Abstract
Paired box 6 (Pax6) is considered to be the master control gene for eye development in all seeing animals studied so far. In vertebrates, it is required not only for lens/retina formation but also for the development of the CNS, olfactory system, and pancreas. Although Pax6 plays important roles in cell differentiation, proliferation, and patterning during the development of these systems, the underlying mechanism remains poorly understood. In the fruit fly, Drosophila melanogaster, Pax6 also functions in a range of tissues, including the eye and brain. In this report, we describe the function of Pax6 in Drosophila eye-antennal disc development. Previous studies have suggested that the two fly Pax6 genes, eyeless (ey) and twin of eyeless (toy), initiate eye specification, whereas eyegone (eyg) and the Notch (N) pathway independently regulate cell proliferation. Here, we show that Pax6 controls eye progenitor cell survival and proliferation through the activation of teashirt (tsh) and eyg, thereby indicating that Pax6 initiates both eye specification and proliferation. Although simultaneous loss of ey and toy during early eye-antennal disc development disrupts the development of all head structures derived from the eye-antennal disc, overexpression of N or tsh in the absence of Pax6 rescues only antennal and head epidermis development. Furthermore, overexpression of tsh induces a homeotic transformation of the fly head into thoracic structures. Taking these data together, we demonstrate that Pax6 promotes development of the entire eye-antennal disc and that the retinal determination network works to repress alternative tissue fates, which ensures proper development of adult head structures.
Keywords: Pax6, eyeless, twin of eyeless, eye-antennal disc, Drosophila
An important question in developmental biology concerns how multiple adult tissues are derived from a single field of cells. For example, the eye, hypothalamus, diencephalon, and telencephalon all arise from adjacent cells within the vertebrate anterior neural plate (1). Similarly, multiple adult head structures of the fruit fly, Drosophila melanogaster, including the compound eyes, ocelli, antennae, maxillary palps, and head epidermis emerge from neighboring cells within a pair of monolayer epithelia called the eye-antennal imaginal discs (2). In both cases competing developmental programs consisting of hundreds of genes are activated in close proximity to one another. To ensure the fidelity of organ specification, mechanisms must be in place within each tissue to ensure that only the desired developmental program is executed. The Drosophila eye-antennal disc is an excellent model system for determining how a single initially uniform field of cells is later subdivided into multiple distinct tissues.
During embryogenesis two groups of cells invaginate from the surface ectoderm and form a pair of eye-antennal discs (3–5). These discs initiate their first cell divisions during the first-larval instar and continue proliferating throughout larval development (4). During the second-larval instar tissue-specific gene regulatory networks (GRNs) are expressed within distinct fields, thereby initiating the subdivision of the eye-antennal disc (6), whereas some GRN members are expressed throughout the entire tissue early in development only to be segregated later (7–12). The molecular battle among GRNs allows for the subdivision of the eye-antennal disc to be maintained within a single continuous cellular field (13–16). Of the GRNs that are known to operate within the eye-antennal disc, the retinal determination (RD) network, which controls eye development, is the best studied (17). At the core of the RD network lie the Paired box 6 (Pax6) genes eyeless (ey) and twin of eyeless (toy), the SIX family member sine oculis (so), the transcriptional coactivator eyes absent (eya), and the Ski/Sno family member dachshund (dac) (17).
Pax6 is an evolutionarily conserved transcription factor that plays a major role in the development of the vertebrate eye and CNS. Mutations in mammalian Pax6 are characterized by eye defects, such as anophthalmia, aniridia, and congenital cataracts, as well as CNS disorders like microcephaly, dysgenesis of both the diencephalon and telencephalon, neuron to glia transformations, and impaired migration of neural crest cells (18–26). Although many studies have demonstrated the importance of Pax6 to vertebrate cell differentiation, proliferation, and patterning (27), the mechanisms by which Pax6 regulates these processes are not fully understood.
The Drosophila Pax6 genes, ey and toy, are activated in the embryonic eye-antennal disc primordium and they, in turn, activate the expression of the other core RD members (7, 9, 28–33). During embryogenesis and the first-larval instar, both genes are expressed throughout the entire eye-antennal disc but by the second-larval instar their expression is restricted to the eye field (7, 9–11). Removal of core RD genes during development causes severe disruptions to eye formation, whereas forced expression in nonocular tissues is sufficient to induce ectopic eyes (17). Thus, these core members are thought to control specification of the developing eye. It has been proposed that other members of the RD network, such as eyegone (eyg) and teashirt (tsh), promote proliferation of the eye field independent of Pax6 and the other core members (12, 34–36). Eyg is similar to the vertebrate Pax6 splice variant Pax6(5a) (11, 35, 37, 38). It is activated by Notch (N) signaling and has been proposed to control tissue proliferation through activation of the JAK/STAT pathway (12, 34, 35, 39). The expression of eyg and its paralog gene twin of eyegone (toe) is initiated throughout the entire eye-antennal disc during embryogenesis, but this activation is thought to be independent of Pax6 (10–12). Tsh and its paralog Tiptop (Tio) are zinc-finger transcription factors (40, 41). Overexpression of either factor leads to massive tissue overgrowth (42–45). Tsh promotes growth of the eye field through participation in a complex that includes Hth (Homothorax) and Yorki (Yki), a Hippo signaling pathway antagonist (36).
A body of evidence suggests that ey and toy also play roles in formation/ development of nonocular structures that are derived from the eye-antennal imaginal disc. Certain Pax6 alleles (toyhdl, eyD) give rise to headless pharate lethal adults (46, 47). However, these mutants are not ideal because the phenotypes are variably penetrant and the mutant loci produce truncated proteins that may function as dominant-negative factors. The combined loss of both Pax6 genes also leads to headless pharate lethal adults (15). The requirement that both genes be simultaneously removed to see effects outside of the retina is consistent with the model in which Ey and Toy function redundantly to each other and together account for the combined activities of vertebrate Pax6 in the eye and CNS (18–26, 48, 49). What is not clear is whether this headless phenotype results from the loss of tissue specification or from a lack of cell proliferation during early eye-antennal disc development, or both. Here we show that Ey and Toy directly promote growth of the entire nascent eye-antennal disc and are later required for eye progenitor cell survival and proliferation. We also show that Pax6 mediates these processes through the activation of tsh, eyg, and N signaling.
The loss of individual Pax6 genes has no effect on eyg expression and this led to the conclusion that Ey/Toy controls eye specification, whereas Eyg independently promotes tissue proliferation (35, 50). We show here that eyg and tsh expression are lost when both Pax6 proteins are simultaneously removed. Expression of tsh or activation of the N signaling pathway can, in the absence of Pax6, partially restore antennal and head epidermis development. Flies that are rescued by tsh expression also display ectopic expression of the Hox gene Antennapedia (Antp) within the eye-antennal disc and a partial homeotic transformation of the rescued head epidermis into thorasic tissue. Our results indicate that Ey and Toy promote proliferation of the eye-antennal disc and specification of the eye, while simultaneously repressing alternate nonocular fates. These results are consistent with a previous report showing that Pax6 is also required for the development of nonocular head structures in the flour beetle, Tribolium castaneum (51), and there is growing evidence in several systems that Pax6 promotes cell proliferation (52–55). These observations suggest that the function of Pax6 has been conserved across 500 million years of animal development.
Results
Function of Pax6 Proteins at Different Developmental Stages in the Eye-Antennal Disc.
A previous study demonstrated that simultaneous expression of RNAi lines targeting ey and toy using the Dorsal Eye (DE)-GAL4 driver results in headless pharate lethal adult flies (15). DE-GAL4 is a GAL4 insertion within the mirror (mirr) locus and mimics its expression pattern (56). During late embryogenesis and the first-larval instar, DE-GAL4 is expressed throughout the entire eye-antennal disc (Fig. S1 A and B), but as development proceeds, expression from the driver is restricted to the dorsal half of the eye as well as a few peripodial cells overlying the antenna (Fig. S1 C–E). The ey RNAi and toy RNAi lines that we are using in this study are efficient at silencing the expression of their respective target genes (Fig. S2 A and B). Knockdown of ey individually with DE-GAL4 does not have a strong effect on the eye-antennal disc or the adult head (Fig. S2 B and E). Similarly, although the knockdown of toy leads to head defects in 11% of animals, the remaining 89% are relatively normal with only slight ocellar and bristle defects (Fig. S2 C and D). Consistent with Wang and Sun (15), in our hands the simultaneous knockdown of both ey and toy results in larvae whose eye-antennal discs are completely missing and headless pharate adult flies that lack all structures derived from the eye-antennal disc (Fig. 1 A–D).
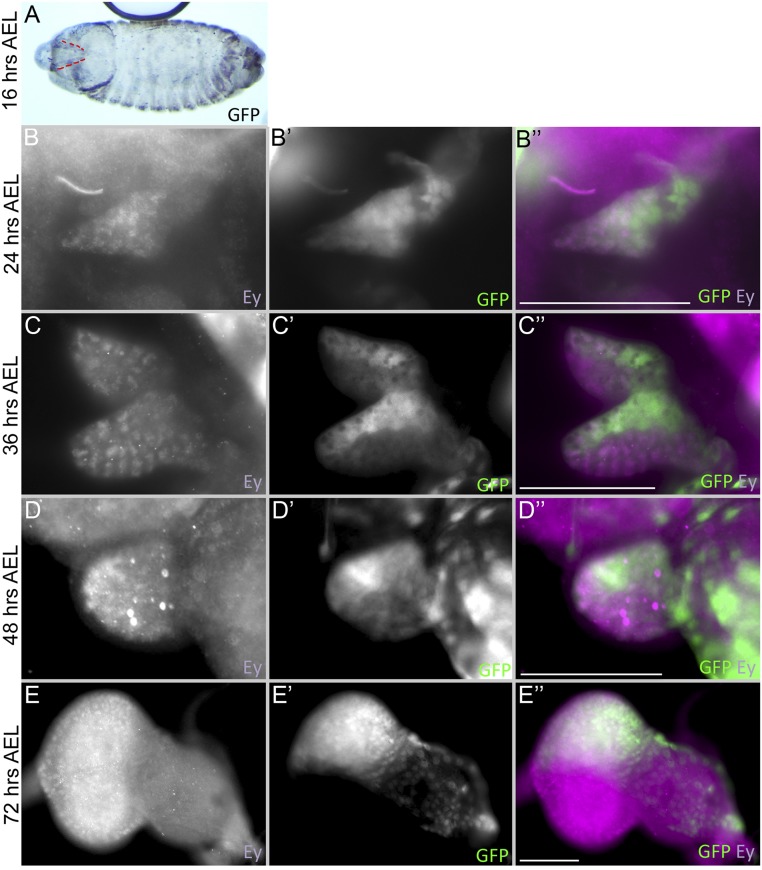
Fig. S1.
