Abstract
Alzheimer’s disease (AD) risk is modified by both genetic and environmental risk factors, which are believed to interact to cooperatively modify pathogenesis. Although numerous genetic and environmental risk factors for AD have been identified, relatively little is known about potential gene-environment interactions in regulating disease risk. The strongest genetic risk factor for late-onset AD is the ε4 allele of apolipoprotein E (APOE4). An important modifiable risk factor for AD is obesity, which has been shown to increase AD risk in humans and accelerate development of AD-related pathology in rodent models. Potential interactions between APOE4 and obesity are suggested by the literature but have not been thoroughly investigated. In the current study, we evaluated this relationship by studying the effects of diet-induced obesity (DIO) in the EFAD mouse model, which combines familial AD transgenes with human APOE3 or APOE4. Male E3FAD and E4FAD mice were maintained for 12 weeks on either a control diet or a Western diet high in saturated fat and sugars. We observed that metabolic outcomes of DIO were similar in E3FAD and E4FAD mice. Importantly, our data showed a significant interaction between diet and APOE genotype on AD-related outcomes in which Western diet was associated with robust increases in amyloid deposits, β-amyloid burden, and glial activation in E4FAD but not in E3FAD mice. These findings demonstrate an important gene-environment interaction in an AD mouse model that suggests that AD risk associated with obesity is strongly influenced by APOE genotype.
Keywords: Alzheimer’s disease, apolipoprotein E, β-amyloid, gliosis, obesity, transgenic
Significance Statement
The ε4 allele of apolipoprotein E (APOE4) is the strongest genetic risk factor for Alzheimer’s disease (AD), but not all APOE4 carriers will develop the disease suggesting that APOE genotype interacts with other factors to modulate Alzheimer’s risk. Here, we show that diet-induced obesity (DIO) interacts with APOE4 genotype to increase Alzheimer’s-like pathology in an Alzheimer’s transgenic mouse model that contains human APOE3 versus APOE4 isoforms. Interestingly, mice with APOE3 do not show diet-induced increases in pathology, suggesting that the adverse effects of obesity on Alzheimer’s risk may be limited to APOE4 carriers. These findings identify an important gene-environment interaction that may have significant impact for understanding Alzheimer’s risk and etiology and promoting development of targeted therapeutic approaches that incorporate both obesity and APOE genotype.
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder, the underlying causes of which are currently incompletely understood. Both genetic and environmental factors are important in determining individual risk for AD. The strongest genetic risk factor for late-onset AD is the ε4 allele of apolipoprotein E (APOE4; Strittmatter et al., 1993; Liu et al., 2013). In the United States, roughly 12% of the population carries the ε4 allele, but its frequency increases to ∼60% in AD patients (Rebeck et al., 1993). APOE4 not only increases risk, but also accelerates the age of onset of AD (Corder et al., 1993; van der Flier et al., 2011). However, since homozygous carriers of APOE4 have a ∼50% lifetime risk of AD, a significant number of APOE4 carriers never develop the disease (Genin et al., 2011). Thus, APOE4 likely interacts with other genetic and or environmental factors to drive AD risk.
A significant modifiable risk factor for dementia is obesity. Obesity has numerous adverse neural effects (Lee and Mattson, 2013) and increases the risk of dementia up to three-fold (Whitmer et al., 2008). Body mass index, a commonly used measure of obesity, has been shown to be associated with AD risk (Profenno et al., 2010) as well as with reduced brain volume in AD patients (Ho et al., 2010). Several studies indicate that obesity may be particularly problematic at midlife (Fitzpatrick et al., 2009; Profenno et al., 2010; Meng et al., 2014; Emmerzaal et al., 2015), suggesting that obesity contributes to the development of AD. Similar relationships have been observed in animal models. In particular, diet-induced obesity (DIO) accelerates AD-related pathology in mouse models of AD (Ho et al., 2004; Julien et al., 2010; Kohjima et al., 2010; Barron et al., 2013; Orr et al., 2014). Further, genetic models of obesity and type 2 diabetes exhibit features of AD-like neuropathology (Kim et al., 2009; Jung et al., 2013; Ramos-Rodriguez et al., 2013).
The extent to which APOE4 and obesity interact to regulate AD risk is unclear. Interestingly, APOE4 carriers can be more sensitive to metabolic consequences associated with obesity (de-Andrade et al., 2000; Kypreos et al., 2009; Niu et al., 2009; Atabek et al., 2012; Zarkesh et al., 2012; Guan et al., 2013). Although some studies do not report an APOE4 bias in obesity-associated AD risk (Profenno and Faraone, 2008; Luchsinger et al., 2012), others have found that AD risk is increased by obesity (Peila et al., 2002; Ghebranious et al., 2011) and diets high in calories and fatty acids (Luchsinger et al., 2002) only in APOE4 carriers. Though the human literature suggests a gene-environment interaction between APOE and obesity in regulating development of AD, this question has not been addressed in experimental models. To study these relationships, we used EFAD transgenic mice, which combine AD transgenes with targeted replacement of mouse APOE with human APOE (Youmans et al., 2012). We compared metabolic and AD-related effects of Western diet in male APOE3 (E3FAD) and APOE4 (E4FAD) mice. Here, we report that DIO increases amyloid pathology and gliosis almost exclusively in E4FAD mice. Our data reveal a gene-environment interaction between APOE genotype and obesity, suggesting that APOE4 carriers may be more susceptible to obesity associated increases in AD risk.
Materials and Methods
Animal procedures
A colony of EFAD mice, which are heterozygous for the 5xFAD transgenes and homozygous for human APOE3 or APOE4 (Youmans et al., 2012), were maintained at vivarium facilities at University of Southern California from breeder mice generously provided by Dr. Mary Jo LaDu (University of Illinois at Chicago). All animals were housed under a 12-hour light/dark cycle with lights on at 6 A.M. and ad libitum access to food and water. At three months of age, male E3FAD and E4FAD mice were randomized to dietary treatment groups (N = 7–11/group): control diet (10% fat, 7% sucrose; #D12450J Research Diets) or Western diet (45% fat, 17% sucrose; #D12451, Research Diets). EFAD mice were maintained on experimental diets for 12 weeks, an exposure period previously established to yield obesity-induced metabolic impairments in APOE mice (Arbones-Mainar et al., 2010; Segev et al., 2016). Body weight and food consumption were recorded weekly.
At the end of the treatment period, mice were anesthetized with inhalant isoflurane and transcardially perfused with ice-cold 0.1 M PBS. The brains were rapidly removed and immersion fixed for 48 h in 4% paraformaldehyde/0.1 M PBS, then stored at 4°C in 0.1 M PBS/0.3% NaN3 until processed for immunohistochemistry. Gonadal and retroperitoneal fat pads were dissected and weighed as a measure of adiposity, and snap frozen for RNA extraction. All animal procedures were conducted under protocols approved by the University of Southern California Institutional Animal Care and Use Committee and in accordance with National Institute of Health standards.
Glucose, cholesterol, and triglyceride measurements
Blood glucose readings were measured after overnight fasting (16 h) every four weeks beginning at week 0 of the 12-week treatment period. Blood was collected from the lateral tail vein and immediately assessed for glucose levels using the Precision Xtra Blood Glucose and Ketone Monitoring System (Abbott Diabetes Care).
Glucose tolerance testing (GTT) was performed at week 11. Fasting, baseline glucose readings were taken after which mice were administered a glucose bolus (2 g/kg body weight) via oral gavage. Blood glucose levels were recorded 15, 30, 60, and 120 min after the glucose bolus was given. Area under the curve (AUC) was calculated.
Plasma cholesterol and triglyceride levels were enzymatically determined at the conclusion of the experiment using commercially available kits (LabAssay Triglycerides #290-63701, Wako Chemicals; Total Cholesterol Colorimetric Assay kit, #K603, BioVision). All samples were run in duplicate according to manufacturer’s instructions.
Thioflavin-S (Thio-S) staining and quantification
Fixed hemi-brains were fully sectioned in the horizontal plane at 40 μm using a vibratome (Leica Biosystems). Every eighth section was stained for Thio-S (#230456, Sigma-Aldrich) using standard methodology. Sections were mounted and allowed to dry overnight, after which they were washed three times in 50% ethanol for 5 min each, then washed in double-distilled H2O before being incubated for 10 min in 1% Thio-S dissolved in H2O. Stained slides were then rinsed in 70% ethanol before being dehydrated and coverslipped in aqueous anti-fade mounting medium (Vector Laboratories). Digital images were captured at 20× magnification using an Olympus BX50 microscope equipped with a DP74 camera and CellSens software (Olympus). The number of spherical thioflavin-positive deposits were counted using NIH ImageJ 1.50i (United States National Institutes of Health) with the cell counter plugin to mark stained plaque-like structures. Thioflavin-positive deposits were counted in entorhinal cortex (three fields/section), subiculum (two fields/section), and hippocampal subfields CA1 (three fields/section) and CA2/3 (three fields/section), across four sections per animal, for a total of ∼44 fields per brain.
Immunohistochemistry
Immunohistochemistry was performed using a standard avidin/biotin peroxidase approach with ABC Vector Elite kits (Vector Laboratories). Aβ immunohistochemistry was performed on every eighth section using sections immediately adjacent to those processed for Thio-S. Briefly, sections were pretreated with 95% formic acid for 5 min, then rinsed in TBS before being treated with an endogenous peroxidase blocking solution for 10 min. After three 10 min washes in 0.1% Triton-X/TBS, sections were incubated for 30 min in a blocking solution consisting of 2% bovine serum albumin in TBS. Blocked sections were incubated overnight at 4°C in primary antibody directed against Aβ (#71-5800, 1:300 dilution, Invitrogen) that was diluted in blocking solution. Next, sections were rinsed and incubated in biotinylated secondary antibody diluted in blocking solution. Immunoreactivity was visualized using 3,3’-diaminobenzidine (Vector Laboratories). Additional sections were similarly immunostained without formic acid pretreatment using IBA-1 (#019-19741, 1:2000 dilution, Wako) and GFAP (#ab7260, 1:1000 dilution, Abcam).
To quantify the percentage area occupied by Aβ immunoreactivity (Aβ load), images of nonoverlapping fields were taken at 20× magnification in entorhinal cortex (three fields/section), subiculum (three fields/section), and hippocampal subfields CA1 (five fields/section) and CA2/3 (three fields/section) across 4 tissue sections, for a total of ∼56 images per brain. Images were digitally captured using an Olympus BX50 microscope and DP74 camera paired with a computer running CellSens software (Olympus). The pictures were converted to grayscale images and thresholded using NIH ImageJ 1.50i to yield binary images separating positive and negative immunostaining. Aβ load was calculated as the percentage of the total area that was positively immunolabeled.
Microglia and astrocyte activation was quantified using live imaging (Olympus BX50, CASTGrid software, Olympus) at 40× magnification. Each cell was categorized as either resting or reactive based on its morphology, as reported in previous studies (Ayoub and Salm, 2003; Wilhelmsson et al., 2006). Specifically, microglia were scored as resting (type 1) if they had spherical cell bodies, with numerous thin, highly ramified processes. Cells were scored as type 2 cells if they exhibited enlarged rod-shaped cell bodies with fewer processes that were shorter and thicker, and scored type 3 cells if they had very few or no processes or several filopodial processes. Both type 2 and type 3 morphologies were considered an activated microglia phenotype. Astrocytes were visualized with GFAP immunostaining and categorized as exhibiting either nonreactive (normally sized cell bodies with a few rather short projections) or reactive (both cell bodies and projections are enlarged) morphology phenotypes. Entorhinal cortex (four fields/section), subiculum (four fields/section), and hippocampal subfields CA-1 (five fields/section) and CA-2/3 (three fields/section) were quantified for both microglia and astrocytes. The number of cells across brain regions scored for each animal averaged ∼700 microglia and ∼600 astrocytes.
RNA isolation and real-time PCR
For RNA extractions, gonadal fat pads and hippocampi were homogenized using TRIzol reagent (Invitrogen), following the manufacturer’s protocol. The RNA pellet was treated with RNase-free DNase I (Epicentre) for 30 min at 37°C, and a phenol/chloroform extraction was performed to isolate RNA. The iScript cDNA synthesis system (Bio-Rad) was used to reverse transcribe cDNA from 1 μg of purified RNA. Real-time quantitative PCR was performed on the resulting cDNA using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and a Bio-Rad CFX Connect Thermocycler. All measurements were performed in duplicates. Quantification of PCR products was conducted by normalizing with a combination of corresponding hypoxanthine-guanine phosphoribosyltransferase (HPRT) and succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial (SDHA) expression levels from the gonadal fat samples, and with β-actin expression levels from hippocampus, using the ΔΔ-CT method to obtain relative mRNA levels. Gonadal fat was probed for levels of cluster of differentiation factor 68 (CD68) and EGF-like module-containing mucin-like hormone receptor-like 1 (F4/80), while hippocampus was probed for β-secretase 1 (BACE1), neprilysin, insulin-degrading enzyme (IDE), CD68, glial fibrillary acidic protein (GFAP), and cluster of differentiation factor 74 (CD74). Primer pair sequences are shown in Table 1.
