Abstract
Establishing a timeline for the evolution of novelties is a common, unifying goal at the intersection of evolutionary and developmental biology. Analyses of gene regulatory networks (GRNs) provide the ability to understand the underlying genetic and developmental mechanisms responsible for the origin of morphological structures both in the development of an individual and across entire evolutionary lineages. Accurately dating GRN novelties, thereby establishing a timeline for GRN evolution, is necessary to answer questions about the rate at which GRNs and their subcircuits evolve, and to tie their evolution to paleoenvironmental and paleoecological changes. Paleogenomics unites the fossil record and all aspects of deep time, with modern genomics and developmental biology to understand the evolution of genomes in evolutionary time. Recent work on the regulatory genomic basis of development in cidaroid echinoids, sand dollars, heart urchins, and other nonmodel echinoderms provides an ideal dataset with which to explore GRN evolution in a comparative framework. Using divergence time estimation and ancestral state reconstructions, we have determined the age of the double-negative gate (DNG), the subcircuit which specifies micromeres and skeletogenic cells in Strongylocentrotus purpuratus. We have determined that the DNG has likely been used for euechinoid echinoid micromere specification since at least the Late Triassic. The innovation of the DNG thus predates the burst of post-Paleozoic echinoid morphological diversification that began in the Early Jurassic. Paleogenomics has wide applicability for the integration of deep time and molecular developmental data, and has wide utility in rigorously establishing timelines for GRN evolution.
Keywords: evolution, evo-devo, euechinoid, cidaroid, gene regulatory networks
The investigation of gene regulatory networks (GRNs) in modern taxa allows for the understanding of evolutionary changes in the regulatory genome that have underpinned the evolution of new morphological structures in deep time (1–4). Establishing a timeline for the rates at which these novel structures arise, and the rate at which the developmental GRNs that encode them evolve, lies at the heart of evolutionary developmental biology (5). In recent years, identifying genetic regulatory differences between diverse organisms has become more feasible with broader phylogenetic sampling of developmental and gene expression data across Metazoa (6, 7). These new data provide insight into genomically encoded developmental programs and the species-specific GRNs that direct animal development in previously unexplored branches of the tree of life. Thus, studies comparing GRN subcircuit wiring in distantly diverged taxa (8, 9) are paving the way for the study of GRN evolution.
The arrival of these new data has introduced new problems, however. Importantly, as comparative studies of developmental GRNs are becoming more commonplace, it is critical to keep in mind that simple pairwise comparisons between taxa violate statistical assumptions of independence and must be carried out in an explicit phylogenetic framework (10). Phylogenetic comparative methods thus provide a rigorous methodology in which to examine gene expression datasets, and ultimately animal body plan evolution (11), within the context of evolutionary time. After genomic novelties underlying differential body plan development have been identified, we can then consider the rates at which these novelties arise, and the rates at which GRNs evolve. Achieving this end requires an explicit timeline in which to explore GRN evolution.
In a phylogenetically informed, comparative framework, it is possible to infer where on a phylogenetic tree and when, in deep time, GRN innovations are likely to have first arisen. Using a paleogenomic approach (12, 13), it is possible to incorporate deep time into an analysis of when GRN novelties arose and to infer the regulatory interactions directing the development of extinct organisms, thereby bringing forth a unique understanding of the evolution of GRNs and the body plans that they encode. Paleogenomics allows for the dating of the appearance of apomorphic GRNs, their subcircuits, and particular network linkages using a combination of the fossil record, statistically derived divergence dates, and comparative analyses of robust experimental data from extant organisms. With reliable dates in hand, the task of determining rates of GRN evolution is not far off. We set out to establish a rigorous framework for determining the timeline of GRN evolution, and to test a recently proposed hypothesis (8) concerning the timing of the evolution of a key sea urchin GRN novelty, the double-negative specification of micromeres.
Echinoids, or sea urchins, represent an ideal model system for understanding the mechanistic basis of GRNs in development and for studying the evolution of development (14–16). Research on the early embryonic development of echinoids has revealed the regulatory interactions that compose the circuitry of developmental GRNs driving early development of the purple sea urchin Strongylocentrotus purpuratus (14). Importantly, echinoids also have an excellent fossil record that dates back to Ordovician strata, more than 400 Mya (17). The combination of a robust fossil record and detailed understanding of the early developmental GRNs in numerous species makes echinoids an opportune group in which to implement integrated approaches to understanding GRN evolution.
The echinoid crown group comprises two clades, the euechinoids and the cidaroids (18). The adult body plans of these two clades provide prime examples of differential morphological disparity (19). Euechinoids have evolved numerous diverse morphologies throughout their evolutionary history and include morphologically distinct clades, such as the bilaterally symmetrical sand dollars and heart urchins. In contrast, cidaroids have shown remarkable morphological conservation, and the earliest fossil cidaroids are almost morphologically identical to cidaroids living in the oceans today (20, 21). Comparative studies of GRN architecture between cidaroids and euechinoids have revealed extensive differences in the wiring of their early developmental GRNs (8, 22, 23), and have served to inform our understanding of the genomic underpinning of the myriad morphological differences in their embryonic and larval development (24, 25).
The Double-Negative Gate
A striking example of a GRN subcircuit is the double-negative gate (DNG) that operates in the early development of S. purpuratus (15, 26, 27). This GRN subcircuit uses a double-repression mechanism that spatially localizes genes critical to primary mesenchyme cell (PMC) specification to the micromeres of the 16-cell-stage euechinoid embryo. PMC genes [e.g., alx homeobox 1 (alx1), delta, ets proto-oncogene 1/2 (ets1), tbrain (tbr)] are kept silent in the embryo by Hesc, a globally expressed repressor. Asymmetric localization of maternal regulatory factors at the vegetal pole results in the up-regulation of a repressor of Hesc, paired-class micromere anti-repressor 1 (pmar1), specifically in the micromere lineage (14, 26). Cells that express pmar1 also express PMC-specific genes and are fated to become PMCs, which later construct the larval skeleton of the embryo.
Although the DNG was first identified in S. purpuratus, its activity has widely been regarded to occur throughout the euechinoid order Camarodonta (28). The now-classic experiment in which ectopic overexpression of pmar1 converts all embryonic cells to a skeletogenic fate (26) has been duplicated in numerous camaradont euechinoids, including Hemicentrotus pulcherrimus (29), Paracentrotus lividus (30), and Lytechinus variegatus (31), suggesting that DNG function and circuitry are conserved in these taxa. Furthermore, whole-mount in situ hybridization data from the camarodonts H. pulcherrimus (32) and L. variegatus (33) reveal spatial distributions of hesc and its downstream targets that are consistent with the existence of the DNG in these euechinoids.
