Abstract
Sea stars and sea urchins are model systems for interrogating the types of deep evolutionary changes that have restructured developmental gene regulatory networks (GRNs). Although cis-regulatory DNA evolution is likely the predominant mechanism of change, it was recently shown that Tbrain, a Tbox transcription factor protein, has evolved a changed preference for a low-affinity, secondary binding motif. The primary, high-affinity motif is conserved. To date, however, no genome-wide comparisons have been performed to provide an unbiased assessment of the evolution of GRNs between these taxa, and no study has attempted to determine the interplay between transcription factor binding motif evolution and GRN topology. The study here measures genome-wide binding of Tbrain orthologs by using ChIP-sequencing and associates these orthologs with putative target genes to assess global function. Targets of both factors are enriched for other regulatory genes, although nonoverlapping sets of functional enrichments in the two datasets suggest a much diverged function. The number of low-affinity binding motifs is significantly depressed in sea urchins compared with sea star, but both motif types are associated with genes from a range of functional categories. Only a small fraction (∼10%) of genes are predicted to be orthologous targets. Collectively, these data indicate that Tbr has evolved significantly different developmental roles in these echinoderms and that the targets and the binding motifs in associated cis-regulatory sequences are dispersed throughout the hierarchy of the GRN, rather than being biased toward terminal process or discrete functional blocks, which suggests extensive evolutionary tinkering.
Keywords: Tbrain, echinoderm, binding site affinity, ChIP-seq
One of the most striking revelations that arose from the emergence of the field of “EvoDevo” in the late 1980s was the concept of functional equivalence [i.e., that orthologous transcription factors (TFs) from vastly different species will functionally compensate for each other when experimentally substituted in vivo]. This concept implies that the function of these orthologous proteins has remained essentially unchanged over hundreds of millions of years. The experimental observations that demonstrated this concept (e.g., refs. 1 and 2) fit neatly with theoretical predictions that TFs, which regulate many target genes in multiple spatiotemporal contexts, will be highly constrained, and therefore any change in function will have wide-sweeping changes that are unlikely to pass the filter of natural selection (3). Thus, it is argued, changes in developmental gene regulatory networks (GRNs) must arise from alterations to the DNA of the cis-regulatory targets bound by these factors. In other words, the biochemical function of a TF (i.e., the preferred sequence motif to which the factor binds) remains conserved, whereas its developmental function, the genes that are regulated, evolves more rapidly through cis-DNA evolution of the target genes.
Modern and genome-wide assessments have largely borne out the results of these earlier experiments and theoretical predictions (4, 5). However, there has been a recent and growing interest in understanding how TFs might evolve changed biochemical functions (6). Conceptually, this investigation must entail understanding how these proteins evolve changes that limit the dramatic effects of pleiotropy and can include evolving differences with small phenotypic effects and independently changing individual subsets, or modules, of functions (7).
A recently recognized, and surprising, source of modularity has been DNA binding itself. The potential prevalence of this source of variation was realized only after protein-binding microarray technology, which universally assesses DNA-binding (8, 9), provided a high-resolution description of binding-site preferences. Studies using this technology have shown that many TF binding motif preferences can be described by multiple position weight matrices (PWMs). These PWMs are commonly called primary and secondary motifs, where the primary is the most preferred and often higher-affinity motif. An increasing number of studies are showing that low-affinity secondary sites can execute particular functions, including providing specificity, controlling the timing of gene expression, and even mediating activation vs. repressive regulation (10–12). Recent work from our laboratory has shown that Tbrain (Tbr), a T-box TF, has evolved modular preferences for binding sequence recognition (13). We showed that the primary motif recognized by the sea star (Patiria miniata) and sea urchin (Strongylocentrotus purpuratus) Tbr proteins were extraordinarily similar, and this similarity extended even to the mouse ortholog Eomesodermin. Sea stars and sea urchins are representatives of two classes of echinoderms, which last shared an ancestor >450 million years ago in the early Ordovician (14), whereas the common ancestor of echinoderms and mice sits at the base of all deuterostomes. Our finding, therefore, is in keeping with the theoretical prediction that highly pleiotropic factors are constrained from evolving changed motif preferences over even immense evolutionary distances. Importantly, however, we also showed that the factors from mouse and sea star recognized different secondary motifs, whereas the sea urchin protein showed no preference for any secondary motif. These results therefore demonstrate that these proteins have evolved a modular change of preference for a low-affinity binding motif.
The next challenge is to understand the developmental consequences of this modular use of evolvable binding motifs in these taxa. Sea urchin and sea star tbr genes have been partially functionally characterized (15–17), but we have no genome-wide understanding of their function. Therefore, we first determined the developmental function of Tbr in these taxa and asked whether these factors are indeed highly pleiotropic, whether they share developmental functions, and how these functions might fit within a developmental GRN. As a measure of developmental function, we assessed the binding of Tbr factors genome-wide in both the sea star and sea urchin at comparable developmental stages and identified genes likely to be regulated by Tbr at these sites. We then sought to determine the distribution of primary and secondary motif use associated with these taxa and the functions of identified genes. This approach allowed us to assess the evolution of the developmental role of Tbr in these taxa and to understand how this developmental function then interfaces with the evolution of the biochemical function of motif use.
Results
Tbr function in sea urchins is reasonably well understood, because the factor acts within the GRN for specification of the primary mesenchyme lineage, which is considered the most well-described GRN for the specification of any cell type (15). In the sea urchin, tbr is localized exclusively to this lineage (ref. 18 and Fig. 1). However, there has been no systematic assessment of Tbr binding sites, and there is no cis-regulatory module (CRM) yet known to be directly bound by Tbr. These data are needed to understand the intersection of binding site and developmental function of Tbr. Therefore, we used ChIP-sequencing (ChIP-seq) to identify S. purpuratus Tbr (SpTbr) binding sites genome-wide. Whole-mount immunofluorescence (IF) using our specific antibody (13) revealed that SpTbr is localized to primary mesenchyme cells (PMC) at 24 h (Fig. 1B), which is the same cell population that expresses the Sp-tbr transcript (18), although the transcripts are known to be maternally deposited. ChIP-seq was performed by using this antibody to specifically enrich SpTbr-bound chromatin at this stage. Resulting reads were mapped to the genome, and peaks were called by MACS2 (Materials and Methods; see Fig. S1 for experimental workflow). In total, we found 3,149 peak regions that were enriched by SpTbr ChIP across the genome compared with the chromatin-only input control.
Fig. 1.
Anti-SpTbr antibody validation. (A) WMISH for Sp-tbr in S. purpuratus at 24 hpf showing expression localized to the ingressed primary mesenchyme cell population. (B) IF localization of SpTbr in an S. purpuratus embryo; staining is also observed in the primary mesenchyme cells. (C) Example data from the SpTbr ChIP-seq experiments showing the Sp-Nrl locus. The top track shows regions called as SpTbr ChIP peaks by MACS2 analysis, where the color intensity of the box corresponds to peak fold enrichment. The two tracks in gray are bedgraph data output from MACS2 showing sequence tag pileups from ChIP and local lambda from input chromatin datasets. The blue bed track represents consensus ATAC-seq peaks for three replicate 24-hpf datasets (hosted at Echinobase.org), and the bottom track is the annotated S. purpuratus gene models. The target gene is highlighted by a red line. (Scale bar: 10 kb.) The area surrounding the detected ChIP peaks, indicated by a square bracket, is expanded in D to show finer resolution of ChIP and ATAC intersection. (Scale bar: 1 kb.)
Fig. S1.