DE-GAL4 drives expression dynamically within the eye-antennal disc. (A) Sixteen-hour AEL embryo (stage 17) from DE-GAL4 > GFP flies. The red dash lines mark expression of the GAL4 driver in the eye-antennal disc primordium. (B–E) Eye-antennal discs from DE-GAL4 > GFP larvae at (B) 24 h AEL (early-L1), (C) 36 h AEL (mid-L1), (D) 48 h AEL (early-L2), and (E) 72 h AEL (early-L3) stained with Ey (Purple) and GFP (green). Anterior is to the right. (Scale bars, 50 µm.)
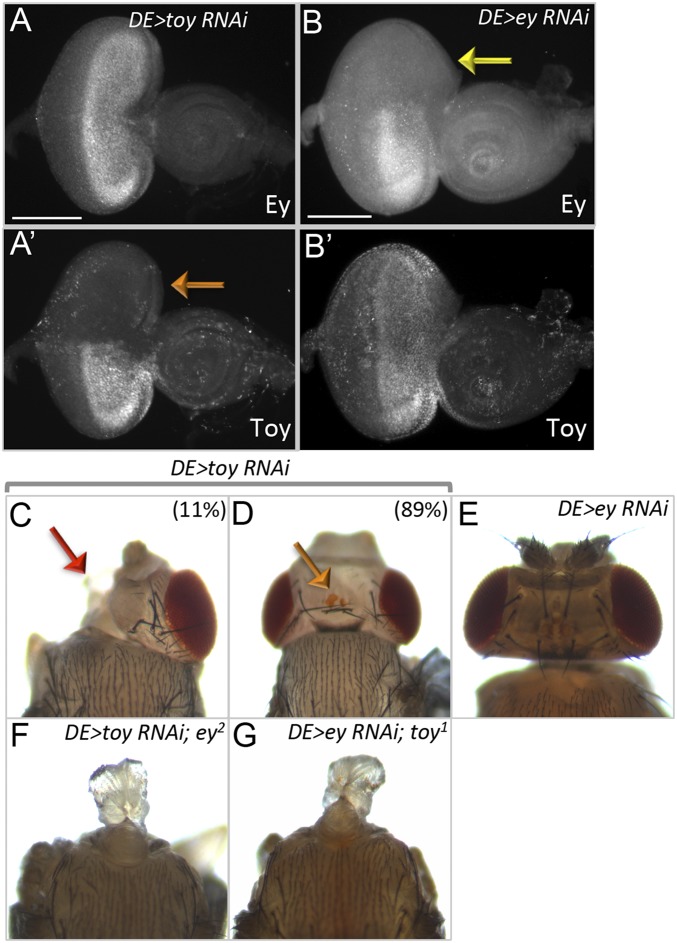
Fig. S2.
Eye antennal discs and adult phenotypes from Pax6 knockdown animals. (A and B) Third-instar larval eye-antennal discs from DE-GAL4 > toy RNAi and DE-GAL4 > ey RNAi larvae at 25 °C. The orange and yellow arrows mark the dorsal half of the disc where DE-GAL4 is expressed and Toy/Ey proteins are absent. Adult heads from (C and D) DE-GAL4 > toy RNAi, (E) DE-GAL4 > ey RNAi, (F) DE-GAL4 > toy RNAi; ey2, and (G) DE-GAL4 > ey RNAi; toy1. Red arrow marks the loss of one half of the head and orange arrow marks the fusion of two ocelli in the DE > toyRNAi flies. (Scale bars, 100 µm.)
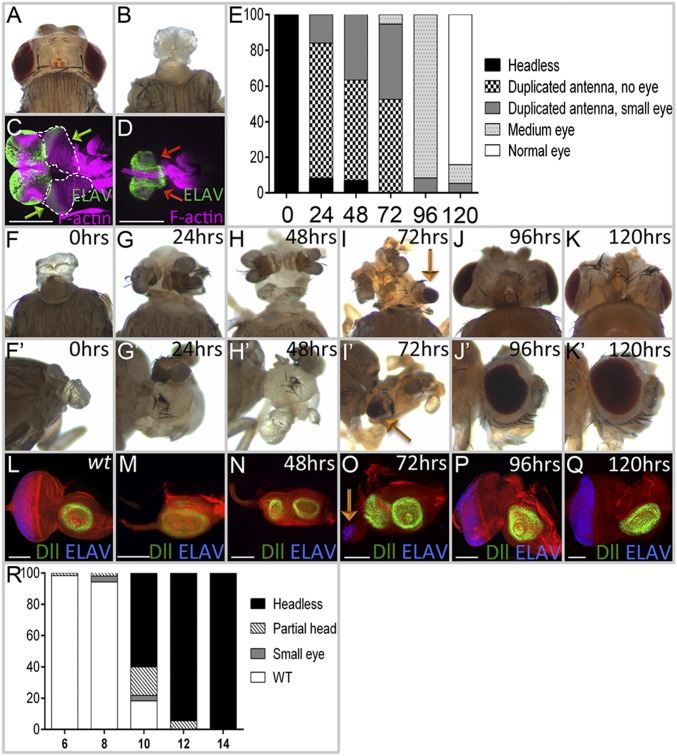
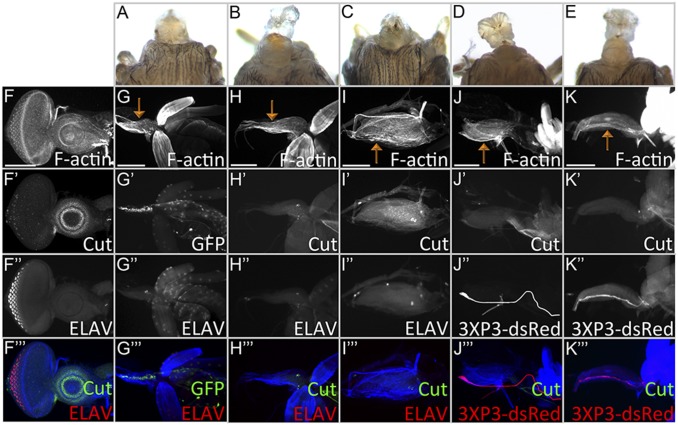
Fig. 1.
Transient removal of Pax6 proteins reveals their roles in the development of all head structures derived from the eye-antenna disc. (A) Wild-type adult head. (B) DE-GAL4 > toy RNAi + ey RNAi flies lack all head structures derived from the eye-antennal discs (100%, n = 30). (C) In wild-type larvae, eye-antennal discs (green arrows) are found between the mouth hooks and brain. (D) Eye-antennal discs are absent in DE > toy RNAi + ey RNAi flies (red arrows, 100%, n = 27). (E–R) Flies are of the following genotype: tub-GAL80ts; DE-GAL4 > toy RNAi + ey RNAi. (E) Quantification of phenotypes observed when animals are transferred from 18 °C to 30 °C to remove Ey and Toy (n = 45, 50, 30, 38, 48, and 28, respectively). x axis = hours AEL at 18 °C. y axis = phenotype percent. (F–K) Light-microscope images of adult heads. Embryos/larvae were kept at 18 °C AEL for 0, 24, 48, 72, 96, and 120 h before being transferred to 30 °C. (L–Q) Corresponding third-instar eye-antennal discs of larvae from F–K. (R) Quantification of phenotypes observed when animals are transferred from 30 °C to 18 °C to restore Ey and Toy (n = 57, 53, 55, 55, and 50, respectively). x axis = hours AEL at 30 °C; y axis = phenotype percent. (Scale bars, 100 µm.)
To rule out RNAi off-target effects, we overexpressed the toy RNAi line in an ey2 mutant and again observed headless pharate adults (Fig. S2F). We also observed headless pharate lethal adults when we expressed an ey RNAi line within the eye-antennal discs of a toy-null mutant that we generated using the CRISPR/Cas9 genome editing system (toy1) (Figs. S2G and S3A). Ey protein is detected in toy1 eye discs (Fig. S3 B–E), which is consistent with a prior report showing that ey expression remains when a toy RNAi line is used to knock down toy expression, as well as with our own analysis of toy RNAi lines (Fig. S2A) (57). The toy1-null mutant animals die as pharate adults and display head defects that vary in severity and penetrance (Fig. S3 F–I). Only when both Pax6 genes are removed together is the headless phenotype seen in 100% of animals (Fig. 1 A and B). These experiments indicate that both Pax6 genes are required and redundant to each other for the development of the entire eye-antennal disc.
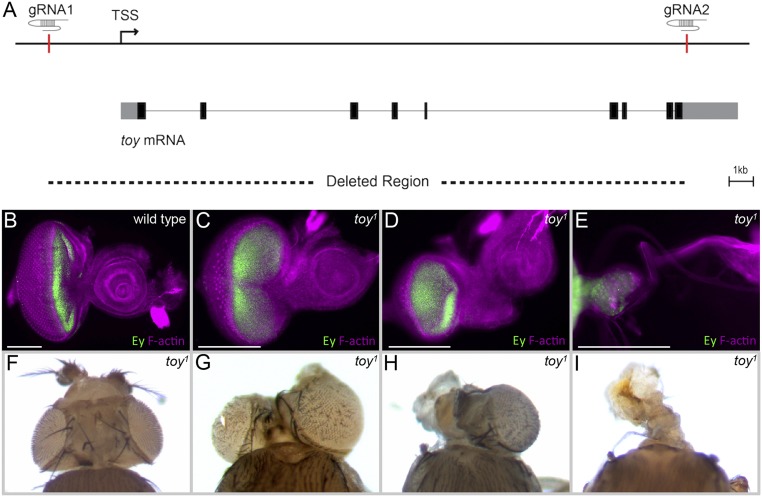
Fig. S3.
ey continues to be expressed in the eye-antennal discs of toy1-null mutants. (A) Schematic depicting the deletion of the toy locus using CRISPR. We are calling the resulting mutant toy1. (B–E) Third-instar eye-antennal discs from wild-type flies (B) and toy1-null mutants (C–E) stained with Ey (green) and F-actin (purple). Anterior is to the right. (F–I) Adult heads from toy1-null mutants. (Scale bars, 100 µm.)
We set out to determine the developmental roles of Ey/Toy at different stages of eye-antennal disc development. To do this, we used a temperature-sensitive GAL80 protein (58) to modulate the activity of DE-GAL4 and therefore gain temporal control of RNAi expression. At 18 °C (permissive temperature), GAL80 is active and interferes with GAL4 activity, thereby preventing RNAi lines from being expressed. At 30 °C (nonpermissive temperature) GAL80 is nonfunctional, GAL4 is active, the RNAi lines are expressed, and the levels of ey/toy are knocked down. If tub-GAL80[ts]; DE-GAL4, UAS-ey RNAi, UAS-toy RNAi animals are kept at 30 °C continuously after a short egg laying period (after egg lay, AEL), then the animals die as pharate adults and are headless (Fig. 1 E and F). If the same animals are kept at 18 °C instead, then the animals have normal compound eyes and heads. By toggling back and forth between these two temperatures, we can control the timing of ey/toy knockdown.
