ABSTRACT
Poxviruses use a complex strategy to escape immune control, by expressing immunomodulatory proteins that could limit their use as vaccine vectors. To test the role of poxvirus NF-κB pathway inhibitors A52, B15, and K7 in immunity, we deleted their genes in an NYVAC (New York vaccinia virus) strain that expresses HIV-1 clade C antigens. After infection of mice, ablation of the A52R, B15R, and K7R genes increased dendritic cell, natural killer cell, and neutrophil migration as well as chemokine/cytokine expression. Revertant viruses with these genes confirmed their role in inhibiting the innate immune system. To different extents, enhanced innate immune responses correlated with increased HIV Pol- and Gag-specific polyfunctional CD8 T cell and HIV Env-specific IgG responses induced by single-, double-, and triple-deletion mutants. These poxvirus proteins thus influence innate and adaptive cell-mediated and humoral immunity, and their ablation offers alternatives for design of vaccine vectors that regulate immune responses distinctly.
IMPORTANCE Poxvirus vectors are used in clinical trials as candidate vaccines for several pathogens, yet how these vectors influence the immune system is unknown. We developed distinct poxvirus vectors that express heterologous antigens but lack different inhibitors of the central host-cell signaling pathway. Using mice, we studied the capacity of these viruses to induce innate and adaptive immune responses and showed that these vectors can distinctly regulate the magnitude and quality of these responses. These findings provide important insights into the mechanism of poxvirus-induced immune response and alternative strategies for vaccine vector design.
KEYWORDS: NF-κB, NYVAC, T cell immunity, adaptive immunity, human immunodeficiency virus, immunomodulation, innate immunity, poxvirus, vaccines, vaccinia virus
INTRODUCTION
Several vaccine strategies have been developed to improve immune responses to heterologous antigens expressed by attenuated poxvirus vectors (1). Given the limited effectiveness of the ALVAC poxvirus vector in the RV144 phase III HIV/AIDS clinical trial (2), the need remains to improve poxvirus vector capacity as an immunogen, to increase protection levels, and to understand how innate and adaptive immune responses can be regulated.
The attenuated poxvirus strain NYVAC (New York vaccinia virus) has been used as a vaccine vector in HIV clinical trials (3, 4). Studies in nonhuman primates showed that NYVAC expressing Env and/or the Gag-Pol-Nef (GPN) clade C HIV antigens elicited a balanced CD4/CD8 T cell response (5) or robust T cell immunity (6). In clinical trials in healthy volunteers and in chronically HIV-infected patients, NYVAC showed clear immunogenic potential to induce expansion of preexisting T cell responses as well as the appearance of newly detected polyfunctional CD8 T cell responses (7).
Deletion of vaccinia virus (VACV) immunomodulatory genes that encode inhibitors of the Toll-like receptor (TLR) pathway is a common vaccine optimization strategy (8–10). VACV binding induces TLR2 and TLR4 homo- or heterodimerization (11, 12); this is followed by recruitment of Toll/interleukin-1 receptor (IL-1R) (TIR) domain-containing adaptor proteins such as myeloid differentiation factor 88 (MyD88) and MyD88-adaptor-like (MAL) or TIR domain-containing adapter inducing beta interferon (IFN-β) (TRIF) and TRIF-related adaptor molecule (TRAM). MyD88 mediates activation of tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) by phosphorylation of IL-1R-associated kinases (IRAK) 1, 2, and 4; TRIF interacts directly with TRAF6. TRAF6 recruits transforming growth factor beta (TGF-β)-activated kinase (TAK1) as well as TAK1-binding proteins 1 (TAB1) and 2 (TAB2) to phosphorylate TAK1 and activate the IκB kinase (IKK) complex (13).
A52, B15 (corresponding to Western Reserve strain B14), and K7 are VACV inhibitors of the TLR pathway (14). A52 and K7 block IRAK2 and TRAF6 activation (15, 16), and B15 inhibits IκBα phosphorylation and proteasome degradation (17), thus avoiding release of the NF-κB complex into the cell nucleus. NYVAC infection limits NF-κB binding to the κB site and transcription of proinflammatory cytokine and chemokine genes (18).
