Abstract
Growing interest in microbial contributions to human health and disease has increasingly led investigators to examine the microbiome in both healthy skin and cutaneous disorders, including acne, psoriasis and atopic dermatitis. The need for common language, effective study design, and validated methods are critical for high-quality, standardized research. Features, unique to skin, pose particular challenges when conducting microbiome research. This review discusses microbiome research standards and highlights important factors to consider, including clinical study design, skin sampling, sample processing, DNA sequencing, control inclusion, and data analysis.
Introduction
The relationship between host and cutaneous microbes has been of great clinical scientific interest, often studied with traditional cultivation methods and focused on a single/few bacteria (Evans et al., 1950; Kligman et al., 1976; Lai et al., 2010; Marples, 1965; Nizet et al., 2001). Reduced costs and increased access to high-throughput sequencing have enabled global examination of the skin microbiome, broadly defined as skin microbiota with their genomes and surrounding environmental conditions (Marchesi and Ravel, 2015).
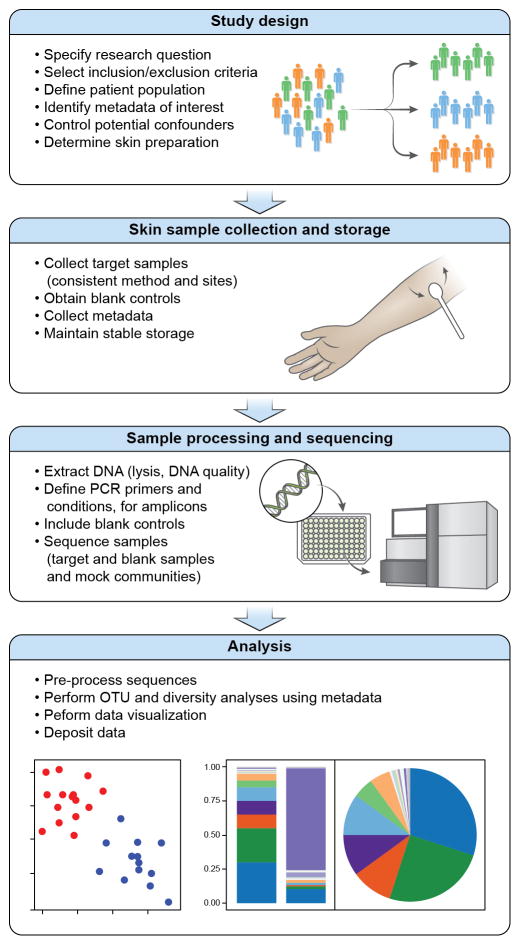
Early skin microbiome studies described healthy human skin microbial communities as more diverse than previously recognized through cultivation methods (Dekio et al., 2005; Gao et al., 2007; Grice et al., 2008a) and unique to skin (Costello et al., 2009; Human Microbiome Project, 2012a, b). Several reviews (Clavel et al., 2016; Goodrich et al., 2014; Huttenhower et al., 2014) have outlined important elements of high-quality microbiome studies. The unique aspects of skin, including low microbial biomass, high contamination risk (Salter et al., 2014), accessibility and diversity of cutaneous habitats, site-specific microbiota, and a distinct immune system (Naik et al., 2012; Watanabe et al., 2015) necessitate important considerations for conducting skin microbiome studies (Figure 1). Several reviews have summarized skin microbiome literature (Edmonds-Wilson et al., 2015; Jo et al., 2016b; Schommer and Gallo, 2013; Zeeuwen et al., 2013).
Figure 1.
Steps for Performing a Skin Microbiome Study. The multiple elements of a skin microbiome study begin with study design, followed by skin sample collection and storage, sample processing and sequencing, and analysis.
In emerging fields, studies to identify optimal methodologies are often performed, and several include elements related to the skin microbiome (Human Microbiome Project, 2012a, b). Study design for skin microbiome research is multifaceted and integral to all downstream steps. Published studies examined skin sampling methods (Chng et al., 2016; Grice et al., 2008a), sample storage (Lauber et al., 2010), controls and contamination sources (Salter et al., 2014), sequencing biases (Meisel et al., 2016), and possible quantitation (Gao et al., 2010). The current review integrates this combined expertise and focuses on methodology and challenges of factors important for skin microbiome research to promote reliability and comparability (Figure 1). Of note, we primarily discuss 16S rRNA gene amplicon sequencing, as the most widely used method.
Potential Pitfalls
Similar to other interdisciplinary fields, multiple factors are important in conducting or assessing a skin microbiome study.
Study Design: Consistent metadata collection; considering potential confounding factors
Skin Sample Collection/Storage: Standardized collection/handling of samples
Sample Processing/Sequencing: DNA extraction; PCR conditions, Primer selection
Process Controls: Negative/blank controls; mock community comparison
Analysis Methods: Pipeline description; sequencing data availability with associated metadata
Study design
Since the skin microbiome is comprised of different microbes including bacteria, fungi, and viruses, whether the scientific focus is on one particular kingdom or all microbes will influence study design, sequencing, costs, and analysis. Bacteria have been the main focus, but several have used sequencing methods to examine skin fungal (Findley et al., 2013; Jo et al., 2016a; Paulino et al., 2006; Zhang et al., 2011), viral (Foulongne et al., 2012; Hannigan et al., 2015; Oh et al., 2014; Oh et al., 2016; Wylie et al., 2014) and archaeal communities (Probst et al., 2013).