Table 1.
Gene targets for the PCR analyses are listed with their corresponding oligonucleotide sequences for the forward and reverse primers
| Target gene | Sequence |
|---|---|
| CD68 | Forward: 5’-TTCTGCTGTGGAAATGCAAG-3’ Reverse: 5’-AGAGGGGCTGGTAGGTTGAT-3’ |
| F4/80 | Forward: 5’-TGCATCTAGCAATGGACAGC-3’ Reverse: 5’-GCCTTCTGGATCCATTTGAA-3’ |
| HPRT | Forward: 5’-AAGCTTGCTGGTGAAAAGGA-3’ Reverse: 5’-TTGCGCTCATCTTAGGCTTT-3’ |
| SDHA | Forward: 5’-ACACAGACCTGGTGGAGACC-3’ Reverse: 5’-GGATGGGCTTGGAGTAATCA-3’ |
| Neprilysin | Forward: 5’-GAGAAAAGCCCACTTGCTTG-3’ Reverse: 5’-GAAAGACAAAATGGGGCAGA-3’ |
| BACE1 | Forward: 5’-TCGCTGTCTCACAGTCATCC-3’ Reverse: 5’-AACAAACGGACCTTCCACTG-3’ |
| IDE | Forward: 5’-TGTTTCCACACACAGGCAAT-3’ Reverse: 5’-ACCTGTGAAAAGCCGAGAGA-3’ |
| CD74 | Forward: 5’-CAAGTACGGCAACATGACCC-3’ Reverse: 5’-GCACTTGGTCAGTACTTTAGGTG-3’ |
| GFAP | Forward: 5’-AACGACTATCGCCGCCAACTG-3’ Reverse: 5’-CTCTTCCTGTTCGCGCATTTG-3’ |
| β-Actin | Forward: 5’-AGCCATGTACGTAGCCATCC-3’ Reverse: 5’-CTCTCAGCTGTGGTGGTGAA-3’ |
Statistical analyses
For the analysis of body weight and glucose tolerance data, two-way repeated measures ANOVAs were run using the Statistical Package for Social Sciences (SPSS; version 23, IBM). All other data were analyzed by two-way ANOVA using Prism (version 5, GraphPad Software). In the case of significant main effects, planned comparisons between groups of interest were made using the Bonferroni correction. All data are presented as the mean ± SEM. Significance was set at a threshold of p < 0.05. Statistical results are presented in Tables 2, 3.
Table 2.
Statistical table
| Figure | Kolmogorov-Smirnov test for normality (p value) | Statistical significance |
|---|---|---|
|
Figure 1A
body weight |
All groups at all time points are normally distributed (p > 0.05). | Genotype: F(1,29) = 0.10, p = 0.759 diet: F(1,29) = 10.51, p = 0.003 interaction: F(1,29) = 2.68, p = 0.112 |
|
Figure 1B
plasma cholesterol |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,29) = 2.86, p = 0.103 diet: F(1,29) = 1.58, p = 0.221 interaction: F(1,29) = 2.60, p = 0.119 |
|
Figure 1C
plasma triglycerides |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,29) = 0.56, p = 0.46 diet: F(1,29) = 2.87, p = 0.102 interaction: F(1,29) = 1.91, p = 0.179 |
|
Figure 1D
gonadal fat weight |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,29) = 0.18, p = 0.673 diet: F(1,29) = 37.04, p < 0.001 interaction: F(1,29) = 5.01, p = 0.033 |
|
Figure 1E
CD68 |
E3FAD CTL N/A E3FAD WD > 0.10 E4FAD CTL = 0.004 E4FAD WD > 0.10 |
Genotype: F(1,21) = 0.90, p = 0.353 diet: F(1,21) = 11.54, p = 0.003 interaction: F(1,21) = 0.85, p = 0.366 |
|
Figure 1F
F4/80 |
E3FAD CTL N/A E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,21) = .09, p = .768 diet: F(1,21) = 7.02, p = 0.015 interaction: F(1,21) = 1.19, p = 0.288 |
|
Figure 1G
glucose (GTT) |
All groups at all time points are normally distributed (p > 0.05), except: E4FAD CTL 0 min = 0.002 E4FADWD 15 min = 0.025 E3FAD WD 30 min = 0.011 E4FAD WD 30 min = 0.008 | Genotype: F(1,29) = 0.02, p = 0.886 diet: F(1,29) = 5.03, p = 0.033 interaction: F(1,29) = 0.10, p = 0.750 |
|
Figure 1H
GTT AUC |
E3FAD CTL = 0.07 E3FAD WD = 0.097 E4FAD CTL > 0.10 E4FAD WD = 0.033 |
Genotype: F(1,29) = .06, p = 0.817 diet: F(1,29) = 5.73, p = 0.023 interaction: F(1,29) = 0.12, p = 0.737 |
|
Figure 1I
percent glucose change |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,29) = .83, p = 0.371 diet: F(1,29) = 3.84, p = 0.059 interaction: F(1,29) = 0.90, p = 0.352 |
|
Figure 2B
Thio-S: entorhinal cortex |
E3FAD CTL > 0.10 E3FAD WD = 0.049 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,29) = 50.30, p < 0.001 diet: F(1,29) = 6.62, p = 0.016 interaction: F(1,29) = 4.09, p = 0.053 |
|
Figure 2C
Thio-S: subiculum |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,29) = 59.40, p < 0.001 diet: F(1,29) = 2.98, p = 0.095 interaction: F(1,29) = 9.75, p = 0.004 |
|
Figure 2D
Thio-S: CA1 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,29) = 80.58, p < 0.001 diet: F(1,29) = 4.95, p = 0.034 interaction: F(1,29) = 8.41, p = 0.007 |
|
Figure 2E
Thio-S: CA2/3 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 46.39, p < 0.001 diet: F(1,29) = 7.41, p = 0.011 interaction: F(1,29) = 7.32, p = 0.011 |
|
Figure 3B
Aβ load: entorhinal cortex |
E3FAD CTL > 0.10 E3FAD WD = 0.002 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 21.38, p < 0.001 diet: F(1,29) = 7.83, p = 0.009 interaction: F(1,29) = 4.91, p = 0.035 |
|
Figure 3C
Aβ load: subiculum |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 25.40, p < 0.001 diet: F(1,29) = 11.19, p = 0.002 interaction: F(1,29) = 0.11, p = 0.742 |
|
Figure 3D
Aβ load: CA1 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD = 0.036 |
Genotype F(1,29) = 37.66, p < 0.001 diet: F(1,29) = 2.91, p = 0.099 interaction: F(1,29) = 2.71, p = 0.110 |
|
Figure 3E Aβ load: CA2/3 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 47.27, p < 0.001 diet: F(1,29) = 10.36, p = 0.003 interaction: F(1,29) = 4.48, p = 0.043 |
|
Figure 4B
microglia number: entorhinal cortex |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,27) = 9.78, p = 0.004 diet: F(1,27) = 2.31, p = 0.141 interaction: F(1,27) = 1.05, p = 0.316 |
|
Figure 4C
microglia number: subiculum |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,27) = 42.77, p < 0.001 diet: F(1,27) = 4.20, p = 0.050 interaction: F(1,27) = 4.75, p = 0.038 |
|
Figure 4D
microglia number: CA1 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,27) = 51.42, p < 0.001 diet: F(1,27) = 10.78, p = 0.003 interaction: F(1,27) = 7.97, p = 0.009 |
|
Figure 4E
microglia number: CA2/3 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,27) = 21.64, p < 0.001 diet: F(1,27) = 1.97, p = 0.172 interaction: F(1,27) = 1.90, p = 0.180 |
|
Figure 4F
microglia reactivity: entorhinal cortex |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,27) = 109.10, p < 0.001 diet: F(1,27) = 1.64, p = 0.212 interaction: F(1,27) = 5.52, p = 0.027 |
|
Figure 4G
microglial reactivity: subiculum |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL = 0.07 E4FAD WD < 0.001 |
Genotype F(1,27) = 19.70, p < 0.001 diet: F(1,27) = 0.00, p = 0.995 interaction: F(1,27) = 0.51, p = 0.480 |
|
Figure 4H
microglial reactivity: CA1 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD = 0.04 |
Genotype F(1,27) = 78.70, p < 0.001 diet: F(1,27) = 5.00, p = 0.034 interaction: F(1,27) = 11.58, p = 0.002 |
|
Figure 4I
microglial reactivity: CA2/3 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,27) = 165.70, p < 0.001 diet: F(1,27) = 21.04, p < 0.001 interaction: F(1,27) = 32.66, p < 0.001 |
|
Figure 5B
astrocyte number: entorhinal cortex |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 3.82, p = 0.060 diet: F(1,29) = 0.29, p = 0.593 interaction: F(1,29) = 0.41, p = 0.528 |
|
Figure 5C
astrocyte number: subiculum |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 9.95, p = 0.004 diet: F(1,29) = 4.79, p = 0.037 interaction: F(1,29) = 1.04, p = 0.316 |
|
Figure 5D
astrocyte number: CA1 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 5.88, p = 0.022 diet: F(1,29) = 3.55, p = 0.069 interaction: F(1,29) = 0.49, p = 0.489 |
|
Figure 5E
astrocyte number: CA2/3 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 1.82, p = 0.188 diet: F(1,29) = 4.26, p = 0.048 interaction: F(1,29) = 0.02, p = 0.894 |
|
Figure 5F
astrocyte reactivity: entorhinal cortex |
E3FAD CTL > 0.10 E3FAD WD = 0.004 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 46.97, p < 0.001 diet: F(1,29) = 5.75, p = 0.023 interaction: F(1,29) = 4.82, p = 0.036 |
|
Figure 5G
astrocyte reactivity: subiculum |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL = 0.045 E4FAD WD > 0.10 |
Genotype F(1,29) = 27.72, p < 0.001 diet: F(1,29) = 3.13, p = 0.088 interaction: F(1,29) = 0.00, p = 0.989 |
|
Figure 5H
astrocyte reactivity: CA1 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 87.49, p < 0.001 diet: F(1,29) = 23.82, p < 0.001 interaction: F(1,29) = 2.08, p = 0.160 |
|
Figure 5I
astrocyte reactivity: CA2/3 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype F(1,29) = 11.68, p = 0.002 diet: F(1,29) = 7.83, p = 0.009 interaction: F(1,29) = 2.405, p = 0.132 |
Table 3.
Relative gene expression in hippocampus
| Gene | Mean ± SEM | Kolmogorov-Smirnov test for normality (p value) | Statistical significance |
|---|---|---|---|
| BACE1 | E3FAD CTL = 1 ± N/A E3FAD WD = 1.53 ± 0.31 E4FAD CTL = 1.32 ± 0.19 E4FAD WD = 1.76 ± 0.41 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,28) = 1.10, p = 0.304 diet: F(1,28) = 3.44, p = 0.074 interaction: F(1,28) = 0.03, p = 0.874 |
| Neprilysin | E3FAD CTL = 1 ± N/A E3FAD WD = 1.61 ± 0.79 E4FAD CTL = 0.94 ± 0.30 E4FAD WD = 1.79 ± 0.63 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,28) = 0.02, p = 0.902 diet: F(1,28) = 2.49, p = 0.126 interaction: F(1,28) = 0.06, p = 0.802 |
| IDE | E3FAD CTL = 1 ± N/A E3FAD WD = 1.27 ± 0.39 E4FAD CTL = 1.30 ± 0.39 E4FAD WD = 1.12 ± 0.35 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL = 0.01 E4FAD WD > 0.10 |
Genotype: F(1,28) = 0.08, p = 0.785 diet: F(1,28) = 0.00, p = 0.955 interaction: F(1,28) = 0.49, p = 0.489 |
| CD68 | E3FAD CTL = 1 ± N/A E3FAD WD = 1.21 ± 0.29 E4FAD CTL = 1.74 ± 0.30 E4FAD WD = 2.30 ± 0.29 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,28) = 10.75, p = 0.003 diet: F(1,28) = 1.91, p = 0.178 interaction: F(1,28) = 0.40, p = 0.532 |
| GFAP | E3FAD CTL = 1 ± N/A E3FAD WD = 1.02 ± 0.11 E4FAD CTL = 1.56 ± 0.21 E4FAD WD = 2.70 ± 0.04 |
E3FAD CTL > 0.10 E3FAD WD > 0.10 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,28) = 14.26, p < 0.001 diet: F(1,28) = 0.23, p = 0.634 interaction: F(1,28) = 0.14, p = 0.712 |
| CD74 | E3FAD CTL = 1 ± N/A E3FAD WD = 1.28 ± 0.28 E4FAD CTL = 3.32 ± 0.62 E4FAD WD = 5.04 ± 1.30 |
E3FAD CTL > 0.10 E3FAD WD = 0.01 E4FAD CTL > 0.10 E4FAD WD > 0.10 |
Genotype: F(1,28) = 16.98, p < 0.001 diet: F(1,28) = 1.86, p = 0.184 interaction: F(1,28) = 0.96, p = 0.335 |
Data are presented as mean fold differences (±SEM) relative to E3FAD mice on a control diet. The Kolmogorov-Smirnov test for normality was performed, with p > 0.05 indicating a normal distribution. Genes related to β-amyloid production (BACE-1) and clearance (neprilysin, IDE) showed no significant changes with either diet or genotype, while genes related to glial activation (CD68, GFAP, and CD74) were increased in E4FAD mice on both control and Western diets.