Flowchart describing filtering processes applied to ChIP-seq datasets. PmTbr ChIP and input alignments were filtered to remove any that overlapped predicted repetitive elements, and then alignments were used as input to MACS2 for peak detection. MACS parameters were empirically optimized for each dataset. Resultant SpTbr peaks were further filtered to include only peaks within 75 kb of an annotated gene and overlapping consensus ATAC-seq peaks from 24 hpf. PmTbr peaks were filtered based on proximity to genes found to be sensitive to PmTbr knockdown. Finally, the set of orthologous genes where the ortholog in each species is associated with a peak in the respective peak set were defined.
Targets of Tbr in sea urchin have been detected by performing quantitative PCR and whole-mount in situ hybridization (WMISH) to analyze the effects of Sp-tbr morpholino antisense oligonucleotide (MASO) knockdown (15, 16). Known targets include Sp-Nrl, Sp-FoxN2/3, Sp-FoxB, Sp-Erg, and Sp-Msp130, which are curated within the PMC-GRN, as well as Sp-Nebnph, Sp-Lisp1, Sp-C-lectin/PMC, SPU_018403, and Sp-Hypp_2998. For 8 of these 11 targets, SpTbr ChIP peaks are detected within 75 kb of the gene (Table 1). In several cases, multiple genes that are clustered in the genome share a single peak. Peaks associated with several genes also overlap with assay for transposase-accessible chromatin-sequencing (ATAC-seq) chromatin accessibility peaks from similar-stage embryos (Table 1, Fig. 1 C and D, and Fig. S2) (see below). Although there is a MACS called peak on the same genomic scaffold as Sp-FoxB, it is 281 kb away from the body of the gene and, therefore, is less likely to represent a functional interaction between the binding site and the gene. This finding supports an indirect effect of SpTbr on Sp-FoxB expression, as suggested by the data supporting the GRN model. The only gene from the developmental GRN without an SpTbr ChIP-seq peak detected on the same scaffold is Sp-Erg. The identification of significantly enriched peaks near known SpTbr target genes supports the conclusion that the ChIP-seq protocol is capable of detecting genomic targets of SpTbr binding.
Table 1.
ChIP-seq peaks near predicted targets of SpTbr
| Glean ID | Gene name | Peak_ID | ATAC overlap | Distance, kb |
| SPU_013698* | Sp-Nrl | tbr_macs_peak_2546 | Yes | 72 |
| SPU_015243* | Sp-FoxN2/3 | tbr_macs_peak_2415 | Yes | 63 |
| SPU_002088* | Sp-Msp130 | tbr_macs_peak_1696 | No† | 36 |
| SPU_013821* | Sp-Msp130_1 | 34 | ||
| SPU_004551* | Sp-FoxB | (tbr_macs_peak_3011) | (No) | (281) |
| SPU_018483* | Sp-Erg | — | — | — |
| SPU_009123‡ | Sp-Nebnph | tbr_macs_peak_2337 | No | 48 |
| SPU_009124‡ | Sp-Lisp1 | 28 | ||
| SPU_018403‡ | None | tbr_macs_peak_977 | Yes | 18 |
| SPU_018407‡ | Sp-Hypp_2998 | 35 | ||
| SPU_027906‡ | Sp-C-lectin/PMC1 | — | — | — |
Genes predicted to be targets of SpTbr either from the curated sea urchin developmental GRN dataset or from Rafiq et al. (16) were surveyed to determine whether a SpTbr ChIP peak was detected on the same scaffold. If a detected SpTbr ChIP peak was detected, the presence of an overlapping ATAC-seq peak was scored, and the distance from the peak to the body of the gene is reported. In several cases, multiple genes shared a single peak (e.g., Sp-Msp130 and Sp-Msp130_1 or Sp-Nebnph and Sp-Lisp1). These genes are grouped together by like shading; the peak ID is reported once, and the distance from the peak to each gene is indicated. Genes in this set for which no peak is detected are marked with —. The peak associated with Sp-FoxB is indicated in parentheses because its extreme distance to the body of the gene—281 kb—makes it unlikely that this peak is involved in the direct regulation of Sp-FoxB.
SpTbr targets predicted from PMC-GRN.
Although there is no ATAC-seq peak overlapping the SpTbr peak at the Sp-Msp130 locus, there is an ATAC peak immediately adjacent, as shown in Fig. S2.
SpTbr targets predicted by Rafiq et al. (16).
Fig. S2.
SpTbr ChIP-seq peaks in regions proximal to known targets of SpTbr. All loci shown represent those summarized in Table 1. For each locus, the top track shows regions called as SpTbr ChIP peaks by MACS2 analysis, where the color intensity of the box corresponds to peak fold-enrichment. The two tracks in gray are bedgraph data output from MACS2 showing sequence tag pileups from ChIP and local lambda from input chromatin datasets. The blue bed track represents consensus ATAC-seq peaks for three replicate 24-hpf datasets (hosted at Echinobase), and the bottom track is the annotated S. purpuratus gene models. The target gene is highlighted by a red line. (Scale bar: 10 kb.) The area surrounding the detected ChIP peaks, indicated by a square bracket, is expanded to the right to show finer resolution of ChIP and ATAC intersection. (Scale bar: 1 kb.)
To reduce potential false-positive peaks from our analysis, we additionally restricted our peak set to those associated with an annotated gene. It is not uncommon, in the sea urchin genome, for characterized CRMs to be 10 kb or more from the coding gene, both upstream and downstream of the transcription start (e.g., refs. 19 and 20). Given that the majority of predicted SpTbr targets for which a ChIP peak is detected are observed within 75 kb of that peak (Table 1), we elected to further filter the SpTbr ChIP peaks to those within 75 kb of a gene annotation. This distance corresponds to approximately three times the average intergenic distance predicted for the sea urchin genome (21). This method further filtered the set of SpTbr peaks to 1,952 (61.9% of all peaks) (Fig. S1). This approach may also eliminate genuinely functional peaks, but because our goal is to associate peaks with putative function, those not obviously associated with a gene are not required in our later analyses.
To further corroborate these peaks, we used unpublished, but publicly accessible, ATAC-seq chromatin accessibility data. The ATAC assay identifies open chromatin based on the propensity for in vitro transposase integration genome-wide. These data are available as JBrowse tracks at Echinobase.org (22), representing MACS2 called peaks of ATAC-based chromatin accessibility data from three replicate measurements of embryos at 24 hours post fertilization (hpf). We used the regions common to all three replicate ATAC samples and filtered the detected 3,149 peaks for only those that overlap the consensus region of ATAC sensitivity. A total of 1,542 SpTbr peaks (49.0%) overlap ATAC peaks. This fraction of ChIP overlap with chromatin accessibility is comparable to what has been reported in the literature for other organisms (e.g., ref. 23). In total, 1,492 SpTbr peaks both overlap an ATAC peak and are positioned within 75 kb of an annotated gene (Fig. S1). These peaks now represent the highest-stringency set that we use for further analyses.
