Abstract
Rice yield is subjected to severe losses due to adverse effect of a number of stress factors. The most effective method of controlling reduced crop production is utilization of host resistance. Recent technological advances have led to the improvement of DNA based molecular markers closely linked to genes or QTLs in rice chromosome that bestow tolerance to various types of abiotic stresses and resistance to biotic stress factors. Transfer of several genes with potential characteristics into a single genotype is possible through the process of marker assisted selection (MAS), which can quicken the advancement of tolerant/resistant cultivars in the lowest number of generations with the utmost precision through the process of gene pyramiding. Overall, this review presented various types of molecular tools including MAS that can be reasonable and environmental friendly approach for the improvement of abiotic and biotic stress resistant rice with enhanced quality.
Keywords: gene pyramiding, genome mapping, phenotype traits, physiological traits, molecular markers, marker assisted selection, rice
Introduction
Rice, which is the world's most important food crop, is a key source of carbohydrate (Narciso and Hossain, 2002). The cultivation of rice is a principal activity and source of income around the world, and several Asian and African countries depend on rice as a basis of earnings (Khan et al., 2009). However, there is severe yield loss in rice cultivation due to a number of abiotic and biotic stresses (Ramegowda and Senthil-Kumar, 2015). Several environmental factors have threatened sustainable agricultural production in emerging countries, with the main variables affecting the future of agricultural production being higher incidence of extreme weather and a number of environmental problems (Joneydi, 2012).
Biotic stresses including pathogens, pests and weeds and abiotic stresses such as drought and periodic cycles of submergence, extreme cold, soil salinity, affects rice cultivation throughout the world. Crop losses caused by major biotic stressors such as bacterial blight and blast disease, and due to insect pests are quite high (Hasan et al., 2015). The occurrence of new stresses necessitates development of highly improved and novel approaches to enhance the capability of various rice varieties that can survive attacks caused by several pathogens at once while also surviving in unfavorable environments with high level of grain quality. In conventional breeding techniques, along with the desired genes other unwanted genes also continue to a long term in next generations, even with several backcross generations and it is not possible to screen unwanted genes, using conventional breeding. Despite their limitations, conventional approaches are also important for conserving wild germplasms, sexual hybridization between contrasting parental lines, novel genetic variants, and mutations (Werner et al., 2005). A variety of methods are used in conventional breeding, such as backcrossing, recurrent selection, and mutation breeding methods. However, using molecular tools such as markers that flank a target gene, can minimize the number of backcross generations (Hasan et al., 2015).
Molecular marker techniques are currently the most advanced method available for the transfer of desired gene in desired rice variety with required combination. The improvement of breeding programs using the most widely used molecular techniques, and their application is a novel prospect for enhancement of rice yields. Hence, the present study was conducted to briefly review the adverse effects of various types of biotic and abiotic stresses on production of rice and to improve the resistance with higher grain quality of rice through application of various types molecular tools especially the MAS.
Marker assisted selection (MAS): an advanced molecular tool in rice breeding
MAS is a process in which a marker is used for indirect selection of a genetic determinant or determinants of a trait of interest, i.e., abiotic stress tolerance, disease resistance, productivity, and/or quality (Prabhu et al., 2009). This method involves selection of plants carrying genomic regions that are involved in the expression of traits of interest through the application of molecular markers. The development and availability of an array of molecular markers and dense molecular genetic maps in crop plants has made application of MAS possible for traits governed by major genes and QTLs (Choudhary et al., 2008). The success of MAS depends on several factors, including the number of target genes to be transferred and the distance between the flanking markers and the target gene (Perumalsamy et al., 2010). MAS is gaining considerable importance as it can improve the efficiency of plant breeding through precise transfer of genomic regions of interest and acceleration of the recovery of the recurrent parent genome (Wijerathna, 2015).
Marker and QTL identification for MAS
The identification of DNA markers, genes and quantitative trait loci (QTLs) associated with particular traits is accomplished through QTL mapping. As a result, QTL mapping signifies the basis of development of molecular markers for MAS. However, there are several aspects that affect the accuracy of QTL mapping, such as replication levels of phenotypic information, population sizes and types, genotyping errors, and environmental effects. Genes or QTLs can be detected relative to a linkage map, by means of statistical methods such as single-marker analysis or interval mapping to identify associations between DNA markers and phenotypic data (Kearsey and Farquhar, 1998). Using DNA markers for identification of QTLs was a breakthrough in the characterization of quantitative traits. In plants, the identification of genomic regions related to quantitative traits has mostly been attained through QTL mapping (Borba et al., 2010).
Advantages of MAS for the improvement of stress resistant rice
With the application of MAS, individual plants can be selected based on their genotype during the selection procedure. For most traits, homozygous and heterozygous plants cannot be distinguished by conventional phenotypic screening. MAS can be used to assist selection of parents, increasing the effectiveness of backcross breeding and improving sex-limited traits (Zhou et al., 2007). MAS can be used to investigate heterosis for hybrid crop production (Reif et al., 2003), and there is the potential for use of DNA marker data along with phenotypic data to select hybrids (Jordan et al., 2003).
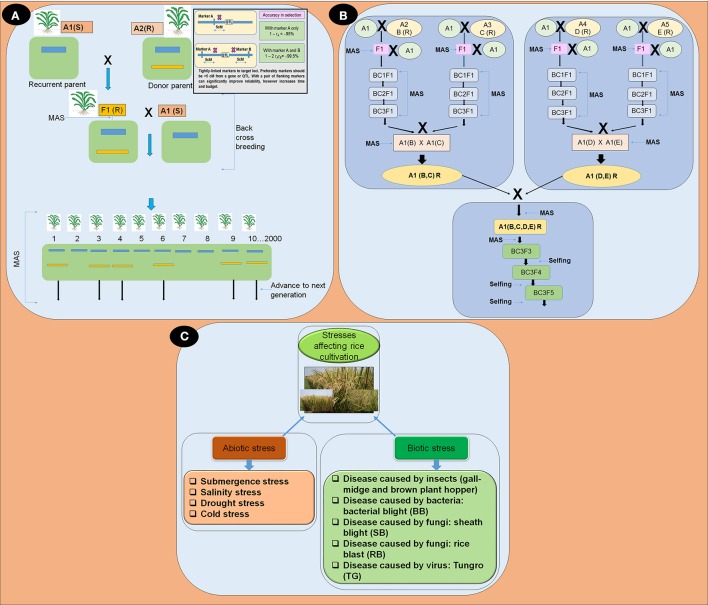
There are various advantages of using MAS in rice breeding. For example, it may be simpler than phenotypic screening; therefore, it can reduce time, effort, and resources. Selection of quality traits in rice generally requires expensive screening procedures that are made feasible through MAS (Figure 1A). Additionally, MAS selection can be conducted at the seedling stage and undesirable plant genotypes can quickly be eliminated (Khan et al., 2015). The advantages associated with the use of markers includes speed, consistency, efficiency, biosafety, and the ability to skew the odds in our favor, even while dealing with complex traits.
