Abstract
Disorders of pain neural systems are frequently chronic and, when recalcitrant to treatment, can severely degrade the quality of life. The pain pathway begins with sensory neurons in dorsal root or trigeminal ganglia and the neuronal subpopulations that express the TRPV1 ion channel transduce sensations of painful heat and inflammation, and play a fundamental role in clinical pain arising from cancer and arthritis. In the present study we elucidate the complete transcriptomes of neurons from the TRPV1 lineage and a non-TRPV1 neuro-glial population in sensory ganglia through the combined application of next-gen deep RNA-Seq, genetic neuronal labeling with fluorescence-activated cell sorting, or neuron-selective chemoablation. RNA-Seq accurately quantitates gene expression, a difficult parameter to determine with most other methods especially for very low and very high expressed genes. Differentially expressed genes are present at every level of cellular function from the nucleus to the plasma membrane. We identified many ligand receptor pairs in the TRPV1 population suggesting that autonomous presynaptic regulation maybe a major regulatory mechanism in nociceptive neurons. The data define, in a quantitative, cell population specific fashion, the molecular signature of a distinct and clinically important group of pain-sensing neurons and provide an overall framework for understanding the transcriptome of TRPV1 nociceptive neurons.
Keywords: pain, nociception, RNA-Seq, transcriptome, dorsal root ganglion, C-fibers, Aδ fibers, TRPV1, capsaicin, resiniferatoxin, neuropeptides, autocrine, paracrine, GPCR, TRP-channels, bradykinin, Cre, glutamate, kainate, AMPA receptors, metabotropic glutamate receptors, cholinergic receptors, potassium channels, maxi K+ channels, natriuritic polypeptide B, Nppb, galanin, somatostatin, GDNF, BDNF, NGF, MCP1, CCL1, CCL2, TrkA, TrkB, TrkC, c-kit, c-ret, synaptoneurin, Piezo, Vglut1, P2X3, neurexophillin, neurexin, cerebellin, P2Y receptors, P2X3 receptors, ectonucleotidases, adenosine receptors, purinergic receptors, CDK5, tyrosine hydroxylase, hippocalcin, NAV1.7, NAV1.8, NAV1.9, Hox genes, homeobox
INTRODUCTION
Detailed investigations of the molecular biology of nociceptive neurons are key elements for advancing basic knowledge of pain-sensing neural circuits and translational investigations for pain control, but their complete molecular repertoire, their transcriptome, has not been fully ascertained. The subset of nociceptive Aδ and C-fiber primary afferents that express the TRPV1 ion channel transduce signals arising from exposure to painful heat, inflammatory and chemical stimuli, and low pH.63,91 Chemoablation of these neurons and/or their axonal projections using resiniferatoxin (RTX), a potent vanilloid agonist, rapidly and effectively eliminates nociceptive input.39,68 Loss of TRPV1 neuronal input yields analgesia to acute experimental nociceptive stimuli in rodents or bone cancer pain from osteosarcoma in companion dogs.8 In humans, intrathecal administration of RTX is being evaluated in intractable cancer pain, <http://clinicaltrials.gov/ct2/show/NCT00804154> and preliminary results suggest sustained improvement in pain symptoms.29,31,54 Based on their pivotal role, the precise molecular signature of TRPV1-expressing DRG neurons is hypothesized to be critical for understanding the molecular biology of nociception and the ability to respond to tissue damage and inflammatory clinical pain.
RNA-Seq presents several unique attributes for the investigation of molecular interrelationships,28 although methods including subtraction cloning, SAGE, and microarrays have been evaluated.6,46,103 We examine the TRPV1 neuronal transcriptome by combining RNA-Seq with genetic and chemoablative strategies for enriching/deleting TRPV1 neurons from sensory ganglia. Neurons from bacterial artificial chromosome (BAC)-transgenic, TRPV1-promoter-Cre recombinase mice expressing red fluorescent protein were isolated by fluorescence activated cell sorting (FACS). In the adult, this has the advantage of broadly including the majority of nociceptive modalities, itch, and several other modalities.1,59–62,77,79,104 To provide a contrasting transcriptome, the TRPV1-lineage neurons were eliminated by expressing diphtheria toxin (DTA) using the same BAC-TRPV1-promoter line of mice.62 The cells remaining in the DRG were then subjected to RNA-Seq. To confirm the mouse genetic manipulations, we injected rat trigeminal ganglion with RTX to remove TRPV1-expressing neurons.39 As with the DTA lesion, the ganglionic cells remaining after the RTX lesion were subjected to RNA-Seq. These manipulations yield three differentially-expressed populations of DRG transcripts: (1) TRPV1-lineage (TRPV1L) or (2) TRPV-depleted (TRPV1D) populations, and (3) those equally expressed/common to both groups. These RNA-Seq data outline the beginnings of a “nociceptome” and define critical molecular interrelationships for experimental and in silico analyses.
Here we report multiple new transcripts in the TRPV1L population such as the hippocalcin family of calcium binding proteins and receptor subunits related to nociceptive or analgesic processes.2,74 We also identify interrelated gene families hypothesized to be involved in potential autocrine or paracrine presynaptic molecular microcircuits which can control nerve terminal excitability. For example, transcripts coding for a purinergic ion channel –> adenosine generating enzyme –> G-protein coupled receptor cascade are abundantly present in TRPV1L neurons and may sequentially increase and then decrease nociceptor excitability following tissue damage and ATP release.107 Individually, all three elements in this cascade have been targets of analgesic manipulations or drug development efforts.27,42,89,107 Potential autocrine regulatory loops are revealed by co-expression of neuropeptide-receptor pairs suggesting multiple mechanisms for dynamic, autonomous regulation of nociceptive nerve terminal excitability. These data highlight molecular pathways and mechanisms involved in acute and neuropathic pain, neurological disorders affecting peripheral nerve, and potential routes to new therapies.
METHODS
Animal Studies
Animals were maintained with water and food ad libitum and housed in groups of 2–3 on a 12 hr light-dark cycle. The procedures were approved by the Institutional Animal Care and Use Committee and performed in accordance with the NIH guidelines for the care and use of laboratory animals and the International Association for the Study of Pain.
TRPV1-Lineage DRG Labeling and Isolation
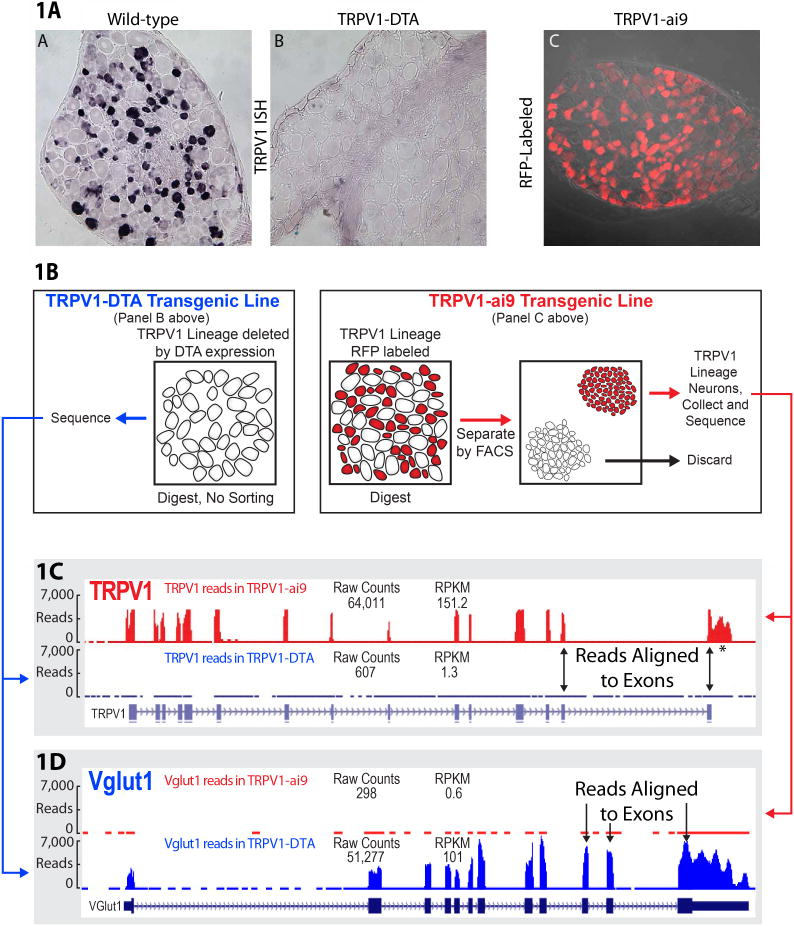
Transgenic mice were engineered to express Cre recombinase under control of the TRPV1 promoter.62 These TRPV1-Cre animals were crossed with the promoter line ai9 ROSA-stop-tdTomato creating TRPV1-ai9 mice whose dorsal root ganglia (DRG) and trigeminal ganglia selectively contained neurons marked with the red fluorescent protein tdTomato (Figure 1A, Panel C). DRG tissue depleted of the TRPV1-lineage population was obtained by Cre-mediated excision of a floxed transcriptional stop 5′ to the diphtheria toxin fragment A coding sequence.62 TRPV1-Cre dependent excision activates DTA expression resulting in the specific ablation of Cre-expressing TRPV1-lineage neurons. Figure 1A panels A and B show TRPV1 in situ hybridization in the wild type and diphtheria toxin expressing ganglion. The TRPV1 lineage neurons have been removed after diphtheria toxin expression (Figure 1A, panel B). Figure 1A panel C shows the red fluorescent protein expression in the TRPV1-lineage neurons. The selectivity of Cre expression has been confirmed using double label in situ hybridization.62 To prepare the material for RNA-Seq (Figure 1B), dorsal root ganglia were dissected from two to three groups of 10 mice from each of the two genotypes (TRPV1-DTA and TRPV1-ai9). The DRGs were dissociated enzymatically, and, for the TRPV1-ai9 mice, the cell suspension was subjected to cell sorting on a FACS Vantage SE (BD Biosciences) at 18 psi. DAPI staining was used for exclusion of dead cells. RFP+ neurons were sorted to >97% purity and were collected and extracted for total RNA. For the TRPV1D mice, the dissociated cells were directly extracted for total RNA. This procedure is shown diagrammatically in Figure 1B. Figure 1C and 1D illustrate two things: read enrichment and read alignment. For each gene in panels C and D, the exon structure is shown below the read panels. Note that the histogram-like accumulation of reads aligns to the exon structure. Figure 1C shows the enrichment of TRPV1 sequences in the RFP-sorted, TRPV1-lineage neurons and the lack of TRPV1 reads in the TRPV1-DTA preparation. In contrast, Figure 1D shows the enrichment of Vglut1 reads in the DTA preparation and the lack Vglut1 reads in the RFP-sorted, TRPV1-lineage neurons. Both of these genes were picked for illustrative purposes because they were very strongly differentially expressed and Vglut1 is a marker for large diameter proprioceptive neurons. For example, in the TRPV1L neuron, 63,000 reads from the deep RNA sequencing runs align with the TRPV1 exons. An exception is indicated with an asterisk, which denotes a 3′ extension and this is discussed further in relationship to Figure 6. These aspects are considered further in the DRG RNA-Seq metrics section.
Figure 1. Assessment of TRPV1 Enrichment Manipulation.
1a. Results of in situ hybridization for TRPV1 are shown for (a) wild type mouse DRG, (b) TRPV1D (TRPV1-DTA), and (c) TRPV1L (TRPV1-ai9) ganglia. Figure 1 B illustrates the procedure for cell deletion using the TRPV1-DTA transgenic mouse line and isolation of the RFP TRPV1L neurons. The panels of Figure 1C and 1D show alignments of raw reads for TRPV1 and Vglut1 (vesicular glutamate uptake, Slc17a7) mRNAs to their exon structures in the TRPV1L neurons and the TRPV1D preparation. Reference sequences are shown below the reads (the asterisk denotes a 3′ extension of the TRPV1 transcript which is analyzed further in Figure 6). Both the alignment and in situ hybridization show that the TRPV1 BAC-transgenic RFP labeling and FACS strongly enriched for TRPV1 and that the DTA lesion is thorough, as almost no TRPV1 transcripts are detectable in ganglia from the TRPV1D mice. Both of these genes were picked for illustrative purposes because they were very strongly differentially expressed. Vglut1 is a marker for large diameter proprioceptive neurons.10 These aspects are considered further in the DRG RNA-Seq metrics section.
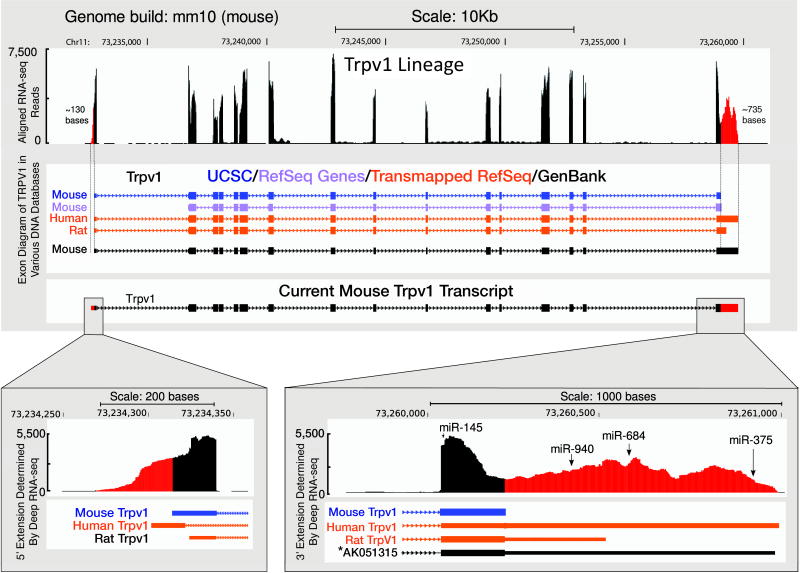
Figure 6. Illustration of Mouse 5′ and 3′ UTR Annotation.
The region in red corresponds to the 5′ and 3′ UTR extensions using the TRPV1L deep sequencing data. The location of three evolutionarily conserved miRNA binding sites are identified in the 3′extension. The region of the mouse genome (mm10) was extracted and aligned with the corresponding region in rat (rn5) and human (hg19) genomes using the UCSC genome table browser. The aligned sequence was used as input by TargetScan to predict the miRNA binding sites. Please note that NCBI Mus musculus Annotation Release 104 now corresponds to the extensions which we illustrate and the exact sequence in SFig3 includes these data.
Intratrigeminal resiniferatoxin injection
The potent TRPV1 agonist resiniferatoxin (RTX) was used to deplete the adult rat sensory ganglion of TRPV1-expressing neurons. RTX activates the TRPV1 ion channel and produces a cytoplasmic calcium overload that selectively kills TRPV1-expressing primary afferent neurons. RTX was stereotaxically injected into the trigeminal ganglion as described earlier (Karai 2004).39 Briefly, the rat is placed in a stereotaxic frame; the coordinates for the intratrigeminal injection were 2.5 mm posterior and 1.5 mm lateral to bregma; the depth is determined during the course of needle placement. The main technical considerations are (a) use of a shallow bevel needle (45° custom ordered from Hamilton), and (b) once the needle gently contacts the base of the skull it is retracted 0.5 mm so the opening of the needle bore is embedded completely within the ganglionic tissue. Ganglia were removed at day 10 after microinjection.
Technical Caveats
Some alterations in gene expression may have occurred as a result of genetic manipulations and/or tissue preparation steps. The acute dissection of ganglion and enzymatic liberation of cell followed by FACS may have induced some genes. The FACS sorting is a relatively brief procedure (10 minutes) although the pressure for running the sorter is a critical variable. For example PSIs above 20 tended to lyse the cells and reduce or completely eliminate intact RNA. The microinjection may also have resulted in some alterations related to the lesion that were not fully resolved by day 10 when we sampled the tissue.
RNA Isolation
Rats were euthanized by CO2 exposure. The trigeminal ganglia were removed from the base of the skull and the L4 and L5 DRGs were removed after laminectomy. Tissues were frozen immediately in dry ice, and stored at −80°C until processed. Samples were homogenized in l ml Qiazol reagent (Life Technologies, Carlsbad, CA) using a Polytron homogenizer. The RNeasy Mini kit (Qiagen Inc., Valencia, CA, USA) plus an additional step of DNase treatment was used to isolate total RNA. The RNA integrity was assessed by gel electrophoresis on Agilent Bioanalyzer. Samples with a RNA integrity number (RIN) above 8.5 were sequenced.
Sample Preparation and Next-Gen Sequencing
Poly A+ isolation, size fragmentation, cDNA synthesis, size selection, ligation of Illumina (San Diego, CA) paired end adaptors and next-gen sequencing were performed at the NIH Intramural Sequencing Center (NISC) according to their standard protocols. Briefly, Poly-A selected mRNA libraries were constructed from 1 μg total RNA using the Illumina TruSeq RNA Sample Prep V2 Kits according to manufacturer’s instructions except where noted. The cDNAs were fragmented to ~275 bp using a Covaris E210 machine. Amplification was performed using 10 cycles to minimize the risk of over-amplification. Unique barcode adapters were applied to each library. Equal volumes of individual libraries were pooled and run on a MiSeq Illumina. The libraries were then repooled based on the MiSeq demultiplexing results. The pooled libraries (N=4 per pool) were sequenced on two lanes of an Illumina HiSeq2000 using version 3 chemistry. The data were processed using RTA version 1.13.48 and CASAVA 1.8.2. For the TRPV1L and D samples, we sequenced to a depth of approximately 166 and 178 million paired end reads of 100 bases each on the HiSeq 2000. An earlier run on an Illumina Genome Analyzer (GAIIX) provided an average of 36.7 million reads of 35 bases each; the correlation coefficients from the two platforms (after removing unannotated genes and genes expressed below 1 RPKM) were 0.93 and 0.94 (Figure S1).
