Significance
Hsp70 is a protein nanometer-sized molecular machine operating similar like a macroscopic engine: Combustion of fuel (ATP in the case of Hsp70) triggers mechanical motion for its function. However, in contrast to macroscopic engines, protein machines face additional limitations imposed by thermal fluctuations and by the low stability of the individual components. Using single-molecule force spectroscopy, we drive the passive machine through its mechanical states that allows us to detect its relevant parts. Applying this approach for the substrate binding domain (SBD) of Hsp70, we identified mechanical subunits and hinges of the SBD. Interestingly, these parts are involved in an ATP-fueled biological conformational change of Hsp70. In summary, single-molecule manipulation gives insight into the operating principles of nanoscale molecular machines.
Keywords: laser trapping, parallel pathways, elasticity, force, protein extension
Abstract
Owing to the cooperativity of protein structures, it is often almost impossible to identify independent subunits, flexible regions, or hinges simply by visual inspection of static snapshots. Here, we use single-molecule force experiments and simulations to apply tension across the substrate binding domain (SBD) of heat shock protein 70 (Hsp70) to pinpoint mechanical units and flexible hinges. The SBD consists of two nanomechanical units matching 3D structural parts, called the α- and β-subdomain. We identified a flexible region within the rigid β-subdomain that gives way under load, thus opening up the α/β interface. In exactly this region, structural changes occur in the ATP-induced opening of Hsp70 to allow substrate exchange. Our results show that the SBD’s ability to undergo large conformational changes is already encoded by passive mechanics of the individual elements.
When looking at protein structures at atomic resolution, it is often tempting to use macroscopic mechanical analogies to describe their function as molecular machines. However, such analogies are often misleading because boundaries between independently stable subdomains cannot often be determined from structures, owing to the high cooperativity of protein folding and structural transitions. Single-molecule protein nanomechanics have emerged as a tool to force biomolecules through their conformational space and, hence, identify hinges, breaking points, and mechanically stable subdomains (1–3).
A prominent example of a protein machine undergoing large conformational change during its functional cycle is the ATP-regulated Hsp70 chaperone DnaK—a central molecular chaperone of the protein quality control network in a cell (4–6). Once ATP is bound to the nucleotide binding domain (NBD, blue-yellow; Fig. 1A) of DnaK, the initially closed substrate binding domain (SBD) opens its binding cleft by engaging the β-subdomain to the NBD. In doing so, it undergoes a dramatic ∼10 Å displacement of its lid subdomain (Fig. 1A; refs. 7 and 8) to allow exchange of substrates (9). Several crystal structures of the isolated SBD (in which the NBD is absent) have been solved (10–15). In these structures, the absence or presence of peptide clients or nonnatural ligands induce no significant structural changes in the closed conformation. There is no indication in the crystal structures of the huge conformational change of the lid domain of the SBD, seen in the ATP form of the full-length two-domain DnaK. Therefore, the large conformational change of the SBD is only observed in the two-domain DnaK after ATP binding. Thus, although the crystal structures provide us with valuable insights into the 3D arrangement of individual atoms, the thermodynamic and mechanical stability of individual substructures are difficult to predict based on this information alone. Here, we ask how the large ATP-induced changes of the SBD, as seen in the two-domain DnaK, are mirrored in the subdomain integrity and nanomechanics of the closed form of the SBD and how this integrity is affected by peptide (client) binding.
Fig. 1.
