ABSTRACT
The human respiratory syncytial virus (hRSV) fusion (F) protein is considered a major target of the neutralizing antibody response to hRSV. This glycoprotein undergoes a major structural shift from the prefusion (pre-F) to the postfusion (post-F) state at the time of virus-host cell membrane fusion. Recent evidences suggest that the pre-F state is a superior target for neutralizing antibodies compared to the post-F state. Therefore, for vaccine purposes, we have designed and characterized a recombinant hRSV F protein, called Pre-F-GCN4t, stabilized in a pre-F conformation. To show that Pre-F-GCN4t does not switch to a post-F conformation, it was compared with a recombinant post-F molecule, called Post-F-XC. Pre-F-GCN4t was glycosylated and trimeric and displayed a conformational stability different from that of Post-F-XC, as shown by chemical denaturation. Electron microscopy analysis suggested that Pre-F-GCN4t adopts a lollipop-like structure. In contrast, Post-F-XC had a typical elongated conical shape. Hydrogen/deuterium exchange mass spectrometry demonstrated that the two molecules had common rigid folding core and dynamic regions and provided structural insight for their biophysical and biochemical properties and reactivity. Pre-F-GCN4t was shown to deplete hRSV-neutralizing antibodies from human serum more efficiently than Post-F-XC. Importantly, Pre-F-GCN4t was also shown to bind D25, a highly potent monoclonal antibody specific for the pre-F conformation. In conclusion, this construct presents several pre-F characteristics, does not switch to the post-F conformation, and presents antigenic features required for a protective neutralizing antibody response. Therefore, Pre-F-GCN4t can be considered a promising candidate vaccine antigen.
IMPORTANCE Human respiratory syncytial virus (RSV) is a global leading cause of infant mortality and adult morbidity. The development of a safe and efficacious RSV vaccine remains an important goal. The RSV class I fusion (F) glycoprotein is considered one of the most promising vaccine candidates, and recent evidences suggest that the prefusion (pre-F) state is a superior target for neutralizing antibodies. Our study presents the physicochemical characterization of Pre-F-GCN4t, a molecule designed to be stabilized in the pre-F conformation. To confirm its pre-F conformation, Pre-F-GCN4t was analyzed in parallel with Post-F-XC, a molecule in the post-F conformation. Our results show that Pre-F-GCN4t presents characteristics of a stabilized pre-F conformation and support its use as an RSV vaccine antigen. Such an antigen may represent a significant advance in the development of an RSV vaccine.
KEYWORDS: hydrogen/deuterium exchange mass spectrometry, pre-F conformation, respiratory syncytial virus F protein, vaccine antigen
INTRODUCTION
Human respiratory syncytial virus (hRSV) is an enveloped single-stranded RNA virus belonging to the Paramyxoviridae family. It is one of the major causes of lower respiratory tract infections in very young children (1). The hRSV disease spectrum includes respiratory signs such as rhinitis, otitis, pneumonia, and bronchiolitis. Among these, pneumonia and bronchiolitis are the major causes of hRSV-associated morbidity and mortality (2, 3). In the United States, data collected from 1993 to 2008 for infants less than 1 year of age showed that hRSV was annually responsible for 2,350 per 100,000 severe respiratory infections needing hospitalization (4). The incidence of hRSV is higher in children born prematurely or those with cardiopulmonary diseases (5, 6). These populations are considered particularly at risk for severe hRSV infections. In addition, hRSV is increasingly recognized as an important cause of morbidity and mortality in the elderly, causing influenza-like illnesses (7–11).
There is currently no licensed vaccine to protect against hRSV despite multiple attempts and persistent efforts during the last 50 years. The only licensed prophylactic treatment currently available is palivizumab (12), a neutralizing monoclonal antibody given to infants considered at higher risk for hRSV infection (13–15). Therefore, the development of an efficacious and safe hRSV vaccine is still a priority. Several vaccine candidates are currently under development (recently reviewed in references 16–18), but a commercial vaccine against hRSV has yet to be licensed.
Induction of potent neutralizing antibodies is a major goal for a hRSV vaccine, as the presence of serum neutralizing antibodies has been linked with protection against hRSV infection (19–22). In this regard, the hRSV F protein is probably the most appealing vaccine antigen candidate. Indeed, this antigen was shown to be the target of most of the RSV-neutralizing activity in human serum (23, 24) and is known to be the target of palivizumab (12). In addition, its amino acid sequence presents a high degree of similarity between the two known hRSV subtypes, subtypes A and B (25, 26).
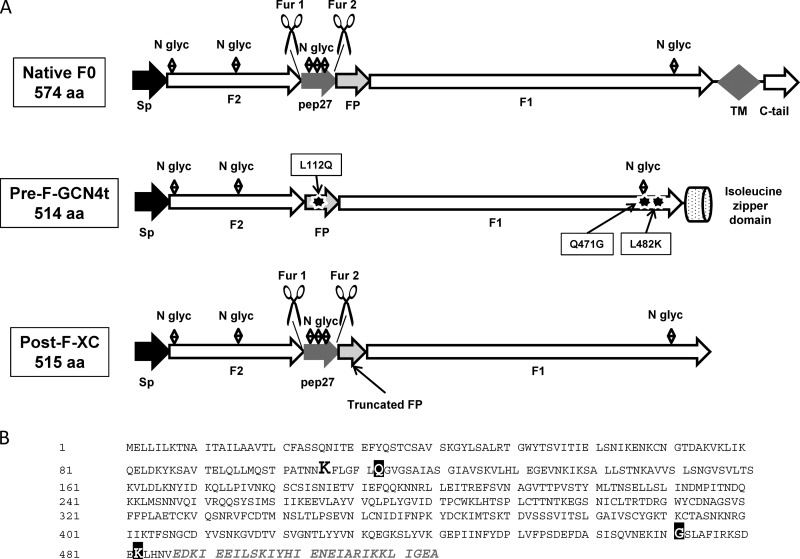
The F protein is a trimeric glycoprotein that mediates viral entry into host cells through the fusion of the viral envelope and cell membrane (27–29). The monomeric precursor peptidic chain, called F0, is subdivided into two main segments, F1 and F2. These two segments are separated by an intervening peptide, designated peptide 27 (pep27), flanked by two furin sites (Fig. 1A). During maturation, F0 is activated through proteolytic cleavage at the two furin sites, leading to the removal of pep27. F1 and F2 remain bound together by two disulfide bridges.
FIG 1.
(A) Comparison between the structures of native RSV F0 and the Pre-F-GCN4t and Post-F-XC monomers. Sp, signal peptide; N glyc, N-glycan; FP, fusion peptide; Fur 1 and Fur 2, furin cleavage sites; TM, transmembrane sequence, C-tail, C-terminal tail; aa, amino acids. F1 and F2 are the two main domains of RSV F. Black stars represent the added mutations L112Q, Q471G, and L482K. (B) Full amino acid sequence of Pre-F-GCN4t. The lysine residue (K) that remains after the deletion of pep27 is highlighted in boldface type and a larger font. The three point mutations are shown with white letters on a black background. The GCN4t domain replacing the transmembrane domain and C-terminal tail is shown in gray italics. The rest of the sequence is unchanged compared with the native one.
To allow host cell-viral membrane fusion, the F protein undergoes a conformational switch from a prefusion (pre-F) state, a transiently stabilized or metastable state, to a more stable postfusion (post-F) state (30). During this transition, two regions of F1, heptad repeat A (HRA) and HRB, are subjected to a dramatic and irreversible rearrangement resulting in the formation of a thermodynamically favored 6-helix bundle (6HB), characteristic of the post-F conformation. The pre-F and post-F conformations were shown to be structurally and antigenically distinct (31). Some antibodies specifically recognize the pre-F conformation (32), others are directed against the post-F-specific 6HB structure (33), and the others bind both pre-F and post-F structures, such as palivizumab (34–37). Importantly, data suggested that a hRSV subunit vaccine based on pre-F triggers a broad protective antibody response, as it has been shown that most of the neutralizing antibodies produced during natural infection are directed against the pre-F conformation (24, 38, 39). In addition, it was reported previously that monoclonal neutralizing antibodies directed against the pre-F state can prevent the conformational changes leading to the post-F state (32), thereby inhibiting virus-cell fusion. The generation of a soluble prefusion F protein has long been a challenging task due to the transient nature of the prefusion state and its spontaneous rearrangement into the postfusion conformation when produced as a soluble recombinant protein. Nevertheless, several stabilized pre-F proteins were described recently (40–45). We have also designed and produced a molecule with pre-F characteristics, called Pre-F-GCN4t, that has now reached clinical development.
In this study, we present the design and characterization of Pre-F-GCN4t. We present data showing that Pre-F-GCN4t can be produced as a soluble, stable, and homogeneous protein trimer. Particularly, Pre-F-GCN4t was analyzed in parallel with Post-F-XC, an F molecule in the postfusion conformation, in order to dissect their differences and to ensure that Pre-F-GCN4t did not spontaneously rearrange into the postfusion structure. Finally, we evaluated the ability of Pre-F-GCN4t to bind palivizumab, to deplete hRSV-neutralizing antibodies from a human immunoglobulin preparation, and to bind the pre-F-specific D25 monoclonal antibody and compared these activities to those of Post-F-XC.
RESULTS
Pre-F-GCN4t and Post-F-XC design.
Two constructs based on the hRSV RSS2 sequence were produced in this study: (i) Pre-F-GCN4t, an RSV F vaccine candidate stabilized in the prefusion conformation, and (ii) Post-F-XC, a post-F conformer made for comparison purposes.