Mapping and Assembly of RNA-Seq Reads
FastQC (version 0.10.0) was used to run preliminary QC analyses. All reads had an average minimum per base sequence quality score above 28, indicative of a high quality sequencing run, and did not require trimming before being mapped. RNA-Seq unified mapper program (RUM v2.05) was selected to align the 100 base reads to the appropriate genome because of the accuracy and speed of this algorithm over other popular aligners for next-generation sequencing.26 Mouse mm10 and rat rn4 assemblies were used for mapping by RUM yielding three groups of reads: unique, non-unique, and not mapped. A summary of unique and non-unique mapped reads can be viewed in Table S1. The mapped reads were assembled using gene models from RefSeq, UCSC, and Ensembl databases. The UCSC table browser was used to convert the various database accession ids (used as ids in RUM) to gene symbols. A perl script was used to select one instance for each gene from the different isoforms and/or gene models. The selection was based on the number of reads mapped to the gene model, and the database source of the gene model. Preference was given to the model that accounted for the most reads mapped and RefSeq and UCSC databases were given preference over Ensembl. By selecting the gene instance with the highest read count, the major isoform of each gene or the most well supported gene structure was considered for differential expression. While we did not perform extensive analysis of alternate splicing events, we looked at isoforms of the genes in Table S2 (TRPV1L 5X or greater fold difference), and confirmed that, overwhelmingly, the assignment of the gene to either the TRPV1 lineage or depleted group was not affected by the choice of the isoform. There are 269 genes in Table S2 and of those 51 have one or more isoforms. From the genes that have isoforms, only 2 genes (3%) had an isoform with an expression level above 1 RPKM that was opposite from the selected major isoform.
Technical Caveats
The RPKM expression values presented within this manuscript are determined by the mapping tool used and the selected gene model; minor differences in values are expected across different RNA-Seq mapping tools and with the use of different gene models. With poly-A selection, we mainly detect protein coding transcripts. It is noteworthy that while the mouse genome is very well annotated, increasing amounts of RNA-Seq data make genome re-annotation a fluid and continuously evolving process, thus remapping of the RNA-Seq data may result in some new genes being discovered.
Immunohistochemistry: animals were deeply anesthetized with ketamine or pentobarbital and perfused transcardially with cold phosphate buffered saline (PBS) followed by 4% paraformaldehyde. For double labeling, tissues were cryoprotected in 20% sucrose, frozen, and blocks were sectioned (10μ) by Histoserv, Inc (Germantown, MD). Slide mounted sections were washed 3×1 min in HEPES Immunostaining Buffer (HIB) containing 145mM NaCl, 5mM KCl, 1.8mM CaCl2, 0.8mM MgCl2 and 10mM HEPES, pH 7.4, followed by washing 3×1 min in dH2O. Sections were subjected to antigen retrieval by microwaving for 2 min at 300 W, cooling for 1 min, microwaving for an additional 4 min at 180 W and then cooling for 30 min followed by washing 3×1 min in HIB then 3×1 min in dH2O. Sections were blocked with 10% normal goat serum for 1 hr at RT and then washed 3×1 min in dH2O. The sections were then incubated at RT for 1 hr with rabbit anti-TRPV1 antibody in HIB (1:1000, BioReagents). Sections were washed 3×1 min in HIB then 3×1 min in dH2O prior to incubating for 1 hr at RT with Alexa Fluor 647 donkey anti-rabbit antibody in HIB (1:150, Abcam). Sections were washed 3×1 min in HIB then 3×1 min in dH2O prior to coverslipping and imaged using a Zeiss Axio Imager.Z2. Slides were incubated overnight in HIB at 4°C to remove coverslip. To remove the initial primary and fluorescent antibodies, sections were subjected to a second round of antigen retrieval and stained as above with rabbit anti-hippocalcin antibody (1:10,000, Abcam) followed by donkey anti-rabbit FITC (1:150, Jackson). Images from both stainings were aligned based on DAPI to determine co-localization.56
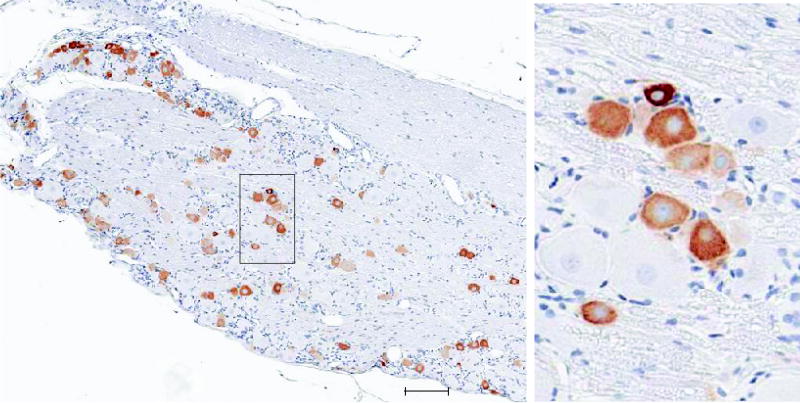
For TRPV1 alone, six micron sections were cut from a paraffin block containing the L4 and L5 dorsal root ganglia. Following deparaffinization and epitope unmasking with Target Retrieval Solution (S1700, Dako, Carpinteria, CA) at 95° C for 20 min, sections were blocked with 10% normal goat serum (Vector Laboratories, Inc., Burlingame, CA) and incubated overnight at 4° C with the primary TRPV1 antibody. Antibody detection was performed using Vectastain Elite Rabbit IgG and Peroxidase Substrate Kits (SK-4700 and SK-4100, Vector Laboratories, Inc., Burlingame, CA). Sites of peroxidase activity were visualized using 3,3-diaminobenzidene tetrahydrochloride (DAB); sections were then counterstained with hematoxylin. Control specimens for the assessment of non-specific binding were processed in an identical way, except for omission of the primary antibody. Histological sections were visualized by scanning with a 20X objective in an Aperio ScanScope slide scanner. A typical section from whole DRG is shown with a subsection enlarged.
5′ and 3′ UTR extension
The region corresponding to the mouse genome (mm10) that was covered by at least 50 bases was extracted and aligned with the corresponding region in rat (rn5) and human (hg19) genomes using the UCSC genome table browser.40 The aligned sequences were used as input into the TargetScan algorithm47 to report evolutionary conservation of miRNA seed regions.
RESULTS
DRG RNA-Seq Metrics
RNA-Seq and Level of Expression
RNA-Seq data are derived from sequencing of nucleic acid and an alignment of the sequence to a reference genome which results in accurate quantitation and unambiguous assignment of gene identity; the process yields highly reproducible results.51,57,66 The data reported here derive from 21 samples (Supplemental Table 1) in four different cohorts, (a) four sequencing runs averaging 34 and 172 million paired end reads of the TRPV1L and TRPV1D mouse Bac transgenic manipulations (Figure 1A), (b) intratrigeminal rat resiniferatoxin cell deletion (N=7, 4 controls and 3 lesioned animals), (c) basal rat DRG (N=9), and (d) a rat “mixed tissue” preparation consisting of equal amounts of total RNA from cerebral cortex, cerebellum, midbrain, hypothalamus, hindbrain, spinal cord, retina, pituitary, heart, liver, lung, kidney, skeletal muscle, small intestine and adrenal gland (113 million paired end reads) for a total of ~2.5 billion individual reads. The treatments and total reads per tissue sequenced are supplied in Supplemental Table 1. The methodological pipeline and criteria used for mapping and filtration are detailed in Supplemental Figure 1 and frequency histograms comparing level of gene expression to number of genes in these different preparations can be found in Supplemental Figure 2. This latter figure allows an appreciation of the overall distribution and number of genes, out of the total of ~12,500 ganglionic genes that are expressed at low, medium, and high levels.
RNA-Seq Unified Mapper (RUM) was used for alignment of sequence reads to the genome; the number of mapped reads is expressed as reads per kilobase of transcript per million bases sequenced (RPKM);26 an example of a typical alignment is seen in Figure 1C and 1D. Genes with >20 raw reads are considered expressed and those with an RPKM of ≥1 are analyzed in this paper. In instances where families of genes are analyzed, low expressed genes are not excluded from discussion. It is recognized that ≥ 1 RPKM is a stringent criterion. A more relaxed criterion would include a portion of the ~3,300 genes expressed between 0.1 and 1.0 RPKM a portion of which are included in a supplemental table (STable7). Behavioral and drug studies suggest that genes with relatively low expression are capable of having a functional impact on nociceptive processes with pharmacological implications. An example is the bradykinin 2 receptor GPCR (Bdkrb2), which is expressed at 1.3 RPKM. Bradykinin is a well-known peptide algesic mediator, and Bdkrb2 antagonists have been tested as analgesic agents.76,87 Thus, a recurrent theme in this paper is that the absolute level of expression may not be a full predictor of functional impact and application of additional genetic targets and labeling will be required to obtain a complete transcriptional matrix of nociceptive neuronal populations.
Mouse DRG
Mapping the 170 million read mouse TRPV1L and TRPV1D ganglionic gene expression data with RUM yielded ~25,000 distinct transcripts. Functionally unannotated and non-coding transcripts were separated out, at which point ~16,200 gene transcripts met the inclusion criterion of 20 raw reads or more. Out of this group, 12,683 transcripts had a RPKM of one or greater, our main criterion for further analysis. RNA-Seq to a depth of 36.7 million unpaired reads on the Illumina GaIIx yielded 12,583 expressed transcripts. The correspondence between the number of transcripts is consistent with data from chicken99 and observations that ultra-high sequencing depth mainly aids detection of non-coding, low-expression RNA’s.93 Taken together, our data indicate that cells of the DRG express about 12,500 well expressed, functionally annotated genes transcripts coding for defined proteins.
Rat DRG
The rat sequencing consisted of 40 ± 5.6 million paired end reads (N=9) and yielded 10,992 distinct transcripts that met the eligibility criteria of >20 raw reads, ≥1 RPKM and annotated in either the Refseq, UC Santa Cruz, or Ensembl databases. Examination of the data after filtration and annotation exclusion showed an apparent lower number of genes (~1500) expressed in rat which reflects the more complete annotation of the mouse genome rather than absence of the transcript in the rat.
Assessment of TRPV1 Enrichment and Depletion
Results of in situ hybridization for TRPV1 are shown in Figure1A for (a) wild-type, (b) TRPV1D, (TRPV1-DTA) and (c) TRPV1L (TRPV1-ai9) mouse DRG. The use of FACS to isolate RFP-labeled neurons yielded strong differential expression: a 100-fold enrichment of TRPV1 transcripts was observed compared to the ganglia in which TRPV1-lineage neurons are eliminated. It is important to note that, because of the sorting, most of the transcripts in the TRPV1L preparation are of neuronal origin. Throughout this paper, we use the terms “differentially” or “predominantly” to mean a higher level of expression in one population vs. another, but frequently the difference in expression is not absolute.
The DTA lesion is thorough and no TRPV1 neurons are detectable in ganglia from these mice. The resulting transcriptome analysis shows that TRPV1 mRNA was nearly absent in the DTA lesioned transcriptome, while other genes are strongly differentially expressed in the TRPV1D ganglia (Figure 1C, 1D). Vglut1 (Slc17a7) a vesicular glutamate transporter is 170-fold differentially expressed in TRPV1D ganglia compared to the TRPV1L neurons (Figure 1D) and immunocytochemistry shows it is mainly localized to large diameter proprioceptive DRG neurons.10 Osteopontin (Spp1) is also strongly differentially expressed in the TRPV1D ganglia and is highly expressed in non-nociceptive muscle spindle afferents as seen with immunocytochemistry.33 Population specific expression extends to developmentally important transcription factors such as short stature homeobox 2 (shox2), which shows a 38-fold enriched expression in the TRPV1D population and is a developmental determinant for a subpopulation of TrkB expressing DRG neurons,82 consistent with TrkB expression in the TRPV1D population (Figure 2). While the neuronal components of the TRPV1D transcriptome are mainly non-nociceptive, this preparation also contains satellite cells, Schwann cells and blood vessels. The same cellular heterogeneity applies to intact ganglia. Thus, in whole rat ganglia and TRPV1D mouse preparation we detect substantial amounts of myelin protein zero and proteolipid myelin protein 22 transcripts from Schwann cells, and even hemoglobin transcripts (Hbb-a1 and -a2) from reticulocytes (Figure S2).
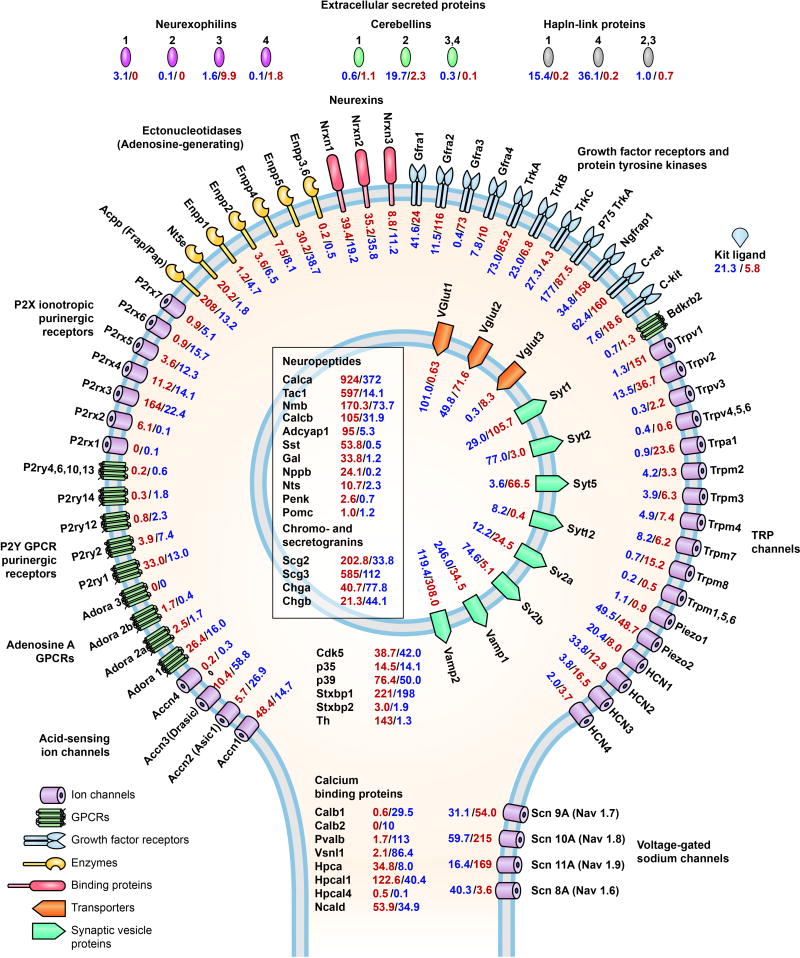
Figure 2. Primary Afferent Nerve Ending.
This figure illustrates a primary afferent nerve ending and shows levels of transcripts coding for families of ion channels, enzymes, GPCRs, growth factor receptors, intracellular and vesicular proteins, neuropeptides, and extracellular linking proteins placed in the cellular locations of the corresponding proteins. The quantitative values for the TRPV1L neurons are in red and the TRPV1D population in blue. In the figure, the outside rim is formed by the plasma membrane and a round synaptic vesicle (light blue) is in the interior. The RPKM values for adenosine, P2Y purinergic/pyrimidine GPCRs, P2X3 purinergic ionotropic receptors, as well ectonucleotidases that can convert ATP, ADP or AMP to adenosine are arrayed on the left side. Neurexins, neurexophilins, cerebellins, and the Halpn extracellular link proteins found in perineuronal nets are at the top. Growth factor tyrosine kinase receptors, TRPA1, the TRPV and TRPM families, putative mechanosensitive Piezo 1 and 2, and the hyperpolarization activated cyclic nucleotide-gated potassium channel (HCN) family of ion channels are on the right side. At the bottom are several voltage gated sodium channels, the families of parvalbumin and hippocalcin calcium binding proteins, and proteins related to cyclin-dependent protein kinase 5 (Cdk5).97 [Gene symbols/names- Nt5e: 5′-ectonucleotidase; Enpp[1,2,4,5,6]: ectonucleotide pyrophosphatase/phosphodiesterase; Gfra[1,2,3,4]: glial cell line derived neurotrophic factor family receptor alpha; TrkA: Ntrk1, neurotrophic tyrosine kinase, receptor, type 1; TrkB: Ntrk2, neurotrophic tyrosine kinase, receptor, type2; TrkC: Ntrk3, neurotrophic tyrosine kinase, receptor, type 3; p75 TrkA: nerve growth factor receptor; Ngfrap1: nerve growth factor receptor associated protein 1; Bdkrb2: bradykinin receptor B2; HCN[1,2,3,4]: hyperpolarization-activated, cyclic nucleotide-gated K+ channel; Vglut1: Slc17a7, solute carrier family 17 (vesicular glutamate transporter), member 7; Vglut2: Slc17a6, solute carrier family 17 (vesicular glutamate transporter), member 6; Vglut3: Slc17a8, solute carrier family 17 (vesicular glutamate transporter), member 8; Syt[1,2,5,12]: Synaptotagmin; Sv2[a,b]: synaptic vesicle glycoprotein 2; Vamp[1,2]: synaptobrevin, vesicle-associated membrane protein; Calca, Calcb: Cgrp[alpha, beta], calcitonin gene-related polypeptide; Tac1: tachykinin 1; Nmb: neuromedin B; Adcyap1: adenylate cyclase activating polypeptide 1 (PACAP); Sst: somatostatin; Gal: galanin; Nppb: natriuretic peptide B; Nts: neurotensin; Penk: preproenkephalin; Pomc: proopiomelanocortin; Scg[2,3]: secretogranin; Chga: chromogranin A; Chgb: chromogranin B (also called secretogranin 1); Cdk5: cyclin-dependent kinase 5; p35: Cdk5 regulatory subunit 1; p39: Cdk5 regulatory subunit 2; Stxbp1: Munc18-1, syntaxin binding protein 1; Stxbp2: Munc18b, syntaxin binding protein 2; Th: tyrosine hydroxylase; Calb[1,2]: calbindin; Pvalb: parvalbumin; Vsnl1: visinin-like 1; Hpca: hippocalcin; Hpcal[1,4]: hippocalcin-like; Ncald: neurocalcin delta
TRPV1L Transcripts: Molecular Signatures of Nociceptive Neurons
Quantitative Aspects and Guide to the Tables
In the following sections, two aspects of gene expression are considered: the absolute level of expression (RPKM) and the fold-differential expression; both facets are captured in Table 1 and Figure 2. We mainly focus on transcripts coding for known or potential mediators of nociceptive transduction or analgesic drug actions. All genes in Table 1 and the supplemental tables are listed by their gene symbol as found in NCBI gene database. However, in the literature they may be identified by many aliases or common names, thus, finding the entry may require accessing the gene menu of NCBI < www.ncbi.nlm.nih.gov/gene/. Additionally, the accession id is provided in the supplemental tables to identify the exact gene isoform that was used in the analysis, which in almost all cases is the main splice variant (see methods). Genes expressed below 1 RPKM are included in a separate tabulation (Table S7). If a particular gene of interest is not found, it either (a) may not be expressed or (b) the annotation may not yet be validated in the current mouse genome build; for mouse the latter is less likely. To facilitate gene searches we supply supplemental Table 8 which is alphabetized based on gene symbol. This table is particularly useful for looking up gene families.