A “study population” may refer to individuals with/without a particular disease, in a specific age range (Capone et al., 2011; Costello et al., 2013; Dominguez-Bello et al., 2010; Oh et al., 2012; Ying et al., 2015), or in a geographic region (Blaser et al., 2013; Clemente et al., 2015). Studies have demonstrated some interindividual differences in the skin microbiome even when matched for body site and sexual maturity, highlighting the need for the study design (e.g. sample size) to account for a certain degree of heterogeneity in the skin microbiome of a target study population; however, many features of the skin microbiome can be commonly observed (i.e. sebaceous sites hosting lipophilic bacteria). Skin bacterial communities in neonates, infants, and young children are notably distinct from those in sexually mature children and adults, particularly at certain skin sites (Capone et al., 2011; Costello et al., 2013; Dominguez-Bello et al., 2010; Jo et al., 2016a; Oh et al., 2012; Ying et al., 2015). The skin microbiomes in patients with different cutaneous and general medical conditions show distinctive patterns of skin microbiomes, but heterogeneity in the experimental study designs highlights the challenges in comparing results between studies and emphasizes a need for minimal standards.
Screening subjects involves collecting demographic data, obtaining detailed history on prior and/or current medical conditions and topical/systemic medications, performing clinical examinations, and considering diagnostic criteria. Explicit criteria for defining healthy individuals are important (Aagaard et al., 2013). Disease phenotyping (validated diagnostic criteria, severity scoring, and clinical photography) enables more accurate comparison of subpopulations within a particular disorder.
A typical exclusion criterion for healthy individuals is prior systemic antibiotic usage, based on antibiotic use within the last 12 months (Grice et al., 2009a), 6 months (Costello et al., 2009; Findley et al., 2013; Human Microbiome Project, 2012b), or 1 month (Gao et al., 2007). For individuals with skin disorders, prior usage of topical and systemic medications can be used as exclusion criteria or defining metadata which can affect analyses (Kong et al., 2012). Other medications may influence the skin microbiome, and collecting a complete medication history is desirable.
Clinical metadata documentation is critical for downstream analyses and may help explain differences within/between studies. Commonly collected metadata include age, sex, antibiotic use, and sampling sites. Some factors such as pet ownershp (Song et al., 2013), deodorant usage (Callewaert et al., 2013), physical activities (Meadow et al., 2013), season, time of day, country of birth, race/ethnicity, mode of delivery and diet may influence the skin microbiome.
Calculating sample sizes for skin microbiome studies can be difficult without pre-existing data for estimating effect sizes. A few methods have been proposed to calculate sample sizes (Kelly et al., 2015; La Rosa et al., 2012), including a web-based tool called Evident (https://github.com/biocore/Evident). With growing numbers of skin microbiome studies, pre-existing data for estimating potential effect sizes are increasingly available for use in designing well-powered studies.
Skin preparation
Questions often arise regarding factors to control in skin microbiome studies. Standardizing controllable factors reduces confounders, maximizing ability to determine the experimental variable responsible for any observed difference. Factors such as washing can alter the bacterial communities present on hands (Fierer et al., 2008) and after experimental exposure to non-skin bacteria (Costello et al., 2009; Two et al., 2016). Due to potential fluctuations in skin microbial communities in the hours after bathing/showering, several studies have utilized a minimum number of hours after baths/showers before sampling; the minimum time since last washing has ranged from 12 hours (Human Microbiome Project, 2012a, b) to 24 hours (Grice et al., 2009a; Oh et al., 2016; Oh et al., 2012). Skin hygiene (e.g. soap and shampoo practices) can also alter the skin microbial communities in some circumstances (Perez Perez et al., 2016) but not in others (Two et al., 2016). Use of emollients can influence the skin microbiome (Seite et al., 2014); therefore, the restrictions on emollient use in studies has varied between 1 to 7 days of avoidance prior to sampling (Human Microbiome Project, 2012a, b; Kong et al., 2012). The effects of topical medications, e.g. steroids, on the skin microbiome can persist for up to 7 days (Kong et al., 2012).
Skin Sampling Methods
Multiple methodologies have been documented and validated for skin microbiome sampling, including swabs, biopsies, surface scrapes, cup scrubs and tape strips (Chng et al., 2016; Costello et al., 2009; Findley et al., 2013; Gao et al., 2007; Grice et al., 2008b; Nakatsuji et al., 2013; Oh et al., 2014; Zeeuwen et al., 2012). Each method varies in biomass yield, human DNA contribution, sampling depth and discomfort level. Method should be based on scientific question, study design, study population, and sampling sites. The skin microbiome composition at the skin surface can differ from deeper layers of the skin (Grice et al., 2008b; Nakatsuji et al., 2013; Zeeuwen et al., 2012) and between skin sub-compartments and structures. Thus, consistency is key for skin microbiome sampling methods.
