Abstract
Background:
Inflammatory bowel diseases (IBDs), including ulcerative colitis (UC) and Crohn's disease (CD), are chronic inflammatory disorders of the gastrointestinal tract. A combination of environmental factors and interactions with a genetic predisposition are suggested to play an important role in the etiology and pathogenesis of the IBD. Glutathione S-transferases (GSTs) are multifunctional enzymes involved in the cellular oxidative stress handling. Possible associations between GSTs gene polymorphisms and susceptibility to UC and CD have been reported in different population. The relationship between GSTM1 and GSTT1 deletion polymorphisms and susceptibility to UC and CD were investigated in the Iranian population.
Materials and Methods:
The study was performed in 106 IBD patients and 243 age- and sex-matched healthy Iranian controls consulting the IBD registry center of the Motahari Clinic, Shiraz University of Medical Sciences, Shiraz, Iran, between 2011 and 2013. GSTM1 and GSTT1 genotyping were performed using multiplex polymerase chain reaction and differences in the distribution of gene polymorphisms were analyzed statistically between the studied groups.
Results:
Statistically significant higher frequency of GSTM1 null genotype was observed in IBD patients (P = 0.01) and in the subgroup of patients with UC (P = 0.04) compared to healthy controls, whereas this was not true for CD patients. No significant association was found between GSTT1 gene polymorphism and UC or CD.
Conclusions:
Absence of GSTT1 functional gene does not play an important role in the pathophysiology and development of IBD, UC, and CD in Iranian population whereas GSTM1 null genotype could be considered as a possible genetic predisposing factor for more susceptibility to IBD and UC.
Keywords: Crohn's disease, genetic polymorphism, glutathione S-transferases, inflammatory bowel disease, ulcerative colitis
Introduction
Inflammatory bowel diseases (IBD) are chronic inflammatory disorders of the gastrointestinal tract, characterized by uncontrolled immune responses and severe intestinal inflammation. Ulcerative colitis (UC) and Crohn's disease (CD) are two major subtypes of IBD that have distinct clinical and histopathological characteristics.[1,2] Although the exact etiology of IBD is still unknown, genetic and environmental factors are believed to have important roles in disease predisposition and pathogenesis.[3,4] Epidemiological data have revealed that no single gene or distinct mechanism can explain all the aspects of IBD and genetic predisposition to UC and CD depends on the contribution of several candidate genes, such as IL23R, IL12B, NKX2-3, XRCC1, CARD15, and HLA. Genetic factors seem to be more involved in the pathogenesis and clinical course of CD than UC.[5,6,7,8,9]
Highly reactive molecules, such as reactive oxygen species (ROS) and imbalanced immune responses are known to be one of the major risk factors of intestinal inflammation.[10,11] ROS might be products of bacterial fermentation and endogenous metabolism of xenobiotics within the intestinal lumen. They might also be produced by activated immune cells.[12,13,14] Biotransformation enzymes such as glutathione S-transferases (GSTs), epoxide hydrolases and cytochrome P450s play a key role in the metabolism of ROS and xenobiotics. Allelic polymorphism in genes encoding these enzymes could change their activities, resulting in defective detoxification and finally, might confer susceptibility to inflammatory diseases or even cancers.[12,15,16,17]
GSTs are a family of multi-functional enzymes having detoxifying properties which catalyze the conjugation of mutagenic electrophiles and peroxidized lipids with nucleophilic glutathione, yielding hydrophilic, and less toxic metabolites.[2,18] GSTs consist of three major families: Cytosolic, microsomal, and mitochondrial. Human cytosolic GSTs are divided into eight classes (alpha, zeta, theta, mu, pi, sigma, kappa, and omega) of which mu (GSTM), theta (GSTT), pi (GSTP), and alpha (GSTA) families display significant gene polymorphisms.[19,20] An important genetic polymorphism in GSTM1 coding gene, located on chromosome 1p13.3, has been described as partial gene deletion (GSTM1 null genotype) leading to complete absence of enzyme activity. A deletion polymorphism was also reported for GSTT1 gene, located on chromosome 22q11.2 (GSTT1 null genotype).[9,17] As recently reviewed, the prevalence of GSTM1 null allele ranges from 22% in Africans to 62% among Europeans, whereas GSTT1 null status differs from 20% to 60% in Caucasian and Asian people, respectively.[21,22] Due to the important role of GSTs in oxidative stress and detoxifying carcinogens, GSTM1 and GSTT1 allelic polymorphisms may result in less effective removal of intestinal toxic metabolites, thus modifying the susceptibility to UC and CD.[2,14,22,23]
This study aimed to evaluate the frequency of GSTM1 and GSTT1 genetic polymorphisms in IBD patients and normal controls to explore the association between GSTs genotypes and prevalence of UC and CD in the south of Iran. This is the first report describing the GSTs gene polymorphism in the Iranian IBD patients.