Table 1.
Molecular Signature of TRPV1L Neurons.
| Gene Symbol | Gene Name | Lineage RPKM | Depleted RPKM | Adjusted Fold Change |
|---|---|---|---|---|
| Ligand and Voltage Gated Ion Channels, Channel Subunits and Channel Interacting proteins | ||||
| Scn11a | sodium channel, voltage-gated, type XI, alpha; Nav1.9 | 169.2 | 16.4 | 10.3 |
| P2rx3 | purinergic receptor P2X, ligand-gated ion channel, 3 | 163.6 | 22.4 | 7.3 |
| Trpv1 | transient receptor potential cation channel, subfamily V, member 1 | 151.2 | 1.3 | 105.3 |
| Kcnip4 | a-type potassium channel modulatory protein 4 | 133 | 19.6 | 6.8 |
| Grik1 | glutamate receptor, ionotropic, kainate 1 | 130.8 | 8.6 | 15.1 |
| Kcnip2 | a-type potassium channel modulatory protein 2 | 102.2 | 14.8 | 6.9 |
| Scn3b | sodium channel, voltage-gated, type III, beta | 39.9 | 7.8 | 5.1 |
| Trpc3 | transient receptor potential cation channel, subfamily C, member 3 | 38.9 | 4.8 | 7.9 |
| Kcnh6 | potassium voltage-gated channel, subfamily H (eag-related), member 6, KV11.2 | 30.6 | 2.5 | 11.9 |
| Chrna6 | cholinergic receptor, nicotinic, alpha polypeptide 6 | 30.3 | 4 | 7.4 |
| Trpa1 | transient receptor potential cation channel, subfamily A, member 1 | 23.6 | 0.9 | 22.6 |
| Kcnmb1 | potassium large conductance calcium-activated channel, subfamily M, BKbeta1, Maxi K β1 | 16.3 | 3.6 | 5 |
| Trpm8 | transient receptor potential cation channel, subfamily M, member 8 | 15.2 | 0.7 | 18.4 |
| Kcnc2 | potassium voltage gated channel, Shaw-related subfamily, member 2, KV3.2 | 9.9 | 0.7 | 12.6 |
| Kcnk12 | potassium channel, subfamily K, member 12, THIK2, tandem pore domain K+ channel | 8.9 | 0.8 | 9.9 |
| G-protein Coupled Receptors and Associated Proteins | ||||
| Rgs4 | regulator of G-protein signaling 4 | 606 | 125.2 | 5 |
| Rgs10 | regulator of G-protein signaling 10 | 222.9 | 28.6 | 7.8 |
| Mrgprd | MAS-related GPR, member D | 125.4 | 4.2 | 29.1 |
| Gnb4 | guanine nucleotide binding protein (G protein), beta 4 | 68 | 8.1 | 8.3 |
| F2rl2 | coagulation factor II (thrombin) receptor-like 2; PAR3 | 57.5 | 10.1 | 5.6 |
| Gna14 | guanine nucleotide binding protein, alpha 14 | 53 | 4.9 | 10.6 |
| Lpar3 | lysophosphatidic acid receptor 3 | 45.8 | 1.9 | 22.8 |
| Mrgpra3 | MAS-related GPR, member A3 | 37.4 | 6.3 | 5.8 |
| Rgs3 | regulator of G-protein signaling 3 | 37.4 | 6.5 | 5.7 |
| Mrgprx1 | MAS-related GPR, member X1 | 28.6 | 1.3 | 21.3 |
| Rgs8 | regulator of G-protein signaling 8 | 22.6 | 0.8 | 26 |
| Ptgdr | prostaglandin D receptor | 20 | 1.4 | 13.3 |
| Mrgprb5 | MAS-related GPR, member B5 | 17.3 | 0.7 | 21.9 |
| Gpr35 | G protein-coupled receptor 35 | 17.3 | 1 | 15.5 |
| Npy1r | neuropeptide Y receptor Y1 | 17.3 | 2.4 | 7 |
| Npy2r | neuropeptide Y receptor Y2 | 14.6 | 1.4 | 10 |
| Agtr1a | angiotensin II receptor, type 1a | 13.3 | 1.6 | 8 |
| Lpar5 | lysophosphatidic acid receptor 5 | 13.1 | 0.9 | 13.1 |
| Sstr2 | somatostatin receptor 2 | 11.2 | 0.3 | 28.5 |
| Neuropeptides and Cytokines | ||||
| Calca* | calcitonin-related polypeptide alpha; CGRP | 924.2 | 372.1 | 2.5 |
| Tac1 | tachykinin 1; substance P | 597.7 | 14.1 | 42 |
| Calcb* | calcitonin-related polypeptide beta; CGRP2 | 105.4 | 31.9 | 3.3 |
| Adcyap1 | adenylate cyclase activating polypeptide 1; PACAP | 95 | 5.3 | 17.6 |
| Cartpt | CART prepropeptide | 64.7 | 2.1 | 29 |
| Sst | somatostatin; growth hormone release-inhibiting factor | 53.8 | 0.5 | 97.3 |
| Gal | galanin | 33.8 | 1.2 | 25.7 |
| Nppb | natriuretic peptide precursor type B | 24.1 | 0.2 | 86.9 |
| Ccl1 | chemokine (C-C motif) ligand 1 | 7.8 | 0.3 | 22.5 |
| Nts* | neurotensin | 10.7 | 2.3 | 4.4 |
| Synaptic Vesicle Proteins and Associated Synaptic Proteins | ||||
| Scg3 | secretogranin III | 585.8 | 111.5 | 5.2 |
| Scg2 | secretogranin II; secretoneurin | 202.8 | 33.8 | 6 |
| Synpr | synaptoporin | 184.9 | 14 | 13.1 |
| Mal2 | mal, T-cell differentiation protein 2 | 124.8 | 6.4 | 19.1 |
| Syt7 | synaptotagmin VII | 75.1 | 9.3 | 8 |
| Nptx2 | neuronal pentraxin 2 | 73.8 | 10.4 | 7 |
| Syt9 | synaptotagmin IX | 66.9 | 6.2 | 10.7 |
| Syt5 | synaptotagmin V | 66.5 | 3.8 | 17.3 |
| Nptxr | neuronal pentraxin receptor | 66.3 | 12.6 | 5.2 |
| Nrsn1 | neurensin 1 | 65.4 | 11.5 | 5.6 |
| C1ql4 | complement component 1, q subcomponent-like 4 | 31.8 | 1.4 | 21.9 |
| Syt16 | synaptotagmin XVI | 3.6 | 0.3 | 8.7 |
| Transcription Factors, mRNA-binding proteins and Transcription Related and Splicing Factors | ||||
| Prdm12 | PR domain zinc finger protein 12 | 136.4 | 21.8 | 6.2 |
| Nbl1 | neuroblastoma, suppression of tumorigenicity 1 | 132.9 | 22.6 | 5.9 |
| Tshz2 | teashirt zinc finger family member 2 | 115.8 | 19.3 | 6 |
| Ldb2 | LIM domain binding 2 | 103.5 | 5 | 20.4 |
| Carhsp1 | calcium regulated heat stable protein 1 | 66.7 | 7.9 | 8.3 |
| Fos | FBJ osteosarcoma oncogene; cFos | 59.6 | 1.7 | 34.1 |
| Junb | Jun-B oncogene | 55.8 | 9.8 | 5.6 |
| Celf6 | bruno-like 6, RNA binding protein (Drosophila) | 41.5 | 1.1 | 35.4 |
| Runx1 | runt related transcription factor 1 | 35.2 | 2.4 | 14 |
| Egr1 | early growth response 1; Krox-1; nerve growth factor-induced protein A | 30 | 4.3 | 6.9 |
| Klf5 | Kruppel-like factor 5; basic transcription element-binding protein 2 | 27.5 | 3 | 8.9 |
| Paqr5 | progestin and adipoQ receptor family member V | 27.4 | 1 | 24.9 |
| Bex1 | brain expressed gene 1 | 26.7 | 1 | 24.9 |
| Zfp36 | zinc finger protein 36 | 21.7 | 3.4 | 6.2 |
| Growth Factors, Matrix and Glycoproteins, Cell Adhesion, Semaphorins, and Cytokine Receptors and Modulators | ||||
| Cd24a | CD24a antigen; nectadrin | 924.2 | 83.6 | 11 |
| Fgf13 | fibroblast growth factor 13, microtubule regulation | 395.2 | 76.1 | 5.2 |
| Cd55 | CD55 antigen; complement decay-accelerating factor | 226.6 | 39.6 | 5.7 |
| sgigsf | cell adhesion molecule 1; Cadm1 | 158.4 | 22.4 | 7 |
| Gfra2 | glial cell line neurotrophic factor family receptor alpha 2; neurturin receptor | 115.7 | 11.5 | 9.9 |
| Ly86 | lymphocyte antigen 86 | 109.5 | 13.7 | 8 |
| Cd44 | CD44 antigen; hyaluronate receptor | 81.4 | 5.2 | 15.3 |
| Plxnc1 | plexin C1; semaphorin receptor | 77.6 | 10.3 | 7.4 |
| Gfra3 | glial cell line derived neurotrophic factor family receptor alpha 3 | 73.5 | 0.4 | 148.6 |
| Stac1 | src homology three (SH3) and cysteine rich domain | 47.2 | 6.2 | 7.5 |
| Ccdc68 | cutaneous T-cell lymphoma-associated antigen | 28 | 2.1 | 12.8 |
| Cgref1 | cell growth regulator with EF hand domain 1 | 24.9 | 4.7 | 5.2 |
| Osmr | oncostatin M receptor | 23.6 | 1.8 | 12.8 |
| Il31ra | interleukin 31 receptor A | 22 | 0.6 | 33.9 |
| Socs3 | suppressor of cytokine signaling 3 | 14.6 | 2.2 | 6.5 |
| Membrane, Structural and Rho/Rac Proteins | ||||
| Tmem233 | transmembrane protein 45b | 461.8 | 55.5 | 8.3 |
| Tmem45b | transmembrane protein 45B | 178.1 | 6.6 | 26.8 |
| Tubb2b | tubulin, beta 2a | 254.2 | 37.1 | 6.8 |
| Rasgrp1 | RAS guanyl releasing protein 1 | 90.5 | 2.5 | 34.8 |
| Ctxn3 | cortexin 3 | 81.3 | 4.2 | 18.8 |
| Ctxn1 | cortexin 1 | 62.8 | 8.8 | 7.1 |
| Rarres1 | retinoic acid receptor responder (tazarotene induced) 1 | 60.1 | 9.1 | 6.5 |
| Rasgrp | RAS guanyl releasing protein 1 | 51.1 | 1.5 | 31.8 |
| Cpne2 | copine II | 48.5 | 7.2 | 6.7 |
| Nrn1l | neuritin 1-like, Cpg15-2 | 46.2 | 6.7 | 6.8 |
| Tmem158 | transmembrane protein 158 | 45.7 | 2.1 | 20.8 |
| Rab27b | RAB27b, member RAS oncogene family | 22.6 | 4.4 | 5.1 |
| Arhgap26 | Rho GTPase activating protein 26 | 21.6 | 3.4 | 6.3 |
| Hspa1b | 70kDa heat shock protein 1B; chaperone protein | 16.8 | 3 | 5.4 |
| Ms4a3 | membrane-spanning 4-domains, subfamily A, member 3 | 6.6 | 0.7 | 8.3 |
| Kinases, Phosphatases, Proteases, Mmps, Phosphodiesterases and Other Enzymes and Enzyme inhibitors | ||||
| Lxn | latexin; endogenous carboxypeptidase inhibitor | 328.5 | 66.3 | 5 |
| Prkcd | protein kinase C, delta | 224.8 | 24.4 | 9.2 |
| Acpp | acid phosphatase, prostate; PAP | 207.6 | 13.2 | 15.6 |
| Plcb3 | phospholipase C, beta 3 | 202.2 | 28.4 | 7.1 |
| Dusp26 | MAP kinase phosphatase 8 | 169.3 | 14.4 | 11.6 |
| Th | tyrosine 3-hydroxylase | 142.8 | 1.3 | 103.7 |
| Dgkh | diacylglycerol kinase, eta | 91.9 | 12.3 | 7.4 |
| Gpx3 | glutathione peroxidase 3 | 90.6 | 6.9 | 13 |
| Camk2a | calcium/calmodulin-dependent protein kinase II alpha | 79.7 | 9 | 8.8 |
| Ppp1r1a | protein phosphatase 1, regulatory (inhibitor) subunit 1A | 51.3 | 7.3 | 7 |
| Hs6st2 | heparan sulfate 6-O-sulfotransferase 2 | 42.8 | 6.4 | 6.6 |
| Prkcq | protein kinase C, theta | 40.3 | 3.1 | 12.7 |
| Mmp25 | matrix metallopeptidase 25 | 35.7 | 4.1 | 8.5 |
| A3galt2 | alpha 1,3-galactosyltransferase 2 (isoglobotriaosylceramide synthase) | 33.5 | 2 | 15.7 |
| Dgki | diacylglycerol kinase, iota | 28.9 | 5.5 | 5.2 |
| Transporters, Trafficking and Interacting Proteins | ||||
| Fxyd2 | FXYD ion transport regulator 2; ATPase subunit gamma | 2196.7 | 252.1 | 8.7 |
| Aqp1 | aquaporin 1 | 152.6 | 21.9 | 6.9 |
| Atp2b4 | ATPase, Ca++ transporting, plasma membrane 4 | 18.5 | 2.1 | 8.4 |
| Slc4a11 | solute carrier family 4, sodium bicarbonate transporter-like, member 11 | 15.8 | 2.2 | 6.8 |
| Slc45a3 | solute carrier family 45, member 3 | 15.2 | 2.6 | 5.7 |
| Slc37a1 | solute carrier family 37 (glycerol-3-phosphate transporter), member 1 | 10.1 | 1.9 | 5.2 |
| Slc10a6 | solute carrier family 10 (sodium/bile acid cotransporter family), member 6 | 9.2 | 0.5 | 15.5 |
| Slc17a8 | sodium-dependent inorganic phosphate cotransporter, member 8 | 8.3 | 0.3 | 22.9 |
| Slc16a12 | monocarboxylic acid transporters, member 12 | 6.3 | 0.5 | 10.6 |
| Slc47a2 | solute carrier family 47, member 2 | 3.7 | 0.5 | 6.1 |
| Slc7a3 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 | 3 | 0.3 | 7.8 |
Genes in the TRPV1L preparation with a differential expression level of 5X and greater above values in the TRPV1D preparation were grouped into functional categories. Each category was sorted based on quantitative expression (high to low) from the deep RNA-Seq datasets. The table contains the top ~10–20 TRPV1L highest expressed genes within each category; this is only a sampling and the full listing is found in STable2. The adjusted fold change values are color coded: Red: >20 fold, Blue: 20-10 fold, and Green: < 10 Fold difference.
Calca, Calcb, and Nts, while not having a differential above 5X, are included because of their relevance to nociception.
Table 1 and Supplemental Table 2 provide the molecular signature of the TRPV1L transcriptome based on differential expression of 5-fold or greater. The transcripts are placed into nine functional categories (10–19 per category) listed from high to low based on quantitative transcript levels (RPKM). It is notable that our cutoff of ≥ 5-fold enrichment is arbitrary and many familiar genes associated with nociceptive processes are not on this list. This in no way minimizes or impugns their role in nociception, but simply reflects lower differential expression and/or performance of a function common to both TRPV1L and D DRG neurons. The theme of differential expression is continued in the remaining supplemental tables (STable3-STable7) which tabulate the remaining differentially expressed genes in TRPV1L and D populations and genes common to both. Together these transcriptome data greatly expand the nodes for connecting molecular processes with our understanding of nociception, its modulation and the differentiation of DRG neuronal functions. Obviously, many but by no means all, of the well expressed DRG genes have been the subjects of pain research and the fact that they are enriched is consistent with the methodological focus on nociceptive neurons.
Potassium Channels
A striking feature of the TRPV1L population is the high representation of differentially expressed potassium channels: out of the 30 ligand and voltage gated ion channels in the 5-fold or greater group, 12 (40%) are K+ channel subunits or modulatory proteins (Tables 1, S2 and S3). In the TRPV1L and D preparations, ~115 K+ channels and interacting proteins are identified and ~90 are expressed at ≥ 1 RPKM. Transcripts for two A-type channel modulatory proteins Kcnip (KChIP) 4 and 2 were the highest expressed (>100 RPKM) and both contain an EF hand calcium binding motif (Table 1). While these have not been studied in DRG, the proteins can affect K+ channel trafficking49 and knockout of Kcnip2 increased susceptibility to limbic seizures, suggesting that Kcnip2 may also influence DRG neuronal excitability.69 The TRPV1L neurons also differentially express both β1 and β2 subunits of the high conductance MaxiK (BK, Kcnmb) channel which is calcium regulated. The potential colocalization of multiple K+ channels and modulators suggests a system for membrane potential control that may be triggered by calcium entry subsequent to activation of TRPV1. Numerous efforts have been aimed at developing K+ “channel opener” compounds, mainly as antiepileptics,67 and some have been partially explored for analgesic activity.14,95 Implementation of the present transcriptome data may facilitate fine-tuning of K+ channel isoform targeting for analgesic therapeutic purposes.