The most established collection method is premoistened swabbing (Aagaard et al., 2013; Human Microbiome Project, 2012a, b; Paulino et al., 2006). Dry swabbing (Schowalter et al., 2010) has not been as widely utilized, due to reduced biomass collection. Tape stripping provides a reproducibly high amount of biomass (Chng et al., 2016) with stratum corneum depth analysis in a defined collection area (Zeeuwen et al., 2012). Tape stripping is not suitable for all body sites because of its dimensions and sample acquisition time. Scrapes increase biomass collected and could be useful for studies of low abundance microbes, e.g. fungi (Findley et al., 2013; Grice et al., 2008b). Cup scrubbing was pioneered for culture-based studies (Williamson and Kligman, 1965) and can also be used for microbiome studies (Chng et al., 2016). Skin punch biopsies enable analysis of microbial DNA potentially deeper in skin. In addition to tissue homogenization for total DNA collection, a portion of skin biopsies can be processed for other analyses, including laser capture microscopy (Grice et al., 2008b; Nakatsuji et al., 2013). However, biopsies are invasive with reduced ability to sample multiple sites in patients.
Most major bacterial taxa are similarly identified in swabs, scrapes, and biopsies, but a tree-based analysis could segregate sampling methods (Grice et al., 2008b). Comparing swabs, tape strips, and cup scrubs for 16S rRNA gene or shotgun metagenomic sequencing provided high concordance when analyzing relative abundance (Chng et al., 2016). While highlighting that multiple methods can be used, it is important to maintain a consistent standardized approach for sample collection throughout a study.
Other aspects of sample collection have been emphasized in prior studies, i.e. location, sampling frequency, and use of controls. Skin microbial communities exhibit striking site-specific differences and notable topographical diversity over the human skin surface, even within the same individual. Yet, regions of skin with common physiological characteristics - sebaceous, moist, dry - share some similarities in the composition of skin microbial communities (Costello et al., 2009; Grice et al., 2009a; Oh et al., 2014; Oh et al., 2012). Consistent sampling of the same anatomic area in the entire study cohort reduces confounders and maximizes the ability to identify microbiome differences (Costello et al., 2009; Grice et al., 2009a). Sampling body sites with bilateral symmetry has also been used to confirm that consistency is observed with low intra-individual variability (Chng et al., 2016; Grice et al., 2009a). Frequency of sampling has varied in different studies from a single timepoint to repeated sampling. Longitudinal sampling of skin has demonstrated that skin microbiomes are highly individualized (Flores et al., 2014; Oh et al., 2016), which suggests that repeated sampling can provide an internal control, and help to increase the statistical power for analyzing changes in a chronic skin disorder (Kong et al., 2012).
Sample storage
For sample handling, most studies have used the standard method of immediate freezing after sample collection, followed by storage at −80°C (Blaser et al., 2013; Costello et al., 2009; Grice et al., 2009a). No major differences in the bacterial communities were found in skin and fecal samples collected from 2 individuals and stored at different temperatures for two weeks (Lauber et al., 2010). However, storage at −80°C is generally advised (Goodrich et al., 2014). Studies have demonstrated that freeze-thaw cycles can alter the microbial composition observed in samples and should be avoided (Cuthbertson et al., 2015; Sergeant et al., 2012).
Sample Processing
DNA extraction
During early steps of sample preparation, bias can easily be introduced (Brooks et al., 2015; Yuan et al., 2012), especially during the lysis of microbial cells (Yuan et al., 2012). Mechanical lysis (with or without enzymatic treatment) provides more comprehensive profiles of Gram-positive bacterial populations and fungi than enzymatic treatment alone, and is thus strongly advisable (Albertsen et al., 2015; Findley et al., 2013; Santiago et al., 2014; Sergeant et al., 2012; Walker et al., 2015; Yuan et al., 2012). After lysis, DNA can be purified following different approaches, e.g. by alcohol precipitation or via binding to affinity columns. Downstream applications will dictate genomic DNA requirements: higher standards in terms of fragment size, amount, and purity of target DNA are often required for some sequencing methods. Therefore, consistent documentation of the quality and quantity of DNA obtained (e.g. fluorometry, UV spectroscopy) is important (Olson and Morrow, 2012) and performing gel electrophoresis on high yield samples will help to characterize fragment size distribution.
Library construction: Amplicons
PCR amplification introduces biases in datasets. Use of PCR replicates, controls, appropriate PCR primers and PCR cycle conditions help control library quality. PCR replicates (at least duplicates; if possible triplicates) that are pooled in downstream steps (e.g. during library purification) can be prepared to limit effects of early amplification bias (Acinas et al., 2005).
Primer choice significantly influences microbial profiles as PCR efficiency can vary between organisms (Hiergeist et al., 2016; Meisel et al., 2016; Walker et al., 2015). The most commonly used gene in amplicon metagenomics studies is the 16S rRNA gene. Several sets of 16S rRNA gene-specific primers have been designed, targeting different variable (V) regions and amplicon sizes (Klindworth et al., 2013). Multiple studies have evaluated primer combinations and compared the utility with different sample types. Primer region selection depends on the scientific question and potential bacterial taxa of interest (Human Microbiome Project, 2012a). Staphylococcus and Streptococcus spp. are major skin bacteria best distinguished using V1 and V2 regions (Conlan et al., 2012). A recent study showed that products spanning V1–3 were more accurate in classifying common skin bacteria and were more similar to results obtained using shotgun metagenomic sequencing of a mock community, as compared to V4 (Meisel et al., 2016). The currently most used region for analysis of skin bacterial communities is V1–3. Consistent use of a single target region facilitates comparison between studies yet may limit differentiation of bacteria optimally sequenced with other primer regions. Fungal sequencing studies have targeted a few different regions associated with the 5.8S, 18S, 28S, and ITS1/2 ribosomal genes (Findley et al., 2013; Paulino et al., 2006; Wang et al., 2015; Zhang et al., 2011).