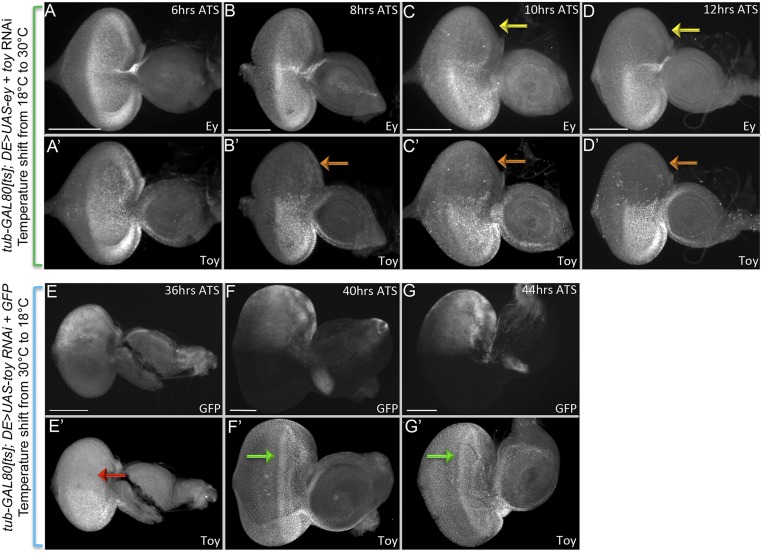
We first determined how long it takes for endogenous Ey/Toy proteins to be cleared once expression of each RNAi line is initiated. To do this, tub-GAL80[ts]; DE-GAL4, UAS-ey RNAi, UAS-toy RNAi animals are kept at 18 °C until the third-larval instar and then shifted to 30 °C. After the shift to 30 °C, it takes ∼10 h for Ey and 8 h for Toy proteins to be cleared from the dorsal half of the retina (Fig. S4 A–D). Flies kept at 30 °C develop twice as fast as those kept at 18 °C; therefore, we combined this developmental difference with the time it takes to clear endogenous proteins to calculate when the two Pax6 genes are required. tub-GAL80[ts]; DE-GAL4, UAS-ey RNAi, UAS-toy RNAi animals are kept at 18 °C AEL and shifted to 30 °C at different times. We scored adult head features (Fig. 1 E–K) and eye-antennal discs for photoreceptor specification (anti-ELAV) and antennal development (anti-Dll) in the late third-instar disc (Fig. 1 L–Q). If both Pax6 proteins are removed from the eye-antennal disc during the first-larval instar then a significant portion of the head epidermis remains, the antenna is partially duplicated, but in most cases the compound eyes fail to form (Fig. 1 E and G–I). In the discs the antennal duplication can be detected by a second zone of Dll expression, whereas the loss/reduction of ELAV indicates the loss of eye development (Fig. 1 M–O). Removal of Ey/Toy after the late second-larval instar or at the beginning of the third-larval instar has a less-severe impact on eye development and the antennal duplications are no longer observed (Fig. 1 E, J, K, P, and Q). These data suggest that by the third-larval instar Ey/Toy no longer contribute to the specification of the compound eyes, despite their continued expression ahead of the morphogenetic furrow. Our findings suggest that the critical window for Pax6 in controlling growth of the entire disc is during the late embryonic/first-larval instar, and the important period for eye specification is during the second-larval instar. Our timings are consistent with the critical window for eye specification that was proposed in an earlier study (8).
Fig. S4.
Pax6 knockdown and restoration after RNAi treatment in the time-course experiment. (A–D) Third-instar larval eye-antennal discs from tub-Gal80[ts]; DE-GAL4 > ey RNAi+ toy RNAi flies. Animals were kept at 18 °C AEL until the third-larval instar stage and were then transferred to 30 °C. Discs were dissected and immunostained to detect Ey and Toy proteins at 6, 8, 10, and 12 h ATS. Yellow arrows show the loss of Ey and orange arrows show loss of Toy in the dorsal compartment. (E–G) Third-instar larval eye-antennal discs from tub-Gal80[ts]; DE-GAL4 > toy RNAi + UAS-GFP flies. Animals were kept at 30 °C AEL until the third-larval instar stage, then being transferred to 18 °C. Discs were dissected and immunostained to detect Toy protein and GFP expression at 36, 40, and 44 h ATS. Red arrow indicates that Toy protein has not been restored at 36 h ATS. Green arrows indicate that at 40 h ATS, Toy expression has been restored in the dorsal half of the eye. Anterior is to the right. (Scale bars, 100 µm.)
We also did the opposite experiment by first keeping flies of the same genotype at 30 °C for varying periods of time and then shifting them to 18 °C. When flies were kept at 30 °C AEL for 10 h and then shifted to 18 °C, the headless phenotype predominated (Fig. 1R). The double-stranded RNA interfering constructs are stable and it takes nearly 40 h at 18 °C for Toy protein levels to be restored back to wild-type (Fig. S4 E–G). Based on this calculation, the point at which Pax6 protein is restored to normal levels is during the mid-first–larval instar stage. We conclude that if Ey and Toy are continually removed before this stage, then the restoration of normal Pax6 protein levels later in development is insufficient to rebuild the eye-antennal disc.
Ey and Toy Proteins Are Essential for Survival and Proliferation of Retinal Progenitor Cells.
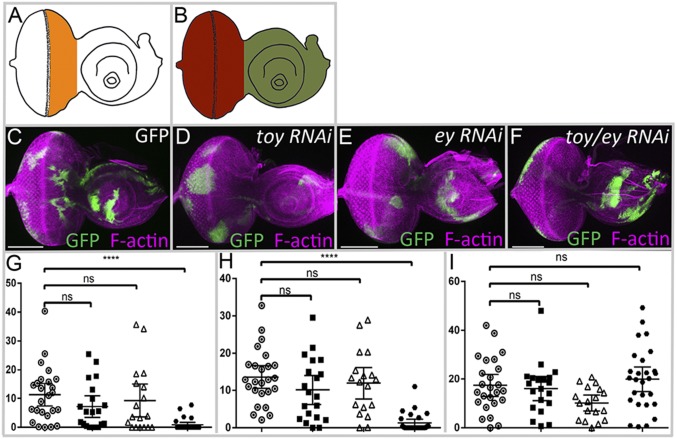
We generated flp-out clones that express ey RNAi and toy RNAi lines either individually or simultaneously and then monitored the growth of the clone when Ey or Toy are eliminated. Clones were induced during the early first-larval instar and their size was determined 72-h later. We measured clones that reside within three zones of the eye-antennal disc: the eye progenitor region (Fig. 2A, orange), the complete eye field region (Fig. 2B, red), and the antennal region (Fig. 2B, green). Wild-type GFP-expressing clones are recovered in all three zones (Fig. 2 C and G–I). Knocking down ey or toy alone does not significantly alter the size of the clones within these three regions, indicating that the loss of either gene individually has little to no effect on growth (Fig. 2 D, E, and G–I). In contrast, clones that lack both Pax6 genes and lie within either the complete eye field or just the eye progenitor zone are significantly smaller and mostly disappear compared with wild-type and single RNAi clones (Fig. 2 F–H). Neither Pax6 gene is expressed within the antennal region after the first-larval instar. Hence, as expected, the size of either single- or double-mutant clones in the antenna is not different from wild-type clones (Fig. 2 D–F and I). The observed inhibition on growth using clonal analysis is consistent with our results using DE-GAL4 to drive the RNAi lines throughout the nascent eye-antennal disc. Our findings also indicate that Ey and Toy are functionally redundant.
Fig. 2.
Toy and Ey are required for cell survival in the eye disc. (A and B) Diagrams of third-larval instar eye-antennal discs demarcating the eye progenitor region (orange), the entire eye region (red), and the antennal region (green). (C–F) Light-microscope images of third-instar eye-antennal discs containing flp-out clones of GFP, toy RNAi, ey RNAi, and ey RNAi + toy RNAi constructs. Purple represents F-actin; green represents GFP. Anterior is to the right. (G–I) y axis = percent clonal area expressing either GFP alone or GFP and RNAi lines within different regions of the eye-antennal disc (n = 25, 18, 20, and 27, respectively). (G) Eye progenitor region, (H) eye field, (I) antenna. Open circle represents GFP, black square represents ey RNAi, open triangle represents toy RNAi, black circle represents ey RNAi + toy RNAi Error bars represent 95% confident interval (CI). ****P ≤ 0.0001. (Scale bars, 100 µm.)
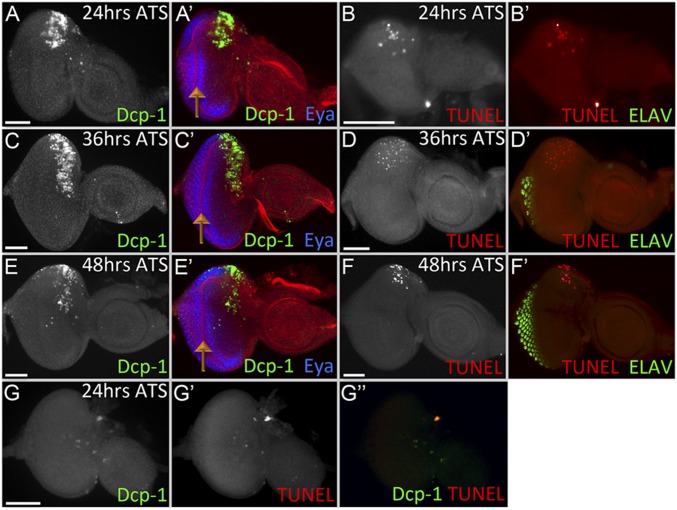
We next set out to determine if the underlying cause of the headless phenotype or the disappearance of double knockdown clones is activation of apoptosis because elevated cell death levels are observed when other retinal determination genes are removed (16, 59, 60). To examine apoptosis in the developing eye-antennal disc, we induced the loss of Ey/Toy using the tub-GAL80[ts]/DE-GAL4 system. tub-GAL80[ts]; DE-GAL4, UAS-ey RNAi, UAS-toy RNAi animals were kept at 18 °C for 96 h AEL to grow until the second-larval instar before being transferred to 30 °C. The eye-antennal discs were dissected and assayed for cell death levels using an antibody against Death caspase-1 (Dcp-1) (61) and TUNEL staining at 24, 36, and 48 h after the shift in temperature (ATS) (Fig. 3 A–F). At this point both Pax6 genes are expressed just within the eye field ahead of the morphogenetic furrow (7, 9, 62). Apoptosis is detected ahead of the furrow in the dorsal half of the eye field (where Ey/Toy are removed) at all time points analyzed (Fig. 3 A–F). However, apoptosis remains suppressed behind the morphogenetic furrow where Ey/Toy are no longer expressed (marked by the presence of Eya) (Fig. 3 A–F). Cell death is significantly higher when both Pax6 proteins are removed than what is reported for single Pax6 gene knockdowns (33, 49). We also detected apoptosis in the flp-out clones expressing both ey and toy RNAi and the surrounding wild-type cells (Fig. S5), which is likely a nonautonomous induction of apoptosis resulting from cell competition (63). Another possibility is that there is a transient knockdown of ey/toy in the shadow RNAi clones when using the FLP-out GAL4 system (64). Taken together, our data suggests that Ey/Toy are required for the survival of retinal progenitor cells.