For the pre-F construct, both the transmembrane and C-terminal domains of F0 were replaced by the engineered Saccharomyces cerevisiae GCN4 trimerization domain (46); the furin cleavage sites and pep27 sequences were removed with a single lysine residue remaining at this position, between F2 and the fusion peptide (FP). To convert the HRB C terminus into a canonical left-handed trimeric coiled-coil and create a network of oppositely charged side chains (47), a leucine (L) residue was mutated into a lysine (K) residue (residue 482 in the Pre-F-GCN4t sequence) (Fig. 1B). Two additional mutations were introduced to increase protein homogeneity and stability (data not shown). The original design of the pre-F protein was N-glycosylated at three different sites: N27, N70, and N470 (corresponding to N500 in the native sequence). SDS-PAGE analysis supported by liquid chromatography-mass spectrometry (LC-MS) investigation indicated partial N-glycosylation at position N470 (data not shown). Modification of the NQS N-glycosylation site by the NGS sequence (Q471G mutation) increased the N-glycosylation yield and protein expression levels in transiently transfected Chinese hamster ovary (CHO) cells. Further analyses identified the hydrophobic region of the FP as a region potentially susceptible to degradation. Therefore, the leucine residue at position 112 was replaced by a glutamine residue (L112Q), resulting in a reduction in proteolysis as well as an unexpected improvement in expression levels (data not shown). These two mutations that were added to the initial construct led to the final design of the pre-F molecule, identified as Pre-F-GCN4t. Sequence modifications of Pre-F-GCN4t, compared to the unmodified hRSV F protein and Post-F-XC, are summarized in Fig. 1A.
Comparative primary and quaternary structure analysis of Pre-F-GCN4t and Post-F-XC.
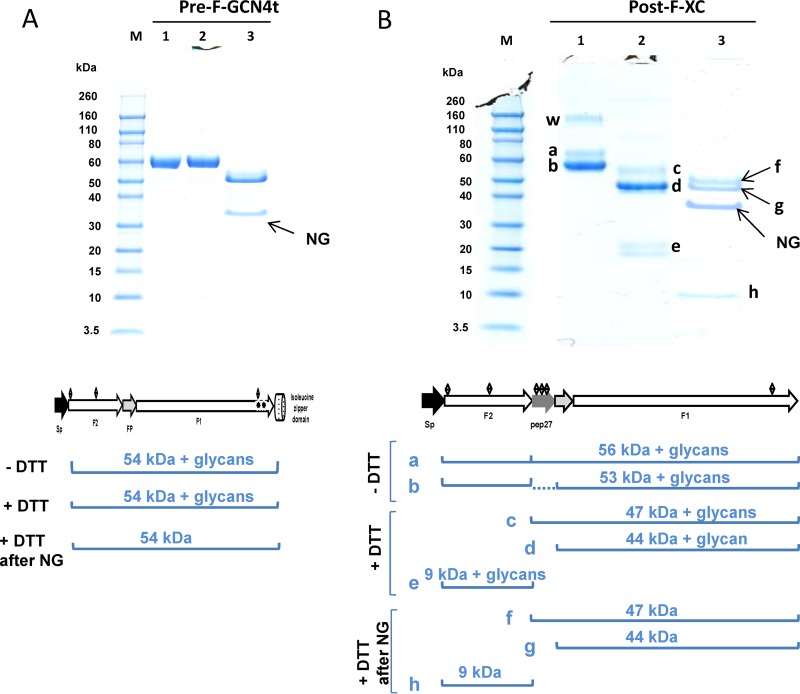
After purification, the trimeric nature of Pre-F-GCN4t and Post-F-XC was confirmed by analytical ultracentrifugation (AUC) (data not shown). It was observed in both cases that more than 90% of the protein adopted a trimeric structure. Both molecules were also analyzed by SDS-PAGE under nonreducing and reducing conditions. For Pre-F-GCN4t (Fig. 2A, lanes 1 and 2, respectively), band patterns were similar under both conditions, indicating that Pre-F-GCN4t consisted of a single polypeptide. Moreover, after N-glycosidase treatment, the observed protein band shifted to a lower molecular weight (MW), consistent with the removal of glycan moieties (Fig. 2A, lane 3), which is in line with the N-glycosylation pattern of the native protein.
FIG 2.
SDS-PAGE analysis of Pre-F-GCN4t (A) and Post-F-XC (B) under reducing (with dithiothreitol [+DTT]) and nonreducing (without dithiothreitol [−DTT]) conditions. Lane 1, nonreducing conditions; lane 2, reducing conditions; lane 3, N-glycosidase-treated sample under reducing conditions. Band a, polypeptide with incomplete cleavage at the second furin site; band b, fully processed polypeptide; bands c and f, F1 with pep27; bands d and g, F1 without pep27; bands e and h, F2; band w, higher-order protein structure; M, molecular mass markers. The band at 35 kDa represents the N-glycosidase F enzyme (NG). Expected molecular masses (with and without glycosylations) are shown at the bottom of each gel photograph.
On the other hand, the band profile of purified Post-F-XC was more complex. Under nonreducing conditions, three bands (bands a, b, and w) were detected (Fig. 2B, lane 1). LC-tandem MS (LC-MS/MS) analysis (data not shown) revealed that band a is the polypeptide resulting from incomplete cleavage at the second furin site, whereas band b corresponds to the fully processed protein. This result is consistent with furin site 1 being the site most susceptible to proteolytic cleavage upon the preparation of the anchorless fusion protein (48). Band w represents a higher-order protein structure. Under reducing conditions, the gel profile was different, because of the dissociation of the F1 and F2 subunits. F1 was present as a doublet migrating at 50 to 70 kDa (bands c and d). These bands represent the F1 polypeptide with (band c) and without (band d) pep27. F2 migrated as a doublet at 20 kDa (band e), which we attributed to glycosylation heterogeneity. This pattern was confirmed by the presence of a single band (band h) after N-glycosidase treatment, migrating near the theoretical molecular mass of F2, i.e., 10 kDa. After N-glycosidase treatment, the F1 band bearing pep27 (band f) showed a shift in the migration pattern due to the removal of up to 4 saccharide moieties, which was not observed for F1 lacking pep27, as it harbors only a single N-glycosylation site (band g). The sequence identity of electrophoresed Post-F-XC bands was confirmed by LC-MS/MS (data not shown). Besides partial cleavage of pep27, this gel profile reflected band patterns expected from the designed molecule.
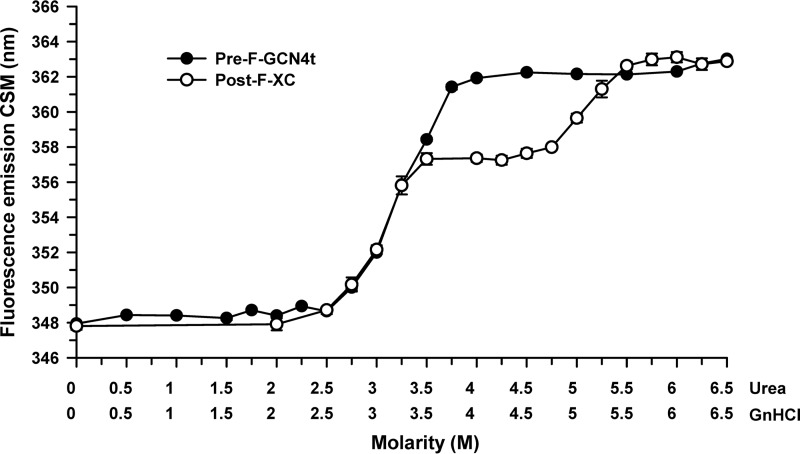
Chemical denaturation.
Pre-F-GCN4t and Post-F-XC polypeptide chain unfolding was assessed by monitoring the intrinsic fluorescence intensity under denaturing conditions. Thermodynamic analysis of protein denaturation profiles can provide information on the stability and domain organization of large protein molecules. Both constructs contain the same number of tryptophans, since the GCN4 coiled-coil added to the pre-F structure is devoid of tryptophans. Because neither urea nor guanidine hydrochloride (GnHCl) treatment was able to completely denature these two proteins (data not shown), a series of urea-GnHCl equimolar mixtures with increasing concentrations was used (Fig. 3). Pre-F-GCN4t and Post-F-XC started unfolding at similar denaturant concentrations and reached comparable plateau values. However, under these conditions, the Pre-F-GCN4t structure changed in what appeared to be a one-step process, with unfolding starting at 3.1 M urea–3.1 M GnHCl. In contrast, Post-F-XC unfolded in a two-step process, with the first unfolding step at 3.0 M urea–3.0 M GnHCl, thus being similar to the single unfolding step of Pre-F-GCN4t, and the second unfolding step at 5.1 M urea–5.1 M GnHCl. This two-step unfolding profile suggests that Post-F-XC consists of at least two domains with different thermodynamic stabilities.
FIG 3.
Chemical unfolding of Pre-F-GCN4t and Post-F-XC. Chemical unfolding profiles of Pre-F-GCN4t and Post-F-XC were monitored by the change in the fluorescence emission center of spectral mass (CSM) upon incubation with a solution containing various equimolar concentrations of urea and guanidine-HCl. Samples were excited at 295 nm.
Electron microscopy.