A key PCR parameter is the number of cycles used for amplification. An increase in the number of amplification cycles can result in lower diversity, and can skew bacterial profiles towards detection of low abundant taxa (Acinas et al., 2005; Bonnet et al., 2002). Hence, it is advisable that PCR cycles are kept to the lowest number that still delivers reproducible DNA amounts across a wide range of samples while also avoiding overamplification and ensuring that negative controls do not yield a product. Several groups have used ~30 cycles of PCR (Dekio et al., 2005; Gao et al., 2007; Grice et al., 2009a; Human Microbiome Project, 2012a, b; Smeekens et al., 2013). The quality of the amplicon libraries produced should be documented, e.g. by controlling for the presence of unspecific bands using high-sensitivity electrophoresis.
Sequencing
Different sequencing platforms are available with various chemistries and technologies, and selection of the platform will be dependent on sequencer availability, amplicon size, sequencing depth, sequencing accuracy, and/or budget. With increased sequencing capacity, it has become feasible to routinely carry out shotgun metagenomic sequencing from human skin samples (Chng et al., 2016; Kang et al., 2015; Oh et al., 2014; Oh et al., 2016). Some advantages of this strategy include reduced amplification bias, generation of multikingdom genetic information, strain identification, and detailed genomic coverage for prediction of functional capacity (Chng et al., 2016; Human Microbiome Project, 2012b; Oh et al., 2014; Oh et al., 2016). Challenges with whole metagenomic sequencing are associated with data analysis complexity and low biomass from human skin samples, typically in the pg to ng range, which can make sequencing library construction difficult and increases sensitivity to contamination with mammalian DNA and microorganisms from the environment and laboratory reagents.
Use of Blank and Control Samples
For the low biomass of skin samples, blank and control samples must be collected and run in parallel to target samples (Costello et al., 2009; Grice et al., 2009b; Paulino et al., 2006). Blank samples are controls intended to contain no biological starting material of interest (e.g. only collection/storage buffers), including mock samples collected during patient sampling (e.g. premoistened swab fanned in the air) and DNA extraction processes, and PCR/sequencing blanks (containing only purified PCR-grade water as template). Blanks are processed with target samples to identify extraction contaminants and amplification artefacts, cross-contamination during library preparation, and contaminants entering during sequencing.
Mock communities (artificial mixtures of known target microorganisms) and reference samples of known composition are critical for benchmarking sample processing and sequence analysis (D’Amore et al., 2016; Jumpstart Consortium Human Microbiome Project Data Generation Working, 2012; Yuan et al., 2012). A mock community used with each sequencing run or across sequencing centers can promote standardization (Sinha et al., 2015) and should include multiple taxa important to the ecosystem of interest. A widely available synthetic DNA mock community (BEI Resources HM-276D) has been developed (Jumpstart Consortium Human Microbiome Project Data Generation Working, 2012); this includes bacterial species important to skin (P. acnes, S. aureus, and S. epidermidis).
Additionally, when studying a patient cohort, it would be ideal to leverage data generated from healthy volunteers in previous small and larger studies. While in principle this increases the power of the study, collection and analysis of data from some healthy volunteers alongside the patient population of interest would control for factors specific to the study.
Analysis Methods
The quality of analysis is dependent on consistent and thorough documentation of clinical metadata, sample collection/storage, DNA processing of target samples, negative controls, mock community positive controls, and sequencing methods (Goodrich et al., 2014). The metadata allow researchers to delineate potential causes/associations with sequencing results. Analysis pipelines, e.g. QIIME (Caporaso et al., 2010) mothur (Schloss et al., 2009), and IMNGS (Lagkouvardos et al., 2016), have tutorials and best practices with parameters that should be evaluated for appropriate use with a particular dataset. There are many approaches to analyzing 16S rRNA gene sequencing data and the following overview briefly highlights most commonly used methods.
The 16S rRNA gene sequence analysis pipeline is comprised of three main components: pre-processing sequences, constructing OTU (Operational Taxonomic Units, similar to bacterial taxa or species) tables, and annotating OTU tables. Sequence pre-processing is used to remove low quality sequences prior to OTU table construction. Chimeric sequences generated during the PCR amplification process also need to be identified and removed from the dataset (Haas et al., 2011). The appropriate analysis methods and parameters are dependent on the sequencing method and amplicon region (D’Amore et al., 2016).
After pre-processing, the OTU table is constructed by clustering (grouping) similar sequences based on a defined similarity threshold. There are several approaches to clustering (Chen et al., 2013). The reference clusters used and the similarity threshold significantly affect the clustering results (He et al., 2015; Kopylova et al., 2016; Schloss, 2016). Normalization or rarefaction is used to address issues due to sequencing depth differences (McMurdie and Holmes, 2014).