Materials and Methods
Patients and controls population
A total of 106 consecutive patients with IBD (94 with UC and 12 with CD) consulting the IBD registry center of the Motahari Clinic, affiliated by the Gastroenterohepatology Research Center, Shiraz University of Medical Sciences, Shiraz, Iran, were included in this study. Case–control study was performed on subjects recruited between 2011 and 2013. The diagnosis of IBD was confirmed by gastroenterology attending, based on clinical, endoscopic, and histological findings in accordance with Lennard-Jones criteria. Patients without pathologic proof were not included. A total of 243 age- and sex-matched healthy controls were selected from healthy blood donors referred to the National Blood Transfusion Organization, Shiraz, Iran. Informed consent was obtained from all participants for being included in the study. All procedures followed were in accordance with the ethical standards of the Shiraz University of Medical Sciences committee on human experimentation and with the Helsinki declaration of 1975, as revised in 2008.
DNA extraction
Two milliliters of venous blood samples were obtained from each IBD patient and also from controls and stored in vacuum tubes containing EDTA at −20°C before examination. Genomic DNA was extracted from the peripheral blood leukocytes using Accuprep Genomic DNA Extraction Kit (Bioneer, Korea) according to the manufacturer's instructions.
Genotyping of glutathione S-transferases genes polymorphisms
Genetic polymorphisms in GSTM1 (rs 366631) and GSTT1 (rs 17856199) genes were determined using multiplex polymerase chain reaction (PCR) followed by gel electrophoresis. The PCR reactions were performed in a final volume of 25 μl, containing 100 ng genomic DNA, 30 pmol of each primers, 0.3 mM dNTP, 1.5 mM MgCl2 and 1.5 U Taq DNA polymerase (Fermentas). The appropriate fragments of the GSTM1 and GSTT1 genes were amplified using the following specific primers: GSTM1 forward primer, 5’-GAACTCCCTGAAAAGCTAAAGC-3’ and reverse primer 5’-GTTGGGCTCAAATATACGGTGG-3’; GSTT1 forward primer, 5’-TTCCTTA CTGGTCCTCACATCTC-3’ and reverse primer 5’-TCACCGGATCATGGCCAGCA-3’.[2] Beta-globin gene amplification was also used as an internal control in the multiplex PCR, for which the primers were as follows: Forward primer 5′-CAACTTCATCCACGTTCACC-3′ and reverse primer 5′-GAAGAGCCAAGGACAGGTAC-3′.
The amplification conditions consisted of an initial denaturation at 95°C for 5 min, followed by 35 cycles at 94°C for 1 min, 64.5°C for 1 min and 72°C for 1 min, with a final extension at 72°C for 5 min. PCR products were separated by electrophoresis on 2% agarose gel and visualized by UV illumination using ethidium bromide staining [Figure 1].
Figure 1.
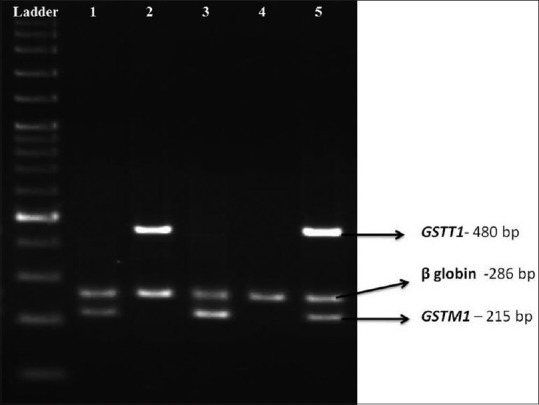
Polymerase chain reaction amplification of the GSTM1, GSTT1 and internal control beta-globin genes, separated by electrophoresis on 2% agarose gel. Lane 5 shows GSTM1 (+)/GSTT1 (+) sample, Lanes 1 and 3 contain GSTM1 (+)/GSTT1 (null) samples, Lane 2 GSTM1 (null)/GSTT1 (+) and Lane 4 shows GSTM1 (−)/GSTT1 (−) sample
Statistical analysis
Statistical significant differences in the distribution of polymorphisms between groups were analyzed using Chi-square test. Odds ratios with 95% confidence intervals (95% CI) were also calculated. In all analyses, a two-tailed P < 0.05 was considered statistically significant. All statistical analyses were conducted using SPSS software, version 18 (SPSS Inc., Chicago, IL, USA).