Fig. 3.
Ey and Toy are required to prevent cell apoptosis in the eye progenitor domain. (A–F) Light-microscope images of third-instar eye-antennal discs from tub-Gal80[ts]; DE > ey RNAi + toy RNAi. Animals were kept at 18 °C AEL for 96 h before being transferred to 30 °C. Cell death is observed in the dorsal half of the retina at 24 h (A and B), 36 h (C and D), and 48 h (E and F) ATS. (G) tub-Gal80[ts]; DE > UAS-p35 + ey RNAi + toy RNAi. Animals were kept at 18 °C for 96 h before being shifted to 30 °C. Discs were dissected 24 h ATS. Cell death was detected with an antibody against Dcp-1 (A, C, and E) and TUNEL labeling (B, D, and F). Anterior is to the right. Orange arrows mark the position of the morphogenetic furrow (Scale bars, 50 μm.)
Fig. S5.
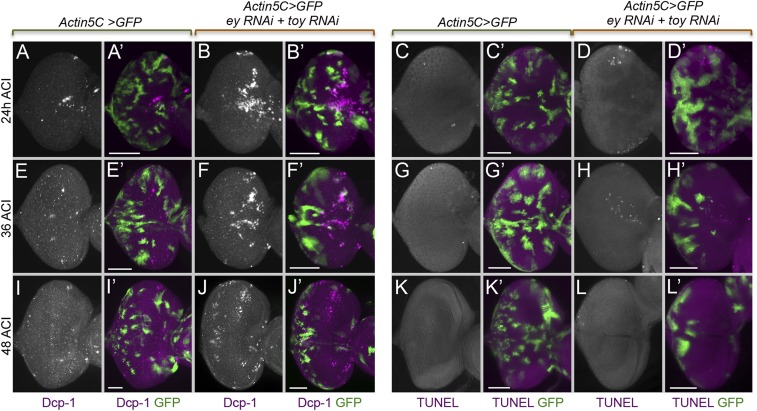
Ey and Toy are required for cell survival in the progenitor region. Light-microscope images of third-instar eye discs containing flp-out clones expressing either GFP alone (A, C, E, G, I, and K) or GFP + ey RNAi + toy RNAi constructs (B, D, F, H, J, and L). Flp-out clones were induced at 48 h AEL and apoptosis was detected using antibodies against Dcp-1 (A, B, E, F, I, and J) and the TUNEL assay (C, D, G, H, K, and L) at 24 h (A–D), 36 h (E–H), and 48 h (I–L) after clonal induction (ACI). Anterior is to the right. (Scale bars, 50 μm.)
We attempted to rescue the headless phenotype of ey/toy double-knockdown flies by overexpressing Death-associated inhibitor of apoptosis 1 (Diap-1) and baculovirus P35, two potent inhibitors of cell death (65, 66). When P35 is overexpressed during the time-shift experiment, cell death in the dorsal eye field is inhibited significantly (Fig. 3G). However, at 25 °C, if Pax6 is knocked down throughout the development, flies expressing Diap-1 or P35 still die as headless pharate adults, lack all structures derived from the eye-antennal disc, and show no morphological difference compared with flies expressing a UAS-GFP control transgene (Fig. 4 A–C). Similarly, overexpression of P35 does not rescue the headless phenotype of ey-GAL4, UAS- ey RNAi; toy1 flies (Fig. 4 D and E). In all genotypes, the tissue that remains between the mouth hook and the brain shows neither Cut nor ELAV protein, indicating a near complete loss of the eye-antennal disc (Fig. 4 F–K). The failure to rescue the headless phenotypes through expression of Diap-1 or P35 suggests that the increased cell death levels is not the proximate cause for the loss of the eye-antennal disc in ey/toy double-knockdown flies.
Fig. 4.
Inhibition of cell death is insufficient to rescue the headless phenotype of ey/toy double mutants. Flies of (A and G) DE > UAS-GFP + ey RNAi + toy RNAi, (B and H) DE > UAS-Diap1 + ey RNAi + toy RNAi, (C and I) DE > UAS-P35 + ey RNAi + toy RNAi, (D and J) ey-Gal4 > UAS-GFP, ey RNAi; toy1, and (E and K) ey-Gal4 > UAS-P35, ey RNAi; toy1 are headless (100%, n = 27, 22, 35, 35, and 28, respectively). (F) Wild-type: Cut expression is observed within the antennal disc and ELAV is seen in developing photoreceptors. (G–K) Blocking cell death with either Diap1 or P35 does not restore eye-antennal disc development. Orange arrows indicate remnants of the eye-antennal disc. Anterior is to the right. (Scale bars, 100 μm.)
We also found that the expression of a 3XP3-dsRed transgene, which marks the Bolwig nerve, aligns along the margin of the remaining tissue in the ey/toy double-knockdown flies (Fig. 4 J and K). A recent study shows that the dorsal portion of the retina develops first with the Bolwig nerve running along one edge of the disc (67). By monitoring DE-GAL4 expression, we have confirmed that all cells of the early disc are dorsally fated (Fig. S1 A and B). As development proceeds, cells fated to develop into the ventral half divide rapidly until the dorsal and ventral halves are of similar sizes. By the late second-larval instar, the Bolwig nerve runs across the middle of the eye-antennal disc along the dorsal-ventral axis (68). Thus, the position of the Bolwig’s nerve in the mutant tissue indicates that loss of disc development may be a result of the loss of cell proliferation.
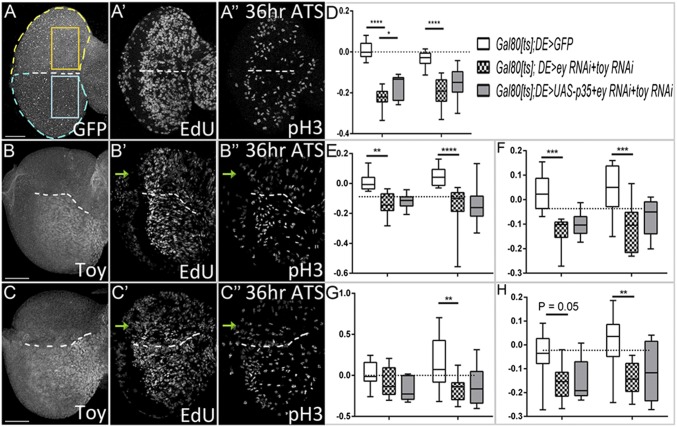
To test our hypothesis, we used the GAL80[ts]/DE-GAL4 system to knockdown ey/toy during the second-larval instar stage and then tested if cell proliferation levels are reduced. First, we compared the number of cells in the dorsal and ventral domains (Fig. 5A, yellow and blue dashed lines outline dorsal and ventral regions) at 24 and 36 h ATS. When Ey/Toy are removed using DE-GAL4, cell numbers in the dorsal domain are significantly lower than in the ventral compartment (Fig. 5 B and D; P ≤ 0.0001). To rule out the effect of apoptosis, we overexpressed P35 while knocking down both Pax6 genes (Fig. 5C). At 24 h ATS, there are more cells in the dorsal domain with P35 overexpression compared with the double knockdown group (Fig. 5D) (P ≤ 0.05). However, as the disc continues to grow there is no significant difference between these two groups (Fig. 5D), indicating that cell proliferation is the major cause of tissue growth loss. Next, we detected cells in S phase and M phase in the dorsal and ventral eye progenitor regions (Fig. 5A, orange and blue boxes) using an EdU assay and a pH3 antibody. When Ey/Toy are removed from the dorsal eye field, the density of cells labeled with EdU and the intensity of EdU florescence in the dorsal compartment is significantly lower than the ventral domain at both 24 and 36 h ATS (Fig. 5 B, E, and F). Similarly, the density of cells labeled with pH3 and the fluorescence intensity of pH3 is significantly lower in the dorsal domain compared with the ventral compartment at 36 h ATS (Fig. 5 B, G, and H). Blocking cell death fails to rescue these phenotypes (Fig. 5 C and E–H), thus our results directly demonstrate that both Ey and Toy are required for eye progenitor cell proliferation.
Fig. 5.
Toy and Ey are required for cell proliferation in the developing eye disc. (A–C) Third-instar eye discs from tub-Gal80[ts]; DE>UAS-GFP (A), tub-Gal80[ts]; DE > ey RNAi + toy RNAi (B), and tub-Gal80[ts]; DE>UAS-P35 + ey RNAi + toy RNAi (C). Animals were kept at 18 °C AEL for 96 h before being transferred to 30 °C. Eye-antennal discs were dissected at 24 and 36 h ATS to detect cells in S phase (EdU) and M phase (pH3). (D–H) y axis in all panels = change index [(dorsal − ventral)/(dorsal + ventral)]. (D) Total number of cells in the dorsal and ventral eye fields (yellow and blue outlines of disc in A). (E–H) Progenitor domains shown in orange and blue boxes in A. (E) EdU+ cell density, (F) EdU fluorescence intensity, (G) pH3+ cell density, (H) pH3 fluorescence intensity (A, yellow and blue boxes) are quantified [for each genotypes at 24 h (Left grouping) and 36 h (Right grouping) ATS, n = 8, 10, 7, 8, 10, and 11 respectively]. Error bars represent 95% CI. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. (Scale bars, 50 μm.)
Ey and Toy Regulate Eye-Antennal Disc Proliferation Through Eyg and Tsh.