Figure 1.
(A) Overview of marker assisted backcross breeding program; (B) Flow diagram depicting the gene pyramiding of multiple stress resistance (R) genes into a single line using marker assisted backcross breeding; and (C) Flow diagram of stresses affecting rice productivity.
Molecular markers used in rice breeding
The development of novel molecular tools has significantly influenced rice breeding programs, permitting application of advanced molecular breeding techniques in rice that leads to the improvement of novel rice varieties through advanced biotechnology, which majorly includes MAS and genetic engineering.
Several researches shown that, in plants, single-nucleotide polymorphisms (SNPs) in addition to insertions and deletions (InDels) are extremely abundant and distributed throughout the genome (Batley et al., 2003). In plant genomes the abundance of polymorphisms makes the SNP marker system a smart tool for marker-assisted selection in breeding program (Hayashi et al., 2004). 1.7 million SNPs have been identified in rice, by comparative analysis of the draft genomic sequences of cv. Nipponbare (Kurokawa et al., 2016). Using NGS (Next-generation sequencing) technologies transcriptome resequencing permits fast and reasonable SNP detection within genes and avoids extremely repetitive sections of a genome (Mammadov et al., 2012). In the rice genome, SNPs are the most frequent polymorphism. In recent times, SNP arrays have been developed for rice by breeders, which are designed to cover a wide range of cross combinations in rice. For separating “linkage drag” the tightly linked set of SNPs can be used. The SNP array can also be readily used to construct an NIL (Near isogenic line) with a very minor introgressed chromosome segment from the donor parent, if a large segregating population is existing (Kurokawa et al., 2016). The employment of SNPs as molecular markers for breeding is becoming an actual possibility.
Advancement of molecular techniques in rice breeding has significantly extended the applicability of identification of genes and pyramiding valuable genes (Wijerathna, 2015). In gene pyramiding for a variety MAS not only shortened the breeding period but also removed the extensive trait assessment involved. There are many rice varieties improved by MAS (Table 1) (Rao et al., 2014). The steps of gene pyramiding using marker assisted selection (MAS) is displayed in (Figures 1A,B). There are many stress resistance genes and genes related to quality of rice which are tightly linked SNPs, SSRs, and STS markers are available (Table 1). All DNA marker techniques are designed to detect and exploit naturally occurring DNA polymorphisms. The main considerations for the use of DNA markers in MAS are as follows: particular markers should be tightly linked to target loci, preferably less than 5 cM genetic distance (Figure 1A).
Table 1.
Selected lists of genotypes improved by MAS; selected abiotic and biotic stress resistance genes/QTLs and linked markers; selected stresses, resistance genes/QTLs, and their donor parents.
| List of genotypes improved by MAS | |||
|---|---|---|---|
| Genotypes | Rice variety improved by MAS | Traits and resistant genes | References |
| Pusa basmati I | Improved Pusa Basmati I | Bacterial blight (xa13 + Xa21) | Kottapalli et al., 2010 |
| Samba Mahsuri (BPT-5204) | Improved Samba Mahsuri | Bacterial blight (xa5 + xa13 + Xa21) | Kottapalli et al., 2010 |
| Swarna | Swarna sub1 | Submergence (Sub1) | Nandi et al., 1997 |
| IR64 | IR64 Sub1 | Submergence (Sub1) | Reddy et al., 2009 |
| Pusa RH 10 | PRR78/IRBB60 | Rice blast (Pi54 + Piz5) | Singh et al., 2011 |
| Pusa RH 10 | Pusa 6A, Pusa 6B | Bacterial blight (xa13 + Xa21) | Singh et al., 2011 |
| KMR3 Restorer | KMR3/IRBB60 | Xa4 + xa5 + xa13 + Xa21 | Shanti et al., 2010 |
| Lalat | Improved Lalat | Bacterial blight (Xa4 + xa5 + xa13 + Xa21) | Dokku et al., 2013 |
| Tapaswini | Improved Tapaswini | Bacterial blight (Xa4 + xa5 + xa13 + Xa21) | Dokku et al., 2013 |
| Mangeumbye | Improved Mangeumbye | Bacterial blight (Xa4 + xa5 + Xa21) | Suh et al., 2013 |
| PRR78 | Improved Pusa RH10 | Rice blast (Piz5 + Pi54) | Singh et al., 2013 |
| Wuyujing 3 | K01, K04 | Low-amylose content gene (Wx-mq) | Tao et al., 2016 |
| List of selected abiotic and biotic stress resistance and quality genes/QTLs and linked markers | |||
| Stress/Disease/Traits | Resistance genes/QTLs | Available linked markers | References |
| Submergence | Sub1 | SUB1BC2, RM464A, RZ698, C1232, RG381 and RG345 | Das and Rao, 2015 |
| Salinity | Saltol | RM8094, RM140, RM10745, RM10772 | Nejad et al., 2008, 2010; Das and Rao, 2015 |
| Drought | QTLs | RM212, RM319, RM316, RM537 | Lin et al., 2007 |
| Cold | qPSST-3, qPSST-7, qPSST-9, qSCT1a, qSCT2 | RM231, RM1377, RM24545, RM3602 RM1211 | Jena et al., 2010; Kim et al., 2014 |
| Heat | qHTSF4.1 | M4 | Ye et al., 2015; Nogoy et al., 2016 |
| Gall midge | Gm1, Gm2, Gm4 | RM444, RM316, and RM219, RG476, RG329, RM547 | Biradar et al., 2004; Das and Rao, 2015 |
| Hopper burn | Bph-1 and Bph-10 (t), Bph-3, Bph-17, Bph-18, Bph-20, Bph21, Bph25, Bph26 | XNpb248 and RG457, RM589, RM5953, RM6217, BP-20-2, B121,RM6273, RM6775, RM5479 | Singh et al., 2011; Kurokawa et al., 2016 |