The resulting OTU table is annotated based on representative OTU taxonomic and phylogenetic relatedness, using specific databases (Conlan et al., 2012; Schloss, 2010). A number of different methods are used to classify 16S rRNA gene sequences, e.g. reference-based clustering, sequence similarity (e.g. BLAST), K-mer based methods (e.g. RDP), and phylogenetic placement (Nguyen et al., 2016). Phylogenetic trees are commonly used in diversity metrics (e.g. UniFrac) or for data visualization.
Analysis of shotgun metagenomic data is challenging due to the orders of magnitude larger amounts of data generated and requires filtering high percentages of human sequences (Chng et al., 2016; Oh et al., 2014; Oh et al., 2016). Limitations of microbial sequencing analyses relate to the need for more reference genomes and for fundamental research on the function of genes identified in metagenomic sequencing. Analytical methods will continue to evolve to utilize both reference-based and reference-free mapping strategies (Human Microbiome Project, 2012b; Ma et al., 2014; Oh et al., 2014; Wylie et al., 2014). For microbiome and other genomic data, depositing data with relevant metadata in public repositories is a standard requirement for many funding agencies and allows acceleration of research through data sharing, re-analysis, validation, and compilation.
Guidelines and Future Needs
Maintaining a standardized format for reporting sample handling and processing with a common scientific language (Marchesi and Ravel, 2015) is important to promote reproducibility and advance science (Ravel and Wommack, 2014). Minimal standards/guidelines are often developed to further these goals, e.g. MIQE for quantitative real-time PCR (Bustin et al., 2009), MIAME for microarray studies (Knudsen et al., 2005), CONSORT for clinical trials (Begg et al., 1996), and MIMARKS and MIxS for genomics (Yilmaz et al., 2011). Research utilizing large DNA sequence datasets requires significant data analysis with a unique set of criteria for reproducibility (Peng, 2011). Depositing primary data into public databases enables independent cross-evaluation and comparison with previous studies, but requires access to raw data, metadata, software, and code used to process the data (Sandve et al., 2013; Stodden et al., 2014). The Genomic Standards Consortium has promoted minimum information standards and provides checklists for a broad range of genomic studies (Yilmaz et al., 2011). Adapting existing guidelines to skin microbiome studies, key areas that are important to clearly describe in manuscripts and attach (as metadata) to sequencing data:
Study design: Include clinical protocol information, study population, inclusion/exclusion criteria, medical and medication history, clinical phenotyping/validated tools for diagnosis and/or severity assessment, documented clinical metadata, and skin preparation regimen.
Sample collection/storage: Include description of sampling methods, skin sites, sample storage, and controls.
Sample processing/sequencing: Include detailed lysis methods, DNA purification, PCR primers, PCR conditions, processing controls, and sequencing.
Process controls: Include negative controls during sample collection, processing, and sequencing and a mock community.
Analysis pipeline: Include software (version), any specific commands, parameter settings, statistical tests, scripts for new pipelines (if applicable), and reference databases (version, access date, and modifications).
Data deposition: Genomic data sharing is often required with publication in many journals. Note submission of raw sequences to a publicly available database should be accompanied by specific relevant metadata, e.g. collection date, skin site/location, clinical features (Yilmaz et al., 2011).
In addition, several areas for growth could facilitate advancement towards mechanistic studies that better elucidate host-microbial interactions. Skin microbiome studies are an important window into our microbial communities and enable formulation of hypotheses, but cause-or-effect relationships can be difficult to untangle. To test functional hypotheses, conducting studies that progress beyond DNA sequence data to include clinical isolates is key. Genomic and biological differences are well-recognized at the microbial species and strain level, e.g. Staphylococcus aureus strains with or without methicillin-resistance (Greenblum et al., 2015; Oh et al., 2014). Thus, strains of a particular species available from biorepositories may or may not function in the same manner as the strains directly obtained in parallel from subjects studied as part of a microbiome analysis. Cultivating and curating skin-associated strains from human studies and making these available would improve sequencing analyses and provide live and highly relevant microbes for biological experiments. To isolate new reference strains, it is important to optimize methods for capturing a diverse set of skin microbes that reflects the complexity of the microbial communities as has been done for other body sites (Browne et al., 2016). Single cell microbial sequencing could be a future option (Lasken and McLean, 2014), which would still benefit from reference genomes that take advantage of the relative ease of culturing most skin-associated bacterial strains.
Current methods to study the skin microbiome are based on relative abundance of microbes. Determination of bioburden and quantification of skin microbes is relevant to understanding the bioburden of microbes the skin typically harbors and whether it increases in certain diseases. One could perform quantitative PCR of the 16S rRNA gene to determine bacterial recovery in a given sample. A major issue is normalization; does one normalize to total DNA, which would include both human and microbial and could alter with disease state? Or does one normalize to surface area sampled, which might be affected by pressure applied and thus layers of skin released? In parallel with sequencing, developing microscopy methods to improve visualization of skin bacterial communities would greatly advance our ability to understand the structure and potential interactions within microbial communities.