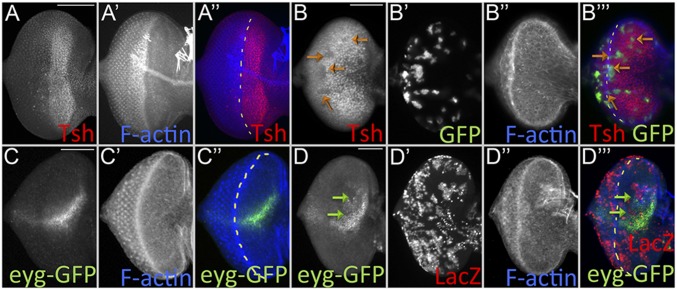
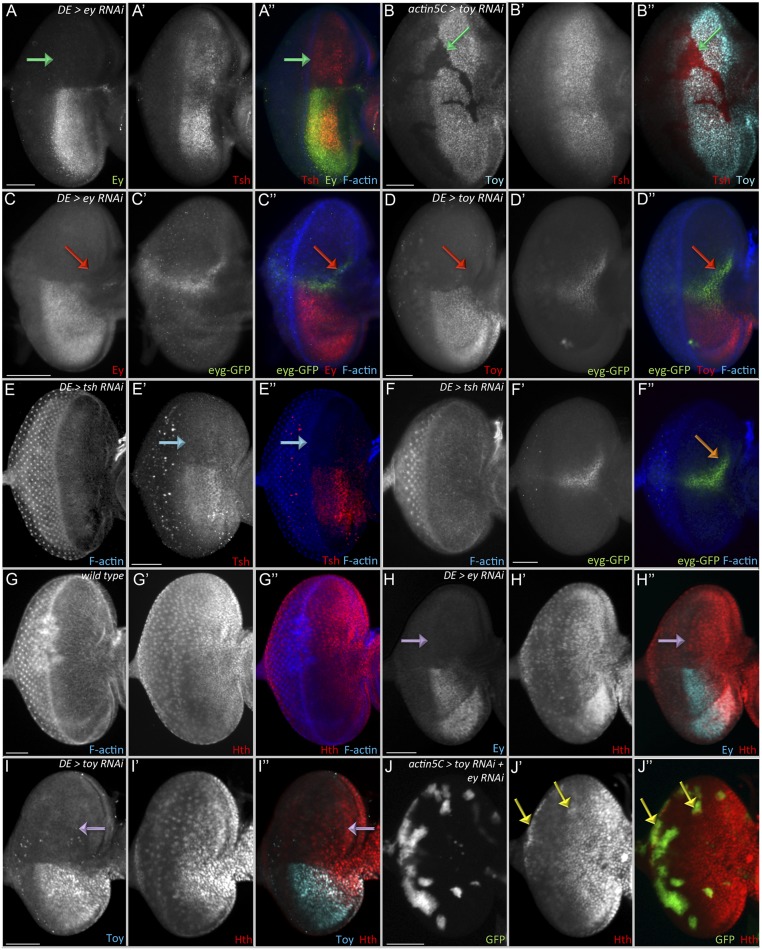
To understand the mechanism by which Ey and Toy promote early eye-antennal disc proliferation, we turned our attention to retinal determination genes that are known to promote progenitor cell survival and proliferation in the eye-antennal disc. Eya, as part of the So–Eya complex, is sufficient to promote cell proliferation and inhibit cell death (16, 59, 60, 69, 70); however, it is not activated until the second-larval instar, and its expression is limited to the eye field. On the other hand, eyg and tsh initiate expression during embryogenesis and the first-larval instar, respectively (12, 34–36, 71). Tsh promotes growth of the eye through formation of a complex with Homothorax (Hth) and Yki (36, 42). Along with ey and toy, tsh is expressed ahead of the morphogenetic furrow (Fig. 6A); therefore, we tested the idea that tsh might lie downstream of Ey/Toy. tsh expression is unaffected when either Pax6 protein is removed individually (Fig. S6 A and B, green arrows). However, the simultaneous removal of both Pax6 proteins leads to a reduction in Tsh levels (Fig. 6B, orange arrows). hth expression is not changed in Pax6 single knockdown eye-antennal discs (Fig. S6 G–I, purple arrows). In some double-knockdown clones there is actually a slight up-regulation of hth expression (Fig. S6J, yellow arrows).
Fig. 6.
Pax6 regulates the expression of tsh and eyg. Light-microscope images of third-instar eye discs. (A) tsh expression in the wild-type eye. (B) tsh expression is lost in clones lacking both Ey and Toy (clones are marked with GFP) 36 h after induction (orange arrows). (C) eyg-GFP expression in the wild-type eye. (D) eyg-GFP expression is lost in clones (green arrows) lacking both Ey and Toy (clones are marked with LacZ) 24 h after induction. Anterior is to the right. The white dashed lines mark the morphogenetic furrow. (Scale bars, 50 µm.)
Fig. S6.
Regulation of Tsh, Eyg, and Hth by Pax6. (A–J) Light-microscope images of third-instar eye discs. Removal of either Ey alone (A: DE-GAL4 > ey RNAi) or Toy alone (B: hs-flp; Actin5C-GAL4 > toy RNAi) has no effect on tsh expression (green arrows). Removal of either Ey alone (C: DE-GAL4 > ey RNAi) or Toy alone (D: DE-GAL4 > toy RNAi) has no effect on eyg-GFP expression (red arrows). Removal of Tsh (E: DE-GAL4 > tsh RNAi, aqua arrow) has no effect on eyg-GFP expression (F, orange arrow). (G) Hth expression pattern in the wild-type eye-antennal disc. Removal of Ey alone (H: DE-GAL4 > ey RNAi) or Toy alone (I: DE-GAL4 > toy RNAi) has no effect on the hth expression pattern (purple arrows) (J: hs-flp; Actin5C-GAL4 > toy RNAi + ey RNAi). hth expression is not repressed in the ey/toy double-knockdown clones, but actually increases in some.(yellow arrows). Anterior is to the right. (Scale bars, 50 μm.)
Earlier studies have suggested that Ey/Toy and Eyg/N regulate independent branches of the RD network and that their expression is independent of each other (12, 35). However, in these early studies eyg expression was examined only in ey mutants; Toy protein is still present and might compensate for Ey by activating eyg. Indeed, eyg-GFP expression is not affected when ey or toy is knocked down individually using DE-GAL4 (Fig. S6 C and D, red arrows). However, it is lost in flp-out clones expressing both ey RNAi and toy RNAi ahead of morphogenetic furrow (Fig. 6 C and D, green arrows), thereby confirming our proposal that Ey/Toy activate eyg expression in the eye-antennal disc and are functionally redundant. We asked if Tsh genetically controls eyg-GFP expression but see no effect on eyg-GFP levels when Tsh is removed from the dorsal half of the retina (Fig. S6 E and F, blue and orange arrows).
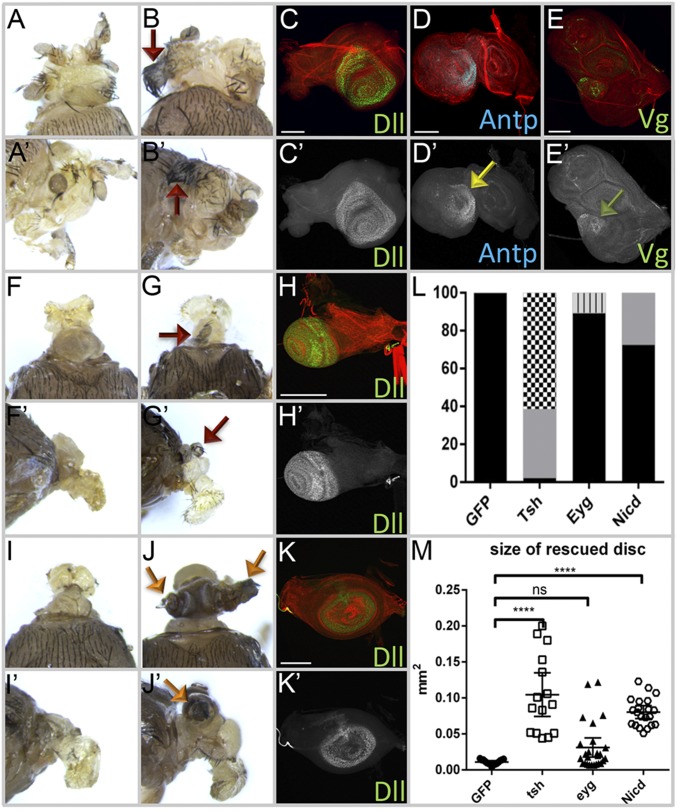
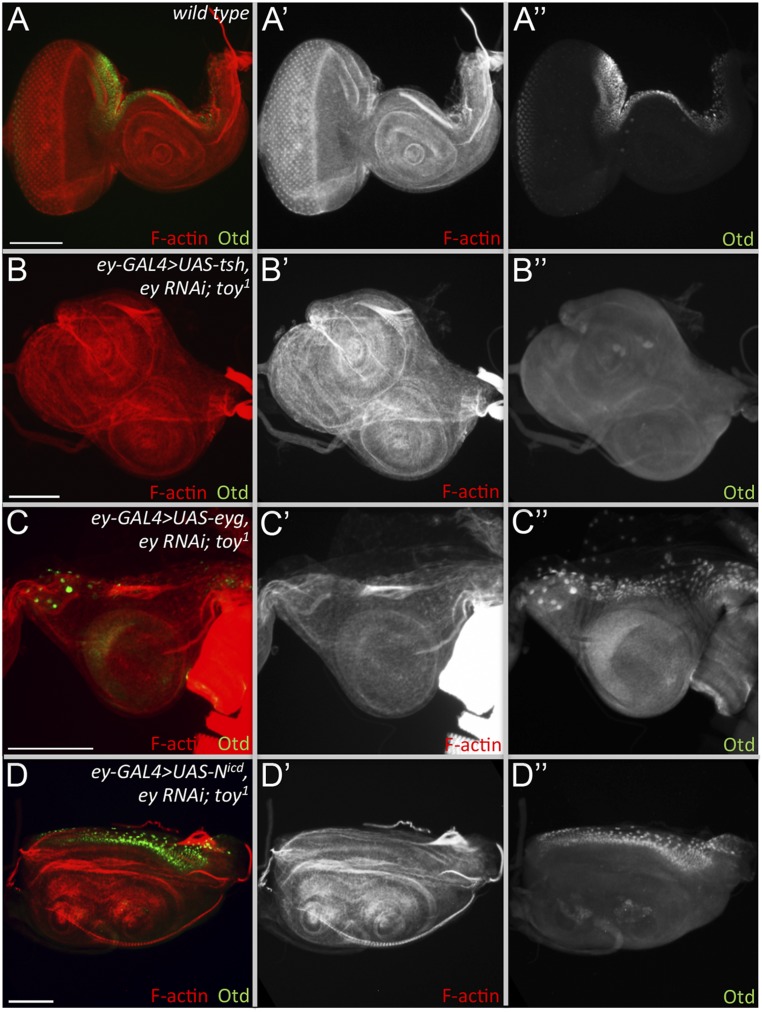
Next, we tested whether overexpression of either eyg or tsh can rescue the headless phenotype. Expression of either gene using DE-GAL4 results in embryonic and larval lethality; therefore, we used ey-Gal4 for these rescue experiments. Overexpression of tsh gives strong recovery of nonocular head structures, including the antenna (Fig. 7 A, B, and L), with Dll expression being restored to third-larval instar discs (Fig. 7C). Interestingly, the rescued head epidermis shows thoracic epidermis transformation. Ectopic wing tissue is also found in some rescued fly heads (Fig. 7 B and L), which is possibly because of the ectopic activation of Antp and vestigial (vg) in the rescued disc (Fig. 7 D and E). Orthodenticle (otd) expression, which marks a portion of the dorsal head epidermis, however, is not detected in the rescued eye-antennal discs (Fig. S7 A and B), which might mean that either the entire head epidermis has been transformed into thoracic tissue or that the rescued head epidermis is fated from an otd− portion of the disc. The size of the rescued eye-antennal disc size is significantly larger than the UAS-GFP overexpression control (Fig. 7M).