It also has been shown that the DNG appears to be present in a number of euechinoid echinoid taxa outside the camaradonts (28, 32). Micro1, a pmar1 ortholog, localizes to the 16-cell-stage micromeres of the clypeasteroid sand dollar Scaphechinus mirabilis (32) and the spatangoid heart urchin Echinocardium cordatum (28). Furthermore, hesc shows complementary yet nonoverlapping expression patterns with alx1, ets1, and delta at the 10th cleavage stage in S. mirabilis (32), and roughly complementary expression with alx1, tbr, and ets1 at the blastula stage in E. cordatum and the stomopneustoid Glyptocidaris crenularis (28).
Recent work has suggested that the DNG is absent in two species of cidaroid echinoids, Eucidaris tribuloides (8) and Prionocidaris baculosa (34). No evidence supporting the presence of pmar1 was found in the E. tribuloides genome, and both organisms show variable, overlapping expression patterns of hesc and its downstream targets in S. purpuratus, tbr, and ets1 (8). These data strongly suggest that the DNG is absent in cidaroid echinoids. In addition, both pmar1 and DNG circuitry are absent from asteroids (35) and ophiuroids (36), although the latter study identified a potential ortholog to pmar1, pplx, that lacked its function as a repressor in the ophiuroid Amphiura filiformis. Here we present data on the expression of hesc from the holothurian Parastichopus parvimensis that also suggests that the DNG is absent in the sister group to echinoids (37). The wide phylogenetic sampling of data indicating the presence or absence of the DNG in numerous euechinoid and cidaroid echinoids, as well as in other echinoderms, makes it an ideal candidate subcircuit to study the tempo of GRN evolution.
Previously, the initial evolution of the DNG was associated with the divergence of cidaroid and euechinoid echinoids, at least 268 Mya (8, 21). However, this conclusion was reached outside the context of a phylogenetic comparative framework and was based on a comparison of the differential GRN circuitry responsible for micromere specification in regular euechinoid and cidaroid echinoids given the most recent date at which euechinoid and cidaroid echinoids could have diverged (21). There are multiple evolutionary scenarios pertaining to the timing of the evolution of the DNG that could explain the presence of this circuitry in regular euechinoids like S. purpuratus and its absence in cidaroids. By analyzing the presence or absence of the DNG within an explicit phylogenetic framework, it is possible to estimate the age of particular nodes in deep time. We can then estimate the probability that the DNG was present or absent at ancestral nodes using ancestral state reconstruction to determine the support for particular hypotheses explaining the course of GRN evolution.
To determine when, on evolutionary timescales, the DNG likely evolved, we set out to establish a framework for rigorously investigating a timeline of GRN evolution. We used comparative phylogenetic estimates of echinoid relationships, ancestral state reconstructions, the fossil record, and a wide array of experimental data concerning the presence or absence of the DNG in numerous echinoid taxa and other echinoderm outgroups. This paleogenomic approach allowed us to estimate the age of the DNG and to establish a timeline for GRN evolution in echinoids.
Results
Holothurians Appear to Lack the DNG.
The holothurian P. parvimensis was developed as an experimental developmental system only recently, once improved methods for spawning animals were established (38, 39). Those earlier studies revealed that P. parvimensis has an alx1-expressing PMC-like cell lineage that produces a small larval spicule. This lineage arises in the vegetal pole, and, before ingression, alx1 is coexpressed with holothurian tbr, ets1, and erythroblast transformation specific ets-related gene (erg) orthologs (38). The expression of hesc, and indeed any earlier mechanisms that might lead to the specification of this lineage at the vegetal pole of P. parvimensis, were not determined, however.
For the present study, we blasted hesc and pmar1 sequences obtained from multiple species of echinoderms against the P. parvimensis transcriptomes (https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP012968). These transcriptomes were obtained from RNAs of early gastrula and larval stages (38). This identified a single predicted hesc transcript (E values of 9e-13 for SpHesc and 6e-15 for PmHesc). Phylogenetic analyses (SI Appendix, Sensitivity Analyses) showed that this transcript is the direct ortholog of the sea star and sea urchin hesc genes, and thus we named this gene Pp-hesc. No sequences with significant similarity to pmar1 or the brittle star pplx were identified, but we cannot discount the possibility that additional sequence coverage, especially of earlier embryonic stages, might reveal an ortholog.
We next used whole-mount in situ hybridization to determine the spatial expression of Pp-hesc. Transcripts are localized in the vegetal plate from hatching through to early invagination (SI Appendix, Fig. S1), and in a spotted pattern throughout the animal ectoderm. hesc continues to be expressed in the archenteron, and by the larval stage, transcripts are localized to the top of the archenteron and diffusely across the ectoderm (SI Appendix, Fig. S1F). Expression of Pp-hesc is very reminiscent of the expression of the orthologous gene in the sea star Patiria miniata (shown for comparison in SI Appendix, Fig. S1). Thus, these data cannot support a role for Hesc as a dominant repressor of tbr or erg in P. parvimensis. Because Hesc in P. parvimensis does not repress tbr, a key downstream target of Hesc repression in the DNG, we coded the DNG as absent in holothurians in our ancestral state reconstructions.
Divergence Time Estimation.
To constrain the age of the DNG with respect to evolutionary time, and also the age of particular nodes onto which we reconstructed ancestral states, we integrated fossil and molecular data using a fossil-calibrated relaxed molecular clock (40) to estimate times of divergence. Because genome-scale data are lacking for echinoids, we instead decided to explicitly account for topological and branch length uncertainty in our downstream analyses. We thus estimated divergence times using 15 alternative topologies (13 of which reached convergence and are discussed herein) resulting from the resolution of polytomies existing in our consensus tree inferred from Bayesian phylogenetic analyses in Phylobayes 4.1 (41). We also estimated divergence times on the best maximum likelihood (ML) tree, which we used to perform a number of sensitivity analyses (SI Appendix, Sensitivity Analyses).