To predict the function of SpTbr genome-wide, we next performed Gene Ontology (GO) term enrichment analysis based on the annotation of the 1,052 genes that are closest to the 1,492 SpTbr peaks. The most significantly enriched terms among these genes were found to be “nucleic acid binding TF activity” (63 genes, P = 2.3e−22), “MAP kinase phosphatase activity” (5 genes, P = 1.3e−4), “proteoglycan binding” (3 genes, P = 0.02), and “regulation of signal transduction” (10 genes, P = 0.04) (Fig. 2 and Dataset S1). These enrichments highlight the known roles of SpTbr in regulating both gene expression and cell signaling, both central mechanisms of information flow through GRNs. Indeed, the genes identified are significantly enriched for genes present in the current GRN model (49 genes, hypergeometric P = 2.3e−45). The MAPK pathway, in particular, is known to be critical for PMC ingression and patterning during the stage sampled here (24, 25), although to our knowledge Tbr has not been previously been implicated as a direct regulator of this pathway. It has been demonstrated, however, that Tbr is required for the epithelial–mesenchymal transition associated with PMC ingression (26). These data suggest the potential for regulatory feedback between MAPK signaling and PMC-GRN regulatory genes, where, for example, SpTbr may regulate aspects of the MAPK pathway that, in turn, have been shown to phosphorylate and regulate SpEts1 at the top of the network and act as a regulatory input to SpTbr in PMCs (25, 26).
Fig. 2.
GO term enrichments for Tbr target genes. The significantly enriched GO terms for each set of targets is reported as a point where the color intensity corresponds to the corrected hypergeometric P value for that enrichment, and the area corresponds to the percent of all genes annotated with that term identified among the target set. (A) GO term enrichments for targets of SpTbr peaks, including all peaks as well as the subsets of peaks containing primary motifs (1° peak) or secondary motifs (2° peak). (B) GO term enrichments for PmTbr targets, including genes that are differentially expressed after Tbr knockdown (All DEG), DEGs that are up- or down-regulated (DEG [up] and DEG [down], respectively), DEGs that have a PmTbr peak assigned (DEG + peak), and DEGs with assigned peaks that contain primary motifs (1° peak) or secondary motifs (2° peak). (C) GO term enrichments for the 108 orthologous target genes found to be regulated by Tbr in both sea urchin and sea star datasets. These are subsetted to indicate whether the assigned peak in each dataset contains primary motifs (SpTbr 1° peak and PmTbr 1° peak) or secondary motifs (SpTbr 2° peak and PmTbr 2° peak).
Having defined a set of high-confidence SpTbr binding sites genome-wide and associations with adjacent genes in sea urchins, we next shifted our focus to ascertaining the targets of sea star Tbr (P. miniata Tbr; PmTbr). Gastrulation in the sea star begins with the invagination of the vegetal plate (there is no ingressing primary mesenchyme) at ∼30 hpf (27). At this stage, PmTbr is localized throughout endomesoderm, as well as at lower levels throughout the ectoderm (13), and therefore the ortholog is distributed far more broadly within the sea star than the sea urchin embryo.
The sea star anti-Tbr antibody was generated against a peptide specific to PmTbr, as described in ref. 13. Our initial analysis of PmTbr ChIP-seq peaks revealed a statistical overrepresentation of predicted repetitive elements (details in SI Materials and Methods). Sequencing reads that aligned to predicted RTE-2–like elements, which conspicuously and significantly overlapped with the initial peak set, were removed before subsequent peak detection. Ultimately, 13,977 peaks were called by MACS2 using the filtered read set, of which 9,164 (65.6%) were within 75 kb of an annotated gene. We detected more peaks in the PmTbr ChIP-seq than in the SpTbr ChIP-seq (9,164 vs. 1,952), although a similar percentage were associated with a gene (65.6% vs. 62.0%). This observation is consistent with the larger domain of PmTbr in sea stars, throughout the endomesoderm and ectoderm, compared with the highly restricted localization of SpTbr to the small number of PMCs in sea urchins.
The only previously known direct target of PmTbr is Pm-Otx (28). The previously functionally characterized CRM is completely overlapped by a significant MACS-called ChIP-seq peak (Fig. 3A). Furthermore, Pm-Delta has previously been shown to be sensitive to PmTbr morpholino knockdown, and in the ChIP-seq data, we found a significant MACS-called PmTbr peak 11 kb away from the Pm-Delta gene (Fig. 3B). Although there are fewer known targets for PmTbr in sea star compared with sea urchin, we found ChIP peaks proximal to both known targets, indicating that the PmTbr ChIP-seq dataset is capable of detecting genomic binding sites of PmTbr.
Fig. 3.
Known PmTbr targets have proximal ChIP peaks. For both the Pm-Otx locus (A) and Pm-Delta locus (B), the top track shows regions called by MACS2 as PmTbr ChIP peaks, and corresponding ChIP sequence tag pileup and local lambda pileup (input) are shown as bedgraph tracks in gray. The bottom track shows annotated genes in the locus with the known target of PmTbr indicated by a red line. For each locus, an expansion of the area around the detected peak, indicated by a square bracket, is shown to the right of the vertical black line. The position of the previously described Pm-Otx CRM is indicated by the red box.
In the absence of available sea star ATAC-seq data to corroborate peaks, and given the limited known functions of sea star Tbr, we performed RNA-sequencing (RNA-seq) on Pm-tbr knockdowns to enable further filtering of PmTbr peaks for functionality. We used RNA collected at early gastrula from Tbr antisense morpholino injected embryos (anti-Tbr MASO) compared with control morpholino injected (control). Genes significantly differentially expressed [false discovery rate (FDR) < 0.05] between control and anti-Tbr MASOs in biological triplicate were detected. In total 2,562 genes (9.3% of all expressed genes) were found to be significantly differentially expressed and 1,165 of these (45.5%) were found to be within 75 kb of a PmTbr ChIP-seq peak (1,105 peaks) (Fig. 4 and Fig. S1). This fraction of differentially expressed genes (DEGs) associated with ChIP peaks is within the range of what has been observed in other model systems (29, 30). Importantly, when we considered the union of DEGs with an associated peak we found both up- and down-regulated genes; 676 of 1,521 (44.4%) significantly up-regulated genes and 489 of 1,041 (47.0%) significantly down-regulated genes had an associated ChIP peak. This finding is strong evidence that PmTbr has the potential to act as both a direct repressor and an activator. This activity is consistent with known dual repressor/activator functions in related Tbox genes from other taxa (31, 32).
Fig. 4.
Identification of targets of PmTbr by integration of ChIP-seq and RNA-seq data. Each point represents a gene detected in the Pm-Tbr MASO RNA-seq experiment, and the fold change for each gene is plotted against the calculated FDR. Significantly, DEGs are further annotated to indicate whether a corresponding ChIP peak was detected in the PmTbr ChIP-seq dataset (red points), where the intensity of the point color corresponds to the FDR of the associated peak; color scale is indicated on bottom key. Finally, any significant DEG that has a sea urchin ortholog with a peak detected within 75 kb is indicated as a black point. Points where both the sea star and sea urchin ortholog have an associated peak are colored red with a black point in the center. Several genes of interest from this category are labeled.
We examined the previously characterized targets of PmTbr (i.e., Pm-Otx and -Delta) and found that neither were significantly differentially expressed. Pm-Otx was found to be 1.3-fold down relative to the control, and Pm-Delta was found to be 1.7-fold down. The directionality of both of these genes was consistent with previous data, even though the changes were not statistically significant by RNA-seq. Thus, our RNA-seq analysis at this time point may miss some target genes we expect are regulated by PmTbr, but we chose to use the DEGs to help filter detected peaks because, again, we sought to minimize the potential for false positives among identified peaks.
