Significance
Organelle contact sites are specialized intracellular zones called membrane contact sites (MCS), in which two distinct suborganelles are closely apposed in eukaryotic cells. The nucleus–vacuole junction (NVJ) is the first identified interorganellar MCS in the budding yeast Saccharomyces cerevisiae, and its formation depends on the nuclear membrane protein Nvj1p and vacuolar membrane protein Vac8p. We present the crystal structure of Vac8p–Nvj1p complex at 2.4-Å resolution. Based on the structure, we propose a molecular mechanism in which Vac8p competitively recognizes Nvj1p or Atg13p and present a model showing how Vac8p facilitates NVJ formation, mediates piecemeal microautophagy of the nucleus, and participates in the cytoplasm-to-vacuole targeting pathway.
Keywords: nucleus–vacuole junction, Vac8p, Nvj1p, membrane contact sites, crystal structure
Abstract
Formation of the nucleus–vacuole junction (NVJ) is mediated by direct interaction between the vacuolar protein Vac8p and the outer nuclear endoplasmic reticulum membrane protein Nvj1p. Herein we report the crystal structure of Vac8p bound to Nvj1p at 2.4-Å resolution. Vac8p comprises a flexibly connected N-terminal H1 helix followed by 12 armadillo repeats (ARMs) that form a right-handed superhelical structure. The extended 80-Å-long loop of Nvj1p specifically binds the highly conserved inner groove formed from ARM1−12 of Vac8p. Disruption of the Nvj1p–Vac8p interaction results in the loss of tight NVJs, which impairs piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Vac8p cationic triad (Arg276, Arg317, and Arg359) motifs interacting with Nvj1p are also critical to the recognition of Atg13p, a key component of the cytoplasm-to-vacuole targeting (CVT) pathway, indicating competitive binding to Vac8p. Indeed, mutation of the cationic triad abolishes CVT of Ape1p in vivo. Combined with biochemical data, the crystal structure reveals a Vac8p homodimer formed from ARM1, and this self-association, likely regulated by the flexible H1 helix and the C terminus of Nvj1p, is critical for Vac8p cellular functions.
Membrane contact sites (MCSs) between subcellular compartments play pivotal roles in cellular processes such as cooperative lipid biosynthesis, ion homeostasis, and interorganellar trafficking of molecules in eukaryotic cells (1–3). MCSs are physically formed through dynamic and direct interactions between proteins that are located in two distinct subcompartments. The nucleus–vacuole junction (NVJ), one of the best-characterized MCSs, is a physical contact site between perinuclear and vacuolar membranes and mediates essential cellular processes such as piecemeal microautophagy of the nucleus (PMN) and lipid biosynthesis in yeast (4, 5). PMN is a selective autophagic recycling process stimulated by carbon or nitrogen starvation through the target of rapamycin signaling pathway in Saccharomyces cerevisiae. Upon nutrient starvation, the region of the nucleus in the vicinity of NVJs invaginates into the vacuolar lumen and forms a bleb-like structure, which is released as a vesicle and eventually degraded by vacuolar hydrolases (5, 6). Because PMN occurs at the NVJ sites, their formation is essential for the initiation of the PMN process (7). NVJs are also involved in lipid metabolism by recruiting the two lipid-modifying enzymes, oxystereol-binding proteins homology (Osh1p) involved in nonvesicular lipid trafficking and the enoyl-CoA reductase Tsc13p that mediates the synthesis of very-long-chain fatty acids (8–11).
Previous studies revealed that NVJs are generated by forming tight interactions between Vac8p, a vacuolar membrane protein, and Nvj1p, a nuclear membrane protein (12). Vac8p was initially found as a key player in vacuole inheritance by cooperating with Vac17p, actin, profilin, and Myo2p (13). In addition, Vac8p has a crucial role in mediating the processing of aminopeptidase I through interaction with Atg13p in the cytoplasm-to-vacuole targeting (CVT) pathway during this selective autophagy (13, 14). Sequence analyses of Vac8p predicted several armadillo repeat (ARM) domains, each consisting of 40 residues that fold into three characteristic short helices tandemly organized into a superhelical structure that serves as a protein-binding platform for interaction with other physiologically relevant molecules (15). Whereas Vac8p is involved in various processes by providing a protein–protein interaction module in the vacuole, Nvj1p is found exclusively in the circumscribed area of NVJs at the nuclear membrane (4, 10, 12, 16). It has been suggested that Nvj1p acts as a scaffold to physically and functionally connect NVJs to lipid metabolism by interacting with both Vac8p and Tsc13p or Osh1p.
Despite the biological importance of the Vac8p–Nvj1p interaction for the formation of NVJs and the PMN process, the underlying mechanism by which Vac8p recognizes Nvj1p and exactly how the complex is organized to form NVJs and carry out PMN remain largely unknown. In this study, we determined the 2.4-Å resolution crystal structure of Vac8p bound to Nvj1p. Together with biochemical analyses and in vivo functional studies using the budding yeast S. cerevisiae, the structure provides fundamental insight into the structural and multifunctional roles of Vac8p in the formation of NVJs and physiological processes such as PMN and CVT pathways at the molecular level.
Results
Vac8p Directly Interacts with Nvj1p.
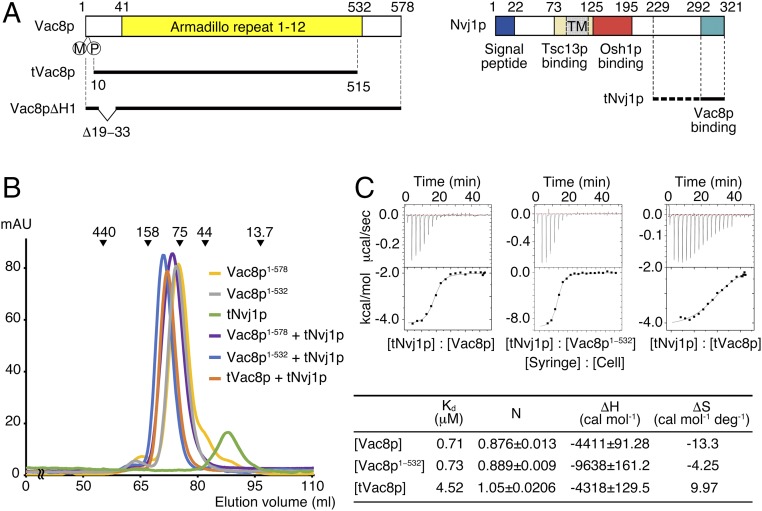
Vac8p is composed of 578 residues and is predicted to have several ARM repeats spread across the whole sequence (Fig. 1A and Fig. S1). The N-terminal residues (1–10) are modified by lipidation in the form of myristoylation and palmitoylation and are anchored into vacuolar lipid bilayers (13), whereas the remaining regions are localized to the cytosol. Nvj1p comprises 321 amino acids and is a typical type I membrane protein consisting of a single transmembrane domain that passes through the nucleus and endoplasmic reticulum (ER) membranes in this case, whereas the C terminus is localized to the cytosol (Fig. 1A). Previous studies using yeast two-hybrid analysis and coimmunoprecipitation revealed that the C-terminal domain (residues 281–321) of Nvj1p that is exposed to the cytosol binds to Vac8p (12). To further examine whether this interaction is direct, we prepared full-length recombinant Vac8p and truncated Nvj1p comprising residues 229–321 (subsequently referred to as tNvj1p) by overexpression and purification using Escherichia coli and analyzed their binding using size-exclusion chromatography (SEC). Fig. 1B shows that purified Vac8p directly interacts with purified tNvj1p, because the Vac8p-tNvj1p complex was eluted from the gel-filtration column at a volume corresponding to the molecular size of the expected heterodimer. Furthermore, we used isothermal titration calorimetry (ITC) to measure the binding affinity between the two proteins, and the results showed that Vac8p bound to tNvj1p with a Kd value of 0.71 μM (Fig. 1C). Both SEC and ITC indicated that Vac8p binds to Nvj1p in a 1:1 molar ratio.
Fig. 1.
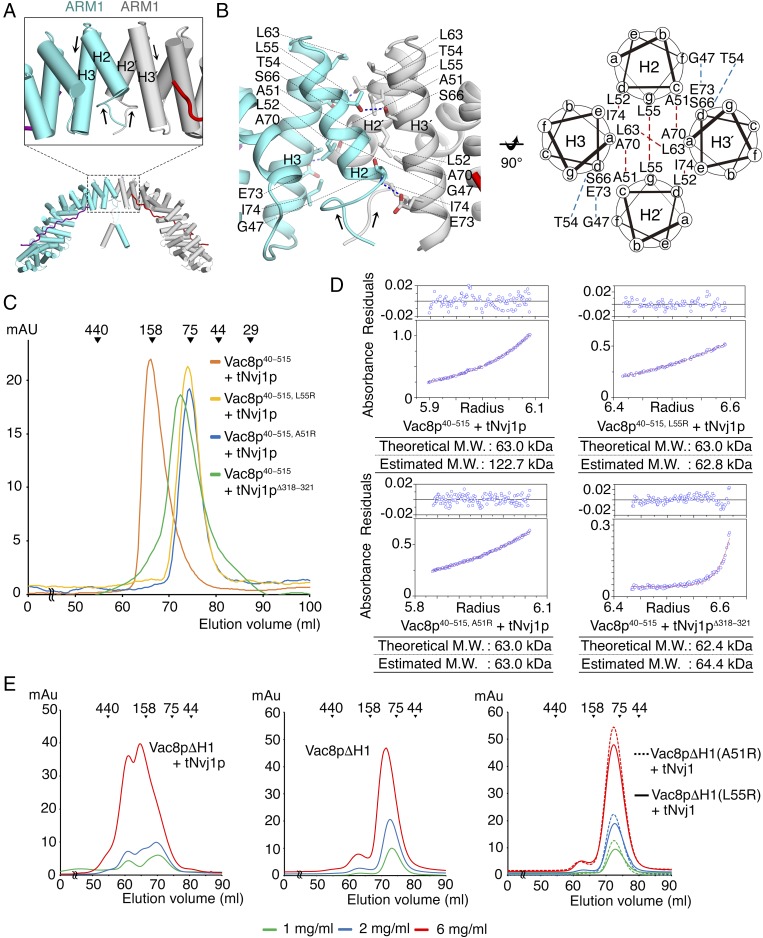
Vac8p directly interacts with Nvj1p. (A) Schematic diagram showing the domain structure of yeast Vac8p (Left) and Nvj1p (Right) truncation constructs used in this study. TM, transmembrane domain. (B) Analysis of the direct interaction between Vac8p and Nvj1p by SEC. Full-length Vac8p (residues 1−578), Vac8p1−532 (residues 1−532), tNvj1p (residues 229−321), Vac8p-tNvj1p, Vac8p1−532-tNvj1p, and the tVac8p–tNvj1p complex were injected on a Superdex 200 column equilibrated with buffer containing 25 mM Tris⋅HCl, 150 mM NaCl, and 5 mM DTT (pH 7.5). The molecular masses of the standards are indicated above the chromatogram curves to indicate the relative molecular masses of the samples. (C) ITC measurements for tNvj1p binding to full-length Vac8p, Vac8p1−532, and truncated tVac8p. Thermodynamic parameters measured by ITC experiments are tabulated below the thermograms.