The 95% credible intervals (CIs) and mean posterior divergence times for nodes that are informative for tracing the evolution of the DNG on one of our 13 alternative topologies are shown in Fig. 1, and mean ages and 95% CIs for all other divergences are shown in SI Appendix, Figs. S7–S19 and Tables S8–S20. Because some dated nodes are not present across all trees owing to topological variation, mean ages and 95% CIs for nodes not explicitly discussed in the main text are provided in SI Appendix, Figs. S7–S19 and Tables S8–S20. Across all trees, 95% CIs on nodes that were directly calibrated by fossils are all relatively narrow, whereas 95% CIs on nodes not calibrated by fossils, such as the Irregularia and Camarodonta, are wider. Of particular interest for questions regarding GRN evolution are the ages of particular nodes onto which we have reconstructed the presence or absence of the DNG. All of our analyses show a Carboniferous-Permian (∼265–342 Mya) divergence for the euechinoids and cidaroids, and a Triassic to Early Jurassic diversification of basal euechinoid lineages. The mean divergence time of the Camarodonta is at the oldest in the Triassic and at the youngest in the Late Cretaceous, dependent on the topology used to estimate divergence times (SI Appendix, Figs. S7–S19). The mean divergence time of the Irregularia is approximately Jurassic in most analyses, although the 95% CI on this node in all analyses is wide (>100 Ma). The node representing the most recent common ancestor (MRCA) of all analyzed cidaroids has a 95% CI between ∼8 and ∼230 Mya, putting the divergence of the analyzed extant cidaroids in the Cenozoic or Mesozoic. This node was not directly calibrated by fossils, which likely explains the wide 95% CI. The mean age for the node representing the most recent common ancestor of all extant euechinoids is Middle Triassic in all alternative topologies, whereas the 95% CI ranges from Early Triassic or Latest Permian (∼252 Mya) to Late Triassic (∼220 Mya) on all topologies. The mean estimated divergence times for the divergence between the echinothurioids (Araeosoma + Asthenosoma) and (aspidodiadematoids + pedinoids) + diadematoids are in the Late Triassic, with 95% CIs from the Late Triassic (∼211 Mya) to Middle Triassic (∼246 Mya) on all 13 topologies. Furthermore, the mean ages and CIs for the divergences between (aspidodiadematoids + pedinoids) and diadematoids and aspidodiadematoids and pedinoids reside within the Late Triassic. Results from analyses using the ML topology are broadly comparable (SI Appendix, Fig. S3) and are generally insensitive to changes in model parameters and tree topology (SI Appendix, Figs. S4 and S6).
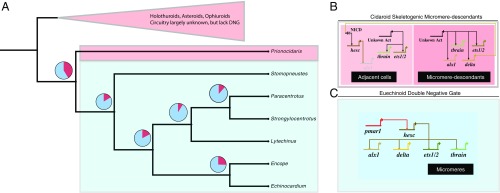
Fig. 1.
Fossil-calibrated time tree showing age of clades of crown group echinoids based on one of the 15 alternative topologies resulting from the Bayesian phylogenetic analysis and showing the probability of the presence or absence of the DNG on nodes of interest. Blue bars are 95% CIs on divergence times for nodes. Pie charts show mean PPs that the DNG was present (blue) or absent (red) at particular ancestral nodes in ancestral state reconstructions on 1,300 alternative topologies under priors on transition rates of U(0, 0.01). The taxa in blue have been demonstrated experimentally to use the DNG for micromere specification, whereas the taxa in red do not use the DNG. Taxonomic names represent higher-level echinoid taxonomic groupings. PPs for ancestral state reconstructions are presented in SI Appendix, Table S8. *Indicates Stomopneustoida + Arbacioida. The geological timescale was created in strap (62). Illustrations of spatangoid silhouette, by Hans Hillewaert (vectorized by T. Michael Keesey); cidaroid silhouette, by Didier Descouens (vectorized by T. Michael Keesey); and regular euechinoid, from Frank Förster (based on a picture by Jerry Kirkhart, modified by T. Michael Keesey), are available for use under a Creative Commons Attribution-ShareAlike 3.0 Unported license (https://creativecommons.org/licenses/by-sa/3.0/). Clypeasteroid image, from Michael Site, is available for use under a Creative Commons Attribution-NonCommercial 3.0 Unported license (https://creativecommons.org/licenses/by-nc/3.0/). All images are from Phylopic (www.phylopic.org).
Ancestral State Reconstruction.
Recent paleogenomic studies have used the fossil record to date the appearance of particular GRNs in evolutionary time by determining the first appearance of the morphological structures that are specified by these GRNs in the fossil record (12). There is no fossil record of embryonic sea urchins; thus, we used ancestral state reconstructions (42) to determine the probability that the DNG was present or absent in the last common ancestor of particular clades. We analyzed two datasets for our ancestral state reconstructions. One of these datasets maximized taxon sampling and included many more taxa for which experimental evidence was unknown than known. This dataset is based on 1,300 time-scaled trees (Divergence Time Estimation), with 100 trees of differing branch lengths sampled at random from the posterior distribution of each of the 13 divergence time estimation analyses run using alternative topologies. In this way, we explicitly account for phylogenetic uncertainty in our ancestral state reconstructions. The other dataset was limited to taxa for which direct experimental evidence concerning the DNG was available, and the topology onto which this ancestral state reconstruction was run reflected the ML estimates of topological relationships (Fig. 2 and SI Appendix, Fig. S5).
Fig. 2.
Ancestral state reconstruction of the DNG. (A) Pruned analyses showing reconstructed ancestral presence or absence of the DNG at select nodes on the ML tree using ML estimates of branch lengths. Pie charts show the mean PPs, with red sections representing the probability of absence of the DNG and blue sections representing the reconstructed probability of the DNG being present at a given ancestral node. (B) Wiring diagram for extant cidaroid echinoids showing the probable interactions of key genes involved in the DNG in S. purpuratus. There is no double-negative repression, because there is no Pmar1 gene, and hesc does not repress tbrain or ets1, as it does in euechinoids. (C) Wiring diagram showing the interactions of genes involved in the DNG in S. purpuratus and other examined euechinoids.