Cholinergic Receptors
A second example of data mining related to analgesics is the acetylcholine receptors. The discovery of the analgesic actions of the nicotinic cholinergic agonist epibatidine emphasized a role for acetylcholine receptors at the ganglionic and/or spinal levels.3,96 Table 1 shows that the α6 subunit is highly expressed in the TRPV1L population (30.3 RPKM in TRPV1L vs 4.0 RPKM in TRPV1D), consistent with expression and colocalization studies.19,23 Despite these data, the α6 subunit appears to have been largely ignored in favor of experiments using the α7 subunit. Interestingly, the α7 subunit is also well expressed in DRG but in an inverse fashion: mainly in the TRPV1D population (5.3 RPKM for TRPV1L vs 30.8 for TRPV1D). Transcriptome analysis disclosed a strong expression of the nicotinic β2 subunit (common to both populations 16.3 and 20.2 RPKM for TRPV1L vs D) and a strong differential expression (5.3 vs. 0.1 RPKM) of the β3 subunit in the TRPV1L neurons. These quantitative considerations suggest that differential α6, β3 or β2 subunit expression may be key determinants for development of nociceptive neuron specific analgesic cholinergic agents.
RGS Proteins
An examination of the G-protein coupled receptors and associated proteins category in Table 1 and Supplemental Table 2 reveal an overrepresentation of Regulators of G Protein Signaling (RGS proteins, n=5) compared to the TRPV1D population (n=1) (Tables S4A and B). The RGS proteins are multifunctional modulators of GPCRs that generally increase the GTPase activity of active Gα subunits thereby reducing the amplitude and time course of GPCR signaling.85 The order of expression levels in TRPV1L neurons is RGS4>10≫3>8≫14 with RGS4 and RGS10 exhibiting very high expression (606 and 222.9 RPKM, respectively). Several studies have implicated RGS proteins in DRG nociceptive functions related to partial axotomy-induced neuropathy15,88 and regulation of opioid and metabotropic glutamate receptor function.16,94 Most opioid receptor-RGS interaction studies have focused on spinal and supraspinal sites and investigated either analgesia or addiction22 but no work has directly focused on the DRG. Knock-in of a RGS insensitive Gα was shown to increase morphine analgesia and the effect was partially attributed to enhanced opioid mediated disinhibitory actions in the periaqueductal gray. However, given the high concentrations of RGS proteins in the TRPV1L population, actions on the “input function” coming from DRG afferents need further consideration and targeting of the TRPV1L RGS isoforms may be a route to modulation of opioid receptor action44 in peripheral neurons.
Mrg Receptors
Four Mrg receptors are expressed ≥ 5 fold in the TRPV1L neurons. This is consistent with their roles in nociception and itch. MAS-related GPR, member D (Mrgprd) is the most highly expressed GPCR in the DRG (125.4 RPKM) and exhibits a 29-fold differential expression compared to TRPV1D. Mouse behavioral studies suggest that Mrgprd alone does not affect responsiveness to noxious thermal stimuli. However, dual ablation of DRG neurons expressing Mrgprd and TRPV1 produces a large decrease in sensitivity to noxious thermal stimuli and dual ablation of Mrgprd- and TRPM8-coexpressing neurons reduces responses to extreme cold.77 These data are consistent with the idea that Mrgprd neurons are polymodal nociceptors that are activated indirectly by the consequences of extreme temperature. In fact Mrgprd-cell deficient mice have profoundly reduced responses to ATP, as well as reduced sensitivity to mechanical stimuli, which is not surprising given the complement of ligand gated and mechanosensitive ion channels expressed by these neurons (Figure 2). 80 MAS-related GPR, member A3 (Mrgpra3) and MAS-related GPR, member X1 (Mrgprx1) receptors respond to various chemical puritogens and appear to be markers of itch sensory neurons.50,75 In mouse, Mrg genes are greatly expanded.106 We detected 16 Mrg receptors in murine DRG and 11 were differentially expressed in the TRPV1L population (between 1.3 to 125.4 RPKM). By comparison rat DRG only expresses the Mrgpra1, b4, d, e, r, h and x1 paralogs, the RPKM values are 9.3, 0.8, 21.3, 21.2, 4.6, 1.6, 14.4, respectively.
Taken together the above transcriptome analyses show that a detailed quantitative and cell-specific understanding of expression levels of paralogs and receptor subunits can have a profound effect on integrating molecular and neuronal functions and ultimately circuit operations for nociception and analgesia. This theme is further explored in Figures 2, 4A, and 4B, which illustrate the molecular “micro-circuit relationships” that can be extracted from the TRPV1L transcriptome.
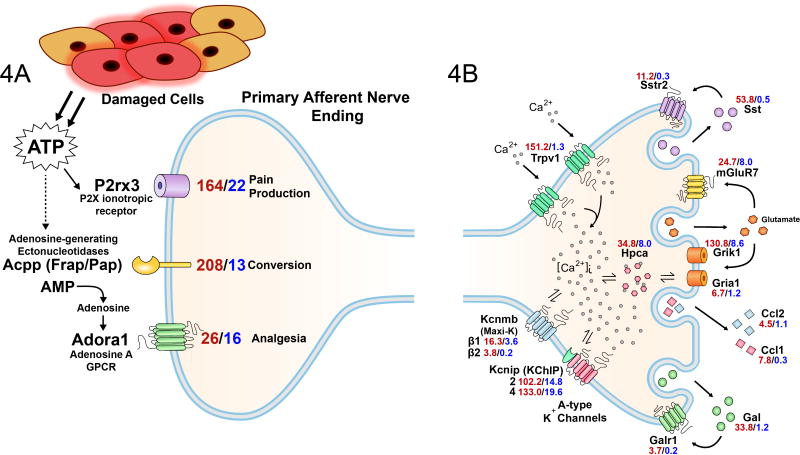
Figure 4. Integratory Molecular Circuits in TRPV1L Neurons.
Diagram showing how expression of linked signaling pathways and transmitter-receptor pairs can generate (A) paracrine and (B) potential autocrine signaling pathways to control the excitatory and inhibitory status of the peripheral nerve endings. (A) Purinergic nociceptive signaling can be modified by co-expressed ATP metabolizing ectonucleotidases and subsequent activation of adenosine A receptors to generate an analgesic component [P2rx3: P2X3, purinergic receptor, ligand-gated ion channel, 3; Acpp: prostatic acid phosphatase; Adora1: adenosine A1 G-protein coupled receptor]. (B) Depiction of several potential autocrine and calcium-dependent processes. Levels of expression receptors; all show higher expression (3- to15-fold) in the TRPV1L population and can support excitatory and inhibitory feedback by binding released glutamate. Neuropeptide receptor pairings for the Somatostatin receptor 2 (Sstr2) and somatostatin (Sst) and galanin receptor 1 (Galr1) and galanin (Gal) support a presynaptic autocrine inhibitory role. We identify several new calcium regulated process in the TRPV1L population based on differential expression of the hippocalcins and the potassium channel interacting proteins Kcnip2 and 4, all of which contain EF hand calcium binding motifs. The β1 and β2 MaxiK channel are also Ca++-regulated and can modulate nerve terminal hyperpolarization. Hippocalcin (Hpca) is known to play a regulatory role for AMPA receptor function (see text). The overall goals of the paracrine, autocrine and calcium binding proteins are hypothesized to mediate autonomous regulation of nerve terminal excitability and [Ca++]i under conditions of acute and chronic tissue damage or inflammation. [Gene symbols/names- mGluR7: Grm7, metabotropic glutamate receptor 7; Grik1: glutamate receptor, ionotropic, kainate 1; Gria1: glutamate receptor, ionotropic, AMPA 1; Ccl2: MCP-1, chemokine (C-C motif) ligand 2; Ccl1: chemokine (C-C motif) ligand 1; Kcnmb: potassium large conductance calcium-activated channel, subfamily M, beta member]
TRPV1L Transcripts: Purines, Peptides, Cytokines and Paracrine-Autocrine Communication
General orientation
The following sections explore in more detail: (a) interpretation of the expression data in terms of neuronal heterogeneity even within the TRPV1 population (Figure 3), (b) some of the relationships devolving from Figure 2 and the tables (mainly Table 1) that integrate several signaling pathways into multifactorial presynaptic regulatory networks (Figure 4A, B), and (c) an example of co-localization of a calcium binding proteins (the hippocalcins) identified by RNA-Seq within TRPV1 neurons (Figure 5).
Figure 3. TRPV1 Expression in DRG.
Immunocytochemical staining for TRPV1 shows a gradient of expression in adult rat DRG neurons. The heterogeneity of TRPV1 expressing neurons ranges from small densely stained cell bodies, more lightly stained medium sized neurons and a range of very faintly stained neurons of various sizes. Many large unstained neurons are also visible. This can be seen in the panoramic view and appreciated better at the high magnification. This range of TRPV1 expression is subsumed in the TRPV1 lineage population thus the level of expression of genes other than TRPV1 can also vary. The large unstained neurons are contained in the TRPV1D preparation. Scale bar: 100μ
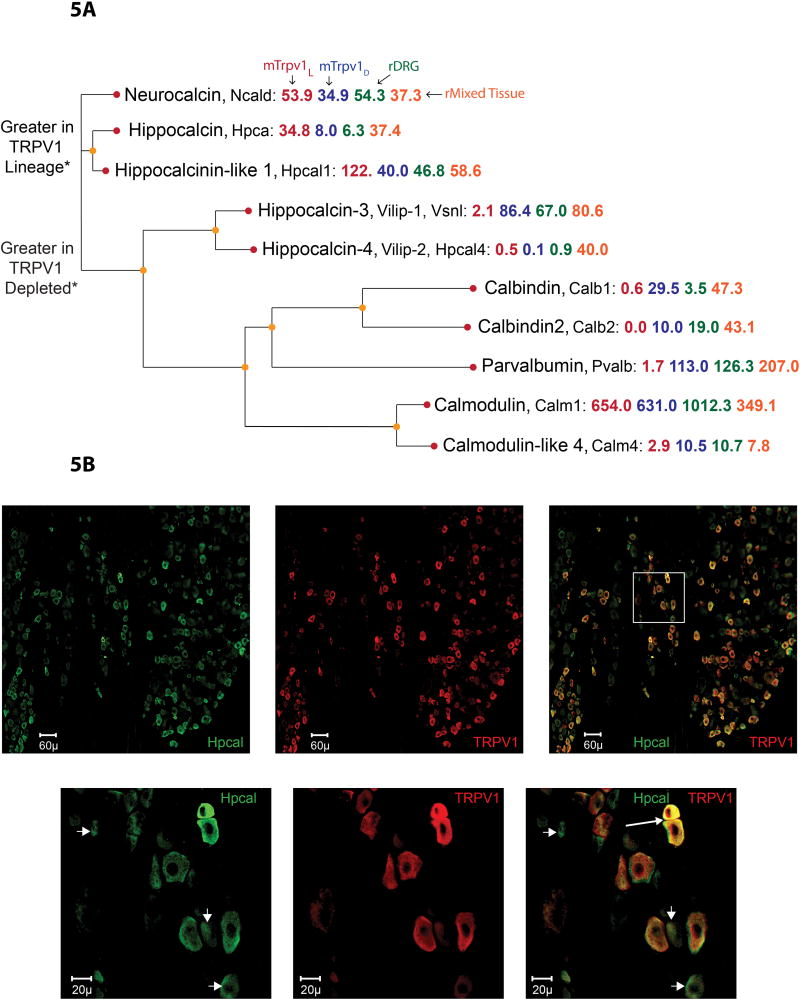
Figure 5. Hippocalcin in DRG.
(A) Molecular relationships, determined by protein sequence alignment in Clustal Omega, between the EF hand gene family members is depicted in this dendrogram. RNA-Seq data for TRPV1L, TRPV1D, whole rat DRG, and rat mixed tissue are given in red, blue, green and orange, respectively. Sequence alignment of hippocalcin and hippocalcin like 1 proteins reveals them to be nearly identical (not shown) and the carboxy-terminal directed antibody used for immunocytochemistry will detect both proteins. (B) Double label immunocytochemistry shows colocalization of hippocalcin (green) and TRPV1 (red). Panels on the right show merged images with many double labeled cells. In lower right panel the arrow points to two very strong coexpressing neurons and the arrowheads point to neurons that predominantly express only hippocalcin. The whole tissue sample was acquired by scanning at three emission wavelengths as a series of fields at 40X with excitation emission filters for DAPI, Alexa 488 and Alexa 594. Consistent with the transcriptional expression data showing major expression of hippocalcin in the TRPV1L neurons, but also detectable expression in the TRPV1D preparation, a range of dual and single labeled neurons are seen for either TRPV1, hippocalcin or both.
Figure 2 illustrates a primary afferent nerve ending and the cellular locations of proteins coding for families of ion channels, enzymes, GPCRs, growth factor receptors, intracellular and vesicular proteins, neuropeptides, and extracellular linking proteins. The corresponding transcript levels are placed next to the protein. Several gene families are portrayed in detail such that most members of a family are listed, even if the values do not reach the ≥ 1 RPKM cutoff (e.g., Trpm1, 5 and 6). Proteins are identified by the gene symbols from NCBI. Full protein names for many of the genes indicated are in the figure legend. It is important to note that the molecules depicted in Figure 2 are not necessarily expressed in a single nerve ending; rather the respective peripheral nerves collectively express these molecules. In addition, as depicted, Figure 4 shows various molecules in the same nerve ending. Such co-localization of a ligand and receptor, for example somatostatin and Somatostatin receptor 2 (Sst and Sstr2) in TRPV1L neurons, needs further verification by anatomical co-localization studies, although somatostatin is known to inhibit DRG neurons via Sstr2. It is also important to note that a paracrine relationship may apply if the ligand and its receptors are in adjacent nerve endings.
Heterogeneity within the TRPV1 population
Neuronal labeling through the developmental TRPV1 lineage allows us to more broadly sample neuronal populations related to nociceptive modalities. At this stage of exploration, we felt that the lineage strategy was more advantageous than to more narrowly focus on the TRPV1 population as it exists in the adult, which is also heterogeneous as shown in Figure 3. The heterogeneity in cell size and intensity of staining is evident in the immunohistochemical staining for TRPV1. Variation in Trpv1 staining intensity also can be seen in Figure 5b and in many publications that mainly use immunofluorescence64,83 and in part reflects expression in a subpopulation of Aδ neurons.64 Thus, the absolute level of expression for each gene needs to be interpreted in terms of a neuronal population within which variation in expression level can occur.
In terms of nociceptive modalities, our data, as well as results from others, dating back to the early 1970’s, support the hypothesis that the adult DRG is likely a mixture of polymodal nociceptors and more refined subpopulations of modality specific nociceptors and non-nociceptive neurons.5,11,18,41,64,72 Figure 3, in which multiple populations of TRPV1 expressing cells can be seen, emphasizes this fact. In addition, in a recent paper,64 we show that injection of resiniferatoxin in the periphery (hind paw) causes a profound loss to both noxious heat and to thermal stimuli that produce tissue damage. This observation, which is obtained at a high dose of RTX, is consistent with a range of TRPV1 expression within a range of nociceptive subtypes. Additional labeling and sorting techniques will be required to isolate and further define these specific subpopulations to obtain a deeper molecular-level understanding of neuronal populations in the DRG.
Purine, Pyrimidine and ASIC receptors and Conversion of ATP to Adenosine
The TRPV1L neuronal lineage shows strong expression of the acid-sensing (proton-gated) ion channel 2, Accn1 (Asic2), the Adenosine 2A and P2Y1 GPCRs, the P2X3 ATP-gated ion channel, and the ectonucleotidases FRAP (fluoride resistant acid phosphatase, Acpp, PAP) and Nt5e (5′ ectonucleotidase). The high expression of Accn1/Asic2 suggests that this paralog is a major acid sensing ion channel subtype responding to an acidic inflammatory environment in the TRPV1L population. Accn3/Drasic exhibits the inverse pattern, although expression overlaps substantially. When the ectonucleotidases are included, the quantitative relationships support the idea that a cascade of transmitter ATP production and enzymatic processing occurs upon tissue damage (Figure 4A). We hypothesize, that ATP released from damaged cells activates TRPV1L neurons through the P2X3 ionotropic receptor (the major P2X subtype on the TRPV1L population). At the same time, high concentrations of extracellular, locally tethered ectonucleotidases can convert ATP to adenosine, which can activate the adenosine A1 GPCR to reduce afferent excitability.107 In this paracrine fashion the nerve ending converts an algesic substance to an analgesic substance. It is known through in vitro and in vivo studies with RTX that calcium overload can produce a long-term inactivation of nociceptive endings.39, 63,68,72 While we use this inactivation property for therapeutic purposes (<http://clinicaltrials.gov/ct2/show/NCT00804154>), in situations of tissue damage, pain sensitivity and the functional integrity of the peripheral nerve ending must be preserved until the damaging stimuli are terminated or removed. RNA-Seq also identifies a new set of intracellular calcium binding molecules, the hippocalcins (Hpca, Hpcal1) that are 3–4 fold enriched in Trpv1L neurons and may reduce free internal calcium levels resulting from excessive or prolonged P2X3 or TRPV1 activation. Thus, multiple molecules and mechanisms may be engaged to maintain the physiological and structural integrity of nociceptive nerve endings in conditions of massive tissue damage or prolonged chronic pain (Figure 4A,B).