Results
Obesity-related outcomes of Western diet
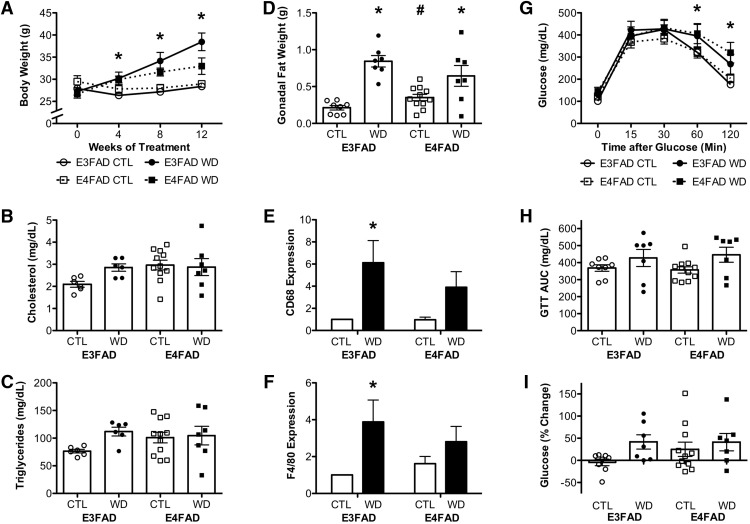
To begin investigating whether there are gene X environment interactions between APOE and Western diet, we first compared measures of DIO in E3FAD versus E4FAD mice following the 12-week exposure to control and Western diets. The control diet was associated with <1% gain in body weight in both E3FAD and E4FAD mice, whereas Western diet yielded a 39 ± 7.7% increase in body weight in E3FAD and a 24 ± 7.21% increase in E4FAD mice (Fig. 1A), such that the effects of diet did not vary significantly across genotypes (p = 0.112; Fig. 1A; Table 2). A 2 × 2 repeated measures ANOVA revealed a significant main effect of diet on body weight (F = 10.51, p = 0.003; Fig. 1A) in which Western diet was associated with increased weight. APOE genotype did not significantly affect body weight (p = 0.759; Fig. 1A). Between group comparisons revealed that E3FAD mice fed a Western diet weighed significantly more than E3FAD mice fed a control diet at 4, 8, and 12 weeks (p < 0.05). There were no statistically significant differences in body weights at any time point between control and Western diet groups in E4FAD mice.
Figure 1.
Metabolic outcomes associated with DIO in E3FAD and E4FAD mice. A, Body weights in male E3FAD and E4FAD mice maintained on control (CTL) and Western (WD) diets taken at baseline (week 0) and four-week intervals across the 12-week experimental period. Plasma levels of cholesterol (B) and triglyceride levels (C) in E3FAD and E4FAD mice on control and Western diets at the end of the experimental period. D, Weight of the gonadal fat pads across groups. Relative mRNA expression of macrophage markers (E) CD68 and (F) F4/80 in gonadal fat, as determined by real time PCR. Data show fold differences relative to the E3FAD + control diet group. G, GTT showing blood glucose levels over time after a glucose bolus. H, AUC for the GTT. I, Percentage change in fasting blood glucose levels relative to baseline after 12-weeks of control or Western diet. Data are presented as mean (±SEM) values; n = 7–11/group. E3FAD mice are shown as circles, E4FAD mice are shown as squares; control diet groups are indicated as open symbols or bars, whereas Western diet groups are filled symbols or bars. *, p < 0.05 relative to genotype-matched mice in control diet condition. #, p < 0.05 relative to E3FAD mice in same diet condition.
We next examined plasma levels of cholesterol and triglycerides as measures of adverse effects of Western diet. We found that plasma cholesterol levels were significantly affected by neither genotype (p = 0.103) nor diet (p = 0.221), and we did not find an interaction effect (p = 0.119; Fig. 1B; Table 2). Likewise, there were no effects of either genotype (p = 0.46) or diet (p = 0.102), or an interaction effect (p = 0.179) on plasma triglyceride levels (Fig. 1C).
Because metabolic impairments associated with obesity have been linked to adiposity, we assessed fat deposition across groups. We observed a significant interaction effect (F = 5.01, p = 0.033; Table 2), such that on the control diets, E4FAD mice had more gonadal fat than E3FADs (p = 0.027), but there was no difference between E3FAD and E4FAD mice on Western diet (p = 0.230; Fig. 1D). Additionally, there was a significant main effect of diet (F = 37.04, p < 0.001) on weight of the gonadal fat pads, so that both E3FAD and E4FAD mice had increased fat pads with Western diet (Fig. 1D). Parallel findings were observed in the retroperitoneal fat pads (data not shown). Because inflammation is an established hallmark of obesity, we examined gene expression of the macrophage markers CD68 and F4/80 by PCR in the adipose tissue. We found a significant main effect of diet on CD68 expression (F = 11.54, p = 0.003), although this effect reached statistical significance only in E3FAD but not in E4FAD mice (Fig. 1E). There was no statistically significant effect of genotype (p = 0.353), nor was there an interaction between diet and genotype (p = 0.366) on CD68 expression. Diet had a main effect on adipose F4/80 expression (F = 7.02, p = 0.015), and again, this effect reached statistical significance only in E3FAD mice (Fig. 1F). There was no statistically significant effect of genotype (p = .768), and no interaction effect (p = 0.288) on F4/80 expression (Table 2).
In addition to increasing body weight and adiposity, Western diet can induce metabolic impairments including dysregulation of glucose homeostasis. When examining glucose clearance in the GTT, we found a significant main effect of diet (F = 5.03, p = 0.033), such that both E3FAD and E4FAD mice fed a Western diet were impaired at clearing glucose (Fig. 1G; Table 2). There was no main effect of genotype (p = 0.886), or interaction effect between diet and genotype (p = 0.750) on glucose clearance. We also calculated the area under the curve (AUC) for GTT, and found that there was a significant main effect of diet (F = 5.73, p = 0.023), but not of genotype (p = 0.817) on GTT AUC (Fig. 1H). However, the effect of diet failed to reach statistical significance when examined separately in E3FAD and E4FAD mice. There was no interaction between genotype and diet on GTT AUC (p = 0.737). Changes in fasting glucose levels over the diet treatment period showed a trend toward a main effect of diet (F = 3.84, p = 0.059; Fig. 1I). There was no effect of genotype (p = 0.371) nor was there an interaction between diet and genotype (p = 0.352) on changes in glucose levels (Table 2).
Western diet increases β-amyloid deposition in E4FAD but not in E3FAD mice
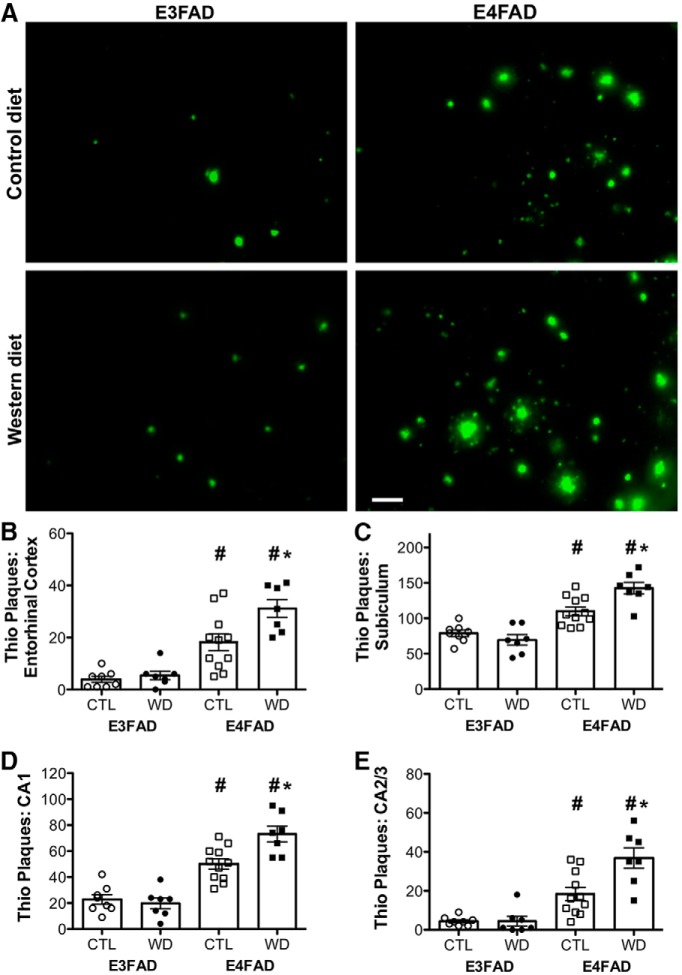
The primary AD-related neuropathological change in EFAD mice at this age is accumulation of β-amyloid protein, largely in the form of extracellular deposits, many of which exhibit positive Thio-S staining that is indicative of amyloid. Thus, to begin assessing AD-related neuropathology, Thio-S positive plaques were counted in entorhinal cortex and in subregions of the hippocampus. Visual inspection of stained sections qualitatively showed not only the expected increase in amyloid deposits in E4FAD mice, but also the surprising finding that Western diet increased Thio-S positive plaques only in E4FAD mice (Fig. 2A). Specifically, there were significant interaction effects between genotype and diet on Thio-S positive plaques in subiculum (F = 9.75, p = 0.004; Fig. 2C), CA1 (F = 8.41, p = 0.007; Fig. 2D), and CA2/3 (F = 7.32, p = 0.011; Fig. 2E), and a nonsignificant trend toward an interaction in entorhinal cortex (F = 4.09, p = 0.053; Fig. 2B; Table 2). Further analyses revealed that diet significantly increased Thio-S positive plaque counts in E4FAD but not E3FAD males across all brain regions sampled (p < 0.01). Additionally, there was a significant main effect of genotype even in the absence of diet, such that E4FAD mice had a greater number of Thio-S positive plaques in entorhinal cortex (F = 50.30, p < 0.001; Fig. 2B), subiculum (F = 59.40, p < 0.001; Fig. 2C), CA1 (F = 80.58, p < 0.001; Fig. 2D), and CA2/3 (F = 46.39, p < 0.001; Fig. 2E), than did E3FAD mice.
Figure 2.
Accumulation of amyloidogenic deposits assessed by Thio-S staining in E3FAD and E4FAD mice across dietary treatments. A, Representative images of Thio-S staining in the subiculum of E3FAD and E4FAD males fed control and Western diets. Scale bar, 50 µm. Numbers of Thio-S positive plaque numbers in E3FAD and E4FAD mice maintained on control and Western diets were quantified in (B) entorhinal cortex, and hippocampal subregions (C) subiculum, (D) CA1, and (E) CA2/3. Data are presented as mean (±SEM) values; n = 7–11/group. E3FAD mice are shown as circles, E4FAD mice are shown as squares; control diet groups are indicated as open symbols, and Western diet groups as filled symbols. *, p < 0.05 relative to genotype-matched mice in control diet condition. #, p < 0.05 relative to E3FAD mice in same diet condition.