To assess GO term enrichment to explore the function of PmTbr genome-wide, GO annotations were mapped from sea urchin genes to predicted sea star orthologs. First, likely sea star orthologs of sea urchin genes were predicted by using a reciprocal best blast hits (BBH) method. We examined term enrichments among the sea urchin orthologs to genes of interest from the sea star dataset—both the 2,562 genes that significantly changed after Pm-tbr knockdown, as well as the subset of 1,165 DEGs that also have a PmTbr ChIP peak detected within 75 kb. In general, GO term enrichments from the sea star dataset were less robust, owing to the fact that only a fraction of the genes examined (35–41%) have a reciprocal BBH mapping that informs the annotation of the sea star genes. Nonetheless, these enrichments suggest both conserved and divergent functions for Tbr in these two species. For example, the set of DEGs proximal to PmTbr ChIP peaks were enriched for some of the same ontology terms as was found in the sea urchin set—including “sequence-specific DNA binding TF activity” (17 genes, P = 0.069). The MAP kinase-associated terms were not found to be enriched in the sea star set, and there is no evidence to suggest that the MAP kinase pathway is used during sea star gastrulation. Additionally, there were several other terms enriched in the sea star set, not found in the sea urchin set, including “scavenger receptor activity” (six genes, P = 0.023), “apoptotic process” (six genes, P = 0.14), “cell adhesion” (six genes, P = 0.35), and “aminoacyl-tRNA ligase activity” (seven genes, P = 0.069) (Fig. 2 and Dataset S1). These terms are all more significantly enriched (all P < 0.05) among genes that were up-regulated by Pm-tbr knockdown (i.e., those genes that PmTbr would be predicted to repress), whereas there were no significant term enrichments for genes down-regulated by the Pm-tbr MASO knockdown. Thus, genes up-regulated by Pm-tbr knockdown are more functionally coherent, further supporting the hypothesis that PmTbr functions as a repressor.
We next compared binding site motif utilization in peaks from both species. We have previously defined the PWMs for Tbr primary and secondary site motifs in vitro. Although the sea star and sea urchin primary site PWM are quite similar, only sea star Tbr has an enhanced preference and affinity for the defined secondary site motif. We used the Finding Individual Motif Occurrences (FIMO) tool from the MEME-suite to scan both peak sets for the presence of the primary and secondary site motifs, and overlapping motif occurrences were filtered such that only the most significant motif was reported for each position. This filtering step is important because the primary and secondary site PWMs are partially overlapping, and frequently we would find both primary and secondary sites called at one position within a peak, although one with a much higher score and associated P value. Each peak was then classified based on the presence or absence of each motif. Although the SpTbr protein has a very low affinity for the secondary motif in vitro, the protein is nonetheless apparently capable of binding to sites with this motif in vivo, because we found several hundred SpTbr peaks in which only this motif was detected (Fig. S6). However, the proportion of sea urchin peaks with a secondary motif-containing site was significantly lower than the proportion of sea star peaks with a secondary site (χ2 P = 1.8e−4) (Fig. S6). The proportion of peaks with primary sites, conversely, was not significantly different between the two datasets (χ2: P = 0.069). Therefore, even though the secondary motif was present in the sea urchin dataset, it was present in significantly fewer of the detected peaks, suggesting a bias against use of this lower-affinity motif. We found minimal differences among the GO term enrichments for peaks with primary vs. secondary site motifs. In the sea urchin, primary site-containing peaks were near genes enriched in the MAPK pathway and thyroid hormone receptor activity, whereas secondary site peaks were near genes enriched for growth factor activity. Sea star primary site-containing peaks were near genes enriched for developmental processes and tRNA ligase activity, whereas targets near secondary site peaks were enriched for cell death (Fig. 2 and Dataset S1). There was no association between motif use and up- or down-regulation among the sea star target genes.
Fig. S6.
Distribution of Tbr primary and secondary motifs among detected peaks. For each Venn diagram, the number of peaks containing primary, secondary, or both motifs are shown. The numbers below each circle represent the total number of peaks with that motif detected. (A) The SpTbr peaks set are those 1,492 peaks that both overlap an ATAC-seq peak and are within 75 kb of an annotated gene. Of the 1,492 SpTbr peaks, 793 have at least one primary site, 690 have at least one secondary site, 360 of which have both a primary and a secondary site, and 369 have no detectable motif. (B) The PmTbr peaks set are the 1,105 peaks that are within 75 kb of a DEG. Of the 1,105 PmTbr peaks, 628 have at least one primary site, 594 have at least one secondary site, 351 have both, and 234 have no detectable Tbr motifs below the P value cutoff. The conserved targets sets are the peaks associated with the 108 genes found to be targets of both SpTbr and PmTbr. The numbers of peaks with motifs found among the associated SpTbr peaks (C) and the PmTbr peaks (D) are shown. For PmTbr peaks associated with these genes, 57 peaks have a primary site, 64 have a secondary site, and 32 have both primary and secondary sites, whereas 19 have no motif detected. For the SpTbr peaks in this set, 49 have primary sites, 45 have secondary sites, 19 have both primary and secondary sites, and 33 have no motifs detected.
We were next interested to understand which, if any, orthologous genes and processes were regulated by Tbr in both sea urchin and sea star. Although multiple studies have compared gene function by assessing specific differential target gene expression in knockdowns vs. controls (e.g., refs. 33–36), this study provides the opportunity to perform an unbiased genome-wide survey, as well as to compare potential direct regulation through comparison of our ChIP-seq datasets. There is no previous knowledge to provide an expectation of conservation and divergence between these echinoderms. Estimates of binding site conservation across other taxa show a wide range of target conservation. Experiments in yeast (37) and mammals (5, 38–40) reported low levels (e.g., 5–40%) of target conservation for several TFs examined, whereas studies of the mesodermal regulator Twist in Drosophila species revealed as high as 60–80% target conservation across the six species tested (41). Thus, the level of target conservation may be factor- and clade-dependent.
Using the reciprocal BBH mappings, we sought to identify orthologous genes that have proximal peaks in both sea urchin and sea star datasets. Given the differences in genome assembly completeness, and our preference toward eliminating false positives over maintaining data, we cannot make strong conclusions about missing associations (i.e., those genes that have a peak in one dataset but are missing a peak in the other) because there is a likelihood for false-negative associations in the sea star dataset, where the genome is assembled into generally smaller scaffolds (20) (Fig. S5). Therefore, we restrict the following analyses to associations between genes and peaks found in both datasets. Of the 4,444 sea urchin genes detected within 75 kb of a SpTbr peak, 995 had an orthologous sea star gene that also had a proximal PmTbr peak. A total of 108 of these genes (10.9%) were also differentially expressed in the Pm-Tbr knockdown RNA-seq dataset (Dataset S2). The GO terms enriched in the set of 108 genes with orthologous genes in both species that had peaks included “protein phosphorylation” (eight genes, P = 0.007), aminoacyl-tRNA ligase activity (three genes, P = 0.02), and “TF activity, sequence-specific DNA binding” (five genes, P = 0.057) (Fig. 2 and Dataset S1).
Fig. S5.
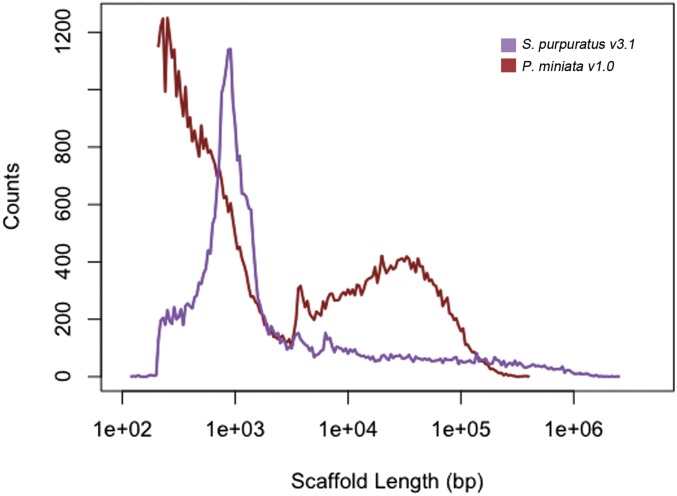
Comparison of genome assembly for S. purpuratus and P. miniata. For each genome, a histogram of scaffold lengths is shown. The maximum scaffold length in the S. purpuratus assembly is approximately an order of magnitude larger than that in the P. miniata genome assembly.