Fig. 7.
Expression of tsh, eyg, and the Notch pathway rescues the headless phenotype of ey/toy mutants. (A) Of ey-GAL4 > UAS-tsh + ey RNAi; toy1 flies, 46.7% have a small head with antenna. (B) About 50% of ey-GAL4 > UAS-tsh + ey RNAi; toy1 flies showed rescued head with thoracic tissue transformation. Red arrows mark ectopic wing tissue on the head. (C–E) Dll, Antp (yellow arrow), and Vg (green arrow) are present in the eye-antennal disc of tsh rescued flies. (F and G) ey-GAL4 > UAS-eyg + ey RNAi; toy1 flies are pharate adult lethal headless. (H) Activation of dll is detected in few eye-antennal discs rescued by overexpression of eyg. (I) Of ey-GAL4 > UAS-Nicd + ey RNAi; toy1, 73% are still headless. (J) Of flies of ey-GAL4 > UAS-Nicd + ey RNAi; toy1, 27% have restored antenna and some head epidermis development. (K) dll is activated in the eye-antennal disc rescued by overexpression of Nicd. (L) Phenotype quantification of the rescued flies (n = 35, 35, and 44, respectively). Black represents headless, gray represents small head with antennal tissue, black/white checkered represents ectopic thorax tissue, gray/black bar represents small antennal tissue. (M) Size of the rescued eye-antennal disc quantification (n = 15, 15, 26, and 21, respectively). Error bars represent 95% CI. ****P ≤ 0.0001. (Scale bars, 100 µm.)
Fig. S7.
Otd expression in the rescued eye-antennal discs. (A) Expression of otd in wild-type eye-antennal discs. (B and C) Otd protein levels are not restored in the rescued eye-antennal discs in which either tsh (B) or eyg (C) is forcibly expressed in the absence of Pax6. (D) Activation of the Notch pathway partially restores otd expression to the antennal disc.
Overexpression of eyg did not rescue the headless phenotype (Fig. 7 F, G, and L). The eye-antennal disc is still absent and only a few discs show activation of dll in the restored antennal field (Fig. 7H). Otd protein is also absent (Fig. S7C), which further indicates a lack of rescue by Eyg. Compared with flies rescued by Tsh, the eye-antennal discs that express eyg are not significantly different in size from UAS-GFP–expressing control flies (Fig. 7M). This result suggests that Eyg must work cooperatively with other factors to promote disc growth. N signaling is required for the activation of eyg expression and it promotes growth of the early eye field (34, 35, 72–74). We activated the N pathway by expressing the intracellular domain of the N receptor (Nicd) in the ey/toy double-knockdown flies. Although most of the mutant flies are still headless, about 27% of the mutant flies have restored antennae and head epidermal tissue (Fig. 7 I, J, and L). This result was confirmed by activation of dll and otd in the antennal primordium (Fig. 7K and Fig. S7D). The size of discs in which N signaling is activated is significantly larger than those in which eyg is expressed (Fig. 7M). In ey/toy knockdown animals that are rescued by Tsh or Notch signaling, we find that the antenna is restored more often than any other tissue. The head epidermis is restored frequently as well. However, we never observed a restoration of photoreceptor specification, suggesting that the reintroduction of Eyg, Tsh, or N signaling is not suitable or sufficient to substitute for Pax6 in the context of eye specification. Our data indicate that downstream of Ey/Toy, proliferation is controlled by N signaling, Eyg, and Tsh, whereas other RD proteins, such as So, Eya, and Dac, control specification (Fig. 8).
Fig. 8.
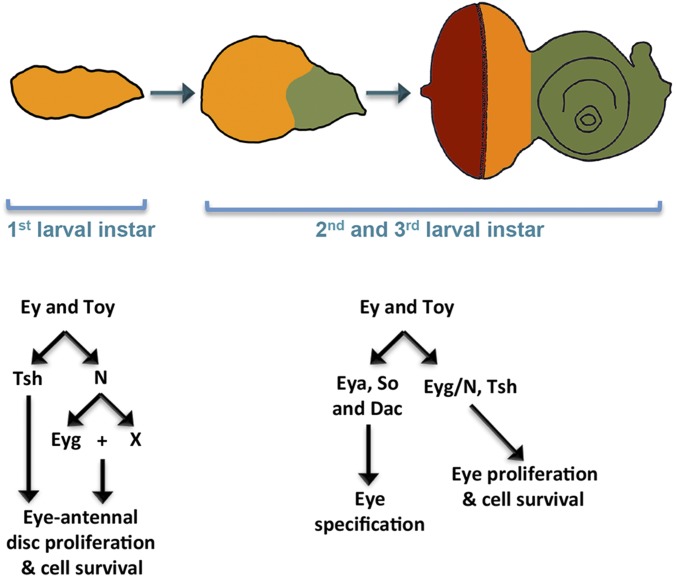
A model illustrating the roles of Pax6 in the eye-antennal disc development. During embryogenesis and the first-larval instar, Ey and Toy promote growth of the entire eye-antennal disc (orange region) through tsh, eyg, and N signaling. During later stages of development, the expression of ey and toy are restricted to the undifferentiated cells of the eye disc (orange region). Here, proliferation continues to be promoted through tsh, eyg, and N signaling. In addition, Ey/Toy promote eye specification through other RD genes, such as so, eya, and dac.
Discussion
In contrast to vertebrates that have a single Pax6 gene, the Drosophila genome contains two Pax6 homologs, ey and toy. Both genes are expressed broadly throughout the entire eye-antennal disc but are later limited to a far more restricted domain within the undifferentiated cells of the eye field. Whereas most studies on Pax6 in the eye-antennal disc have focused on the developing compound eye, several reports have hinted at a role for both genes outside of the eye (15, 46, 75–77). However, the underlying mechanism of how Ey/Toy promote eye-antennal disc development has been elusive. This is, in part, because of the use of single Pax6 mutants to study development. The phenotypes associated with individual mutants are variable and often restricted to the eye. Several studies have suggested that Ey and Toy function redundantly to each other (46, 48, 49). This finding most likely explains the variability of phenotype severity and penetrance. Thus, the combined loss of both Ey/Toy may be a more accurate reflection of the effect that Pax6 loss has on Drosophila development. Indeed, this appears to be the case as it is reported that the combined loss of both ey and toy leads to the complete loss of all head structures that are derived from the eye antennal disc (15). In this report we have attempted to determine the mechanism by which Ey/Toy support eye-antennal disc development.
Previous studies in the fly eye proposed that Pax6 is concerned solely with eye specification, whereas Notch signaling and other retinal determination proteins, such as Eyg, Tsh, and Hth, control cell proliferation and tissue growth (35, 36, 42, 50). Here we propose an alternate model in which Ey/Toy are in fact required for cell survival and proliferation in addition to eye specification. Our data indicate that Ey/Toy regulate growth of the eye-antennal disc through Tsh, N/Eyg, and additional N-dependent proliferation promoting genes (Fig. 8, Left). We propose that on simultaneous removal of Ey and Toy the eye-antennal disc fails to develop, in part, because the expression of eyg and tsh is lost in complete absence of Pax6. Expression of tsh and activation of the N pathway are sufficient to restore tissue growth to the eye-antennal disc. Support for our model linking Ey/Toy to cell proliferation via Eyg and Tsh comes from studies showing that eyg loss-of-function mutants display a headless phenotype identical to that seen in the ey/toy double knockdowns, that cells lacking eyg do not survive in the eye disc, and overexpression of Tsh causes overproliferation (12, 36).
Our results also show that the combined loss of Ey and Toy affects the number of cells that are in S and M phases of the cell cycle. This observation directly supports our model that Ey/Toy control growth of the eye-antennal disc and is consistent with studies in vertebrates that demonstrate roles for Pax6 in the proliferation of neural progenitors within the brain (53, 78–80). Earlier studies observed cells undergoing apoptosis in Pax6 single-mutant eye-antennal discs and showed that blocking cell death alone can partially rescue the head defects of the eyD and toyhdl mutants (46). Although we show that retinal progenitor cells lacking both Pax6 proteins undergo even greater levels of apoptosis, blocking cell death does not restore the eye-antennal disc. What accounts for the differences in the two experiments? In the eyD and toyhdl rescue experiments, each genotype contained wild-type copies of the other Pax6 paralog, but here we have knocked down both Pax6 genes simultaneously. Another possible difference is that we are reducing Pax6 levels while the eyD and toyhdl mutants are likely functioning as dominant negatives. We conclude from our results that a reduction in cell proliferation but not elevated apoptosis levels is the proximate cause for the complete loss of the eye-antennal disc.
Although the activation of Tsh and the Notch pathway can restore antennal and head epidermal development, neither factor is capable of restoring eye development to the ey/toy double-knockdown discs. This is most likely because both Pax6 genes are also required for the specification of the eye. In particular, Ey/Toy are required for the activation of several other retinal determination genes, including so, eya, and dac (17). Thus, our results suggest that Notch signaling, Eyg, and Tsh can restore nonocular tissue growth to the eye field but cannot compensate for the Pax6 requirement in eye specification (Fig. 8, Right).
Finally, our results using the double knockdown of ey/toy are consistent with the dosage effects that are seen in mammalian Pax6 mutants. Although mutations in ey have just eye defects (81), the combined loss of ey/toy lacks all head structures (15). Mice that are heterozygous for Pax6 mutations have small eyes, whereas those that are homozygous completely lack eyes, have severe CNS defects, and die prematurely (21). Similarly, human patients carrying a single mutant copy of Pax6 suffer from aniridia, whereas newborns that are homozygous for the mutant Pax6 allele have anophthalmia, microcephaly, and die very early as well (20, 22). As a master control gene of eye development, Pax6 appears to initiate both retinal specification and proliferation. Our data demonstrate that the functions of Ey and Toy in the eye-antennal disc are redundant and dependent upon gene dosage, thereby making the roles of Pax6 in the Drosophila similar to what is observed in vertebrates where Pax6 controls both specification and proliferation of the brain and retina in a dosage-sensitive manner (18–26, 48, 49).
Materials and Methods
Fly Stocks.
The following fly stocks were used: (i) DE-GAL4 (Georg Halder, VIB-KU Leuven Center for Cancer Biology, Leuven, Belgium); (ii) hsFLP22 [Bloomington Drosophila Stock Center (BDSC)]; (iii) Actin5C > y+>GAL4, UAS-GFP S65T (BDSC); (iv) y1 w*; Actin5C > y+>GAL4, UAS-lacZ (BDSC); (v) UAS-ey RNAi (BDSC); (vi) UAS-toy RNAi (BDSC); (vii) tub-GAL80[ts] (BDSC); (viii) UAS-NICD (Kevin Moses, Emory University, Atlanta); (ix) UAS-GFP (BDSC); (x) UAS-cycE (BDSC); (xi) UAS-tsh (Amit Singh, University of Dayton, Dayton, OH); (xii) UAS-eyg; (xiii) UAS-p35 (BDSC); (xiv) UAS-Diap1 (BDSC); (xv) UAS-cut (Craig Micchelli, Washington University in St. Louis, St. Louis); (xvi) eyg-GFP (BDSC); (xvii) ey-GAL4 (BDSC); (xviii) vasa-Cas9 (Kate O’Connor-Giles, University of Wisconsin–Madison, Madison, WI). All crosses were conducted at 25 °C except for time-course experiments, which were conducted at 18 °C or 30 °C.