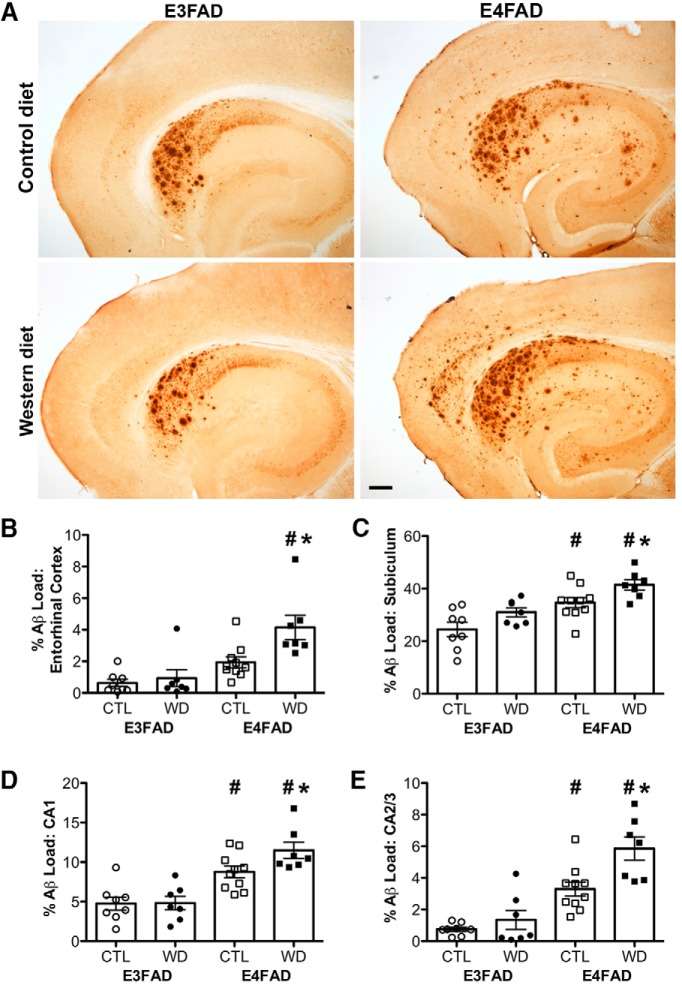
As a second measure of AD-like pathology, we assessed total β-amyloid burden by immunohistochemistry. This provides a measure of complete β-amyloid, as the antibody recognizes intra- and extracellular accumulations of Aβ, even those that have not progressed to Thio-S positive amyloid deposits. Results repeated the same general pattern observed with Thio-S staining. That is, (1) E4FAD mice exhibit greater β-amyloid burden, and (2) E4FAD but not E3FAD mice show increased β-amyloid accumulation with Western diet (Fig. 3A). We found significant interaction effects between genotype and diet in entorhinal cortex (F = 4.91, p = 0.035; Fig. 3B) and in CA2/3 (F = 4.48, p = 0.043; Fig. 3E), but not in subiculum (F = 0.11, p = 0.742; Fig. 3C) or in CA1 (F = 2.71, p = 0.110; Fig. 3D; Table 2). Bonferroni post hoc tests showed that Western diet significantly increased Aβ load in E4FAD but not in E3FAD mice across all brain regions surveyed (p < 0.05). There was a significant main effect of genotype with E4FAD mice having greater Aβ load than E3FAD mice in entorhinal cortex (F = 21.38, p < 0.001; Fig. 3B), subiculum (F = 25.40, p < 0.001; Fig. 3C), CA1 (F = 37.66, p < 0.001; Fig. 3D), and CA2/3 (F = 47.27, p < 0.001; Fig. 3E).
Figure 3.
Accumulation of β-amyloid deposits assessed by immunohistochemistry in E3FAD and E4FAD mice across dietary treatments. A, Representative images of β-amyloid immunoreactivity in entorhinal cortex and hippocampus in E3FAD and E4FAD males maintained on control and Western diets. Scale bar, 100 µm. β-Amyloid burden was quantified as immunoreactivity load in E3FAD and E4FAD mice in control and Western diets groups in (B) entorhinal cortex, and hippocampal subregions (C) subiculum, (D) CA1, and (E) CA2/3. Data are presented as mean (±SEM) values; n = 7–11/group. E3FAD mice are shown as circles, E4FAD mice are shown as squares; control diet groups are indicated as open symbols, and Western diet groups as filled symbols. *, p < 0.05 relative to genotype-matched mice in control diet condition. #, p < 0.05 relative to E3FAD mice in same diet condition.
Western diet increases gliosis more strongly in E4FAD than in E3FAD mice
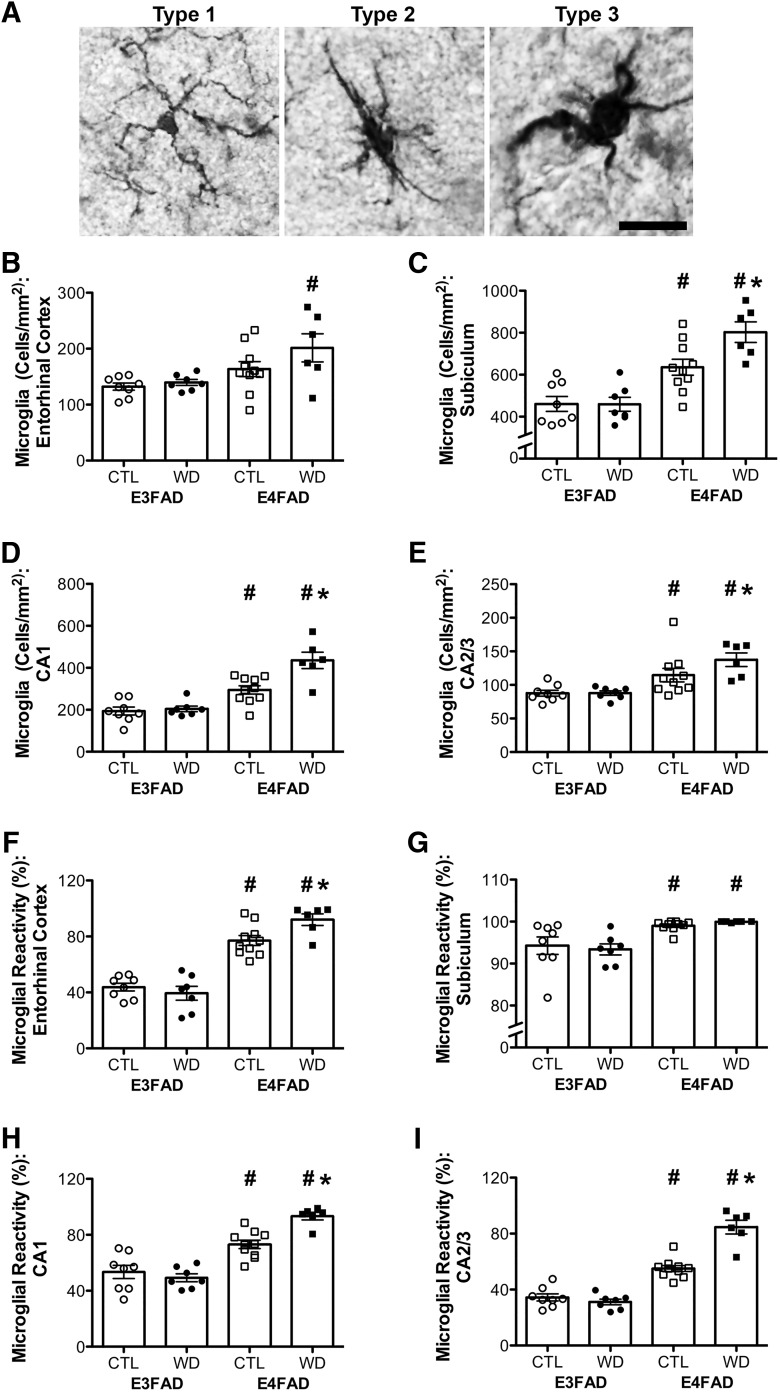
Gliosis is an important neuropathological feature of AD that is also associated with both obesity and APOE4. To assess gliosis, we compared both the relative cell numbers and morphologic activation state of microglia and astrocytes across groups. We found that, in comparison to E3FAD mice, E4FAD mice consistently had a higher total number of glial cells as well as a higher percentage of glial cells with reactive versus resting phenotypes. Moreover, the effects of diet on glial number and reactivity were stronger in E4FAD than in E3FAD mice.
We first examined microglia number and morphology by IBA-1 staining. Figure 4A shows a resting microglial cell with thin, ramified processes (type 1), and activated cells with rod-shaped cell bodies and fewer, thicker processes (type 2), and amoeboid cells (type 3). We found significant interactions between genotype and diet when examining the total number of microglia per mm2 in subiculum (F = 4.75, p = 0.038; Fig. 4C) and in CA1 (F = 7.97, p = 0.009; Fig. 4D), with Bonferroni post hoc tests showing that Western diet increased microglia number in E4FAD but not in E3FAD mice in these brain regions (p < 0.05; Table 2). There were no interaction effects on microglia number in entorhinal cortex (p = 0.316; Fig. 4B), or in CA2/3 (p = 0.180; Fig. 4E). There was a significant effect of genotype on the total number of microglia per mm2 in entorhinal cortex (F = 9.78, p = 0.004; Fig. 4B), subiculum (F = 42.77, p < 0.001; Fig. 4C), CA1 (F = 51.42, p < 0.001; Fig. 4D), and CA2/3 (F = 21.64, p < 0.001; Fig 4E), such that E4FAD mice had a greater total number of microglia across these brain regions than did E3FAD mice. However, in entorhinal cortex, the effect of genotype was significant only in animals on a Western diet.
Figure 4.
Microglia number and morphologic status assessed by IBA-1 immunohistochemistry in E3FAD and E4FAD mice across dietary treatments. A, Representative images of microglial morphology associated with resting (type 1) and reactive (types 2 and 3) phenotypes. Scale bar, 40 µm. B–E, Densities (cells/mm2) of IBA-1-immunoreactive cells in E3FAD and E4FAD mice on control and Western diets were quantified in (B) entorhinal cortex, and hippocampal subregions (C) subiculum, (D) CA1, and E) CA2/3. F–I) Percentages of all IBA-1-immunoreactive cells scored as having reactive phenotype (types 2 and 3) were quantified in (F) entorhinal cortex, and hippocampal subregions (G) subiculum, (H) CA1, and (I) CA2/3. Data are presented as mean (±SEM) values; n = 7–11/group. E3FAD mice are shown as circles, E4FAD mice are shown as squares; control diet groups are indicated as open symbols, and Western diet groups as filled symbols. *, p < 0.05 relative to genotype-matched mice in control diet condition. #, p < 0.05 relative to E3FAD mice in same diet condition.
Measures of microglial reactivity showed similar results as microglial number. Significant interaction effects between genotype and diet were observed in entorhinal cortex (F = 5.52, p = 0.027; Fig. 4F), CA1 (F = 11.58, p = 0.002; Fig. 4H), and CA2/3 (F = 32.66, p < 0.001; Fig. 4I), but not in subiculum (p = 0.480; Fig. 4G; Table 2). Bonferroni post hoc tests revealed that Western diet increased the percentage of reactive microglia in entorhinal cortex, CA1, and CA2/3 of E4FAD, but not E3FAD, male mice. There was a significant main effect of genotype even in the absence of diet, such that E4FAD mice had a greater percentage of reactive microglia than E3FAD mice in entorhinal cortex (F = 109.10, p < 0.001; Fig. 4F), subiculum (F = 19.70, p < 0.001; Fig. 4G), CA1 (F = 78.70, p < 0.001; Fig. 4H), and CA2/3 (F = 165.70, p < 0.001; Fig. 4I).
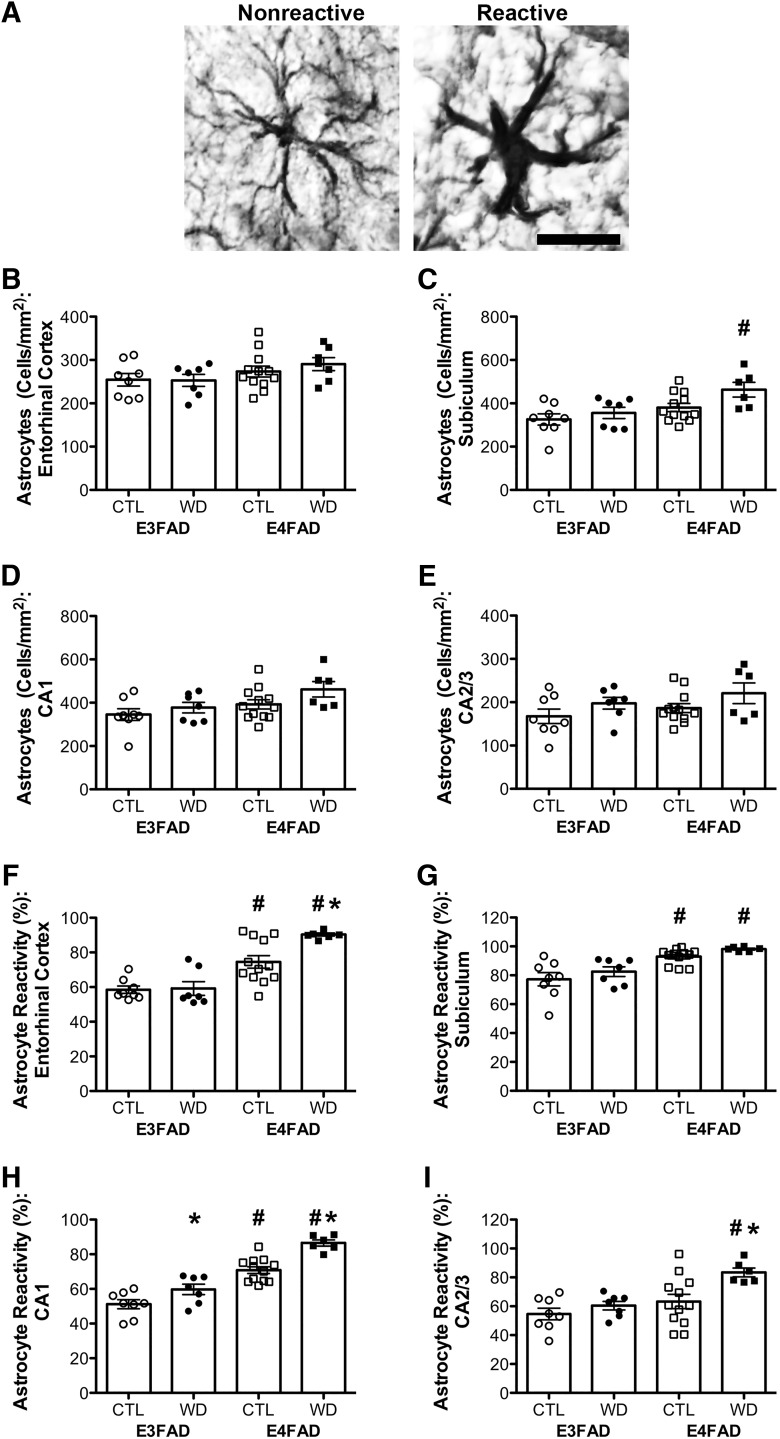
We next examined astrocyte number and activation by GFAP staining. Figure 5A shows examples of a nonreactive astrocyte with a normally sized soma versus a reactive phenotype with enlarged soma and projections. For the measure of astrocyte number, the effects of diet did not differ across genotype for any of the brain regions sampled (Table 2). We found significant main effects of genotype on the total number of astrocytes in subiculum (F = 9.95, p = 0.004; Fig. 5C), although this effect was only statistically significant in animals on a Western diet. There was a main effect of genotype on astrocyte number in CA1 (F = 5.88, p = 0.022; Fig. 5D), but this did not reach statistical significance when examined separately in control and Western diet-fed animals. There was a trend toward a significant effect of genotype in entorhinal cortex (F = 3.82, p = 0.060; Fig. 5B), but no effect in CA2/3 (p = 0.188; Fig. 5E). Diet had significant main effects on astrocyte number in subiculum (F = 4.79, p = 0.037; Fig. 5C), and CA2/3 (F = 4.26, p = 0.048; Fig. 5E), with a trend toward a main effect in CA1 (F = 3.55, p = 0.069; Fig. 5D), although this effect did not reach statistical significance when examined separately in E3FAD and E4FAD mice in any brain region. There was no effect of diet on astrocyte number in entorhinal cortex (p = 0.593; Fig. 5B).