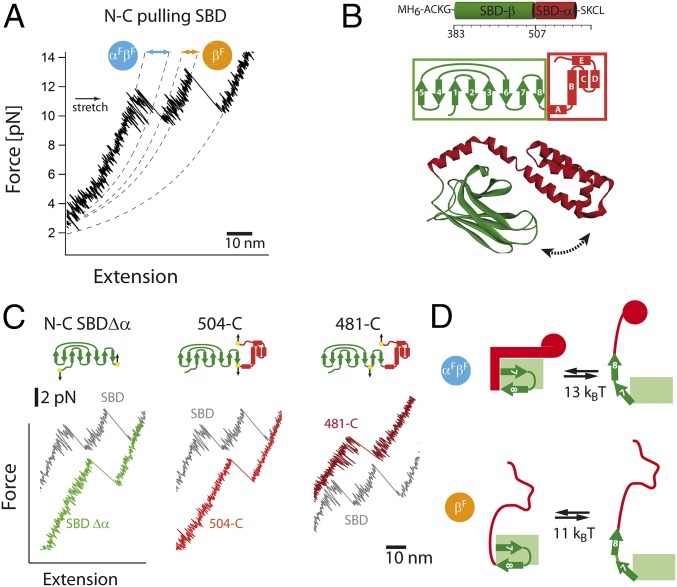
Single-molecule force experiments of the SBD of the Hsp70 chaperone. (A) Three-dimensional structure of the Hsp70 chaperone DnaK in the ADP/apo state (PDB ID code 2KHO; ref. 30) and the ATP state (PDB ID code 4B9Q; ref. 8). The protein consists of two domains: the nucleotide binding domain, which is further divided into lobe I (blue) and lobe II (red), and the substrate binding domain, which is divided into the α-subdomain and the β-subdomain. The figures were prepared by using DS Viewer (Accelrys Software). (B) Optical trap assay. The SBD (green/red surfaces) is tethered to the beads (gray spheres) by two DNA handles, and beads are trapped in highly focused laser beams (red cones). The connection between the DNA and protein is realized by the modification of the two cysteine residues of the protein by the single-stranded DNA-maleimide oligonucleotide complementary to the DNA-handle overhang (for details, see ref. 16). One of the beams is reflected by a steerable mirror, which enables pulling and stretching of a single protein. The conditions were 10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, and 137 mM NaCl (PBS buffer), pH 7.5 at 30 °C. (C) Shown is a sequence of force-extension curves of a single SBD domain. The protein-DNA construct was stretched several times (black curves, 1 kHz filtered) at constant velocity of 200 nm/s. In every individual trace, two unfolding events of different contour-length increase are visible. The dashed lines correspond to worm-like chain (WLC) fits. (D) The order of the individual unfolding events varies. Pathway I corresponds to unfolding of the larger fragment first, followed by the shorter SBD fragment. For pathway II, the order of unfolding events is reversed. (E) Force extension curves of the SBDΔα construct at 200 nm/s (1 kHz). (F) Scatter plot of unfolding forces and corresponding contour length increases with respective distributions for pathway I and pathway II (red, α-helical part; green, β-subdomain). We found significant fluctuations at forces > 9 pN (Fig. 2A), hence the worm-like chain analysis was performed within the 0–6 pN force range. (G) Summary of bifurcating unfolding pathways of the SBD.
Generally, a folded protein that undergoes large conformational changes (such as the SBD) faces a seemingly contradictory demand; a high folding stability and cooperativity ensures that a protein is in a functional state, but at the same time it increases the energy penalty for disrupting the interactions in the native state during conformational transition. In this study, we show that to balance these contradictory demands, the SBD decreases the unfolding cooperativity by mechanical destabilization of the critical part of the structure β7–β8 and the interface between the subdomains (C-terminal β-strands), which are involved in the allosteric ATP-induced conformational change. These regions have been also identified in silico by coarse-grained steered molecular dynamics simulations using the self-organized polymer (SOP) model (16). In previous studies, various parts of the SBD have been implicated in a hinge function, based on local structural heterogeneity with a possible impact on allosteric signaling pathways in DnaK. Although such thermally accessible states can be readily populated on fast timescales, in our approach, force is used to drive the closed SBD structure toward the open state by providing a large energy bias to investigate collective dynamics at much slower timescales.
Results
Nanomechanical Units of the SBD During N- and C-Terminal Pulling Experiments.
To scrutinize the mechanical integrity of the SBD of DnaK, we carried out single molecule force experiments by using an optical tweezer setup (Fig. 1B). A wild-type SBD (residues 393–607, for sequence and biochemical characterization, see SI Appendix, Fig. S1) construct was prepared for optical tweezers by adding N- and C-terminal cysteine residues, assembled and assayed in single-molecule force experiments by using optical tweezers, as described (17). Consecutive force-extension curves using a constant velocity of 200 nm/s at a 1 kHz sampling rate showed two distinct unfolding transitions of different lengths during a single pull corresponding to unfolding of longer and shorter protein portions (Fig. 1C). The order of unfolding changed from trace to trace. We classified these different unfolding patterns as pathway I and pathway II. In pathway I, the larger portion of the SBD unfolds first, followed by the shorter part, whereas the order is reversed in pathway II. The pathways were stochastically distributed; 65 of 96 unfolding events followed pathway II and 31 of 96 events followed pathway I. The analysis using the Worm-like chain model yielded contour lengths approximately 20–24 nm and approximately 40–44 nm for shorter and longer protein portions, respectively. The total contour length for both patterns ranged between 61 and 64 nm, slightly less than the expected contour length of 68 nm as estimated based on the number of amino acids (aa) in the folded part of the SBD (398–603, 0.35 nm per aa) minus the N- and C-terminal distance in the folded state (approximately 4 nm). To interpret these unfolding transitions in structural terms, we calculated the number of amino acids involved in the particular unfolding transition using contour-length increases. We calculated that 120 aa are involved in the longer unfolding transition, which matches well with the number of amino acids in the β-subdomain of the SBD (117 aa shown in green, Fig. 1B). For the shorter transition, we determined that approximately 80 aa (5.8 nm now being the N-C distance, after the unfolding of the larger fragment) are involved, which is slightly less than the size of the α-helical portion (90 aa, helices A–E shown in red, Fig. 1B). To corroborate this preliminary structural assignment, we deleted the α-helical subdomain completely and investigated a truncated construct, SBDΔα, in optical tweezers experiments (Fig. 1E). As expected, force extension curves showed only a single unfolding transition with contour length approximately 42 nm and a mean unfolding force distribution of 10.5 pN, which matches the longer unfolding transition seen in WT SBD. In summary, the WT SBD comprises two mechanically and energetically distinct domains corresponding to an α-helical subdomain (red) and a β-subdomain (shown in green).
