Abstract
Background:
Chronic idiopathic urticaria (CIU) is a complicated skin disease with unknown pathophysiology. MicroRNAs (miRNA) have been shown to be active in cellular regulation. The goal of this pilot study was to examine whether miRNAs may be involved in the regulation of CIU or as biomarkers for CIU.
Methods:
Four groups of three patients each were selected: patients with either active hives or no hives and with positive or negative chronic urticaria (CU) index results. MiRNAs were isolated from patient plasma and analyzed by using miRNA microarray technology to determine the amount of each of the 2567 known human miRNAs.
Results:
A total of 16 miRNAs were found to be differentially expressed in patients with active hives. Among them, five (2355–3p, 4264, 2355–5p, 29c-5p, and 361–3p) were significantly increased in samples with positive CU index results, which could be useful biomarkers for patients with chronic autoimmune urticaria. The miRNA data bases were used to find the targets of these selected miRNA sequences. These potential targets were then compared against a list of 154 urticaria-related genes. Twenty-five genes were found to match. These included eight that were significantly downregulated and eight that were significantly upregulated; however, seven of the eight downregulated genes (FBXL20, OPHN1, YPEL2, STARD9, EZH1, KLHL24, ING4) and five of the eight upregulated genes (BYSL, PNO1, ADAMTS9, STEAP4, SRGN) have no reported roles in signaling. For the 13 genes with reported roles in signaling, the following pathways were found: transforming growth factor beta signaling pathway (NRC31, KITLG, THBS1, CCL2), glucocorticoid receptor signaling pathway (NR3C1, SELE, CCL2), p53 signaling pathway (CCNG2, THBS1, CCL2), p21-activated kinase pathway (PAK1IP1, KITLG, CCL2), phosphoinositide-3 kinase protein kinase B signaling pathway (KITLG, CHRM, THBS1), and neuroactive ligand-receptor interaction (NRC31, HRH1, CHRM), which could play important roles in CIU.
Conclusion:
A better understanding of those genes with undefined function and simultaneous quantitation of both miRNAs and messenger RNAs are needed to fully understand CIU disease.
Keywords: CIU, CAU, microRNA, target mRNA, signal pathway, and cellular function
Chronic idiopathic urticaria (CIU) is a complicated skin disease. It is defined by ≥6 weeks with itchy red bumps and/or welts, which could be associated, in some patients, with a positive autologous serum skin test result, thyroid autoimmunity, and profound basopenia.1 Current findings indicate that one subgroup of chronic autoimmune urticaria (CAU) is due to circulating immunoglobulin G autoantibodies against Fc ε receptor 1 (FcεR1) or immunoglobulin E.2 In fact, chronic spontaneous urticaria is represented in a majority of patients with CIU, and more than half of these patients with chronic spontaneous urticaria might have CAU.3
MicroRNAs (miRNA) represent an important class of small noncoding RNAs.4 MiRNAs usually induce gene silencing by binding to target sites in the 3′untranslated region of the targeted messenger RNA (mRNA), which results in disruption of normal gene expression and protein production.5,6 MiRNAs are highly conserved throughout evolution across species.7 At least 2567 unique mature miRNAs have been identified for humans in miRBase 20.8 A given miRNA can have thousands of potential mRNA targets because the miRNA seed sequence is very short. Conversely, a given mRNA can be targeted by many miRNAs. Overall, ∼60% of the human genome could be targeted by the known miRNAs7,9 However, the effect on protein expression by transfected miRNAs is often mild.10 Extracellular miRNAs are found in all body fluids. They are relatively stable in biologic samples and have been reported as promising biomarkers for cancer and other nonmalignant diseases.11 But whether they play significant clinical roles remain to be evaluated.12
The etiology of CIU is unknown. Studies were performed that tried to link stomach ulcers caused by Helicobacter pylori infection to CIU, but it has not been substantiated.13 Alterations in miRNA expression have been reported to be associated with several diseases, including asthma and allergic rhinitis,14–17 but it is unknown in CIU. A recent review indicates that miRNAs may be involved in skin disorders by influencing regulatory mechanisms of inflammation.18 In this study, we investigated whether the anticipated differential expression of miRNAs in CIU are similar or different from that shown in allergy and asthma.15,17 After identifying the differentially expressed (DE) miRNAs in the patients with CIU and with active hives, the next approach was to find out which mRNAs could be affected by these miRNAs. We relied on miRNA data bases to identify potential targets.19,20
Next, for validation of these potential miRNA targets, we compared them with reported urticaria-related genes. From “Gene expression profiles in chronic idiopathic (spontaneous) urticaria,” by Patel et al.,21 and other sources,22,23 we created an urticaria gene panel, which consisted of 154 genes. We then compared these two gene groups with identified candidate mRNAs in CIU during active hives. Also, these selected candidate genes were subjected to several online gene function search engines such as the KEGG (Kanehisa Laboratories, Kyoto, Japan) gene pathway data base24 and PathCards (Weizmann Institute of Science, Rehovot, Israel)25 to examine their possible roles in pathways and cellular functions. This approach allowed us to look at how these DE miRNAs could be involved in patients with CIU and with active hives.
METHODS
Patients were recruited, during office visits, as part of a CIU study approved by Kaiser Permanente Institutional Review Board. Hive status was diagnosed at the time. Patients consented to have additional blood drawn for a chronic urticaria (CU) index test (a functional anti-FcεR test that supports an autoimmune basis for the disease) and other tests for further investigation. Subjects with no active hives and with negative CU index were invited to participate in the study as volunteers. The patients were divided into four groups, those with (1) active hives and with positive CU index results; (2) active hives and with a negative CU index; (3) a positive CU index but no active hives; and (4) normal, negative CU index and with no active hives. Three samples from each of the four groups were used in this pilot study. The plasma was separated from blood samples and kept frozen at −70°C. Isolation of miRNAs was performed by using Qiagen miRNeasy serum/plasma kit (Qiagen, Germantown, MD) in an RNase free environment. The isolated miRNAs were quantitated by using a Qubit microRNA assay kit (Molecular Probes by Life Technologies, Eugene, OR). The miRNA samples were stored at −70°C immediately and were processed by LC Sciences, LLC (Houston, TX) by using microarray technology for quantitation of the 2567 known human miRNAs.