Figure 5.
Astrocyte number and morphologic status assessed by GFAP immunohistochemistry in E3FAD and E4FAD mice across dietary treatments. A, Representative images of astrocyte morphology associated with resting and reactive phenotypes. Scale bar, 50 µm. B–E, Densities (cells/mm2) of GFAP-immunoreactive cells in E3FAD and E4FAD mice on control and Western diets were quantified in (B) entorhinal cortex, and hippocampal subregions (C) subiculum, (D) CA1, and (E) CA2/3. F–I, Percentages of all GFAP-immunoreactive cells scored as having reactive phenotype (type 2) were quantified in (F) entorhinal cortex, and hippocampal subregions (G) subiculum, (H) CA1, and (I) CA2/3. Data are presented as mean (±SEM) values; n = 7–11/group. E3FAD mice are shown as circles, E4FAD mice are shown as squares; control diet groups are indicated as open symbols, and Western diet groups as filled symbols. *, p < 0.05 relative to genotype-matched mice in control diet condition. #, p < 0.05 relative to E3FAD mice in same diet condition.
When examining astrocyte reactivity, we found similar trends as with microglial reactivity. That is, there was a significant interaction effect between genotype and diet on astrocyte reactivity in entorhinal cortex (F = 4.82, p = 0.036; Fig. 5F), with Western diet increasing reactivity only in E4FAD mice (Table 2). There were no significant interaction effects between genotype and diet in subiculum (p = 0.989; Fig. 5G), CA1 (p = 0.160; Fig. 5H), or CA2/3 (p = 0.132; Fig. 5I). Moreover, in the absence of diet, genotype had a significant effect on astrocyte reactivity, with E4FAD mice having a greater percentage of reactive astrocytes in entorhinal cortex (F = 46.97, p < 0.001; Fig. 5F), subiculum (F = 27.72, p < 0.001; Fig. 5G), CA1 (F = 87.49, p < 0.001; Fig. 5H), and CA2/3 (F = 11.68, p = 0.002; Fig. 5I). In CA2/3 the effect of genotype was only significant in Western diet-fed animals. Furthermore, Western diet significantly increased astrocyte reactivity in CA1 (F = 23.82, p < 0.001; Fig. 5H), and CA2/3 (F = 7.83, p = 0.009; Fig. 5I), although this effect was only significant in E4FAD mice in CA2/3. There was a nonsignificant trend toward an effect of diet in subiculum (F = 3.13, p = 0.088; Fig. 5G).
E4FAD mice have increased gene expression of inflammatory markers
To begin addressing possible mechanisms underlying the interactive effects of APOE4 and Western diet, we examined hippocampal gene expression of several markers related to Aβ production and clearance, as well as inflammation. Overall, our results indicate that gene expression of factors involved in Aβ clearance and production are not significantly altered by genotype or diet, and that inflammatory gene expression is increased in E4FAD mice, without being altered by Western diet (Table 3).
For BACE1, relative mRNA levels did not show evidence of an interaction between the diet and APOE genotypes (p = 0.874). There was no significant main effect genotype (p = 0.304), but there was a nonsignificant trend of increased BACE1 levels with Western diet (p = 0.074). Expression of the Aβ clearance factor neprilysin was not significantly affected by genotype (p = 0.902) or diet (p = 0.126), and there was no interaction between genotype and diet (p = 0.802). Likewise, gene expression of IDE was not altered by genotype (p = 0.785), diet (p = 0.955), or the interaction between genotype and diet (p = 0.489).
In assessing gene expression of inflammatory markers we found that E4FAD mice had significantly greater levels of the microglial markers CD68 (F = 10.75, p = 0.003), the astrocyte marker GFAP (F = 14.26, p < 0.001), and the innate immune marker CD74 (F = 16.98, p < 0.001), than did E3FAD mice. However, there were no significant effects of diet on levels of CD68 (p = 0.178), GFAP (p = 0.634), or CD74 (p = 0.184). Moreover, there were no significant interactions between genotype and diet on levels of CD68 (p = 0.532), GFAP (p = 0.712), or CD74 (p = 0.335).
Discussion
The goal of this study is to examine whether APOE genotype and obesity interact to promote AD pathogenesis. Comparing E3FAD and E4FAD mice maintained on standard versus Western diets, we demonstrate a significant gene-environment interaction whereby DIO drives AD-related pathology primarily in APOE4 mice. Our results are consistent with previous findings in humans (Fitzpatrick et al., 2009; Profenno et al., 2010), and confirm studies in rodent models (Ho et al., 2004; Julien et al., 2010; Kohjima et al., 2010; Barron et al., 2013) that obesity increases risk for development of AD. Similarly, our findings replicate prior rodent data (Fryer et al., 2005; Castellano et al., 2011; Youmans et al., 2012; Rodriguez et al., 2014; Cacciottolo et al., 2016) that model the human observation that APOE4 increases the risk and or accelerates the onset of AD pathology (Corder et al., 1993; Saunders et al., 1993; Strittmatter et al., 1993; Morris et al., 2010; Jack et al., 2015). Importantly, our data indicate that the effects of DIO and APOE4 are not strictly additive. Although APOE4 status is associated with greater AD-like pathology on both control and Western diets, obesity increased AD-like pathology in E4FAD but not E3FAD mice. Our finding that E3FAD mice did not show a diet-induced increase in AD-related pathology is similar to null findings in some rodent models of obesity (Zhang et al., 2013; Knight et al., 2014; Niedowicz et al., 2014), suggesting that deleterious effects of obesity can be regulated by genetic factors besides APOE4. Thus, these data suggest an important gene X environment interaction in which APOE4 carriers are more susceptible to the AD-promoting effects of obesity.
How neural outcomes in human populations are impacted by the relationship between APOE genotype and metabolic risk factors remains incompletely defined. Many studies simply control for APOE genotype rather than considering its potential moderating role in the relationship between obesity and AD risk (Vanhanen et al., 2006; Luchsinger et al., 2012). When APOE status has been considered as a modulator of AD risk associated with metabolic factors, the results have been mixed. In some studies, APOE4 carriers showed significantly more cognitive impairment in association with adverse metabolic conditions including atherosclerosis, peripheral vascular disease, type 2 diabetes (Haan et al., 1999), and high systolic blood pressure at midlife (Peila et al., 2001). Further, levels of senile plaques and neurofibrillary tangles were highest in obese men that were also APOE4 carriers (Peila et al., 2002). However, several other studies reported that the AD risk associated with obesity and metabolic syndrome is stronger in APOE3 carriers (Dixit et al., 2005; Leiva et al., 2005; Singh et al., 2006; Profenno and Faraone, 2008).
An important consideration in interpreting these seemingly discordant findings is the potential role of sex differences. Although the impact of sex differences in the interactions among obesity, APOE, and AD risk has not been thoroughly addressed, AD is characterized by numerous sex differences (Li and Singh, 2014; Pike, 2017). Further, the AD-associated risk of APOE4 appears to disproportionately affect women (Payami et al., 1994; Farrer et al., 1997; Altmann et al., 2014). Additionally, there are sex differences in various aspects of obesity (Lovejoy et al., 2009; Mauvais-Jarvis, 2015; Moser and Pike, 2016), including observations that women exhibit relative protection against obesity until menopause (Meyer et al., 2011; Sugiyama and Agellon, 2012; Bloor and Symonds, 2014). Given that sex differences have been found in each of these factors, future studies should address sex as a possible mediator in the relationship between APOE4 and obesity. Ongoing projects in our lab have begun to address this issue using female E3FAD and E4FAD mice.
How obesity and APOE interact to regulate AD pathogenesis remains to be determined. One candidate mechanism linked to both factors is metabolic impairment. Obesity is strongly associated with development of impaired glucose and insulin metabolism (Kahn et al., 2006; Singla et al., 2010), which are also characteristic of AD patients and have been proposed as possible mechanisms driving AD pathogenesis (Craft, 2005; Martins et al., 2006; Craft, 2009). Notably, APOE genotype affects metabolic responses to diet (Snook et al., 1999; Barberger-Gateau et al., 2011), and several studies show that APOE4 carriers are at increased risk for a number of metabolic disturbances (de-Andrade et al., 2000; Oh and Barrett-Connor, 2001; Elosua et al., 2003; Marques-Vidal et al., 2003; Sima et al., 2007; Kypreos et al., 2009; Niu et al., 2009; Atabek et al., 2012; Zarkesh et al., 2012; Guan et al., 2013), although some studies find no effect of APOE genotype on metabolic outcomes (Meigs et al., 2000; Ragogna et al., 2011). Our findings suggest that E3FAD mice may be more susceptible to some metabolic effects of Western diet, although E4FAD mice trend toward metabolic disturbances even in the absence of a Western diet. Specifically, relative to E4FAD mice, E3FAD mice showed greater diet-induced body weight gain, gonadal fat inflammatory cytokine expression, and higher glucose levels on Western diet. Conversely, E4FAD mice had higher gonadal fat pad weight and a trend toward higher fasting glucose levels than E3FAD mice under the control diet condition. These findings are consistent with several previous reports showing that mice with human APOE3 gain more weight in response to a high fat diet than mice with either human APOE4 (Arbones-Mainar et al., 2008; Segev et al., 2016) or mouse APOE (Karagiannides et al., 2008). It is important to note that the Western diet used in this study has elevated levels of saturated fats, cholesterol, and sucrose, all of which have been independently associated with increased AD-related pathology (Refolo et al., 2000; Oksman et al., 2006; Cao et al., 2007; Takechi et al., 2010). Understanding how APOE genotype interacts with various dietary components should be one target of future studies. Though metabolic factors may have a role in AD pathogenesis, our findings that metabolic outcomes of DIO were greater in E3FAD than E4FAD mice argue against the possibility that metabolic impairment significantly contributes to the observed APOE4 bias in diet-induced increases in AD-like pathology.