Next, we focused on analysis of the differences between the individual subdomains after they unfold in different pathways. Importantly, the first unfolding event of either α- or β-subdomain disrupts the interface between them, which gives an additional length increase. To investigate the role breaking the interface plays, we assembled scatter plots of unfolding force vs. contour length for the α-helical and β-subdomains for pathway I (Fig. 1F, closed symbols) and II separately (Fig. 1F, open symbols). We found significant pathway-dependent differences in contour-length increases. For the α-helical subdomain (n = 110), we found contour-length increases of 22.0 ± 0.1 nm vs. 23.4 ± 0.1 nm for pathway I versus pathway II. For the β-subdomain (n = 86), we found contour-length increases of 43.8 ± 0.1 nm vs. 42.5 ± 0.1 nm for pathway I versus pathway II. We analyzed the contour-length increases by using two-sample t test and found that the differences between pathway I and pathway II are statistically significant (P < 0.001). Thus, the domain that unfolds first shows an increased contour length.
Force Separates the SBD Subdomains by Detaching the Terminal β-Strands.
To detect possible fast fluctuations, we analyzed force-extension curves at 10 kHz resolution (see details in Fig. 2A). Note that because unfolding of subdomains occurs stochastically, some of our constant velocity traces showed unfolding of the subdomains below the average unfolding force and some traces showed unfolding well above the average force. We observed that in force-extension traces after reaching a critical force of approximately 9 pN, rapid equilibrium fluctuations—exchanges between upper and lower state—appeared and gradually vanished as force was increased. We termed the observed rapid fluctuations as αFβF fluctuations because they are observed only if the folded α-domain and the folded β-subdomain are present. Further experiments revealed pathway-dependent differences; after unfolding of the α-domain (pathway II), we still observed fluctuations, however with a shorter length. Because in this situation the β-subdomain is still folded, we named these fluctuations as type βF. In pathway I, after unfolding of the β-subdomain, we did not observe any fluctuations and, thus, both longer and shorter fluctuations require the folded β-subdomain.
Fig. 2.
High-resolution single-molecule force experiments reveal fluctuations between subdomains. (A) A magnification of the SBD force-extension trace at 10 kHz. Before unfolding of the α- and β-subdomain, significant fluctuations are observed (αFβF). After unfolding of α-subdomain, much shorter fluctuations are observed (βF). The dashed lines correspond to worm-like chain (WLC) fits. (B) Structure and secondary structure topology of the SBD. (C) A magnification of the force-extension trace at 10 kHz for the SBD along different pulling directions. (D) Structural interpretation (helical part in red, β-subdomain in green) of upper and lower states and associated free energies with αFβF and βF fluctuations. For details, see Results.
A worm-like chain fit to these fluctuations gave estimates of the contour-length increases of 12 nm and 8 nm for αFβF and βF, respectively (see contour-length transformation in SI Appendix, Fig. S2). The fluctuations gradually vanish with increasing force and vanish completely at forces above 13 pN. To estimate the free energy costs of these fluctuations, we take into account all energetic contributions, including the beads in a harmonic trapping potential well and the worm-like chain elasticity of the DNA tether and protein (SI Appendix). Fluctuations are assumed as a two-state transition between upper and lower state associated with a contour-length increase (12 nm for αFβF and 8 nm for βF). Using this model, force vs. distance curves were fitted to obtain free energy changes (18). We calculated a free energy difference at zero force of 13 (±1) and 11 (±1) kBT for the αFβF and βF fluctuations, respectively. Unfortunately, a quantitative estimation of microscopic rates in constant force experiments was not possible because, at loads necessary to induce fluctuations, the protein will rapidly proceed to further unfolded states.