Cytokine and chemokine proinflammatory signals are involved in recruiting several cell types that act as the first cell line of defense in the innate immune response (19), including monocytes, dendritic cells (DC), natural killer (NK) cells, B cells, and neutrophils, which generate adaptive immunity (20). Distinct poxvirus vectors induce substantial differences in chemokine and cytokine profiles (21), which influence the adaptive immune responses. Whereas DCs and neutrophils transport VACV antigens to lymphoid organs and directly induce antigen-specific T cell responses (22, 23), NK cells can generate the environment necessary to induce recruitment of activated T cells (20) or cross talk with DCs for the generation of antigen-specific T cell immunity (24).
We prepared several NYVAC deletion mutants that express HIV-1 Env and GPN clade C antigens (NYVAC-C), but lack the A52R, K7R, and B15R genes that encode TLR NF-κB inhibitors. The effect of their concomitant ablation on increased neutrophil migration and HIV-specific CD8 T cell responses has been studied (18). Here we used NYVAC-C (18) to define the specific role of single, double, and triple deletion of these genes in innate and adaptive HIV-specific cell-mediated and humoral responses. We show that deletion of these genes increases cytokine/chemokine secretion and innate immune responses at the infection site. This enhanced innate immune response correlates with increased HIV-specific adaptive cell-mediated and humoral immunity.
RESULTS
Deletion of NF-κB inhibitory proteins in NYVAC leads to increased innate immune responses.
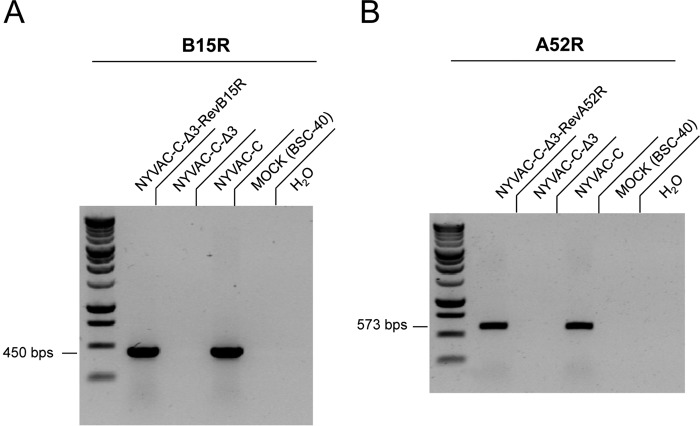
To define the immunomodulatory role of the VACV NF-κB inhibitory A52, K7, and B15 proteins, we used mutants with single, double, and triple deletions of these genes on a backbone NYVAC-C vector expressing HIV-1 clade C antigens Env (gp120) as a cell-released product and Gag-Pol-Nef (GPN) as an intracellular polyprotein (25). BALB/c mice were infected by intraperitoneal (i.p.) injection of 107 PFU of the sucrose-purified NYVAC-C and NYVAC-C deletion mutants NYVAC-C ΔΒ15R, NYVAC-C ΔΑ52R, NYVAC-C ΔΚ7R, NYVAC-C ΔΒ15R ΔΚ7R, NYVAC-C ΔΑ52R ΔΒ15R, NYVAC-C ΔΑ52R ΔΚ7R, and NYVAC-C ΔΑ52R ΔΒ15R ΔΚ7R (here termed the NYVAC-C Δ3 or triple-deletion mutant). Reverse transcription-PCR (RT-PCR) of BSC-40 cells infected with the different viruses confirmed the lack of A52R, K7R, and B15R gene transcription following gene deletion (Fig. 1A to C).
FIG 1.