There are several other mechanisms besides metabolic impairment that may contribute to the observed interactions among obesity, APOE, and AD-like pathology. One established consequence of obesogenic diets is pro-amyloidogenic alteration in the expression and or activity of factors that regulate generation and clearance of Aβ, including BACE1, neprilysin, and IDE (Standeven et al., 2010; Maesako et al., 2012; Brandimarti et al., 2013; Wei et al., 2014; Maesako et al., 2015). Although we cannot exclude a significant role of such pathways in our observations, we did not observe that mRNA levels of BACE1, neprilysin, and IDE were significantly altered by either the simple or interactive effects of Western diet and APOE. Another compelling candidate mechanism is neuroinflammation, which is widely implicated as a significant regulator of AD risk and development of AD pathology (Glass et al., 2010; Wyss-Coray and Rogers, 2012; Heneka et al., 2015). Notably, both obesity and APOE4 are associated with increased inflammation in brain and systemically. For example, obesity is linked with increased immune cell infiltration into brain (Buckman et al., 2014), as well as increased glial activation (Koga et al., 2014; Dorfman and Thaler, 2015; Douglass et al., 2017). In addition, obesity increases inflammation in peripheral organs including adipose tissue (Weisberg et al., 2003; Zeyda and Stulnig, 2009) and liver (Park et al., 2010). APOE4 is also associated with greater levels of inflammation in the brain (Ophir et al., 2005; Vitek et al., 2009) and throughout the body (Colton et al., 2004; Gale et al., 2014). Moreover, stimulating innate inflammation in the presence of apoE4 increases cell death and damage in macrophages (Cash et al., 2012), and in microglia and neurons (Maezawa et al., 2006a; 2006b). In the context of AD pathology, APOE4 is associated with greater glial activation in EFAD mice (Rodriguez et al., 2014). Similarly, we found that both the total number and the relative level of morphologic activation of microglia and astrocytes were higher in E4FAD than E3FAD mice. Further, we observed that E4FAD mice expressed significantly higher mRNA levels of glial markers than E3FAD mice under both control and Western diets. These glial markers were significantly increased across several brain regions in response to DIO in E4FAD but not E3FAD mice. Perhaps in contrast to our results, middle-aged female APOE4 mice showed higher levels of neuroinflammation in hippocampus under control diet but decreased neuroinflammation with high-fat diet, relative to age- and sex-matched wild-type mice (Janssen et al., 2016). Though the presence of familial AD transgenes and Aβ pathology in the EFAD model may account for these divergent findings, there may also be age and sex differences in inflammatory responses to both diet and APOE4. Further, because reactive astrocytes and microglia are associated with Aβ plaques, the changes in gliosis we observe with APOE4 and DIO may be a consequence of, rather than a contributor to, Aβ pathology. Thus, additional research is needed to directly assess the potential mechanistic role of gliosis in the interaction between APOE4 and obesity in AD.
To our knowledge, this is the first experimental investigation examining the interaction between APOE4 and obesity in the context of AD. Interactions among genetic risk factors like APOE4 and environmental and modifiable lifestyle risk factors in AD have thus far not been well studied, although there are some epidemiological studies consistent with this possibility (Dufouil et al., 2000; Hanson et al., 2013; Rajan et al., 2014; Wirth et al., 2014; Ishioka et al., 2016; Zheng and Li, 2016). Our findings suggest that APOE genotype affects the relationship between obesity and AD, such that APOE4 carriers may be more susceptible to obesity-associated risks than APOE3 carriers. This illustrates an important gene-environment interaction and points to the need for additional research exploring such relationships in the context of AD, as well as identifying underlying mechanisms. Additionally, these findings identify a large population that may be at increased risk of AD, but whose chance of developing the disease may be reduced by preventative lifestyle changes.
Acknowledgments
Acknowledgements: EFAD mice to generate a colony were generously provided by Dr. Mary Jo LaDu (University of Illinois at Chicago). We thank Dr. Amy Christensen, Antoine Ganivet, and Daniella Lent-Schochet for technical assistance.
Synthesis
Reviewing Editor: Liisa Galea, University of British Columbia
Decisions are customarily a result of the Reviewing Editor and the peer reviewers coming together and discussing their recommendations until a consensus is reached. When revisions are invited, a fact-based synthesis statement explaining their decision and outlining what is needed to prepare a revision will be listed below. The following reviewer(s) agreed to reveal their identity: Gregory Cole.
This is a well written manuscript that presents data illustrating interactions between ApoE genotype and the effectiveness of 3 months of a high fat "Western" diet in 3 month old male human ApoE3 or E4 TR (Sullivan) x 5xFAD (Vassar) "EFAD" mice. The Western diet leads to predictable weight gain (obesity) and metabolic (glucose) regulatory changes that are similarly significant with human ApoE3 and ApoE4, but with a selective increase in amyloid plaques and gliosis in the E4FAD mice. The findings are useful in explaining some discrepant results in the literature as there are robust but somewhat inconsistent epidemiological evidence for diet and lifestyle modification of ApoE4 genetic AD risk, the selective effect of high fat Western diet on E4FAD amyloid deposits and gliosis is an important observation from this study. As the authors note, the results are limited to relatively young male mice and the increased gliosis may simply track the amyloid. The reviewers have only a few relatively minor comments and agreed that this will be a nice addition to the literature.
Statistics:
1) A lot of the data are actually not normally distributed, as the authors indicate in Table 1. Although in practice it often makes little difference to the analysis, frequency and ratio variables (i.e., counting plaques and expressing amyloid load as % area) have a Poisson distribution, and are sometimes better treated as nonparametric variables (although these tests are not preferred due to their own limitations). However, normality assumptions can be checked and reported, and this is usually fine when it's done; if not, the data can be converted to an approximately normal distribution (for example, by a logarithmic transformation). A lot of times, this all works out the same way, but sometimes it doesn't. The reason this may be important is that the more interesting outcomes in this paper are interactions, and these in particular are sometimes vulnerable to violations of the underlying ANOVA assumptions. The authors need to at least check these assumptions.
2) Due to the number of significant interactions, it is important to acknowledge these significant interactions first and that when interactions exist it is important to describe these exclusively even in the face of significant main effects. This order is particularly notable for Thio-S results.
3) The results section is very through and perhaps to cut down when there is no significant interaction or main effect the authors could cut out the F and df and simply report the p values if they chose.
4) Bonferroni corrections are very conservative and the authors suggest they use them when analyzing significant main effects (p 8, last line)- but in fact because there are only two groups pairwise comparisons are not needed ( ie 2 genotypes and 2 diets). If Bonferroni were used with significant interactions they authors may consider using Tukey's instead for posthoc comparisons.
5) Were there any effects of WD on APP expression? This would seem important to take into consideration, as this could be the source of changes in amyloid deposition.
6) Were there any differences in peripheral cholesterol in the different mouse lines, either with or without Western Diet?
7) The authors could consider a figure for type 1,2,and 3 morphologies for microglia and astrocytic activation - this could be incorporated into figure 4 or 5.
8) It could be worthwhile to examine PS1 and BACE expression. In many cases of dietary and/or obesity studies we see effects on AD-related pathology, but get little insight into the underlying mechanism. The authors do hint at the problems with establishing mechanism, but don't delve too deeply into the problem. This could also be acknowledged in the discussion.
Discussion
9) The authors point might be helped by incorporating into the discussion more papers that show minimal effects of diet and obesity on amyloid pathology. For example, Niedowicz et al (Acta Neuropathologica Comm, 2014) found no effects on amyloid pathology in a db/db mouse crossed with an APPxPS1 mouse. In another example, Zhang et al (BBA: Mol Basis of Dis, 2013) found little effect of western diet of amyloid in a model of CAA. These papers and others like it - including this one - show that the interactions between metabolism, obesity, and amyloid pathology are more complex than many investigators realize.
10) In this study there is no isolation of the type of fat involved as the diet is high in cholesterol, saturated fat, calories from fat and calories. This invites further study, but does not negate the significance of the main observations and could be added into the discussion on future experiments.
Minor: "APOE4 is associated with greater glial activation EFAD mice (Rodriguez et al., 2014)." Needs "in" EFAD
Minor: Provide a reference for the use of 12 weeks of exposure to the diet.
References
- Altmann A, Tian L, Henderson VW, Greicius MD; Alzheimer's Disease Neuroimaging Initiative Investigators (2014) Sex modifies the APOE-related risk of developing Alzheimer disease. Ann Neurol 75:563–573. 10.1002/ana.24135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbones-Mainar JM, Johnson LA, Altenburg MK, Maeda N (2008) Differential modulation of diet-induced obesity and adipocyte functionality by human apolipoprotein E3 and E4 in mice. Int J Obes Relat Metab Disord 32:1595–1605. 10.1038/ijo.2008.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbones-Mainar JM, Johnson LA, Altenburg MK, Kim HS, Maeda N (2010) Impaired adipogenic response to thiazolidinediones in mice expressing human apolipoproteinE4. FASEB J 24:3809–3818. 10.1096/fj.10-159517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atabek ME, Özkul Y, Eklioğlu BS, Kurtoğlu S, Baykara M (2012) Association between apolipoprotein E polymorphism and subclinic atherosclerosis in patients with type 1 diabetes mellitus. J Clin Res Pediatr Endocrinol 4:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub AE, Salm AK (2003) Increased morphological diversity of microglia in the activated hypothalamic supraoptic nucleus. J Neurosci 23:7759–7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberger-Gateau P, Samieri C, Féart C, Plourde M (2011) Dietary omega 3 polyunsaturated fatty acids and Alzheimer's disease: interaction with apolipoprotein E genotype. Curr Alzheimer Res 8:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron AM, Rosario ER, Elteriefi R, Pike CJ (2013) Sex-specific effects of high fat diet on indices of metabolic syndrome in 3xTg-AD mice: implications for Alzheimer's Disease. PLoS One 8:e78554. 10.1371/journal.pone.0078554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloor ID, Symonds ME (2014) Sexual dimorphism in white and brown adipose tissue with obesity and inflammation. Horm Behav 66:95–103. 10.1016/j.yhbeh.2014.02.007 [DOI] [PubMed] [Google Scholar]
- Brandimarti P, Costa JJM, Ferreira SM, Protzek AO, Santos GJ, Carneiro EM, Boschero AC, Rezende LF (2013) Cafeteria diet inhibits insulin clearance by reduced insulin-degrading enzyme expression and mRNA splicing. J Endocrinol 219:173–182. 10.1530/JOE-13-0177 [DOI] [PubMed] [Google Scholar]
- Buckman LB, Hasty AH, Flaherty DK, Buckman CT, Thompson MM, Matlock BK, Weller K, Ellacott KLJ (2014) Obesity induced by a high-fat diet is associated with increased immune cell entry into the central nervous system. Brain Behav Immun 35:33–42. 10.1016/j.bbi.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciottolo M, Christensen A, Moser A, Liu J, Pike CJ, Smith C, LaDu MJ, Sullivan PM, Morgan TE, Dolzhenko E, Charidimou A, Wahlund L-O, Wiberg MK, Shams S, Chiang GC-Y, Finch CE; Alzheimer's Disease Neuroimaging Initiative (2016) The APOE4 allele shows opposite sex bias in microbleeds and Alzheimer's disease of humans and mice. Neurobiol Aging 37:47–57. 10.1016/j.neurobiolaging.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Lu H, Lewis TL, Li L (2007) Intake of sucrose-sweetened water induces insulin resistance and exacerbates memory deficits and amyloidosis in a transgenic mouse model of Alzheimer disease. J Biol Chem 282:36275–36282. 10.1074/jbc.M703561200 [DOI] [PubMed] [Google Scholar]