| BB | Xa21, xa13, Xa10, xa5, Xa4, Xa3, Xa1, Xa7 | pTA248, AB9, RG103, Xa13p, RG136, O072000,CDO365, RG556, XNpb181, R1506-S12886, MP, XNpb181, Y5212L, C600, Y5212R, 16PFXa1/EcoRV, M5 | Ma et al., 1999; Porter et al., 2003; Shin et al., 2007; Singh et al., 2011; Kottearachchi, 2013; Kurokawa et al., 2016 |
| SB (sheath blight) | qShB1, qShB2-1, qSB5, qShB6, qShB9-2 | RM104, RM341, RM13, RM190, RM245 | Liu et al., 2009 |
| RB (rice blast) | Pi-1, Pi 2, Pi 4(t), Pi 5, Piz-5, Pi 5(t), Pi 7 (t), Pi 10 (t), Pi-b, Pi54, Pi21, Pia | RZ536, RG64, RG869, S04G03, AP4007, AP5930 RG498, RG788, RG103A, RG16, RRF6, RZ213, RZ123, G1234, RM206, Os04g0401000, YCA72 | Cho et al., 2007; Singh et al., 2011; Kottearachchi, 2013 |
| TG (Tungro) | RTSV replicase gene | RZ262 | Khondker et al., 2005 |
| Deep roots | QTLs on chromosomes 1, 2, 7 and 9 | RFLP and SSR markers | Hasan et al., 2015 |
| Root traits C Aroma | QTLs on chromosomes 2, 7, 8, 9 and 11 | RFLP and SSR markers | Hasan et al., 2015 |
| Heading date | QTLs for heading date (Hd1,Hd4, Hd5, or Hd6) | RFLP, STS, SSR, CAPS, dCAPs | Hasan et al., 2015 |
| Quality | Waxy | RFLP | Hasan et al., 2015 |
| Eating quality | Amylose content gene | RM190 | Jairin et al., 2009 |
| Fragrance | Fragrance gene | BO3_127.8 | Jairin et al., 2009 |
| High yield | Gn1a/OsCKX2, APO1, WFP/OsSPL14 | Os01g0197700, Os06g0665400, Os08g0509600 | Ashikari et al., 2005; Kurokawa et al., 2016 |
| Seed shape | GW2, GS3, qSW5 | Os02g0244100, Os03g0407400, Os05g0187500 | Song et al., 2007; Shomura et al., 2008; Takano-Kai et al., 2009; Kurokawa et al., 2016 |
| List of selected stresses, resistance genes/QTLs and their donor parents | |||
| Disease/Stress | Resistance genes/QTLs | Donor parents | References |
| BIOTIC STRESS | |||
| Bacterial blight | Xa1, Xa2, Xa3, Xa4, Xa5, Xa10, Xa13, Xa21 | Kogyku, Tetep, Chogoku 45, IR20, IR1545-339, CAS209, O. longistaminata, Pusa 1460 | Swamy et al., 2006; Singh et al., 2011 |
| Rice blast | Pi-1(t), Pi-2(t), Pi-4(t), Pi-5(t), Pi-zh, Pi2, Pi9, Piz-5, Pi54, Piz, pi21 | LAC23, 5173, Tetep, IRAT13, Moroberekan, Zhiyeqing, C1O1A51, O. minuta derivative, Pusa 1602, IRBLZ5-a, DHMAS-70 Q164-2a,z2143,z1671, Os04g0401000 | Hayashi et al., 2004; Deng et al., 2006; Fukuoka et al., 2009; Singh et al., 2011; Kurokawa et al., 2016 |
| Gall midge | Gm1, Gm2, Gm4(t), Gm5(t), Gm6(t), Gm7(t), Gm8(t), Gm9(t), Gm10(t), Gm11(t) | Kavya, Siam 29, Abhaya, ARC5984, Duokang #1, Bhumansan, NHTA 8, Banglei | Jain et al., 2004; Kumaravadivel et al., 2006; Dubey and Chandel, 2010 |
| RTSV and GLH | Replicase genes, Glh-1, Glh-2, Glh-3, Glh-4, Glh-5, Glh-6, Glh-7, Glh-8, Grh4, Grh2 | Taipei 309-147.4 and Taipei 309-147.8, ARC11554, Pankhari 203, ASD7, IR8, Ptb8, Tightly linked to XNpb144, Tightly linked to G1465 | Khondker et al., 2005; Kurokawa et al., 2016 |
| BPH | Bph1, Bph2, Bph3, Bph4, Bph5, Bph6, Bph7, Bph8, Bph9, Bph10(t),Bph20(t), Bph21(t) Bph12(t), Bph13(t), Bph14 (Qbp1) and Bph15 (Qbp2) | Mudgo, ASD7, Rathu Heenati, Babawee, ARC10550, Swarnalata, T12, Chin Saba, pokkali, O. australiensis, Oryza latifolia, Oryza eichingeri, Oryza officinalis | Sun et al., 2006; Yadavalli et al., 2011 |
| ABIOTIC STRESS | |||
| Drought | Dreb1 | Nagina 22 | Reddy et al., 2009 |
| Submergence | Sub1 | FR13A, Swarna sub1, IR64 sub1, FR43B, Kurkurappan and Thavalu | Endang et al., 2009, 2012 |
| Salinity | Saltol | FL496, FL478, FL378, Pokkali, SR26B, Patnai 23, Vytilla 1 | Reddy et al., 2009; Nejad et al., 2010 |
| QUALITY | |||
| Eating quality | Low-amylose content gene Wx-mq | Kanto 194 | Tao et al., 2016 |
By exploring the diverse molecular techniques and advanced genomic technologies such as genome sequencing, SNPs array, genome-wide association mapping, and transcriptome profiling, the molecular mechanism and their relation between the genotypes and phenotypic traits leading to development of improved rice varieties can be realized (McCouch et al., 2016; Peng et al., 2016). Currently SSRs (second-generation markers) are widely used markers in MAS due to the easy availability and comparatively cheaper than others and they require a comparatively simple technique with a higher polymorphism rate (Gao et al., 2016).
Molecular markers for phenotype and physiological traits
The QTL analysis is an essential step to phenotyping of mapping of the population for their resistance to biotic and tolerance to abiotic stress. By pyramiding resistance genes through MAS, more durable resistance can be achieved via the identification of additional QTLs. Generally, biotic stress resistance is monitored using a specific scale value. Alternative methods such as quantification of the infected area can be used or tested for it. Phenotyping for abiotic stress is also essential to reducing the gap between genotype and phenotype, particularly for quantitative traits, which are the prime factors of abiotic stress resistance (Boopathi, 2012). The future of plant breeding depends on the complete knowledge regarding genetic control of physiological traits and the linkage of these physiological characteristics to molecular markers on chromosomes, and eventually the genes underlying the traits. Molecular markers have been rapidly adopted by researchers worldwide as an effective and appropriate tool for primary and practical studies addressing physiological traits. An important way of linking marker loci to a particular plant phenotype is through the use of genetic linkage maps (Graham et al., 2008).