Results
The average age was 36.6 ± 12.9 years in UC patients and 35 ± 13.3 in CD patients. The patients’ and controls’ demographic characteristics are presented in Table 1. Genotypes of GSTM1 and GSTT1 were screened at the genome level by PCR. The PCR products of GSTM1 (215 bp) and GSTT1 (480 bp) were detected in normal samples but absent in individuals who carry only the null allele [Figure 1]. The frequencies for GSTM1 and GSTT1 deletion polymorphisms were compared between IBD patients and control group. The genotype frequencies were also compared between UC and CD patients with the control group, separately. There was a significantly higher rate of GSTM1 null genotype in IBD (including both UC and CD) patients compared to controls (61.32% vs. 47.32%, P = 0.01, OR = 1.76, 95% CI = 1.10–2.80) [Table 2].
Table 1.
Demographic characteristics of inflammatory bowel diseases patients and healthy controls
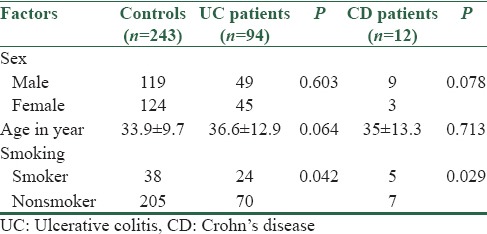
Table 2.
Frequency distribution of GSTM1 gene polymorphisms (rs 366631) in inflammatory bowel diseases patients and healthy controls
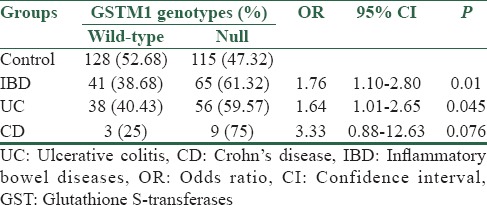
In UC patients, a significantly higher frequency of GSTM1 null genotype was observed compared to healthy controls (59.57% vs. 47.32%. P =0.045, OR = 1.64, 95% CI = 1.01–2.65). On the contrary, no significant difference was observed between control and patient groups regarding GSTT1 gene polymorphism (27.98% vs. 20.21%. P = 0.146, OR = 0.65, 95% CI = 0.36–1.16) [Tables 2 and 3].
Table 3.
Frequency distribution of GSTT1 gene polymorphisms (rs 17856199) in inflammatory bowel diseases patients and healthy controls
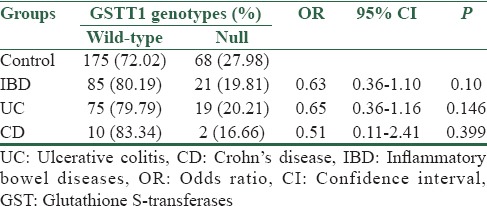
In CD patients, no significant differences in the frequency of GSTM1 and GSTT1 null genotypes were observed between patient and control groups (75% vs. 47.32%, P = 0.076, OR = 3.33, %95 CI = 0.88–12.63 and 16.66% vs. 27.98%, P = 0.399, OR = 0.51, %95 CI = 0.11–2.41, respectively) [Tables 2 and 3].