- Cash JG, Kuhel DG, Basford JE, Jaeschke A, Chatterjee TK, Weintraub NL, Hui DY (2012) Apolipoprotein E4 impairs macrophage efferocytosis and potentiates apoptosis by accelerating endoplasmic reticulum stress. J Biol Chem 287:27876–27884. 10.1074/jbc.M112.377549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, Goate AM, Bales KR, Paul SM, Bateman RJ, Holtzman DM (2011) Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med 3:89ra57. 10.1126/scitranslmed.3002156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton CA, Needham LK, Brown C, Cook D, Rasheed K, Burke JR, Strittmatter WJ, Schmechel DE, Vitek MP (2004) APOE genotype-specific differences in human and mouse macrophage nitric oxide production. J Neuroimmunol 147:62–67. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261:921–923. [DOI] [PubMed] [Google Scholar]
- Craft S (2005) Insulin resistance syndrome and Alzheimer's disease: age- and obesity-related effects on memory, amyloid, and inflammation. Neurobiol Aging 26:65–69. 10.1016/j.neurobiolaging.2005.08.021 [DOI] [PubMed] [Google Scholar]
- Craft S (2009) The role of metabolic disorders in Alzheimer disease and vascular dementia: two roads converged. Arch Neurol 66:300–305. 10.1001/archneurol.2009.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de-Andrade FM, Larrandaburu M, Callegari-Jacques SM, Gastaldo G, Hutz MH (2000) Association of apolipoprotein E polymorphism with plasma lipids and Alzheimer's disease in a Southern Brazilian population. Braz J Med Biol Res 33:529–537. 10.1590/S0100-879X2000000500007 [DOI] [PubMed] [Google Scholar]
- Dixit M, Bhattacharya S, Mittal B (2005) Association of CETP TaqI and APOE polymorphisms with type II diabetes mellitus in North Indians: a case control study. BMC Endocr Disord 5:7 10.1186/1472-6823-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman MD, Thaler JP (2015) Hypothalamic inflammation and gliosis in obesity. Curr Opin Endocrinol Diabetes Obes 22:325–330. 10.1097/MED.0000000000000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass JD, Dorfman MD, Thaler JP (2017) Glia: silent partners in energy homeostasis and obesity pathogenesis. Diabetologia 60:226–236. 10.1007/s00125-016-4181-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufouil C, Tzourio C, Brayne C, Berr C, Amouyel P, Alpérovitch A (2000) Influence of apolipoprotein E genotype on the risk of cognitive deterioration in moderate drinkers and smokers. Epidemiology 11:280–284. [DOI] [PubMed] [Google Scholar]
- Elosua R, Demissie S, Cupples LA, Meigs JB, Wilson PWF, Schaefer EJ, Corella D, Ordovas JM (2003) Obesity modulates the association among APOE genotype, insulin, and glucose in men. Obes Res 11:1502–1508. 10.1038/oby.2003.201 [DOI] [PubMed] [Google Scholar]
- Emmerzaal TL, Kiliaan AJ, Gustafson DR (2015) 2003-2013: a decade of body mass index, Alzheimer's disease, and dementia. J Alzheimers Dis 43:739–755. 10.3233/JAD-141086 [DOI] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM (1997) Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278:1349–1356. [PubMed] [Google Scholar]
- Fitzpatrick AL, Kuller LH, Lopez OL, Diehr P, O'Meara ES, Longstreth WT, Luchsinger JA (2009) Midlife and late-life obesity and the risk of dementia: cardiovascular health study. Arch Neurol 66:336–342. 10.1001/archneurol.2008.582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JD, Simmons K, Parsadanian M, Bales KR, Paul SM, Sullivan PM, Holtzman DM (2005) Human apolipoprotein E4 alters the amyloid-beta 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J Neurosci 25:2803–2810. 10.1523/JNEUROSCI.5170-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale SC, Gao L, Mikacenic C, Coyle SM, Rafaels N, Murray Dudenkov T, Madenspacher JH, Draper DW, Ge W, Aloor JJ, Azzam KM, Lai L, Blackshear PJ, Calvano SE, Barnes KC, Lowry SF, Corbett S, Wurfel MM, Fessler MB (2014) APOε4 is associated with enhanced in vivo innate immune responses in human subjects. J Allergy Clin Immunol 134:127–134. 10.1016/j.jaci.2014.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, Bullido MJ, Engelborghs S, De Deyn P, Berr C, Pasquier F, Dubois B, Tognoni G, Fiévet N, Brouwers N, Bettens K, Arosio B, Coto E, Del Zompo M, Mateo I, et al. (2011) APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol Psychiatry 16:903–907. 10.1038/mp.2011.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebranious N, Mukesh B, Giampietro PF, Glurich I, Mickel SF, Waring SC, McCarty CA (2011) A pilot study of gene/gene and gene/environment interactions in Alzheimer disease. Clin Med Res 9:17–25. 10.3121/cmr.2010.894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH (2010) Mechanisms underlying inflammation in neurodegeneration. Cell 140:918–934. 10.1016/j.cell.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J, Zhao H-L, Sui Y, He L, Lee H-M, Lai FMM, Tong PCY, Chan JCN (2013) Histopathological correlations of islet amyloidosis with apolipoprotein E polymorphisms in type 2 diabetic Chinese patients. Pancreas 42:1129–1137. 10.1097/MPA.0b013e3182965e6e [DOI] [PubMed] [Google Scholar]
- Haan MN, Shemanski L, Jagust WJ, Manolio TA, Kuller L (1999) The role of APOE epsilon4 in modulating effects of other risk factors for cognitive decline in elderly persons. JAMA 282:40–46. [DOI] [PubMed] [Google Scholar]
- Hanson AJ, Bayer-Carter JL, Green PS, Montine TJ, Wilkinson CW, Baker LD, Watson GS, Bonner LM, Callaghan M, Leverenz JB, Tsai E, Postupna N, Zhang J, Lampe J, Craft S (2013) Effect of apolipoprotein E genotype and diet on apolipoprotein E lipidation and amyloid peptides. JAMA Neurol 70:972–980. 10.1001/jamaneurol.2013.396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka MT, Carson MJ, El Khoury J, Landreth GE, Brosseron F, Feinstein DL, Jacobs AH, Wyss-Coray T, Vitorica J, Ransohoff RM, Herrup K, Frautschy SA, Finsen B, Brown GC, Verkhratsky A, Yamanaka K, Koistinaho J, Latz E, Halle A, Petzold GC, et al. (2015) Neuroinflammation in Alzheimer's disease. Lancet Neurol 14:388–405. 10.1016/S1474-4422(15)70016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho AJ, Raji CA, Becker JT, Lopez OL, Kuller LH, Hua X, Lee S, Hibar D, Dinov ID, Stein JL, Jack CR Jr, Weiner MW, Toga AW, Thompson PM (2010) Obesity is linked with lower brain volume in 700 AD and MCI patients. Neurobiol Aging 31:1326–1339. 10.1016/j.neurobiolaging.2010.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Qin W, Pompl PN, Xiang Z, Wang J, Zhao Z, Peng Y, Cambareri G, Rocher A, Mobbs CV, Hof PR, Pasinetti GM (2004) Diet-induced insulin resistance promotes amyloidosis in a transgenic mouse model of Alzheimer's disease. FASEB J 18:902–904. [DOI] [PubMed] [Google Scholar]
- Ishioka YL, Gondo Y, Fuku N, Inagaki H, Masui Y, Takayama M, Abe Y, Arai Y, Hirose N (2016) Effects of the APOE ε4 allele and education on cognitive function in Japanese centenarians. Age (Dordr) 38:495–503. 10.1007/s11357-016-9944-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Wiste HJ, Weigand SD, Knopman DS, Vemuri P, Mielke MM, Lowe V, Senjem ML, Gunter JL, Machulda MM, Gregg BE, Pankratz VS, Rocca WA, Petersen RC (2015) Age, sex, and APOE ε4 effects on memory, brain structure, and β-Amyloid across the adult life span. JAMA Neurol 72:511–519. 10.1001/jamaneurol.2014.4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen CIF, Jansen D, Mutsaers MPC, Dederen PJWC, Geenen B, Mulder MT, Kiliaan AJ (2016) The effect of a high-fat diet on brain plasticity, inflammation and cognition in female ApoE4-knockin and ApoE-knockout mice. PLoS One 11:e0155307. 10.1371/journal.pone.0155307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien C, Tremblay C, Phivilay A, Berthiaume L, Emond V, Julien P, Calon F (2010) High-fat diet aggravates amyloid-beta and tau pathologies in the 3xTg-AD mouse model. Neurobiol Aging 31:1516–1531. 10.1016/j.neurobiolaging.2008.08.022 [DOI] [PubMed] [Google Scholar]
- Jung H-J, Kim Y-J, Eggert S, Chung KC, Choi KS, Park SA (2013) Age-dependent increases in tau phosphorylation in the brains of type 2 diabetic rats correlate with a reduced expression of p62. Exp Neurol 248:441–450. 10.1016/j.expneurol.2013.07.013 [DOI] [PubMed] [Google Scholar]
- Kahn SE, Hull RL, Utzschneider KM (2006) Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444:840–846. 10.1038/nature05482 [DOI] [PubMed] [Google Scholar]
- Karagiannides I, Abdou R, Tzortzopoulou A, Voshol PJ, Kypreos KE (2008) Apolipoprotein E predisposes to obesity and related metabolic dysfunctions in mice. FEBS J 275:4796–4809. 10.1111/j.1742-4658.2008.06619.x [DOI] [PubMed] [Google Scholar]
- Kim B, Backus C, Oh S, Hayes JM, Feldman EL (2009) Increased tau phosphorylation and cleavage in mouse models of type 1 and type 2 diabetes. Endocrinology 150:5294–5301. 10.1210/en.2009-0695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight EM, Martins IVA, Gümüsgöz S, Allan SM, Lawrence CB (2014) High-fat diet-induced memory impairment in triple-transgenic Alzheimer's disease (3xTgAD) mice is independent of changes in amyloid and tau pathology. Neurobiol Aging 35:1821–1832. 10.1016/j.neurobiolaging.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga S, Kojima A, Kuwabara S, Yoshiyama Y (2014) Immunohistochemical analysis of tau phosphorylation and astroglial activation with enhanced leptin receptor expression in diet-induced obesity mouse hippocampus. Neurosci Lett 571:11–16. 10.1016/j.neulet.2014.04.028 [DOI] [PubMed] [Google Scholar]
- Kohjima M, Sun Y, Chan L (2010) Increased food intake leads to obesity and insulin resistance in the Tg2576 Alzheimer's disease mouse model. Endocrinology 151:1532–1540. 10.1210/en.2009-1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypreos KE, Karagiannides I, Fotiadou EH, Karavia EA, Brinkmeier MS, Giakoumi SM, Tsompanidi EM (2009) Mechanisms of obesity and related pathologies: role of apolipoprotein E in the development of obesity. FEBS J 276:5720–5728. 10.1111/j.1742-4658.2009.07301.x [DOI] [PubMed] [Google Scholar]
- Lee EB, Mattson MP (2013) The neuropathology of obesity: insights from human disease. Acta Neuropathol 127:3–28. 10.1007/s00401-013-1190-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiva E, Mujica V, Orrego R, Prieto M, Arredondo M (2005) Apolipoprotein E polymorphism in type 2 diabetic patients of Talca, Chile. Diabetes Res Clin Pract 68:244–249. 10.1016/j.diabres.2004.09.017 [DOI] [PubMed] [Google Scholar]
- Li R, Singh M (2014) Sex differences in cognitive impairment and Alzheimer's disease. Front Neuroendocrinol 35:385–403. 10.1016/j.yfrne.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-C, Kanekiyo T, Xu H, Bu G (2013) Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 9:106–118. 10.1038/nrneurol.2012.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy JC, Sainsbury A; Stock Conference 2008 Working Group (2009) Sex differences in obesity and the regulation of energy homeostasis. Obes Rev 10:154–167. 10.1111/j.1467-789X.2008.00529.x [DOI] [PubMed] [Google Scholar]
- Luchsinger JA, Cheng D, Tang MX, Schupf N, Mayeux R (2012) Central obesity in the elderly is related to late-onset Alzheimer disease. Alzheimer Dis Assoc Disord 26:101–105. 10.1097/WAD.0b013e318222f0d4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchsinger JA, Tang M-X, Shea S, Mayeux R (2002) Caloric intake and the risk of Alzheimer disease. Arch Neurol 59:1258–1263. [DOI] [PubMed] [Google Scholar]
- Maesako M, Uemura K, Kubota M, Kuzuya A, Sasaki K, Hayashida N, Asada-Utsugi M, Watanabe K, Uemura M, Kihara T, Takahashi R, Shimohama S, Kinoshita A (2012) Exercise is more effective than diet control in preventing high fat diet-induced β-amyloid deposition and memory deficit in amyloid precursor protein transgenic mice. J Biol Chem 287:23024–23033. 10.1074/jbc.M112.367011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maesako M, Uemura M, Tashiro Y, Sasaki K, Watanabe K, Noda Y, Ueda K, Asada-Utsugi M, Kubota M, Okawa K, Ihara M, Shimohama S, Uemura K, Kinoshita A (2015) High fat diet enhances β-Site cleavage of amyloid precursor protein (APP) via promoting β-Site APP cleaving enzyme 1/adaptor protein 2/clathrin complex formation. PLoS One 10:e0131199. 10.1371/journal.pone.0131199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa I, Maeda N, Montine TJ, Montine KS (2006a) Apolipoprotein E-specific innate immune response in astrocytes from targeted replacement mice. J Neuroinflammation 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa I, Zaja-Milatovic S, Milatovic D, Stephen C, Sokal I, Maeda N, Montine TJ, Montine KS (2006b) Apolipoprotein E isoform-dependent dendritic recovery of hippocampal neurons following activation of innate immunity. J Neuroinflammation 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Vidal P, Bongard V, Ruidavets J-B, Fauvel J, Hanaire-Broutin H, Perret B, Ferrières J (2003) Obesity and alcohol modulate the effect of apolipoprotein E polymorphism on lipids and insulin. Obes Res 11:1200–1206. 10.1038/oby.2003.165 [DOI] [PubMed] [Google Scholar]