Genome-wide association mapping
In a large germplasm collection, an alternative QTL mapping approach is conducting association analysis, known as association mapping. In a population, association mapping is based on linkage disequilibrium (LD) or the non-independence of alleles. Association mapping studies using sparse SSR and SNP markers have been proven to be effective when identifying marker-trait associations in rice (Qiu et al., 2015; McCouch et al., 2016). Genome-wide association studies (GWASs) by means of high density markers have become increasingly popular in rice genetics with the development of high-throughput sequencing and SNP chip techniques (Abe et al., 2010; Yu et al., 2014). GWAS is a powerful strategy to clearly understand the genetic basis of complex traits that has been especially productive for rice (Qiu et al., 2015).
Gene pyramiding using MAS: a smart approach to breeding programs
Gene pyramiding in rice is the transfer or pyramiding more than one resistance/tolerance genes/QTLs into a single rice genotype (Figure 1B) (Collard and Mackill, 2008). Pyramiding of resistance genes into a single line for each disease or stress is a novel strategy in rice breeding to prevent the breakdown of resistance against specific disease or stress. Pyramiding of genes/QTLs that confer resistance to biotic stresses and tolerance against various types of abiotic stresses is now feasible because of advancements in molecular markers (Das and Rao, 2015). MAS has been found to work efficiently for transferring genes from pyramided lines into new plants and into the improved varieties (Magar et al., 2014).
Breeders have used marker-assisted selection to “pyramid” resistance conferred by several separate resistance genes/QTLs with the help of closely linked markers against diseases such as BB, RB, and gall midge in rice, leaf rust resistance and powdery mildew resistance in wheat, and insect pest resistance in cotton, as well as for several abiotic stresses such as submergence, salinity, drought, and cold stress (Table 1) into rice, wheat, and cotton (Das and Rao, 2015; Pradhan et al., 2015; Suh et al., 2015; Shamsudin et al., 2016).
To get the desired population with required gene combinations without unwanted genes, backcrossing with the recurrent parent is required. The use of molecular markers which were unlinked to the assembled genes/QTLs, for back ground selection enhances the proportion of recovery of the recipient genome (Figures 1A,B). The gene pyramiding scheme can be distinguished into two parts, development of a pedigree, which is designed to accumulate all target genes in a single genotype known as the root genotype, and a fixation step, which is intended to fix the target genes into a homozygous state to derive the ideal genotype from one single genotype (Figure 1B).
Prospects of molecular methods managing stresses affecting rice productivity
Abiotic stress
Abiotic and biotic (Table 1, Figure 1C) stresses are the two major constraints accountable for the declination of growth and productivity of rice varieties (Wani and Sah, 2014). Abiotic stress (Table 1, Figure 1C) influence the distribution of plant species, survival rate, and the yields of the crop (Lee et al., 2005). Each year, about 25% of the rice croplands worldwide are submerged by flash floods (Loo et al., 2015). With a number of minor QTLs, submergence tolerance in rice is controlled by a single major QTL on chromosome 9 that provides complete submergence tolerance for up to 14 days or more (Septiningsih et al., 2009).
Sensitivity of rice to salt-stress changes throughout the lifecycle, but the effects are most severe in the seedling and reproductive stages (Thitisaksakul et al., 2015). Several salt-resistant rice varieties have been produced by expressing salt-responsive genes. QTLs associated with salt tolerant rice varieties can be mapped using microsatellite markers (Singh et al., 2007). Use of molecular breeding techniques have been shown to be the most efficient tools for development of improved varieties tolerable to salt (Mondal et al., 2013). In the salinity-tolerant cultivar NonaBokra, mapping of SKC1 on chromosome1 was a breakthrough that preserved K+ ion homeostasis under salinity conditions (Ren et al., 2005; Das et al., 2015).
Significant improvements have been made in mapping QTLs for drought resistance traits in rice; however, few have been effectively used in marker-assisted breeding (Prince et al., 2015). A number of drought tolerant QTLs for rice have been identified (Huang et al., 2014). DREB transcription factors play a major role in induction of the expression of genes involved in drought stress, and the genes encoding DREB transcription factors exhibit significant enhancement of the response of plants to drought stress (Udvardi et al., 2007). Several approaches have been improved to enable identification of low temperature stress resistant rice varieties. Cold tolerance of rice in the seedling stage is controlled by multiple genes and several QTLs (Zhang et al., 2014). In 2014, Xu and Cai reported that the Ran gene, OsRAN1, is necessary for the development of cold tolerant rice varieties. Pyramiding of cold resistance QTLs using MAS is useful for improvement of new rice cultivars with cold tolerance (Shinada et al., 2014).
Biotic stress
Biotic stresses in rice (Table 1, Figure 1C) are caused by insects, including the gall midge and brown plant hopper, and by diseases including bacterial blight, blast, and sheath blight. Asian rice Gall-midge (GM), Orseolia oryzae (wood-mason), is a serious pest of rice in China, India, Sri Lanka, and several other countries (Katiyar et al., 2000). So far 11 GM resistance genes have been acknowledged in various rice varieties, Gm1, Gm2, gm3, Gm4, Gm5, Gm6, Gm7, Gm8, Gm9, Gm10, and Gm11(t) (Dutta et al., 2014; Das and Rao, 2015; Hasan et al., 2015; Bentur et al., 2016). The brown plant hopper (BPH), Nilaparvata lugens, has been one of the most devastating pests to rice crops in Vietnam and Asia. There is successful report of the use of SSR and STS markers in pyramiding two BPH resistance genes Bph14 and Bph15 into three elite japonica varieties Shengdao 15, Shengdao 16, Xudao 3 using marker assisted backcross breeding program (Xu, 2013).
Rice cultivation across tropical and semi-tropical regions of the world is affected by bacterial blight (BB) disease caused by Xanthomonas oryzae pv. oryzae (Xoo). A total of 38 R genes of BB have been identified in rice (Khan et al., 2014). Resistant cultivars with one or two major resistant genes are unsustainable in the field and the only way to delay such a breakdown of BB resistance is to pyramid many resistance genes using MAS (Rafique et al., 2010).
Rice sheath blight disease caused by Rhizoctonia solani Kuhn reduces trivial yield in rice-growing areas around the globe (Yellareddygari et al., 2014; Yadav et al., 2015). Genetic studies have shown that SB resistance can be controlled by polygenic QTLs. Using MAS it is possible to pyramid SB resistance QTLs into rice varieties. Rice blast disease (RB), is generally considered as the most important rice disease worldwide (Divya, 2013; Miah et al., 2013). RB is caused by a filamentous heterothallic ascomycetous fungus, Pyricularia grisea, which is known as Magnaporthe grisea (Hebert) Barr. in its sexual state (Divya, 2013). Rice tungro (RT) disease consists of a spherical RNA virus (RTSV) and a DNA bacilliform virus (RTBV). The green leafhopper (GLH), Nephotettix virescens, is the most proficient vector of rice RT virus disease.