Our Bayesian ancestral state reconstructions using the taxonomically inclusive dataset were run under a variety of priors on the transition rates for each state. Given that the presence or absence of the DNG is unknown for many of the taxa in our tree, with more uninformative priors (uniform distribution with bounds 0 and 100), the probability of reconstructed ancestral states at all nodes in the phylogeny is 0.5, with an equal probability of presence or absence of the DNG at each ancestral node. Using narrower priors, which may be more appropriate for modeling the presence or absence of the DNG, which is unlikely to have evolved and unevolved numerous times throughout the evolutionary history of echinoids, resulted in more informative reconstruction of ancestral states. ML reconstruction of these ancestral states on all but four of the 1,300 trees resulted in transition rate parameters of <0.001 (Dataset S1), which is in line with the more informative reconstructed ancestral states resulting from smaller priors on transition rates. Using a uniform prior from 0 to 0.01 on transition rates resulted in relatively high mean posterior probabilities (PPs) in favor of the presence of the DNG in the last common ancestor of camarodonts (0.76), of Irregularia (0.76), and of Strongylocentrotus and Paracentrotus (0.97) (Fig. 1).
The DNG was present in the MRCA of camarodonts and the Irregularia, with a mean PP of 0.79; in the MRCA of diadematoids and camarodonts, with a mean PP of 0.76; and in the MRCA of camarodonts and stomopneustoids, with a mean PP of 0.78. The MRCA of echinoids appears to have used the DNG, with a mean PP of 0.65, whereas the MRCA of euechinoids appears to have used the DNG, with a mean PP of 0.76; however, the MRCA of extant cidaroids appears to have not used the DNG, with a mean PP of 0.95. These probabilities of the reconstructed presence and absence of the DNG at select nodes are plotted in Fig. 1. When using a slightly more diffuse prior [U(0, 0.05)], the mean PPs of the presence or absence of the DNG at all reconstructed ancestral nodes approached 0.5. Under this prior, the mean PPs of the presence or absence of the DNG of most reconstructed ancestral nodes were between 0.45 and 0.55 The exceptions to these less informative reconstructions are the node representing the MRCA of extant analyzed cidaroids, where the DNG was absent with a mean PP of 0.72, and the node representing the MRCA of Strongylocentrotus and Paracentrotus, which likely used the DNG with a mean PP of 0.75.
Many of our mean PP values derived using the dataset that maximized taxon sampling are systematically close to 0.50, resulting from the large amount of taxa included in the analysis for which no data on the DNG are available. This is especially the case when wider priors are used. This is likely because the tips with missing data are treated as having an equal probability of the presence or absence of the DNG in ancestral state reconstructions (42). Using a pruned dataset (Fig. 2), which included only taxa from orders for which direct experimental evidence regarding the presence or absence of the DNG exists, resulted in PPs farther from 0.50 and more representative of the current state of knowledge with regard to taxonomic sampling of developmental data. This analysis was run using the ML branch lengths as opposed to a time-scaled tree, which was able to more adequately resolve ancestral states with a wider prior [U(0, 100)]. This dataset had high mean PPs (Fig. 2 and SI Appendix, Table S3), and showed that the MRCA of the camarodonts used the DNG, with a mean PP of 0.91, and that the DNG was present in the MRCA of the Irregularia + Camadodonta, with a mean PP of 0.82, and also in the MRCA of the Carinacea, with a mean PP of 0.83. The last common ancestor of echinoids showed a mean PP of 0.41 and 0.59, respectively, in favor of the absence or presence of the DNG. Using a tree topology based on morphology (18) with this pruned dataset also revealed similarly high posterior mean PPs, supporting the presence of the DNG in the MRCAs of the camarodonts, the Carinacea, the Irregularia, and the Camarodonta + Stomopneustidae (SI Appendix, Table S3 and Fig. S5).
Discussion
The Antiquity of the DNG.
Our data strongly favor a euechinoid origin of the DNG, which appears to have evolved at the latest sometime before the Late Triassic (∼220–230 Mya depending on the topology used). Support is greatest for the presence of the DNG in the MRCA of camarodonts and in the MRCA of Paracentrotus and Strongylocentrotus, which is in line with other recently published studies showing marked conservation in the timing of gene expression between camarodonts (43). Our results also favor the presence of the DNG over its absence in the last common ancestor of early branching euechinoid clades. This indicates that the DNG was likely used in the MRCA of extant euechinoids. The pruned dataset in particular shows strong support for a pre-Carinacea age of the DNG; however, this dataset provides limited data concerning the non-Carinacea euechinoids, given the absence of direct experimental evidence from these early diverging euechinoid taxa. Future experimental work in these euechinoids, such as the diadematoids, will augment phylogenetic sampling and further resolve the age of the DNG in the euechinoid lineage. In the last common ancestor of echinoids, our Bayesian analyses show a mean PP of 0.65 for the presence of the DNG at this node under our smallest prior. Our analyses using the ML topology also show mean PPs slightly favoring the presence of the DNG over its absence at this node.
Because mean PPs at this node showed the lowest support among all nodes examined for the presence of the DNG, we can only tentatively interpret our findings as indicating that the DNG was used in the MCRA of all extant echinoids. Given the much higher mean PPs at the MRCA of extant euechinoids, however, it seems more likely that the DNG evolved at some point between the divergence of euechinoids and cidaroids in the Carboniferous-Permian and the MRCA of extant euechinoids in the Triassic. We also note that that our results at the more basal euechinoid nodes are more sensitive to prior choice compared with those at more recently branching nodes (SI Appendix, Table S21), and broader phylogenetic sampling in the future should decrease the sensitivity of results at these nodes to prior choice.
Given the low support for either the presence (mean PP, 0.65) or absence (mean PP, 0.35) in the node representing the divergence of euechinoids and cidaroids, we are unable to confidently attribute the evolution of the DNG to the divergence of the euechinoids and cidaroids as we have done in the past (8, 21). Previous authors have characterized the DNG as an evolutionary novelty unique to all euechinoid echinoids (1, 8, 21), and our ancestral state reconstructions have revealed mean PPs that strongly identify the DNG as a novelty unique to all extant euechinoids. Nonetheless, our analysis also fails to demonstrate unambiguously that the last common ancestor of echinoids (including cidaroids) did not use the DNG, and instead used the mechanism present in extant cidaroids (8, 21). Our results instead slightly favor the alternative hypothesis, that the DNG was used in the MRCA of extant echinoids. We are hesitant about endorsing this interpretation, however, given that PPs for both the presence and absence of the DNG at this node are closer to 0.5 than at any other nodes examined.