- Martins IJ, Hone E, Foster JK, Sünram-Lea SI, Gnjec A, Fuller SJ, Nolan D, Gandy SE, Martins RN (2006) Apolipoprotein E, cholesterol metabolism, diabetes, and the convergence of risk factors for Alzheimer's disease and cardiovascular disease. Mol Psychiatry 11:721–736. 10.1038/sj.mp.4001854 [DOI] [PubMed] [Google Scholar]
- Mauvais-Jarvis F (2015) Sex differences in metabolic homeostasis, diabetes, and obesity. Biol Sex Differ 6:14. 10.1186/s13293-015-0033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meigs JB, Ordovas JM, Cupples LA, Singer DE, Nathan DM, Schaefer EJ, Wilson PW (2000) Apolipoprotein E isoform polymorphisms are not associated with insulin resistance: the Framingham Offspring Study. Diabetes Care 23:669–674. 10.2337/diacare.23.5.669 [DOI] [PubMed] [Google Scholar]
- Meng X-F, Yu J-T, Wang H-F, Tan M-S, Wang C, Tan C-C, Tan L (2014) Midlife vascular risk factors and the risk of Alzheimer's disease: a systematic review and meta-analysis. J Alzheimers Dis 42:1295–1310. 10.3233/JAD-140954 [DOI] [PubMed] [Google Scholar]
- Meyer MR, Clegg DJ, Prossnitz ER, Barton M (2011) Obesity, insulin resistance and diabetes: sex differences and role of oestrogen receptors. Acta Physiol (Oxf) 203:259–269. 10.1111/j.1748-1716.2010.02237.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, Mintun MA (2010) APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol 67:122–131. 10.1002/ana.21843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser VA, Pike CJ (2016) Obesity and sex interact in the regulation of Alzheimer's disease. Neurosci Biobehav Rev 67:102–118. 10.1016/j.neubiorev.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedowicz DM, Reeves VL, Platt TL, Kohler K, Beckett TL, Powell DK, Lee TL, Sexton TR, Song ES, Brewer LD, Latimer CS, Kraner SD, Larson KL, Ozcan S, Norris CM, Hersh LB, Porter NM, Wilcock DM, Murphy MP (2014) Obesity and diabetes cause cognitive dysfunction in the absence of accelerated β-amyloid deposition in a novel murine model of mixed or vascular dementia. Acta Neuropathol Commun 2:64. 10.1186/2051-5960-2-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Qi Y, Qian Y, Gao P, Zhu D (2009) The relationship between apolipoprotein E e2/e3/e4 polymorphisms and hypertension: a meta-analysis of six studies comprising 1812 cases and 1762 controls. Hypertens Res 32:1060–1066. 10.1038/hr.2009.164 [DOI] [PubMed] [Google Scholar]
- Oh JY, Barrett-Connor E (2001) Apolipoprotein E polymorphism and lipid levels differ by gender and family history of diabetes: the Rancho Bernardo Study. Clin Genet 60:132–137. 10.1034/j.1399-0004.2001.600207.x [DOI] [PubMed] [Google Scholar]
- Oksman M, Iivonen H, Hogyes E, Amtul Z, Penke B, Leenders I, Broersen L, Lütjohann D, Hartmann T, Tanila H (2006) Impact of different saturated fatty acid, polyunsaturated fatty acid and cholesterol containing diets on beta-amyloid accumulation in APP/PS1 transgenic mice. Neurobiol Dis 23:563–572. 10.1016/j.nbd.2006.04.013 [DOI] [PubMed] [Google Scholar]
- Ophir G, Amariglio N, Jacob-Hirsch J, Elkon R, Rechavi G, Michaelson DM (2005) Apolipoprotein E4 enhances brain inflammation by modulation of the NF-κB signaling cascade. Neurobiol Dis 20:709–718. 10.1016/j.nbd.2005.05.002 [DOI] [PubMed] [Google Scholar]
- Orr ME, Salinas A, Buffenstein R, Oddo S (2014) Mammalian target of rapamycin hyperactivity mediates the detrimental effects of a high sucrose diet on Alzheimer's disease pathology. Neurobiol Aging 35:1233–1242. 10.1016/j.neurobiolaging.2013.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Lee JH, Yu G-Y, He G, Ali SR, Holzer RG, Österreicher CH, Takahashi H, Karin M (2010) Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 140:197–208. 10.1016/j.cell.2009.12.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payami H, Montee KR, Kaye JA, Bird TD, Yu C-E, Wijsman EM, Schellenberg GD (1994) Alzheimer's disease, apolipoprotein E4, and gender. JAMA 271:1316–1317. [PubMed] [Google Scholar]
- Peila R, Rodriguez BL, Launer LJ (2002) Type 2 diabetes, APOE gene, and the risk for dementia and related pathologies: the Honolulu-Asia Aging Study. Diabetes 51:1256–1262. [DOI] [PubMed] [Google Scholar]
- Peila R, White LR, Petrovich H, Masaki K, Ross GW, Havlik RJ, Launer LJ (2001) Joint effect of the APOE gene and midlife systolic blood pressure on late-life cognitive impairment: the Honolulu-Asia aging study. Stroke 32:2882–2889. [DOI] [PubMed] [Google Scholar]
- Pike CJ (2017) Sex and the development of Alzheimer's disease. J Neurosci Res 95:671–680. 10.1002/jnr.23827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Profenno LA, Faraone SV (2008) Diabetes and overweight associate with non-APOE4 genotype in an alzheimer's disease population. Am J Med Genet 147B:822–829. 10.1002/ajmg.b.30694 [DOI] [PubMed] [Google Scholar]
- Profenno LA, Porsteinsson AP, Faraone SV (2010) Meta-analysis of Alzheimer’s disease risk with obesity, diabetes, and related disorders. Biol Psychiatry 67:505–512. 10.1016/j.biopsych.2009.02.013 [DOI] [PubMed] [Google Scholar]
- Ragogna F, Lattuada G, Ruotolo G, Luzi L, Perseghin G (2011) Lack of association of apoE ε4 allele with insulin resistance. Acta Diabetol 49:25–32. 10.1007/s00592-011-0255-3 [DOI] [PubMed] [Google Scholar]
- Rajan KB, Skarupski KA, Rasmussen HE, Evans DA (2014) Gene-environment interaction of body mass index and apolipoprotein E ε4 allele on cognitive decline. Alzheimer Dis Assoc Disord 28:134–140. 10.1097/WAD.0000000000000013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Rodriguez JJ, Ortiz O, Jimenez-Palomares M, Kay KR, Berrocoso E, Murillo-Carretero MI, Perdomo G, Spires-Jones T, Cozar-Castellano I, Lechuga-Sancho AM, Garcia-Alloza M (2013) Differential central pathology and cognitive impairment in pre-diabetic and diabetic mice. Psychoneuroendocrinology 38:2462–2475. 10.1016/j.psyneuen.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Rebeck GW, Reiter JS, Strickland DK, Hyman BT (1993) Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron 11:575–580. [DOI] [PubMed] [Google Scholar]
- Refolo LM, Malester B, LaFrancois J, Bryant-Thomas T, Wang R, Tint GS, Sambamurti K, Duff K, Pappolla MA (2000) Hypercholesterolemia accelerates the Alzheimer's amyloid pathology in a transgenic mouse model. Neurobiol Dis 7:321–331. 10.1006/nbdi.2000.0304 [DOI] [PubMed] [Google Scholar]
- Rodriguez GA, Tai LM, LaDu MJ, Rebeck GW (2014) Human APOE4 increases microglia reactivity at Aβ plaques in a mouse model of Aβ deposition. J Neuroinflammation 11:111 10.1186/1742-2094-11-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ (1993) Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology 43:1467–1472. [DOI] [PubMed] [Google Scholar]
- Segev Y, Livne A, Mints M, Rosenblum K (2016) Concurrence of high fat diet and APOE gene induces allele specific metabolic and mental stress changes in a mouse model of Alzheimer’s disease. Front Behav Neurosci 10:170 10.3389/fnbeh.2016.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima A, Iordan A, Stancu C (2007) Apolipoprotein E polymorphism–a risk factor for metabolic syndrome. Clin Chem Lab Med 45:1149–1153. 10.1515/CCLM.2007.258 [DOI] [PubMed] [Google Scholar]
- Singh PP, Naz I, Gilmour A, Singh M, Mastana S (2006) Association of APOE (Hha1) and ACE (I/D) gene polymorphisms with type 2 diabetes mellitus in North West India. Diabetes Res Clin Pract 74:95–102. 10.1016/j.diabres.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Singla P, Bardoloi A, Parkash AA (2010) Metabolic effects of obesity: a review. World J Diabetes 1:76–88. 10.4239/wjd.v1.i3.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook JT, Park S, Williams G, Tsai Y-H, Lee N (1999) Effect of synthetic triglycerides of myristic, palmitic, and stearic acid on serum lipoprotein metabolism. Eur J Clin Nutr 53:597–605. [DOI] [PubMed] [Google Scholar]
- Standeven KF, Hess K, Carter AM, Rice GI, Cordell PA, Balmforth AJ, Lu B, Scott DJ, Turner AJ, Hooper NM, Grant PJ (2010) Neprilysin, obesity and the metabolic syndrome. Int J Obes Relat Metab Disord 35:1031–1040. 10.1038/ijo.2010.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD (1993) Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA 90:1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama MG, Agellon LB (2012) Sex differences in lipid metabolism and metabolic disease risk. Biochem Cell Biol 90:124–141. 10.1139/o11-067 [DOI] [PubMed] [Google Scholar]
- Takechi R, Galloway S, Pallebage-Gamarallage MMS, Wellington CL, Johnsen RD, Dhaliwal SS, Mamo JCL (2010) Differential effects of dietary fatty acids on the cerebral distribution of plasma-derived apo B lipoproteins with amyloid-beta. Br J Nutr 103:652–662. 10.1017/S0007114509992194 [DOI] [PubMed] [Google Scholar]
- van der Flier WM, Pijnenburg YA, Fox NC, Scheltens P (2011) Early-onset versus late-onset Alzheimer's disease: the case of the missing APOE ε4 allele. Lancet Neurol 10:280–288. 10.1016/S1474-4422(10)70306-9 [DOI] [PubMed] [Google Scholar]
- Vanhanen M, Koivisto K, Moilanen L, Helkala EL, Hänninen T, Soininen H, Kervinen K, Kesäniemi YA, Laakso M, Kuusisto J (2006) Association of metabolic syndrome with Alzheimer disease: a population-based study. Neurology 67:843–847. 10.1212/01.wnl.0000234037.91185.99 [DOI] [PubMed] [Google Scholar]
- Vitek MP, Brown CM, Colton CA (2009) APOE genotype-specific differences in the innate immune response. Neurobiol Aging 30:1350–1360. 10.1016/j.neurobiolaging.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ke B, Zhao Z, Ye X, Gao Z, Ye J (2014) Regulation of insulin degrading enzyme activity by obesity-associated factors and pioglitazone in liver of diet-induced obese mice. PLoS One 9:e95399. 10.1371/journal.pone.0095399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr(2003) Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112:1796–1808. 10.1172/JCI19246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmer RA, Gustafson DR, Barrett-Connor E, Haan MN, Gunderson EP, Yaffe K (2008) Central obesity and increased risk of dementia more than three decades later. Neurology 71:1057–1064. 10.1212/01.wnl.0000306313.89165.ef [DOI] [PubMed] [Google Scholar]
- Wilhelmsson U, Bushong EA, Price DL, Smarr BL, Phung V, Terada M, Ellisman MH, Pekny M (2006) Redefining the concept of reactive astrocytes as cells that remain within their unique domains upon reaction to injury. Proc Natl Acad Sci USA 103:17513–17518. 10.1073/pnas.0602841103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth M, Villeneuve S, La Joie R, Marks SM, Jagust WJ (2014) Gene-environment interactions: lifetime cognitive activity, APOE genotype, and beta-amyloid burden. J Neurosci 34:8612–8617. 10.1523/JNEUROSCI.4612-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T, Rogers J (2012) Inflammation in Alzheimer disease-a brief review of the basic science and clinical literature. Cold Spring Harb Perspect Med 2:a006346 10.1101/cshperspect.a006346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youmans KL, Tai LM, Nwabuisi-Heath E, Jungbauer L, Kanekiyo T, Gan M, Kim J, Eimer WA, Estus S, Rebeck GW, Weeber EJ, Bu G, Yu C, LaDu MJ (2012) APOE4-specific changes in amyloid-beta accumulation in a new transgenic mouse model of Alzheimer disease. J Biol Chem 287:41774–41786. 10.1074/jbc.M112.407957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkesh M, Daneshpour MS, Faam B, Hedayati M, Azizi F (2012) Is there any association of apolipoprotein E gene polymorphism with obesity status and lipid profiles? Tehran Lipid and Glucose Study (TLGS). Gene 509:282–285. 10.1016/j.gene.2012.07.048 [DOI] [PubMed] [Google Scholar]
- Zeyda M, Stulnig TM (2009) Obesity, inflammation, and insulin resistance–a mini-review. Gerontology 55:379–386. 10.1159/000212758 [DOI] [PubMed] [Google Scholar]
- Zhang L, Dasuri K, Fernandez-Kim SO, Bruce-Keller AJ, Freeman LR, Pepping JK, Beckett TL, Murphy MP, Keller JN (2013) Prolonged diet induced obesity has minimal effects towards brain pathology in mouse model of cerebral amyloid angiopathy: implications for studying obesity-brain interactions in mice. Biochim Biophys Acta 1832:1456–1462. 10.1016/j.bbadis.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Li Q (2016) Impact of apolipoprotein E gene polymorphism and additional gene-obesity interaction on type 2 diabetes risk in a Chinese Han old population. Obes Res Clin Pract pii:S1871-403X(16)30010-2. [DOI] [PubMed] [Google Scholar]