Next, we focused on the structural interpretation of these fluctuations and whether they are linked to the specific secondary-structure elements of the SBD subdomains (Fig. 2B). As expected, deletion of the α-helical part in the variant SBDΔα resulted in only the βF fluctuations being seen (Fig. 2C, for biochemical characterization, see SI Appendix, Fig. S1 and Table S1). Hence, the fluctuations originate from the part of the β-sheets that reversibly attaches and detaches. Observation of the αFβF fluctuations depended on the presence of both folded domains. To identify the substructures involved in these fluctuations, we applied force across specific parts of the SBD while keeping the rest of the structure unperturbed. To this end, we removed the N-terminal cysteine and introduced a cysteine residue at location 504 (lid opening variant 1, Fig. 2C) and 481 (lid opening variant 2, Fig. 2C) with the second cysteine retained at the C terminus. In lid opening variant 1, the pulling force is applied to the residues from 504 through the C terminus and does not include any β-sheet. In lid opening variant 2, force is applied between residues 481 through the C terminus including β7 and β8. The sites for attachment were selected such that the overall pulling force vector had a similar orientation in both constructs. In variant 1, we did not detect any sign of fluctuations of either type during constant velocity cycles. In the second variant (481-C-terminal pulling, Fig. 2C), both types of fluctuations, αFβF and βF, appeared. These results indicate that both fluctuations are structurally interrelated.
Based on these observations from different variants, we propose a model where the closed form of the SBD undergoes opening and closing at the expense of significant energy necessary for the displacement of β-sheets 7 and 8 and breaking the interface (Fig. 2D). In this model, the upper state in the αFβF fluctuations is a fully closed SBD (with completely folded α/β subdomains), whereas the lower state represents a protein with a folded β-core (presumably folded β1-β5/6) and with β7-β8 detached. After unfolding of the α-part, the increase in length causes the force to drop and, as a consequence, β7-β8 folds back to the β-subdomain. Thus, for the βF fluctuations, the upper state is a fully folded β-subdomain (β1-β8) and the lower state is again the residual β-core with detached β7-β8. Thus, the free energy calculation from the analysis of βF fluctuations gives us an estimate for the energy costs of the unfolding of the β7-β8 part (11 kBT) and length gain of 8 nm. For αFβF fluctuations, the total energy costs (13 kBT) are higher because, in addition to the unfolding of the β7-β8 strands, the disruption of the interface between the folded subdomains also takes place. Subdomain reorientation after physical separation and unfolding of helix A contributes to the higher length gain of αFβF fluctuations.
Peptide Binding Couples Strongly the Subdomains’ Interface, Stabilizes the β-Core.
Having structurally characterized αFβF and βF fluctuations, we returned to the WT SBD to investigate the effect of the peptide binding (19) on protein stability. For our experiments, we used so-called σ32 (19) and NR peptide (20). However, the NR peptide showed better behavior in optical tweezers assays and, hence, was used in our experiments (Fig. 3A). Constant velocity cycles of the WT SBD at 200 nm/s in the presence of 250 μM NR peptide are shown in Fig. 3B. In some of the traces (gray arrows, Fig. 3B), significantly higher unfolding forces for the β-core were observed, up to 30 pN, indicating that the peptide binds to the β-core stabilizing it (for statistics, see the gray squares in Fig. 3C, n = 87). Unfolding force distribution was bimodal with two maxima at 13 and 23 pN. In analogy to the experiments in Fig. 2, we studied the influence of substrate binding on the αFβF and βF fluctuations (Fig. 3 D and E). We picked only those traces that showed high unfolding forces (≥18 pN) of the β-subdomain as a fingerprint for the substrate bound state (gray arrows, Fig. 3B). In these events, the midpoint of the αFβF transition shifts from 11 pN to 12.5 pN, whereas only a minor shift is observed, 10.8–11.4 pN, for βF fluctuations. We calculated the free energies to be 17 kBT and 12 kBT for αFβF and βF fluctuations, respectively. In the case of the peptide-bound state, the subdomain interface is stabilized by approximately 5 kBT free energy. Because β7-β8 can be detached from the remaining core, we identify the reduced β-core (1–6) as a potentially new peptide-binding minidomain.
Fig. 3.
Peptide binding increases mechanical stability of the subdomain and tightens subdomain interface. (A) Three-dimensional structure of the SBD with the bound NR peptide (PDB ID code 4EZW; ref. 11). Schematic below illustrates the variant used for single-molecule experiments. (B) Consecutive force extension pulling curves of the SBD in the presence of 250 µM NR peptide and corresponding distribution of unfolding forces. (C) Scatter plot of unfolding forces vs. contour lengths of the β-subdomain for the apo state (empty symbols) and in the presence of the NR peptide (gray). (D and E) Block-averaged mean of the contour-length increase as a function of force for αFβF and βF type of fluctuations in the absence (open symbols) or presence (filled symbols) of the NR peptide. Solid lines are corresponding fits of a two-state open/close equilibrium model. (F) Structural interpretation (helical part in red, β-subdomain in green) of the upper (closed state) and lower states (force open state) and associated free energies with αFβF and βF fluctuations in the presence peptides.
Molecular Simulations of the Forced Unfolding of the SBD Pinpoint Mechanically Flexible Regions.