Antibodies/Microscopy.
The following antibodies were used: (i) rat anti-ELAV [1:100; Developmental Studies Hybridoma Bank (DSHB)], (ii) mouse anti-Ey (1:100; DSHB), (iii) mouse anti-Wg (1:800; DSHB), (iv) mouse anti-Cut (1:100; DSHB), (v) mouse anti-Eya (1:5; DSHB), (vi) mouse anti-Dac (1:100; DSHB), (vii); chicken anti–β-gal (1:800; Abcam), (viii) rabbit anti-GFP (1:1,000; Invitrogen), (ix) rabbit anti–Dcp-1 (1:100; Cell Signaling Technologies), (x) rabbit anti-Tsh (1:3,000; Stephen Cohen, University of Copenhagen, Copenhagen), (xi) rabbit anti-Hth (1:1,000; Richard Mann, Columbia University, New York), (xii), guinea pig anti-Toy (1:500; Henry Sun, Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan), (xiii) mouse anti-Dll (1:500; Diana Duncan, Washington University in St. Louis, St. Louis), and (xiv) guinea pig anti-Otd (1:650; Tiffany Cook, Wayne State University, Detroit). Fluorophore-conjugated secondary antibodies and phalloidin-fluorophore conjugates were obtained from Jackson Immuno Research Laboratories and Life Technologies. TUNEL assay (Roche) was performed as per the manufacturer’s instructions. Imaginal discs were prepared as described previously (82). Eye-antennal discs were photographed on a Zeiss Axioplan II compound microscope and Leica SP5 Scanning Confocal. Adult flies were viewed with a Zeiss Discovery light microscope and Leica M205FA Stereo Microscope.
Generation of toy-Null Mutant.
toy1 was generated using CRISPR/Cas9-mediated homology-directed repair (83). CRISPR target sites were selected by CRISPR Optimal Target Finder to delete the coding region and ∼5.7 kb upstream of the TSS (tools.flycrispr.molbio.wisc.edu/targetFinder/). Guide RNAs (gRNAs) were designed and cloned into the pU6-BbsI-gRNA plasmid (Kate O’Connor-Giles, University of Wisconsin–Madison, Madison, WI, flycrispr.molbio.wisc.edu/protocols/gRNA). Homology arms were cloned into the pHD-DsRed-attP donor plasmid (Kate O’Connor-Giles, University of Wisconsin–Madison, Madison, WI). The deleted genomic region was replaced with 3XP3-DsRed flanked by LoxP sites (flycrispr.molbio.wisc.edu/protocols/pHD-DsRed-attP). The injection mix of gRNA plasmids (100 ng/µL each) and the donor plasmid (500 ng/µL) were injected into Drosophila embryos carrying vasa-cas9. Mutants were selected based on the expression of DsRed in the eye and were further verified by DNA sequencing. 3XP3-DsRed was later removed by Cre-Lox recombination.
gRNA 1.
Sense oligo: 5′-CTT CGC ATT CCA CTT ACC CAT CTA-3′;
Antisense oligo: 5′-AAA CTA GAT GGG TAA GTG GAA TGC-3′.
gRNA 2.
Sense oligo: 5′-CTT CGA ATG TTT GGA ACT TAA AAA-3′;
Antisense oligo: 5′-AAA CTT TTT AAG TTC CAA ACA TTC-3′.
Homology arm 1.
F primer: 5′-ATA ATA CAC CTG CAA AAT CGC ATC ATC ACC GGC ACA CG-3′;
R primer: 5′-ATA ATA CAC CTG CAA AAT TAT CAT GTG TTT TTT TAA TCA ATT TAA AGT GTA TG-3′.
Homology arm 2.
F primer: 5′-ATA ATA GCT CTT CTT ATT TCC TGA TCT GCT AAG ATA GGT TAA AGT AT-3′;
R primer: 5′-ATA ATA GCT CTT CAT ATA CGC CGA CAT GGT CTA AAG AG-3′.
Clonal Induction and Analysis.
Flp-out overexpression clones were induced with UAS-ey RNAi or UAS-toy RNAi. Embryos were collected for 2 h at 25 °C and then heat-shocked at 37 °C for 10 min during the early first-larval instar (about 24-h AEL) or at early second instar (about 48-h AEL). Larvae were cultured at 25 °C and dissected at times specified in figures. Adobe Photoshop CC 2015 was used to outline and measure the area of the clones induced in the eye-antenna disc (in pixels). Statistical significance was calculated using one-way ANOVA with GraphPad Prism.
Temperature Shifts.
tub-Gal80[ts]; DE-GAL4> ey RNAi + toy RNAi embryos were collected for 2 h at 25 °C and then kept either at 18 °C or 30 °C before being shifted to the opposite temperature. Eye-antennal discs were dissected either as wandering third-instar larvae or at defined time points after shifts in temperature.
Cell Proliferation Analysis.
S-phase cells were detected using the Click-iT EdU Alexa Fluor 555 imaging Kit (Invitrogen). Eye-antennal discs were dissected in PBS and incubated in 50 µM EdU PBS for 20 min, fixed, and then washed in 0.1% Triton-X PBS, 3% (wt/vol) BSA in PBS. Next, eye-antennal discs were incubated with the Click-iT reaction mixtures per the manufacturer’s instructions before standard immunostaining with pH3 antibody (M phase). Finally, samples were stained with Hoechst 33342 (Invitrogen) to label DNA. Eye-antennal discs were imaged using Leica SP5 confocal. Total cell numbers in the dorsal/ventral eye field, EdU+ and pH3+ cell density, and fluorescence intensity in the dorsal/ventral eye progenitor region were measured using Imaris (Bitplane). Statistical significance was calculated using the Holm–Sidak’s multiple comparisons test followed by two-way ANOVA.
Acknowledgments
We thank Amit Singh, Steve Cohen, Georg Halder, Richard Mann, Craig Micchelli, Kevin Moses, Kate O’Connor-Giles, Henry Sun, Diana Duncan, Jim Powers (Indiana Light Microscopy Imaging Center), Tiffany Cook, the Bloomington Drosophila Stock Collection, and the Developmental Studies Hybridoma Bank for gifts of fly stocks, antibodies, and plasmids; Luke Baker for the original observation that knocking down toy and ey simultaneously yields headless flies; Lena Weber for showing that eyg-GFP is unchanged when either toy or ey is knocked down individually; and members of the J.P.K. Laboratory for comments on the manuscript. This work is supported by National Eye Institute Grant R01 EY014863 (to J.P.K.).
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Gene Regulatory Networks and Network Models in Development and Evolution,” held April 12–14, 2016, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Gene_Regulatory_Networks.
This article is a PNAS Direct Submission. E.V.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610614114/-/DCSupplemental.
References
- 1.Speman H. Embryonic Development and Induction. Hafner; New York: 1962. [Google Scholar]
- 2.Ferris GF. External morphoogy of the adult. In: Demerec M, editor. Biology of Drosophila. John Wiley and Sons; New York: 1950. pp. 368–419. [Google Scholar]
- 3.Garcia-Bellido A, Merriam JR. Cell lineage of the imaginal discs in Drosophila gynandromorphs. J Exp Zool. 1969;170(1):61–75. doi: 10.1002/jez.1401700106. [DOI] [PubMed] [Google Scholar]
- 4.Madhaven MM, Schneiderman HA. Histological analysis of the dynamics of growth of imaginal discs and histoblast nests during the larval development of Drosophila melanogaster. Wilhelm Roux Archive. 1977;183:269–305. doi: 10.1007/BF00848459. [DOI] [PubMed] [Google Scholar]
- 5.Newby WW, Thelander RP. Early development of the head in normal and tumorous-head Drosophila melanogaster. Drosoph Inf Serv. 1950;24:89. [Google Scholar]
- 6.Domínguez M, Casares F. Organ specification-growth control connection: New in-sights from the Drosophila eye-antennal disc. Dev Dyn. 2005;232(3):673–684. doi: 10.1002/dvdy.20311. [DOI] [PubMed] [Google Scholar]
- 7.Czerny T, et al. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3(3):297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- 8.Kumar JP, Moses K. EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell. 2001;104(5):687–697. doi: 10.1016/s0092-8674(01)00265-3. [DOI] [PubMed] [Google Scholar]
- 9.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the small eye gene in mice and aniridia in humans. Science. 1994;265(5173):785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 10.Jones NA, Kuo YM, Sun YH, Beckendorf SK. The Drosophila Pax gene eye gone is required for embryonic salivary duct development. Development. 1998;125(21):4163–4174. doi: 10.1242/dev.125.21.4163. [DOI] [PubMed] [Google Scholar]
- 11.Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc Natl Acad Sci USA. 1998;95(23):13720–13725. doi: 10.1073/pnas.95.23.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang CC, et al. Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development. 2003;130(13):2939–2951. doi: 10.1242/dev.00522. [DOI] [PubMed] [Google Scholar]
- 13.Anderson AM, Weasner BM, Weasner BP, Kumar JP. Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development. 2012;139(5):991–1000. doi: 10.1242/dev.077255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev Cell. 2003;5(3):403–414. doi: 10.1016/s1534-5807(03)00243-0. [DOI] [PubMed] [Google Scholar]
- 15.Wang CW, Sun YH. Segregation of eye and antenna fates maintained by mutual antagonism in Drosophila. Development. 2012;139(18):3413–3421. doi: 10.1242/dev.078857. [DOI] [PubMed] [Google Scholar]
- 16.Weasner BM, Kumar JP. Competition among gene regulatory networks imposes order within the eye-antennal disc of Drosophila. Development. 2013;140(1):205–215. doi: 10.1242/dev.085423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar JP. Retinal determination the beginning of eye development. Curr Top Dev Biol. 2010;93:1–28. doi: 10.1016/B978-0-12-385044-7.00001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoykova A, Fritsch R, Walther C, Gruss P. Forebrain patterning defects in small eye mutant mice. Development. 1996;122(11):3453–3465. doi: 10.1242/dev.122.11.3453. [DOI] [PubMed] [Google Scholar]
- 19.Matsuo T, et al. A mutation in the Pax-6 gene in rat small eye is associated with impaired migration of midbrain crest cells. Nat Genet. 1993;3(4):299–304. doi: 10.1038/ng0493-299. [DOI] [PubMed] [Google Scholar]
- 20.Jordan T, et al. The human PAX6 gene is mutated in two patients with aniridia. Nat Genet. 1992;1(5):328–332. doi: 10.1038/ng0892-328. [DOI] [PubMed] [Google Scholar]
- 21.Hill RE, et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354(6354):522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 22.Glaser T, et al. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7(4):463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 23.Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet. 1992;2(3):232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 24.Grindley JC, Hargett LK, Hill RE, Ross A, Hogan BL. Disruption of PAX6 function in mice homozygous for the Pax6Sey-1Neu mutation produces abnormalities in the early development and regionalization of the diencephalon. Mech Dev. 1997;64(1-2):111–126. doi: 10.1016/s0925-4773(97)00055-5. [DOI] [PubMed] [Google Scholar]
- 25.Holm PC, et al. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol Cell Neurosci. 2007;34(1):99–119. doi: 10.1016/j.mcn.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Warren N, Price DJ. Roles of Pax-6 in murine diencephalic development. Development. 1997;124(8):1573–1582. doi: 10.1242/dev.124.8.1573. [DOI] [PubMed] [Google Scholar]