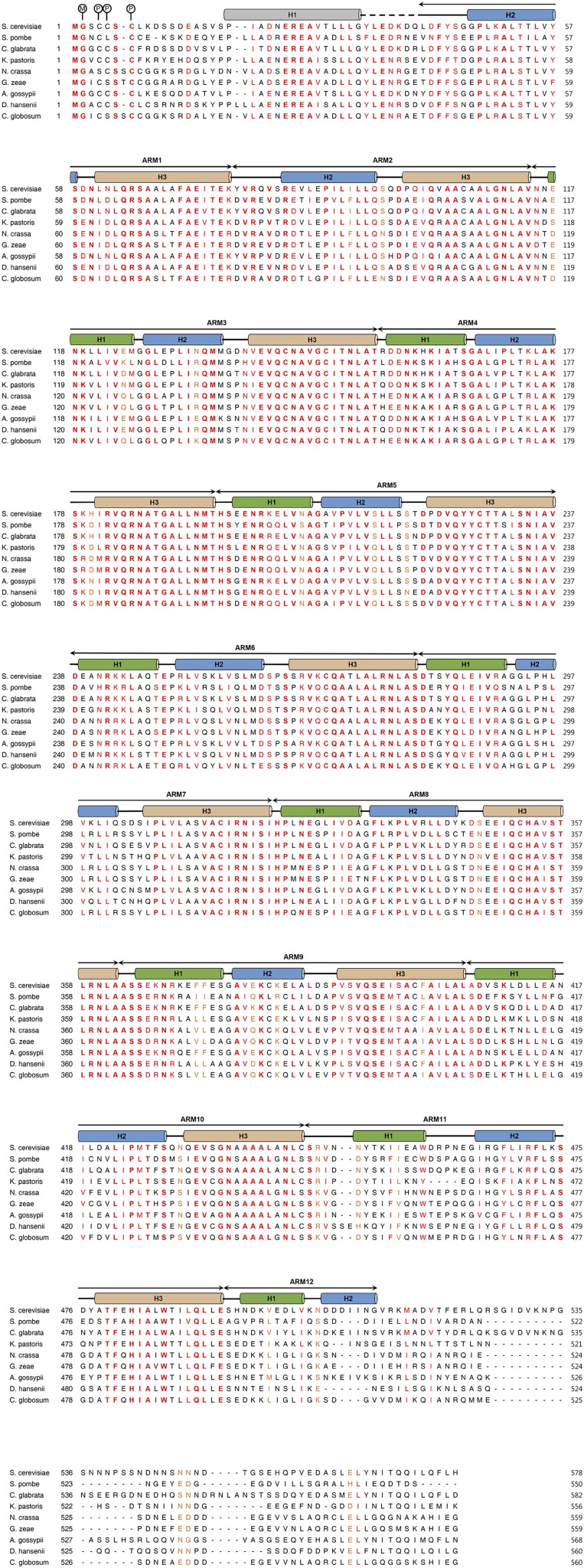
Fig. S1.
Sequence alignment of yeast Vac8p orthologs. Sequence alignment of full-length Vac8p orthologs in fungi. The N terminus of Vac8p is known to be modified by lipidation. The circled M and P indicate N-myristoyl glycine and S-palmitoyl cysteine, respectively. Secondary structural elements are indicated above the sequences with helices, β-stands, loops, and disordered regions depicted as cylinders, arrows, solid lines, and dashed lines, respectively. The crystal structure revealed that Vac8p is composed of an N-terminal helix (gray) followed by 12 ARMs. The three helices forming each ARM repeat are colored differently. Absolutely conserved, highly similar, and similar sequences are highlighted in bold red, red, and orange, respectively. UniProt entries for the sequences are as follows: S. cerevisiae (P39968), Schizosaccharomyces pombe (O43028), Candida glabrata (Q6FJV1), Komagataella pastoris (Q5EFZ4), Neurospora crassa (Q7RXW1), Gibberella zeae (Q4I1B1), Aphis gossypii (Q757R0), Debaryomyces hansenii (Q6BTZ4), and Chaetomium globosum (Q2GW27).
Crystal Structure of the Vac8p–Nvj1p Complex.
To gain structural insight into the recognition of Nvj1p by Vac8p we crystallized the Vac8p–Nvj1p complex. Attempts to obtain crystals from full-length Vac8p were unsuccessful, but we were instead able to crystallize a truncated version (residues 10–515, referred to as tVac8p) in complex with the Vac8p-binding domain of Nvj1p comprising residues 229–321 (referred to as tNvj1p). The N- and C-terminal truncated variant of Vac8p showed reduced but still substantial binding affinity for tNvj1p, suggesting that the truncation did not profoundly affect the overall structure and function of Vac8p (Fig. 1C). Crystals of the tVac8p–tNvj1p complex belong to the P21 space group and diffracted to a maximum of 2.4-Å resolution using synchrotron radiation. The structure was determined by single-wavelength dispersion (SAD) using Se-Met derivatized crystals, and the final model was refined with native data to 2.4-Å resolution (Fig. S2A). X-ray crystallographic data and refinement statistics are listed in Table S1.
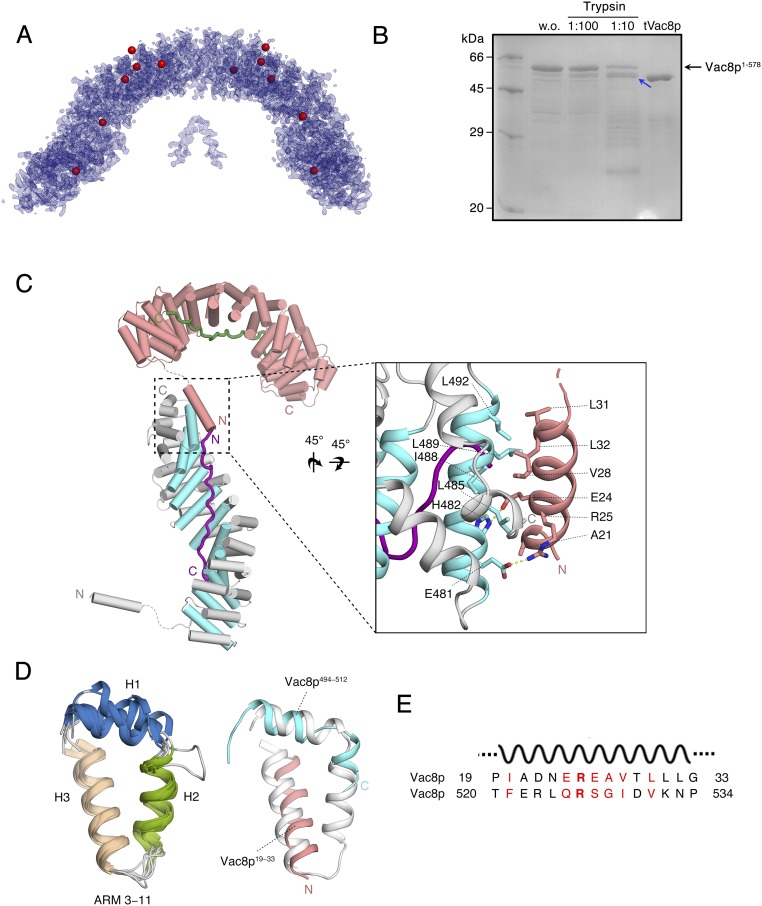
Fig. S2.
The H1 helix of tVac8p is critically involved in crystal packing. (A) Experimental electron density map (calculated using Se-SAD phasing at 2.4-Å resolution and contoured at 1.5σ) of the crystallographic asymmetric unit of the tVac8p–tNvj1p complex. Selenium atoms found by the PHENIX program (37) are represented by red spheres. (B) Trypsin digestion of full-length Vac8p. Vac8p was mixed with trypsin at the indicated trypsin:Vac8p ratio for 1 h at 4 °C. Reactions were stopped by adding 1 mM PMSF and SDS sample buffer, and the products were separated by SDS/PAGE. Sequencing analysis identified that the N terminus of the protease-resistant fragment, indicated by the blue arrow, is residue number 10 of Vac8p. SDS/PAGE shows comparison of the relative molecular massess between protease-resistant product and tVac8p (residues 10−515). (C) Cartoon representation of the tVac8p interface showing crystal contacts and highlighting the flexibly connected H1 helix from the neighboring molecule (pink) placed in the H3 position of the incomplete ARM12 lacking the H3 helix due to truncation. The 12 intact ARM repeats are also included. Residues involved in this interaction are shown in stick representation in the zoom view on the right. (D) Structural alignment of tVac8p ARM repeats 3–11 (Left). To compare the structural role of the N-terminal H1 helix, the intact ARM11 (gray) and the composite ARM12 comprising H1 and H2 (blue) together with H1 (residues 19−33) from the neighboring molecule (pink) are superimposed (Right). (E) Sequence alignment of Vac8p residues 19–33 corresponding to the H1 helix and residues 520–534 that might correspond to the H3 helix of ARM12. Conserved residues are colored red. The identical and similar residues between two sequences were highlighted in bold font and red text, respectively. w.o., without.
Table S1.
Data collection and refinement statistics
| Dataset | Native | Se-SAD |
| X-ray source | Beamline 7A, PAL | Beamline 7A, PAL |
| Temperature, K | 100 | 100 |
| Space group | P21 | P21 |
| Cell parameters a, b, c, Å | 84.082, 115.630, 84.131 | 84.116, 115.500, 84.631 |
| α, β, γ, ° | 90.00, 114.677, 90.00 | 90.00, 114.896, 90.00 |
| Data processing | ||
| Wavelength, Å | 0.97933 | 0.97934 |
| Resolution, Å | 50.0–2.40 | 50.0–2.40 |
| Rmerge,* % | 12.8 (61.9) | 9.9 (60.0) |
| I/σ | 13.3 (1.90) | 16.2 (2.21) |
| Completeness, % | 98.8 (97.4) | 98.7 (99.7) |
| Redundancy | 2.9 (2.6) | 3.1 (3.2) |
| Measured reflections | 166,859 | 176,447 |
| Unique reflections | 56,806 | 57,011 |
| Refinement statistics | ||
| Data range, Å | 31.9–2.40 | |
| Reflections | 57,022 | |
| Nonhydrogen atoms | 8,212 | |
| Water molecules | 181 | |
| Rms ∆ bonds,† Åb | 0.004 | |
| Rms ∆ angles,† ° | 1.118 | |
| R-factor,‡ % | 19.30 | |
| Rfree,ठ% | 21.72 | |
| Ramachandran plot, residues in | ||
| Most favored regions, % | 98.4 | |
| Additional allowed regions, % | 1.6 | |
| Generously allowed regions, % | 0.0 | |
| Disallowed regions, % | 0 |
Highest-resolution shell is shown in parentheses.
Rmerge = 100 x ∑h∑i | Ii(h) − <I(h)> |/∑h<I(h)>, where Ii(h) is the ith measurement and <I(h)> is the weighted mean of all measurement of I(h) for Miller indices h.
Root-mean-squared deviation (Rms ∆) from target geometries.
R-factor = 100 x ∑|FP – FP(calc)|/∑ FP.
Rfree was calculated with 5% of the data.