Synaptogranins and Cytokines
Subpopulation-enriched expression was also observed for synaptic vesicle proteins. The intra-vesicular proteins secretogranin-2 and -3 are highly and predominantly expressed in the TRPV1L population (Figure 2 center). Neuropeptides are concentrated in the TRPV1L neurons and are known to be stored in large dense core vesicles, which co-store the granins. While members of the granins are expressed in all neuroendocrine tissues, the 33 amino acid peptide secretoneurin, derived from secretogranin-2, has been reported to play a role in neuroimmune interactions mediated by DRG neurons.38 Although a distinct receptor had not been identified for this peptide, it has been shown to be cleaved from the precursor, stored in C-fiber dense core vesicles, released peripherally in certain pathological states, such as rheumatoid and osteoarthritis and has macrophage chemoattractant properties.86
Two other macrophage chemoattractant molecules are preferentially expressed in the TRPV1L population, CCL1 and CCL2 (monocyte chemoattractant peptide 1, MCP-1). Both have been investigated for neuroimmune interactions in several studies, especially MCP-1.37,102 The receptors for these cytokine molecules are not expressed in sensory ganglionic tissue. These data raise the possibility that vesicles of primary afferents store one or both cytokines and that nociceptive nerve endings play an active role in monocyte recruitment. Interestingly, leukocyte diapedesis appears to occur at both the peripheral and central ends of the primary afferents. In models of nerve injury, monocyte infiltration from peripheral blood has been demonstrated in the dorsal horn,20 and peripherally, monocytes make up a second wave of infiltrating leukocytes into inflamed tissue, the first wave being predominantly neutrophils. These data are consistent with the idea that the primary afferent ending can shape nociceptive responses to tissue injury and local regulation of specific leukocyte populations at primary afferent nerve termination zones. Both CCL1 and CCL2/MCP-1 contain signal peptides consistent with processing in the regulated secretory pathway and vesicular storage, however, the co-localization of CCL1, CCL2 and secretoneurin is illustrative, and whether they are in the same vesicle requires further studies (Figure 4B).
Neuropeptides
Transcripts coding for neuropeptides display more prominent expression in the TRPV1L population (Tables 1, S2, and S3). None of the classical neuropeptides are differentially expressed in the TRPV1D population (Table S4A). Again, it is important to point out that the word “differential” does not equate to absolute, and we include in Table 1 the Calca and Calcb transcripts encoding CGRPα and β which are highly expressed, but the expression overlaps into both TRPV1L and TRPV1D cellular populations. This is consistent with many co-localization studies which indicate a broad array of DRG neurons express CGRP. In contrast, many other neuropeptide transcripts (preprotachykinin (Tac1), pituitary adenylate cyclase activating peptide (Adcyap1), cocaine and amphetamine regulated peptide (Cartpt), somatostatin (Sst), Galanin (Gal), and natriuritic polypeptide B [Nppb]) are strongly differentially expressed in the TRPV1L population (avg. of 50-fold).
Potential Autocrine Communication by Neuropeptides
Examination of the RNA-Seq data for neuropeptide-receptor pairs suggested the potential for autocrine or paracrine communication. Cases where both ligand and receptor are made by the same set of neurons include somatostatin (Sst) and galanin (Gal) (Figure 4B). In the mouse, strong differential expression of transcripts coding for Sst (97-fold) and Gal (25-fold) is seen in the TRPV1L population, which also primarily expresses somatostatin receptor 2 (Sstr2) and galanin receptor 1 (Galr1). Sst inhibits primary afferent nociceptive functions and gene knockouts identify Sstr2 as the receptor mediating analgesic actions.35 Similarly, galanin has been shown to inhibit the efferent local effector functions activated by RTX or capsaicin-sensitive peripheral nerve terminals in a fashion consistent with a presynaptic site of action.24 These data support an inhibitory presynaptic autocrine function for both Sst/Sstr2 and Gal/Galr1 peptide-receptor pairs and we speculate, as with the purinergic paracrine circuit, an autocrine mechanism that functions to regulate excitability of afferent endings in situations of prolonged tissue damage and chronic pain. Pharmacologically this can be manifested as analgesia, but in pathophysiological conditions we hypothesize that the inhibitory functions participate in preserving the physiological and anatomical integrity of the primary afferent ending.
Potential Autocrine Communication by Glutamate
The excitatory amino acid glutamate may also participate in autocrine/paracrine communication via the kainate and AMPA subtypes of glutamatergic receptors. Expression of the kainate subtype (Grik1, 130.8 RPKM) far exceeds that of the AMPA receptor Gria1, 6.7 RPKM). Ionotropic glutamate receptors are generally heteromeric and the other subunits are also expressed to varying degrees (Tables S3, S4 and S5). Expression of kainate and AMPA receptors may represent an excitatory presynaptic “opponent process” to the above analgesic neuropeptide scenarios (Figure 4B) and kainic acid is known to excite peripheral nerve endings.105 Metabotropic glutamate receptors are also expressed, the highest levels were for mGluR7 (Grm7: TRPV1L 24.7 and TRPV1D 8.0 RPKM), the expression levels for the other isoforms were below 1.5 RPKM in the TRPV1L neurons; several other mGLuRs are made in the TRPV1D population (Tables S4A, B). These quantitative measures suggest that mGluR7 is the main metabotropic glutamate receptor paralog performing a presynaptic function in murine nociceptive neurons. As such, mGluR7 also fits the “transmitter-receptor pairing” criterion to form an autocrine molecular circuit. In this case an analgesic function may be engaged since local injection of a mGluR7 allosteric agonist attenuated carrageenan-induced and post-surgical hyperalgesia.17
Other receptor families
Table 1 and Table S2 contain the GPCRs Lpar3 and 5 that bind lysophaphatidic acid, an algesic mediatior and a target for therapeutic anti-lysophaphatidic acid antibodies.25 Two receptors for neuropeptide Y, Npy1r and 2r are expressed in TRPV1L neurons, however, while NPY is not well expressed in basal state ganglion, it is strongly induced following axotomy.45,98 Thus, it is possible that newly synthesized NPY and the two preexisting NPY receptors could form an autocrine loop following nerve injury. Growth factor receptors are also highly differentially expressed with the strongest TRPV1L transcripts being glial cell line derived growth factors 3 and 1 (Gfra3, Gfra1, Figure 2). Genes for most of the tyrosine kinase growth factor receptor ligands (GDNF, artemin, neurturin, persephin, neurotrophin 3, and NGF) are not expressed in DRG. Exceptions are BDNF (9.7/5.2), CNTF (4.4/46.7), and C-kit ligand (5.8/21.3); values are RPKM in TRPV1L/TRPV1D populations. Roles for NGF and BDNF have been reviewed.58,70 For c-kit both the ligand and receptor are expressed in the TRPV1L and D populations and developmentally are involved in early DRG differentiation and neurite growth cone extension.30 In the adult, intrathecal injection of c-kit ligand is reported to produce sensitization in a variety of pain tests.90,92 Additionally, the inhibition of c-kit with monoclonal antibodies has been reported to reduce heat and cold pain sensitivity in humans.12
RTX Lesion of Rat Trigeminal TRPV1 Neurons
Stereotaxic microinjection of RTX into rat trigeminal ganglion to remove TRPV1-expressing neurons at 10 days provided a validation of the mouse TRPV1 manipulation and allowed the mouse dataset to be extended to the rat. We analyzed gene expression in the cell remaining in the ganglion after RTX treatment. The trigeminal ganglion and the dorsal root ganglion share a very similar overall gene expression pattern.21 We examined the similarity between TG and lumbar DRG using the quantitative rat data for genes in Figure 2 as a test transcript set. The resultant DRG:TG correlation coefficient was significant with a R2 value of 0.88 (Figure S4, panel A). From our more global analysis of ganglionic transcriptomes, one difference we noted was that, out of 18 Hox genes present in the lumbar DRG, Hoxd1 is the only Hox gene also expressed in the trigeminal ganglion (Figure S4, panel B).
Table S6 contains the comparison of the rat RTX data with the TRPV1L 5X and above differentially expressed genes (Tables 1 and S2). 85% of the TRPV1L genes expressed at or above 3 RPKM in the functional groups of “Ligand and Voltage Gated Ion Channels,” “Transporters and Channel Interacting Proteins,” and “GPCRs and Associated Proteins” were significantly down regulated after intraganglionic injection of RTX. Unlike the genetic expression of diphtheria toxin, the microinjection generally leaves some low expressing TRPV1 neurons intact, which can attenuate the degree of decrease (Karai et al., 2004).39 The RTX dataset also indicates the polymodal nature of some neurons in the TRPV1L population: loss of TRPV1-expressing neurons was accompanied by significant decreases in TRPM8 (cold), Trpa1 (chemo-nociception), TRPC3 (light touch), Mrgprd (pinch and high threshold stimuli), and Mrgprx1 (itch and nociception).79 The overlap between the mouse genetic labeling/ablation and rat RTX chemoablation suggest a core “nociceptome” that is key to understanding the first steps in pain processing. These genes, in turn, are embedded in a supporting transcriptional network that can also be exploited for further understanding of nociception.
Species Differences
Tyrosine Hydroxylase
We made a comparison between rat and mouse DRGs. The vast majority of transcripts showed similar levels of expression. One conspicuous species differences occurred for TH, the first enzyme in the catecholamine biosynthetic pathway. TH is highly expressed in mouse DRG TRPV1L neurons (143 RPKM) compared to whole rat DRG (0.7 RPKM). In rat, TH immunostaining shows infrequent, sporadic staining of DRG neurons,78 which matches the low transcript abundance. The comprehensive nature of RNA-Seq indicates that the catecholamine synthetic pathway apparently ends with TH in both mouse and rat. Transcripts coding for aromatic amino acid decarboxylase, dopamine β-hydroxylase, and vesicular dopamine or norepinephrine uptake were either not detectable or were below 1.0 RPKM, consistent with the earlier neuroanatomical studies.9 Thus, it is tempting to speculate that L-3,4-dihydroxyphenylalanine (DOPA), the end product of TH, has a unique role in mouse C-fiber low-threshold mechano-sensory neurons. However, deep sequencing showed that Moxd1, a dopamine β-hydroxylase paralog,13 is well expressed (19.1 RPKM) in the TRPV1L neurons suggesting that β-hydroxyL-3,4-dihydroxyphenylalanine may also be an enzymatic end product and potential transmitter.
Neuropeptides
Several neuropeptides also exhibit strong differential species expression, notably the cocaine and amphetamine regulated transcript (CART, Cartpt), galanin (Gal), natriuretic peptide precursor b (Nppb), and neurotensin (Nts). CART, Gal, Nppb and Nts and are mainly expressed in the TRPV1L population (RPKM: CART 64.7 vs 2.1; Gal 33.8 vs 1.2; Nppb 24.1 vs 0.2; Nts 10.7 vs 2.3; enriched vs depleted, respectively, see Table 2 and Figure 2), but in whole rat ganglion these peptides are expressed at much lower levels (CART 1.1, Gal 3.2, Nppb 0.7, and Nts 0.6 RPKM). Nonetheless, such low levels can translate into a measureable signal in immunocytochemical studies. In rat, CART-positive neurons only account for ~1.26% of > 25,000 trigeminal ganglion neurons,36 and, based on additional colocalization markers, some may be TRPV1-expressing Aδ neurons.63 Most of the other familiar neuropeptides (Calcitonin gene related peptides (Calca, Calcb), preprotachkinin precursor (substance P, Tac1), somatostatin (Sst) and pituitary adenylate cyclase activating polypeptide 1 (Adcyap1, PACAP) are well expressed in both mouse and rat DRG.
Hippocalcin Expression and New Transcript Annotations Resulting from Deep Sequencing
Immunocytochemical Localization of Hippocalcin (Hpca) in Rat DRG
The calcium binding proteins calbindin D28, parvalbumin and calretinin have been used to label subpopulations of DRG neurons.32,84 Up till now the presence of the hippocalcins in DRG has not been appreciated. RNA-Seq transcriptome data indicate that hippocalcin (Hpca) and hippocalcin-like protein 1 (Hpcal1) are highly expressed in the TRPV1L population. Amino acid sequence comparisons show that the two hippocalcins are very similar and diverge from the parvalbumins and calbindins (Figure 5A). Immunofluorescent staining of DRG reveals that Hpca immunoreactive neurons are in the small to medium size range consistent with the nociceptive class of DRG neurons. Immunofluorescent double labeling shows colocalization of TRPV1 with hippocalcin, consistent with predominant expression in TRPV1L neurons (Figure 5B). The hippocalcin calcium binding proteins provide new markers for the TRPV1 population of DRG neurons and may play key roles in regulating nerve terminal free calcium in conditions of acute and chronic pain. This potential role is illustrated as part of Figure 4B.
5′ and 3′ Untranslated Regions (5′ and 3′-UTRs) and MicroRNA Binding Sites in the TRPV1 Gene
Earlier studies on RNA-Seq have been used for genomic re-annotations and studying alternate splicing events.28 Deep transcriptome sequencing can also be used to provide a more complete assessment of 5′ and 3′ UTRs and the information content therein. In the present report, using TRPV1 as an example, we analyzed the Refseq entries of mouse, rat and human. Deep sequencing showed that the 5′ and 3′ mouse UTRs are extended approximately 130 and 735 bases, respectively (Figure 6 and S3). The 5′ sequence reported herein extends beyond the Refseq entries for mouse, rat, and human and suggests the start site for transcription is upstream from the current database annotation. This is significant because the genetic control elements for transcription are at the 5′ end and accurately determining the start site allows more precise mapping of transcriptional control elements and a better understanding of molecular transcription mechanisms for TRPV1 expression or any other genes that need reannotation. The 3′ extension brings the mouse sequence into register with the human and rat annotations and provides a new consensus 3′-UTR. (Note: the above predicted 5′ and 3′ UTR extensions have been recently incorporated into the TRPV1 Refseq gene model and now correspond fairly well to extensions seen in our RNA-Seq data). It has recently been observed that 3′-UTRs frequently contain binding sites for regulatory microRNAs.4 We identified 3 consensus sites that were conserved between mouse, rat and human for the microRNAs miR-940, miR-684, miR-375 (Figure 6). The conservation of the miRNA binding sequences between species implies potential direct roles for miRNAs in TRPV1 regulation in DRG neurons. However, none of the early studies of miRNA in DRG after peripheral inflammation or nerve injury examined the above three miRNAs.7,43,48 This suggests more comprehensive bioinformatic and experimental approaches to the analysis of miRNAs in DRG are necessary to guide experimentation and to delineate the roles of miRNAs in pain models.
DISCUSSION
The present sets of experiments were conducted to probe the pain transcriptome in primary afferent neurons. Our understanding of nociceptive molecular biology is rudimentary at best and has not yet been dissected with a strategy that combines genetic labeling with comprehensive transcriptome sequencing. Using such an approach we have elucidated the complete transcriptome of an important and clinically relevant population of nociceptors: those that express TRPV1. We make the assertion of clinical relevance based on our previous and current work in animals and humans using the TRPV1 neuronal/nerve terminal deletion reagent resiniferatoxin, which is presently being evaluated as a pain treatment in patients with advanced cancer and has been shown to be effective to treat pain in canine osteosarcoma.29,31,54
Molecular Elements for Integrative Communication
We examine many aspects of nociceptive processes and pharmacology within the TRPV1L population. One of the more important observations to emerge from this research is the identification of several new potential autocrine and paracrine molecular communication and control pathways as illustrated in Figure 4. The presence of multiple molecular microcircuits in dorsal root ganglion highlights the crucial role for autonomous, presynaptic regulatory mechanisms. The representation of both inhibitory and excitatory autocrine/paracrine loops suggests, at least at the peripheral terminals, that the nociceptive sensory ending has elaborated a diverse set of control mechanism for maintaining sensitivity in chronic pain states. It is known that excessive excitation can produce calcium cytotoxicity and “deactivate” the nerve ending. Such a deactivation would make the nerve terminal insensitive to the local nociceptive environment thereby losing the protective function of pain. Thus, maintaining excitability in persistent pain states is a critical requirement of nociceptive nerve endings and both peptidergic and metabotropic glutamate receptors as well as ectonucleotidases are hypothesized to reduce excitation. Interestingly excitatory glutamatergic kainite and AMPA receptors are present apparently to maintain a balance. In addition, these processes are bolstered by multiple intracellular calcium regulatory mechanisms involving calcium binding proteins, such as the hippocalcins and calcium regulated potassium channel interacting proteins. These data quantitatively identify a rich repertoire for combinatorial approaches to pain control.
This paper mainly examines ion channels, receptors and transmitters but RNA-Seq transcriptome extends the fingerprint to all aspects of neuronal function, this in turn provides many new ways to investigate nociceptive neurobiology. Indeed further in silico transcriptome data mining can take a wide variety of directions. Molecular circuits, immunocytochemistry, and 5′ and 3′ analyses provide templates for integration into intra- and intercellular communication networks. These aspects are somewhat tangentially addressed here. The most functionally abundant groups are enzymes, and certain regulatory pathways are highly enriched in the TRPV1L neurons. In Table 1, five protein kinases are well expressed and show a 5.2 to 12.7 fold differential expression in the TRPV1L population. This suggests that a specific phosphoprotein signature can be elaborated from substrates in the RNA-Seq transcriptome that contain the specific phosphorylation motifs for these kinases. This type of reasoning can be applied to many different protein families within the datasets presented allowing a more detailed and heuristic model of nociceptive neuronal function to be elaborated.