Relative levels of miRNAs between the hive and the nonhive groups or among the four different groups of patients were determined and analyzed by using analysis of variance. DE miRNAs were selected based on the significance of the p value and at least twofold changes in the group mean. The potential targets of these identified miRNA sequences were then investigated by using various data bases, such as TargetScanHuman (Whitehead Institute for Biomedical Research, Cambridge, MA)19 and miRDB (Washington University, School of Medicine, St. Louis, MO).20 In general, hundreds to thousands of potential targets were proposed for each miRNA. Therefore, we used a more stringent selection criterion, such as three or more selected miRNAs that target a certain mRNA to consider it as a likely target. These potential targets were further narrowed to known CIU-related genes. Based on the report by Patel et al.21 of gene expression profiles in CIU that identified significant upregulation of 506 genes and downregulation of 51 genes in the CIU lesions and other online resources,22,23 we were able to generate a highly reliable urticaria-related gene panel, which consisted of 154 genes.
Finally, by matching these miRNA targets and urticaria-related genes, a candidate gene set for CIU in active hives was found. These candidate genes were then subjected to online search engines such as KEGG pathway24 and PathCards25 to explore what pathways and cellular functions could be involved in CIU with hives. In addition, the following three application programs were evaluated: (1) Diana Tool's mirPath v.3 (Diana Lab, Athens, Greece),26 which allows direct pathways search from a set of miRNAs; (2) Qiagen's Ingenuity Pathway Analysis (IPA),27 which uses its miRNA target filter to analyze miRNA data, prioritize mRNA targets, and examine miRNA–mRNA pairings to explore canonical pathways; and (3) Advaita Bioinformatics iPathwayGuide (Advaita Bioinformatics, Plymouth, MI),28 which performs meta-analyses of genes for predicted miRNA, pathways, and gene ontology (GO) analysis, to simulate the connection between the selected miRNAs, mRNAs, and their downstream effects on signal pathways and cellular functions. This approach may provide useful information that links miRNAs to possible causes or effects in CIU.
RESULTS
DE miRNAs During Active Hives
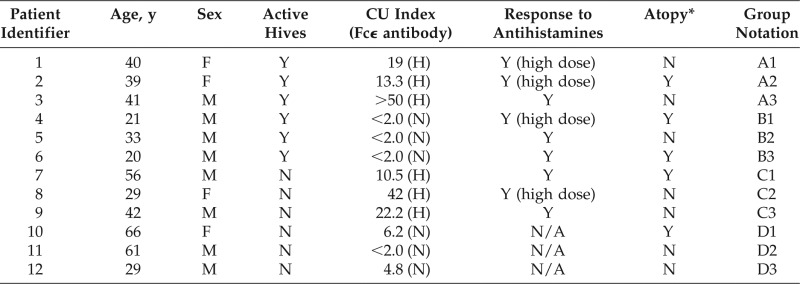
Our primary goal was to identify which miRNAs in circulating blood were DE in patients with CIU and with active hives. Twelve subjects were included in the study, divided into four groups as described in the Methods section. The demographic details of these patients are shown in Table 1. DE miRNAs could be useful biomarkers for patients with active hives. To evaluate this, we compared blood samples from these 12 patients by using a microarray platform against the latest 2567 known mature human miRNAs. The relative miRNA level results are illustrated in the heat map in Fig. 1, which shows that miR-4649–5p and miR-6769a-5p differed the most between patients with hives and patients without hives with a p value of <0.01. We further analyzed these data by using analysis of variance to take the actual fold changes into consideration.
Table 1.
Demographics of patients with CIU
CIU = Chronic idiopathic urticaria; CU = chronic urticaria; Fcϵ = Fc region of immunoglobulin E; Y = yes; (H) = high; N = no; (N) = normal; N/A = not available; A1, A2, A3 = subjects 1, 2, and 3 in group A; B1, B2, B3 = subjects 1, 2, and 3 in group B; C1, C2, C3 = subjects 1, 2, and 3 in group C; D1, D2, D3 = subjects 1, 2, and 3 in group D.
*Indicates patients who had a history of either allergic rhinitis or food allergy with a previous positive allergy testing result.
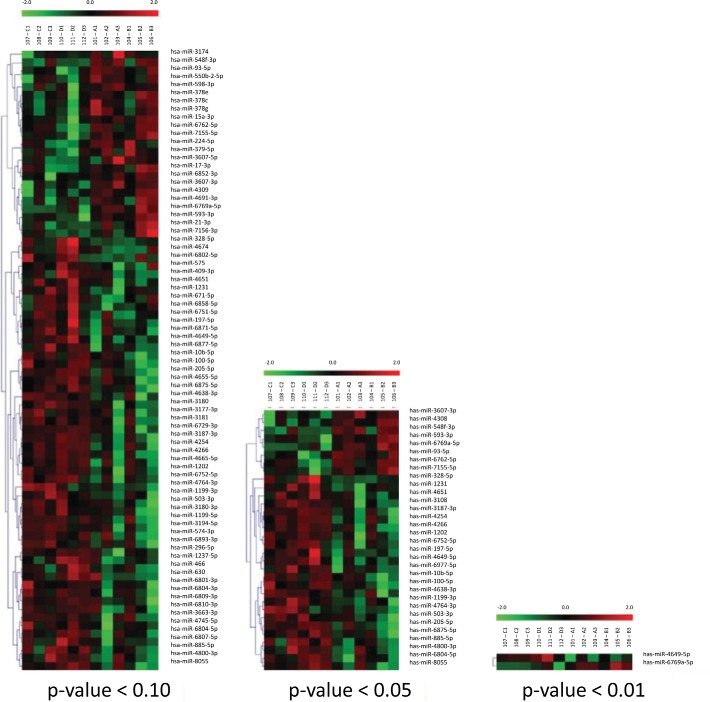
Figure 1.
Heat map of differentially expressed microRNAs (miRNA) by hives status. Only those miRNAs whose level of expression differed at a given p value are shown. The first six columns represent patients who were negative for hives; the last six columns represent patients with active hives. Different miRNAs are clustered according to the degree of similarity in the level of expression. Red represents miRNAs with up to two-fold or greater increased expression; green represents miRNAs with up to two-fold or greater decreased expression; and different levels of shading (from red to black, or green to black, with black indicating no difference) indicate values between these extremes. The average for all 12 patients is taken as the reference value. Each colored column is labeled with the sample number as well as the group and patient identity corresponding to Table 1.