Discussion
ROS and xenobiotics are of the most important factors affecting inflammation and tissue injuries in the mucosa. Imbalance between production and detoxification of these toxic compounds may play an important role in the etiology of intestinal inflammatory diseases.[24,25] Antioxidant defenses have an important role in the metabolism and removal of toxic molecules and polymorphism in the genes related to their activities could increase the susceptibility to several diseases, especially cancers.[12,16,22]
GSTs are detoxifying enzymes with high expression in the liver, gonads and colon.[2,26] GSTM1 and GSTT1 are two GSTs main subclasses in human involved in the metabolism of different toxins such as polycyclic aromatic hydrocarbons, epoxides, methane, and ethylene oxide.[2] Deletion polymorphism in GSTM1 and GSTT1 genes could result in nonfunctional (null) alleles and loss of enzymatic activities. It has been shown that individuals carrying the GSTM1 and GSTT1 null genotypes may have an impaired detoxifying ability and, therefore, be at increased risk of different cancers, including lung,[27,28] breast,[29] bladder[30] and colorectal cancers.[19,21] A significant association between GSTM1 and GSTT1 gene polymorphisms and greater susceptibility to UC and CD were also reported for different ethnic groups.[2,14,22]
In the present research, we studied the GSTM1 and GSTT1 deletion polymorphisms in Iranian UC and CD patients as well as healthy controls. The prevalence of GSTM1 and GSTT1 null genotypes in controls (47.32% and 27.98%, respectively) were similar to previously reported results.[12,14,22] Our finding showed a significantly higher frequency of GSTM1 null genotype in all IBD (P = 0.01) and also in UC subgroup (P = 0.045) patients relative to controls, but not in the CD patients (P = 0.076). However, no significant differences were observed between the frequency of GSTT1 null genotype in the IBD patients (P = 0.10), UC (P = 0.146) and CD (P = 0.399) subgroups, compared to the controls. The observed higher frequency of the GSTM1 null genotype in IBD and UC Iranian patients relative to the controls is in consistent with previous studies had done on Asian populations. These studies have shown an important role of GSTM1 gene deletion in the development of UC.[2,14,23] Ye et al. reported a positive association of GSTM1 and GSTT1 null alleles with susceptibility to UC in central China population and suggested a predominant role of GSTM1 to influence the UC.[2] An association between the null genotype of GSTM1 with UC (but not CD) was also reported in Indian IBD patients.[23] Conversely, an Israeli study found a lower GSTM1 null frequency in IBD and CD Arab Moslem patients than controls.[22]
The GSTT1 null genotype has been correlated with the increased risk of UC and CD in different populations,[2,14,22,23] however in some studies such a relationship was not found.[12,13,31] In our study, the GSTT1 null frequencies did not differ significantly between the UC and CD patients compared to controls. This is in agreement with the results of de Jong et al and Ernst et al., who showed no association between GSTT1 gene deletion and occurrence of UC and CD in the Netherlands and Danish population, respectively.[12,31] Overall, it could be concluded that GSTM1 and GSTT1 null alleles may confer more susceptibility to intestinal inflammatory disorders, although these conflicting results may be due to confounding variables, such as ethnicity, different genetic background and additional susceptibility genes.
Our results indicate that absence of the GSTM1 and GSTT1 functional genes does not play an important role in the pathophysiology of CD in Iranian population. However, it should be noted that making a conclusion based on the results obtained from a small CD patients group (n = 12) might not be very exact (the limitation of this study). More comprehensive studies regarding larger sample sizes are needed for the more definite conclusion.