TRPV1 Lineage and Polymodality
The idea of a molecular fingerprint for nociceptive neurons is in part realized by the combined genetic and sequencing approaches in this study. It is of note that the early developmental stage genetic labeling strategy we used yields a transcriptional profile that is applicable to multiple sensory modalities and may be informative for pathological pain states. In the mouse, TRPV1 and, consequently, Cre recombinase is expressed early in DRG development; as a result, expression of red fluorescent protein labels neurons in which TRPV1 expression is persistent and others in which TRPV1 expression is transient. Therefore, the labeling strategy encompasses a broader range of sensory modalities in the adult than the final expression pattern of TRPV1.59,62 Behavioral studies suggest the presence of “labeled lines” specific for modalities such as noxious heat, cool and mechanosensations.11 However, in species from the mouse to monkey, the presence of C-fiber polymodal nociceptors that are responsive to touch, pressure, pinch, noxious heat and cold5,11,18 suggest that developmental segregation of nociceptive modalities into distinct neuronal populations may only occur in a partial fashion in mammals. We observe, for example, that TRPM8 is significantly reduced in the RTX-treated rat trigeminal ganglion consistent with a subpopulation of polymodal thermo-nociceptors in rat (Table S6). Mouse TRPV1-lineage DRG neurons abundantly and predominantly express the TH and Vglut3 genes and these neurons appear to comprise a distinct population of low-threshold C-fiber mechanoreceptors. Despite their low threshold and apparent hedonic valence this group of neurons are observed to synapse centrally in spinal cord lamina I and inner lamina II (substantia gelatinosa); regions typically associated with nociceptive processing.1 In this regard, deletion of the Vglut3 gene, which causes an inability to store transmitter glutamate, was reported to block the establishment of mechano-hyperalgesia following nerve injury,83 although recent data suggest the participation of these afferents may not be so clear.53 Itch-responsive neurons are also subsumed in the TRPV1 lineage: genetic or chemical ablation of TRPV1 neurons blocks the response to puritogens34 and TRPV1 modulates transmission of itch sensations.52 It is evident from this discussion that, for the TRPV1 lineage population, enriched gene expression can present in several ways at the cellular level. For example, expression of a particular gene can be distributed across several modalities of nociceptive neurons or more exclusively expressed in a particular subpopulation. Thus, further genetic labeling strategies, complementary to those employed here will fine tune the explicit nature of these co-expression matrices.
High and Low Levels of Gene Expression
Throughout this paper the quantitative nature of RNA-Seq has been emphasized, at the same time, we want to dispel any impression that only highly expressed genes are important: the strategic location of the protein product from a low expressed gene can have a profound effect on neuronal function. Emerging aspects of protein function continue to be revealed such as biased agonism and combinatorial heterodimerization of GPCRs.100 In this regard, quantitative cell-specific transcriptomics can play a fundamental role in guiding experimental design and determining, a priori, which combinations to investigate. Comprehensive data are particularly relevant for single or combinatorial molecular interventions aimed at complex networks such as second messenger signaling pathways or transcriptional control and lay a foundation for a systems biology approach to nociception.
Concluding Remarks
An enormous literature has evolved generally lamenting the inability of a candidate drug’s performance in animal models to predict efficacy in human clinical trials.55,65,101 We document several important differences related to the translational potential of observations made in model species. One implication is that expression of molecular targets or complete signaling pathways in model organisms needs to be verified in humans prior to drug development efforts. Much of the translational difficulties are blamed on lack of physiological correspondence between the human and the model, although more recently, renewed emphasis is being given to large animal and human models.8,71 The present study suggests that establishment of gene expression correspondence may offset some of the problems related to target selection. Genetic manipulations of higher order species (e.g., marmosets) combined with RNA-Seq may further inform human ganglionic gene expression for translational research.81 Our data support the idea that specification of expression to the nociceptive neuronal subpopulation and also the transcript pool common to both populations, as well as proper identification of the precise paralogs are additional critical factors for programs aimed at peripherally-acting analgesics. It will also be extremely informative to incorporate RNA-Seq results from neuron-specific, glial-specific and activation- or lesion-induced expression into archives of searchable databases. The present transcriptome data presage a more fundamental paradigm shift as more RNA-Seq information is accumulated, where experiments in the planning stage include preliminary consultation of one or more transcriptome RNA-Seq databases.
Supplementary Material
The top figure outlines the RNA-Seq filtering process after reads are mapped to the appropriate genome by RUM. Multiple records for the same gene are filtered by selecting the instance which constitutes the most mapped reads, thereby selecting either the major isoform or the most supported gene model. RNA-Seq records with raw counts below 20 are filtered out, as well as those without annotations or annotations corresponding to small non-coding RNA transcripts. For the most part, genes expressed above 1 RPKM are analyzed within the paper. The lower panels analyze the RNA-Seq technical replicates (Illumina HiSeq vs Illumina GaIIx) for the TRPV1L and TRPV1D populations within the R statistical programming language. The correlation coefficients for both datasets are ~ 0.9 supporting the strong reproducibility of the data across platforms.
The table lists the total number of reads and mapping statistics, as generated by RUM, for each dataset included in the study.
This table contains the molecular signature of TRPV1L neurons based on level of expression. The table lists all 263 TRPV1L functionally well annotated transcripts with a differential of 5X and above (there is partial overlap with Table 1). The genes have been grouped into functional categories according to known annotations and publications. Within the groups, the genes have been sorted on expression (high to low) and only genes with an expression level above 1 RPKM are included. The adjusted fold change values are color coded: Red: >20 fold, Blue: 20-10 fold, and Green: < 10 fold difference. The accession id provides the exact reference to the gene isoform analyzed within this study.
This table lists all functionally well annotated TRPV1L genes with a differential above 1.5X and below 5X. The table only includes genes with an expression level above 1 RPKM.
Table 4A is a partial sampling of the TRPV1D genes with a differential of 5X and above. The genes have been grouped into functional categories according to known annotations and publications. Within the groups, the genes were sorted on expression (high to low) and only the top five most highly expressed genes within each group are included. (4B) The table lists all the genes in the TRPV1D population that exhibit differential expression above 1.5X fold difference and are expressed above 1 RPKM. Entries are arranged alphabetically.
This table lists all functionally well annotated genes whose expression is common to both the TRPV1L and D preparations. All entries have a fold difference equal to or below 1.5 when comparing TRPV1L to TRPV1D. Only genes with an RPKM above 1 are included. Entries are arranged alphabetically.
Top panels: The complete histogram showing the distribution of gene expression based on RPKM for TRPV1L neurons: 14% are expressed between 0–1 RPKM, approximately 12% are expressed between 1–3 RPKM, the majority of TRPV1L genes are expressed between 3–20 RPKM, (~37%,), and 20–200 (~34%). The high expressing group, between 200–3600 RPKM contains ~3% of the expressed genes. Thus, a number like 200 RPKM means that gene is in pretty rare company with only 150 other genes in that range or higher; 3 to 30 RPKM means it is expressed at the general level of most cellular genes. The two panels in the second row show the most highly expressed category (200 to highest RPKM) for TRPV1 depleted and common categories. Note that the neurofilament genes (Nefl and Nefm) are highly expressed by the large DRG neurons that comprise the TRPV1D population but these are also expressed in the TRPV1L population but just not to the same extent. The same consideration of differential vs. absolute also applies to a highly expressed TRPV1L gene synuclein γ (Sncg) and, in fact, to nearly all the genes. The remaining panels show the full histogram for low, medium and high expressing groups for whole rat DRG and for a rat mixed tissue preparation (equal amounts of total RNA from cerebral cortex, cerebellum, midbrain, hypothalamus, hindbrain, spinal cord, retina, pituitary, heart, liver, lung, kidney, skeletal muscle, small intestine and adrenal gland). Note the overall similarity between the rat whole DRG and the mouse histograms. For all transcriptomes illustrated in SFigure 2, the high end of each histogram can form a general molecular “fingerprint” of that tissue. For example, familiar transcripts associated with TRPV1 neurons are calcitonin gene related peptide a (Calca) and peripherin (Prph), both are highly expressed in the TRPV1L neurons. A less familiar member of the fingerprint is Cd24A, which codes for a GPI-linked sialoglycoprotein that interacts with contactin 1 and 2 and L1Cam, a system implicated in tissue remodeling subsequent to nerve injury and axotomy (Lieberoth et al., 2009; Yamanaka et al., 2007). The transcriptional fingerprint of the rat mixed tissue reflects the presence of highly specialized cells such as photoreceptors, pituicytes and muscle fibers and includes functionally relevant genes with dedicated functions such as rhodopsin (Rho), growth hormone (Gh1), prolactin (prl), myosin (myh4) and albumin (Alb). Based on overlap and within gene family complementarity (e.g., the calmodulin paralogs) reliable molecular fingerprinting of neuronal subpopulations will likely require inclusion of more transcripts.
Abbreviations: Actb: β actin, Prph: peripherin, Cd24: nectadrin, Calca: α-type CGRP, Tppp3: tubulin polymerization-promoting protein 3, Hsp90ab1: 90kDa, αB1 heat shock protein, S100a6: S100 calcium binding protein A6, Tuba1a: α1 tubulin, Fxyd2: γ Na(+)/K(+) ATPase subunit, Sncg: γ synuclein, Calm3: calmodulin 3, Ywhag: 14-3-3 protein γ, Pcp4: Purkinje cell protein 4, Mbp: myelin basic protein, Nefh: neurofilament, heavy polypeptide, Mtap1b: microtubule-associated protein 1B, Atp1a1: α1 ATPase, Na+/K+ transporting polypeptide, Ndufa4: NADH dehydrogenase 1α, subcomplex4, Pmp22: peripheral myelin protein 22, Apoe: apolipoprotein E, Nefm: medium neurofilament, Nefl: light neurofilament, Mpz: myelin protein zero, Ywhah: 14-3-3 η, Fstl1: follistatin-e3like 1, Anxa2: annexin A2, Atpif1: ATPase inhibitory factor 1, Tceb2: transcription elongation factor B2, Rplp1: ribosomal protein, large, P1, Calm1: calmodulin 1, Calm2: calmodulin 2, Stmn2: stathmin-like 2, Fth1: ferritin, heavy polypeptide 1, Ndrg4: NDRG family member 4, Ubb: ubiquitin B, Cox6a1: cytochrome c oxidase subunit, Aldoa: aldolase A, Tubb3: β3 tubulin, Tmsb4x: β4 thymosin, Uchl1: ubiquitin thiolesterase, Thy1: θ thymus cell antigen 1, Hba-a2: hemoglobin α2, Pmp22: peripheral myelin protein 22, Uchl1: ubiquitin thiolesterase, Scd: stearoyl-CoA desaturase, Atp1a3: α3 ATPase, Na+/K+ transporting polypeptide, Prl: prolactin, Ckm: creatine kinase, Myh4: myosin, heavy polypeptide 4, Atp5b: β ATP synthase, Acta1: α1 actin, Rho: rhodopsin, Alb: albumin, Plp1: myelin proteolipid protein, and Gh1: growth hormone 1.
The extended TRPV1 annotation sequence is provided for the 5′ (top) and 3′ (bottom) UTR sequence. We designated the end of the selected segment at 50 mapped reads. The extended segment is given in red. Please note that since the submission of this manuscript, RNA-Seq data has been incorporated into the Refseq annotation pipeline. Thus Refseq TRPV1 (NC_000077.6) 5′ UTR now corresponds to our 5′UTR extension; the Refseq 3′ UTR, extends beyond our 3′ UTR extension. The reannotation of genomes is a fluid process. As the RNA-Seq databases evolve and grow, the precise UTR locations become more precisely defined. Attempts were made using several promoter prediction tools, including Berkeley Neural Network Promoter Prediction tool (version 2.3) < http://www.fruitfly.org/seq_tools/promoter.html> to predict a new promoter upstream of the start of the newly extended 5′ UTR. However, none of the tools predicted a conventionally defined sequence close to the new start. RNA-Seq data for rat dorsal root ganglion showed that the TRPV1 5′ and 3′UTR regions could also be similarly extended in rat. The human TRPV1 gene was well annotated and did not require modifications.
A. Genes for rat DRG and TG were ranked according to descending expression value within their respective tissue transcriptome. The rat TG and DRG contain approximately 12,000 genes and the x and y axes reflect this number. Genes from Figure 2 are plotted as a test set for assessing similarity between the TG and DRG. The plot includes genes that are expressed greater than 1 RPKM in both TRPV1L neurons and the TRPVD ganglion preparation. The correlation coefficient for relative expression is 0.88. B. When a broader analysis of TG and DRG is performed, Hoxd1, a member of the homeobox gene family is noticeably different. Hoxd1 is expressed in TG and DRG whereas the other 17 homeobox genes are only expressed in DRG to varying degree. In the context of the Hox gene family, it is important to note that only lumbar rat DRGs were dissected in the present experiments. The mean ± SEM values are provided in the table.
Lists all the transcripts from Tables 1 and S2 (TRPV1L with a fold difference above or equal to 5) that were significantly (padj < .05) down regulated after RTX. DESeq was used for the statistical analysis according to standard protocol (Anders and Huber, 2010).
This table lists all the functionally well annotated genes (12,098) in the mouse lineage experiment that are expressed at or below 1 RPKM. The number of raw reads ranged from 0 to 2,807 in the TRPV1L population and were distributed as following: 0 = 5,614; 1–2 = 1,216; 3–19 = 2,983; 20–99 = 1,329; 100–499 = 861; 500–2807 = 95. To give some interpretation, 200 reads, for example, can cover all the exons of particular gene and may allow for assembly of several full length transcripts. However one has to take into account the number of neurons in the ganglion. Depending on how the reads are distributed, one neuron could have several full length transcripts or raw reads might be present in multiple cell types in which case a full length transcript is, unlikely to be assembled.
Table S8: Expressed Mouse DRG Gene Database. This table lists in alphabetical order all the genes/transcripts in the mouse lineage experiment that are expressed above 20 raw counts. Few duplicate entries may exist due to aliases in the “Functionally Not Well Annotated Genes” section. The table is provided to facilitate looking up of gene families and genes of interest. This full coverage table also contains many non-protein coding genes (i.e pseudogenes, small nuclear ribonucleic acid [snRNA], and non-coding RNA [ncRNA]) however the table is not meant to be an exhaustive list of all transcripts within the mouse transcriptome. Our poly-A selection was not designed to comprehensively capture non-polyadenylated, non-coding RNAs, such as microRNA [miRNA], long intergenic non-coding RNAs [lincRNA], piwi-interacting RNA [piRNA], or small nucleolar RNA [snoRNA], and we removed the incidental transcripts that were captured in these categories. Similarly, the table does include unannotated transcripts, but as the NCBI database evolves more entries can be added in the future.
Highlights.
Next-gen RNA-Seq and genetic labeling identifies all genes expressed in pain-sensing neurons
Provides an in silico resource for planning future studies that is low cost, animal sparing, and fast
Suggests molecular microcircuits for autonomous control of afferent nerve terminal excitability
Provides molecular basis for more detailed, heuristic models in pain systems biology
PERSPECTIVE.
Next-gen RNA-Seq, combined with molecular genetics, provides a comprehensive and quantitative measurement of transcripts in TRPV1 lineage neurons and a contrasting transcriptome from non-TRPV1 neurons and cells. The transcriptome highlights previously unrecognized protein families, identifies multiple molecular circuits for excitatory or inhibitory autocrine and paracrine signaling, and suggests new combinatorial approaches to pain control.
Acknowledgments
We thank Catalina Dumitrascu for help with the immunocytochemistry.
Footnotes
There are no actual or potential conflicts of interest in relation to this article.