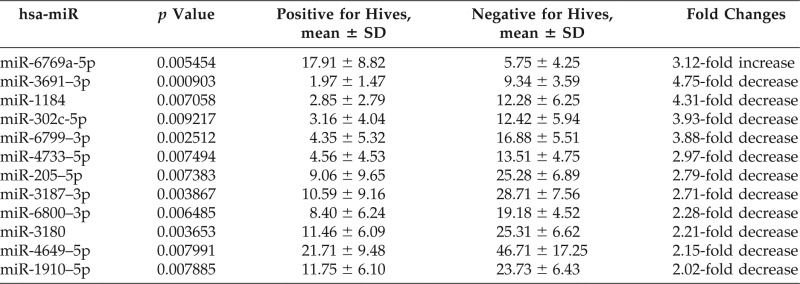
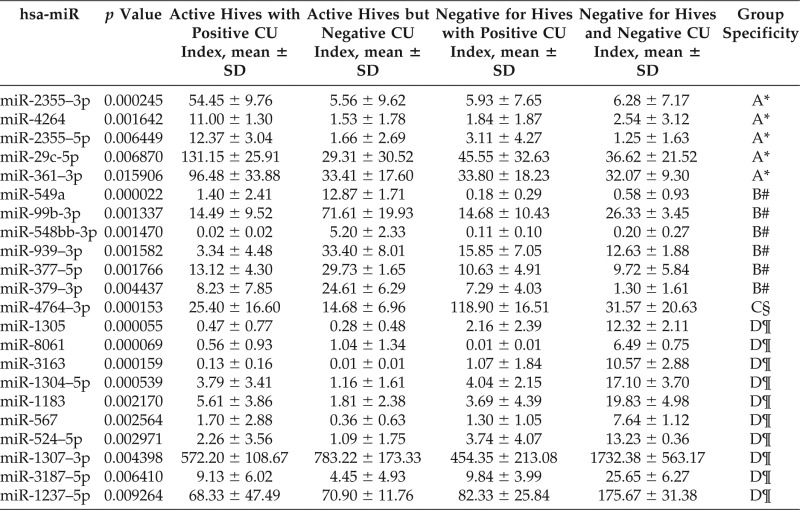
Twelve DE miRNAs were found to have at least twofold changes in miRNA expression between active hives and no hives. Among these, only miR-6769–5p showed an increase of more than twofold, whereas the other 11 miRNAs all showed decreases of more than twofold, as shown in Table 2. This was somewhat unexpected because, for miRNAs to act as inhibitors of mRNA function, these would have been expected to be increased in patients with CIU and with hives. Next, we compared the differential expression of miRNAs among the four groups to specifically evaluate the active hives with a positive CU index (i.e., CAU) group. When this CAU group was compared with the other groups, we found five DE miRNAs: 2355–3p, 2355–5p, 4264, 29c-5p, and 361–3p were all increased between 3- to 10-fold in mean values, as listed in Table 3. This result implied that genes targeted by these five miRNAs could be greatly inhibited and that they could be useful as biomarkers for the pathophysiology of CAU.29
Table 2.
Differential expression of miRNAs between patients with active hives and patients negative for hives by ANOVA analysis
miRNA = MicroRNA; ANOVA = analysis of variance; hsa-miR = homo sapiens related miRNA; SD = standard deviation.
Table 3.
Differential expression of miRNAs in different groups of patients by ANOVA analysis
miRNA = MicroRNA; ANOVA = analysis of variance; hsa-miR = homo sapiens related miRNA; CU = chronic urticaria; SD = standard deviation.
*Active hives, positive CU index.
#Active hives, negative CU index.
§Negative for hives, positive CU index.
¶Normal, negative for hives and negative CU index.
Potential mRNA Targets by the Selected DE miRNAs
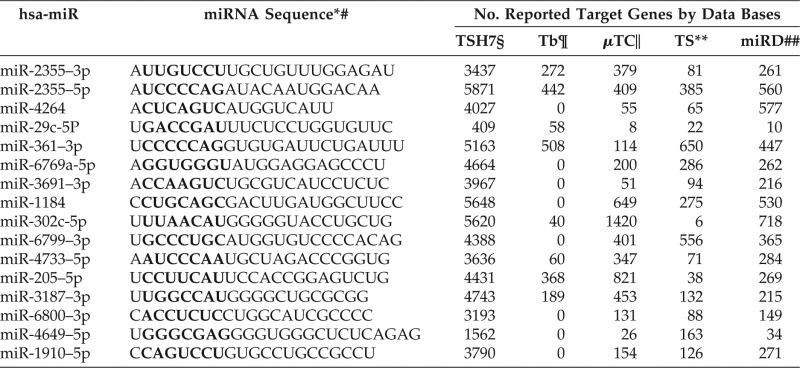
Next, we investigated potential genes targeted by the miRNAs characteristic of active hives. Several miRNA target data bases, such as TargetScanHuman 7.1, TarBase, Diana-microT-CD, miRDB, were found by using a Wikipedia (The Free Encyclopedia, Wikimedia Foundation, San Francisco, CA) search.30 The number of potential targets identified by each databases differed, as shown in Table 4. We elected to use the TargetScanHuman 7.1 miRNA data base because it was used by Qiagen's IPA simulation program and because it shows the largest number of possible targets for each miRNA. Because miRNA data bases predict targets based on sequence alone, there is some element of chance involved. To avoid this pitfall, we decided to consider only those potential targets that were reported to be related to CIU, as shown in Table 5, with the gene names and references listed for those matched gene symbols.
Table 4.
Sequence of selected miRNAs and the number of proposed target genes in various miRNA data bases
miRNA = MicroRNA; hsa-miR = homo sapiens related miRNA; mRNA = messenger RNA.
*miRBase (from Ref. 20).
#The seed sequences for mRNA binding are shown in bold.
§TargetScan Human 7.1 (from Ref. 19).
¶Tarbase (from Ref. 26).
‖microT-CDS (from Ref. 26).
**TargetScan (from Ref. 26).
##miRDB (from Ref. 20).