Conclusions
There are several abiotic and biotic stresses that affect the productivity of rice cultivation throughout the world. To meet the demands of the growing population, there is urgent need to protect rice plants from various abiotic and biotic stresses that reduce yield and quality. Presently, the conventional breeding of rice is rapidly advancing due to the integration of molecular markers and MAS techniques. Through the application of molecular markers with the help of MAS in gene pyramiding, multiple stress resistant genes could be incorporated into a single rice variety with high yield, abiotic stress tolerance and biotic stress resistance with enhanced nutritional quality.
Author contributions
GD and JP collected literature and wrote the manuscript. KB edited the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant from the Systems and Synthetic Agro-biotech Center through the Next-Generation BioGreen 21 Program (PJ011117), Rural Development Administration, South Korea.
References
- Abe Y., Mieda K., Ando T., Kono I., Yano M., Kitano H., et al. (2010). The Small and round seed1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes Genet. Syst. 85, 327–339. 10.1266/ggs.85.327 [DOI] [PubMed] [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. 10.1126/science.1113373 [DOI] [PubMed] [Google Scholar]
- Batley J., Barker G., O'Sullivan H., Edwards K. J., Edwards D. (2003). Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol. 132, 84–91. 10.1104/pp.102.019422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentur J. S., Rawat N., Divya D., Sinha D. K., Agarrwal R., Atray I., et al. (2016). Rice-gall midge interactions: battle for survival. J. Insect Physiol. 84, 40–49. 10.1016/j.jinsphys.2015.09.008 [DOI] [PubMed] [Google Scholar]
- Biradar S. K., Sundaram R. M., Thirumurugan T., Bentur J. S., Amudhan S., Shenoy V. V., et al. (2004). Identification of flanking SSR markers for a major rice gall midge resistance gene Gm1 and their validation. Theor. Appl. Genet. 109, 1468–1473. 10.1007/s00122-004-1763-0 [DOI] [PubMed] [Google Scholar]
- Boopathi M. (2012). Genetic Mapping and Marker Assisted Selection: Basics, Practice and Benefits. Dordrecht; London: Springer Science and Business Media. [Google Scholar]
- Borba T. C. D. O., Brondani R. P. V., Breseghello F., Coelho A. S. G., Mendonça J. A., Rangel P. H. N., et al. (2010). Association mapping for yield and grain quality traits in rice (Oryza sativa L.). Genet. Mol. Biol. 33, 515–524. 10.1590/S1415-47572010005000065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. C., Kwon S. J., Choi I. S., Lee S. K., Jeon J. S., Oh M. K., et al. (2007). Identification of major blast resistance genes in Korean rice varieties (Oryza sativa L.) using molecular markers. J. Crop Sci. Biotechnol. 10, 265–276. [Google Scholar]
- Choudhary K., Choudhary O. P., Shekhawat N. S. (2008). Marker assisted selection: a novel approach for crop improvement. Am. Eurasian J. Agric. 1, 26–30. [Google Scholar]
- Collard B. C. Y., Mackill D. J. (2008). Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 557–572. 10.1098/rstb.2007.2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G., Rao G. J. N. (2015). Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 6:698. 10.3389/fpls.2015.00698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P., Nutan K. K., Singla-Pareek S. L., Pareek A. (2015). Understanding salinity responses and adopting ‘omics-based’ approaches to generate salinity tolerant cultivars of rice. Front. Plant Sci. 6:712. 10.3389/fpls.2015.00712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Zhu X., Shen Y., He Z. (2006). Genetic characterization and fine mapping of the blast resistance locus Pigm(t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety Theor. Appl. Genet. 113, 705–713. 10.1007/s00122-006-0338-7 [DOI] [PubMed] [Google Scholar]
- Divya K. (2013). Study of genetic diversity in Karnataka rice (Oryza Sativa) landraces using trait specific Simple Sequence Repeat (SSR) Markers. Int. J. Thesis Projects Disser. 1, 45–70. [Google Scholar]
- Dokku P., Das K. M., Rao G. J. N. (2013). Genetic enhancement of host plant-resistance of the Lalat cultivar of rice against bacterial blight employing marker-assisted selection. Biotechnol. Lett. 35, 1339–1348. 10.1007/s10529-013-1212-8 [DOI] [PubMed] [Google Scholar]
- Dubey M., Chandel G. (2010). In silico survey and characterization of resistance gene analogues (rgas) in the genomic regions encompassing gall midge resistance genes gm4 and gm5 in rice (Oryza sativa L.). Plant Omics J. 3, 140–148. [Google Scholar]
- Dutta S. S., Divya D., Durga R. C. V., Reddy D. T., Visalakshmi V., Cheralu C., et al. (2014). Characterization of gall midge resistant rice genotypes using resistance gene specific markers. J. Exp. Biol. Agric. Sci. 2, 339–446. [Google Scholar]
- Endang M. S., Alvaro M. P., Darlene L. S., Chirravuri N. N., Georgina V. V., Sigrid H., et al. (2009). Development of submergence-tolerant rice cultivars: the Sub1 locus and beyond. Ann. Bot. Lond. 103, 151–160. 10.1093/aob/mcn206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endang M. S., Darlene L. S., Singh N., Pamella M. D., Pamplona S. A. M., Heuer S., et al. (2012). Identifying novel QTLs for submergence tolerance in rice cultivars IR72 and Madabaru. Theor. Appl. Genet. 124, 867–874. 10.1007/s00122-011-1751-0 [DOI] [PubMed] [Google Scholar]
- Fukuoka S., Saka N., Koga H., Ono K., Shimizu T., Ebana K., et al. (2009). Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325, 998–1001. 10.1126/science.1175550 [DOI] [PubMed] [Google Scholar]
- Gao L., Jia J., Kong X. (2016). A SNP-based molecular barcode for characterization of common wheat. PLoS ONE 11:e0150947. 10.1371/journal.pone.0150947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J., Ratnaparkhe M. B., Powell W. (2008). Molecular Mapping and Breeding of Physiological Traits. Scottish Crop Research Institute; Division of Plant Sciences, University of Missouri-Columbia, Dundee.