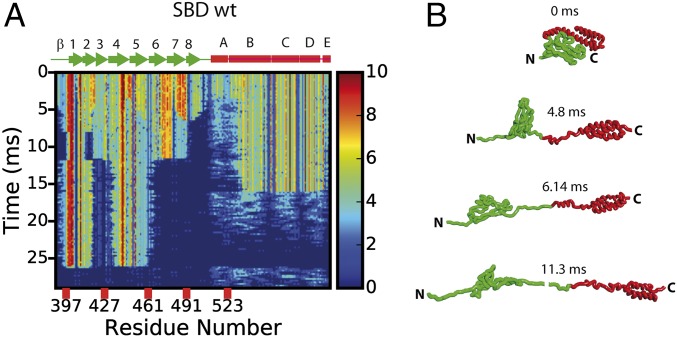
We set up SOP model (on graphics processing unit) simulations of the SBD in close correspondence to force experiments and monitored the behavior of the WT SBD and its variants. Our simulations with a pulling speed of 2,394 nm/s showed (Fig. 4) a clear hierarchy of the unfolding events of the SBD substructures. First, helix A unfolds and the loop connecting α/β subdomains stretches out, resulting in the separation of the folded subdomains (Fig. 4). Next, β8 breaks from the β-subdomain followed by the detachment of the residues 464–490 (β6 and β7 and the loop connecting them to the β-core). The remaining substructures unfold in two steps: first the α-helical part (helices B to E) then the rest of the β-core. The observed order of unfolding events in simulations resembles experimental unfolding pathway II (Movie S1). In some simulations (e.g., 1 of 25 trajectories for WT SBD; SI Appendix, Table S4), we observed unfolding of the β-core before unfolding of the α-helical part, thus the two domains in SBD have comparable stability, because either one can unfold first.
Fig. 4.
Forced unfolding simulations of the SBD at 2,394 nm/s. (A) Contact map evolution for the unfolding of the SBD (SI Appendix). (B) Conformational snapshots of intermediate structures at 0, 4.8, 6.14, and 11.3 ms. Shown is a gradual unfolding of the weakest structural elements.
The increase in contour length between the first two events of ∼14 nm corresponds to the stretching of β6–β8 and the first half of helix B. This increase is close to the 12 nm reported above from optical trap experiments and, thus, we assign it to the αFβF fluctuations. SI Appendix, Fig. S6A shows a similar ∼14 nm contour length increase for the unraveling events at the start of the run, i.e., the signature of the αFβF fluctuations. Details of the unraveling processes leading to these fluctuations from time evolution of contacts between strands in the β-subdomain are presented in SI Appendix, Fig. S8 and discussed in SI Appendix, SI Methods. Using the SBDΔα ending at 504 (SBDΔα-504) construct, we found (SI Appendix, Fig. S6B) that there is a ∼9-nm increase in contour length corresponding to the stretching of the 461–490 region (Movie S2). We identified this region as the βF fluctuations, based on the similarity in contour-length increase (8 nm) with our experiments. Steps leading to these fluctuations are presented in the time evolution of contacts between strands in SI Appendix, Fig. S9. We found a similar behavior for the SBDΔα-507 construct (SI Appendix, Fig. S13). Moreover (SI Appendix, Fig. S6D), we found evidence of the αFβF fluctuations in our simulations of the 481 to C-terminal variant, but no fluctuations in the 504 to C-terminal one (SI Appendix, Fig. S6C) in accordance with experimental results. For the 481 to C-terminal variant, steps leading to the αFβF fluctuations are detailed in SI Appendix, Figs. 6D and S10. Importantly, when pulling on the SBD-lidopen, i.e., the SBD structure from the ATP-bound DnaK state (SI Appendix, Table S4 and Fig. S14), we found evidence of the αFβF fluctuations seen in the WT SBD case, but at a lower critical force for the unraveling of the β-subdomain (SI Appendix, SI Methods).
In our simulations with the peptide, we observed only one pathway (SI Appendix, Fig. S7 and Movie S3): first the separation between the β-sandwich and the α-helical lid, accompanied by the stretching of the connector loop and unfolding of helix A. Next, β8 unfolds. In contrast to the simulation in the absence of peptide, unfolding of the helical lid occurs before the detachment of β6–β7. Finally, the β-sandwich breaks into the β6, β7, and the β-core, followed by its complete unfolding. Compared with the case without peptide, we found an increase in the critical forces for the major peaks (unfolding of α-helical lid and of the β-core), indicative of the stabilizing role of the peptide (SI Appendix, Figs. S7, S11, and S12 and Table S2). Importantly, our simulations show (SI Appendix, Fig. S12) that the peptide acts through changes in the flexibility of loops in the β-subdomain, corroborating results from earlier NMR-based studies (13, 21).
We conclude that the Achilles’ heel in the SBD is the C-terminal β-strands of the β-subdomain and helix A.
Discussion
Bifurcating Unfolding Pathways for SBD and Mechanical Hinge Regions.