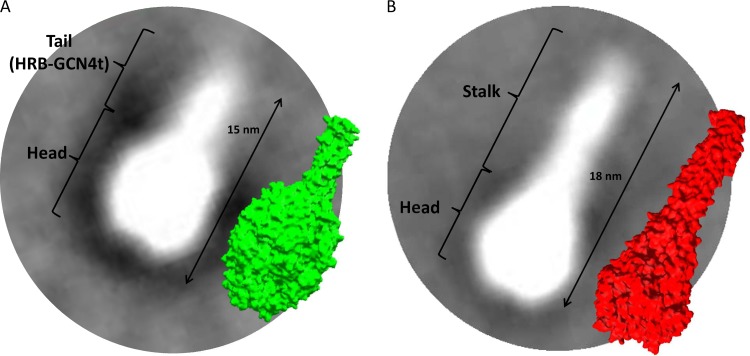
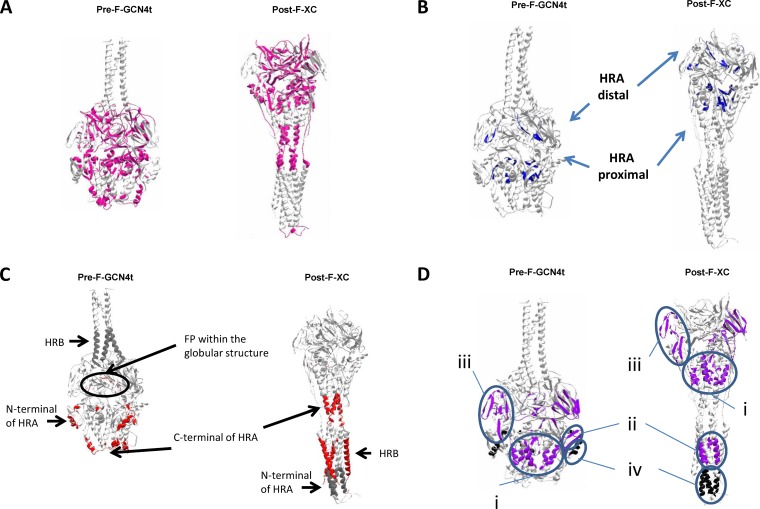
We used electron microscopy (EM) and single-particle analysis to determine whether Pre-F-GCN4t and Post-F-XC adopt conformations similar to those reported previously (24, 36, 37). Figure 4 presents two-dimensional (2D) class-average images generated from representative particle populations (representing between 70 and 80% of the total) and surface representations derived from structural models of both proteins for comparison (PDB accession no. 4JHW for the pre-F structure and 3RKI for the post-F structure). In order to adjust the molecules to scale, protein modeling and distance measurements were performed with Swiss-PdbViewer software. A Pre-F-GCN4t homology model was generated by using a combination of the recently reported structure of the RSV pre-F ectodomain (32) and the structure of the GCN4 trimeric coiled-coil region added at the C-terminal end. Representative 2D class-average images agreed well with the respective molecular surface of the Pre-F-GCN4t model (Fig. 4, green) and post-F (red). It was observed that the Pre-F-GCN4t adopts a lollipop-like structure, with a size and shape similar to those of the model. In approximately 13% of these particles, the head portion appeared to have an indentation in the center.
FIG 4.
Superimposition of representative averaged electron micrograph images with structural models for Pre-F-GCN4t and Post-F-XC at the same scale. (A) Superimposition of a representative image from the Pre-F-GCN4t electron micrograph with the structural model reported under PDB accession no. 4JHW; (B) superimposition of a representative image from the Post-F-XC electron micrograph with the structural model reported under PDB accession no. 3RKI.
Post-F-XC appeared as an elongated conical structure, consistent with post-F structures described previously (36, 37, 49). 2D class-average images also suggested a 3-nm difference in length between Pre-F-GCN4t and Post-F-XC, consistent with the predictions from the models.
Hydrogen/deuterium exchange mass spectrometry.
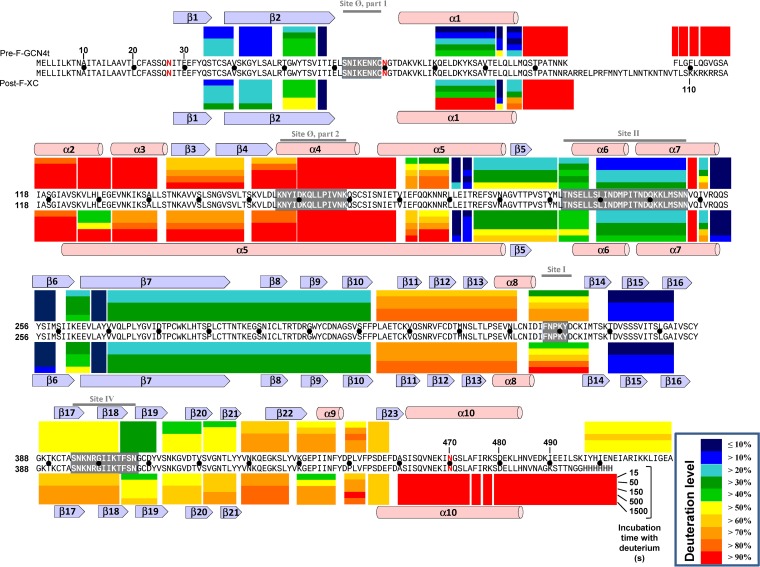
Overall, hydrogen/deuterium exchange mass spectrometry (HDX-MS) patterns of Pre-F-GCN4t and Post-F-XC were very similar, indicating that the dynamic properties of the two proteins are comparable despite the expected structural differences (Fig. 5). This is in line with the fact that a sizeable portion of their secondary and tertiary structures is superimposable (32). Since Pre-F-GCN4t and Post-F-XC share most of the sequence, a direct comparison of the two HDX-MS patterns was possible. There were 37 proteolytic segments observed in both constructs. Among the 37 common segments, 24 segments did not show a significant difference (<10% change) in the average deuteration levels between the two constructs (Fig. 6A). Average deuteration levels at five time points for each segment in Pre-F-GCN4t and Post-F-XC correlated very well with a coefficient of determination (R-squared value) of 0.87. This implies that most of the dynamic regions in Pre-F-GCN4t are dynamic in Post-F-XC and that most of the rigid regions in Pre-F-GCN4t remain rigid in Post-F-XC.
FIG 5.
Color representation of data from HDX-MS analysis of Pre-F-GCN4t and Post-F-XC. HDX-MS results for each pepsin-released peptide at each time point (15, 50, 150, 500, or 1,500 s) are represented as colored bars above and below the sequences of Pre-F-GCN4t and Post-F-XC, respectively. Exchange values are presented in the key. Secondary structure motifs are specified above and below the HDX-MS results for Pre-F-GCN4t and Post-F-XC, respectively. Antibody binding sites are highlighted in gray with white amino acid letters in the sequences, and their names are shown in gray above the secondary structure motifs. N-glycosylation sites are indicated in red in the sequences. To facilitate residue counting, a black dot is inserted between the two sequences every 10 residues.
FIG 6.
Categorized HDX-MS results on the modeled Pre-F-GCN4t and Post-F-XC structures. (A) Segments with similar deuterium incorporation (difference in average deuterium incorporation of <10%). (B) Segments with no or little deuterium incorporation (slow-exchanging segments; shown in dark blue). (C) Segments with high deuterium incorporation (fast-exchanging segments). Red, shared by both structures; dark gray, exclusive to each structure. (D) Segments which show significant change in deuterium incorporation (difference in average deuterium incorporation of >10%) between Pre-F-GCN4t and Post-F-XC. Purple, slower exchange for Pre-F-GCN4t; black, slower exchange for Post-F-XC.
Six segments, spanning positions 57 to 58, 201 to 202, 203 to 206, 252 to 259, 267 to 269, and 370 to 382, showed very low levels of deuterium incorporation in both proteins (average deuteration level of <15% in both states). Such behavior is typical of protein core regions, since the minimal local fluctuation protects the amide hydrogens from exchanging with the surrounding deuteriums. The first core region included five segments: the C terminus of β2 (segment spanning positions 57 to 58), the C terminus of α5 (segments spanning positions 201 to 202 and 203 to 206), the N terminus of β6 (segment spanning positions 252 to 259), and the N terminus of β7 (segment spanning positions 267 to 269). According to model prediction, these segments are part of the core of the globular head in the HRA-proximal region (Fig. 6B). The second region was around β15 (segment spanning positions 370 to 382). This region involves an antiparallel β-hairpin structure, which is part of a three-stranded β-sheet located within the HRA-distal subunits. It should be noted that these rigid regions are easily superimposable when both conformation models are compared, with a calculated α-carbon root mean square deviation (RMSD) of less than 2 Å, indicating that they remain in the core of the globular structure and do not account for the conformational differences between Pre-F-GCN4t and Post-F-XC.
There were four very dynamic regions in Pre-F-GCN4t with deuteration levels at 15 s of >90% (Fig. 5 and 6C). They are the C-terminal region of F2 (segment spanning positions 98 to 107), the FP (segments spanning positions 108 to 109, 110 to 111, and 112 to 117), the N-terminal region of HRA (positions 126 to 132 and 133 to 141), and the C-terminal region of HRA (segment spanning positions 170 to 189). Similarly, there were three very dynamic regions in Post-F-XC: the C-terminal region of F2 (segment spanning positions 98 to 107), the C terminus of HRA (segment spanning positions 170 to 189), and α10 of HRB (segments spanning positions 460 to 473, 474 to 475, 476 to 477, and 478 to 502). All of these very dynamic regions experience a drastic conformational change between the pre-F structure and the post-F structure.
There were 11 segments that increased average deuteration levels by 10% or more (becoming more dynamic or less stable) in Post-F-XC compared to Pre-F-GCN4t. These 11 segments are part of three regions. The first region (Fig. 6Di) lies in the HRA-proximal part of Pre-F-GCN4t and includes the N-terminal side of β2 (segment spanning positions 50 to 56), α1 (segments spanning positions 81 to 92 and 95 to 97), and α5 (segments spanning positions 192 to 193 and 194 to 200). The removal of the HRB region from the HRA-proximal part upon the transition from the pre-F state to the post-F state makes the HRA-proximal part thinner. This may weaken the hydrogen bond network and give more flexibility to this part of the molecule. The second region (Fig. 6Dii) is also in the HRA-proximal parts of Pre-F-GCN4t and covers β3-β4 (segment spanning positions 144 to 158). It sustains a complete transition from β3-β4 in Pre-F-GCN4t to α5 in Post-F-XC upon a structural switch and participates in the formation of the HRA coiled coil buried in the center of the 6HB. The third region adjacent to the pre-F HRB inverted pyramid (Fig. 6Diii) consists of β17-β18 (segment spanning positions 389 to 404), β19 (segment spanning positions 405 to 410), β20 (segment spanning positions 413 to 420), β21 (segment spanning positions 421 to 427), and β22 (segment spanning positions 429 to 437). Here, the parallel strand β22 sustains a large-scale rearrangement in order to accommodate the movement of HRB toward HRA, which translates into more flexible structures. The higher stability of Pre-F-GCN4t in the pre-F state may result from this additional beta-strand reinforcing the hydrogen bond network of this particular beta-sheet.