Finally, we asked whether the gene targets that are regulated in common (108 genes) (Fig. 4 and Dataset S2) had any notable binding site motif attributes. We found a similar decrease in the number of secondary motifs present in SpTbr peaks associated with the 108 genes in the overlapping ortholog set, compared with the total peak dataset (Fig. S6). The proportion of secondary sites in the sea urchin peaks in this subset was significantly lower (χ2: P = 0.014), whereas there was no significant difference in the proportion of primary site peaks (χ2: P = 0.34). A total of 17 of these genes were associated with peaks that had the same combination of primary and secondary motifs in both organisms (Dataset S2). Here, again, we found limited functional differences for genes associated with either primary vs. secondary site motif-containing peaks (Fig. 2 and Dataset S1). Therefore, primary and secondary motif use is not biased toward any of the functional categories tested here.
SI Materials and Methods
ChIP.
Approximately 100,000 embryos (1 × 108 cells) were cultured in artificial seawater at 15 °C until mesenchyme blastula stage (S. purpuratus; ∼24 hpf) or hatched blastula stage (P. miniata; ∼30 hpf). Embryo samples were fixed in 1% formaldehyde in PBS for 10 min at room temperature, and fixation was stopped by the addition of 0.125 M glycine. Samples were pelleted and flash-frozen. Chromatin was extracted by using standard protocols (51), except that shearing was achieved by enzymatic digestion rather than sonication. Micrococcal nuclease (Cell Signaling Technology) and digestion were performed according to the manufacturer’s SimpleChIP instructions. ChIP was performed by using standard protocols (51) with 50 µg of fragmented chromatin and 10 µg of custom antibody for each Tbr ortholog.
ChIP-Seq Analysis.
High-quality reads were mapped to either the S. purpuratus v3.1 genome or the P. miniata v1.0 genome assemblies [Bowtie (Version 2.2.2; ref. 52)], and nonredundant unique alignments were used for peak detection [MACS2 (Version 2.1.0; ref. 53)]. MACS parameters were empirically optimized for each read set based on the generated MACS models. Applying the same parameters to the analysis of these two datasets, which were generated by using different antibodies, was found to be suboptimal for each. For the sea urchin dataset, an mfold of [5,50] and a q-value cutoff of 0.05 were used. For the sea star dataset, an mfold of [10,50] and a P value cutoff of 1e−5 were used. Finally, bedtools suite applications (Version 2.26.0; ref. 54) were used to identify peaks overlapping ATAC-seq peaks and/or identify the closest gene to each detected peak.
Masking Repeat Regions.
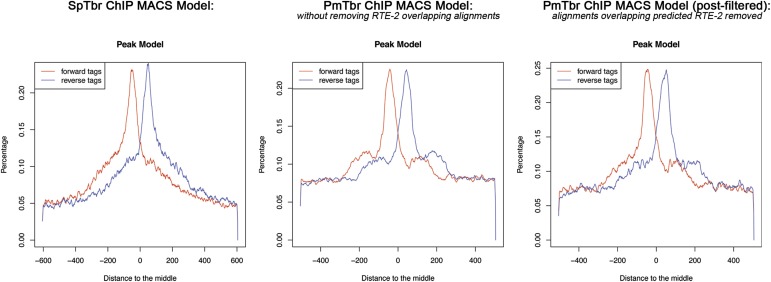
P. miniata repeat predictions, accessible as JBrowse tracks at Echinobase.org, were generated by Susan Ernst, Tufts University, Medford, MA and Andrew Cameron, California Institute of Technology, Pasadena, CA, based on similarities to repetitive elements present in other genomes. The predicted RTE-2–like retroelements were found to be statistically enriched in the initial peak set relative to the whole genome. In total, 23.4% of this initial peak set was found to overlap an RTE-2–like element (hypergeometric P < 1e−113). Peaks were abundant on highly repetitive regions and scaffolds (e.g., an extreme example in Scaffold119; Fig. S3). We expect that these peaks do not reflect a true enrichment, but, rather, artefactual enrichment owing to chromatin features of these repeat regions. To minimize the potential for false-positive binding sites, sequencing reads that overlapped predicted RTE-2–like elements were removed before peak detection. This filtering reduced the number of peaks detected by 10,049 and also, importantly, improved the shape of the derived MACS2 model (Fig. S4).
Fig. S3.
Example of a highly repetitive region from the P. miniata genome. Initial peak detection called hundreds of PmTbr peaks along Scaffold119 (top track), many of which were found to overlap the RTE-2–like elements predicted in this region. IP and input alignments were filtered to remove any alignments overlapping these elements, and after filtering no MACS peaks were called.
Fig. S4.
MACS models generated from the SpTbr and PmTbr ChIP-seq datasets. For the PmTbr data, the models generated from the data both prefiltering and postfiltering of alignments that overlap predicted repeat elements are shown. After filtering out repetitive elements, the shoulders present in the MACS model are reduced and resemble more closely the peak model generated from the SpTbr ChIP dataset.
PmTbr Knockdown.
P. miniata zygotes were injected with 600 µM Tbr MASO or standard control MASO (Gene Tools) as described (1, 2, 49, 50). Embryos were sorted on the basis of rhodamine tracer and morphology to ensure that only healthy injected embryos were processed for RNA. Embryos that arrested in early development or that exhibited aberrant development were discarded. Embryos were cultured in artificial seawater at 15 °C until hatching, ∼30 hpf for standard control injected and typically 36 hpf for Tbr MASO injected.
RNA-Seq Analysis.
RNA-seq reads were trimmed of residual adapter sequences and low-quality bases [Trimmomatic (Version 0.32; ref. 55)]. High-quality reads were mapped to the P. miniata (Version 1.0) genome assembly [Tophat (Version 2.0.12; ref. 56)]. Reads uniquely mapping to transcripts from an in-house Cufflinks transcriptome assembly generated from several RNA-seq datasets (bouzouki.bio.cs.cmu.edu/Echinobase/Pmin_transcripts.gtf) were counted [HTSeq-count (Version 0.6.1p1; ref. 57)). Read counts were normalized, and any gene with more than two reads per million reads in at least three different samples was included in further analyses. Differential expression was assessed by using a generalized linear model quasilikelihood F test [edgeR (58, 59)], controlling for sample batch, and genes with a calculated FDR < 0.05 were considered differentially expressed. Raw and processed sequencing reads have been deposited into the NCBI Gene Expression Omnibus (accession no. GSE89863), and analysis scripts are available upon request.
Ortholog Mapping.
Orthologous genes between the sea star and sea urchin transcriptome were detected by using a reciprocal BBH script generated in-house. Briefly, the Cufflinks-assembled sea star transcriptome was searched against a custom-built sea urchin database generated from the SPU_Nucleotides.fasta (Echinobase) using tblastx at an E-value cutoff of 1e−5 (60), and the sea urchin genes were searched against the sea star transcriptome database. The top-scoring match for each sea star and sea urchin gene was determined, and for gene pairs with the reciprocal best match, the pair was defined as putative orthologs. A total of 35.3% of genes found to be expressed in the sea star transcriptome (9,761 of 27,630) had a reciprocal BBH mapping to a sea urchin gene.