- Hasan M. M., Rafii M. Y., Ismail M. R., Mahmood M., Rahim H. A., Alam M. A., et al. (2015). Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol. Biotechnol. Equip. 29, 237–254. 10.1080/13102818.2014.995920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Hashimoto N., Daigen M., Ashikawa I. (2004). Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor. Appl. Genet. 108, 1212–1220. 10.1007/s00122-003-1553-0 [DOI] [PubMed] [Google Scholar]
- Huang L., Zhang F., Zhang F., Wang W., Zhou Y., Fu B., et al. (2014). Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genomics 15:1026. 10.1186/1471-2164-15-1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Kumar A., Srivastava M. N., Nair S. (2004). Tagging and mapping of a rice gall midge resistance gene, Gm8, and development of SCARs for use in marker-aided selection and gene pyramiding. Theor. Appl. Genet. 109, 1377–1384. 10.1007/s00122-004-1774-x [DOI] [PubMed] [Google Scholar]
- Jairin J., Teangdeerith S., Leelagud P., Kothcharerk J., Sansen K., Yi M., et al. (2009). Development of rice introgression lines with brown planthopper resistance and KDML105 grain quality characteristics through marker-assisted selection. Field Crops Res. 110, 263–271. 10.1016/j.fcr.2008.09.009 [DOI] [Google Scholar]
- Jena K. K., Kim S. M., Suh J. P., Kim Y. G. (2010). Development of Cold-Tolerant Breeding Lines Using QTL Analysis in Rice. Second Africa Rice Congress; Innovation and Partnerships to Realize Africa's Rice Potential, Bamako.
- Joneydi M. S. (2012). Factors affecting in sustainability of agricultural production systems in Iran. Ann. Biol. Res. 3, 4578–4583. [Google Scholar]
- Jordan I. K., Rogozin I. B., Glazko G. V., Koonin E. V. (2003). Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19, 68–72. 10.1016/S0168-9525(02)00006-9 [DOI] [PubMed] [Google Scholar]
- Katiyar S. K., Chandel G., Tan Y., Zhang Y., Huang B., Nugaliyadde L., et al. (2000). Biodiversity of Asian rice gall midge (Orseolia oryzae Wood Mason) from five countries examined by AFLP analysis. Genome 43, 322–332. [PubMed] [Google Scholar]
- Kearsey M. J., Farquhar A. G. L. (1998). QTL analysis in plants, where are we now? Heredity 80, 137–142. 10.1046/j.1365-2540.1998.00500.x [DOI] [PubMed] [Google Scholar]
- Khan M. A., Awan I. U., Zafar J. (2009). Energy requirement and economic analysis of rice production in western part of Pakistan. Soil Environ. 28, 60–67. [Google Scholar]
- Khan M. A., Naeem M., Iqbal M. (2014). Breeding approaches for bacterial leaf blight resistance in rice (Oryza sativa L.), current status and future directions. Eur. J. Plant Pathol. 139, 27–37. 10.1007/s10658-014-0377-x [DOI] [Google Scholar]
- Khan M. H., Dar Z. A., Dar S. A. (2015). Breeding strategies for improving rice yield-a review Agric. Sci. 6, 467–478. 10.4236/as.2015.65046 [DOI] [Google Scholar]
- Khondker S., Sebastian L. S., Pastor H. M., Cruz A. A., Tabanao D. A. (2005). Phenotyping backcrossed rice with pyramided transgenes for resistance to bacterial blight & tungro diseases. Philip. J. Crop Sci. 30, 29–35. [Google Scholar]
- Kim Y. S., Kim S., Shin S. Y., Park T. H., Park H. M., Kim Y. H., et al. (2014). Overexpression of dehydroascorbate reductase confers enhanced tolerance to salt stress in rice plants (Oryza sativa L. japonica). J. Agron. Crop Sci. 200, 444–456. 10.1111/jac.12078 [DOI] [Google Scholar]
- Kottapalli K., Lakshmi N. M., Jena K. (2010). Effective strategy for pyramiding three bacterial blight resistance genes into fine grain rice cultivar, Samba mahsuri, using sequence tagged site markers. Biotechnol. Lett. 32, 989–996. 10.1007/s10529-010-0249-1 [DOI] [PubMed] [Google Scholar]
- Kottearachchi N. S. (2013). Utility of DNA Markers in rice breeding. Eur. Int. J. Sci. Technol. 2, 111–122. [Google Scholar]
- Kumaravadivel N., Uma M. D., Saravanan P. A., Suresh H. (2006). Molecular marker-assisted selection and pyramiding genes for gall midge resistance in rice suitable for Tamil Nadu Region, in ABSTRACTS- 2nd International Rice Congress (New Delhi: ), 257. [Google Scholar]
- Kurokawa Y., Nodaa T., Yamagatac Y., Angeles-Shima R., Sunoharaf H., Ueharaa K., et al. (2016). Construction of a versatile SNP array for pyramiding useful genes of rice. Plant Sci. 242, 131–139. 10.1016/j.plantsci.2015.09.008 [DOI] [PubMed] [Google Scholar]
- Lee S. C., Lee M. Y., Kim S. J., Jun S. H., An G., Kim S. R. (2005). Characterization of an abiotic stress-inducible dehydrin gene, OsDhn1, in rice (Oryza sativa L.). Mol.Cells 19, 212–218. [PubMed] [Google Scholar]
- Lin R., Zhao W., Meng X., Wang M., Peng Y. (2007). Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci. 172, 120–130. 10.1016/j.plantsci.2006.07.019 [DOI] [Google Scholar]
- Liu G., Jia Y., Correa-Victoria F. J., Prado G. A., Yeater K. M., McClung A., et al. (2009). Mapping quantitative trait loci responsible for resistance to sheath blight in rice. Phytopathology 99, 1078–1084. 10.1094/PHYTO-99-9-1078 [DOI] [PubMed] [Google Scholar]
- Loo Y. Y., Billa L., Singh A. (2015). Effect of climate change on seasonal monsoon in Asia and its impact on the variability of monsoon rainfall in Southeast Asia. Geosci. Front. 6, 817–823. 10.1016/j.gsf.2014.02.009 [DOI] [Google Scholar]
- Ma B. J., Wang W. M., Zhao B., Zhou Y. L., Zhu L., Zhai W. (1999). Studies of PCR marker for the rice bacterial blight resistance gene Xa-4. Hereditas 21, 9–12. [Google Scholar]
- Magar M. M., Rani C. V. D., Anuradha G. (2014). Marker assisted selection for bacterial leaf blight resistance in segregating populations of Cottondora sannalu. Int. J. Appl. Sci. Biotechnol. 2, 229–237. 10.3126/ijasbt.v2i3.10570 [DOI] [Google Scholar]