To identify individual mechanical parts of the SBD in an unbiased approach, we applied single molecule force spectroscopy using optical tweezers. We found that the SBD consists of two largely independent stable nanomechanical units, consisting of the structural α- and β-subdomains, and a stable hinge within the β-subdomain formed by the terminal β-sheets. Both α and β subdomains as mechanical units impose roughly similar energy barriers against unfolding, which result in bifurcating pathways. Hence, after applying the force at the N and C terminus of the SBD, the protein unfolds through parallel unfolding reactions starting with either the β-subdomain (68% pathway I) or the α (32%, pathway II). The largely independent subdomains and bifurcating pathways indicate that the subdomain interface contributes negligibly to the overall SBD stability. This behavior contrasts with the N-terminal domain of DnaK, the NBD, which unfolds highly cooperatively because its subdomain interface is strongly stabilized by a C-terminal helix. In the case of the SBD, stable subdomains are connected only by a weak interface that easily breaks, leading to splitting subdomains and, hence, to lower unfolding cooperativity. We speculate that a high unfolding cooperativity in the SBD with a strong connecting interface between subdomains would inhibit large conformational motions that are crucial for its function. Additionally, independently stable subdomains of the SBD are necessary to secure a properly folded structure in the open state, in which the subdomains are far apart.
During mechanical separation of the subdomains, the protein undergoes rapid equilibrium fluctuations (αFβF and βF) between closed and a force-induced open state. Uncovering the regions of the protein involved in these fluctuations under load is not straightforward because force propagates through the entire protein structure. A key advantage of single molecule force spectroscopy is its potential to select specific regions of force application. The comparison of three different variants with different attachment points allowed us to unequivocally identify β7–β8 strands as the critical hinge elements involved in the fluctuations. In our SOP simulations, an additional unfolding of β6 was observed, which we cannot completely rule out from happening in our pulling experiments as well (see SI Appendix, Figs. S5 and S6 for further discussion and comparison for the case of WT SBD). In such a case, this strand plays a passive role during the unfolding of the critical hinge elements formed by β7–β8.
We found that under force, the C-terminal strands can reversibly attach and detach from the β-core at a significant energy cost of 12 kBT. After the unraveling of these β-strands, the β-core behaves like a single cooperative unit. Such dynamics within the β-subdomain are unexpected given the compact appearance of 3D structure of the SBD. In the presence of the folded α-domain, breaking the interface between subdomains requires 2 kBT of free energy (Fig. 5). This “interface free energy” may be stored in the folded helix A and/or in direct interactions between the subdomains. Thus, this energy is a measure of the interaction between the folded subdomains in the closed state of the SBD. Using the interface free energy, we can calculate a relative occurrence of the corresponding states at zero force: the fully closed state is populated to approximately 88%, whereas the state with a disturbed subdomain interface is populated to approximately 12%. It indicates that the later state (presumably the “open state”) is populated even at equilibrium, which might be detectable by sensitive methods. Single-molecule FRET experiments (22) showed that the open SBD form is present (8–11%) under ADP conditions, supporting our results. A weak interaction between the α/β-folded subdomains and physical separation of subdomains was also observed in native-state molecular simulations by using the replica exchange method or following the dynamics of DnaK over tens of nanoseconds of equilibrium molecular dynamics (23, 24). As pointed out above, the subdomain interface is easily perturbed allowing the opening of the lid, whereas β7–β8 are still attached to the β-core, which reduces energy costs.
Fig. 5.
Schematics of nucleotide and force-induced conformational changes of the SBD in the absence or presence of peptide substrate. The Hsp70 chaperone DnaK undergoes large ATP-dependent conformational changes leading to the open state of the SBD (green color corresponds to the β-subdomain, red color corresponds to the α-subdomain). During this transition, the interface breaks and α/β subdomains move apart. In our approach, the interface is mechanically broken and subdomains are physically separated, which leads to the force-induced open states. The states shown in squared brackets []* are not observed directly but can be postulated based on the reference states obtained from αFβF and βF fluctuations.
Force Triggers Transitions Between High- and Low-Affinity States of the Single SBD Molecule.
Switching between low- and high-affinity states for substrate peptides is key for the function of Hsp70 chaperones (9). The closed state binds firmly to peptide clients to prevent aggregation, whereas the open state must release the peptides to allow them to fold (5). The free energy stabilization of the closed vs. open lidless state should be directly associated with the differences in peptide affinity of the closed state and ATP-induced open state in bulk assays (25, 26) (1 and 86 µM, respectively). In fact, the ratio of those affinities:
agrees with our analysis (5 kBT). Assuming that the closed states are identical, this analysis indicates that the absolute KD value for the force-open state is similar to the ATP-induced open state. Obviously, the peptide secures the lid domain to the β-core, which results in a decrease of the relative population of the open state to a negligible level.