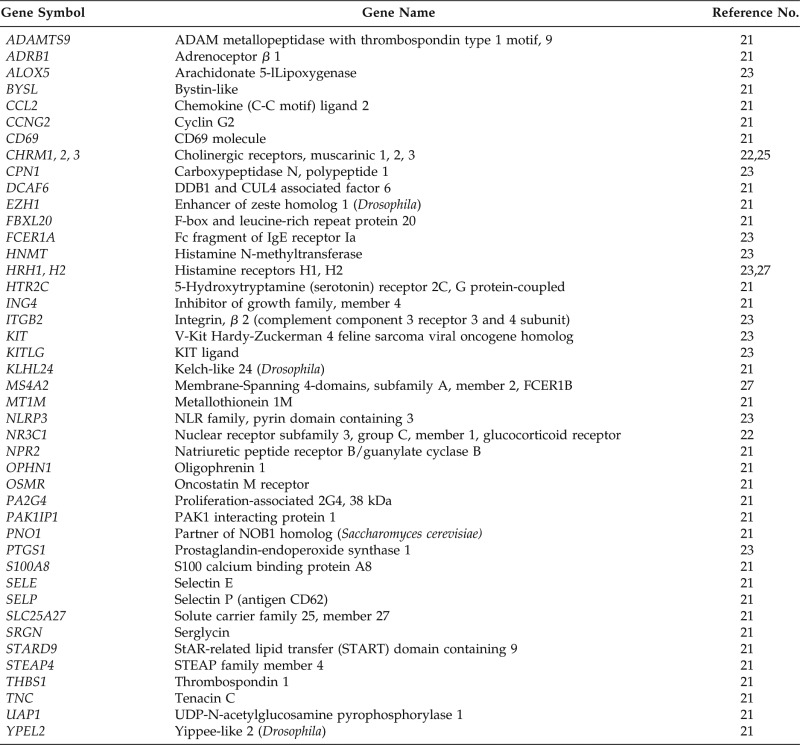
Table 5.
A list of 154 CIU-related genes with gene names shown for those potential microRNA targets*
CIU = Chronic idiopathic urticaria.
Other CIU-related genes not listed are the following: ACE, ADORA3, ATR, BCL2, CCL11 (Eotaxin), CCL13, CCL17 (TARC), CCL22 (MDC), CCL24 (Eotaxin-2), CCL26 (Eotaxin-3), CCL5 (RANTES), CCL8 (MCP-2), CCR1, CDKN1A, CH25H, CISH, CLC, CLEC4A, CMA1, CSF1, CSF2 (GM-CSF), CTSL1, CYP2C9, CysLTR1, E2F4, FARSB, FCGR2A, GARS, GNL3, GPR44 (CRTH2), HDAC2, HSP90AA1, HSPA5, HSPA8, HSPA9, HSPD1, IARS, ICAM1, ICOS, IFNAR, IFNAR2, IFNF (IL10), IFNGR1, IFNGR2, IGHE, IL-12A, IL-12B, IL-13, IL-17A, IL-18, IL1R1, IL1RL1, IL-21, IL-25 (IL-17E), IL-3, IL-31, IL-33, IL-4, IL4R, IL-5, IL-9, IPO5, IRF1, JAK3, KPNA1, KPNA2, KPNA4, KPNB1, LARS, LTC4S, MAP3K14, MARS, MARS2, MCEE, MMP12, MT2A, MYC, MYD88, NFKB1, PDCD1, PMCH, PTGER4, PTPN2, PTPN22, RAN, RANBP1, RANBP2, RNASE3, SGK1, SMARCA2, SOCS2, SOCS3, SRA1, STAT1, STAT3, TARS, TGFB1, TGFBR1, TGFβ1, TNFAIP6, TNFRSF1A, TNF-α, TNSF4 (OX40L), TSLP, VCAM1, WARS, YARS, and YPEL2NPR2.
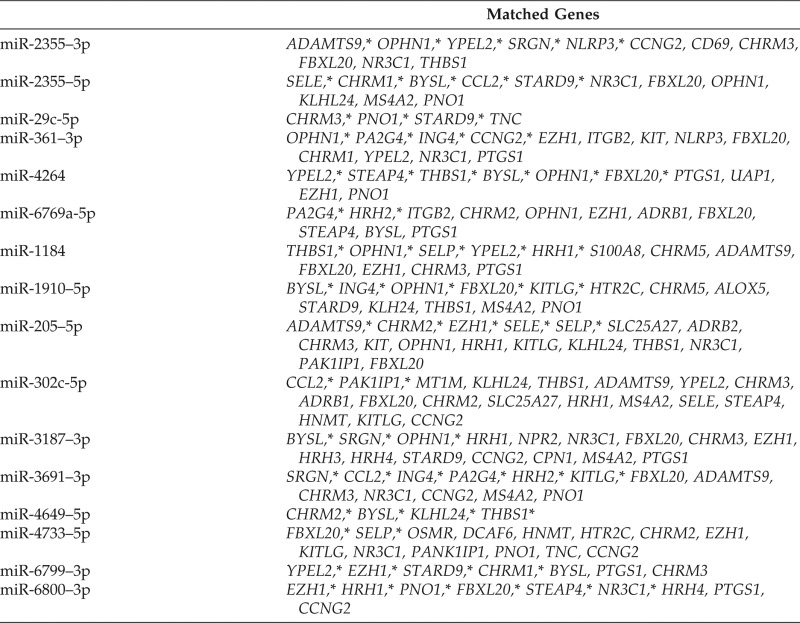
For each of the selected 16 DE miRNAs, we first created a gene set of potential targets found in the TargetScanHuman 7.1 miRNA data base.19 We then compared them with the 154 urticaria-related genes. The matched genes, in rank order by the miRNA data base, are shown in Table 6. To weigh the importance of these selected targets, we counted the number of times that these genes were targeted by the selected DE miRNAs. The result is shown in Fig. 2. Among them, FBXL20 shows the highest count and is targeted by 13 of the 16 DE miRNAs, followed by EZN1 and OPHN1 as targets of 9 of the 16 DE miRNAs.
Table 6.
Matched candidate genes between CIU-related genes and miRNA targets for each of the selected miRNAs in active hives
CIU = Chronic idiopathic urticaria; miRNA = microRNA.
*Higher-ranked genes.
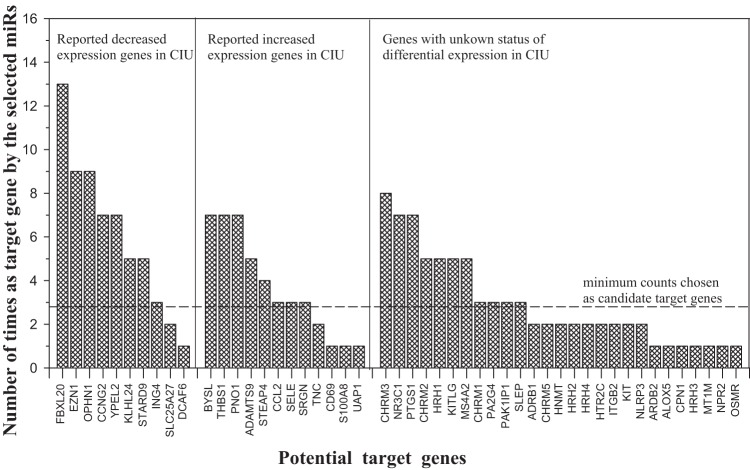
Figure 2.