While skin microbiome studies may identify differences between affected and unaffected subjects, correlation-versus-causation questions remain. Longitudinal data across multiple timepoints can provide insights into the natural history of diseases and dynamics of ecological succession of the skin microbial communities. Another method to gather further evidence supporting causation is to identify colonization with a specific microbe or community of microbes prior to disease manifestation. Given the interest in microbial education of the immune system, birth cohort study designs provide an opportunity to obtain multiple skin samples prior to disease development, similar to published gut studies (Bokulich et al., 2016; Vatanen et al., 2016; Yassour et al., 2016). Meta-transcriptomics would also offer valuable information regarding the expression of microbial genes during the natural course of disease.
Conclusion
Studies of skin microbiome research have the potential to improve our understanding of host-microbial interactions. A byproduct of the expansion in the number of published skin microbiome studies is the need to understand how studies interrelate. Several scientific communities have developed minimal standards to improve the overall quality of different fields of research (Yilmaz et al., 2011). Minimal standards will contribute to the development of robust studies in skin microbiome research.
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Dr. Jay-Hyun Jo and Mr. Ethan Tyler for assistance with figures and Clay Deming for discussions. Certain commercial equipment, instruments, or materials are identified in this paper only to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the NIST, nor is it intended to imply that the materials or equipment are necessarily the best available for the purpose. Official contribution of NIST; not subject to copyrights in USA.
Abbreviations
- rRNA
ribosomal RNA
- ITS
internal transcribed spacer
Footnotes
Conflict of Interest
The authors state no conflict of interest.
References
- Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013;27:1012–22. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl Environ Microbiol. 2005;71:8966–9. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen M, Karst SM, Ziegler AS, Kirkegaard RH, Nielsen PH. Back to Basics--The Influence of DNA Extraction and Primer Choice on Phylogenetic Analysis of Activated Sludge Communities. PLoS One. 2015;10:e0132783. doi: 10.1371/journal.pone.0132783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg C, Cho M, Eastwood S, Horton R, Moher D, Olkin I, et al. Improving the quality of reporting of randomized controlled trials. The CONSORT statement. JAMA. 1996;276:637–9. doi: 10.1001/jama.276.8.637. [DOI] [PubMed] [Google Scholar]
- Blaser MJ, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Estrada I, et al. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J. 2013;7:85–95. doi: 10.1038/ismej.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet R, Suau A, Dore J, Gibson GR, Collins MD. Differences in rDNA libraries of faecal bacteria derived from 10- and 25-cycle PCRs. Int J Syst Evol Microbiol. 2002;52:757–63. doi: 10.1099/00207713-52-3-757. [DOI] [PubMed] [Google Scholar]
- Brooks JP, Edwards DJ, Harwich MD, Jr, Rivera MC, Fettweis JM, Serrano MG, et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–6. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Callewaert C, Kerckhof FM, Granitsiotis MS, Van Gele M, Van de Wiele T, Boon N. Characterization of Staphylococcus and Corynebacterium clusters in the human axillary region. PLoS One. 2013;8:e70538. doi: 10.1371/journal.pone.0070538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol. 2011;131:2026–32. doi: 10.1038/jid.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010:7. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang CK, Cheng Y, Zhang S, Zhao H. A Comparison of Methods for Clustering 16S rRNA Sequences into OTUs. PLoS ONE. 2013;8:e70837. doi: 10.1371/journal.pone.0070837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng KR, Tay ASL, Li C, Ng AHQ, Wang J, Suri BK, et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat Microbiol. 2016 doi: 10.1038/nmicrobiol.2016.106. [DOI] [PubMed] [Google Scholar]
- Clavel T, Lagkouvardos I, Hiergeist A. Microbiome sequencing: challenges and opportunities for molecular medicine. Expert Rev Mol Diagn. 2016:1–11. doi: 10.1080/14737159.2016.1184574. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015:1. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan S, Kong HH, Segre JA. Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PLoS One. 2012;7:e47075. doi: 10.1371/journal.pone.0047075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Carlisle EM, Bik EM, Morowitz MJ, Relman DA. Microbiome assembly across multiple body sites in low-birthweight infants. MBio. 2013;4:e00782–13. doi: 10.1128/mBio.00782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Rogers GB, Walker AW, Oliver A, Hoffman LR, Carroll MP, et al. Implications of multiple freeze-thawing on respiratory samples for culture-independent analyses. J Cyst Fibros. 2015;14:464–7. doi: 10.1016/j.jcf.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amore R, Ijaz UZ, Schirmer M, Kenny JG, Gregory R, Darby AC, et al. A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiling. BMC Genomics. 2016;17:1–20. doi: 10.1186/s12864-015-2194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekio I, Hayashi H, Sakamoto M, Kitahara M, Nishikawa T, Suematsu M, et al. Detection of potentially novel bacterial components of the human skin microbiota using culture-independent molecular profiling. J Med Microbiol. 2005;54:1231–8. doi: 10.1099/jmm.0.46075-0. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds-Wilson SL, Nurinova NI, Zapka CA, Fierer N, Wilson M. Review of human hand microbiome research. J Dermatol Sci. 2015;80:3–12. doi: 10.1016/j.jdermsci.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Evans CA, Smith WM, Johnston EA, Giblett ER. Bacterial flora of the normal human skin. J Invest Dermatol. 