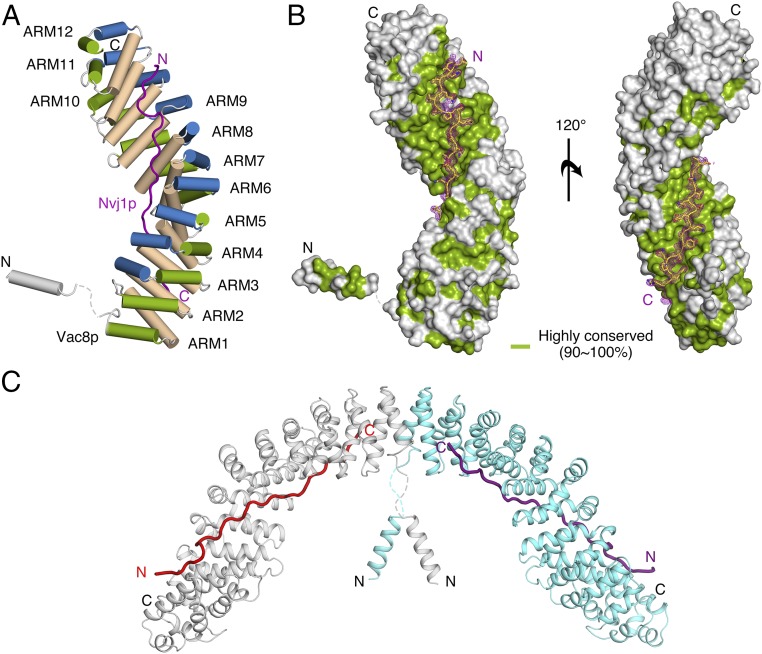
Fig. 2 A and B show the overall structure of the tVac8p–tNvj1p complex. Crystals contained two tVac8p–tNvj1p complexes arranged by twofold internal symmetry in the asymmetric unit (Fig. 2C). No structural differences were found between the two complexes, which had an rmsd of 0.8 Å for all Cα atoms. Each complex consists of one tVac8p and one tNvj1p (1:1 stoichiometry), consistent with the biochemical results (Fig. 1 B and C). Overall, the tVac8p–tNvj1p complex resembles a serpent column in which tNvj1p represents the axis and tVac8p represents the snake. tVac8p has an all α-helical topology with an elongated superhelical structure of 40 × 50 × 120 Å in size. The two structural elements consisting of (i) the N-terminal amphipathic helix spanning residues 19–33 (referred to as helix H1 throughout) and (ii) the central core domain of 12 ARM repeats (residues 41–512) are organized into a right-handed superhelical structure. This is a common structural feature in most ARM-containing proteins such as β-catenin and importin α (Fig. 2A) (15). Notably, the ARM repeats start from residue 41, and the two structural elements are connected with flexible loops comprising residues 34–40 that are disordered in the crystal structure. The truncated tVac8p construct used for crystallization ends at residue 515, which might remove part of H2 and all of H3 of the last of the ARM repeats (ARM12). Indeed, H2 and H3 of ARM12 were vulnerable to proteolytic degradation using limited proteolysis (Fig. S2B). The C-terminal region (residues 533–578) of Vac8p was also sensitive to digestion by trypsin, suggesting that this region is structurally disordered. Interestingly, the N-terminal amphipathic helix H1 from a neighboring tVac8p molecule in the crystal lattice occupies the ARM12 H3 position and may play a similar role to the H3 helix in ARM repeats (Fig. S2 C and D).
Fig. 2.
Overall structure of the tVac8p–tNvj1p complex. (A) Cartoon representation showing the overall structure of tVac8p bound to tNvj1p. tVac8p comprises two structural elements that contain an N-terminal H1 helix and 12 ARM domains. The extended loop of tNvj1p (magenta) binds in an antiparallel manner to the inner groove generated by the 12 ARMs of Vac8p. The three helices characterizing each ARM are shown in a different color. The crystal structure was determined by Se-SAD phasing and refined with native data to 2.4-Å resolution. (B) Surface representation of tVac8p based on the degree of sequence conservation from nine Vac8p orthologs generated using the Consurf website (consurf.tau.ac.il) (34). tNvj1p bound to tVac8p is shown in stick representation. The Fo − Fc difference electron density map (magenta, contoured at 3σ) was calculated in the absence of tNvj1p at 2.4-Å resolution. (C) Ribbon diagram showing two copies of the tVac8p–tNvj1p complex in the asymmetric unit. The two tVac8p molecules are colored cyan and gray, and tNvj1p molecules are colored magenta and red.
The 30-amino acid tNvj1p forms an extended straight structure. Even though the construct used for crystallography starts at residue 229, only residues bound to Vac8p were clearly visible in the electron density, and residues 229–291 are disordered.
Interfaces Between tVac8p and tNvj1p.
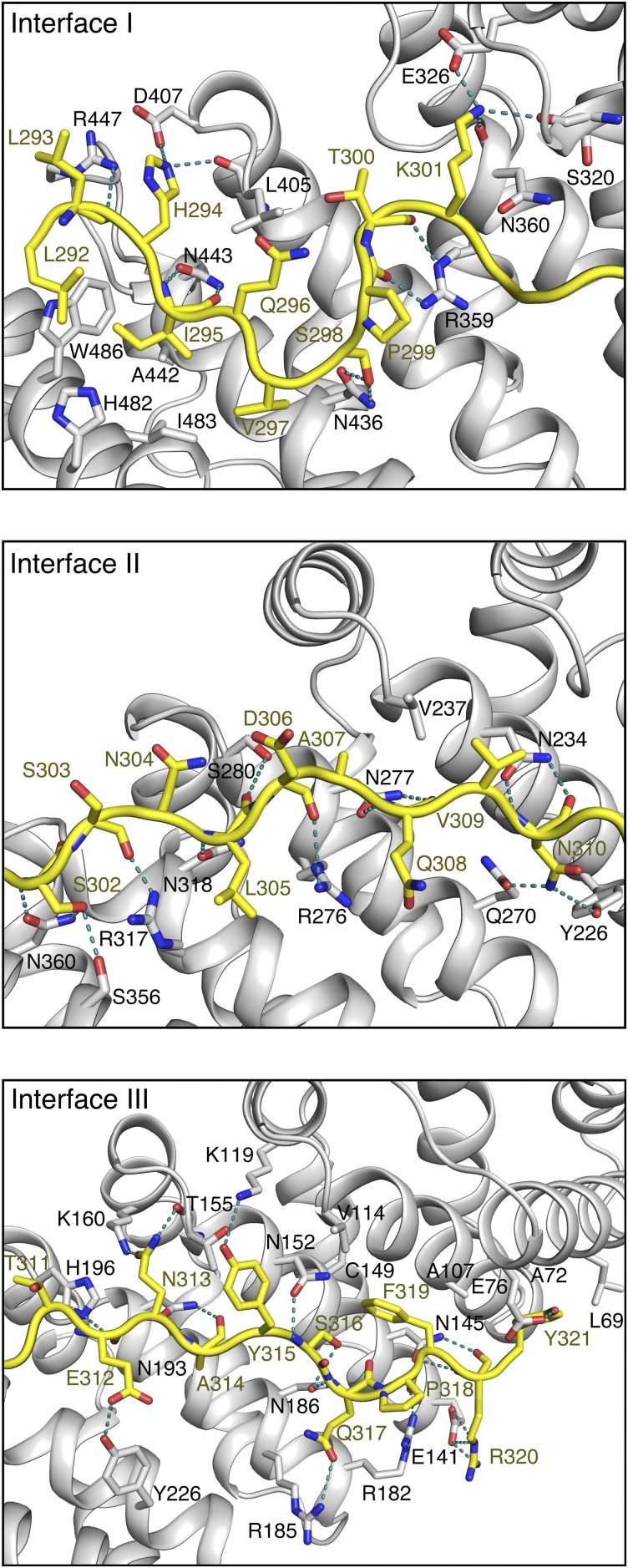
The extended tNvj1p loop of 80 Å in length binds to tVac8p via the highly conserved inner groove formed by all H3 helices from the 12 ARM repeats (Fig. 2 A and B). tNvj1p runs through tVac8p in an antiparallel fashion, and ∼2,300 Å2 of surface area is buried within the complex. The two tNvj1p fragments bound to tVac8p in the asymmetric unit could be superimposed precisely with an rmsd of 0.4 Å for all 30 Cα atoms, suggesting that the tVac8p–tNvj1p complex is mediated by specific interactions rather than random recognition. We divided the contact sites into three main regions (interface I–III) ordered from the N to the C terminus of tNvj1p (Fig. 3A and Fig. S3).
Fig. 3.
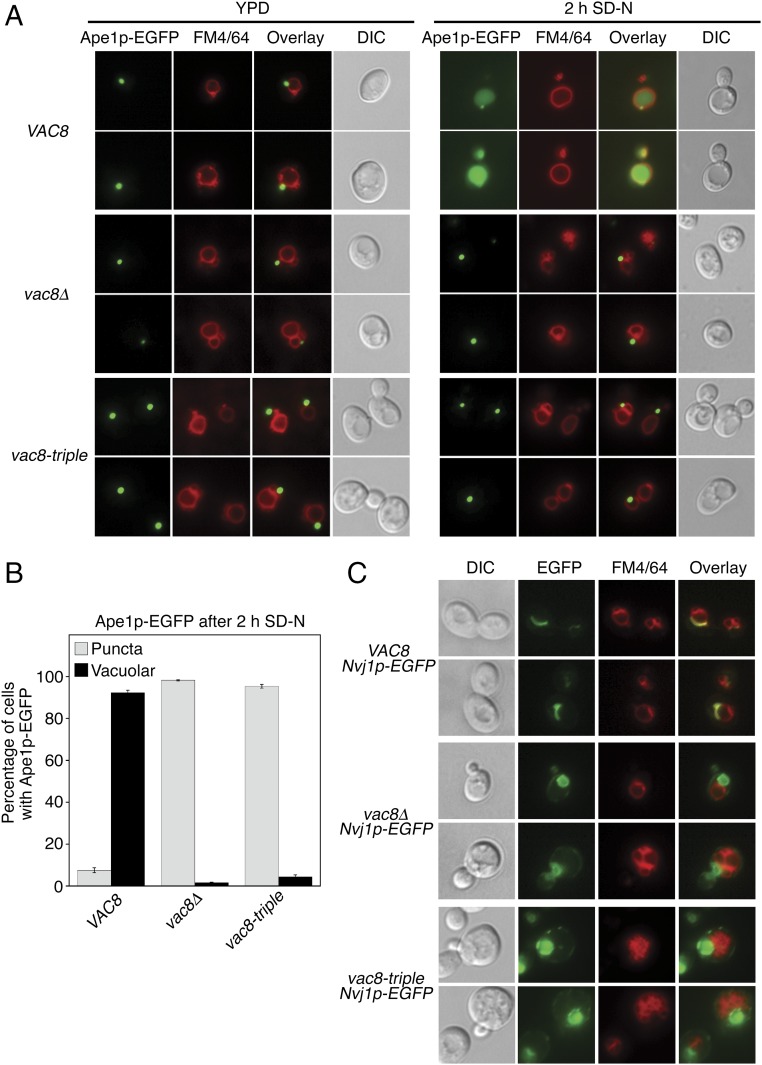
Binding interface between tVac8p and tNvj1p. (A) Schematic diagram showing the tVac8p (blue)–tNvj1p (black line) binding interface (Fig. S3 for more details). Red and blue dotted lines indicate hydrophobic contacts and intermolecular hydrogen bonds, respectively. tVac8p residue labels are boxed. (B) Pull-down experiment demonstrating the direct tVac8p–tNvj1p interaction in solution. The wild type and mutant of his-tagged tNvj1p were coexpressed with GST-tVac8p and immobilized on Ni-NTA resin. Proteins were analyzed by 15% SDS/PAGE and Coomassie blue staining. The asterisk indicates the residues included in the triple mutant. (C) The Nvj1p mutants, Nvj1p-F319E and Nvj1p-triple, do not support the formation of a tight junction between the nucleus and the vacuole. Cells expressing wild-type Nvj1p-EGFP, Nvj1p-EGFP (F319E), or Nvj1p-EGFP (triple) were labeled with FM4/64, a vacuole-specific lipophilic dye, in YPD media for 2 h at 30 °C. After free dye was removed, cells were resuspended in fresh media and allowed to grow for 2 h at 30 °C. GFP fluorescence for wild-type or mutant Nvj1p was then analyzed by fluorescence microscopy. Representative images from each cell type are shown. (Magnification: 100×.) (D) PMN was largely impaired in cells expressing Nvj1p-F319E or Nvj1p-triple. Experiments were carried out as in C except for treatment with 0.2 μM rapamycin for 6 h to induce PMN. (Magnification: 100×.) GFP fluorescence for Nvj1p and red fluorescence for vacuoles were analyzed by fluorescence microscopy. Representative images (Top) of each cell type with or without rapamycin, and quantification (Bottom) of cells with PMN blebs or PMN vesicles is shown. The yeast strains lacked the major vacuolar protease Pep4p to block the degradation of PMN blebs or vesicles in the vacuolar lumen. White arrows indicate nuclear membrane-derived fragments in the vacuole. DIC, differential interference contrast.
Fig. S3.
Interactions between Nvj1p and Vac8p. Close-up view of the tNvj1p and tVac8p interface. The three interfaces of tVac8p (gray) and tNvj1p (yellow) described in Fig. 3A are shown in cartoon representation. Oxygen and nitrogen atoms are colored red and blue, respectively. Blue dotted lines indicate H-bonds.