The potential number of microRNA (miRNA) that targets each gene. Genes are grouped as either differentially decreased or differentially increased in expression in chronic idiopathic urticaria (CIU) (reported in Ref. 21). The last panel is of genes not known to be differentially expressed in CIU. The dashed line indicates the cutoff for selection of potential targets that are targeted by at least three different selected miRNAs.
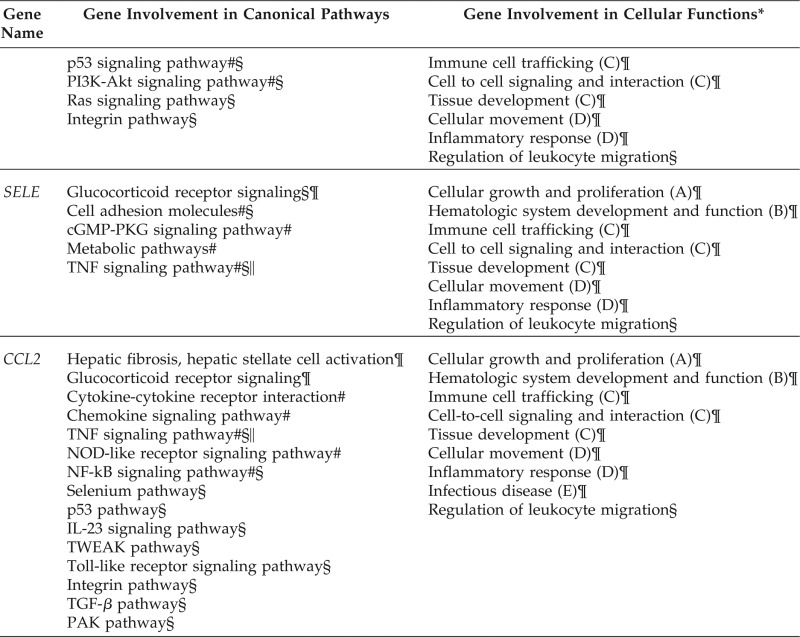
These three genes, along with five other genes (CCNG2, YPEL2, KLHL24, STARD9, and ING4), which were targeted by at least three different DE miRNAs, were reported as the most differentially downregulated genes in CIU.21 Three genes (BYSL, THBS1, and PNO1) were found to be targeted by seven DE miRNAs, followed by five other genes (ADAMTS9, STEAP4, CCL2, SELE, and SRGN), which were targeted by at least three different DE miRNAs were reported by Patel et al.21 as the most differentially upregulated genes in CIU. In addition, 11 genes not known to be DE in CIU were also targeted by at least three different DE miRNAs. In all, we found 25 (when considering CHRM1, CHRM2, CHRM3 as one CHRM family) of 49 targets as common targets by three or more selected DE miRNAs. These were considered as essential CIU candidate genes in active hives.
Role of Candidate Genes Targeted by the Selected DE miRNAs in Signal Pathways and Cellular Functions
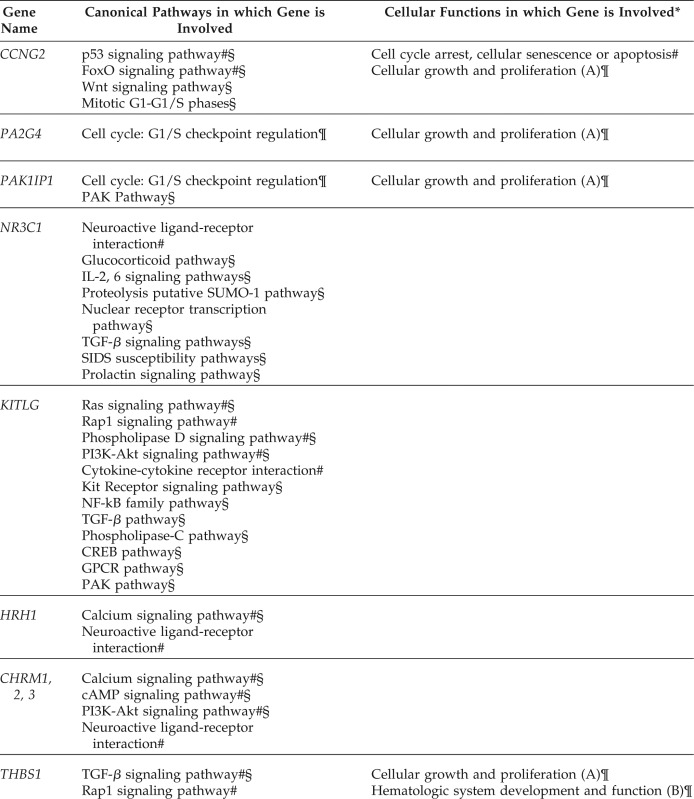
With having found these 25 candidate genes for active hives, the question arises regarding what their roles are in signal pathways and cellular functions, and whether they can shed some light on understanding the mechanism of CIU. We first looked into the bioinformatics of each of these genes by using the KEGG pathway data base24 and the PathCards data base.25 Surprisingly, seven of the eight reported downregulated genes (FBXL20, EZH1, OPHN1, YPEL2, KLHL24, STARD9, ING4) and five of the eight upregulated genes (BYSL, PNO1, ADAMTS9, STEAP4, SRGN) reported by Patel et al.21 have no known roles in signaling pathways and, hence, are unknown as to their subsequent cellular functions. The missing information about these 12 genes is a major setback for understanding the cause and mechanism of CIU because they are not only 12 of 16 most differentially regulated genes in CIU but, also, 9 of these 12 genes account for 18 of the most common targets by at least five or more DE miRNAs.
Only one significantly downregulated gene, CCNG2, and three significantly upregulated genes, THBS1, SELE, and CCL2, are known to be involved in certain signaling pathways that may be relevant to immune pathophysiology of CIU, which, in turn, are related to a variety of cellular functions, as shown in Table 7. However, 9 of the remaining 25 candidate genes (PA2G4, PAK1IP1, NR3C1, KITLG, HRH1, CHRM, MS4A2, PTGS1, SELP), which are uncertain of their differential expression status, are known to be associated with certain pathways, but most of their roles in cellular functions remain uncharacterized.