1950;15:305–24. doi: 10.1038/jid.1950.105. [DOI] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17994–9. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–70. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15:531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Perez-Perez GI, Chen Y, Blaser MJ. Quantitation of major human cutaneous bacterial and fungal populations. J Clin Microbiol. 2010;48:3575–81. doi: 10.1128/JCM.00597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2927–32. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, et al. Conducting a microbiome study. Cell. 2014;158:250–62. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblum S, Carr R, Borenstein E. Extensive strain-level copy-number variation across human gut microbiome species. Cell. 2015;160:583–94. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009a;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science. 2009b;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, Blakesley RW, et al. A diversity profile of the human skin microbiota. Genome Res. 2008a;18:1043–50. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, Blakesley RW, et al. A diversity profile of the human skin microbiota. Genome Research. 2008b;18:1043–50. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ, et al. The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. MBio. 2015;6:e01578–15. doi: 10.1128/mBio.01578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Caporaso JG, Jiang X-T, Sheng H-F, Huse SM, Rideout JR, et al. Stability of operational taxonomic units: an important but neglected property for analyzing microbial diversity. Microbiome. 2015;3:1–10. doi: 10.1186/s40168-015-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiergeist A, Reischl U. Priority Program Intestinal Microbiota Consortium/ quality assessment participants, Gessner A (2016) Multicenter quality assessment of 16S ribosomal DNA-sequencing for microbiome analyses reveals high inter-center variability. Int J Med Microbiol. doi: 10.1016/j.ijmm.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project C. A framework for human microbiome research. Nature. 2012a;486:215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012b;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Knight R, Brown CT, Caporaso JG, Clemente JC, Gevers D, et al. Advancing the microbiome research community. Cell. 2014;159:227–30. doi: 10.1016/j.cell.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J-H, Deming C, Kennedy EA, Conlan S, Polley EWN, et al. Diverse human skin fungal communities in children converge in adulthood. J Invest Dermatol. 2016a doi: 10.1016/j.jid.2016.05.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo JH, Kennedy EA, Kong HH. Research Techniques Made Simple: Bacterial 16S Ribosomal RNA Gene Sequencing in Cutaneous Research. J Invest Dermatol. 2016b;136:e23–7. doi: 10.1016/j.jid.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One. 2012;7:e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Shi B, Erfe MC, Craft N, Li H. Vitamin B12 modulates the transcriptome of the skin microbiota in acne pathogenesis. Sci Transl Med. 2015;7:293ra103. doi: 10.1126/scitranslmed.aab2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BJ, Gross R, Bittinger K, Sherrill-Mix S, Lewis JD, Collman RG, et al. Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics. 2015;31:2461–8. doi: 10.1093/bioinformatics/btv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kligman AM, Leyden JJ, McGinley KJ. Bacteriology. J Invest Dermatol. 1976;67:160–8. doi: 10.1111/1523-1747.ep12513007. [DOI] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic acids research. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen TB, Daston GP, Teratology S. MIAME guidelines. Reprod Toxicol. 2005;19:263. doi: 10.1016/j.reprotox.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–9. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Navas-Molina JA, Mercier C, Xu ZZ, Mahé F, He Y, et al. Open-Source Sequence Clustering Methods Improve the State Of the Art. mSystems. 2016:1. doi: 10.1128/mSystems.00003-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa PS, Brooks JP, Deych E, Boone EL, Edwards DJ, Wang Q, et al. Hypothesis testing and power calculations for taxonomic-based human microbiome data. PLoS One. 2012;7:e52078. doi: 10.1371/journal.pone.0052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, Joseph D, Kapfhammer M, Giritli S, Horn M, Haller D, et al. IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep. 2016;6:33721. doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol. 2010;130:2211–21. doi: 10.1038/jid.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken RS, McLean JS. Recent advances in genomic DNA sequencing of microbial species from single cells. Nature reviews Genetics. 2014;15:577–84. doi: 10.1038/nrg3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol Lett. 2010;307:80–6. doi: 10.1111/j.1574-6968.2010.01965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3:31. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marples M. The Ecology of the Human Skin. Charles C Thomas, Bannerstone House; Springfield, Ill: 1965. [Google Scholar]