In interface I, residues 292–301 of tNvj1p adopt a thunderbolt-like structure containing two sharp bends. Residues 293–297 run antiparallel to the axis of the H3 helices of ARM10 and ARM11. Residues 297–300 form a loop that binds perpendicular to the H3 helices of ARM8–10, followed by residues 301 and 302 that run antiparallel to the axis of helix H3 of ARM8. The main chain atoms of Leu293, Ile295, and Gln296 are recognized by the side chains of Arg447 and Asn443 of tVac8p through intermolecular H-bonds, and two hydrophobic residues (Ile295 and Val297) of tNvj1p make nonpolar contacts with hydrophobic side chains Trp486, Ala442, His482, and Ile483 of tVac8p (Fig. 3A and Fig. S3).
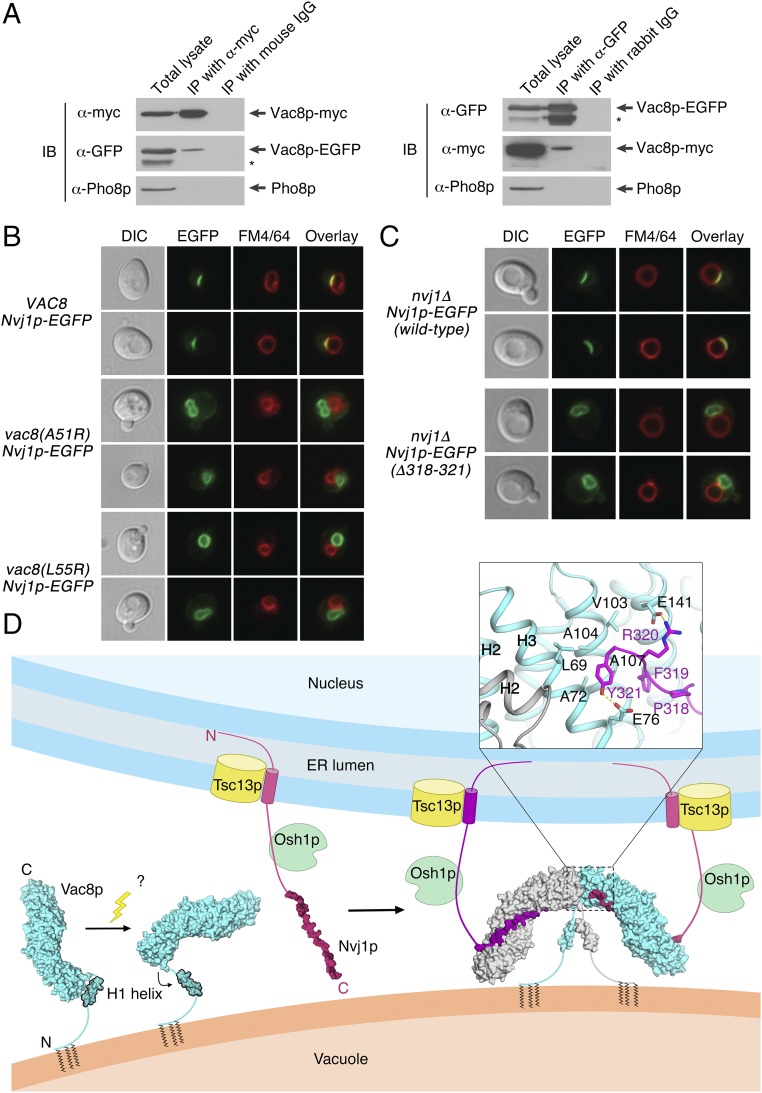
In interface II (residues 302–310) and interface III (residues 311–321), tNvj1p forms straighter β-strand–like structures. A slight bend formed from Thr311 and Glu312 of tNvj1p at the boundary between interfaces II and III facilitates the tight and extended interactions with the inner groove of tVac8p. The nine-residue loop in interface II runs through ARM5–9 of tVac8p in a diagonal direction relative to the axes of the H3 helices and makes extensive contacts with H3 helices from the tVac8p repeats. In interface III (residues 311–321), the main chain atoms of tNvj1p residues 313–317 form hydrogen bonds with the side chains of Asn193, His196, and Asn152 from the H3 helices of ARM2 to ARM7 of tVac8p. Additionally, tNvj1p Arg320 forms a salt bridge with tVac8p Glu141 (Fig. 3A and Fig. S3). There are also hydrophobic contacts in interfaces II and III. Particularly, Ala307 and Val309 of tNvj1p make hydrophobic interactions with Val237 of tVac8p in interface II. Finally, the aromatic residues Phe319 and Tyr321 of tNvj1p make hydrophobic contacts with Cys149, Val114, Ala107, Ala72, and Leu69 of tVac8p.
To verify whether the residues involved in the tVac8p–tNvj1p interactions in the crystal are important in solution, we made a series of N-terminally hexa-histidine-tagged mutants of tNvj1p and tested their ability to interact with GST-tVac8p using histidine tag pull-down assays. Fig. 3B shows that the binding affinity of I295R, K301E, and A307R of tNvj1p for tVac8p was reduced between two- and threefold compared with the native protein. More surprisingly, the F319E single mutant lost almost all ability to interact with tVac8p. Finally, the triple substitution mutant (I295R, A307R, and F319E, referred to as tNvj1p-triple) lost all ability to interact with tVac8p.
Disruption of the Vac8p–Nvj1p Interface Damages NVJ Formation and PMN in Vivo.
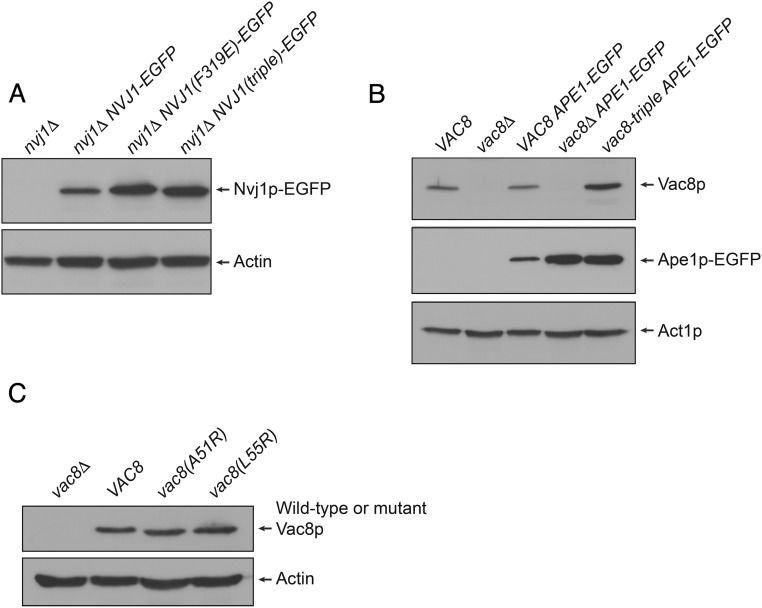
Next, we used S. cerevisiae to examine the functional relevance of the interaction interfaces between Vac8p and Nvj1p in vivo. Because Vac8p–Nvj1p interactions are largely responsible for the formation of NVJs that rely on the tight apposition of a region of the nuclear envelope with the vacuolar membrane, Nvj1p is enriched in the contact region between the nucleus and the vacuole in wild-type yeast cells (12). Consistent with this, when EGFP-conjugated wild-type Nvj1p was expressed in nvj1Δ yeast (Table S2), nearly all Nvj1p-EGFP proteins were localized to normal NVJs (Fig. 3C). By contrast, when Nvj1p-EGFP (F319E), which has a reduced affinity for Vac8p, was expressed, the mutant Nvj1p became dispersed almost evenly along the entire outer nuclear membrane region (Fig. 3C and Fig. S4A). Similarly, Nvj1p-EGFP (triple), which has no affinity for Vac8p, was also distributed over the nuclear envelope, instead of being confined to NVJs (Fig. 3C and Fig. S4A). Furthermore, substitution of wild-type Nvj1p with the triple mutant seemed to result in the loosening of the tight apposition between the nucleus and the vacuole.
Table S2.
Yeast strains used in this study
| Strain | Genotype | Source |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Ref. 43 |
| BY4742 nvj1Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 nvj1Δ::KanMX4 | Ref. 43 |
| BY4742 vac8Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 vac8Δ::KanMX4 | Ref. 43 |
| BY4742 pep4Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pep4Δ::KanMX4 | Ref. 43 |
| BY-Nvj1p-EGFP | BY4742 nvj1Δ with pYJ406-NVJ1-EGFP | This study |
| BY-Nvj1p(F319E)-EGFP | BY4742 nvj1Δ with pYJ406-NVJ1(F319E)-EGFP | This study |
| BY-Nvj1p(triple)-EGFP | BY4742 nvj1Δ with pYJ406-NVJ1(I295R/A307R/F319E)-EGFP | This study |
| BY-Nvj1p(Δ318–321)-EGFP | BY4742 nvj1Δ with pYJ406-NVJ1(Δ318–321)-EGFP | This study |
| BY-Vac8p(triple) | BY4742 vac8Δ with pRS316-VAC8((R276E/R317E/R359E) | This study |
| BY-VAC8-N1-EGFP | BY4742 with pYJ406-NVJ1-EGFP | This study |
| BY-vac8Δ-N1-EGFP | BY4742 vac8Δ with pYJ408-NVJ1-EGFP | This study |
| BY-vac8(t)-N1-EGFP | BY-Vac8p(triple) with pYJ408-NVJ1-EGFP | This study |
| BY-vac8(A51R)-N1-EGFP | BY4742 vac8Δ with pRS316-VAC8(A51R) and pYJ408-NVJ1-EGFP | This study |
| BY-vac8(L55R)-N1-EGFP | BY4742 vac8Δ with pRS316-VAC8(L55R) and pYJ408-NVJ1-EGFP | This study |
| BY-pep4Δ-N1-EGFP | BY4742 pep4Δ with pYJ406-NVJ1-EGFP | This study |
| BY-pep4Δ-N1a-EGFP | BY4742 pep4Δ with pYJ406-NVJ1(F319E)-EGFP | This study |
| BY-pep4Δ-N1b-EGFP | BY4742 pep4Δ with pYJ406-NVJ1(I295R/A307R/F319E)-EGFP | This study |
| BY-Ape1p-EGFP | BY4742 with pYJ408-APE1-EGFP | This study |
| BY-vac8(t)-Ape1p-EGFP | BY-Vac8p(triple) with pYJ408-APE1-EGFP | This study |
| BY-vac8Δ-Ape1p-EGFP | BY4742 vac8Δ with pYJ408-APE1-EGFP | This study |
| BY-Vac8p-EGFP/Vac8p-myc | BY4742 vac8Δ with pYJ406-VAC8-EGFP and pYJ408-VAC8-myc | This study |
Fig. S4.
Expression of Vac8p and Nvj1p mutants. (A) Expression of EGFP-conjugated wild-type and mutant Nvj1p. The expression of wild-type Nvj1p-EGFP, Nvj1p(F319E)-EGFP, and Nvj1p(triple)-EGFP was analyzed by immunoblotting. Actin was used as a loading control. (B) The expression of Vac8p-triple is comparable to that of wild-type Vac8p. The expression of wild-type Vac8p and the Vac8p-triple mutant in Ape1p-EGFP-expressing cells was analyzed by immunoblotting. Actin (Act1p) was used as a loading control. (C) The expression of Vac8p(A51R) and Vac8p(L55R) is comparable to that of wild-type Vac8p. The expression of wild-type Vac8p, Vac8p(A51R), and Vac8p(L55R) was analyzed by immunoblotting. Actin was used as a loading control.