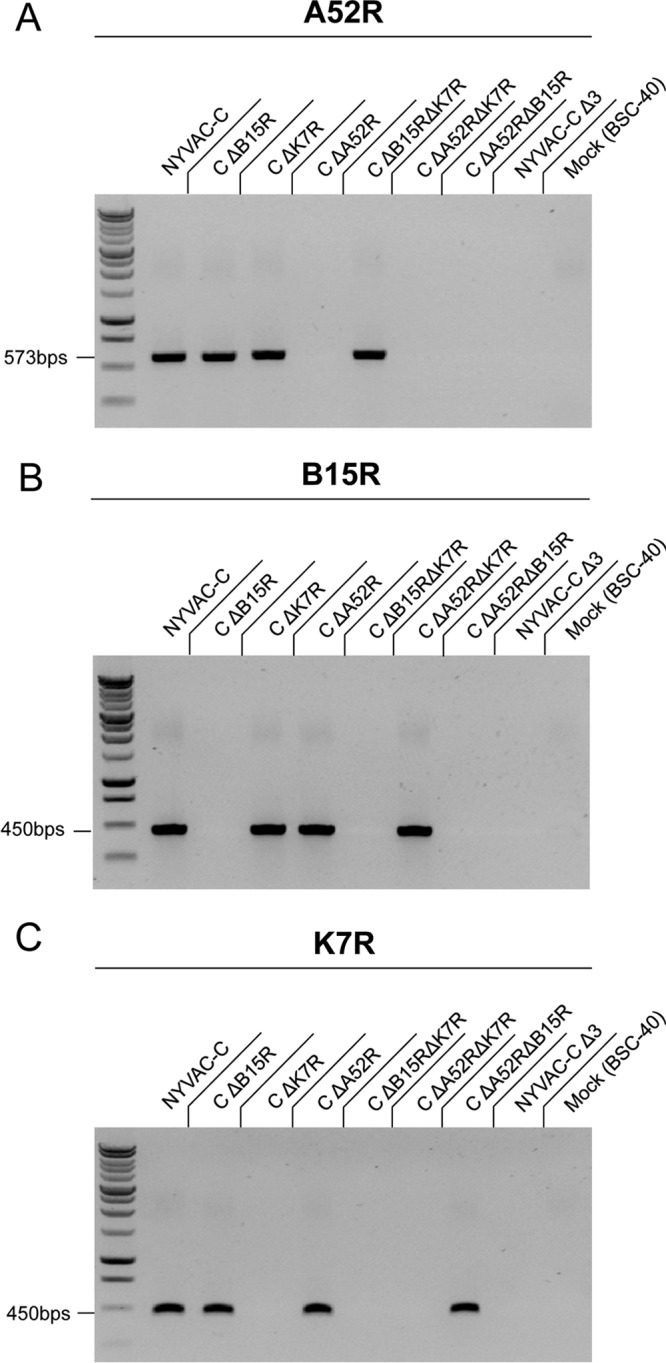
Lack of transcription of the A52R, B15R, and K7R genes in NYVAC deletion mutant-infected cells. BSC-40 cells were infected with NYVAC-C and NYVAC deletion mutant viruses (1 PFU per cell, 24 h). RT-PCR analysis of infected cells confirms the deletion of the A52R (A), B15R (B), and K7R (C) genes by loss of RNA expression.