DISCLOSURES:
This research was supported by the Intramural Research Programs of the National Center for Complementary and Alternative Medicine, the National Institute of Dental and Craniofacial Research, and the Department of Perioperative Medicine, Clinical Center of the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abraira VE, Ginty DD. The sensory neurons of touch. Neuron. 2013;79:618–639. doi: 10.1016/j.neuron.2013.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amici M, Doherty A, Jo J, Jane D, Cho K, Collingridge G, Dargan S. Neuronal calcium sensors and synaptic plasticity. Biochemical Soc Trans. 2009;37:1359–1363. doi: 10.1042/BST0371359. [DOI] [PubMed] [Google Scholar]
- 3.Badio B, Daly JW. Epibatidine, a potent analgetic and nicotinic agonist. Mol Pharmacol. 1994;45:563–569. [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beitel RE, Dubner R. Response of unmyelinated (C) polymodal nociceptors to thermal stimuli applied to monkey’s face. J Neurophysiol. 1976;39:1160–1175. doi: 10.1152/jn.1976.39.6.1160. [DOI] [PubMed] [Google Scholar]
- 6.Bourane S, Mechaly I, Venteo S, Garces A, Fichard A, Valmier J, Carroll P. A SAGE-based screen for genes expressed in sub-populations of neurons in the mouse dorsal root ganglion. BMC Neurosci. 2007;8:97. doi: 10.1186/1471-2202-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandenburger T, Castoldi M, Brendel M, Grievink H, Schlosser L, Werdehausen R, Bauer I, Hermanns H. Expression of spinal cord microRNAs in a rat model of chronic neuropathic pain. Neurosci Lett. 2012;506:281–286. doi: 10.1016/j.neulet.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 8.Brown DC, Iadarola MJ, Perkowski SZ, Erin H, Shofer F, Laszlo KJ, Olah Z, Mannes AJ. Physiologic and antinociceptive effects of intrathecal resiniferatoxin in a canine bone cancer model. Anesthesiology. 2005;103:1052–1059. doi: 10.1097/00000542-200511000-00020. [DOI] [PubMed] [Google Scholar]
- 9.Brumovsky P, Villar MJ, Hökfelt T. Tyrosine hydroxylase is expressed in a subpopulation of small dorsal root ganglion neurons in the adult mouse. Exp Neurol. 2006;200:153–165. doi: 10.1016/j.expneurol.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Brumovsky P, Watanabe M, Hökfelt T. Expression of the vesicular glutamate transporters-1 and -2 in adult mouse dorsal root ganglia and spinal cord and their regulation by nerve injury. Neuroscience. 2007;147:469–490. doi: 10.1016/j.neuroscience.2007.02.068. [DOI] [PubMed] [Google Scholar]
- 11.Cain DM, Khasabov SG, Simone DA. Response properties of mechanoreceptors and nociceptors in mouse glabrous skin: an in vivo study. J Neurophysiol. 2001;85:1561–1574. doi: 10.1152/jn.2001.85.4.1561. [DOI] [PubMed] [Google Scholar]
- 12.Ceko M, Milenkovic N, Ie Coutre P, Westermann J, Lewin GR. Inhibition of c-Kit signaling is associated with reduced heat and cold pain sensitivity in humans. Pain. 2014;155:1222–1228. doi: 10.1016/j.pain.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Chambers KJ, Tonkin LA, Chang E, Shelton DN, Linskens MH, Funk WD. Identification and cloning of a sequence homologue of dopamine beta-hydroxylase. Gene. 1998;218:111–120. doi: 10.1016/s0378-1119(98)00344-8. [DOI] [PubMed] [Google Scholar]
- 14.Chen SR, Cai YQ, Pan HL. Plasticity and emerging role of BKCa channels in nociceptive control in neuropathic pain. J Neurochem. 2009;110:352–362. doi: 10.1111/j.1471-4159.2009.06138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costigan M, Samad TA, Allchorne A, Lanoue C, Tate S, Woolf CJ. High basal expression and injury-induced down regulation of two regulator of G-protein signaling transcripts, RGS3 and RGS4 in primary sensory neurons. Mol Cell Neurosci. 2003;24:106–116. doi: 10.1016/s1044-7431(03)00135-0. [DOI] [PubMed] [Google Scholar]
- 16.De Blasi A, Conn PJ, Pin J, Nicoletti F. Molecular determinants of metabotropic glutamate receptor signaling. Trends Pharmacol Sci. 2001;22:114–120. doi: 10.1016/s0165-6147(00)01635-7. [DOI] [PubMed] [Google Scholar]
- 17.Dolan S, Gunn MD, Biddlestone L, Nolan AM. The selective metabotropic glutamate receptor 7 allosteric agonist AMN082 inhibits inflammatory pain-induced and incision-induced hypersensitivity in rat. Behav Pharmacol. 2009;20:596–604. doi: 10.1097/FBP.0b013e32832ec5d1. [DOI] [PubMed] [Google Scholar]
- 18.Dubner R, Sumino R, Wood WI. A peripheral “cold” fiber population responsive to innocuous and noxious thermal stimuli applied to monkey’s face. J Neurophysiol. 1975;38:1373–1389. doi: 10.1152/jn.1975.38.6.1373. [DOI] [PubMed] [Google Scholar]
- 19.Dussor GO, Leong AS, Gracia NB, Kilo S, Price TJ, Hargreaves KM, Flores CM. Potentiation of evoked calcitonin gene-related peptide release from oral mucosa: a potential basis for the pro-inflammatory effects of nicotine. Eur J Neurosci. 2003;18:2515–2526. doi: 10.1046/j.1460-9568.2003.02935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Echeverry S, Shi XQ, Rivest S, Zhang J. Peripheral nerve injury alters blood-spinal cord barrier functional and molecular integrity through a selective inflammatory pathway. J Neurosci. 2011;31:10819–10828. doi: 10.1523/JNEUROSCI.1642-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eng SR, Dykes IM, Lanier J, Fedtsova N, Turner EE. POU-domain factor Brn3a regulates both distinct and common programs of gene expression in the spinal and trigeminal sensory ganglia. Neural Dev. 2007;2:3. doi: 10.1186/1749-8104-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garnier M, Zaratin PF, Ficalora G, Valente M, Fontanella L, Rhee MH, Blumer KJ, Scheideler MA. Up-regulation of regulator of G protein signaling 4 expression in a model of neuropathic pain and insensitivity to morphine. J Pharmacol Exp Ther. 2003;304:1299–1306. doi: 10.1124/jpet.102.043471. [DOI] [PubMed] [Google Scholar]
- 23.Genzen JR, Van Cleve W, McGehee DS. Dorsal root ganglion neurons express multiple nicotinic acetylcholine receptor subtypes. J Neurophysiol. 2001;86:1773–1782. doi: 10.1152/jn.2001.86.4.1773. [DOI] [PubMed] [Google Scholar]
- 24.Giuliani S, Amann R, Papini AM, Maggi CA, Meli A. Modulatory action of galanin on responses due to antidromic activation of peripheral terminals of capsaicin-sensitive sensory nerves. Eur J Pharmacol. 1989;163:91–96. doi: 10.1016/0014-2999(89)90399-3. [DOI] [PubMed] [Google Scholar]
- 25.Goldshmit Y, Matteo R, Sztal T, Ellett F, Frisca F, Moreno K, Crombie D, Lieschke GJ, Currie PD, Sabbadini RA, Pebay A. Blockage of lysophosphatidic acid signaling improves spinal cord injury outcomes. Am J Pathol. 2012;181:978–992. doi: 10.1016/j.ajpath.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, Stoeckert CJ, Hogenesch JB, Pierce EA. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27:2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gum RJ, Wakefield B, Jarvis MF. P2X receptor antagonists for pain management: examination of binding and physicochemical properties. Purinergic Signal. 2012;8:41–56. doi: 10.1007/s11302-011-9272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammer P, Banck MS, Amberg R, Wang C, Petznick G, Luo S, Khrebtukova I, Schroth GP, Beyerlein P, Beutler AS. mRNA-seq with agnostic splice site discovery for nervous system transcriptomics tested in chronic pain. Genome Res. 2010;20:847–860. doi: 10.1101/gr.101204.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heiss J, Iadarola M, Cantor F, Oughourli A, Smith R, Mannes A. A Phase I study of the intrathecal administration of resiniferatoxin for treating severe refractory pain associated with advanced cancer. J Pain. 2014;15:S67. [Google Scholar]
- 30.Hirata T, Kasugai T, Morii E, Hirota S, Nomura S, Fujisawa H, Kitamura Y. Characterization of c-kit-positive neurons in the dorsal root ganglion of mouse. Brain Res Dev Brain Res. 1995;85:201–211. doi: 10.1016/0165-3806(94)00205-e. [DOI] [PubMed] [Google Scholar]
- 31.Iadarola MJ, Mannes AJ. The vanilloid agonist resiniferatoxin for interventional-based pain control. Curr Top Med Chem. 2011;11:2171–2179. doi: 10.2174/156802611796904942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichikawa H, Deguchi T, Nakago T, Jacobowitz DM, Sugimoto T. Parvalbumin, calretinin and carbonic anhydrase in the trigeminal and spinal primary neurons of the rat. Brain Res. 1994;655:241–245. doi: 10.1016/0006-8993(94)91620-9. [DOI] [PubMed] [Google Scholar]
- 33.Ichikawa H, Itota T, Nishitani Y, Torii Y, Inoue K, Sugimoto T. Osteopontin-immunoreactive primary sensory neurons in the rat spinal and trigeminal nervous systems. Brain Res. 2000;863:276–281. doi: 10.1016/s0006-8993(00)02126-0. [DOI] [PubMed] [Google Scholar]
- 34.Imamachi N, Park GH, Lee H, Anderson DJ, Simon MI, Basbaum AI, Han SK. TRPV1-expressing primary afferents generate behavioral responses to pruritogens via multiple mechanisms. Proc Natl Acad Sci U S A. 2009;106:11330–11335. doi: 10.1073/pnas.0905605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imhof AK, Glück L, Gajda M, Lupp A, Bräuer R, Schaible HG, Schulz S. Differential antiinflammatory and antinociceptive effects of the somatostatin analogs octreotide and pasireotide in a mouse model of immune-mediated arthritis. Arthritis Rheum. 2011;63:2352–2362. doi: 10.1002/art.30410. [DOI] [PubMed] [Google Scholar]
- 36.Ivanusic JJ, Goulding KE, Kwok MM, Jennings EA. Neurochemical classification and projection targets of CART peptide immunoreactive neurons in sensory and parasympathetic ganglia of the head. Neuropeptides. 2012;46:55–60. doi: 10.1016/j.npep.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Jung H, Bhangoo S, Banisadr G, Freitag C, Ren D, White FA, Miller RJ. Visualization of chemokine receptor activation in transgenic mice reveals peripheral activation of CCR2 receptors in states of neuropathic pain. J Neurosci. 2009;29:8051–8062. doi: 10.1523/JNEUROSCI.0485-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kähler CM, Kaufmann G, Kähler ST, Wiedermann CJ. The neuropeptide secretoneurin stimulates adhesion of human monocytes to arterial and venous endothelial cells in vitro. Regul Pept. 2002;110:65–73. doi: 10.1016/s0167-0115(02)00161-1. [DOI] [PubMed] [Google Scholar]
- 39.Karai L, Brown DC, Mannes AJ, Connelly ST, Brown J, Gandal M, Wellisch OM, Neubert JK, Olah Z, Iadarola MJ. Deletion of vanilloid receptor 1-expressing primary afferent neurons for pain control. J Clin Invest. 2004;113:1344–1352. doi: 10.1172/JCI20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi K, Fukuoka T, Obata K, Yamanaka H, Dai Y, Tokunaga A, Noguchi K. Distinct expression of TRPM8, TRPA1, and TRPV1 mRNAs in rat primary afferent neurons with adelta/c-fibers and colocalization with trk receptors. J Comp Neurol. 2005;493:596–606. doi: 10.1002/cne.20794. [DOI] [PubMed] [Google Scholar]
- 42.Korboukh I, Hull-Ryde EA, Rittiner JE, Randhawa AS, Coleman J, Fitzpatrick BJ, Setola V, Janzen WP, Frye SV, Zylka MJ, Jin J. Orally active adenosine A(1) receptor agonists with antinociceptive effects in mice. J Med Chem. 2012;55:6467–6477. doi: 10.1021/jm3004834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kusuda R, Cadetti F, Ravanelli MI, Sousa TA, Zanon S, De Lucca FL, Lucas G. Differential expression of microRNAs in mouse pain models. Mol Pain. 2011;7:17. doi: 10.1186/1744-8069-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamberts JT, Smith CE, Li MH, Ingram SL, Neubig RR, Traynor JR. Differential control of opioid antinociception to thermal stimuli in a knock-in mouse expressing regulator of G-protein signaling-insensitive Gαo protein. J Neurosci. 2013;33:4369–4377. doi: 10.1523/JNEUROSCI.5470-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landry M, Holmberg K, Zhang X, Hökfelt T. Effect of axotomy on expression of NPY, galanin, and NPY Y1 and Y2 receptors in dorsal root ganglia and the superior cervical ganglion studied with double-labeling in situ hybridization and immunohistochemistry. Exp Neurol. 2000;162:361–384. doi: 10.1006/exnr.1999.7329. [DOI] [PubMed] [Google Scholar]
- 46.Legha W, Gaillard S, Gascon E, Malapert P, Hocine M, Alonso S, Moqrich A. stac1 and stac2 genes define discrete and distinct subsets of dorsal root ganglia neurons. Gene Expr Patterns. 2010;10:368–375. doi: 10.1016/j.gep.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:115–120. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Gibson G, Kim JS, Kroin J, Xu S, van Wijnen AJ, Im HJ. MicroRNA-146a is linked to pain-related pathophysiology of osteoarthritis. Gene. 2011;480:34–41. doi: 10.1016/j.gene.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin L, Sun W, Wikenheiser AM, Kung F, Hoffman DA. KChIP4a regulates Kv4.2 channel trafficking through PKA phosphorylation. Mol Cell Neurosci. 2010;43:315–325. doi: 10.1016/j.mcn.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, Kim A, Ru F, Guan Y, Weng HJ, Geng Y, Undem BJ, Kollarik M, Chen ZF, Anderson DJ, Dong X. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell. 2009;139:1353–1365. doi: 10.1016/j.cell.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S, Lin L, Jiang P, Wang D, Xing Y. A comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic acids Res. 2011;39:578–588. doi: 10.1093/nar/gkq817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Abdel Samad O, Zhang L, Duan B, Tong Q, Lopes C, Ji RR, Lowell BB, Ma Q. VGLUT2-dependent glutamate release from nociceptors is required to sense pain and suppress itch. Neuron. 2010;68:543–556. doi: 10.1016/j.neuron.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lou S, Duan B, Vong L, Lowell BB, Ma Q. Runx1 controls terminal morphology and mechanosensitivity of VGLUT3-expressing C-mechanoreceptors. J Neurosci. 2013;33:870–882. doi: 10.1523/JNEUROSCI.3942-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mannes A, Hughes M, Quezado Z, Berger A, Fojo T, Smith R, Butman J, Lonser R, Iadarola M. Resiniferatoxin, a potent TRPV1 agonist: Intrathecal administration to treat severe pain associated with advanced cancer- case report. Pain. 2010;11:S43. [Google Scholar]
- 55.Mao J. Current challenges in translational pain research. Trends Pharmacol Sci. 2012;33:568–573. doi: 10.1016/j.tips.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maric D, Fiorio Pla A, Chang YH, Barker JL. Self-renewing and differentiating properties of cortical neural stem cells are selectively regulated by basic fibroblast growth factor (FGF) signaling via specific FGF receptors. J Neurosci. 2007;27:1836–1852. doi: 10.1523/JNEUROSCI.5141-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKelvey L, Shorten GD, O’Keeffe GW. Nerve growth factor-mediated regulation of pain signalling and proposed new intervention strategies in clinical pain management. J Neurochem. 2013;124:276–289. doi: 10.1111/jnc.12093. [DOI] [PubMed] [Google Scholar]
- 59.McKemy DD. A spicy family tree. TRPV1 and its thermoceptive and nociceptive lineage. EMBO J. 2011;30:453–455. doi: 10.1038/emboj.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKemy DD. The molecular and cellular basis of cold sensation. ACS Chem Neurosci. 2013;4:238–247. doi: 10.1021/cn300193h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mishra SK, Hoon MA. The cells and circuitry for itch responses in mice. Science. 2013;340:968–971. doi: 10.1126/science.1233765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mishra SK, Tisel SM, Orestes P, Bhangoo SK, Hoon MA. TRPV1-lineage neurons are required for thermal sensation. EMBO J. 2011;30:582–593. doi: 10.1038/emboj.2010.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitchell K, Bates BD, Keller JM, Lopez M, Scholl L, Navarro J, Madian N, Haspel G, Nemenov MI, Iadarola MJ. Ablation of rat TRPV1-expressing Adelta/C-fibers with resiniferatoxin: analysis of withdrawal behaviors, recovery of function and molecular correlates. Mol Pain. 2010;6:94. doi: 10.1186/1744-8069-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitchell K, Lebovitz EE, Keller JM, Mannes AJ, Nemenov MI, Iadarola MJ. Nociception and inflammatory hyperalgesia evaluated in rodents using infrared laser stimulation after Trpv1 gene knockout or resiniferatoxin lesion. Pain. 2014;155:733–745. doi: 10.1016/j.pain.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mogil JS. Animal models of pain: progress and challenges. Nat Rev Neurosci. 2009;10:283–294. doi: 10.1038/nrn2606. [DOI] [PubMed] [Google Scholar]
- 66.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 67.Nardi A, Olesen SP. BK channel modulators: a comprehensive overview. Curr Med Chem. 2008;15:1126–1146. doi: 10.2174/092986708784221412. [DOI] [PubMed] [Google Scholar]
- 68.Neubert JK, Karai L, Jun JH, Kim HS, Olah Z, Iadarola MJ. Peripherally induced resiniferatoxin analgesia. Pain. 2003;104:219–228. doi: 10.1016/s0304-3959(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 69.Norris AJ, Foeger NC, Nerbonne JM. Interdependent roles for accessory KChIP2, KChIP3, and KChIP4 subunits in the generation of Kv4-encoded IA channels in cortical pyramidal neurons. J Neurosci. 2010;30:13644–13655. doi: 10.1523/JNEUROSCI.2487-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Obata K, Noguchi K. BDNF in sensory neurons and chronic pain. Neurosci Res. 2006;55:1–10. doi: 10.1016/j.neures.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 71.Oertel BG, Lötsch J. Clinical pharmacology of analgesics assessed with human experimental pain models: bridging basic and clinical research. Br J Pharmacol. 2013;168:534–553. doi: 10.1111/bph.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olah Z, Szabo T, Karai L, Hough C, Fields RD, Caudle RM, Blumberg PM, Iadarola MJ. Ligand-induced dynamic membrane changes and cell deletion conferred by vanilloid receptor 1. J Biol Chem. 2001;276:11021–11030. doi: 10.1074/jbc.M008392200. [DOI] [PubMed] [Google Scholar]
- 73.Okazawa M, Inoue W, Hori A, Hosokawa H, Matsumura K, Kobayashi S. Noxious heat receptors present in cold-sensory cells in rats. Neurosci Lett. 2004;359:33–36. doi: 10.1016/j.neulet.2004.01.074. [DOI] [PubMed] [Google Scholar]
- 74.Palmer CL, Lim W, Hastie PG, Toward M, Korolchuk VI, Burbidge SA, Banting G, Collingridge GL, Isaac JT, Henley JM. Hippocalcin functions as a calcium sensor in hippocampal LTD. Neuron. 2005;47:487–494. doi: 10.1016/j.neuron.2005.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patel KN, Dong X. An itch to be scratched. Neuron. 2010;68:334–339. doi: 10.1016/j.neuron.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Petho G, Reeh PW. Sensory and signaling mechanisms of bradykinin, eicosanoids, platelet-activating factor, and nitric oxide in peripheral nociceptors. Physiol Rev. 2012;92:1699–1775. doi: 10.1152/physrev.00048.2010. [DOI] [PubMed] [Google Scholar]
- 77.Pogorzala LA, Mishra SK, Hoon MA. The cellular code for mammalian thermosensation. J Neurosci. 2013;33:5533–5541. doi: 10.1523/JNEUROSCI.5788-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Price J, Mudge AW. A subpopulation of rat dorsal root ganglion neurones is catecholaminergic. Nature. 1983;301:241–243. doi: 10.1038/301241a0. [DOI] [PubMed] [Google Scholar]