The use of ancestral state reconstruction provides a sound statistical framework within which to explore the evolution of GRNs. Although we previously made statements regarding GRN evolution from cross-species comparisons in a few taxa (8, 21), here we demonstrate that statistical analysis of GRN evolution within the context of a phylogeny provides a more rigorous framework within which to examine their evolution. Whereas a most parsimonious interpretation of our data would have led us to make binary statements regarding the presence or absence of the DNG at particular ancestral nodes, our approach allows us to assign probabilities to our conclusions while taking into account uncertainty in phylogenetic topology and branch lengths. Surprisingly, within this statistical framework, we cannot definitively determine whether or not the DNG was used in the MRCA of all extant echinoids. This result indicates that inferences following from cross-species comparisons of GRNs are at best one of many evolutionary scenarios, and that caution should be exercised regarding definitive statements about evolutionary changes in gene regulation associated with divergence of taxa. As more data on the utilization of the DNG in diverse echinoid clades become available, and as the echinoid phylogeny is improved on with new phylogenomic techniques, our confidence regarding the phylogenetic origin, and antiquity, of the DNG will improve. At the very least, we are able to say that the DNG is an ancient GRN subcircuit that likely evolved in or before the MRCA of extant euechinoids and has been conserved and used throughout the euechinoid tree of life and across at least 220 My of echinoid evolution.
The DNG and Echinoid Macroevolutionary Trends.
Obtaining age estimates for the DNG allows us to compare this GRN innovation with other events in the macroevolutionary history of echinoids. It was recently shown that crown group echinoids underwent heightened rates of evolution during the Early Jurassic (20), which, given the results of our analysis, postdate the origin of the DNG during or before the Late Triassic. This burst of diversification is tightly linked to the diversification of irregular echinoids and other euechinoid clades that, according to our analysis, likely used the DNG. Because the DNG functions to specify skeletogenic cells in developing embryos, its correct spatiotemporal deployment is required for embryonic and larval skeletogenesis. That such a critical function is dependent on the DNG may have even resulted in the existence of “fail safe” regulatory wiring of blimp1 and hesc, wherein Blimp1 represses hesc in the absence of Pmar1 (44). Given that the DNG appears to have been conserved across ∼220 My of echinoid evolution, it is possible that the DNG is so critical for early indirect development in euechinoids that it has undergone stabilizing selection, although this has yet to be demonstrated empirically.
The conserved role of the DNG in micromere specification, which appears to predate the diversification of irregular euechinoids, also indicates that the regulatory mechanism of micromere specification very early in development has been constrained, whereas aspects of later larval and adult morphology, which show high degrees of disparity (20, 45), have been relatively free to vary. Thus, the euechinoid double-negative specification of micromeres and skeletogenesis offers a striking example of genomically encoded developmental constraint early in development, whereas high degrees of plasticity have prevailed in the evolution of later larval and adult morphology. This is also in stark contrast to the cidaroids, which although exhibiting much more constraint in their adult body plans (20), display high levels of morphological variability in early development. Whereas euechinoids have an invariant four micromeres, cidaroids contain a variable number, even within a single species (8, 24, 25). This apparent differential constraint at different times in development found in these two clades represents an intriguing avenue for future research.
“Fossilized” GRNs.
Paleogenomics allows us to frame hypotheses regarding the nature of embryonic micromere specification in fossil echinoids. Because it has been shown that direct development in echinoids was not widespread until the Cretaceous (46), our analyses indicate that most indirectly developing euechinoids, which compose the majority of the echinoid fossil record since the Triassic, likely used the DNG. When put into the context of environmental and ecological change undergone by echinoids through the Cretaceous-Paleogene mass extinction, which had profound effects on echinoid size, feeding strategy, and biogeographic distribution (47), and further Cenozoic climatic changes (48), our findings suggest that the DNG has been very robust to evolutionary change over the course of the past 220 My. Paleogenomics offers the opportunity to compare presumptive evolutionary changes in GRNs with paleoclimatic and paleoenvironmental changes. We assert that this is a promising and intriguing avenue of future research, and that it is only one of the many doors that can be opened by paleogenomic approaches to studying the evolution of GRNs.
Methods
Phylogenetic Tree Construction.
For our dataset, we used previously published mitochondrial 16s, 18s, and 28s small subunit rRNA sequence data (49–51), which comprises the most comprehensive known molecular dataset with respect to echinoid taxonomic sampling. Although such a dataset for inference of topology has limited use compared with more widely used phylogenomic datasets, we accounted for uncertainty in topology and branch length in our downstream ancestral state reconstructions. We used the concatenated aligned dataset of Smith et al. (51) with gaps removed, leaving 3,230 sites (Datasets S2 and S3). For our ML analysis, we determined the best-fitting substitution model using Modeltest 3.7 (52) in PAUP* (53), which, among the models available in MCMCTREE (which we used to estimate divergence times on our ML topology), identified GTR+Γ as the best fit. The outgroup taxa were constrained to represent the recently supported Asterozoa hypothesis (37). Phylogenetic trees were constructed using RaxMLversion 8 (54). We ran 20 ML analyses, with the tree with the highest likelihood used for ancestral state reconstructions and divergence time estimation. This tree, showing bootstrap support from 1,000 replicates, is shown in SI Appendix, Fig. S2.
Our ML topology differs from previously published estimates (51) and the most up-to-date morphological phylogeny (18), predominantly in the placement of a clade consisting of the stomopneustoids and arbacioids plotting as sister group to a clade of Camarodonta + Irregularia. Because this topology differs with morphology and previous molecular estimates, which either put arbacioids as a sister group to the camarodonts or Arbacioida + Stomopneustoida as a sister group to the camarodonts, we decided to run 20 additional ML analyses using a constrained topology that reflects the most recent morphological phylogeny (figure 5 in ref. 18). We used this topology, shown in SI Appendix, Fig. S4, for sensitivity analyses.
In addition to our ML analyses, we performed a Bayesian analysis in Phylobayes 4.1 (41) using the CAT-GTR+ Γ model (55). Two independent Markov chains were run until both reached convergence. The burn-in for each chain was 2,000 points, and each chain was sampled every 10 points up to 20,000 points. Chains were run until the effective sample size for each parameter was >300 and the relative difference between parameters of each run was <0.1. Convergence of Markov chain Monte Carlo (MCMC) was assessed using the tracecomp and bpcomp commands in Phylobayes.