Table 7.
Role of active hives candidate genes with known canonical signal pathways and cellular functions
CIU = Chronic idiopathic urticaria; NF-κB = nuclear factor kappa light chain enhancer of activated B cells.
*Other CIU related genes which also involved in this cellular function as reported by Patel (21) are: (A) ALOX5, CD69, OSMR, S100A8, SGRN, SLC25A27, TNC; (B) ALOX5, CD69, S100A8; (C)CD69, S100A8; (D) ALOX5, CD69, S100A8; (E) ALOX5, CD69, S100A8, UAP1.
#From Ref. 24.
§From Ref. 25.
¶From Ref. 21.
‖From Ref. 27.
Because every pathway consists of many genes, enzymes, and proteins, we specifically searched for those pathways that engaged at least 3 members of these 13 candidate genes with known functions because it could boost their involvement due to possible synergic effects. Based on this consideration, we found the following pathways in transforming growth factor β (TGF-β) signaling pathway (via NRC31, KITLG, THBS1, CCL2), glucocorticoid receptor signaling pathway (NR3C1, SELE, CCL2), p53 signaling pathway (CCNG2, THBS1, CCL2), p21-activted kinase (PAK) pathway (PAK1IP1, KITLG, CCL2), phosphoinositide-3 kinase and protein kinase B (PI3K-Akt) signaling pathway (KITLG, CHRM, THBS1), and neuroactive ligand-receptor interaction (NRC31, HRH1, CHRM) all fit into this category and could be relevant in the pathophysiology of active hives in CIU.
In addition, we tried three pathway simulation programs:
Qiagen Bioinformatics IPA program.27 We uploaded our selected 16 DE miRNAs into the IPA program and found that the IPA automatically identified MS4A2 and HRH1 genes as the pivotal link for urticaria. This program seems to be ideal for the miRNA-mRNA analysis if both comparable differential expression results for miRNA and mRNA are available.
Advaita's iPathwayGuide.28 This program analyzes RNA genes and predicts pathways based on the KEGG pathway data base24 as well as cellular function assessment based on GO biologic process analysis.31 We uploaded the selected 25 candidate genes and found it only returned limited information, such as SELE and CCL2 as involved in the tumor necrosis factor (TNF) signaling pathway and in the regulation of leukocyte migration. This program will be convenient to show the overall effect of the selected genes if all of them have defined roles in signal pathways and cellular functions.
Diana-miRPathv3.0. This program provides a free tool to directly decipher miRNA function for potential pathways and genes regulated by the selected miRNAs.26 We uploaded the selected five DE miRNAs specific for CAU into the program and found that it returned 965 genes that can be targeted by these miRNAs. It also listed four KEGG pathways and 18 GO categories. Overall, this program was a good start to look for the roles of miRNAs in pathways and cellular functions based on miRNAs alone; however, without knowing their roles within specific genes related to the disease, the results will seem to be too broad to explain the actual mechanisms that define that disease.
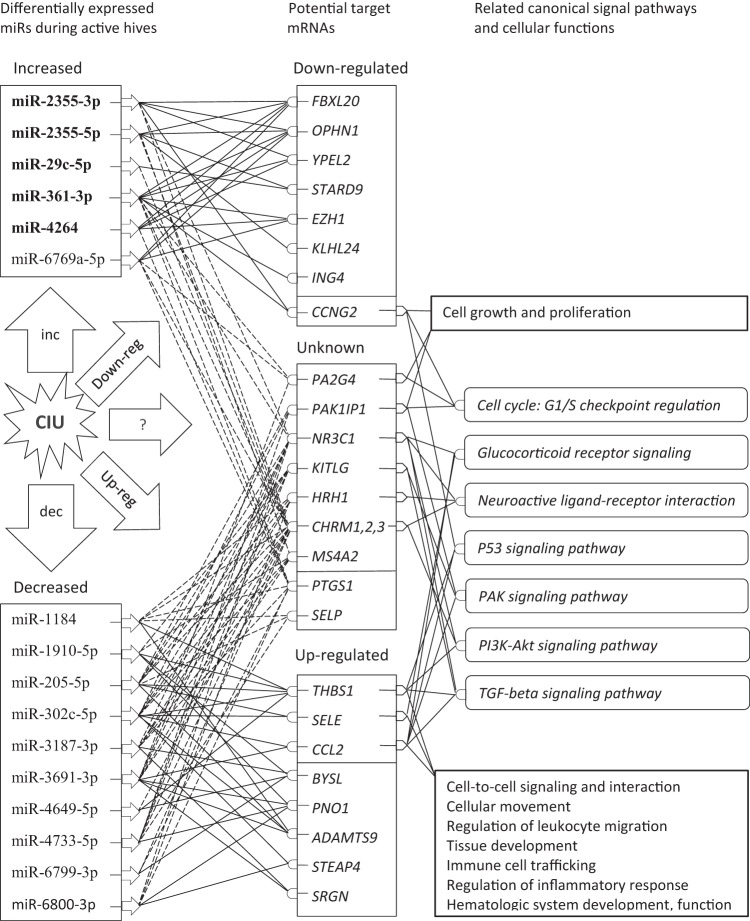
Due to the lack of information about involvement of certain genes in pathways and because we do not have parallel mRNA differential expression results, these simulation programs could not be applied to provide answers about CIU disease in this study. Nevertheless, because of the complexity of the miRNA network, a simplified diagram is shown in Fig. 3 to illustrate the links between the DE miRNAs found in this pilot study and their candidate targets as well as between these mRNAs to their possible roles in signal pathways and cellular functions.
Figure 3.
Simplified diagram, illustrating linkage between selected microRNAs (miRNA) and their potential target messenger RNAs (mRNA) and potential signaling pathways and cell functions. Differentially expressed miRNAs in active hives are placed together in either increased (five miRNAs as potential chronic autoimmune urticaria [CAU] biomarkers are shown in bold) or decreased boxes in the left-hand side. Likewise, the candidate target mRNAs are grouped as either down- or upregulated, or in the unknown boxes in the middle. Due to the complexity of the miRNA-mRNA network, only connections among miRNAs and the chosen candidate genes that were targets of three or more different miRNAs are shown in solid lines if they are in accordance (i.e., increased miRNA expression that corresponded to mRNAs decreased in chronic idiopathic urticaria [CIU], or decreased miRNA expression that corresponds to mRNAs increased in CIU) or in dashed lines if the status between them is unknown. Similarly, only pathways (in round boxes and italic) and possible cellular functions (in rectangular boxes) were shown if they were involved with at least three of the candidate genes with known functions.