NVJs are involved in various cellular functions including mediating PMN, a distinct form of nuclear microautophagy in budding yeast (5). Upon nutrient depletion or rapamycin treatment NVJs proliferate and subsequently undergo engulfment into the lumen of the vacuole, leading to the eventual degradation of the engulfed nuclear membranes (6). To test whether the interaction interfaces between Vac8p and Nvj1p revealed by our tVac8p–tNvj1p crystal structure are also critical for mediating PMN, yeast cells expressing EGFP-conjugated wild-type Nvj1p or mutant Nvj1p were subjected to rapamycin treatment, and PMN-mediated engulfment of Nvj1p-EGFP into the vacuole was analyzed by fluorescence microscopy. As shown in Fig. 3D, PMN blebs or vesicles were readily observed in the vacuoles of pep4Δ yeast cells (in which vacuolar proteases are inactivated and thereby prevented from degrading PMN vesicles in the vacuole) expressing wild-type Nvj1p. By contrast, only a few cells expressing Nvj1p-EGFP (F319E) or Nvj1p-EGFP (triple) exhibited PMN blebs or vesicles upon rapamycin treatment (Fig. 3D). Instead, small nuclear fragments were often found in cells expressing mutant Nvj1p (white arrows), suggesting that another type of nucleophagy occurred independently of NVJ (17, 18). Taken together, these in vivo results clearly demonstrate that the interaction interfaces between Vac8p and Nvj1p revealed by the crystal structure of the tVac8p–tNvj1p complex (Fig. 3A) play a critical role in NVJ formation and their cellular functions.
Competition Between Nvj1p and Atg13p for Vac8p Binding.
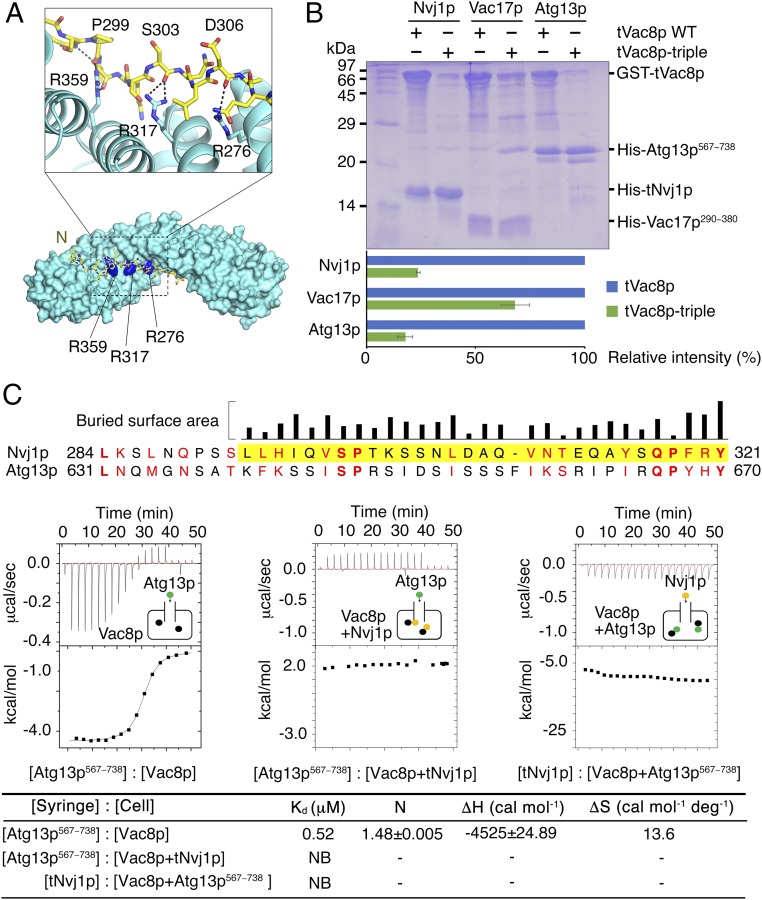
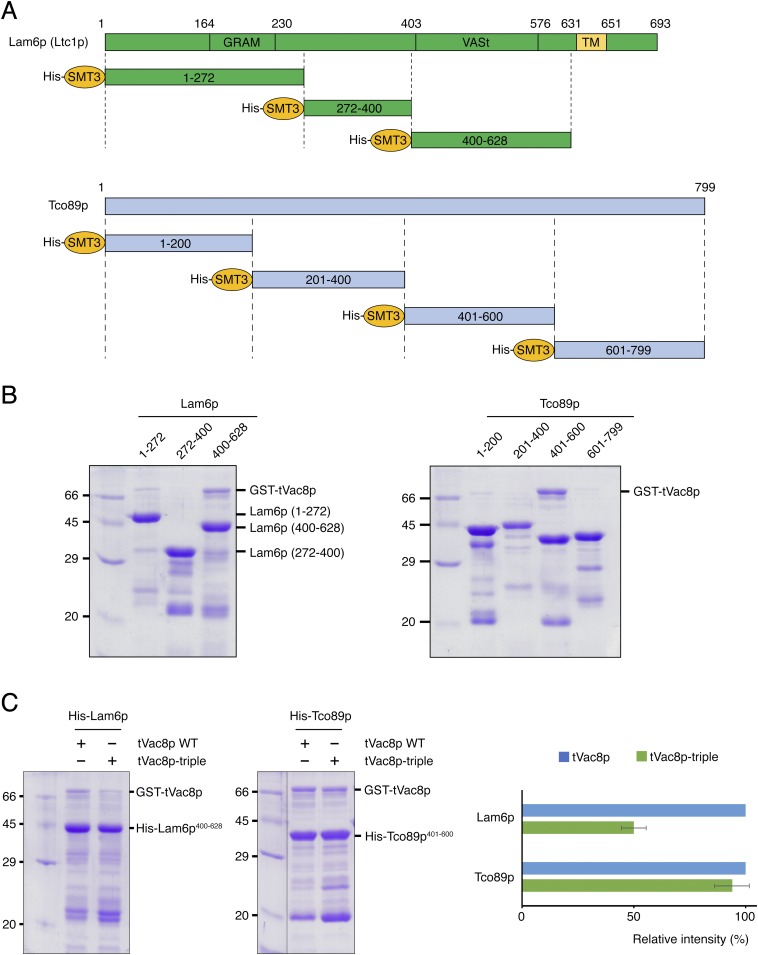
It was previously shown that Vac8p interacts with Vac17p and Atg13p, and these interactions play essential roles in vacuole inheritance and the CVT pathway, respectively (13, 14). Additionally, β-catenin, a structural homolog of Vac8p, shares the typical inner groove formed from the ARM repeats that serves as an interacting platform for their various binding partners (19). We therefore wondered whether Vac8p would also share Nvj1p-binding regions with Vac17p or Atg13p proteins. To test this, we prepared His-tagged tNvj1p, Vac17p (residues 290–380), and Atg13p (residues 567–738) proteins and GST-fused tVac8p and the tVac8p triple mutant (R276E, R317E, R359E, referred to as tVac8p-triple) by coexpression in bacterial cells. According to our structure, a triad of cationic residues (Arg276, Arg317, and Arg359) in the groove of tVac8p seems to be critically involved in the recognition of tNvj1p (Fig. 4A). Fig. 4B shows the results of pull-down experiments revealing interactions between wild-type tVac8p and tNvj1p, Vac17p, and Atg13p, whereas the tVac8p-triple mutant did not interact with tNvj1p or Atg13p. Interestingly, however, the tVac8p-triple retained its ability to interact with Vac17p. Surprisingly, the pull-down data were consistent with the sequence conservation, because regions of Nvj1p involved in Vac8p binding were found to be quite well aligned with those of Atg13p, but not strictly aligned with those of Vac17p (Fig. 4C). Additionally, we found that Lam6p (residues 400–628) and Tco89p (residues 401–600) specifically interact with tVac8p. These interactions were not significantly affected by the tVac8p triple mutation (Fig. S5). Taken together, these results suggest that Atg13p likely binds Vac8p in a similar manner to that shown for the tVac8p–tNvj1p interaction, although additional contacts might also occur between Vac8p and other binding partners including Vac17p, Lam6p, and Tco89p.
Fig. 4.
Nvj1p and Atg13p share a binding interface of Vac8p. (A) Cartoon representation of the cationic triad of tVac8p comprising R276, R317, and R359 that are involved in the interaction with tNvj1p. Critical hydrogen bonds are indicated by dotted lines between important residues shown in stick representation. (B) His-tag pull-down experiments revealing tVac8p interactions with tNvj1p, Vac17p, and Atg13p via the conserved cationic triad. Wild-type and triple mutant (R276E/R317E/R359E) GST-tagged tVac8p were coexpressed with his-tagged tNvj1p, Vac17p (residues 290–380), and Atg13p (residues 567–738) and immobilized on Ni-NTA resin. Copurified proteins were separated by 15% SDS/PAGE and stained with Coomassie blue. Quantitative data are shown below the gel. (C) Sequence alignment of S. cerevisiae Nvj1p (residues 284−321) and Atg13p (residues 631−670). The visible residues of Nvj1p in the structure of tVac8p–tNvj1p is highlighted in yellow. The identical and similar residues between Nvj1p and Atg13p sequences were indicated by bold font and red text, respectively. The alignment suggests that the interaction mode of Vac8p and Atg13p might resemble that of Vac8p and Nvj1p. Upper black bars indicate the relative surface area of Nvj1p buried by tVac8p–tNvj1p complexation calculated using the PISA program (35). The bottom figure shows ITC measurements of Vac8p and binding partners Atg13p567−738 and tNvj1p. Atg13p567−738 or tNvj1p was titrated into Vac8p alone, the Vac8p–tNvj1p complex, and the Vac8p–Atg13p567−738 complex at 25 °C as indicated. The results suggest that Vac8p exclusively and competitively binds to Nvj1p and Atg13p.
Fig. S5.
Particular regions of Lam6p and Tco89p directly interact with tVac8p. (A) Schematic representations showing domain structures of Lam6p and Tco89p. A series of truncated constructs used for pull-down experiments are indicated. All constructs were fused to the His6-SMT3 tag at their N termini. (B) His-tag pull-down experiments showing tVac8p interactions with Lam6p and Tco89p. GST-tagged tVac8p was coexpressed with His6-SMT3-tagged Lam6p or Tco89p and immobilized on Ni2+-NTA resin. Experiments were carried out as described in Fig. 4B. (C) To test whether Vac8p shares the binding interface for Nvj1p with Lam6p or Tco89p, wild-type and triple mutant (R276E/R317E/R359E) GST-tagged tVac8p were coexpressed with his-tagged Lam6p (residues 400–628) or Tco89p (residues 401–600) and immobilized on Ni2+-NTA resin. Quantitative data are shown on the right. TM, transmembrane domain.
Next, we tested whether Nvj1p could compete with Atg13p for interaction with Vac8p using ITC experiments. Atg13p was titrated into Vac8p alone and also into the Vac8p–tNvj1p complex prepared by coexpression. As shown in Fig. 4C, Vac8p alone bound Atg13p with an affinity (Kd) of 0.5 μM, whereas previously liganded Vac8p was unable to further associate with Atg13p. Likewise, Vac8p already bound to Atg13p was not able to interact with tNvj1p, suggesting that Vac8p exclusively and competitively interacts with its specific ligand. Competitive interaction of ligands to ARM-containing proteins has also been reported for β-catenin (20–22), which interacts exclusively with E-cadherin, Tcf-1, and APC in accordance with their molecular functions through a positively charged inner groove formed from H3 ARM helices. Similarly, we suggest that Vac8p interacts exclusively with Nvj1p or Atg13p, in agreement with their known physiological relevance.
Mutation of Vac8p Cationic Residues Disrupts the CVT Pathway.
To examine whether the differential affinity of the Vac8p triple mutant to Atg13p and Vac17p revealed by the in vitro binding experiments is relevant to their in vivo functions, we attempted to observe the CVT pathway of Ape1p and vacuole inheritance in budding yeast cells expressing wild-type or the mutant Vac8p. Atg13p is known to mediate the transport of cytoplasmic Ape1p to the vacuole lumen through the CVT pathway upon starvation (14, 23). In yeast cells expressing wild-type Vac8p, Ape1p-EGFP was dispersed in the lumen of vacuoles upon starvation (Fig. 5A), suggesting that cytoplasmic Ape1p-EGFP was transported to the vacuole via the CVT pathway. By contrast, Ape1p-EGFP formed punctate structures in the cytoplasm of yeast cells lacking Vac8p, indicating a defective CVT pathway (Fig. 5 A and B and Fig. S4B). A similar result was obtained with yeast cells expressing the Vac8p-triple (Fig. 5 A and B and Fig. S4B), suggesting that the triad of cationic residues (Arg276, Arg317, and Arg359) in the groove of Vac8p are also critical for the in vivo interaction between Vac8p and Atg13p, and thus for the CVT pathway.