The majority rule consensus tree resulting from our Bayesian analyses (SI Appendix, Fig. S20) resolved many traditional clades supported by morphology and previous molecular analyses, including the cidaroids, camarodonts, and two clades of irregular echinoids. However, this tree contained a large polytomy in which the relationships of many of these major echinoid clades to one another are unresolved. An analysis in Phylobayes using the same model parameters and convergence criteria was also run using only the 18s and 28s sequence data (Dataset S4). This topology retained most of the same clades as the topology resulting from inference using all three genes, and was equally unresolved (SI Appendix, Fig. S21). We thus chose to use the topology resulting from inference using all three genes as a backbone for topologies used in divergence time estimation and ancestral state reconstruction (Divergence Time Estimation and Ancestral State Reconstruction). Given that we explicitly account for phylogenetic uncertainty in our ancestral state reconstruction, we would not expect inference using polytomy resolutions of the two-gene consensus tree to seriously alter our results. Because the goal of this analysis was not explicitly to reconstruct echinoid phylogeny, but rather to examine the evolution of the DNG within a phylogenetic context, the phylogenetic uncertainty evident from our Bayesian analysis is accounted for in our ancestral state reconstructions (see below).
Divergence Time Estimation.
The ML tree with the highest log-likelihood was used for divergence time estimation in the MCMCTREE package in PAML (56). After removal of the outgroup taxa, a global clock model (clock = 1 in BASEML) with the root of the tree (euechinoid-cidaroid divergence) calibrated was used to estimate a suitable prior for the substitution rate μ in BaseML. We thus used a gamma-Dirichlet distribution with shape parameter α = 1.79 and scale parameter β = 70 (rgene_gamma = 1.79, 70 in MCMCTREE). Parameters α and β of the gamma Dirichlet prior on σ2, specifying the variability of rates across branches, were set to 1 and 2.68, respectively. The birth death priors were set to λ = 1, μ = 1, and ρ = 0 (BDParas= 1 1 0). Divergence time estimation using the ML topology was done with MCMCTREE using the approximate likelihood calculation of dos Reis and Yang (57). We chose to use the independent rates model (58, 59). For our time priors, we used nine uniform constraints, most of which focused on the earlier branching nodes of the tree. MCMCTREE uses soft constraints (40), and for all of our divergence time priors, 2.5% of the distribution served as a soft tail on both the minimum and maximum constraints. Two independent MCMC chains were run for 5,000,000 iterations with a sample drawn every 50 iterations. The first 50,000 runs were discarded as burn in. This resulted in a total of 100,001 samples in each chain. Following analyses, all resultant MCMC samples were examined in Tracer version 1.6 (60) to check for convergence.
Fossil calibrations for analyses in MCMCTREE were compiled and justified with up-to-date absolute dates and the first attempt at using “best practices” (61) to rigorously assemble well-supported fossil calibrations for use in prior construction for divergence time estimation in echinoids. Our compilation is not exhaustive, given our primary goal of rigorously calibrating nodes toward the base of the echinoid tree, where most discussion regarding the origin of the DNG has been focused. A complete list of fossil calibrations with justifications is provided in SI Appendix, Fossil Calibrations for Priors on Divergence Time Estimates and Datasets S5 and S6.
We also ran sensitivity analyses to check the robustness of our results to variations in prior, model choice, and tree topology. Our results were generally robust to changes in model parameters. The results of our sensitivity analyses are shown in SI Appendix, Fig. S6 and discussed in SI Appendix, Sensitivity Analyses.
Because our Bayesian majority rule consensus tree was unresolved, we estimated divergence times using multiple phylogenetic topologies resulting from resolution of our Bayesian majority rule consensus tree. Nodes in the consensus tree with a PP <0.25 were collapsed. After this, all irregular echinoids were constrained to form a clade, based on morphological evidence (18), and divergence times were estimated on the 15 possible resolutions of the resulting tree. Divergence times were estimated using Phylobayes 4.1 (41) under the model of Drummond et al. (58), where the rate on each branch is an independent draw from a gamma distribution, using the CAT-GTR+ Γ model for the substitution process. The birth death priors were set to λ = 1, μ = 1, and ρ = 0. Fossil calibrations were soft and used variable combinations of the same calibrations as were used for divergence time estimation using the ML tree plus three calibrations for divergences outside of echinoids (SI Appendix, Fossil Calibrations for Priors on Divergence Time Estimates and Datasets S7–S20). Thus, each analysis used between 9 and 12 calibrations.
For each topology, two independent chains were run with a burn-in of 1,000 points sampling every 10 points. MCMC convergence was assessed using tracecomp and bpcomp in Phylobayes. Chains were run until the effective sample size for each parameter was >100 and the relative difference between parameters of each run was <0.3. Thirteen of the 15 analyses reached convergence within time available, whereas the two analyses that failed to converge were excluded from ancestral state reconstructions. Those two topologies exhibited relationships widely different from previous molecular and morphological inferences of echinoid topology (18, 51), and thus their exclusion from ancestral state reconstruction likely had no effect on our inferences.
Ancestral State Reconstructions.
Representatives from families (or orders in the case of the Stomopneustoida) and nonechinoid echinoderm classes for which there is experimentally derived evidence in favor or in disfavor of the presence of the double-negative repression of hesc by Pmar1 and of alx1, tbr, or ets1 by Hesc were used to score extant taxa for the presence or absence of the DNG. For instance, the cidaroid P. baculosa expresses hesc in the same cells as tbr, alx1, and ets1 at the late blastula stage, which indicates that Hesc could not be acting as a repressor of these genes, a canonical feature of the DNG, and as such, the DNG is not used in this taxon (34). When experimental data showed precise nonoverlapping expression patterns and experimentally tested GRN linkages mirroring those of S. purpuratus, in which the DNG was first discovered, the DNG was scored as present in that taxon. Given our interest in modeling the evolution of the DNG as it was first described from S. purpuratus, we feel that coding the DNG as present/absent is appropriate for this analysis, because any difference in experimentally derived linkages, especially evidence of nonrepression of alx1, tbr, or ets1 by Hesc or of hesc by Pmar1, allows us to code a taxon as absent and thereby maximize phylogenetic coverage.