A region near the N terminus of HRA (segments spanning positions 126 to 132 and 133 to 141) showed significantly lower deuteration levels in Post-F-XC than in Pre-F-GCN4t (Fig. 6Div, black highlighting). This region is located in the globular head in Pre-F-GCN4t and within the membrane-proximal part of 6HB in Post-F-XC. This is the only region with a much lower deuteration level in Post-F-XC than in Pre-F-GCN4t. We suggest that it contributes to the free energy released upon post-F refolding.
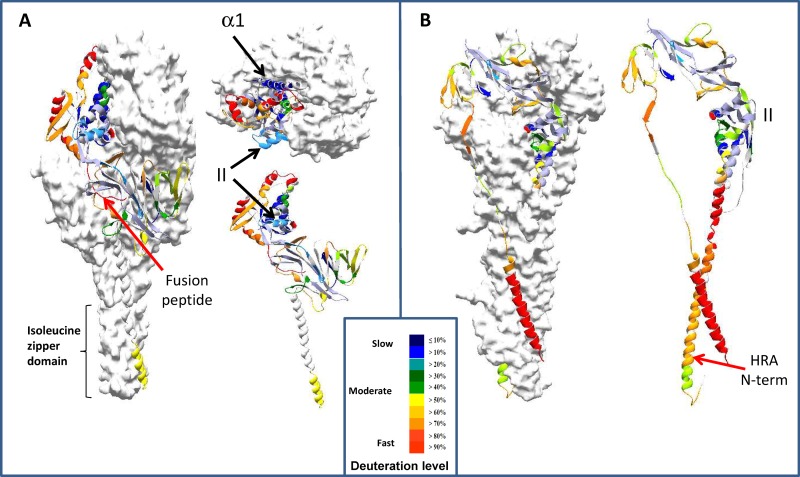
HDX-MS results were mostly consistent with predicted Pre-F-GCN4t and Post-F-XC structures. Figure 7 shows a ribbon backbone model representation of deuteration levels at the 15-s time point. As HDX reactions occur through local and global unfolding events, HDX results should correlate with dynamic properties of each segment, not necessarily with solvent accessibility or distance from the surface (50). However, one can probably expect some secondary correlation between the retardation in HDX reactions and solvent accessibility in such a large polymeric protein. This appears to be the case for most of the segments except for two regions. The first region covered the FP (Fig. 7A), where the deuteration level was >90%, even at 15 s, despite being internally located according to the modeled structure (32). The second region covered site II (the palivizumab binding site) and showed low-level deuterium incorporation despite its location at the surface of both constructs (Fig. 7A and B).
FIG 7.
HDX-MS data transposed on the structural models of Pre-F-GCN4t and Post-F-XC. Two of the Pre-F-GCN4t monomers are shown in a surface representation, while the third is shown in a ribbon representation. Results from the 15-s deuteration time point are shown. Deuteration levels are presented in the key. Red arrows indicate the positions of two peptides lacking correlation between surface accessibility and deuterium incorporation. (A) Exchange kinetics of peptides from Pre-F-GCN4t in longitudinal sections (left and bottom right) and from above (top right); (B) exchange kinetics of peptides from Post-F-XC as seen in longitudinal sections. The positions of α-helix α1 and the palivizumab binding site (site II) are shown by black arrows.
Antigenic characterization.
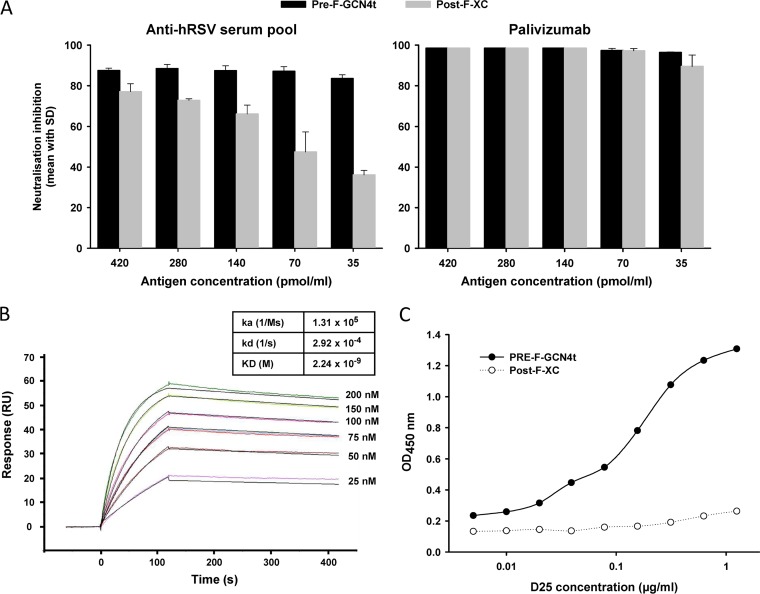
Pre-F-GCN4t and Post-F-XC were compared in a neutralization inhibition experiment using palivizumab and a hRSV-positive human serum pool. As expected, Pre-F-GCN4t was more efficient than Post-F-XC at depleting the neutralizing activity of the seropositive serum pool throughout the range of concentrations tested (Fig. 8A, left) but even more so at the lower competitor concentrations. On the other hand, Pre-F-GCN4t and Post-F-XC were equally efficient at inhibiting palivizumab neutralizing activity (Fig. 8A, right). The affinity of palivizumab for Pre-F-GCN4t was measured by surface plasmon resonance in a Biacore device (Fig. 8B). The KD (equilibrium dissociation constant) for the interaction was 2.24 nM, which is similar to data reported previously for a C-terminally truncated RSV A2 F protein (22, 51).
FIG 8.
Pre-F-GCN4t antigenicity. (A) Pre-F-GCN4t or Post-F-XC at decreasing concentrations was used to inhibit the neutralizing activity of a convalescent-phase serum pool (left) or palivizumab (right) in a neutralization inhibition assay. (B) Biacore sensorgrams of Pre-F-GCN4t injected at different concentrations onto captured palivizumab. Association and dissociation constants (ka, kd, and KD) are indicated in the table. (C) Binding of the D25 antibody to Pre-F-GCN4t and Post-F-XC. D25 at increasing concentrations was used to bind immobilized Pre-F-GCN4t or Post-F-XC in a microtiter plate. D25 binding was revealed by peroxidase-labeled anti-human IgG Fc antibody followed by a peroxidase substrate. OD, optical density.
To confirm the structural and antigenic relationships between the present constructs and the expected pre-F and post-F structures, the binding of the two antigens to the D25 antibody was compared by an enzyme-linked immunosorbent assay (ELISA) (Fig. 8C). We demonstrated the binding of D25 to Pre-F-GCN4t, but in contrast to what was observed with palivizumab (Fig. 8A, right), D25 bound Post-F-XC only minimally (Fig. 8C).
DISCUSSION
The field of recombinant RSV F vaccine candidate development has recently been successful at stabilizing both pre- and post-F states, and both approaches present opportunities for innovative RSV vaccine development. In this report, we present the characterization of a recombinant RSV F antigen that was specifically designed to adopt a pre-F conformation. Modification of hRSV F was initially inspired by a structural study of the parainfluenza virus 5 (PIV5) F protein in a prefusion form (52, 53). We focused on well-established structural and functional features of the RSV F sequence and modified some of them to stabilize the protein in the pre-F conformation, which led to the Pre-F-GCN4t design. The major characteristics of Pre-F-GCN4t were the lack of F2-F1 processing in the absence of a linker region and the C-terminal fusion with an engineered GCN4 coiled-coil region instead of the foldon domain. More particularly, the two furin sites and pep27 were deleted. As a result, the FP remains attached to the C terminus of F2, preventing a structural translocation of approximately 100 Å that would normally be required for the formation of the post-F 6HB. In line with results obtained by others, the removal of pep27 allowed the folding of the protein in its trimeric form (42, 44, 54). We have shown that the presence of pep27 and the two furin sites in the context of the Post-F-XC construct did not necessarily lead to a complete processing of the peptide. Replacement of the C-terminal transmembrane and endodomain with GCN4 was meant to contribute to the trimerization and stability of the soluble ectodomain in the pre-F conformation. The C-terminal GCN4 fusion was designed to compensate for HRB instability in the absence of the transmembrane region, preventing membrane interaction and aggregation and thus allowing secretion. Moreover, the presence of the C-terminal GCN4 triple-helix coiled-coil structure should prevent HRB translocation and formation of the post-F 6HB. N-glycosylated Pre-F-GCN4t was purified from CHO cell culture supernatants with the expected soluble quaternary structure. Finally, the L112Q, Q471G, and L482K substitutions were introduced in the context of Pre-F-GCN4t to improve the stability, homogeneity, and production yield, which are critical parameters for large-scale recombinant vaccine production. For comparison purposes, a recombinant F protein adopting a post-F conformation, called Post-F-XC, was also produced based on data from a previous study (37). AUC analysis confirmed that Post-F-XC was purified in the expected oligomeric state, and EM data also confirmed adequate post-F morphological characteristics. In EM images, Post-F-XC appeared as an elongated conical structure, consistent with post-F structures described previously (36, 37, 49). In addition, Post-F-XC did not bind D25, which was a further indication of its post-F conformation.