GO Set Enrichment.
GO term annotations for sea urchin genes were extracted from the PostgreSQL database dump from build 7 of the S. purpuratus genome (Version 3.1) available through Echinobase. Term enrichment testing was performed by using the GOstats Bioconductor package (61) by building a custom GeneSetCollection from the mapping of GO IDs to sea urchin genes. For sea urchin enrichment tests, the entire set of annotations was used as the universe, whereas for the sea star set, this set was limited to only the sea urchin genes with an identified BBH ortholog in the sea star. We used both the 2,562 significantly differentially expressed following Pm-tbr knockdown and the subset of 1,165 DEGs that also had a PmTbr ChIP peak detected within 75 kb; 963 and 481 of these genes, respectively, were found to have a BBH orthology mapping to a sea urchin gene enabling GO term enrichment analysis. Statistical assessment of term enrichment was determined by the included hypergeometric test function (hyperGTest), and hypergeometric P values were adjusted to compensate for errors associated with multiple testing using the Benjamini–Hochberg correction (62).
Motif Detection.
To ensure the probability of finding a motif was not biased by peak widths, we used the 500 bp surrounding each MACS-defined peak summit to search for motif occurrences. Consistently sized peak regions were generated by extending peaks 250 bp upstream and 250 bp downstream of each MACS-defined peak summit. These 500-bp fragments were used as input along with the 12-bp Tbr primary and secondary motif PWMs to the FIMO program (63), part of the MEME suite of motif discovery and analysis tools (64). Motif occurrences below a P value threshold of 1e−3 were reported. Overlapping motif occurrences were filtered so that only the most significant motif at each position within a peak was reported. This step is important because the primary and secondary site PWM are partially overlapping, and frequently we would find both primary and secondary sites called at one position within a peak, although one with a much higher score and associated P value. Finally, each peak was classified based on the presence or absence of each motif.
Discussion
Echinoderms are a powerful model system for understanding deep divergence in developmental GRNs and the consequences that these changes have for the evolution of morphology. Despite extensive comparative studies (33, 34, 36, 42), there has been no direct and unbiased whole-genome comparison of the role of orthologous factors in these taxa. Put simply, we are as yet completely ignorant of the scale of conservation and change that might occur among these classes. Indeed, although there are many studies among classes of chordates (5), such comparisons are essentially unknown in any other animal taxa at this level of evolutionary comparison. Echinoderms diverged into separate classes ∼450–500 Mya in the Ordovician, but the radiation is considered to have occurred extremely rapidly, with five separate classes emerging in as few as 5 million years (14, 43, 44). During this rapid radiation, the classes underwent dramatic morphological change, and yet their body plans, both as adults and larvae, have remained remarkably stable since. This stability contrasts with vertebrate classes, which have undergone multiple waves of morphological radiations. Recently, this deep evolutionary separation of echinoderm classes has also been shown to present an opportunity to understand how TF proteins might evolve changed biochemical functions (13). Specifically, our previous study showed that the TF Tbr has evolved changed preferences for a low-affinity motif among echinoderm classes, and these secondary motifs were more sensitive to changing levels of Tbr. However, the role that such differences in motif utilization might have for the structure and function of the GRN is unknown in any taxa.
ChIP-seq provides the only method to assess direct binding and to reveal CRMs genome-wide, and it is therefore the only current technology that can provide a holistic comparison of direct regulatory connections and motif utilization. This approach has only recently become feasible in echinoderms because of the improvements made to the assembled genomes for these species (21). The high number of scaffolds in these genomes, however, still limits our ability to collect all associations between factor binding and target genes. In using these genomic techniques, a key goal has been to minimize false positives by stringently filtering the data in various ways. These aspects of our analysis restrict our ability to make conclusions about missing associations because they may be missed due to the limitations of the genome or by the application of stringent filters.
Accepting the nature of these limitations, our study has shown that Tbr in these species is highly pleiotropic, and especially so in sea stars, where PmTbr has 9,164 predicted binding sites compared with only 1,952 in sea urchins. The expression of tbr has been well characterized in many species from different groups of echinoderms. Whereas in euechinoid sea urchins (e.g., S. purpuratus), tbr expression is restricted to skeletogenic mesoderm, orthologs in other echinoderm groups are expressed more broadly throughout the endomesoderm [e.g., as has been shown in sea stars (13), brittle stars (45), cidaroid sea urchins (46), and sea cucumbers (34)]. Parsimony, therefore, suggests that the euechinoid sea urchins have relatively recently lost this broader domain of tbr expression. The reduced number of Tbr binding sites identified in the sea urchin supports this hypothesis. This finding also suggests that our dataset should identify processes associated with a dramatic loss of target genes in sea urchin. Indeed, our data highlight functions found to be significantly enriched among sea star target genes that are lost in the sea urchin set, specifically those related to scavenger receptor activity and apoptotic processes suggesting a specific loss of these functions. Intriguingly, sea star Tbr appears to act as a repressor of these genes, implying that sea urchin Tbr has specifically lost repressor activities relative to the sea star. Both the sea urchin and sea star factors have predicted roles in regulating genes at all levels of the GRN hierarchy (i.e., other regulatory genes as well as genes involved in differentiation, cellular processes, and morphogenesis, which are found at the termini of the GRN). Therefore, alterations in function have not occurred solely through loss of terminal processes, but also through changes to regulation of other TFs and, hence, we predict, to GRN topology.
Even though the sea urchin and sea star proteins have very different domains of expression, and show a marked difference in numbers and types of regulated genes, they nonetheless share ∼10% of their targets. Because we have only two taxa for comparison, we cannot know to what extent these are evolutionarily maintained and thus genuinely homologous, or are converged upon independently. However, this study reveals the overlap in targets that may occur in these taxa and is within the low range of similar comparisons of yeast and vertebrate lineages (5, 37–40). GO analyses and close inspection of the target genes in common indicate that protein kinases and other regulatory genes are commonly regulated, again pointing to a role for Tbr within the body of the GRN topology. This finding implies that GRN structure can change dramatically, while maintaining some regulatory connections. A future direction will be to more carefully dissect the GRN circuitry surrounding these conserved nodes to understand the types of motifs and network functions that can be maintained in the face of such dramatic rewiring.
We found peaks containing secondary binding motifs in sea urchin as well as sea star, which shows that, although it has very low affinity for these motifs in vitro, in vivo SpTbr is able to bind to these sites. This finding suggests that some combination of CRM characteristics must act to stabilize binding to these low-affinity sites in vivo. For example, CRMs may contain multiple Tbr binding sites or potential cofactor binding sites that could mitigate the effect of a single low-affinity site. Importantly, we show that secondary motifs are significantly underrepresented in sea urchins compared with sea stars, which implies that there are indeed fewer permissive contexts, or an increase in the requirements for functional utilization, for these motifs in sea urchin CRMs. This finding has important implications for how changes to TF binding affinity can restructure GRNs during evolution. Genes regulated by CRMs containing only a single low-affinity binding site (i.e., a secondary motif) will be prone to loss of regulation as the TF evolves a further reduced affinity, such as we found for sea urchin Tbr. Many CRMs, however, are likely to be regulated by combinations of binding sites, including multiple low-affinity sites, as well as high-affinity motifs and comotifs, and are therefore expected to be relatively insulated from changes to TF binding affinity. Based on the described functions of low-affinity motifs (47), the CRMs most sensitive to a reduction in TF affinity may be those that regulate genes expressed at low levels, those that provide highly specific expression (for example in the presence of multiple related TFs), those that mediate repressive vs. activator functions, or possibly those that affect precise developmental expression timing.