- 27.Shaham O, Menuchin Y, Farhy C, Ashery-Padan R. Pax6: A multi-level regulator of ocular development. Prog Retin Eye Res. 2012;31(5):351–376. doi: 10.1016/j.preteyeres.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Ostrin EJ, et al. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16(4):466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pappu KS, et al. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development. 2005;132(12):2895–2905. doi: 10.1242/dev.01869. [DOI] [PubMed] [Google Scholar]
- 30.Punzo C, Seimiya M, Flister S, Gehring WJ, Plaza S. Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development. 2002;129(3):625–634. doi: 10.1242/dev.129.3.625. [DOI] [PubMed] [Google Scholar]
- 31.Niimi T, Seimiya M, Kloter U, Flister S, Gehring WJ. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126(10):2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- 32.Michaut L, et al. Analysis of the eye developmental pathway in Drosophila using DNA microarrays. Proc Natl Acad Sci USA. 2003;100(7):4024–4029. doi: 10.1073/pnas.0630561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halder G, et al. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125(12):2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- 34.Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131(16):3839–3847. doi: 10.1242/dev.01258. [DOI] [PubMed] [Google Scholar]
- 35.Dominguez M, Ferres-Marco D, Gutierrez-Aviño FJ, Speicher SA, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat Genet. 2004;36(1):31–39. doi: 10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- 36.Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: Homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23(19):2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epstein JA, et al. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8(17):2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 38.Yao JG, Sun YH. Eyg and Ey Pax proteins act by distinct transcriptional mechanisms in Drosophila development. EMBO J. 2005;24(14):2602–2612. doi: 10.1038/sj.emboj.7600725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutierrez-Aviño FJ, Ferres-Marco D, Dominguez M. The position and function of the Notch-mediated eye growth organizer: The roles of JAK/STAT and four-jointed. EMBO Rep. 2009;10(9):1051–1058. doi: 10.1038/embor.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fasano L, et al. The gene teashirt is required for the development of Drosophila embryonic trunk segments and encodes a protein with widely spaced zinc finger motifs. Cell. 1991;64(1):63–79. doi: 10.1016/0092-8674(91)90209-h. [DOI] [PubMed] [Google Scholar]
- 41.Laugier E, Yang Z, Fasano L, Kerridge S, Vola C. A critical role of teashirt for patterning the ventral epidermis is masked by ectopic expression of tiptop, a paralog of teashirt in Drosophila. Dev Biol. 2005;283(2):446–458. doi: 10.1016/j.ydbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 2002;16(18):2415–2427. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datta RR, Lurye JM, Kumar JP. Restriction of ectopic eye formation by Drosophila teashirt and tiptop to the developing antenna. Dev Dyn. 2009;238(9):2202–2210. doi: 10.1002/dvdy.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Datta RR, Weasner BP, Kumar JP. A dissection of the teashirt and tiptop genes reveals a novel mechanism for regulating transcription factor activity. Dev Biol. 2011;360(2):391–402. doi: 10.1016/j.ydbio.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh A, Kango-Singh M, Sun YH. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development. 2002;129(18):4271–4280. doi: 10.1242/dev.129.18.4271. [DOI] [PubMed] [Google Scholar]
- 46.Kronhamn J, et al. Headless flies produced by mutations in the paralogous Pax6 genes eyeless and twin of eyeless. Development. 2002;129(4):1015–1026. doi: 10.1242/dev.129.4.1015. [DOI] [PubMed] [Google Scholar]
- 47.Arking R, Putnam RL, Schubiger M. Phenogenetics of the eyeless-dominant mutant of Drosophila melanogaster. J Exp Zool. 1975;193(3):301–311. doi: 10.1002/jez.1401930306. [DOI] [PubMed] [Google Scholar]
- 48.Furukubo-Tokunaga K, Adachi Y, Kurusu M, Walldorf U. Brain patterning defects caused by mutations of the twin of eyeless gene in Drosophila melanogaster. Fly (Austin) 2009;3(4):263–269. doi: 10.4161/fly.10385. [DOI] [PubMed] [Google Scholar]
- 49.Jacobsson L, Kronhamn J, Rasmuson-Lestander A. The Drosophila Pax6 paralogs have different functions in head development but can partially substitute for each other. Mol Genet Genomics. 2009;282(3):217–231. doi: 10.1007/s00438-009-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mann RS. Two Pax are better than one. Nat Genet. 2004;36(1):10–11. doi: 10.1038/ng0104-10. [DOI] [PubMed] [Google Scholar]
- 51.Luan Q, Chen Q, Friedrich M. The Pax6 genes eyeless and twin of eyeless are required for global patterning of the ocular segment in the Tribolium embryo. Dev Biol. 2014;394(2):367–381. doi: 10.1016/j.ydbio.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka-Matakatsu M, Miller J, Du W. The homeodomain of Eyeless regulates cell growth and antagonizes the paired domain-dependent retinal differentiation function. Protein Cell. 2015;6(1):68–78. doi: 10.1007/s13238-014-0101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walcher T, et al. Functional dissection of the paired domain of Pax6 reveals molecular mechanisms of coordinating neurogenesis and proliferation. Development. 2013;140(5):1123–1136. doi: 10.1242/dev.082875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marquardt T, et al. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105(1):43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 55.Sansom SN, et al. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5(6):e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison CM, Halder G. Characterization of a dorsal-eye Gal4 Line in Drosophila. Genesis. 2010;48(1):3–7. doi: 10.1002/dvg.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blaquiere JA, Lee W, Verheyen EM. Hipk promotes photoreceptor differentiation through the repression of Twin of eyeless and Eyeless expression. Dev Biol. 2014;390(1):14–25. doi: 10.1016/j.ydbio.2014.02.024. [DOI] [PubMed] [Google Scholar]
- 58.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302(5651):1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 59.Weasner BM, Weasner BP, Neuman SD, Bashirullah A, Kumar JP. Retinal expression of the Drosophila eyes absent gene is controlled by several cooperatively acting cis-regulatory elements. PLoS Genet. 2016;12(12):e1006462. doi: 10.1371/journal.pgen.1006462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: Genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72(3):379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- 61.Song Z, McCall K, Steller H. DCP-1, a Drosophila cell death protease essential for development. Science. 1997;275(5299):536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- 62.Kumar JP, Moses K. The EGF receptor and notch signaling pathways control the initiation of the morphogenetic furrow during Drosophila eye development. Development. 2001;128(14):2689–2697. doi: 10.1242/dev.128.14.2689. [DOI] [PubMed] [Google Scholar]
- 63.Amoyel M, Bach EA. Cell competition: How to eliminate your neighbours. Development. 2014;141(5):988–1000. doi: 10.1242/dev.079129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bosch JA, Sumabat TM, Hariharan IK. Persistence of RNAi-mediated knockdown in Drosophila complicates mosaic analysis yet enables highly sensitive lineage tracing. Genetics. 2016;203(1):109–118. doi: 10.1534/genetics.116.187062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120(8):2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 66.Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83(7):1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 67.Won JH, et al. Cell type-specific responses to wingless, hedgehog and decapentaplegic are essential for patterning early eye-antenna disc in Drosophila. PLoS One. 2015;10(4):e0121999. doi: 10.1371/journal.pone.0121999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bolwig N. Senses and sense organs of the anterior end of the housefly larvae. Vidensk Medd Dansk Naturh Forenh. 1946;109:81–217. [Google Scholar]
- 69.Jemc J, Rebay I. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev Biol. 2007;310(2):416–429. doi: 10.1016/j.ydbio.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheyette BN, et al. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12(5):977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 71.Pan D, Rubin GM. Targeted expression of teashirt induces ectopic eyes in Drosophila. Proc Natl Acad Sci USA. 1998;95(26):15508–15512. doi: 10.1073/pnas.95.26.15508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cho KO, Choi KW. Fringe is essential for mirror symmetry and morphogenesis in the Drosophila eye. Nature. 1998;396(6708):272–276. doi: 10.1038/24394. [DOI] [PubMed] [Google Scholar]
- 73.Domínguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396(6708):276–278. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- 74.Papayannopoulos V, Tomlinson A, Panin VM, Rauskolb C, Irvine KD. Dorsal-ventral signaling in the Drosophila eye. Science. 1998;281(5385):2031–2034. doi: 10.1126/science.281.5385.2031. [DOI] [PubMed] [Google Scholar]
- 75.Blanco J, Pauli T, Seimiya M, Udolph G, Gehring WJ. Genetic interactions of eyes absent, twin of eyeless and orthodenticle regulate sine oculis expression during ocellar development in Drosophila. Dev Biol. 2010;344(2):1088–1099. doi: 10.1016/j.ydbio.2010.05.494. [DOI] [PubMed] [Google Scholar]
- 76.Brockmann A, Domínguez-Cejudo MA, Amore G, Casares F. Regulation of ocellar specification and size by twin of eyeless and homothorax. Dev Dyn. 2011;240(1):75–85. doi: 10.1002/dvdy.22494. [DOI] [PubMed] [Google Scholar]
- 77.Jiao R, et al. Headless flies generated by developmental pathway interference. Development. 2001;128(17):3307–3319. doi: 10.1242/dev.128.17.3307. [DOI] [PubMed] [Google Scholar]
- 78.Estivill-Torrus G, Pearson H, van Heyningen V, Price DJ, Rashbass P. Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development. 2002;129(2):455–466. doi: 10.1242/dev.129.2.455. [DOI] [PubMed] [Google Scholar]
- 79.Georgala PA, Manuel M, Price DJ. The generation of superficial cortical layers is regulated by levels of the transcription factor Pax6. Cereb Cortex. 2011;21(1):81–94. doi: 10.1093/cercor/bhq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26(7):1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 81.Hoge MA. Another gene in the fourth chromosome of Drosophila. Am Nat. 1915;49:47–49. [Google Scholar]
- 82.Spratford CM, Kumar JP. Dissection and immunostaining of imaginal discs from Drosophila melanogaster. J Vis Exp. 2014;(91):51792. doi: 10.3791/51792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gratz SJ, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196(4):961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]