- Mammadov J., Aggarwal R., Buyyarapu R., Kumpatla S. (2012). SNP Markers and their impact on plant breeding. Int. J. Plant Genomics 2012:728398. 10.1155/2012/728398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch S. R., Wright M. H., Tung C. W., Maron L. G., McNally K. L., Fitzgerald M., et al. (2016). Open access resources for genome-wide association mapping in rice. Nat. Commun. 7:10532 10.1038/ncomms10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miah G., Rafii M. Y., Ismail M. R., Puteh A. B., Rahim H. A., Asfaliza R., et al. (2013). Blast resistance in rice: a review of conventional breeding to molecular approaches. Mol. Biol. Rep. 40, 2369–2388. 10.1007/s11033-012-2318-0. [DOI] [PubMed] [Google Scholar]
- Mondal U., Khanom M. S. R., Hassan L., Begum S. N. (2013). Foreground selection through SSRs markers for the development of salt tolerant rice variety. J. Bangladesh Agril. Univ. 11, 67–72. 10.3329/jbau.v11i1.18215 [DOI] [Google Scholar]
- Nandi S., Subudhi P. K., Senadhira D., Manigbas N. L., Sen-Mandi S., Huang N. (1997). Mapping QTLs for submergence tolerance in rice by AFLP analysis and selective genotyping. Mol. Gen. Genet. 255, 1–8. 10.1007/s004380050468 [DOI] [PubMed] [Google Scholar]
- Narciso J., Hossain M. (2002). In: IRRI World Rice Statistics. Available online at: www.irri.org/science/ricestat
- Nejad G. M., Arzani A., Rezai A. M., Singh R. K., Gregorio G. B. (2008). Assessment of rice genotypes for salt tolerance using microsatellite markers associated with the saltol QTL. Afr. J. Biotechnol. 7, 730–736. [Google Scholar]
- Nejad G. M., Singh R. K., Arzani A., Rezaie A. M., Sabouri H., Gregorio G. B. (2010). Evaluation of salinity tolerance in rice genotypes. Int. J. Plant Prod. 4, 199–208. [Google Scholar]
- Nogoy F. M., Song J. Y., Ouk S., Rahimi S., Kwon S. W., Kang K. K., et al. (2016). Current applicable DNA markers for marker assisted breeding in abiotic and biotic stress tolerance in rice (Oryza sativa L.). Plant Breed. Biotechnol. 4, 271–284. 10.9787/PBB.2016.4.3.271 [DOI] [Google Scholar]
- Peng Y., Hu Y., Mao B., Xiang H., Shao Y., Pan Y., et al. (2016). Genetic analysis for rice grain quality traits in the YVB stable variant line using RAD-seq. Mol. Genet. Genomics 291, 297–307. 10.1007/s00438-015-1104-9 [DOI] [PubMed] [Google Scholar]
- Perumalsamy S., Bharani M., Sudha M., Nagarajan P., Arul L., Sarawathi R., et al. (2010). Functional marker-assisted selection for bacterial leaf blight resistance genes in rice (Oryza sativa L.). Plant Breed. 129, 400–406. 10.1111/j.1439-0523.2009.01705.x [DOI] [Google Scholar]
- Porter B. W., Chittoor J. M., Yano M., Sasaki T., White F. F. (2003). Development and mapping of markers linked to the rice bacterial blight resistance Gene, Xa7. Crop Sci. 43, 1484–1492. 10.2135/cropsci2003.1484 [DOI] [Google Scholar]
- Prabhu A. S., Filippi M. C., Silva G. B., Silva-Lobo V. L., Morais O. P. (2009). An unprecedented outbreak of rice blast on a newly released cultivar BRS Colosso in Brazil, in Advances in Genetics, Genomics and Control of Rice Blast, eds Wang G. L., Valent B. (Dordrecht: Springer Science; ), 257–267. [Google Scholar]
- Pradhan S. K., Nayak D. K., Mohanty S., Behera L., Barik S. R., Pandit E., et al. (2015). Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 8, 1–14. 10.1186/s12284-015-0051-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince S. J., Beena R., Gomez S. M., Senthivel S., Babu R. C. (2015). Mapping consistent Rice (Oryza sativa L.) yield QTLs under drought stress in target rainfed environments. Rice 25, 1–13. 10.1186/s12284-015-0053-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Pang Y., Yuan Z., Xing D., Xu J., Dingkuhn M., et al. (2015). Genome-wide association study of grain appearance and milling quality in a worldwide collection of indica rice germplasm. PLoS ONE 10:e0145577. 10.1371/journal.pone.0145577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafique M. Z., Zia M., Rashid H., Chaudhary M. F., Chaudhry Z. (2010). Comparison of transgenic plant production for bacterial blight resistance in Pakistani local rice (Oryza sativa L.) cultivars. Afr. J. Biotechnol. 9, 1892–1904. 10.5897/AJB09.868 [DOI] [Google Scholar]
- Ramegowda V., Senthil-Kumar M. (2015). The interactive effects of simultaneous biotic and abiotic stresses on plants: mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 176, 47–54. 10.1016/j.jplph.2014.11.008 [DOI] [PubMed] [Google Scholar]
- Rao Y., Li Y., Qian Q. (2014). Recent progress on molecular breeding of rice in China. Plant Cell Rep. 33, 551–564. 10.1007/s00299-013-1551-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy C. S., Babu A. P., Swamy B. P. M., Kaladhar K., Sarala N. (2009). ISSR marker based on GA and AG repeats reveal genetic relationship among rice varieties tolerant to drought, flood, salinity. J. Zhejiang Univ. Sci. B 10, 133–141. 10.1631/jzus.B0820183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif J. C., Melchinger A. E., Xia X. C., Warburton M. L., Hoisington D. A., Vasal S. K., et al. (2003). Use of SSRs for establishing heterotic groups in subtropical maize. Theor. Appl. Genet. 107, 947–957. 10.1007/s00122-003-1333-x [DOI] [PubMed] [Google Scholar]
- Ren Z. H., Gao J. P., Li L. G., Cai X. L., Huang W., Chao D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37, 1141–1146. 10.1038/ng1643 [DOI] [PubMed] [Google Scholar]
- Septiningsih E. M., Pamplona A. M., Sanchez D. L., Neeraja C. N., Vergara G. V., Heuer S., et al. (2009). Development of submergence-tolerant rice cultivars: the Sub1 locus and beyond. Ann. Bot. 103, 151–160. 10.1093/aob/mcn206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsudin N. A. A., Swamy B. P. M., Ratnam W., Cruz M. T. S., Raman A., Kumar A. (2016). Marker assisted pyramiding of drought yield QTLs into a popular Malaysian rice cultivar, MR219. BMC Genet. 17:30. 10.1186/s12863-016-0334-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanti M. L., Shenoy V. V., Devi G. L., Kumar V. M., Premalatha P., Kumar G. N., et al. (2010). Marker-assisted breeding for resistance to bacterial leaf blight in popular cultivar and parental lines of hybrid rice. J. Plant. Pathol. 92, 495–501. 10.4454/jpp.v92i2.194 [DOI] [Google Scholar]
- Shin S. H., Kim K. Y., Shin M. S., Ko J. K., Kim C. K. (2007). Identification of rice varieties containing bacterial blight resistance gene Xa1 by SNP marker (16PFXa). Korean J. Breed. Sci. 39, 63–69. [Google Scholar]