We used two datasets for our analyses, one dataset that maximized phylogenetic coverage but had low proportional coverage of experimentally derived presence and absence data, and a pruned dataset that included only taxa from families (or the Stomopneustoida) for which direct experimental evidence exists regarding the presence or absence of the DNG. The dataset maximizing phylogenetic coverage was used to reconstruct ancestral states across the 1,300 time-calibrated trees resulting from divergence time estimation. In this way, phylogenetic uncertainty with respect to both branch length and topology were accounted for in reconstruction (42) (Dataset S23). We ran these analyses under four different sets of priors on transition rates (SI Appendix, Table 21). We also ran this dataset using the ML branch lengths from the best ML tree inferred in RAxML and the ML estimates of branches constrained on the morphological tree of Kroh and Smith (18) (SI Appendix, Tables S1 and S2). The topology of the pruned dataset was constrained in RaxML before branch length estimation, such that the topological relationships matched those of the ML tree inferred from the more complete dataset. All analyses were run in BayesTraits 2.0 (42) using the MRCA command, which reconstructs probabilities of ancestral states at the node representing the most recent common ancestor of the specified taxa (Datasets S21 and S22). We ran our MCMC analyses for 10,000,000 iterations with a burn-in of 2,000,000 iterations and sampling every 1,000 iterations. All analyses were run under varying priors to assess the sensitivity of analyses to prior choice.
Supplementary Material
Acknowledgments
This research is the outgrowth of an extremely fruitful collaboration between the authors and the late Eric H. Davidson, which was funded by US National Science Foundation Grant IOS1240626 (to D.J.B. and Eric H. Davidson). J.R.T. thanks M. Florence, K. Hollis, D. Levin, and J. Strotman at the US National Museum of Natural History and T. Ewin at the Natural History Museum, London, who provided access to collections that house many of the specimens used in fossil calibration for divergence time estimation. We also thank J. Yu, Director of Computing Services at the University of Southern California’s Department of Earth Sciences, for computing time and assistance. The comments of two anonymous reviewers improved on an earlier version of this manuscript and are much appreciated.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Gene Regulatory Networks and Network Models in Development and Evolution,” held April 12–14, 2016, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Gene_Regulatory_Networks.
This article is a PNAS Direct Submission. D.H.E. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610603114/-/DCSupplemental.
References
- 1.Ettensohn CA. Lessons from a gene regulatory network: Echinoderm skeletogenesis provides insights into evolution, plasticity and morphogenesis. Development. 2009;136(1):11–21. doi: 10.1242/dev.023564. [DOI] [PubMed] [Google Scholar]
- 2.McClay DR. Evolutionary crossroads in developmental biology: Sea urchins. Development. 2011;138(13):2639–2648. doi: 10.1242/dev.048967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144(6):970–985. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peter IS, Davidson EH. Implications of developmental gene regulatory networks inside and outside developmental biology. Curr Top Dev Biol. 2016;117:237–251. doi: 10.1016/bs.ctdb.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311(5762):796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 6.Hejnol A, Lowe CJ. Embracing the comparative approach: How robust phylogenies and broader developmental sampling impacts the understanding of nervous system evolution. Philos Trans R Soc Lond B Biol Sci. 2015;370(1684):20150045. doi: 10.1098/rstb.2015.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin M, et al. The mid-developmental transition and the evolution of animal body plans. Nature. 2016;531(7596):637–641. doi: 10.1038/nature16994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erkenbrack EM, Davidson EH. Evolutionary rewiring of gene regulatory network linkages at divergence of the echinoid subclasses. Proc Natl Acad Sci USA. 2015;112(30):E4075–E4084. doi: 10.1073/pnas.1509845112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci USA. 2003;100(23):13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hejnol A, Dunn CW. Animal evolution: Are phyla real? Curr Biol. 2016;26(10):R424–R426. doi: 10.1016/j.cub.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 11.Dunn CW, Luo X, Wu Z. Phylogenetic analysis of gene expression. Integr Comp Biol. 2013;53(5):847–856. doi: 10.1093/icb/ict068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bottjer DJ, Davidson EH, Peterson KJ, Cameron RA. Paleogenomics of echinoderms. Science. 2006;314(5801):956–960. doi: 10.1126/science.1132310. [DOI] [PubMed] [Google Scholar]
- 13.Bottjer DJ. Geobiology and palaeogenomics. Earth Sci Rev. 2016;164:182–192. [Google Scholar]
- 14.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295(5560):1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 15.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105(16):5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peter IS, Davidson EH. Genomic control of patterning. Int J Dev Biol. 2009;53(5-6):707–716. doi: 10.1387/ijdb.072495ip. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith AB, Savill JJ. Bromidechinus, a new Ordovician echinozoan (Echinodermata), and its bearing on the early history of echinoids. Earth Env Sci T R So. 2001;92:137–147. [Google Scholar]
- 18.Kroh A, Smith AB. The phylogeny and classification of post-Palaeozoic echinoids. J Syst Palaeontology. 2010;8:147–212. [Google Scholar]
- 19.Gao F, et al. Juvenile skeletogenesis in anciently diverged sea urchin clades. Dev Biol. 2015;400(1):148–158. doi: 10.1016/j.ydbio.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 20.Hopkins MJ, Smith AB. Dynamic evolutionary change in post-Paleozoic echinoids and the importance of scale when interpreting changes in rates of evolution. Proc Natl Acad Sci USA. 2015;112(12):3758–3763. doi: 10.1073/pnas.1418153112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JR, et al. Reorganization of sea urchin gene regulatory networks at least 268 million years ago as revealed by oldest fossil cidaroid echinoid. Sci Rep. 2015;5:15541. doi: 10.1038/srep15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erkenbrack EM, et al. Ancestral state reconstruction by comparative analysis of a GRN kernel operating in echinoderms. Dev Genes Evol. 2016;226(1):37–45. doi: 10.1007/s00427-015-0527-y. [DOI] [PubMed] [Google Scholar]
- 23.Erkenbrack EM. Divergence of ectodermal and mesodermal gene regulatory network linkages in early development of sea urchins. Proc Natl Acad Sci USA. 2016;113(46):E7202–E7211. doi: 10.1073/pnas.1612820113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schroeder T. Development of a “primitive” sea urchin (Eucidaris tribuloides): Irregularities in the hyaline layer, micromeres, and primary mesenchyme. Biol Bull. 1981;161:141–151. [Google Scholar]