- 79.Quick K, Zhao J, Eijkelkamp N, Linley JE, Rugiero F, Cox JJ, Raouf R, Gringhuis M, Sexton JE, Abramowitz J, Taylor R, Forge A, Ashmore J, Kirkwood N, Kros CJ, Richardson GP, Freichel M, Flockerzi V, Birnbaumer L, Wood JN. TRPC3 and TRPC6 are essential for normal mechanotransduction in subsets of sensory neurons and cochlear hair cells. Open Biol. 2012;2:120068. doi: 10.1098/rsob.120068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rau KK, McIlwrath SL, Wang H, Lawson JJ, Jankowski MP, Zylka MJ, Anderson DJ, Koerber HR. Mrgprd enhances excitability in specific populations of cutaneous murine polymodal nociceptors. J Neurosci. 2009;29:8612–8619. doi: 10.1523/JNEUROSCI.1057-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sasaki E, Suemizu H, Shimada A, Hanazawa K, Oiwa R, Kamioka M, Tomioka I, Sotomaru Y, Hirakawa R, Eto T, Shiozawa S, Maeda T, Ito M, Ito R, Kito C, Yagihashi C, Kawai K, Miyoshi H, Tanioka Y, Tamaoki N, Habu S, Okano H, Nomura T. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459:523–527. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- 82.Scott A, Hasegawa H, Sakurai K, Yaron A, Cobb J, Wang F. Transcription factor short stature homeobox 2 is required for proper development of tropomyosin-related kinase B-expressing mechanosensory neurons. J Neurosci. 2011;31:6741–6749. doi: 10.1523/JNEUROSCI.5883-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seal RP, Wang X, Guan Y, Raja SN, Woodbury CJ, Basbaum AI, Edwards RH. Injury-induced mechanical hypersensitivity requires C-low threshold mechanoreceptors. Nature. 2009;462:651–655. doi: 10.1038/nature08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi TJ, Xiang Q, Zhang MD, Tortoriello G, Hammarberg H, Mulder J, Fried K, Wagner L, Josephson A, Uhlén M, Harkany T, Hökfelt T. Secretagogin is expressed in sensory CGRP neurons and in spinal cord of mouse and complements other calcium-binding proteins, with a note on rat and human. Mol Pain. 2012;8:80. doi: 10.1186/1744-8069-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sjögren B. Regulator of G protein signaling proteins as drug targets: current state and future possibilities. Adv Pharmacol. 2011;62:315–347. doi: 10.1016/B978-0-12-385952-5.00002-6. [DOI] [PubMed] [Google Scholar]
- 86.Sprott H, Pap T, Rethage J, Wintersberger W, Gay RE, Bradley LA, Uebelhart D, Gay S. Expression of the precursor of secretoneurin, secretogranin II, in the synovium of patients with rheumatoid arthritis and osteoarthritis. J Rheumatol. 2000;27:2347–2350. [PubMed] [Google Scholar]
- 87.Steranka LR, Farmer SG, Burch RM. Antagonists of B2 bradykinin receptors. FASEB J. 1989;3:2019–2025. doi: 10.1096/fasebj.3.9.2545496. [DOI] [PubMed] [Google Scholar]
- 88.Stratinaki M, Varidaki A, Mitsi V, Ghose S, Magida J, Dias C, Russo SJ, Vialou V, Caldarone BJ, Tamminga CA, Nestler EJ, Zachariou V. Regulator of G protein signaling 4 [corrected] is a crucial modulator of antidepressant drug action in depression and neuropathic pain models. Proc Natl Acad Sci USA. 2013;110:8254–8259. doi: 10.1073/pnas.1214696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Street SE, Kramer NJ, Walsh PL, Taylor-Blake B, Yadav MC, King IF, Vihko P, Wightman RM, Millán JL, Zylka MJ. Tissue-nonspecific alkaline phosphatase acts redundantly with PAP and NT5E to generate adenosine in the dorsal spinal cord. J Neurosci. 2013;33:11314–11322. doi: 10.1523/JNEUROSCI.0133-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun YG, Gracias NG, Drobish JK, Vasko MR, Gereau RW, Chen ZF. The c-kit signaling pathway is involved in the development of persistent pain. Pain. 2009;144:178–186. doi: 10.1016/j.pain.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Szolcsányi J, Sándor Z. Multisteric TRPV1 nocisensor: a target for analgesics. Trends Pharmacol Sci. 2012;33:646–655. doi: 10.1016/j.tips.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 92.Takagi K, Okuda-Ashitaka E, Mabuchi T, Katano T, Ohnishi T, Matsumura S, Ohnaka M, Kaneko S, Abe T, Hirata T, Fujiwara S, Minami T, Ito S. Involvement of stem cell factor and its receptor tyrosine kinase c-kit in pain regulation. Neuroscience. 2008;153:1278–1288. doi: 10.1016/j.neuroscience.2008.02.073. [DOI] [PubMed] [Google Scholar]
- 93.Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Traynor J. μ-Opioid receptors and regulators of G protein signaling (RGS) proteins: from a symposium on new concepts in mu-opioid pharmacology. Drug Alcohol Depend. 2012;121:173–180. doi: 10.1016/j.drugalcdep.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsantoulas C, McMahon SB. Opening paths to novel analgesics: the role of potassium channels in chronic pain. Trends Neurosci. 2014;37:146–158. doi: 10.1016/j.tins.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Umana IC, Daniele CA, McGehee DS. Neuronal nicotinic receptors as analgesic targets: It’s a winding road. Biochem Pharmacol. 2013;86:1208–1214. doi: 10.1016/j.bcp.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Utreras E, Keller J, Terse A, Prochazkova M, Iadarola MJ, Kulkarni AB. Transforming growth factor-β1 regulates Cdk5 activity in primary sensory neurons. J Biol Chem. 2012;287:16917–16929. doi: 10.1074/jbc.M111.329979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wakisaka S, Kajander KC, Bennett GJ. Increased neuropeptide Y (NPY)-like immunoreactivity in rat sensory neurons following peripheral axotomy. Neurosci Lett. 1991;124:200–203. doi: 10.1016/0304-3940(91)90093-9. [DOI] [PubMed] [Google Scholar]
- 99.Wang Y, Ghaffari N, Johnson CD, Braga-Neto UM, Wang H, Chen R, Zhou H. Evaluation of the coverage and depth of transcriptome by RNA-Seq in chickens. BMC Bioinformatics. 2011;12 (Suppl 10):S5. doi: 10.1186/1471-2105-12-S10-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Williams JT, Ingram SL, Henderson G, Chavkin C, von Zastrow M, Schulz S, Koch T, Evans CJ, Christie MJ. Regulation of mu-opioid receptors: desensitization, phosphorylation, internalization, and tolerance. Pharmacol Rev. 2013;65:223–254. doi: 10.1124/pr.112.005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Woodcock J, Witter J, Dionne RA. Stimulating the development of mechanism-based, individualized pain therapies. Nat Rev Drug Discov. 2007;6:703–710. doi: 10.1038/nrd2335. [DOI] [PubMed] [Google Scholar]
- 102.Yang HY, Mitchell K, Keller JM, Iadarola MJ. Peripheral inflammation increases Scya2 expression in sensory ganglia and cytokine and endothelial related gene expression in inflamed tissue. J Neurochem. 2007;103:1628–1643. doi: 10.1111/j.1471-4159.2007.04874.x. [DOI] [PubMed] [Google Scholar]
- 103.Yang HY, Wilkening S, Iadarola MJ. Spinal cord genes enriched in rat dorsal horn and induced by noxious stimulation identified by subtraction cloning and differential hybridization. Neuroscience. 2001;103:493–502. doi: 10.1016/s0306-4522(00)00573-x. [DOI] [PubMed] [Google Scholar]
- 104.Zhang J, Cavanaugh DJ, Nemenov MI, Basbaum AI. The modality-specific contribution of peptidergic and non-peptidergic nociceptors is manifest at the level of dorsal horn nociresponsive neurons. J Physiol. 2013;591:1097–1110. doi: 10.1113/jphysiol.2012.242115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou S, Bonasera L, Carlton SM. Peripheral administration of NMDA, AMPA or KA results in pain behaviors in rats. Neuroreport. 1996;7:895–900. doi: 10.1097/00001756-199603220-00012. [DOI] [PubMed] [Google Scholar]
- 106.Zylka MJ, Dong X, Southwell AL, Anderson DJ. Atypical expansion in mice of the sensory neuron-specific Mrg G protein-coupled receptor family. Proc Natl Acad Sci USA. 2003;100:10043–10048. doi: 10.1073/pnas.1732949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zylka MJ, Sowa NA, Taylor-Blake B, Twomey MA, Herrala A, Voikar V, Vihko P. Prostatic acid phosphatase is an ectonucleotidase and suppresses pain by generating adenosine. Neuron. 2008;60:111–122. doi: 10.1016/j.neuron.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The top figure outlines the RNA-Seq filtering process after reads are mapped to the appropriate genome by RUM. Multiple records for the same gene are filtered by selecting the instance which constitutes the most mapped reads, thereby selecting either the major isoform or the most supported gene model. RNA-Seq records with raw counts below 20 are filtered out, as well as those without annotations or annotations corresponding to small non-coding RNA transcripts. For the most part, genes expressed above 1 RPKM are analyzed within the paper. The lower panels analyze the RNA-Seq technical replicates (Illumina HiSeq vs Illumina GaIIx) for the TRPV1L and TRPV1D populations within the R statistical programming language. The correlation coefficients for both datasets are ~ 0.9 supporting the strong reproducibility of the data across platforms.
The table lists the total number of reads and mapping statistics, as generated by RUM, for each dataset included in the study.
This table contains the molecular signature of TRPV1L neurons based on level of expression. The table lists all 263 TRPV1L functionally well annotated transcripts with a differential of 5X and above (there is partial overlap with Table 1). The genes have been grouped into functional categories according to known annotations and publications. Within the groups, the genes have been sorted on expression (high to low) and only genes with an expression level above 1 RPKM are included. The adjusted fold change values are color coded: Red: >20 fold, Blue: 20-10 fold, and Green: < 10 fold difference. The accession id provides the exact reference to the gene isoform analyzed within this study.
This table lists all functionally well annotated TRPV1L genes with a differential above 1.5X and below 5X. The table only includes genes with an expression level above 1 RPKM.
Table 4A is a partial sampling of the TRPV1D genes with a differential of 5X and above. The genes have been grouped into functional categories according to known annotations and publications. Within the groups, the genes were sorted on expression (high to low) and only the top five most highly expressed genes within each group are included. (4B) The table lists all the genes in the TRPV1D population that exhibit differential expression above 1.5X fold difference and are expressed above 1 RPKM. Entries are arranged alphabetically.
This table lists all functionally well annotated genes whose expression is common to both the TRPV1L and D preparations. All entries have a fold difference equal to or below 1.5 when comparing TRPV1L to TRPV1D. Only genes with an RPKM above 1 are included. Entries are arranged alphabetically.
Top panels: The complete histogram showing the distribution of gene expression based on RPKM for TRPV1L neurons: 14% are expressed between 0–1 RPKM, approximately 12% are expressed between 1–3 RPKM, the majority of TRPV1L genes are expressed between 3–20 RPKM, (~37%,), and 20–200 (~34%). The high expressing group, between 200–3600 RPKM contains ~3% of the expressed genes. Thus, a number like 200 RPKM means that gene is in pretty rare company with only 150 other genes in that range or higher; 3 to 30 RPKM means it is expressed at the general level of most cellular genes. The two panels in the second row show the most highly expressed category (200 to highest RPKM) for TRPV1 depleted and common categories. Note that the neurofilament genes (Nefl and Nefm) are highly expressed by the large DRG neurons that comprise the TRPV1D population but these are also expressed in the TRPV1L population but just not to the same extent. The same consideration of differential vs. absolute also applies to a highly expressed TRPV1L gene synuclein γ (Sncg) and, in fact, to nearly all the genes. The remaining panels show the full histogram for low, medium and high expressing groups for whole rat DRG and for a rat mixed tissue preparation (equal amounts of total RNA from cerebral cortex, cerebellum, midbrain, hypothalamus, hindbrain, spinal cord, retina, pituitary, heart, liver, lung, kidney, skeletal muscle, small intestine and adrenal gland). Note the overall similarity between the rat whole DRG and the mouse histograms. For all transcriptomes illustrated in SFigure 2, the high end of each histogram can form a general molecular “fingerprint” of that tissue. For example, familiar transcripts associated with TRPV1 neurons are calcitonin gene related peptide a (Calca) and peripherin (Prph), both are highly expressed in the TRPV1L neurons. A less familiar member of the fingerprint is Cd24A, which codes for a GPI-linked sialoglycoprotein that interacts with contactin 1 and 2 and L1Cam, a system implicated in tissue remodeling subsequent to nerve injury and axotomy (Lieberoth et al., 2009; Yamanaka et al., 2007). The transcriptional fingerprint of the rat mixed tissue reflects the presence of highly specialized cells such as photoreceptors, pituicytes and muscle fibers and includes functionally relevant genes with dedicated functions such as rhodopsin (Rho), growth hormone (Gh1), prolactin (prl), myosin (myh4) and albumin (Alb). Based on overlap and within gene family complementarity (e.g., the calmodulin paralogs) reliable molecular fingerprinting of neuronal subpopulations will likely require inclusion of more transcripts.
Abbreviations: Actb: β actin, Prph: peripherin, Cd24: nectadrin, Calca: α-type CGRP, Tppp3: tubulin polymerization-promoting protein 3, Hsp90ab1: 90kDa, αB1 heat shock protein, S100a6: S100 calcium binding protein A6, Tuba1a: α1 tubulin, Fxyd2: γ Na(+)/K(+) ATPase subunit, Sncg: γ synuclein, Calm3: calmodulin 3, Ywhag: 14-3-3 protein γ, Pcp4: Purkinje cell protein 4, Mbp: myelin basic protein, Nefh: neurofilament, heavy polypeptide, Mtap1b: microtubule-associated protein 1B, Atp1a1: α1 ATPase, Na+/K+ transporting polypeptide, Ndufa4: NADH dehydrogenase 1α, subcomplex4, Pmp22: peripheral myelin protein 22, Apoe: apolipoprotein E, Nefm: medium neurofilament, Nefl: light neurofilament, Mpz: myelin protein zero, Ywhah: 14-3-3 η, Fstl1: follistatin-e3like 1, Anxa2: annexin A2, Atpif1: ATPase inhibitory factor 1, Tceb2: transcription elongation factor B2, Rplp1: ribosomal protein, large, P1, Calm1: calmodulin 1, Calm2: calmodulin 2, Stmn2: stathmin-like 2, Fth1: ferritin, heavy polypeptide 1, Ndrg4: NDRG family member 4, Ubb: ubiquitin B, Cox6a1: cytochrome c oxidase subunit, Aldoa: aldolase A, Tubb3: β3 tubulin, Tmsb4x: β4 thymosin, Uchl1: ubiquitin thiolesterase, Thy1: θ thymus cell antigen 1, Hba-a2: hemoglobin α2, Pmp22: peripheral myelin protein 22, Uchl1: ubiquitin thiolesterase, Scd: stearoyl-CoA desaturase, Atp1a3: α3 ATPase, Na+/K+ transporting polypeptide, Prl: prolactin, Ckm: creatine kinase, Myh4: myosin, heavy polypeptide 4, Atp5b: β ATP synthase, Acta1: α1 actin, Rho: rhodopsin, Alb: albumin, Plp1: myelin proteolipid protein, and Gh1: growth hormone 1.
The extended TRPV1 annotation sequence is provided for the 5′ (top) and 3′ (bottom) UTR sequence. We designated the end of the selected segment at 50 mapped reads. The extended segment is given in red. Please note that since the submission of this manuscript, RNA-Seq data has been incorporated into the Refseq annotation pipeline. Thus Refseq TRPV1 (NC_000077.6) 5′ UTR now corresponds to our 5′UTR extension; the Refseq 3′ UTR, extends beyond our 3′ UTR extension. The reannotation of genomes is a fluid process. As the RNA-Seq databases evolve and grow, the precise UTR locations become more precisely defined. Attempts were made using several promoter prediction tools, including Berkeley Neural Network Promoter Prediction tool (version 2.3) < http://www.fruitfly.org/seq_tools/promoter.html> to predict a new promoter upstream of the start of the newly extended 5′ UTR. However, none of the tools predicted a conventionally defined sequence close to the new start. RNA-Seq data for rat dorsal root ganglion showed that the TRPV1 5′ and 3′UTR regions could also be similarly extended in rat. The human TRPV1 gene was well annotated and did not require modifications.
A. Genes for rat DRG and TG were ranked according to descending expression value within their respective tissue transcriptome. The rat TG and DRG contain approximately 12,000 genes and the x and y axes reflect this number. Genes from Figure 2 are plotted as a test set for assessing similarity between the TG and DRG. The plot includes genes that are expressed greater than 1 RPKM in both TRPV1L neurons and the TRPVD ganglion preparation. The correlation coefficient for relative expression is 0.88. B. When a broader analysis of TG and DRG is performed, Hoxd1, a member of the homeobox gene family is noticeably different. Hoxd1 is expressed in TG and DRG whereas the other 17 homeobox genes are only expressed in DRG to varying degree. In the context of the Hox gene family, it is important to note that only lumbar rat DRGs were dissected in the present experiments. The mean ± SEM values are provided in the table.
Lists all the transcripts from Tables 1 and S2 (TRPV1L with a fold difference above or equal to 5) that were significantly (padj < .05) down regulated after RTX. DESeq was used for the statistical analysis according to standard protocol (Anders and Huber, 2010).
This table lists all the functionally well annotated genes (12,098) in the mouse lineage experiment that are expressed at or below 1 RPKM. The number of raw reads ranged from 0 to 2,807 in the TRPV1L population and were distributed as following: 0 = 5,614; 1–2 = 1,216; 3–19 = 2,983; 20–99 = 1,329; 100–499 = 861; 500–2807 = 95. To give some interpretation, 200 reads, for example, can cover all the exons of particular gene and may allow for assembly of several full length transcripts. However one has to take into account the number of neurons in the ganglion. Depending on how the reads are distributed, one neuron could have several full length transcripts or raw reads might be present in multiple cell types in which case a full length transcript is, unlikely to be assembled.
Table S8: Expressed Mouse DRG Gene Database. This table lists in alphabetical order all the genes/transcripts in the mouse lineage experiment that are expressed above 20 raw counts. Few duplicate entries may exist due to aliases in the “Functionally Not Well Annotated Genes” section. The table is provided to facilitate looking up of gene families and genes of interest. This full coverage table also contains many non-protein coding genes (i.e pseudogenes, small nuclear ribonucleic acid [snRNA], and non-coding RNA [ncRNA]) however the table is not meant to be an exhaustive list of all transcripts within the mouse transcriptome. Our poly-A selection was not designed to comprehensively capture non-polyadenylated, non-coding RNAs, such as microRNA [miRNA], long intergenic non-coding RNAs [lincRNA], piwi-interacting RNA [piRNA], or small nucleolar RNA [snoRNA], and we removed the incidental transcripts that were captured in these categories. Similarly, the table does include unannotated transcripts, but as the NCBI database evolves more entries can be added in the future.