Chemical unfolding and HDX-MS analyses were performed to establish the thermodynamic characteristics of both molecules. The chemical unfolding profile highlighted clear thermodynamic differences. This technique showed that the globular heads of both Pre-F-GCN4t and Post-F-XC are remarkably resistant to treatment, as the complete unfolding of either protein required high concentrations of the urea-GnHCl mixture. Nevertheless, Post-F-XC was shown to be globally more stable than Pre-F-GCN4t. A recent report presented differential scanning fluorimetry (DSF) results for protein F prefusion (DS-Cav1) and postfusion (FΔFP) conformers (55). Those DSF results showed a very high melting temperature (Tm) (90°C) for the postfusion conformer and a slightly lower Tm for the prefusion conformer (61 to 81°C). Such data agree well with our chemical unfolding results showing that Post-F-XC has a higher stability than Pre-F-GCN4t. In our work, the unfolding of Post-F-XC occurred with two main transitions. It is noteworthy that Pre-F-GCN4t unfolding and the first unfolding step of Post-F-XC occurred at about the same denaturant concentrations, which suggests that there is a Post-F-XC domain(s) whose stability is similar to that of Pre-F-GCN4t. The second transition in the Post-F-XC unfolding profile could be attributed to the 6HB structure at first sight, as this structure has already been reported to be highly stable (56). However, as there are no tryptophan residues in the HRA or HRB regions which form the 6HB, the chemical unfolding experiment could not monitor its stability directly. In contrast, HDX-MS data showed that all detectable amide hydrogens in the HRA and HRB regions finished exchanging by the 1,500-s time point, which rather indicates that the 6HB is not stable by itself in Post-F-XC. One hypothesis is that the HRA-proximal subunit in Post-F-XC is more stable than that in Pre-F-GCN4t, while the HRA-distal subunits have the same stability in the two constructs. This would be in line with the fact that the HRA-proximal subunit is fused with the 6HB, implying that the two domains would form a single cooperatively unfolding structural entity. As only the HRA-proximal and the HRA-distal subunits in the globular head include a tryptophan residue(s), it is reasonable to assume that these two subunits unfold at different denaturant concentrations in Post-F-XC. There are two tryptophan residues in the HRA-distal subunit and one in the HRA-proximal subunit. As the red shift of the first chemical unfolding transition is approximately twice as large as that of the second transition, it is more likely that the first unfolding event represents the HRA-distal subunit and that the second one represents the HRA-proximal subunit. Therefore, as expected, the HRA-proximal subunit is most probably stabilized in Post-F-XC compared to Pre-F-GCN4t. This observation may be considered a unique post-F thermodynamic signature. Pre-F-GCN4t does not show this signature, which indicates that this molecule does not adopt a post-F conformation.
EM provided further evidence about the morphological specificity of Pre-F-GCN4t. Indeed, the appearance of Pre-F-GCN4t in EM images was easily distinguishable from that of Post-F-XC. However, the Pre-F-GCN4t lollipop-shaped structure was slightly divergent from the rounded pre-F structure presented in works by others (24, 43). We attributed this morphological difference to the addition of the GCN4 trimeric coiled-coil domain. Our goal to obtain a pre-F molecule supposedly stabilized in the pre-F conformation was almost achieved after the demonstration that the molecule does not reassemble into a post-F conformation. However, we did not yet know to which extent the structural specificities induced by the present stabilization strategy could influence the antigenicity of the molecule.
The HDX-MS data are in line with the fact that Pre-F-GCN4t and Post-F-XC share most of their three-dimensional (3D) coordinates, with most peptides showing similar exchange kinetics in both molecules. This technique revealed subtle dissimilarities between the two constructs. Among the regions presenting different deuterium incorporation between Pre-F-GCN4t and Post-F-XC, two trends are of particular interest. We first observed that some regions were more stable in the context of Pre-F-GCN4t than in the context of Post-F-XC. This local stability may reflect the present design approach, as this molecule was especially designed to be stabilized in the pre-F conformation. The second trend concerns regions undergoing a significant drop in deuterium incorporation in the context of Post-F-XC. In line with the 6HB contributing to the stability of Post-F-XC, such regions are localized mostly in HRA. A particular segment of interest is localized in the N-terminal part of Post-F-XC HRA. This α-helical segment may be an important contributor to the free energy loss driving protein folding into the native form and may be essential for membrane fusion activity.
The HDX-MS results indicated a high deuteration level for the FP in Pre-F-GCN4t, suggesting that this region is highly flexible. This is consistent with the observation that the FP is exposed to degradation from secreted endogenous proteases during in-process stability monitoring (data not shown). On the other hand, this might appear to contradict the pre-F crystal structure, which showed that the FP lies inside a protecting cavity (32). There are two possible explanations for this apparent discrepancy. One possible explanation is that the region around pep27 flanked with two furin sites and the FP is dynamic in the native pre-F conformation. In this way, the two furin sites are susceptible to furin cleavage despite their possible location inside the cavity. The dynamic property of this region was just sustained in the Pre-F-GCN4t construct. The other explanation is that the removal of the two furin cleavage sites and pep27 hampers its folding inside a Pre-F-GCN4t internal cavity. In this case, Pre-F-GCN4t may resemble more the previously reported PIV5 pre-F structure (52, 53), and the surface accessibility of the FP could provide specific antigenic attributes to Pre-F-GCN4t in comparison with previously reported fully processed pre-F structures.
Pre-F-GCN4t was recognized by palivizumab, with the affinity constant in the same range as known palivizumab–pre-F affinity values. The epitope of palivizumab is site II, which is displayed by both the Pre-F-GCN4t and Post-F-XC constructs. Two segments covering this site in HDX-MS analysis, the segments spanning positions 222 to 227 and 230 to 247, showed very similar and relatively low deuteration levels in both Pre-F-GCN4t and Post-F-XC, implying that both constructs present this site in similar ways. This reflects the fact that both molecules were equally efficient at inhibiting palivizumab neutralizing activity. Despite being an interesting site to consider for the antigenicity of Pre-F-GCN4t, site II is not specific to the pre-F conformation, and thus, the binding at site II does not confirm the supposed pre-F conformation of Pre-F-GCN4t. Data from two antigenic assays suggested the pre-F nature of Pre-F-GCN4t. First, Pre-F-GCN4t was more efficient than Post-F-XC at inhibiting the neutralizing activity of a human convalescent-phase serum pool. A major contributor to the human neutralizing antibody response is antigenic site ø, which was initially identified by D25 epitope characterization (32, 38). This conformational epitope consists of two segments, residues 62 to 69 in the F2 fragment and residues 166 to 179 in the HRA region of the F1 fragment. After folding into the post-F state, the F1 segment refolds into the 6HB, so the conformational epitope is lost. Accordingly, we showed in a second antigenic assay that D25 bound to Pre-F-GCN4t but not to Post-F-XC in an ELISA. The latter observation reveals a pre-F-like structure but does not confirm the quaternary structure of the molecule, as D25 is known to also bind monomeric pre-F (54). Nevertheless, the trimeric nature of Pre-F-GCN4t was supported by data from AUC experiments. It may be worth pointing out that site ø resides in a very dynamic region, unlike site II, and that dynamic regions are often involved in conformational changes (see above).
Altogether, these data show that Pre-F-GCN4t has important characteristics of a prefusion conformation. It has the requisite product quality and antigenic attributes desired for an RSV F protein vaccine candidate. Further preclinical, particularly immunogenicity characterization, but also clinical assessments of Pre-F-GCN4t are in progress to demonstrate the ability of this recombinant subunit RSV vaccine candidate to address the unmet RSV vaccine needs.
MATERIALS AND METHODS
Design of genetically modified F proteins.
The design of Pre-F-GCN4t was based on the hRSV RSS2 reference strain (subtype A) F0 protein sequence and the PIV5 pre-F structure (52). Peptide 27, located between the two furin cleavage sites of F0, was deleted. A Lys residue from one of the two furin sites was conserved to keep a positive charge at this position. The transmembrane and C-terminal regions were replaced by the engineered yeast GCN4 isoleucine zipper domain variant EDKIEEILSKIYHIENEIARIKKLIGEA (46). This modified GCN4 stabilizes as a trimer, hence the denomination GCN4t. Additional L112Q, Q471G, and L482K point mutations (amino acid numbering refers to the Pre-F-GCN4t sequence) (Fig. 1B) were introduced by using the QuikChange II site-directed mutagenesis kit (Agilent Technologies).
The construct design of Post-F-XC was based on the RSV reference strain RSS2 F0 protein sequence and the known post-F structure (36, 37). In this design, the following modifications were made to F0: deletion of the transmembrane domain and the C-terminal domain and truncation of the first 9 amino acids of the FP. In addition, a C-terminal His tag was added to facilitate purification.
Synthesis of the genes corresponding to Pre-F-GCN4t and Post-F-XC, with codon optimization favoring Cricetulus griseus species, was performed by GeneArt Gene Synthesis Services (Life Technologies).
Design modifications of Pre-F-GCN4t and Post-F-XC, compared with the native RSS2 F0 sequence, are shown in Fig. 1A.
Cloning and expression of Pre-F-GCN4t and Post-F-XC.
The synthetic Pre-F-GCN4t and Post-XC genes were cloned into the pEE14 mammalian expression vector using HindIII-XbaI restriction sites (Lonza). The pEE14 plasmids expressing PreF-GCN4t or Post-F-XC were introduced into host CHO cells by lipofection using the FreeStyle Max reagent (Life Technologies). Glutamine selection was used to establish stable CHO clones. Transfected cells were grown in serum-free CD-CHO medium (Life Technologies) in shake flasks at 37°C with 5% CO2 and passaged at 2- to 3-day intervals. Protein expression was triggered by shifting the incubation temperature to 29°C and adding 2 mM sodium butyrate. After 6 days, the culture supernatant was harvested, and Pre-F-GCN4t or Post-F-XC proteins were purified.
Purification of F proteins.
For the purification of Pre-F-GCN4t, cell debris were removed by centrifugation at 10,000 × g for 10 min, and the supernatant was filtered on a Sartobran 0.45/0.2 filter (Sartorius), adjusted to pH 6.0 with HCl, and injected into a CM ceramic HyperD ion-exchange chromatography column (Pall) equilibrated in 20 mM sodium phosphate buffer (pH 6.0). Pre-F-GCN4t was eluted by using a 0.3 M NaCl step gradient. Fractions containing Pre-F-GCN4t, as determined by SDS-PAGE and Western blotting with a Pre-F-GCN4t-specific polyclonal antibody (from a rabbit immunized with His-tagged Pre-F-GCN4t–incomplete Freund's adjuvant), were pooled and transferred to ceramic hydroxyapatite type II resin (Bio-Rad) in 50 mM sodium phosphate buffer (pH 7.0), followed by stepwise elution with 0.5 M NaCl. The eluate was directly loaded onto Capto Adhere resin (GE Healthcare) equilibrated in 20 mM sodium phosphate (pH 7.0) containing 500 mM NaCl, washed with 10 mM sodium phosphate buffer (pH 7.0) containing 0.1 M arginine, and eluted with a buffer containing 0.6 M arginine. Finally, a pool of Pre-F-GCN4t-containing fractions was transferred to a size exclusion chromatography column packed with Superdex 200 (GE Healthcare) in 10 mM sodium dipotassium phosphate–160 mM NaCl (pH 6.5). Pooled fractions containing Pre-F-GCN4t were sterile filtered, aliquoted, and stored at −70°C until use.