Fig. 5.
Mutation of the cationic triad in Vac8p causes disruption of the NVJ and CVT pathway. (A) The Vac8p-triple mutant fails to support cytoplasm-to-vacuole transport of Ape1p-EGFP upon starvation. The vacuoles of wild-type, vac8Δ, and vac8-triple mutant cells expressing Ape1p-EGFP were labeled with FM4/64 in YPD media for 2 h at 30 °C. After free dye was removed, cells were resuspended in nitrogen starvation medium (SD-N) and allowed to grow for 2 h at 30 °C. GFP signals for Ape1p and FM4/64 signal in vacuoles were then analyzed by fluorescence microscopy. Representative images from each cell type are shown. (Magnification: 100×.) (B) Graph showing quantitative data for cytoplasmic and vacuolar Ape1p-EGFP obtained as described in A. (C) The Vac8p-triple mutant does not support the formation of a tight junction between the nucleus and the vacuole. Experiments were performed as in A without starvation, and GFP fluorescence for Nvj1p was analyzed by fluorescence microscopy. (Magnification: 100×.) DIC, differential interference contrast.
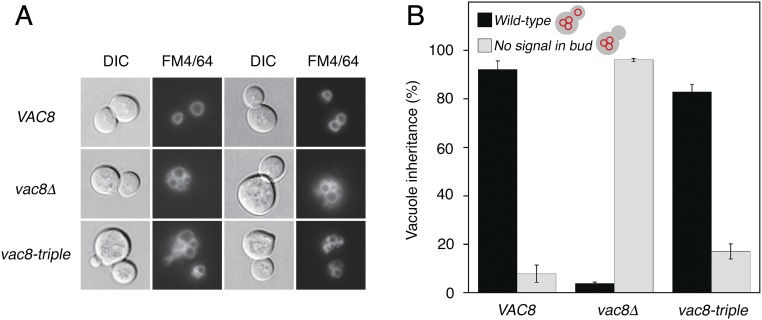
Vac17p interacts simultaneously with Vac8p and Myo2p, the yeast class V myosin motor, and this trimeric complex moves the vacuole to the bud, mediating vacuole inheritance during yeast cell budding (24). To examine whether the Vac8p-triple mutant could support vacuole inheritance, yeast cells expressing wild-type or mutant Vac8p were stained with the vacuole-specific dye FM4/64 and analyzed for vacuole inheritance in newly formed buds. As shown in Fig. S6, no obvious defect in vacuole inheritance was observed in cells expressing the Vac8p-triple mutant, consistent with the finding that Vac8p-triple retained significant affinity for Vac17p (Fig. 4B). However, Vac8p-triple undoubtedly abolished the formation of NVJ as shown in the Nvj1p-triple mutant (Fig. 5C).
Fig. S6.
The Vac8p-triple mutant supports vacuole inheritance. To analyze whether Vac8p-triple affects the vacuole inheritance, the vacuoles of wild-type or Vac8p-triple mutant cells were labeled with FM4/64 for 30 min at 30 °C. After the free dye was removed, cells were resuspended in fresh media and allowed to grow for 3 h at 30 °C. The vacuole morphology and inheritance were analyzed by fluorescence microscopy. Representative images from each cell type (A) and quantitation for vacuole inheritance (B) are shown. (Magnification: 100×.) DIC, differential interference contrast.
Vac8p Self-Associates Through ARM1.
The crystal structure revealed two tVac8p–tNvj1p complexes arranged via twofold rotation symmetry into a four-helix bundle structure comprising H2 and H3 of ARM1 in the asymmetric unit (Fig. 6A). The tVac8p homo-dimeric interface buries 622 Å2 of surface area in the dimeric tVac8p–tNvj1p complex and is mediated by van der Waals contacts between H2 and H3 of ARM1. In particular, the side chains of Ala51, Leu52, and Leu55 from H2 and Leu63, Ala70, and Ile74 from H3 make hydrophobic contacts with the same side chains of the opposing molecule in the homodimer. In addition to hydrophobic interactions, Ser66 and Glu73 make hydrogen bonds with Thr54 and Gly47 from the opposing molecule, respectively (Fig. 6B).
Fig. 6.
The first ARM of tVac8p mediates self-association of Vac8p. (A) Cartoon representation of the tVac8p–tNvj1p complex in the crystal asymmetric unit. Two copies of the tVac8p–tNvj1p heterodimer are organized via twofold symmetry, forming a four-helix bundle from ARM1. The black box highlights the orientation of helices from ARM1 repeats in the tVac8p homo-dimeric interface, and black arrows indicate the direction of the helices. (B) Close-up view of the four-helix bundle in the tVac8p homo-dimeric interface (Left). Blue dotted lines indicate intermolecular H-bonds. (Right) A schematic helical wheel showing the organization of the four helices that form the twofold interface. Blue and red dashed lines indicate hydrogen bonds and hydrophobic contacts, respectively. (C) SEC profiles of Vac8p40−515–tNvj1p, Vac8p40−515, (A51R or L55R)–tNvj1p, and Vac8p40−515–tNvj1pΔ318−321 complexes. The results reveal the behavior of wild-type Vac8p and its variants in solution. Experiments were performed as shown in Fig. 1B. (D) Analytical ultracentrifugation measurement of the molecular mass of wild-type and mutant Vac8p40−515 in complex with tNvj1p using the sedimentation equilibrium method. The upper panel shows the residuals, and the results are summarized on the bottom. (E) Chromatograms comparing the SEC behaviors of Vac8pΔH1-tNvj1p, Vac8pΔH1 alone, and Vac8pΔH1(A51R or L55R)–tNvj1p. Proteins at three different concentrations (1, 2, and 6 mg/mL) were injected into a Superdex 200 column and analyzed as shown in B. M.W., molecular weight.
At first, we assumed that the self-association of tVac8p might be an artifact of crystal contacts and not a biologically relevant interface because the Vac8p–tNvj1p complex migrated as an apparent heterodimer rather than a heterotetramer in gel-filtration chromatography (Fig. 1B). However, the following biochemical data strongly indicated the possibility that Vac8p self-associated in solution. First, the flexibly connected H1 helix was not vulnerable to proteases such as subtilisin or trypsin, and if Vac8p complexed Nvj1p as a monomer this would likely not be the case. However, a dimeric form of Vac8p would likely restrict access of proteases through steric hindrance, as shown in the crystal structure (Fig. 2C). Second, truncated Vac8p (residues 40–515, referred to as Vac8p40−515) that included only the ARM repeats formed a tight heterotetramer based on both gel-filtration chromatography and analytical ultracentrifugation (Fig. 6 C and D). Surprisingly, the single mutation A51R or L55R interfered with the dimer interface to the extent that the mutant forms of Vac8p40−515 were unable to self-associate, suggesting that residues at the dimer interface are important for self-association in solution (Fig. 6 C and D). Collectively, the results strongly suggest that the Vac8p–Nvj1p complex self-oligomerizes in solution into the assembly shown in the crystal structure (Fig. 2C and Fig. 6A), and this may be related to the molecular function. This raises the question of why full-length Vac8p complexed with tNvj1p eluted as a monomer in gel-filtration chromatography. One explanation is that the flexibly linked H1 helix might interact with H2 and H3 of ARM1 from Vac8p to sterically mask the dimerization interface in solution. Indeed, a fold-back mechanism operating through the interaction between ARM repeats on the N- or C-term helices has been previously proposed for β-catenin (25–27). To test this hypothesis, we generated a truncated mutant lacking helix H1 (residues 19−33, referred to as Vac8pΔH1) for the otherwise full-length Vac8p. Because the H1 helix is flexibly connected to ARMs by disordered residues, we predicted that the removal of H1 would not affect the overall structure of Vac8p. Surprisingly, we observed that Vac8pΔH1 complexed with tNvj1p to form a dimer in a concentration-dependent manner. However, Vac8pΔH1 mutants with the A51R or L55R mutation could not form a dimer at all, suggesting that self-association of Vac8p might be regulated by the H1 helix in the presence of tNvj1p (Fig. 6E).
Under appropriate cellular conditions, other (unknown) cellular factor(s) might induce a conformational change of helix H1 that causes it to dissociate from H2 and H3 of ARM1 to promote the self-association of Vac8p through the exposed H2 and H3. Structural analysis showed that the highly conserved final four residues (PFRY) of Nvj1p are directly oriented away from the H2 and H3 helices of tVac8p ARM1 (Fig. 7D). In particular, Tyr321 of tNvj1p forms an H-bond with the conserved Glu76 of tVac8p and also forms hydrophobic contacts with conserved Leu69 and Ala72 of tVac8p. Remarkably, we found that truncated Nvj1p lacking the last four residues (tNvj1pΔ318–321) could still bind to Vac8p40–515 but could not induce the dimerization of Vac8p40–515 (Fig. 6 C and D). We tested whether Nvj1p could affect the self-oligomerization of Vac8p by comparing Vac8p40–515 alone and Vac8p40–515 complexed with tNvj1p using gel-filtration chromatography. We expected that Vac8p40–515 without tNvj1p may elute as a monomer if Nvj1p served as a regulatory factor. However, partially consistent with our expectations, Vac8p40–515 alone was not soluble and aggregated in solution and eventually eluted from the column in the void volume. From this observation, we speculate that H2 and H3 from ARM1 of Vac8p40–515 are highly disordered in the absence of Nvj1p and unable to form the four-helix bundle structure, which exposes the hydrophobic residues from H2 and H3, culminating in aggregation. Consistently, Vac8pΔH1 was unable to form a dimer at even a high concentration when not complexed with Nvj1p (Fig. 6E). Taken together, our results led us to propose that binding of Nvj1p organizes the H2 and H3 helices of Vac8p and induces them to form a four-helix bundle structure when required, rather than directly initiating the self-oligomerization of Vac8p (Fig. 7D).
Fig. 7.
Vac8p forms a homo-dimer in vivo for NVJ formation. (A) Vac8p-EGFP coprecipitates with Vac8p-myc. Vacuoles isolated from BY4742 expressing both Vac8p-EGFP and Vac8p-myc were detergent-solubilized and incubated with either anti-myc antibodies or control mouse IgG, or with either anti-GFP antibodies or control rabbit IgG in the presence of protein A Sepharose. Protein A Sepharose-bound material was then analyzed by immunoblotting using the indicated antibodies. The asterisk indicates degradation products of Vac8p-EGFP. All experiments were performed multiple times with similar results, and the data shown are representative of all results. (B) Normal NVJs do not form in cells expressing the dimerization-defective mutant Vac8p(A51R) or Vac8p(L55R). The experiment was carried out as described in Fig. 5C. (C) The Nvj1p mutant Nvj1p (Δ318−321) fails to support the formation of a tight junction between the nucleus and the vacuole. The experiment was performed as Fig. 3C. (D) Schematic representation showing the putative working model of the self-association of the Vac8p–Nvj1p complex regulated by both the H1 helix and Nvj1p. Unknown factors likely cause dissociation of the flexible H1 helix of Vac8p from the ARM repeats, and binding of Nvj1p might induce the reorganization of helices H2 and H3 of ARM1 to form the four-helix bundle structure at the complex interface. These conformational changes eventually establish a Vac8p–Nvj1p hetero-tetramer that could be functionally critical for NVJ formation and/or PMN signaling. DIC, differential interference contrast; ER, endoplasmic reticulum; IB, immunoblotting; IP, immunoprecipitation.
Vac8p Dimerization Is Required for Normal NVJ Formation.