Peptide-Induced Lid Closure Occurs as Brownian Ratchet Motion.
Large allosteric shape changes of proteins, including action of molecular motors as a prominent example, are often discussed by using energy landscape concepts that distinguish between lever-arm type active motions of the protein and passive so-called Brownian ratchet models. We propose to apply such a picture also to the process of peptide-induced lid closing. In the case of the SBD of DnaK, an active motion would imply that peptide binding actively drives the lid from an open into a closed conformation through rotation of the hinge elements (“lever arm”) (i.e., β7–β8). A ratchet model, however, would imply that peptide binding locks the interface between lid and core as soon as they form physical contact but does not drive the elements β7–β8 involved in the transition actively. Our results favor a ratchet-type closing of the lid because β7–β8 docking energies are independent of bound substrate, hence substrates do not drive active docking elements. This ratchet-type mechanism means that thermal motions agitate the SBD until the lid touches the β-subdomain, and the interface energy locks it in the closed conformation.
Implications for the Energetics of Lid Opening During ATP-Driven Conformational Changes of Hsp70.
How do the states we induce by force relate to the open states populated in the active cycle of Hsp70? In the crystal structure of ATP-bound DnaK, the SBD shows a complete breakdown of the α/β interface and reorientation of β8 (see schematics in Fig. 5). At the same time, our mechanical experiments and SOP simulations identified the subdomain interface and terminal β-strands of the β-subdomain as the most mechanically vulnerable and, hence, easily deformable parts of the SBD. Thus, internal nanomechanics and energetics of the SBD determine its allosteric ATP-induced conformational change in the full-length DnaK (see SI Appendix, SI Methods, Section 2.10. Discussion of Simulation Results). A comparison of the ATP and force-induced open structures shows that the open states are not absolutely identical: In the force-induced open state, strands 7–8, helix A and part of helix B are completely detached and unfolded, whereas in the ATP-induced open state those elements are folded and docked. Despite the differences between the force-induced and ATP-induced open states, the magnitude of the energy penalty for the ATP-induced opening of the SBD can be estimated. In the absence of peptide, the minimum energy penalty is 2 kBT, which is necessary for the disruption of the interface and a maximum penalty 13 kBT, which is required for the complete detachment of the β7–β8 strands. In the presence of the peptide, the energy penalties are higher: 4 kBT for the opening of the closed form and 17 kBT for the complete unraveling of the β7–β8. Thus, peptide binding increases the energy penalty for the conformational change and, thus, opposes the lid-opening reaction that conceptually agrees with the tug-of-war picture (27). From an energy-balance point of view, even the drastic conformational changes with β7–β8 completely detached as seen in force experiments could be driven by ATP hydrolysis. Even the high free-energy input of 17 kBT required in the peptide-bound state is well below the 25 kBT estimated for the energy available from a single ATP hydrolysis cycle under cellular conditions (28).
Methods
Experimental Procedures.
For cloning, protein purification, and design, see SI Appendix, SI Methods. Functionality of the variants were tested by using a σ32 peptide binding assay (SI Appendix, SI Methods). The experimental setup used for optical trapping consists of custom-built high-resolution dual trap optical tweezers with back-focal plane detection as described (18, 29).
SOP Model.
We simulated the mechanical response of the SBD of Hsp70 by using the topology-based SOP model (16) starting from the Cα positions of each amino acid residue in the respective PDB entries [ID codes 2KHO (30) for the apo state, 4EZW for the NR peptide bound state (11), 4B9Q (8) for the lid open ATP state]. Brownian dynamics simulations followed the behavior of the chain under force applied by using constant loading rate conditions. The details are provided in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank the members of our group for suggestions and Dr. Kasia Tych for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft projects SFB 1035/A5 and ZO 291/1-1 projects (to M.R. and G.Ž.) and in part by National Science Foundation Grant MCB-1412183 (to R.I.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619843114/-/DCSupplemental.