It has been shown for many species, including in echinoderms, that there is a high rate of turnover in TF binding sites (48). Thus, there will be a gradual turnover that results in the loss of low-affinity motifs, a switching from high to low affinity, and loss of comotifs. Changes in affinity of these motifs can result from just one SNP, and hence can be quite frequent. Such turnover can lead to scenarios in which a gene is now regulated by a single, or few, low-affinity motifs. In the sea star, where the TF can readily bind these secondary motifs, the gene will maintain TF regulation. The sea urchin, however, may no longer express these genes in response to TF input. At this point, the CRM is no longer under selection for this TF binding and could rapidly acquire further mutations that are deleterious in this context. For example, loss of Tbr regulation might lead to a change in the timing of the regulation of the target, thereby relaxing other features of the CRM that direct function at this stage. Conversely the sea star CRM, being under maintained functional selection, may in the future acquire additional motifs and/or switch back from a low- to a high-affinity site. Thus, the relatively higher-affinity preference for a secondary motif in sea stars should provide an increased robustness to natural turnover in CRM sequences. The sea star has a greater range of functional binding site variants, and thus a larger sequence space for maintained selection. This hypothesis leads to the following predictions: that genes regulated by one or few low-affinity motifs will be more sensitive to TF levels, and that the sea star should have a greater frequency of CRMs under the control of single or few numbers of secondary motifs, whereas the frequency of these should be suppressed in sea urchins. Additionally, we predict greater population-level variation among Tbr-regulated CRMs in sea stars than in sea urchins. Ultimately, it will be essential to consider the consequences of binding site turnover and CRM loss in the context of the broader GRN and recognize that not all losses will manifest equivalent developmental phenotypes.
Materials and Methods
ChIP-Seq Analysis of SpTbr and PmTbr.
ChIP was performed as described (13) (details in SI Materials and Methods). Custom rabbit polyclonal antibodies were produced by Thermo-Fisher against peptides from the nonconserved N terminus of each protein (PmTbr: EQGERYTVSHHGATEDTR; SpTbr: KFQKTTEPEESDKVYEDENLDRD) to ensure high specificity for Tbr over other t-box TFs present in the genome (e.g., Tbx2/3 and Brachyury). Embryos were cultured until mesenchyme blastula stage (S. purpuratus; ∼24 hpf) or hatched blastula stage (P. miniata; ∼30 hpf), at which point they were collected and processed as described (ref. 13 and SI Materials and Methods). One biological replicate each, prepared by pooling chromatin from two or three independently fertilized cultures before immunoprecipitation, was used to prepare sequencing libraries from total (input) and immunoprecipitated chromatin, and Illumina HiSeq 2500 76-bp SR sequencing runs were performed (Yale Center for Genome Analysis). Analysis of ChIP-seq data are described (SI Materials and Methods). Raw and processed sequencing reads have been deposited into the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database (accession no. GSE89862), and analysis scripts are available upon request.
RNA-Seq Analysis of PmTbr Knockdown.
P. miniata zygotes were injected with 600 µM Tbr MASO or standard control MASO (Gene Tools) as described (1, 2, 49, 50). RNA was extracted by using the GenElute Mammalian Total RNA Kit (Sigma-Aldrich). Illumina TruSeq library preparation and HiSeq 2500 50-bp SR sequencing were performed (University of Southern California Epigenome Center). RNA from biological triplicate paired sibling control- and Tbr-MASO injected embryos was analyzed. Analysis of RNA-seq data is described in SI Materials and Methods. Raw and processed sequencing reads have been deposited into the NCBI Gene Expression Omnibus database (accession no. GSE89863), and analysis scripts are available upon request.
Ortholog Mapping, GO Set Enrichment, and Motif Detection.
Orthologous genes between the sea star and sea urchin transcriptome were detected by using a reciprocal BBH script generated in-house (SI Materials and Methods). GO term annotations for sea urchin genes were extracted from annotation files available through Echinobase. Term enrichment testing was performed by using the GOstats Bioconductor package, with statistical assessment determined by hypergeometric test, and reported P values were adjusted to compensate for multiple testing errors using the Benjamini–Hochberg correction (SI Materials and Methods). Motif occurrences within peaks was detected by using the FIMO program (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Pat Leahy and Marinus Scientific for research animals and Andrew Cameron and Susan Ernst for sharing unpublished datasets. This work was supported by National Science Foundation Grants IOS 0844948 and IOS-1557431.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Gene Regulatory Networks and Network Models in Development and Evolution,” held April 12–14, 2016, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Gene_Regulatory_Networks.
This article is a PNAS Direct Submission. D.H.E. is a guest editor invited by the Editorial Board.
Data deposition: The sequence data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (GEO SuperSeries accession no. GSE89865, relating accession nos. GSE89862 and GSE89863).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610611114/-/DCSupplemental.
References
- 1.McGinnis N, Kuziora MA, McGinnis W. Human Hox-4.2 and Drosophila deformed encode similar regulatory specificities in Drosophila embryos and larvae. Cell. 1990;63(5):969–976. doi: 10.1016/0092-8674(90)90500-e. [DOI] [PubMed] [Google Scholar]
- 2.Grens A, Mason E, Marsh JL, Bode HR. Evolutionary conservation of a cell fate specification gene: The Hydra achaete-scute homolog has proneural activity in Drosophila. Development. 1995;121(12):4027–4035. doi: 10.1242/dev.121.12.4027. [DOI] [PubMed] [Google Scholar]
- 3.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8(3):206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 4.Meader S, Ponting CP, Lunter G. Massive turnover of functional sequence in human and other mammalian genomes. Genome Res. 2010;20(10):1335–1343. doi: 10.1101/gr.108795.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328(5981):1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch VJ, Wagner GP. Resurrecting the role of transcription factor change in developmental evolution. Evolution. 2008;62(9):2131–2154. doi: 10.1111/j.1558-5646.2008.00440.x. [DOI] [PubMed] [Google Scholar]
- 7.Cheatle Jarvela AM, Hinman VF. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. Evodevo. 2015;6(1):3. doi: 10.1186/2041-9139-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat Protoc. 2009;4(3):393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger MF, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24(11):1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos AI, Barolo S. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philos Trans R Soc Lond B Biol Sci. 2013;368(1632):20130018. doi: 10.1098/rstb.2013.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rowan S, et al. Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity. Genes Dev. 2010;24(10):980–985. doi: 10.1101/gad.1890410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crocker J, et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell. 2015;160(1-2):191–203. doi: 10.1016/j.cell.2014.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheatle Jarvela AM, et al. Modular evolution of DNA-binding preference of a Tbrain transcription factor provides a mechanism for modifying gene regulatory networks. Mol Biol Evol. 2014;31(10):2672–2688. doi: 10.1093/molbev/msu213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erwin DH, et al. The Cambrian conundrum: Early divergence and later ecological success in the early history of animals. Science. 2011;334(6059):1091–1097. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- 15.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105(16):5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rafiq K, Cheers MS, Ettensohn CA. The genomic regulatory control of skeletal morphogenesis in the sea urchin. Development. 2012;139(3):579–590. doi: 10.1242/dev.073049. [DOI] [PubMed] [Google Scholar]