For the purification of Post-F-XC, the harvested supernatant was filtered, adjusted to pH 6.0, and injected onto a CM ceramic HyperD ion-exchange chromatography column (Pall) equilibrated in 20 mM morpholineethanesulfonic acid (MES) (pH 6.0). After a wash step with 5 mM sodium phosphate–20 mM HEPES (pH 7.0), elution was performed with a NaCl gradient. The target protein was eluted in the range of 300 mM to 1 M. As shown after evaluation by reducing SDS-PAGE, this fraction contained the two fragments corresponding to the expected size of the furin-processed F protein. The fraction was loaded onto His Trap FF crude resin (GE Healthcare) for immobilized-metal ion affinity chromatography and equilibrated in 20 mM Bicine–500 mM NaCl (pH 8.3), and stepwise elution with 250 mM imidazole was performed. The eluate was fractionated by size exclusion chromatography using Superdex 200 26/60 equilibrated in 20 mM Bicine buffer (pH 8.3) containing 0.5 M NaCl. Fractions containing the furin-processed protein, as assessed by SDS-PAGE, were pooled, sterile filtered, aliquoted, and stored at −70°C until use.
SDS-PAGE analysis of deglycosylated proteins.
Protein N-deglycosylation was performed by using an N-glycosidase F deglycosylation kit (catalog no. 11 836 552 001; Roche), according to the manufacturer's instructions. Glycosylated and nonglycosylated proteins were analyzed by SDS-PAGE using a Novex Bis-Tris 4 to 12% gel (Invitrogen) under reducing conditions. The gels were stained with a Coomassie blue-based staining solution.
Analytical ultracentrifugation.
Protein samples in different buffers were centrifuged at 25,000 rpm in an AN-60Ti rotor at 15°C in a Beckman-Coulter ProteomeLab XL-1 analytical ultracentrifuge equipped with an absorbance optical detection system. The absorbance profile at 280 nm was recorded every 5 min. Data were analyzed with Sedfit 14.1 (NIH, Bethesda, MD) using a continuous size distribution, c(S), model (57).
Chemical denaturation.
Chemical unfolding was performed at room temperature and monitored by using fluorescence spectroscopy. Since Pre-F-GCN4t and Post-F-XC do not unfold completely in saturated urea or GnHCl at 25°C, solutions containing equimolar amounts of both urea and GnHCl were used. Buffers ranging from 0.5 M urea–0.5 GnHCl to 6.5 M urea–6.5 M GnHCl were freshly prepared in 20 mM Bicine–150 mM NaCl (pH 8.0). Pre-F-GCN4t and Post-F-XC samples at 2 mg/ml were brought to different concentrations of urea-GnHCl with these buffers and incubated for at least 2 h 30 min at room temperature before analysis. Intrinsic fluorescence was measured by using a Spectramax M5e instrument (Molecular Devices). Samples were excited at 295 nm, which is specific to tryptophan, and fluorescence emission spectra were scanned from 300 to 420 nm with 1.0-nm increments. The center of spectral mass (CSM) was calculated by using the formula CSM = ∑λ × I(λ)/∑I(λ).
Electron microscopy.
Electron microscopy sample preparation and image analyses were performed at NanoImaging Services, Inc. (San Diego, CA, USA). Samples were prepared by using a continuous-carbon-grid method. Nitrocellulose-supported 400-mesh copper grids were prepared by applying the sample suspension to a cleaned grid, blotting with filter paper, and immediately staining the sample with uranyl formate. EM was performed by using an FEI Tecnai T12 electron microscope operating at 120 keV equipped with an FEI Eagle 4,096- by 4,096-physical-pixel charge-coupled-device (CCD) camera. High-magnification images were acquired at nominal magnifications of ×110,000 (0.10 nm/pixel) and ×67,000 (0.16 nm/pixel). The images were acquired at nominal underfocuses of −2 μm to −1 μm (magnification, ×110,000) and −3 μm to −2 μm (magnification, ×67,000) and electron doses of ∼24 to 39 electron/Å2. For 2D averaging analysis, individual particles in the images at a ×67,000 magnification were selected by using automated picking protocols (58), followed by two rounds of reference-free alignment and classification using the XMIPP software package (59).
Model building.
By using Swiss-PdbViewer (also called DeepView) (60), protein sequences of Pre-F-GCN4t and Post-F-XC were aligned with the corresponding structural templates: pre-F (PDB accession no. 4JHW) (32) and post-F (PDB accession no. 3RKI) (37). Modeling of the three-dimensional structures was performed by using the automated Swissmodel homology modeling server (61). A theoretical model of Pre-F-GCN4t fused with GCN4 was finalized by superposing the resulting Pre-F-GCN4t model with the pre-F PIV5 structure, that is, the structured reported under PDB accession no. 2B9B (52). Superposition of the Pre-F-GCN4t model and the GCN4 structure was achieved in accordance with the designed construct. Energy minimization was achieved by using Molecular Operating Environment (MOE) (Chemical Computing Group, Inc.), assisted by the Amber10:EHT force field. Structural analysis was performed and figure representations were generated with Swiss-PdbViewer.
Hydrogen/deuterium-exchange mass spectrometry.
HDX-MS experiments were performed at ExSAR Corporation. Briefly, an exchange reaction was initiated by diluting 5 μl of a Pre-F-GCN4t stock solution (28.2 μM in 10 mM Na2HPO4–150 mM NaCl [pH 6.5]) or 5 μl of a Post-F-XC stock solution (28.2 μM in 20 mM Bicine–500 mM NaCl [pH 8.3]) with 35 μl of deuterated buffer (20 mM Bicine, 500 mM NaCl [pH 8.3]). The reaction mixture was incubated at 0°C for various times (15, 50, 150, 500, and 1,500 s). Control samples consisted of nondeuterated samples (run without deuterated buffers) and fully deuterated samples [prepared by mixing 11 μl of 28.2 μM Pre-F-GCN4t or Post-F-XC with 77 μl of 100 mM tris(2-carboxyethyl)phosphine (TCEP) in D2O (pH 7) and incubating the mixture at 60°C for 3 h]. The reactions were quenched by mixing the solutions with 20 μl of 8 M urea and 1 M TCEP at pH 4.0 at 0°C. The quenched solutions were immediately pumped over a column filled with porcine pepsin (Sigma-Aldrich) immobilized on Poros 20 AL medium (Applied Biosystems) at 30 mg/ml (62) with 0.05% trifluoroacetic acid (TFA) (200 μl/min) for 3 min, with contemporaneous collection of proteolytic products by using a trap column (Poros R2 medium; Applied Biosystems). Subsequently, the peptide fragments were eluted from the trap column and separated by using an analytical column (BioBasic-18, 1-mm inner diameter [ID] by 50 mm, 5 μm, and a 300-Å pore size; Thermo Scientific) with a linear gradient of 13% solvent B to 35% solvent B over 23 min (solvent A, 0.05% TFA in water; solvent B, 95% acetonitrile, 5% water, and 0.0025% TFA) (flow rate of 5 μl/min to 10 μl/min). Mass spectrometric analyses were carried out with a Thermo OrbiTrap XL mass spectrometer (Thermo Scientific) with a capillary temperature at 200°C. Prior to HDX-MS experiments, each pepsin-generated peptide had been identified (63).
The centroids of probe peptide isotopic envelopes were measured by using HDExpress (ExSAR Corporation). The corrections for back-exchange were made by employing methods described previously (64). The deuteration level (percent) was calculated as [m(P) − m(N)]/[m(F) − m(N)] × 100, where m(P), m(N), and m(F) are the centroid values of the partially deuterated peptide, the nondeuterated peptide, and the fully deuterated peptide, respectively.
Neutralization inhibition assay.
Serial dilutions of a reference anti-hRSV adult human serum pool (65) or palivizumab (MedImmune) were mixed with various concentrations of competitor proteins, either Pre-F-GCN4t or Post-F-XC, or no protein, in 50% Dulbecco's modified Eagle's medium (DMEM)–50% 199-H medium containing 0.5% fetal bovine serum, 2 mM glutamine, and 50 μg/ml gentamicin (RSV medium) and incubated for 30 min at 37°C in round-bottom 96-well plates. Sixty microliters of each serial dilution of antibody and protein was mixed with 30 μl of hRSV A Long (ATCC VR-26; 1,000 PFU/ml) and incubated for 20 min at 33°C with 5% CO2. Fifty microliters of the antibody-competitor-virus mixture was then transferred into flat-bottom 96-well plates previously seeded with 1 × 104 Vero cells/well (ATCC CCL-81) and emptied of cell culture medium. Plates were incubated for 2 h at 33°C with 5% CO2. After incubation, the medium was removed, and cells were covered with 0.5% carboxymethylcellulose in RSV medium. The plates were incubated for 3 days at 33°C with 5% CO2 prior to immunofluorescence staining.
Immunofluorescence analysis was performed by using a goat anti-hRSV antibody followed by fluorescein isothiocyanate (FITC)-labeled rabbit anti-goat IgG. Briefly, cell monolayers were washed with phosphate-buffered saline (PBS) and fixed with 1% paraformaldehyde. After blocking with 2% (wt/vol) skim milk in PBS, hRSV-infected cells were detected by using commercial polyclonal goat anti-hRSV (BioDesign, Meridian Life Science) followed by rabbit anti-goat IgG conjugated to FITC (Sigma-Aldrich) diluted in blocking buffer containing a final concentration of 0.01% Evans blue as the counterstain. Following the washing steps, empty plates were read by using an automated fluorescence microscope. Stained foci were counted with automated software (AxioVision v4.7; Carl Zeiss Vision). For human pooled serum and palivizumab, the effective dose (ED60) titer was determined as the serum dilution at which 60% of the virus was neutralized compared to control wells containing virus and competitor protein but no antibody for each competitor-protein concentration. Control wells consisting of virus only (no competitor protein) were included on each plate and had similar numbers of plaques, indicating that the addition of the competitor protein did not impact viral infection. For each competitor-protein concentration, percent neutralization inhibition was calculated as 1 − serum titer in the presence of competitor protein/serum titer in the absence of competitor protein × 100.