Our in vitro experiments (Fig. 6) strongly suggest that Vac8p forms a dimer under the regulatory control of Nvj1p. To confirm whether Vac8p dimerizes in vivo, EGFP-conjugated Vac8p and myc-tagged Vac8p were coexpressed in yeast cells, and their interactions were analyzed by coimmunoprecipitation. As shown in Fig. 7A, Vac8p-EGFP was coprecipitated with Vac8p-myc, suggesting that Vac8p forms a homodimer in vivo. To examine whether Vac8p dimerization is critical for its functions, in particular NVJ formation, Nvj1p-EGFP was expressed in wild-type VAC8 yeast cells or vac8Δ cells expressing Vac8p(A51R) or Vac8p(L55R). Whereas endogenous Vac8p supported the formation of normal NVJs in wild-type VAC8 cells, Nvj1p-EGFP fluorescence was dispersed over the entire nuclear membrane in Vac8p(A51R)- and Vac8p(L55R)-expressing cells (Fig. 7B and Fig. S4C). Consistently, EGFP-conjugated Nvj1pΔ318−321, a mutant that cannot induce Vac8p dimerization, failed to form normal NVJs (Fig. 7C). Thus, these data strongly suggest that Vac8p homodimerization is critical for its function in NVJ formation.
Discussion
In this study we determined the crystal structure of the tVac8p–tNvj1p complex and established the protein organization mediating NVJs at the molecular level. To obtain diffraction-quality crystals of the Vac8p–Nvj1p complex we truncated both the N and C termini of Vac8p. It is noteworthy that truncation of the C terminus of Vac8p was critical to producing crystals because the flexibly connected H1 helix of Vac8p was plausibly located in the H3 helix position of ARM12 of neighboring molecules that stabilized lattice contacts and crystal packing (Fig. S2 C and D). Indeed, the binding affinity of tVac8p for tNvj1p was reduced by around eightfold compared with the native protein. We assumed that the reduced affinity might be because the truncated regions are somewhat involved in the Nvj1p interaction, and/or because the truncations affected protein stability by generating incomplete ARM repeats for ARM12. Structural analysis revealed that ARM12 itself is not directly associated with the Nvj1p interaction. Interestingly, we found that the binding affinity (Kd of 0.7 μM) between tNvj1p and Vac8p (residues 1−532), in which all 12 ARMs are intact, was almost comparable to that of tNvj1p and full-length Vac8p based on ITC measurements (Fig. 1C). This suggests that no additional binding energy would be gained from the presence of the C terminus of Vac8p. Rather, the ARM12 H3 truncation likely affects the stability of tVac8p. From this observation, we suggest that the structural integrity resulting from the correct formation of all 12 ARM repeats in Vac8p might be important for protein stability and interaction with Nvj1p (13).
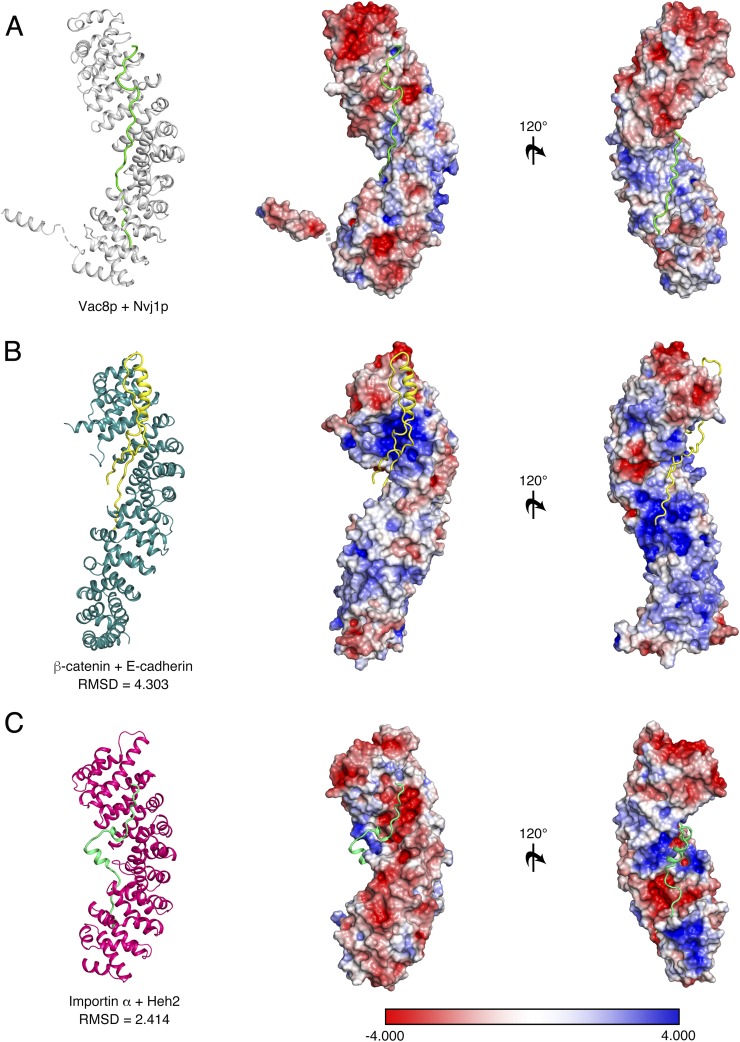
As expected, the overall architecture of the tVac8p–tNvj1p complex resembles the β-catenin/E-cadherin complex (rmsd = 4.3 Å) and the importin α/Heh2 complex (rmsd = 2.4 Å) and has similar structural features, despite the low sequence similarity between them (Fig. S7) (15, 28–30). First, the central domains of Vac8p and β-catenin are composed of 12 tandem ARM repeats that are organized into a right-handed superhelical structure. Second, Vac8p binds Nvj1p in an antiparallel fashion, and β-catenin interacts with E-cadherin, Tcf-1, and APC exclusively or partially in an antiparallel manner. Third, the interacting partners of ARM domain proteins adopt a mainly extended loop structure within the groove formed by the ARM repeats. Additionally, client ligands compete for the same ARM domain-mediated binding platform formed. Finally, the self-interaction referred to as the fold-back mechanism that involves the N- and C-terminal ARM repeats is present in both Vac8p and β-catenin. Because these structural features may be common among all ARM-containing proteins, we propose that the Vac8p–Nvj1p complex might be evolutionarily conserved with β-catenin. Despite the structural similarity, some unique features were apparent. First, the binding affinity of the Vac8p–Nvj1p complex is weaker than that between β-catenin and E-cadherin (22). Second, whereas charged interactions are stronger in the β-catenin/E-cadherin interface, the Vac8p–Nvj1p interface is formed from both H-bonds and nonpolar contacts, and the charged buttons shown in β-catenin are not conserved. Finally, self-association of ARM domains is a unique feature of Vac8p. In summary, although it has been reported that Vac8p and β-catenin do not share all functions, they clearly share some biologically important features in common, specifically the formation of a structural platform through ARM repeats that can interact with other functional molecules and that responds to cellular demands.
Fig. S7.
Comparison of the tVac8p–tNvj1p complex with other ARM-containing proteins. Ribbon and surface charge representations of the tVac8p (gray)–tNvj1p (green) complex (A), β-catenin (dark green)–E-cadherin (yellow) complex (B), and Importin α (magenta)–Heh2 (light green) complex (C) shown in the same orientations. The electrostatic potential of the surface area was calculated using the APBS program (42) and contoured at ± 4 KT/e (blue, positive; red, negative).
The self-association of Vac8p through a specific ARM repeat was unexpected, and the functional implications of this self-association in the working model of Vac8p require further investigation (Fig. 7D). Our accumulated biochemical data suggest that the self-association of Vac8p is dependent on the interaction with Nvj1p, indicating that the oligomerization of Vac8p is associated with a particular function such as forming NVJs or PMN pathway signaling (Fig. 7 B–D). Future work should therefore focus on other potential binding partners such as Atg13p or Vac17p that could drive the self-association of Vac8p.
Recently, several proteins have been suggested to mediate nuclear–vacuolar junctions (31–33). Ltc1p (also known as Lam6p), an ER integral membrane protein, is localized to NVJ via its binding to Vac8p (31, 33). Another integral ER membrane protein, Mdm1p, binds vacuolar phosphatidylinositol-3-phosphate through its PX domain, forming NVJs (32). Although the molecular composition or architecture of NVJs formed by these proteins remains unknown, their contribution to the formation of a tight NVJ is likely to be limited because disruption of the Nvj1p–Vac8p interaction largely impaired the tight apposition between the nucleus and the vacuole (Fig. 3C, Nvj1p-triple). Thus, it is very likely that the NVJ mediated by Nvj1p–Vac8p constitutes the main contact site between the two organelles whereas other NVJs may play a complementary or even a redundant role.
In conclusion, this study provides insight into understanding how Vac8p recognizes Nvj1p, and how the resulting protein complex physically mediates this organelle contact site and promotes the molecular events that occur at these specialized intracellular zones.
Materials and Methods
Cloning.
The full-length Vac8p gene was amplified by PCR using S. cerevisiae genomic DNA and fused into the pGEX-6P-1 vector. DNA fragments encoding S. cerevisiae Nvj1p and Atg13p were cloned into the pCDFDuet-1 vector that had been modified previously to include a tobacco etch virus (TEV) protease cleavage site before the first residues of Nvj1p and Atg13p. Each plasmid was transformed into competent E. coli BL21 (DE3) cells, and protein expression was induced with 0.4 mM isopropyl β-d-1-thiogalactopyranoside when the cell density reached an A600 value of 0.6. After culturing for a further 18 h at 18 °C, cells were harvested by centrifugation at 3,200 × g for 15 min at 4 °C. Cells expressing Vac8p were resuspended in PBS, and cells expressing Nvj1p, Atg13p, or Vac8p–Nvj1p were resuspended in buffer A (25 mM sodium phosphate, pH 7.8, 400 mM sodium chloride, and 10 mM imidazole).
Protein Production.
Nvj1p and Atg13p were purified by Ni2+-immobilized metal affinity chromatography (Ni2+-IMAC). His6-tags were removed by TEV protease during dialysis against buffer B comprising 25 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, and 5 mM β-mercaptoethanol (β-ME). After cleavage, reactions were subjected to subtractive Ni2+-IMAC to remove tags and undigested protein, followed by SEC with a Superdex 200 (16/600) column (GE Healthcare) preequilibrated with buffer C comprising 25 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, and 5 mM DTT. Vac8p proteins were purified by GST affinity chromatography, and the GST tag was removed using PreScission protease. Proteins were further purified using ion-exchange chromatography on a HiTrap Q HP column and gel-filtration chromatography on a Superdex 200 column.
For purification of the tVac8p–tNvj1p complex, plasmids encoding GST-tagged tVac8p and His6-tagged tNvj1p were cotransformed and overexpressed in E. coli BL21 (DE3) cells. Proteins were purified by Ni2+-IMAC (GE Healthcare), and tags were cleaved by TEV and PreScission protease, followed by dialysis overnight against buffer B at 4 °C. Proteins were subsequently subjected to subtractive Ni2+-IMAC and GST affinity chromatography to remove cleaved tags and then further purified using SEC with a Superdex 200 column (GE Healthcare) in buffer C.
Selenomethionine-labeled tVac8p–tNvj1p complex was expressed in E. coli B834 (DE3) cells using M9 minimal media and purified using the same procedure as described above for the native protein. All mutants used in this study were generated by PCR-based site-directed mutagenesis. Mutations were confirmed by DNA sequencing, and proteins were purified as described previously.
Additional methods are described in SI Materials and Methods.
SI Materials and Methods
Crystallization and Structure Determination.