DISCUSSION
miRNA is a new player in the field of molecular biology. Since the first discovery of lin-4 in 1993,32 miRNA research has been booming and the volume of publications has increased greatly. However, although thousands of miRNAs have been found and tens of thousands of miRNA reviews have been reported, the real functions of these small noncoding miRNAs remain largely unsettled. The main reason could be in the difficulty of defining their roles directly and effectively. Nevertheless, deregulation of miRNAs were found to be associated with human diseases, e.g., miR-21 was reported in chronic lymphocytic leukemia33 and allergic diseases.14 The broad posttranscriptional regulation of mRNA was proposed as an alternate for the treatment of various diseases, as shown in ∼369 miRNA studies currently under various stages of clinical trials.34 Two miRNA studies have been reported in asthma and allergic rhinitis,15,35 but, to our knowledge, there is no published miRNA study for CIU to date. We found 16 of 2567 known human miRNAs that were DE with more than twofold changes in patients with active hives. Fourteen of these are mature miRNAs, which were indicated by suffix of -3p or of -5p.4 But only 6 of these 16 miRNAs showed an increase in expression in patients with active hives. Moreover, five of these six differentially increased miRNAs were found in patients with CAU, which implied that these miRNAs (2355–3p, 2355–5p, 29c-5p, 361–3p, and 4264) could be useful biomarkers for the CAU subtype of CIU.
We then screened for potential targets of these 16 DE miRNAs directly by using the TargetScanHuman 7.1 miRNA data bases and then compared the selected potential targets with the 154 CIU-related genes listed in Table 5 and found 49 matches. Furthermore, we found that 25 of these 49 genes were targets by at least 3 of the selected 16 miRNAs, which could be real candidate genes involved in active hives. We then investigated further what was known about these candidate genes. Unfortunately, in half of these 25 candidate genes, it is unclear what their roles are in signaling pathways. The lack of information about these genes presents a drawback in exploring the obscure mechanisms of CIU. Nevertheless, we examined the 13 candidate genes that are known to be involved in signaling pathways to survey what could be concluded in CIU, as shown in Table 7.
For 4 of these 13 candidate genes, the CCNG2 gene was significantly downregulated and the 3 others (THBS1, SELE, CCL2) were significantly upregulated, as reported by Patel et al.21 CCNG2 is one of several genes, including MDM2, COP-1, PIRH-2, WIP1, SIAH1, TP73, that act as a p53 negative feedback regulator; p53 activation is induced by a number of stress signals, including DNA damage and oxidative stress. Because of its role via the p53 pathway, CCNG2 is reported to be associated with cell cycle arrest, cellular apoptosis, cellular growth, and proliferation functions. THBS1 is also involved in p53 signaling pathway; however, it plays a different role in inhibition of angiogenesis and metastasis. Upregulation of THBS1 does not seem to be in conflict with downregulation of CCNG2, which could imply collaboration in their effects on p53 activation. Also, THBS1 is also involved in the TGF-β signaling pathway, Rap1 signaling pathway, and PI3K-Akt signaling pathway. Thus, not only can an miRNA target multiple mRNAs, but a certain mRNA can also participate in multiple signaling pathways. Therefore, for similar reasons, as we reasoned, potential targets should be targeted by three or more selected miRNAs to be a trustworthy candidate gene, a signaling pathway should also comprise ≥3 different genes from these 13 candidate genes to be a trustworthy canonical signaling pathway. With this assumption, p53 signaling seemed to be one of the valid pathways in CIU by means of CCNG2, THBS1, and CCL2 contribution.
SELE is reported to be involved in glucocorticoid receptor signaling, the cyclic 3′,5′-guanosine monophosphate dependent protein kinase G signaling pathway, metabolic pathways, and TNF signaling pathway. CCL2 is also reported to be associated with glucocorticoid receptor signaling, the TNF signaling pathway, chemokine signaling pathway, nucleotide-binding, oligomerization domain-like receptor signaling pathway, NF-κB signaling pathway, p53 signaling pathway, and others. When comparing the roles of SELE and CCL2, we found that two common pathways in glucocorticoid receptor signaling and TNF signaling were shared by these two genes but with different attributes. For example, in the TNF signaling pathway, SELE is related to cell adhesion in the same group with ICAM1, VCAM1; however, CCL2 is related to leukocyte recruitment propelled by CCL5, CCL20, and other chemokine motifs. The other nine selected essential genes (PA2G4, PAK1IP1, NR3C1, KITLG, HRH1, CHRM, MS4A2, PTGS1, and SELP) are unknown regarding their differential expression status in CIU. PA2G4 and PAK1IP1 were reported by Patel et al.21 to be involved in G1/S checkpoint regulation in cell cycle. Along with the CCNG2 gene, which is also involved in mitotic G1-G1/S phases, these three genes are related to cell growth and proliferation.
Three genes (NR3C1, HRH1, CHRM1,2,3) are related to neuroactive ligand-receptor interaction reported in the KEGG pathway data base, but their roles are different. NR3C1 is associated with cortisol in glucocorticoid receptor signaling, HRH1 is associated with histamine, and CHRM1, CHRM2, and CHRM3 are associated with acetylcholine. CHRM1 and CHRM2 are also associated with the PI3K-Akt signaling pathway through the chemokine signaling pathway down to G protein-coupled receptors. The PI3K-Akt signaling pathway is also linked to THBS1 through extracellular matrix receptor interaction and to KITLG, which acts as a growth factor through KIT, a receptor tyrosine kinase. Also, three genes (not shown in Table 7) are the following: PTGS1 is involved in metabolic pathway but has a different role from that of SELE in the metabolic pathway; SELP, like SELE which is shown in Table 7, is a cell adhesion molecule involved in leukocyte migration; and MS4A2 is a lone essential gene that links to the FcεRI signaling pathway, phospholipase D signaling pathway, and sphingolipid signaling pathway.
When taking the 13 candidate genes together, we found the following six pathways in the TGF-β signaling pathway (NRC31, KITLG, THBS1, CCL2), glucocorticoid receptor signaling pathway (NR3C1, SELE, CCL2), p53 signaling pathway (CCNG2, THBS1, CCL2), p21-activted kinase (PAK) pathway (PAK1IP1, KITLG, CCL2), phosphoinositide-3 kinase and protein kinase B (PI3K-Akt) signaling (KITLG, CHRM, THBS1), and neuroactive ligand–receptor interaction (NRC31, HRH1, CHRM) all contain at least 3 of these chosen 13 genes that could play important roles in active hives.