Affinity of palivizumab for Pre-F-GCN4t.
The affinity of palivizumab for Pre-F-GCN4t was analyzed by surface plasmon resonance technology using a Biacore T100 instrument (GE Healthcare). Briefly, 1,500 resonance unit (RU) of a mouse monoclonal anti-human IgG (Fc) antibody (catalog no. BR-1008-39; GE Healthcare) were immobilized on a CM5 sensorchip surface (catalog no. BR-1005-30; GE Healthcare) by N-hydroxysuccinimide–1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride chemistry (amine coupling kit, catalog no. BR-1000-50; GE Healthcare). Each cycle of analysis consisted of the capture of 60 RU of palivizumab followed by the injection of a Pre-F-GCN4t antigen solution prepared in running buffer (phosphate-buffered saline with 0.005% surfactant P20) during 120 s at a 60-μl/min flow rate. The dissociation phase was monitored for 300 s by using the same flow rate. Between cycles, three regeneration cycles of 30 s in glycine (pH 1.5) were performed at a flow rate of 30 μl/min. Background values obtained by antigen binding on the immobilized capture mouse monoclonal antibody were subtracted from each data point. Binding curves at seven different concentrations (0, 25, 50, 75, 100, 150, and 200 nM) of the Pre-F-GCN4t antigen were analyzed with the 1:1 Langmuir binding model using the Biacore BIA evaluation program. Due to the trimeric nature of the antigen, a substantial avidity effect can reduce the off-rate. The apparent dissociation constant (KD) was calculated as a ratio of the two rate constants, koff/kon.
D25 immunoassay.
Ninety-six-well flat-bottom microtiter plates (Thermo Fisher) were coated with 100 ng of either Pre-F-GCN4t or Post-F-XC (2 μg/ml) overnight at 4°C. One hundred microliters of various concentrations of the D25 antibody (in-house produced, from DNA vectors kindly provided by ReiThera SRL, Rome, Italy) was added to each well, and the mixture was incubated for 2 h at 37°C. D25 is a monoclonal antibody that binds and locks the F protein in its prefusion state (32). In the microtiter plates, bound antibodies were detected by incubation with 50 μl of horseradish peroxidase-conjugated mouse anti-human IgG Fc antibody (Life Technologies) for 1 h at 37°C, followed by 50 μl of the SureBlue horseradish peroxidase substrate (KPL) for 30 min at room temperature. Between steps, plates were washed with PBS–0.05% Tween 20. The peroxidase reaction was stopped with 2 N 50 μl H2SO4 and 100 μl acetic acid. Plates were read in a Vmax microplate reader (Molecular Devices) at 450 nm, and results were analyzed with Softmax Pro.
ACKNOWLEDGMENTS
We thank Chantal Molitor, Nathalie Drouin, Pierre Plante, and Jean-Philippe Lièvremont (molecular biology, GSK); Patrick Dumas, Karmen Hodges, and Jean-Philippe Matheise (upstream process development, GSK); Jean-Michel Schauss, Philippe Permanne, Bruno André, and Frédéric Buyse (downstream development, GSK); Xavier Czeszak and Michel Plisnier (analytical development, GSK); Michel Deschuyteneer (electron microscopy, GSK); and Denis Faubert (MS/MS analyses, Institut de Recherches Cliniques de Montréal). We appreciate the input of Jean-Francois Toussaint, Christophe Debacq, and Sonya Cyr (all from GSK at the time of the study). Pascal Cadot (Xpe-Pharma & Science) and Krikor Torossian (GSK) provided scientific writing services, and Ulrike Krause (GSK) provided editorial assistance and coordinated manuscript development.
This work was sponsored and financially supported by GlaxoSmithKline Biologicals SA, which was involved in all stages of study conduct and analysis. The costs associated with the development and publishing of the manuscript, including scientific writing assistance, were also covered by GlaxoSmithKline Biologicals SA.
D.M., G.B., J.-L.R., M.G., N.B., and Y.H. were involved in the conception and/or the design of the study. J.D., M.B., M.G., P.R., V.D., and Y.H. developed the protocol(s) for the study and/or acquired the data. A.-M.S., D.M., J.D., M.B., M.G., N.B., V.D., and Y.H. analyzed and interpreted the results.
A.-M.S., D.M., G.B., J.D., J.-L.R., M.B., M.G., N.B., P.R., and V.D. are, or were at the time of the study, employees of the GSK group of companies. A.-M.S., N.B., and J.-L.R. report ownership of GSK shares and/or restricted GSK shares. A.-M.S., G.B., N.B., and P.R. are listed as inventors on patents owned by the GSK group of companies. M.G. has received consulting fees from GlaxoSmithKline Biologicals SA. Y.H. reports no conflicts of interest.
REFERENCES
- 1.Hall CB. 2012. The burgeoning burden of respiratory syncytial virus among children. Infect Disord Drug Targets 12:92–97. doi: 10.2174/187152612800100099. [DOI] [PubMed] [Google Scholar]
- 2.Leader S, Kohlhase K. 2003. Recent trends in severe respiratory syncytial virus (RSV) among US infants, 1997 to 2000. J Pediatr 143:S127–S132. doi: 10.1067/S0022-3476(03)00510-9. [DOI] [PubMed] [Google Scholar]
- 3.Thorburn K, Harigopal S, Reddy V, Taylor N, van Saene HKF. 2006. High incidence of pulmonary bacterial co-infection in children with severe respiratory syncytial virus (RSV) bronchiolitis. Thorax 61:611–615. doi: 10.1136/thx.2005.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou H, Thompson WW, Viboud CG, Ringholz CM, Cheng P-Y, Steiner C, Abedi GR, Anderson LJ, Brammer L, Shay DK. 2012. Hospitalizations associated with influenza and respiratory syncytial virus in the United States, 1993-2008. Clin Infect Dis 54:1427–1436. doi: 10.1093/cid/cis211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langley GF, Anderson LJ. 2011. Epidemiology and prevention of respiratory syncytial virus infections among infants and young children. Pediatr Infect Dis J 30:510–517. doi: 10.1097/INF.0b013e3182184ae7. [DOI] [PubMed] [Google Scholar]
- 6.Sommer C, Resch B, Simões EAF. 2011. Risk factors for severe respiratory syncytial virus lower respiratory tract infection. Open Microbiol J 5:144–154. doi: 10.2174/1874285801105010144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherukuri A, Patton K, Gasser RA Jr, Zuo F, Woo J, Esser MT, Tang RS. 2013. Adults 65 years and older have reduced numbers of functional memory T cells to respiratory syncytial virus fusion (F) protein. Clin Vaccine Immunol 20:239–247. doi: 10.1128/CVI.00580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 9.Falsey AR, McElhaney JE, Beran J, van Essen GA, Duval X, Esen M, Galtier F, Gervais P, Hwang S-J, Kremsner P, Launay O, Leroux-Roels G, McNeil SA, Nowakowski A, Richardus JH, Ruiz-Palacios G, St Rose S, Devaster J-M, Oostvogels L, Durviaux S, Taylor S. 2014. Respiratory syncytial virus and other respiratory viral infections in older adults with moderate to severe influenza-like illness. J Infect Dis 209:1873–1881. doi: 10.1093/infdis/jit839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClure DL, Kieke BA, Sundaram ME, Simpson MD, Meece JK, Sifakis F, Gasser RA Jr, Belongia EA. 2014. Seasonal incidence of medically attended respiratory syncytial virus infection in a community cohort of adults >=50 years old. PLoS One 9:e102586. doi: 10.1371/journal.pone.0102586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 12.Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang S-C, Dormitzer M, O'Grady J, Koenig S, Tamura JK, Woods R, Bansal G, Couchenour D, Tsao E, Hall WC, Young JF. 1997. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J Infect Dis 176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 13.American Academy of Pediatrics. 2009. Policy statements—modified recommendations for use of palivizumab for prevention of respiratory syncytial virus infections. Pediatrics 124:1694–1701. doi: 10.1542/peds.2009-2345. [DOI] [PubMed] [Google Scholar]
- 14.Geskey JM, Thomas NJ, Brummel GL. 2007. Palivizumab: a review of its use in the protection of high risk infants against respiratory syncytial virus (RSV). Biologics 1:33–43. [PMC free article] [PubMed] [Google Scholar]
- 15.IMpact-RSV Study Group. 1998. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 102:531–537. doi: 10.1542/peds.102.3.531. [DOI] [PubMed] [Google Scholar]
- 16.Graham BS. 2016. Vaccines against respiratory syncytial virus: the time has finally come. Vaccine 34:3535–3541. doi: 10.1016/j.vaccine.2016.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins D, Trujillo C, Keech C. 2016. Advances in RSV vaccine research and development—a global agenda. Vaccine 34:2870–2875. doi: 10.1016/j.vaccine.2016.03.109. [DOI] [PubMed] [Google Scholar]
- 18.Guvenel AK, Chiu C, Openshaw PJM. 2014. Current concepts and progress in RSV vaccine development. Expert Rev Vaccines 13:333–344. doi: 10.1586/14760584.2014.878653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawasaki Y, Hosoya M, Katayose M, Suzuki H. 2004. Role of serum neutralizing antibody in reinfection of respiratory syncytial virus. Pediatr Int 46:126–129. doi: 10.1046/j.1442-200x.2004.01860.x. [DOI] [PubMed] [Google Scholar]
- 20.Piedra PA, Jewell AM, Cron SG, Atmar RL, Glezen WP. 2003. Correlates of immunity to respiratory syncytial virus (RSV) associated-hospitalization: establishment of minimum protective threshold levels of serum neutralizing antibodies. Vaccine 21:3479–3482. doi: 10.1016/S0264-410X(03)00355-4. [DOI] [PubMed] [Google Scholar]
- 21.Shinoff JJ, O'Brien KL, Thumar B, Shaw JB, Reid R, Hua W, Santosham M, Karron RA. 2008. Young infants can develop protective levels of neutralizing antibody after infection with respiratory syncytial virus. J Infect Dis 198:1007–1015. doi: 10.1086/591460. [DOI] [PubMed] [Google Scholar]
- 22.Wu H, Pfarr DS, Tang Y, An L-L, Patel NK, Watkins JD, Huse WD, Kiener PA, Young JF. 2005. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J Mol Biol 350:126–144. doi: 10.1016/j.jmb.2005.04.049. [DOI] [PubMed] [Google Scholar]