- 17.Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci USA. 2003;100(23):13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croce J, Lhomond G, Lozano JC, Gache C. ske-T, a T-box gene expressed in the skeletogenic mesenchyme lineage of the sea urchin embryo. Mech Dev. 2001;107(1-2):159–162. doi: 10.1016/s0925-4773(01)00470-1. [DOI] [PubMed] [Google Scholar]
- 19.Li X-Y, et al. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol. 2011;12(4):R34. doi: 10.1186/gb-2011-12-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Revilla-i-Domingo R, Minokawa T, Davidson EH. R11: A cis-regulatory node of the sea urchin embryo gene network that controls early expression of SpDelta in micromeres. Dev Biol. 2004;274(2):438–451. doi: 10.1016/j.ydbio.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Nam J, Dong P, Tarpine R, Istrail S, Davidson EH. Functional cis-regulatory genomics for systems biology. Proc Natl Acad Sci USA. 2010;107(8):3930–3935. doi: 10.1073/pnas.1000147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cameron RA, Kudtarkar P, Gordon SM, Worley KC, Gibbs RA. Do echinoderm genomes measure up? Mar Genomics. 2015;22:1–9. doi: 10.1016/j.margen.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cameron RA, Samanta M, Yuan A, He D, Davidson E. SpBase: The sea urchin genome database and web site. Nucleic Acids Res. 2009;37(Database issue):D750–D754. doi: 10.1093/nar/gkn887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Z, Ettensohn CA. Signal-dependent regulation of the sea urchin skeletogenic gene regulatory network. Gene Expr Patterns. 2014;16(2):93–103. doi: 10.1016/j.gep.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Röttinger E, Besnardeau L, Lepage T. A Raf/MEK/ERK signaling pathway is required for development of the sea urchin embryo micromere lineage through phosphorylation of the transcription factor Ets. Development. 2004;131(5):1075–1087. doi: 10.1242/dev.01000. [DOI] [PubMed] [Google Scholar]
- 26.Saunders LR, McClay DR. Sub-circuits of a gene regulatory network control a developmental epithelial-mesenchymal transition. Development. 2014;141(7):1503–1513. doi: 10.1242/dev.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinman VF, Nguyen AT, Davidson EH. Expression and function of a starfish Otx ortholog, AmOtx: A conserved role for Otx proteins in endoderm development that predates divergence of the eleutherozoa. Mech Dev. 2003;120(10):1165–1176. doi: 10.1016/j.mod.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Hinman VF, Nguyen A, Davidson EH. Caught in the evolutionary act: Precise cis-regulatory basis of difference in the organization of gene networks of sea stars and sea urchins. Dev Biol. 2007;312(2):584–595. doi: 10.1016/j.ydbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Tong A-J, et al. A stringent systems approach uncovers gene-specific mechanisms regulating inflammation. Cell. 2016;165(1):165–179. doi: 10.1016/j.cell.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cusanovich DA, Pavlovic B, Pritchard JK, Gilad Y. The functional consequences of variation in transcription factor binding. PLoS Genet. 2014;10(3):e1004226. doi: 10.1371/journal.pgen.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakabe NJ, et al. Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Hum Mol Genet. 2012;21(10):2194–2204. doi: 10.1093/hmg/dds034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouimette J-F, Jolin ML, L’honoré A, Gifuni A, Drouin J. Divergent transcriptional activities determine limb identity. Nat Commun. 2010;1:35. doi: 10.1038/ncomms1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCauley BS, Akyar E, Saad HR, Hinman VF. Dose-dependent nuclear β-catenin response segregates endomesoderm along the sea star primary axis. Development. 2015;142(1):207–217. doi: 10.1242/dev.113043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCauley BS, Weideman EP, Hinman VF. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev Biol. 2010;340(2):200–208. doi: 10.1016/j.ydbio.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 35.Koga H, et al. Experimental approach reveals the role of alx1 in the evolution of the echinoderm larval skeleton. PLoS One. 2016;11(2):e0149067. doi: 10.1371/journal.pone.0149067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yankura KA, Koechlein CS, Cryan AF, Cheatle A, Hinman VF. Gene regulatory network for neurogenesis in a sea star embryo connects broad neural specification and localized patterning. Proc Natl Acad Sci USA. 2013;110(21):8591–8596. doi: 10.1073/pnas.1220903110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borneman AR, et al. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317(5839):815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 38.Conboy CM, et al. Cell cycle genes are the evolutionarily conserved targets of the E2F4 transcription factor. PLoS One. 2007;2(10):e1061. doi: 10.1371/journal.pone.0001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42(7):631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 40.Odom DT, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39(6):730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Q, et al. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet. 2011;43(5):414–420. doi: 10.1038/ng.808. [DOI] [PubMed] [Google Scholar]
- 42.Cheatle Jarvela AM, Yankura KA, Hinman VF. A gene regulatory network for apical organ neurogenesis and its spatial control in sea star embryos. Development. 2016;143(22):4214–4223. doi: 10.1242/dev.134999. [DOI] [PubMed] [Google Scholar]
- 43.Smith AB, et al. Testing the molecular clock: Molecular and paleontological estimates of divergence times in the Echinoidea (Echinodermata) Mol Biol Evol. 2006;23(10):1832–1851. doi: 10.1093/molbev/msl039. [DOI] [PubMed] [Google Scholar]
- 44.Smith AB, Zamora S, Álvaro JJ. The oldest echinoderm faunas from Gondwana show that echinoderm body plan diversification was rapid. Nat Commun. 2013;4:1385. doi: 10.1038/ncomms2391. [DOI] [PubMed] [Google Scholar]
- 45.Dylus DV, et al. Large-scale gene expression study in the ophiuroid Amphiura filiformis provides insights into evolution of gene regulatory networks. Evodevo. 2016;7:2. doi: 10.1186/s13227-015-0039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamazaki A, Kidachi Y, Yamaguchi M, Minokawa T. Larval mesenchyme cell specification in the primitive echinoid occurs independently of the double-negative gate. Development. 2014;141(13):2669–2679. doi: 10.1242/dev.104331. [DOI] [PubMed] [Google Scholar]
- 47.Crocker J, Noon EP-B, Stern DL. The soft touch: Low-affinity transcription factor binding sites in development and evolution. Curr Top Dev Biol. 2016;117:455–469. doi: 10.1016/bs.ctdb.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 48.Garfield D, Haygood R, Nielsen WJ, Wray GA. Population genetics of cis-regulatory sequences that operate during embryonic development in the sea urchin Strongylocentrotus purpuratus. Evol Dev. 2012;14(2):152–167. doi: 10.1111/j.1525-142X.2012.00532.x. [DOI] [PubMed] [Google Scholar]
- 49.Cheatle Jarvela AM, Hinman V. A method for microinjection of Patiria miniata zygotes. J Vis Exp. 2014;(91):e51913. doi: 10.3791/51913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci USA. 2007;104(49):19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mortazavi A, Leeper Thompson EC, Garcia ST, Myers RM, Wold B. Comparative genomics modeling of the NRSF/REST repressor network: From single conserved sites to genome-wide repertoire. Genome Res. 2006;16(10):1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lund SP, Nettleton D, McCarthy DJ, Smyth GK. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat Appl Genet Mol Biol. 2012;11(5):8. doi: 10.1515/1544-6115.1826. [DOI] [PubMed] [Google Scholar]
- 60.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23(2):257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 62.Benjamini Y. Hochberg Y controlling the false discovery rate: A practical and powerful approach to multiple testing. JR Stat Soc. 1995;57(1):289–300. [Google Scholar]
- 63.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.