References
- 1.Bauer D, et al. Nucleotides regulate the mechanical hierarchy between subdomains of the nucleotide binding domain of the Hsp70 chaperone DnaK. Proc Natl Acad Sci USA. 2015;112:10389–10394. doi: 10.1073/pnas.1504625112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelz B, Žoldák G, Zeller F, Zacharias M, Rief M. Subnanometre enzyme mechanics probed by single-molecule force spectroscopy. Nat Commun. 2016;7:10848. doi: 10.1038/ncomms10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müller DJ, Dufrêne YF. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat Nanotechnol. 2008;3:261–269. doi: 10.1038/nnano.2008.100. [DOI] [PubMed] [Google Scholar]
- 4.Calloni G, et al. DnaK functions as a central hub in the E. coli chaperone network. Cell Reports. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Clerico EM, Tilitsky JM, Meng W, Gierasch LM. How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J Mol Biol. 2015;427:1575–1588. doi: 10.1016/j.jmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi R, et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol. 2013;20:900–907. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell. 2012;48:863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 9.Nunes JM, Mayer-Hartl M, Hartl FU, Müller DJ. Action of the Hsp70 chaperone system observed with single proteins. Nat Commun. 2015;6:6307. doi: 10.1038/ncomms7307. [DOI] [PubMed] [Google Scholar]
- 10.Stevens SY, Cai S, Pellecchia M, Zuiderweg ER. The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393-507) in complex with the peptide NRLLLTG. Protein Sci. 2003;12:2588–2596. doi: 10.1110/ps.03269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahn M, et al. Structural studies on the forward and reverse binding modes of peptides to the chaperone DnaK. J Mol Biol. 2013;425:2463–2479. doi: 10.1016/j.jmb.2013.03.041. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, et al. NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: A preview of chaperone-protein interaction. Biochemistry. 1998;37:7929–7940. doi: 10.1021/bi9800855. [DOI] [PubMed] [Google Scholar]
- 13.Pellecchia M, et al. Structural insights into substrate binding by the molecular chaperone DnaK. Nat Struct Biol. 2000;7:298–303. doi: 10.1038/74062. [DOI] [PubMed] [Google Scholar]
- 14.Leu JI, Zhang P, Murphy ME, Marmorstein R, George DL. Structural basis for the inhibition of HSP70 and DnaK chaperones by small-molecule targeting of a C-terminal allosteric pocket. ACS Chem Biol. 2014;9:2508–2516. doi: 10.1021/cb500236y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyeon C, Dima RI, Thirumalai D. Pathways and kinetic barriers in mechanical unfolding and refolding of RNA and proteins. Structure. 2006;14:1633–1645. doi: 10.1016/j.str.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Stigler J, Ziegler F, Gieseke A, Gebhardt JC, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 18.Žoldák G, Stigler J, Pelz B, Li H, Rief M. Ultrafast folding kinetics and cooperativity of villin headpiece in single-molecule force spectroscopy. Proc Natl Acad Sci USA. 2013;110:18156–18161. doi: 10.1073/pnas.1311495110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarty JS, et al. Regulatory region C of the E. coli heat shock transcription factor, sigma32, constitutes a DnaK binding site and is conserved among eubacteria. J Mol Biol. 1996;256:829–837. doi: 10.1006/jmbi.1996.0129. [DOI] [PubMed] [Google Scholar]
- 20.Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman ME. Specificity of DnaK-peptide binding. J Mol Biol. 1994;235:848–854. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- 21.Zhuravleva A, Gierasch LM. Substrate-binding domain conformational dynamics mediate Hsp70 allostery. Proc Natl Acad Sci USA. 2015;112:E2865–E2873. doi: 10.1073/pnas.1506692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kudryavtsev V, et al. Combining MFD and PIE for accurate single-pair Forster resonance energy transfer measurements. Chemphyschem. 2012;13:1060–1078. doi: 10.1002/cphc.201100822. [DOI] [PubMed] [Google Scholar]
- 23.Gołaś E, et al. Simulation of the opening and closing of Hsp70 chaperones by coarse-grained molecular dynamics. J Chem Theory Comput. 2012;8:1750–1764. doi: 10.1021/ct200680g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiappori F, Merelli I, Milanesi L, Colombo G, Morra G. An atomistic view of Hsp70 allosteric crosstalk: From the nucleotide to the substrate binding domain and back. Sci Rep. 2016;6:23474. doi: 10.1038/srep23474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buczynski G, Slepenkov SV, Sehorn MG, Witt SN. Characterization of a lidless form of the molecular chaperone DnaK: Deletion of the lid increases peptide on- and off-rate constants. J Biol Chem. 2001;276:27231–27236. doi: 10.1074/jbc.M100237200. [DOI] [PubMed] [Google Scholar]
- 26.Slepenkov SV, Witt SN. Kinetic analysis of interdomain coupling in a lidless variant of the molecular chaperone DnaK: DnaK’s lid inhibits transition to the low affinity state. Biochemistry. 2002;41:12224–12235. doi: 10.1021/bi0263208. [DOI] [PubMed] [Google Scholar]
- 27.Zhuravleva A, Clerico EM, Gierasch LM. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151:1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard J. Mechanics of Motor Proteins and the Cytoskleton. Sinauer Associates; Sunderland, MA: 2001. [Google Scholar]
- 29.von Hansen Y, Mehlich A, Pelz B, Rief M, Netz RR. Auto- and cross-power spectral analysis of dual trap optical tweezer experiments using Bayesian inference. Rev Sci Instrum. 2012;83:095116. doi: 10.1063/1.4753917. [DOI] [PubMed] [Google Scholar]
- 30.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci USA. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.