Conclusion
A significant association was found between the GSTM1 null genotype and greater susceptibility to IBDs and UC in the Iranian population. These results suggest that GSTM1 gene polymorphism could be considered as a possible genetic predisposing factor for IBD and UC in Iranian population.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye X, Jiang Y, Wang H, Chen L, Yuan S, Xia B. Genetic polymorphisms of glutathione S-transferases are associated with ulcerative colitis in central China. Cell Biochem Biophys. 2011;60:323–8. doi: 10.1007/s12013-011-9154-z. [DOI] [PubMed] [Google Scholar]
- 3.Bonen DK, Cho JH. The genetics of inflammatory bowel disease. Gastroenterology. 2003;124:521–36. doi: 10.1053/gast.2003.50045. [DOI] [PubMed] [Google Scholar]
- 4.Molodecky NA, Kaplan GG. Environmental risk factors for inflammatory bowel disease. Gastroenterol Hepatol (N Y) 2010;6:339–46. [PMC free article] [PubMed] [Google Scholar]
- 5.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SW, Kim ES, Moon CM, Park JJ, Kim TI, Kim WH, et al. Genetic polymorphisms of IL-23R and IL-17A and novel insights into their associations with inflammatory bowel disease. Gut. 2011;60:1527–36. doi: 10.1136/gut.2011.238477. [DOI] [PubMed] [Google Scholar]
- 7.Moon CM, Shin DJ, Son NH, Shin ES, Hong SP, Kim TI, et al. Genetic variants in the IL12B gene are associated with inflammatory bowel diseases in the Korean population. J Gastroenterol Hepatol. 2013;28:1588–94. doi: 10.1111/jgh.12214. [DOI] [PubMed] [Google Scholar]
- 8.Bouzid D, Kammoun A, Amouri A, Mahfoudh N, Haddouk S, Tahri N, et al. Inflammatory bowel disease: Susceptibility and disease heterogeneity revealed by human leukocyte antigen genotyping. Genet Test Mol Biomarkers. 2012;16:482–7. doi: 10.1089/gtmb.2011.0132. [DOI] [PubMed] [Google Scholar]
- 9.Tahara T, Shibata T, Nakamura M, Okubo M, Yamashita H, Yoshioka D, et al. Association between polymorphisms in the XRCC1 and GST genes, and CpG island methylation status in colonic mucosa in ulcerative colitis. Virchows Arch. 2011;458:205–11. doi: 10.1007/s00428-010-1038-x. [DOI] [PubMed] [Google Scholar]
- 10.Lih-Brody L, Powell SR, Collier KP, Reddy GM, Cerchia R, Kahn E, et al. Increased oxidative stress and decreased antioxidant defenses in mucosa of inflammatory bowel disease. Dig Dis Sci. 1996;41:2078–86. doi: 10.1007/BF02093613. [DOI] [PubMed] [Google Scholar]
- 11.Rezaie A, Parker RD, Abdollahi M. Oxidative stress and pathogenesis of inflammatory bowel disease: An epiphenomenon or the cause? Dig Dis Sci. 2007;52:2015–21. doi: 10.1007/s10620-006-9622-2. [DOI] [PubMed] [Google Scholar]
- 12.de Jong DJ, van der Logt EM, van Schaik A, Roelofs HM, Peters WH, Naber TH. Genetic polymorphisms in biotransformation enzymes in Crohn's disease: Association with microsomal epoxide hydrolase. Gut. 2003;52:547–51. doi: 10.1136/gut.52.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan H, Swan C, Green J, Jones P, Brannigan K, Alldersea J, et al. Susceptibility to ulcerative colitis and Crohn's disease: Interactions between glutathione S-transferase GSTM1 and GSTT1 genotypes. Clin Chim Acta. 1995;240:53–61. doi: 10.1016/0009-8981(95)06127-4. [DOI] [PubMed] [Google Scholar]
- 14.Buyukgoze O, Osmanoglu N, Arslan S, Sen A. Association of the CYP1A1*2A, GSTT1 null, GSTM1 null, mEPHX*3, and XRCC1-399 genetic polymorphisms with ulcerative colitis. Int J Colorectal Dis. 2013;28:593–5. doi: 10.1007/s00384-012-1507-6. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Closas M, Kelsey KT, Wiencke JK, Xu X, Wain JC, Christiani DC. A case-control study of cytochrome P450 1A1, glutathione S-transferase M1, cigarette smoking and lung cancer susceptibility (Massachusetts, United States) Cancer Causes Control. 1997;8:544–53. doi: 10.1023/a:1018481910663. [DOI] [PubMed] [Google Scholar]
- 16.Sergentanis TN, Economopoulos KP, Choussein S, Vlahos NF. Cytochrome P450 1A1 (CYP1A1) gene polymorphisms and cervical cancer risk: A meta-analysis. Mol Biol Rep. 2012;39:6647–54. doi: 10.1007/s11033-012-1470-x. [DOI] [PubMed] [Google Scholar]