For crystallization of the tVac8p–tNvj1p complex, proteins were concentrated to 10 mg/mL using an Amicon Ultra-15 centrifugal filter (molecular mass 50-kDa cutoff; Merck Millipore). Crystals for both native and Se-Met tVac8p–tNvj1p proteins were grown in 22% PEG 400, 0.1 M bis-Tris propane (BTP), pH 6.5, and 1.5% t-butanol by the hanging-drop vapor diffusion method at 20 °C. Crystals were transferred to a cryoprotectant solution comprising 30% PEG 400, 0.1 M BTP, pH 6.5, and 1.5% t-butanol and frozen in liquid nitrogen. Diffraction data for both native and Se-Met crystals were collected at the Pohang Accelerator Laboratory (7A beamline) and processed using the HKL2000 package (36). Phases were calculated from a SAD dataset collected at a Se peak wavelength of 0.9793 Å using the PHENIX program (37). Structures were built using Coot (38) and refined with PHENIX (37). The final model was refined to R/Rfree values of 0.193/0.217 using native data and included two complexes in the asymmetric unit. Residues 10–18, 35–39, and 513–515 of Vac8p and residues 229–291 of Nvj1p in the first copy, and residues 10–18, 35–39, and 513–515 of Vac8p and residues 229–291 of Nvj1p in the second copy are not visible in the electron density maps and were omitted from the final model. Crystallographic data and refinement results are summarized in Table S1.
SEC Analysis.
To assess the interaction between Vac8p and Nvj1p (as indicated in Fig. 1B) and analyze the relative molecular mass of Vac8p and Vac8p mutants (Vac8p40−515 + tNvj1p, Vac8p40−515, [A51R or L55R] + tNvj1p, and Vac8p40−515 + tNvj1pΔ318−321 in Fig. 6C; Vac8pΔH1 alone and complexed with tNvj1p in Fig. 6E), purified proteins were injected onto a Superdex 200 (16/600) column (GE Healthcare) equilibrated with buffer C. Protein standards used in the experiment were ferritin, 440 kDa; aldolase, 158 kDa; conalbumin, 75 kDa; ovalbumin, 43 kDa; and ribonuclease A, 13.7 kDa.
ITC Analysis.
All protein samples (Vac8p, Vac8p1−532, tVac8p, tNvj1p, Atg13p567−738, Vac8p–tNvj1p, and Vac8p–Atg13p567−738) used in ITC measurements were prepared in buffer B using a HiTrap desalting column (GE Healthcare). ITC experiments were performed using a Microcal ITC200 instrument (GE Healthcare) at 25 °C. The molar concentrations used in the sample cell and the syringe were 40 μM and 0.6 mM, respectively (Fig. 1C and Fig. 4C). Measurements were carried out over 20 injections of 2 μL, with a reference power of 5 μcal/s, and a stirring speed of 1,000 rpm. Titration curves were fitted to the one set of sites model using Origin software provided by the manufacturer.
Analytical Ultracentrifugation.
The absolute molecular mass of Vac8p40−515–tNvj1p, Vac8p40−515 (A51R)–tNvj1p, Vac8p40−515 (L55R)–tNvj1p, and Vac8p40−515–tNvj1pΔ318−321 was measured by sedimentation equilibrium analysis in an XL-A analytical ultracentrifuge (Beckman Coulter) as described previously (39). Protein samples were prepared in a buffer consisting of 25 mM Tris⋅HCl, pH 7.5, 200 mM NaCl, and 5 mM β-mercaptoethanol.
Pull-Down Experiments.
For His-tag pull-down experiments (Fig. 3B), 10 μL of Ni-NTA agarose (Qiagen) was incubated with supernatants from E. coil cells coexpressing GST-tVac8p and wild-type tNvj1p or GST-tVac8p and tNvj1p mutants for 1 h at 4 °C. Beads were washed three times with 500 μL of buffer A supplemented with 50 mM imidazole plus 0.1% Triton X-100 and 0.1% Nonidet P-40. Proteins were eluted with 4× SDS sample buffer, analyzed by 15% SDS/PAGE, and stained with Coomassie blue. To assess the binding of Nvj1p, Atg13p, Lam6p, Tco89p, and Vac17p to tVac8p and the tVac8p-triple mutant, as shown in Fig. 4B and Fig. S5, GST-tVac8p or the GST-tVac8p-triple mutant (R276E/R317E/R359E) was coexpressed with histidine-tagged tNvj1p, Vac17p, and Atg13p and His6-SMT3-tagged Lam6p and Tco89p in E. coli BL21 (DE3) cells. His-tag pull-down experiments were performed as described above.
Yeast Strains and Vacuole Staining with FM4/64.
Yeast strains used in this study are listed in Table S2. Vacuoles of growing yeast cells were labeled with FM4/64 as previously described with minor modifications (40). Briefly, cells were grown at 30 °C to an OD600 of 0.8 in YPD (10 g/L yeast extract, 20 g/L peptone, and 20 g/L dextrose) medium. Then, the cells were incubated with aeration in YPD medium containing 20 μM FM4/64 (Molecular Probes) for 30 min at 30 °C. The cells were then harvested by centrifugation (2,400 × g) at room temperature for 1 min, washed, and resuspended in fresh medium. The cells were then allowed to grow for 3 h at 30 °C. After harvesting by centrifugation, the cells were analyzed by a fluorescence microscope (Nikon Eclipse Ti-U) with a Nikon Plan Apo ×100, 1.45/N.A. oil immersion objective.
PMN and CVT Induction.
Cells were grown overnight at 30 °C with shaking in YPD. Cultures were adjusted to OD600 = 0.5 and 0.5 μM FM4/64 was added to visualize vacuoles. The cells were then allowed to grow for 2 h at 30 °C. For PMN induction, 0.2 μM rapamycin (Sigma-Aldrich) was added and the cells were further incubated for 6 h at 30 °C. For CVT induction, cells grown in YPD were washed with autoclaved water three times and resuspended in nitrogen starvation (SD-N) media. The cells were further allowed to grow for 2 h at 30 °C.
Coimmunoprecipitation.
Yeast vacuoles were isolated as described (41). Vacuoles were resuspended in ice-cold solubilization buffer (PBS containing 0.5% Triton X-100, 1 mM EDTA, 1 mM PMSF, and 10 mM leupeptin) and incubated on ice for 20 min. Detergent-insoluble material was removed by centrifugation at 16,200 × g (10 min, 4 °C). The resulting postcentrifugation supernatants were precleared by incubation with protein A Sepharose (GE Healthcare) at 4 °C for 1 h. Anti-myc antibodies, control mouse IgG, anti-GFP antibodies, or control rabbit IgG were added to the precleared supernatants and incubated 4 °C on a nutator mixer for 2 h. Protein A Sepharose was then added and further incubated for 1 h. Protein A Sepharose beads were collected by centrifugation at 3,000 × g (1 min, 25 °C) and washed three times with ice-cold solubilization buffer. Bound proteins were eluted with SDS sample buffer for SDS/PAGE analysis followed by immunoblotting using anti-myc, anti-GFP, and anti-Pho8p antibodies.
Acknowledgments
We thank staff from beamline 7A at the Pohang Accelerator Laboratory (PAL) for assistance with data collection, Dr. Bill Wickner (Geisel School of Medicine, Dartmouth College) for the generous gift of anti-Vac8p rabbit serum, and So Young Yoon for expert technical assistance. This research was supported by Cell Logistics Research Center Grant 2016R1A5A1007318; National Research Foundation of Korea Grants NRF-2015R1D1A1A01058016, NRF-2014R1A2A1A11051229, and NRF-2015R1A2A1A15055269; and National Research Foundation of Korea Global PhD Fellowship Program Grant NRF-2014H1A2A1020322 (to H.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.H.H. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5XJG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701030114/-/DCSupplemental.
References
- 1.Elbaz Y, Schuldiner M. Staying in touch: The molecular era of organelle contact sites. Trends Biochem Sci. 2011;36:616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helle SC, et al. Organization and function of membrane contact sites. Biochim Biophysica Acta. 2013;1833:2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Roberts P, et al. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kvam E, Goldfarb DS. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007;3:85–92. doi: 10.4161/auto.3586. [DOI] [PubMed] [Google Scholar]
- 6.Krick R, et al. Piecemeal microautophagy of the nucleus: Genetic and morphological traits. Autophagy. 2009;5:270–272. doi: 10.4161/auto.5.2.7639. [DOI] [PubMed] [Google Scholar]
- 7.Dawaliby R, Mayer A. Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear-vacuolar junctions. Mol Biol Cell. 2010;21:4173–4183. doi: 10.1091/mbc.E09-09-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine TP, Munro S. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol Biol Cell. 2001;12:1633–1644. doi: 10.1091/mbc.12.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohlwein SD, et al. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:109–125. doi: 10.1128/MCB.21.1.109-125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kvam E, Goldfarb DS. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J Cell Sci. 2004;117:4959–4968. doi: 10.1242/jcs.01372. [DOI] [PubMed] [Google Scholar]
- 11.Kvam E, Gable K, Dunn TM, Goldfarb DS. Targeting of Tsc13p to nucleus-vacuole junctions: A role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol Biol Cell. 2005;16:3987–3998. doi: 10.1091/mbc.E05-04-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan X, et al. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y-X, Catlett NL, Weisman LS. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott SV, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 15.Tewari R, Bailes E, Bunting KA, Coates JC. Armadillo-repeat protein functions: Questions for little creatures. Trends Cell Biol. 2010;20:470–481. doi: 10.1016/j.tcb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Weisman LS. Organelles on the move: Insights from yeast vacuole inheritance. Nat Rev Mol Cell Biol. 2006;7:243–252. doi: 10.1038/nrm1892. [DOI] [PubMed] [Google Scholar]
- 17.Mochida K, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 18.Mijaljica D, Prescott M, Devenish RJ. A late form of nucleophagy in Saccharomyces cerevisiae. PLoS One. 2012;7:e40013. doi: 10.1371/journal.pone.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, Kimelman D. Mechanistic insights from structural studies of beta-catenin and its binding partners. J Cell Sci. 2007;120:3337–3344. doi: 10.1242/jcs.013771. [DOI] [PubMed] [Google Scholar]
- 20.Hülsken J, Birchmeier W, Behrens J. E-cadherin and APC compete for the interaction with beta-catenin and the cytoskeleton. J Cell Biol. 1994;127:2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Kries JP, et al. Hot spots in beta-catenin for interactions with LEF-1, conductin and APC. Nat Struct Biol. 2000;7:800–807. doi: 10.1038/79039. [DOI] [PubMed] [Google Scholar]
- 22.Choi H-J, Huber AH, Weis WI. Thermodynamics of β-catenin-ligand interactions: The roles of the N- and C-terminal tails in modulating binding affinity. J Biol Chem. 2006;281:1027–1038. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- 23.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 24.Tang F, et al. Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature. 2003;422:87–92. doi: 10.1038/nature01453. [DOI] [PubMed] [Google Scholar]
- 25.Cox RT, Pai L-M, Kirkpatrick C, Stein J, Peifer M. Roles of the C terminus of Armadillo in Wingless signaling in Drosophila. Genetics. 1999;153:319–332. doi: 10.1093/genetics/153.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piedra J, et al. Regulation of β-catenin structure and activity by tyrosine phosphorylation. J Biol Chem. 2001;276:20436–20443. doi: 10.1074/jbc.M100194200. [DOI] [PubMed] [Google Scholar]
- 27.Solanas G, et al. β-Catenin and plakoglobin N- and C-tails determine ligand specificity. J Biol Chem. 2004;279:49849–49856. doi: 10.1074/jbc.M408685200. [DOI] [PubMed] [Google Scholar]
- 28.Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 29.Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 30.Cingolani G, Petosa C, Weis K, Müller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 31.Elbaz-Alon Y, et al. Lam6 regulates the extent of contacts between organelles. Cell Reports. 2015;12:7–14. doi: 10.1016/j.celrep.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henne WM, et al. Mdm1/Snx13 is a novel ER-endolysosomal interorganelle tethering protein. J Cell Biol. 2015;210:541–551. doi: 10.1083/jcb.201503088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murley A, et al. Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J Cell Biol. 2015;209:539–548. doi: 10.1083/jcb.201502033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529-33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krissinel E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 2015;43:W314-9. doi: 10.1093/nar/gkv314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong H, Park J, Lee C. Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex. EMBO Rep. 2016;17:1857–1871. doi: 10.15252/embr.201642706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haas A, Conradt B, Wickner W. G-protein ligands inhibit in vitro reactions of vacuole inheritance. J Cell Biol. 1994;126:87–97. doi: 10.1083/jcb.126.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]