CONCLUSION
From the GO biologic process analysis by IPA and iPathwayGuide simulation programs,27,28 these 13 genes and their affiliated signaling pathways were found to be associated with several biologic functions. For example, the three significantly upregulated genes THBS1, SELE, and CCL2 were reported to be associated with many cellular functions, including cell-to-cell signaling and interaction, cellular movement, tissue development, regulation of leukocyte migration, immune cell trafficking, inflammatory response, and hematologic system development and function, as shown in Fig. 3. Overall, the GO biologic process analysis in the disease of CIU pointed to cellular movement, cell migration, cell-cell interaction, and cell growth of the immune cells; however, it is still unclear as to the exact role in CIU mechanisms because the functions of the 12 most prominent CIU-related genes are still unknown.
ACKNOWLEDGMENTS
We thank John Chase, M.D., for his contribution in recruiting patients and providing the needed patient information for this research project.
Footnotes
Supported by Southern California Permanente Medical Group
The authors have no conflicts of interest to declare pertaining to this article
This work is published and licensed by OceanSide Publications, Inc. The full terms of this license are available at https://www.allergyandrhinology.com/terms and incorporate the Creative Commons License Deed: Attribution – Non-Commercial – NoDerivs 4.0 Unported (CC BY-NC-ND 4.0). By accessing the work you hereby accept the terms. Non-commercial uses of the work are permitted without any further permission from OceanSide Publications, Inc., provided the work is properly attributed. Any use of the work other then as authorized under this license or copyright law is prohibited.
REFERENCES
- 1. Magen E, Zueva E, Mishal J, Schlesinger M. The clinical and laboratory characteristics of acute spontaneous urticaria and its progression to chronic spontaneous urticaria. Allergy Asthma Proc 37:394–399, 2016. [DOI] [PubMed] [Google Scholar]
- 2. Greaves MW. Chronic idiopathic urticaria. Curr Opin Allergy Clin Immunol 3:363–368, 2003. [DOI] [PubMed] [Google Scholar]
- 3. Azkur D, Civelek E, Toyran M, et al. Clinical and etiologic evaluation of the children with chronic urticaria. Allergy Asthma Proc 37:450–457, 2016. [DOI] [PubMed] [Google Scholar]
- 4. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116:281–297, 2004. [DOI] [PubMed] [Google Scholar]
- 5. Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell 115:787–798, 2003. [DOI] [PubMed] [Google Scholar]
- 6. Mazière P, Enright AJ. Prediction of microRNA targets. Drug Discov Today 12:452–458, 2007. [DOI] [PubMed] [Google Scholar]
- 7. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92–105, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42(Database issue):D68–D73, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20, 2005. [DOI] [PubMed] [Google Scholar]
- 10. Baek D, Villén J, Shin C, et al. The impact of microRNAs on protein output. Nature 455:64–71, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ortiz-Quintero B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif 49:281–303, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singh R, Ramasubramanian B, Kanji S, et al. Circulating microRNAs in cancer: Hope or hype? Cancer Lett 381:113–121, 2016. [DOI] [PubMed] [Google Scholar]
- 13. Rasooly MM, Moye NA, Kirshenbaum AS. Helicobacter pylori: A significant and treatable cause of chronic urticaria and angioedema. Nurse Pract 40:1–6, 2015. [DOI] [PubMed] [Google Scholar]
- 14. Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol 132:3–13; quiz 14, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rebane A, Akdis CA. MicroRNAs in allergy and asthma. Curr Allergy Asthma Rep 14:424, 2014. [DOI] [PubMed] [Google Scholar]
- 16. Pua HH, Ansel KM. MicroRNA regulation of allergic inflammation and asthma. Curr Opin Immunol 36:101–108, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Panganiban RP, Wang Y, Howrylak J, et al. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol 137:1423–1432, 2016. [DOI] [PubMed] [Google Scholar]
- 18. Mannucci C, Casciaro M, Minciullo PL, et al. Involvement of microRNAs in skin disorders: A literature review. Allergy Asthma Proc 38:9–15, 2017. [DOI] [PubMed] [Google Scholar]
- 19. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43(Database issue):D146–D152, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patel OP, Giorno RC, Dibbern DA, et al. Gene expression profiles in chronic idiopathic (spontaneous) urticaria. Allergy Rhinol (Providence) 6:101–110, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. MalaCards: Human Disease Database, Weizmann Institute of Science, Rehovot, Israel: Available online at http://www.malacards.org/card/urticaria [Google Scholar]
- 23. GeneAnalytics: Gene set analysis, LifeMap Sciences, Alameda, CA: Available online at http://geneanalytics.genecards.org/ [Google Scholar]
- 24. KEGG: Kyoto Encyclopedia of Genes and Genomes, Kanehisa Laboratories, Kyoto University, Kyoto, Japan: Available online at http://www.genome.jp/kegg/pathway.html [Google Scholar]
- 25. PathCards: Pathway unification database, Weizmann Institute of Science, Rehovot, Israel: Available online at http://pathcards.genecards.org/ [Google Scholar]
- 26. Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res 43:W460–W466, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. IPA, Qiagen, Redwood City, CA: Available online at http://www.ingenuity.com/products/ipa#/?tab=overview [Google Scholar]
- 28. iPathwayGuide, Advaita Bioinformatics, Plymouth, MI: Available online at https://apps.advaitabio.com/oauth-provider/login [Google Scholar]
- 29. Goh CL, Tan KT. Chronic autoimmune urticaria: Where we stand? Indian J Dermatol 54:269–274, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. MicroRNA and microRNA Target Database. Available online at https://en.wikipedia.org/wiki/MicroRNA_and_microRNA_target_database
- 31. Gene Ontology Consortium. Available online at http://www.geneontology.org/
- 32. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854, 1993 [DOI] [PubMed] [Google Scholar]
- 33. Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: How a complex biology gets more complex. Leukemia 29:1004–1017, 2015. [DOI] [PubMed] [Google Scholar]
- 34. U.S. National Institutes of Health. Available online at https://clinicaltrials.gov/ct2/results?term=microRNA&Search=Search
- 35. Suojalehto H, Lindström I, Majuri ML, et al. Altered microRNA expression of nasal mucosa in long-term asthma and allergic rhinitis. Int Arch Allergy Immunol 163:168–178, 2014. [DOI] [PubMed] [Google Scholar]