- Shinada H., Iwata N., Sato T., Fujino K. (2014). QTL pyramiding for improving of cold tolerance at fertilization stage in rice. Breed. Sci. 63, 483–488. 10.1270/jsbbs.63.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., Konishi S., et al. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028. 10.1038/ng.169 [DOI] [PubMed] [Google Scholar]
- Singh A. K., Gopalakrishnan S., Singh V. P., Prabhu K. V., Mohapatra T., Singh N. K., et al. (2011). Marker assisted selection: a paradigm shift in Basmati breeding. Indian J. Genet. 71, 120–128. [Google Scholar]
- Singh R. K., Gregorio G. B., Jain R. K. (2007). QTL mapping for salinity tolerance in rice. Physiol. Mol. Biol. Plants 13, 87–99. [Google Scholar]
- Singh V. K., Singh A. T., Singh S. P., Llur R. A. K. E., Singh D. E., Krishnan S. G. O., et al. (2013). Marker-assisted simultaneous but stepwise backcross breeding for pyramiding blast resistance genes Piz5 and Pi54 into an elite Basmati rice restorer line ‘PRR78.’ Plant Breed. 132, 486–495. 10.1111/pbr.12077 [DOI] [Google Scholar]
- Song X. J., Huang W., Shi M., Zhu M. Z., Lin H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase, Nat. Genet. 39, 623–630. 10.1038/ng2014 [DOI] [PubMed] [Google Scholar]
- Suh J. P., Cho Y. C., Won Y. J., Ahn E. K., Baek M. K., Kim M. K., et al. (2015). Development of resistant gene-pyramided Japonica rice for multiple biotic stresses using molecular marker-assisted selection. Plant Breed. Biotechnol. 3, 333–345. 10.9787/PBB.2015.3.4.333 [DOI] [Google Scholar]
- Suh J. P., Jeung J. U., Noh T. H., Cho Y. C., Park S. H., Park H. S., et al. (2013). Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 6, 1–11. 10.1186/1939-8433-6-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. H., Chun-Ming W., Chang-Chao S. U., Yu-Qiang L. I. U., Hu-Qu Z. H. A. I., Jian-Min W. A. N. (2006). Mapping and marker-assisted selection of a brown planthopper resistance gene bph2 in rice (Oryza sativa L.). Acta Genet. Sin. 33, 717–723. 10.1016/S0379-4172(06)60104-2 [DOI] [PubMed] [Google Scholar]
- Swamy P., Panchbhai A. N., Priti D., Vaishali N., Panchbhai S. D., Zehr U. B., et al. (2006). Evaluation of bacterial blight resistance in rice lines carrying multiple resistance genes and Xa21 transgenic lines. Curr. Sci. 90, 818–824. [Google Scholar]
- Tao C., Hao W., Ya-dong Z., Zhen Z., Qi-yong Z., Li-hui Z., et al. (2016). Genetic improvement of Japonica rice variety Wuyujing 3 for stripe disease resistance and eating quality by pyramiding Stv-bi and Wx-mq. Rice Sci. 23, 69–77. 10.1016/j.rsci.2016.02.002 [DOI] [Google Scholar]
- Takano-Kai N., Jiang H., Kubo T., Sweeney M., Matsumoto T., Kanamori H., et al. (2009). Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182, 1134–1323. 10.1534/genetics.109.103002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitisaksakul M., Tananuwong K., Shoemaker C. F., Chun A., Tanadul O., Labavitch J. M., et al. (2015). Effects of timing and severity of salinity stress on rice (Oryza sativa L.) yield, grain composition, and starch functionality. J. Agric. Food Chem. 63, 2296–2304. 10.1021/jf503948 [DOI] [PubMed] [Google Scholar]
- Udvardi M. K., Kakar K., Wandrey M., Montanari O., Murray J., Andriankaja A., et al. (2007). Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiol. 144, 538–549. 10.1104/pp.107.098061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani S. H., Sah S. K. (2014). Biotechnology and abiotic stress tolerance in rice. J. Rice Res. 2, 1–2. 10.4172/jrr.1000e105 [DOI] [Google Scholar]
- Werner K., Friedt W., Ordon F. (2005). Strategies for pyramiding resistance genes against the barley yellow mosaic virus complex (BaMMV, BaYMV, BaYMV-2). Mol. Breed. 16, 45–55. 10.1007/s11032-005-3445-2 [DOI] [Google Scholar]
- Wijerathna Y. M. A. M. (2015). Marker assisted selection: biotechnology tool for rice molecular breeding. Adv. Crop Sci. Technol. 3:187 10.4172/2329-8863.1000187 [DOI] [Google Scholar]
- Xu J. (2013). Pyramiding of two BPH resistance genes and Stv-bi gene using marker-assisted selection in japonica rice. Crop Breed. Appl. Biotechnol. 13, 99–106. 10.1590/S1984-70332013000200001 [DOI] [Google Scholar]
- Yadav S., Anuradha G., Kumar R. R., Vemireddy L. R., Sudhakar R., Donempudi K., et al. (2015). Identification of QTLs and possible candidate genes conferring sheath blight resistance in rice (Oryza sativa L.). Springer Plus 4:175. 10.1186/s40064-015-0954-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadavalli V., Narwane G. P., Nagarajan P., Bharathi M. (2011). Identification of coupling- and repulsion-phase markers in rice for brown plant hopper resistance genes using F2S of IR 50 X CO 46. J. Plant Breed. Crop Sci. 3, 1–7. [Google Scholar]
- Ye C., Tenorio F. A., Argayoso M. A., Laza M. A., Koh H. J., Redona E. D., et al. (2015). Identifying and confirming quantitative trait loci associated with heat tolerance at flowering stage in different rice populations. BMC Genet. 16:41. 10.1186/s12863-015-0199-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellareddygari S. K. R., Reddy M. S., Kloepper J. W., Lawrence K. S., Fadamiro H. (2014). Rice sheath blight: a review of disease and pathogen management approaches. J. Plant Pathol. Microb. 5:241 10.4172/2157-7471.1000241 [DOI] [Google Scholar]
- Yu H., Xie W., Li J., Zhou F., Zhang Q. (2014). A whole-genome SNP array (RICE6K) for genomic breeding in rice. Plant Biotechnol. J. 12, 28–37. 10.1111/pbi.12113 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Chen Q., Wang S., Hong Y., Wang Z. (2014). Rice and cold stress: methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 7, 1–12. 10.1186/s12284-014-0024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Wang J. K., Yi Q., Wang Y. Z., Zhu Y. G., Zhang Z. H. (2007). Quantitative trait loci for seedling vigor in rice under field conditions. Field Crop. Res. 100, 294–301. 10.1016/j.fcr.2006.08.003 [DOI] [Google Scholar]