- 25.Wray GA, McClay DR. The origin of spicule-forming cells in a “primitive” sea urchin (Eucidaris tribuloides) which appears to lack primary mesenchyme cells. Development. 1988;103(2):305–315. doi: 10.1242/dev.103.2.305. [DOI] [PubMed] [Google Scholar]
- 26.Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol. 2002;246(1):209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- 27.Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci USA. 2007;104(30):12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamazaki A, Minokawa T. Expession patterns of mesenchyme specification genes in two distantly related echinoids, Glyptocidaris crenularis and Echinocardium cordatum. Gene Expr Patterns. 2015;17(2):87–97. doi: 10.1016/j.gep.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki A, Ki S, Kokubo T, Yamaguchi M. Structure-function correlation of micro1 for micromere specification in sea urchin embryos. Mech Dev. 2009;126(8-9):611–623. doi: 10.1016/j.mod.2009.06.1083. [DOI] [PubMed] [Google Scholar]
- 30.Duloquin L, Lhomond G, Gache C. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development. 2007;134(12):2293–2302. doi: 10.1242/dev.005108. [DOI] [PubMed] [Google Scholar]
- 31.Wu SY, McClay DR. The Snail repressor is required for PMC ingression in the sea urchin embryo. Development. 2007;134(6):1061–1070. doi: 10.1242/dev.02805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamazaki A, Furuzawa Y, Yamaguchi M. Conserved early expression patterns of micromere specification genes in two echinoid species belonging to the orders Clypeasteroida and Echinoida. Dev Dyn. 2010;239(12):3391–3403. doi: 10.1002/dvdy.22476. [DOI] [PubMed] [Google Scholar]
- 33.Sharma T, Ettensohn CA. Activation of the skeletogenic gene regulatory network in the early sea urchin embryo. Development. 2010;137(7):1149–1157. doi: 10.1242/dev.048652. [DOI] [PubMed] [Google Scholar]
- 34.Yamazaki A, Kidachi Y, Yamaguchi M, Minokawa T. Larval mesenchyme cell specification in the primitive echinoid occurs independently of the double-negative gate. Development. 2014;141(13):2669–2679. doi: 10.1242/dev.104331. [DOI] [PubMed] [Google Scholar]
- 35.McCauley BS, Weideman EP, Hinman VF. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev Biol. 2010;340(2):200–208. doi: 10.1016/j.ydbio.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 36.Dylus DV, et al. Large-scale gene expression study in the ophiuroid Amphiura filiformis provides insights into evolution of gene regulatory networks. Evodevo. 2016;7:2. doi: 10.1186/s13227-015-0039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Telford MJ, et al. Phylogenomic analysis of echinoderm class relationships supports Asterozoa. Proc Biol Sci. 2014;281(1786):20140479. doi: 10.1098/rspb.2014.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCauley BS, Wright EP, Exner C, Kitazawa C, Hinman VF. Development of an embryonic skeletogenic mesenchyme lineage in a sea cucumber reveals the trajectory of change for the evolution of novel structures in echinoderms. Evodevo. 2012;3(1):17. doi: 10.1186/2041-9139-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le HS, Schulz MH, McCauley BM, Hinman VF, Bar-Joseph Z. Probabilistic error correction for RNA sequencing. Nucleic Acids Res. 2013;41(10):e109. doi: 10.1093/nar/gkt215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z, Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol. 2006;23(1):212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- 41.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25(17):2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 42.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53(5):673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 43.Gildor T, Ben-Tabou de-Leon S. Comparative study of regulatory circuits in two sea urchin species reveals tight control of timing and high conservation of expression dynamics. PLoS Genet. 2015;11(7):e1005435. doi: 10.1371/journal.pgen.1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith J, Davidson EH. Regulative recovery in the sea urchin embryo and the stabilizing role of fail-safe gene network wiring. Proc Natl Acad Sci USA. 2009;106(43):18291–18296. doi: 10.1073/pnas.0910007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wray GA. The evolution of larval morphology during the post-Paleozoic radiation of echinoids. Paleobiology. 1992;18(3):258–287. [Google Scholar]
- 46.Jeffery CH. Dawn of echinoid nonplanktotrophy: Coordinated shifts in development indicate environmental instability prior to the K-T boundary. Geology. 1997;25(11):991–994. [Google Scholar]
- 47.Smith AB, Jeffery CH. Selectivity of extinction among sea urchins at the end of the Cretaceous period. Nature. 1998;392(5):69–71. [Google Scholar]
- 48.Kroh A. Climate changes in the Early to Middle Miocene of the Central Paratethys and the origin of its echinoderm fauna. Palaeogeogr Palaeoclimatol Palaeoecol. 2007;253:169–207. [Google Scholar]
- 49.Littlewood DTJ, Smith AB. A combined morphological and molecular phylogeny for sea urchins (Echinoidea: Echinodermata) Philos Trans R Soc Lond B Biol Sci. 1995;347(1320):213–234. doi: 10.1098/rstb.1995.0023. [DOI] [PubMed] [Google Scholar]
- 50.Stockley B, Smith AB, Littlewood T, Lessios HA, Mackenzie-Dodds JA. Phylogenetic relationships of spatangoid sea urchins (Echinoidea): Taxon sampling density and congruence between morphological and molecular estimates. Zool Scr. 2005;34(5):447–468. [Google Scholar]
- 51.Smith AB, et al. Testing the molecular clock: Molecular and paleontological estimates of divergence times in the Echinoidea (Echinodermata) Mol Biol Evol. 2006;23(10):1832–1851. doi: 10.1093/molbev/msl039. [DOI] [PubMed] [Google Scholar]
- 52.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 53.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods), Version 4. Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- 54.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21(6):1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 56.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 57.dos Reis M, Yang Z. Approximate likelihood calculation on a phylogeny for Bayesian estimation of divergence times. Mol Biol Evol. 2011;28(7):2161–2172. doi: 10.1093/molbev/msr045. [DOI] [PubMed] [Google Scholar]
- 58.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4(5):e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rannala B, Yang Z. Inferring speciation times under an episodic molecular clock. Syst Biol. 2007;56(3):453–466. doi: 10.1080/10635150701420643. [DOI] [PubMed] [Google Scholar]
- 60.Rambaut A, Suchard M, Drummond AJ. 2013 Tracer version 1.6. Available at tree.bio.ed.ac.uk/software/tracer/. Accessed February 6, 2017.
- 61.Parham JF, et al. Best practices for justifying fossil calibrations. Syst Biol. 2012;61(2):346–359. doi: 10.1093/sysbio/syr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bell MA, Lloyd GT. strap: An R package for plotting phylogenies against stratigraphy and assessing their stratigraphic congruence. Palaeontology. 2014;58(2):379–389. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.