- 17.Wan H, Zhou Y, Yang P, Chen B, Jia G, Wu X. Genetic polymorphism of glutathione S-transferase T1 and the risk of colorectal cancer: A meta-analysis. Cancer Epidemiol. 2010;34:66–72. doi: 10.1016/j.canep.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Eaton DL, Bammler TK. Concise review of the glutathione S-transferases and their significance to toxicology. Toxicol Sci. 1999;49:156–64. doi: 10.1093/toxsci/49.2.156. [DOI] [PubMed] [Google Scholar]
- 19.Khan MI, Micheal S, Akhtar F, Ahmed W, Ijaz B, Ahmed A, et al. The association of glutathione S-transferase GSTT1 and GSTM1 gene polymorphism with pseudoexfoliative glaucoma in a Pakistani population. Mol Vis. 2010;16:2146–52. [PMC free article] [PubMed] [Google Scholar]
- 20.Stocco G, Martelossi S, Barabino A, Decorti G, Bartoli F, Montico M, et al. Glutathione-S-transferase genotypes and the adverse effects of azathioprine in young patients with inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:57–64. doi: 10.1002/ibd.20004. [DOI] [PubMed] [Google Scholar]
- 21.Economopoulos KP, Sergentanis TN. GSTM1, GSTT1, GSTP1, GSTA1 and colorectal cancer risk: A comprehensive meta-analysis. Eur J Cancer. 2010;46:1617–31. doi: 10.1016/j.ejca.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Karban A, Krivoy N, Elkin H, Adler L, Chowers Y, Eliakim R, et al. Non-Jewish Israeli IBD patients have significantly higher glutathione S-transferase GSTT1-null frequency. Dig Dis Sci. 2011;56:2081–7. doi: 10.1007/s10620-010-1543-4. [DOI] [PubMed] [Google Scholar]
- 23.Mittal RD, Manchanda PK, Bid HK, Ghoshal UC. Analysis of polymorphisms of tumor necrosis factor-alpha and polymorphic xenobiotic metabolizing enzymes in inflammatory bowel disease: Study from Northern India. J Gastroenterol Hepatol. 2007;22:920–4. doi: 10.1111/j.1440-1746.2006.04538.x. [DOI] [PubMed] [Google Scholar]
- 24.Kruidenier L, Verspaget HW. Review article: Oxidative stress as a pathogenic factor in inflammatory bowel disease – Radicals or ridiculous? Aliment Pharmacol Ther. 2002;16:1997–2015. doi: 10.1046/j.1365-2036.2002.01378.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhu H, Li YR. Oxidative stress and redox signaling mechanisms of inflammatory bowel disease: Updated experimental and clinical evidence. Exp Biol Med (Maywood) 2012;237:474–80. doi: 10.1258/ebm.2011.011358. [DOI] [PubMed] [Google Scholar]
- 26.Ghobadloo SM, Yaghmaei B, Bakayev V, Goudarzi H, Noorinayer B, Rad FH, et al. GSTP1, GSTM1, and GSTT1 genetic polymorphisms in patients with cryptogenic liver cirrhosis. J Gastrointest Surg. 2004;8:423–7. doi: 10.1016/j.gassur.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Stücker I, Hirvonen A, de Waziers I, Cabelguenne A, Mitrunen K, Cénée S, et al. Genetic polymorphisms of glutathione S-transferases as modulators of lung cancer susceptibility. Carcinogenesis. 2002;23:1475–81. doi: 10.1093/carcin/23.9.1475. [DOI] [PubMed] [Google Scholar]
- 28.Zupa A, Sgambato A, Bianchino G, Improta G, Grieco V, LA Torre G, et al. GSTM1 and NAT2 polymorphisms and colon, lung and bladder cancer risk: A case-control study. Anticancer Res. 2009;29:1709–14. [PubMed] [Google Scholar]
- 29.Curran JE, Weinstein SR, Griffiths LR. Polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and breast cancer susceptibility. Cancer Lett. 2000;153:113–20. doi: 10.1016/s0304-3835(00)00361-x. [DOI] [PubMed] [Google Scholar]
- 30.Zeng FF, Liu SY, Wei W, Yao SP, Zhu S, Li KS, et al. Genetic polymorphisms of glutathione S-transferase T1 and bladder cancer risk: A meta-analysis. Clin Exp Med. 2010;10:59–68. doi: 10.1007/s10238-009-0070-0. [DOI] [PubMed] [Google Scholar]
- 31.Ernst A, Andersen V, Østergaard M, Jacobsen BA, Dagiliene E, Pedersen IS, et al. Genetic variants of glutathione S-transferases mu, theta, and pi display no susceptibility to inflammatory bowel disease in the Danish population. Scand J Gastroenterol. 2010;45:1068–75. doi: 10.3109/00365521.2010.490594. [DOI] [PubMed] [Google Scholar]