- 23.Collins PL, Melero JA. 2011. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res 162:80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magro M, Mas V, Chappell K, Vázquez M, Cano O, Luque D, Terrón MC, Melero JA, Palomo C. 2012. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A 109:3089–3094. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson PR, Spriggs MK, Olmsted RA, Collins PL. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci U S A 84:5625–5629. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sullender WM. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin Microbiol Rev 13:1–15. doi: 10.1128/CMR.13.1.1-15.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calder LJ, González-Reyes L, García-Barreno B, Wharton SA, Skehel JJ, Wiley DC, Melero JA. 2000. Electron microscopy of the human respiratory syncytial virus fusion protein and complexes that it forms with monoclonal antibodies. Virology 271:122–131. doi: 10.1006/viro.2000.0279. [DOI] [PubMed] [Google Scholar]
- 28.González-Reyes L, Ruiz-Argüello MB, García-Barreno B, Calder L, López JA, Albar JP, Skehel JJ, Wiley DC, Melero JA. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci U S A 98:9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell CJ, Luque LE. 2006. The structural basis of paramyxovirus invasion. Trends Microbiol 14:243–246. doi: 10.1016/j.tim.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang A, Dutch RE. 2012. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 4:613–636. doi: 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLellan JS. 2015. Neutralizing epitopes on the respiratory syncytial virus fusion glycoprotein. Curr Opin Virol 11:70–75. doi: 10.1016/j.coviro.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palomo C, Mas V, Vázquez M, Cano O, Luque D, Terrón MC, Calder LJ, Melero JA. 2014. Polyclonal and monoclonal antibodies specific for the six-helix bundle of the human respiratory syncytial virus fusion glycoprotein as probes of the protein post-fusion conformation. Virology 460–461:119–127. doi: 10.1016/j.virol.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Graham BS, Modjarrad K, McLellan JS. 2015. Novel antigens for RSV vaccines. Curr Opin Immunol 35:30–38. doi: 10.1016/j.coi.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magro M, Andreu D, Gómez-Puertas P, Melero JA, Palomo C. 2010. Neutralization of human respiratory syncytial virus infectivity by antibodies and low-molecular-weight compounds targeted against the fusion glycoprotein. J Virol 84:7970–7982. doi: 10.1128/JVI.00447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLellan JS, Yang Y, Graham BS, Kwong PD. 2011. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol 85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, Dormitzer PR, Carfi A. 2011. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A 108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ngwuta JO, Chen M, Modjarrad K, Joyce MG, Kanekiyo M, Kumar A, Yassine HM, Moin SM, Killikelly AM, Chuang G-Y, Druz A, Georgiev IS, Rundlet EJ, Sastry M, Stewart-Jones GBE, Yang Y, Zhang B, Nason MC, Capella C, Peeples ME, Ledgerwood JE, McLellan JS, Kwong PD, Graham BS. 2015. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 7:309ra162. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sastre P, Melero JA, García-Barreno B, Palomo C. 2004. Comparison of antibodies directed against human respiratory syncytial virus antigens present in two commercial preparations of human immunoglobulins with different neutralizing activities. Vaccine 23:435–443. doi: 10.1016/j.vaccine.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 40.Boyington JC, Joyce MG, Sastry M, Stewart-Jones GB, Chen M, Kong WP, Ngwuta JO, Thomas PV, Tsybovsky Y, Yang Y, Zhang B, Chen L, Druz A, Georgiev IS, Ko K, Zhou T, Mascola JR, Graham BS, Kwong PD. 2016. Structure-based design of head-only fusion glycoprotein immunogens for respiratory syncytial virus. PLoS One 11:e0159709. doi: 10.1371/journal.pone.0159709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joyce MG, Zhang B, Ou L, Chen M, Chuang GY, Druz A, Kong WP, Lai YT, Rundlet EJ, Tsybovsky Y, Yang Y, Georgiev IS, Guttman M, Lees CR, Pancera M, Sastry M, Soto C, Stewart-Jones GB, Thomas PV, Van Galen JG, Baxa U, Lee KK, Mascola JR, Graham BS, Kwong PD. 2016. Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV. Nat Struct Mol Biol 23:811–820. doi: 10.1038/nsmb.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJM, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, Langedijk JPM. 2015. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun 6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GBE, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang G-Y, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko S-Y, Todd J-P, Rao S, Graham BS, Kwong PD. 2013. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart-Jones GBE, Thomas PV, Chen M, Druz A, Joyce MG, Kong W-P, Sastry M, Soto C, Yang Y, Zhang B, Chen L, Chuang G-Y, Georgiev IS, McLellan JS, Srivatsan S, Zhou T, Baxa U, Mascola JR, Graham BS, Kwong PD. 2015. A cysteine zipper stabilizes a pre-fusion F glycoprotein vaccine for respiratory syncytial virus. PLoS One 10:e0128779. doi: 10.1371/journal.pone.0128779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Widjaja I, Rigter A, Jacobino S, van Kuppeveld FJM, Leenhouts K, Palomo C, Melero JA, Leusen JHW, Haijema BJ, Rottier PJM, de Haan CAM. 2015. Recombinant soluble respiratory syncytial virus F protein that lacks heptad repeat B, contains a GCN4 trimerization motif and is not cleaved displays prefusion-like characteristics. PLoS One 10:e0130829. doi: 10.1371/journal.pone.0130829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harbury PB, Zhang T, Kim PS, Alber T. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 47.Dragan AI, Potekhin SA, Sivolob A, Lu M, Privalov PL. 2004. Kinetics and thermodynamics of the unfolding and refolding of the three-stranded α-helical coiled coil, Lpp-56. Biochemistry 43:14891–14900. doi: 10.1021/bi048365+. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz-Argüello MB, González-Reyes L, Calder LJ, Palomo C, Martín D, Saíz MJ, García-Barreno B, Skehel JJ, Melero JA. 2002. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology 298:317–326. doi: 10.1006/viro.2002.1497. [DOI] [PubMed] [Google Scholar]
- 49.Chaiwatpongsakorn S, Epand RF, Collins PL, Epand RM, Peeples ME. 2011. Soluble respiratory syncytial virus fusion protein in the fully cleaved, pretriggered state is triggered by exposure to low-molarity buffer. J Virol 85:3968–3977. doi: 10.1128/JVI.01813-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Englander SW, Kallenbach NR. 1983. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys 16:521–655. doi: 10.1017/S0033583500005217. [DOI] [PubMed] [Google Scholar]
- 51.Wu H, Pfarr DS, Johnson S, Brewah YA, Woods RM, Patel NK, White WI, Young JF, Kiener PA. 2007. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol 368:652–665. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 52.Yin H-S, Wen X, Paterson RG, Lamb RA, Jardetzky TS. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, Lamb RA. 2012. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci U S A 109:16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swanson KA, Balabanis K, Xie Y, Aggarwal Y, Palomo C, Mas V, Metrick C, Yang H, Shaw CA, Melero JA, Dormitzer PR, Carfi A. 2014. A monomeric uncleaved respiratory syncytial virus F antigen retains prefusion-specific neutralizing epitopes. J Virol 88:11802–11810. doi: 10.1128/JVI.01225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flynn JA, Durr E, Swoyer R, Cejas PJ, Horton MS, Galli JD, Cosmi SA, Espeseth AS, Bett AJ, Zhang L. 2016. Stability characterization of a vaccine antigen based on the respiratory syncytial virus fusion glycoprotein. PLoS One 11:e0164789. doi: 10.1371/journal.pone.0164789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao X, Singh M, Malashkevich VN, Kim PS. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc Natl Acad Sci U S A 97:14172–14177. doi: 10.1073/pnas.260499197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamn equation modeling. Biophys J 78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P-W, Lyumkis D, Potter CS, Carragher B. 2009. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sorzano COS, Marabini R, Velázquez-Muriel J, Bilbao-Castro JR, Scheres SHW, Carazo JM, Pascual-Montano A. 2004. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol 148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 60.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 61.Arnold SR, Wang EEL, Law BJ, Boucher FD, Stephens D, Robinson JL, Dobson S, Langley JM, McDonald J, MacDonald NE, Mitchell I. 1999. Variable morbidity of respiratory syncytial virus infection in patients with underlying lung disease: a review of the PICNIC RSV database. Pediatric Investigators Collaborative Network on Infections in Canada. Pediatr Infect Dis J 18:866–869. doi: 10.1097/00006454-199910000-00006. [DOI] [PubMed] [Google Scholar]
- 62.Hamuro Y, Coales SJ, Southern MR, Nemeth-Cawley JF, Stranz DD, Griffin PR. 2003. Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. J Biomol Tech 14:171–182. [PMC free article] [PubMed] [Google Scholar]
- 63.Hamuro Y, Coales SJ, Molnar KS, Tuske SJ, Morrow JA. 2008. Specificity of immobilized porcine pepsin in H/D exchange compatible conditions. Rapid Commun Mass Spectrom 22:1041–1046. doi: 10.1002/rcm.3467. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Z, Smith DL. 1993. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci 2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang DP, Zielinska E, Quiroz J, Madore D, Rappaport R. 2007. Preparation of a respiratory syncytial virus human reference serum for use in the quantitation of neutralization antibody. Biologicals 35:183–187. doi: 10.1016/j.biologicals.2006.09.004. [DOI] [PubMed] [Google Scholar]