- McMurdie PJ, Holmes S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadow JF, Bateman AC, Herkert KM, O’Connor TK, Green JL. Significant changes in the skin microbiome mediated by the sport of roller derby. PeerJ. 2013;1:e53. doi: 10.7717/peerj.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel JS, Hannigan GD, Tyldsley AS, SanMiguel AJ, Hodkinson BP, Zheng Q, et al. Skin microbiome surveys are strongly influenced by experimental design. J Invest Dermatol. 2016 doi: 10.1016/j.jid.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–9. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N-P, Warnow T, Pop M, White B. A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. Npj Biofilms And Microbiomes. 2016;2:16004. doi: 10.1038/npjbiofilms.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, et al. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–7. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Kong HH, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA. Temporal Stability of the Human Skin Microbiome. Cell. 2016;165:854–66. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome medicine. 2012;4:77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ND, Morrow JB. DNA extract characterization process for microbial detection methods development and validation. BMC Res Notes. 2012;5:668. doi: 10.1186/1756-0500-5-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–41. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng RD. Reproducible Research in Computational Science. Science. 2011;334:1226–7. doi: 10.1126/science.1213847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Perez GI, Gao Z, Jourdain R, Ramirez J, Gany F, Clavaud C, et al. Body Site Is a More Determinant Factor than Human Population Diversity in the Healthy Skin Microbiome. PLoS One. 2016;11:e0151990. doi: 10.1371/journal.pone.0151990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AJ, Auerbach AK, Moissl-Eichinger C. Archaea on human skin. PLoS One. 2013;8:e65388. doi: 10.1371/journal.pone.0065388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel J, Wommack KE. All hail reproducibility in microbiome research. Microbiome. 2014;2:8. doi: 10.1186/2049-2618-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandve GK, Nekrutenko A, Taylor J, Hovig E. Ten Simple Rules for Reproducible Computational Research. PLoS Comput Biol. 2013;9:e1003285. doi: 10.1371/journal.pcbi.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago A, Panda S, Mengels G, Martinez X, Azpiroz F, Dore J, et al. Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol. 2014;14:112. doi: 10.1186/1471-2180-14-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput Biol. 2010;6:e1000844. doi: 10.1371/journal.pcbi.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD. Application of a Database-Independent Approach To Assess the Quality of Operational Taxonomic Unit Picking Methods. mSystems. 2016:1. doi: 10.1128/mSystems.00027-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009:75. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer NN, Gallo RL. Structure and function of the human skin microbiome. Trends Microbiol. 2013;21:660–8. doi: 10.1016/j.tim.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell host & microbe. 2010;7:509–15. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seite S, Flores GE, Henley JB, Martin R, Zelenkova H, Aguilar L, et al. Microbiome of affected and unaffected skin of patients with atopic dermatitis before and after emollient treatment. J Drugs Dermatol. 2014;13:1365–72. [PubMed] [Google Scholar]
- Sergeant MJ, Constantinidou C, Cogan T, Penn CW, Pallen MJ. High-throughput sequencing of 16S rRNA gene amplicons: effects of extraction procedure, primer length and annealing temperature. PLoS One. 2012;7:e38094. doi: 10.1371/journal.pone.0038094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 2015;16:276. doi: 10.1186/s13059-015-0841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens SP, Huttenhower C, Riza A, van de Veerdonk FL, Zeeuwen PL, Schalkwijk J, et al. Skin Microbiome Imbalance in Patients with STAT1/STAT3 Defects Impairs Innate Host Defense Responses. Journal of innate immunity. 2013 doi: 10.1159/000351912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stodden V, Leisch F, Peng RD. Implementing reproducible research. CRC Press; 2014. [Google Scholar]
- Two AM, Nakatsuji T, Kotol PF, Arvanitidou E, Du-Thumm L, Hata TR, et al. The cutaneous microbiome and aspects of skin antimicrobial defense system resist acute treatment with topical skin cleansers. J Invest Dermatol. 2016 doi: 10.1016/j.jid.2016.06.612. [DOI] [PubMed] [Google Scholar]
- Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:842–53. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome. 2015;3:26. doi: 10.1186/s40168-015-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Clavaud C, Bar-Hen A, Cui M, Gao J, Liu Y, et al. Characterization of the major bacterial-fungal populations colonizing dandruff scalps in Shanghai, China, shows microbial disequilibrium. Exp Dermatol. 2015;24:398–400. doi: 10.1111/exd.12684. [DOI] [PubMed] [Google Scholar]
- Watanabe R, Gehad A, Yang C, Scott LL, Teague JE, Schlapbach C, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med. 2015;7:279ra39. doi: 10.1126/scitranslmed.3010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson P, Kligman AM. A new method for the quantitative investigation of cutaneous bacteria. J Invest Dermatol. 1965;45:498–503. doi: 10.1038/jid.1965.164. [DOI] [PubMed] [Google Scholar]
- Wylie KM, Mihindukulasuriya KA, Zhou Y, Sodergren E, Storch GA, Weinstock GM. Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol. 2014;12:71. doi: 10.1186/s12915-014-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8:343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol. 2011;29:415–20. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S, Zeng DN, Chi L, Tan Y, Galzote C, Cardona C, et al. The Influence of Age and Gender on Skin-Associated Microbial Communities in Urban and Rural Human Populations. PLoS One. 2015;10:e0141842. doi: 10.1371/journal.pone.0141842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeuwen PL, Boekhorst J, van den Bogaard EH, de Koning HD, van de Kerkhof PM, Saulnier DM, et al. Microbiome dynamics of human epidermis following skin barrier disruption. Genome Biol. 2012;13:R101. doi: 10.1186/gb-2012-13-11-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeuwen PL, Kleerebezem M, Timmerman HM, Schalkwijk J. Microbiome and skin diseases. Curr Opin Allergy Clin Immunol. 2013;13:514–20. doi: 10.1097/ACI.0b013e328364ebeb. [DOI] [PubMed] [Google Scholar]
- Zhang E, Tanaka T, Tajima M, Tsuboi R, Nishikawa A, Sugita T. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol Immunol. 2011;55:625–32. doi: 10.1111/j.1348-0421.2011.00364.x. [DOI] [PubMed] [Google Scholar]