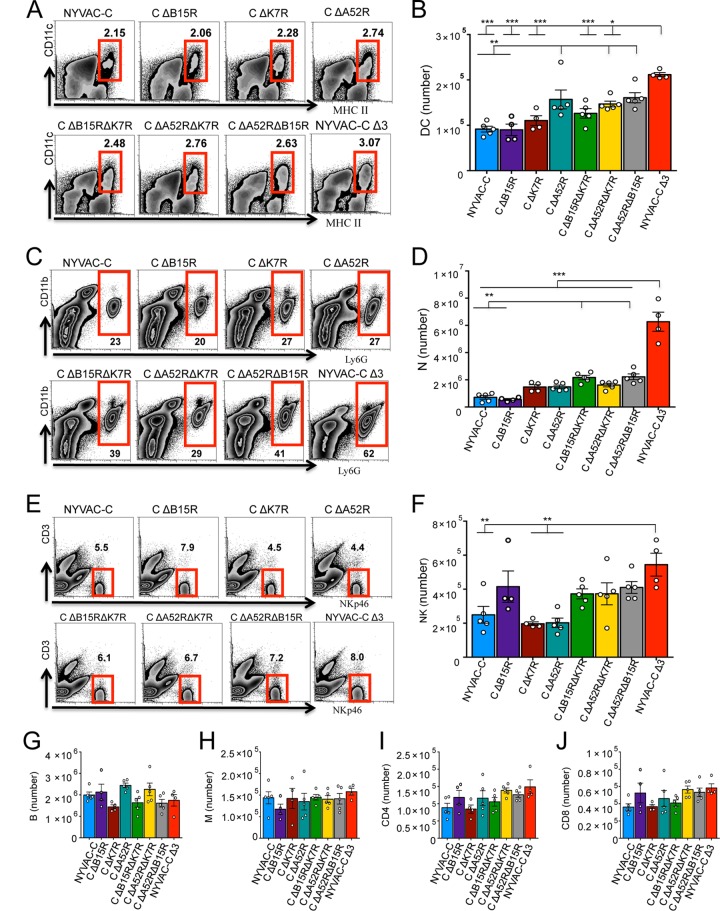
Peritoneal exudate cells (PECs) were collected 6 h postinfection to study the innate immune response. Mice infected with NYVAC-C ΔA52R, NYVAC-C ΔA52R ΔK7R, or NYVAC-C ΔA52R ΔB15R showed an increase in the percentages of CD11c+ major histocompatibility complex class II (MHC-II)+ DCs compared to NYVAC-C-infected mice (Fig. 2A). Absolute DC numbers in these groups were significantly enhanced (Fig. 2B). There were no differences in DC migration between mice that received K7R or B15R single or combined deletion and mice that received the parental virus (Fig. 2B). Mice injected with NYVAC-C Δ3 showed a significant increase in DC numbers compared to those that received the NYVAC-C ΔA52R ΔK7R or NYVAC-C ΔB15R ΔK7R double-deletion mutants (Fig. 2B).
FIG 2.
Deletion of the A52R, B15R, and K7R genes influences peritoneal cell migration. Shown are the percentages and absolute numbers of CD11c+ MHC-II+ dendritic cells (DCs) (A and B), Ly6G+ CD11b+ neutrophils (N) (C and D), NKp46+ CD3− natural killer cells (NK) (E and F), CD19+ B cells (B) (G), F4/80low CD11blow monocytes (M) (H), CD4+ CD3+ T cells (CD4) (I), and CD8+ CD3+ T cells (CD8) (J) in the peritoneal cavity of BALB/c mice at 6 h postinjection of 107 PFU of NYVAC-C or the NYVAC-C deletion mutants. Graphs show the mean ± standard error of the mean (SEM); each point represents an individual mouse. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Compared to the parental and single-deletion mutants, NYVAC-C ΔA52R ΔB15R and NYVAC-C ΔB15R ΔK7R showed an increase in the percentages of neutrophils in the peritoneal cavity (Fig. 2C). NYVAC-C Δ3 further enhanced neutrophil migration (Fig. 2C) and significantly increased absolute neutrophil numbers compared to all other groups (Fig. 2D).
The percentages and absolute numbers of NK cells in NYVAC-C Δ3-injected mice were also higher than in NYVAC-C-, NYVAC-C ΔA52R- or NYVAC-C ΔK7R-injected mice (Fig. 2E and F). There were no significant differences between single-deletion mutants and NYVAC-C, between double- and single-deletion mutants, or between triple- and double-deletion mutants in total numbers of monocytes or B cells or CD4 and CD8 T cells (Fig. 2G to J).
These data indicate that A52R single gene deletion partially influences the innate immune response, as indicated by DC migration. Double gene deletion had a clear effect on DC and neutrophil migration, and concomitant A52R, B15R, and K7R deletion increased DC, neutrophil, and to some extent, NK cell migration.
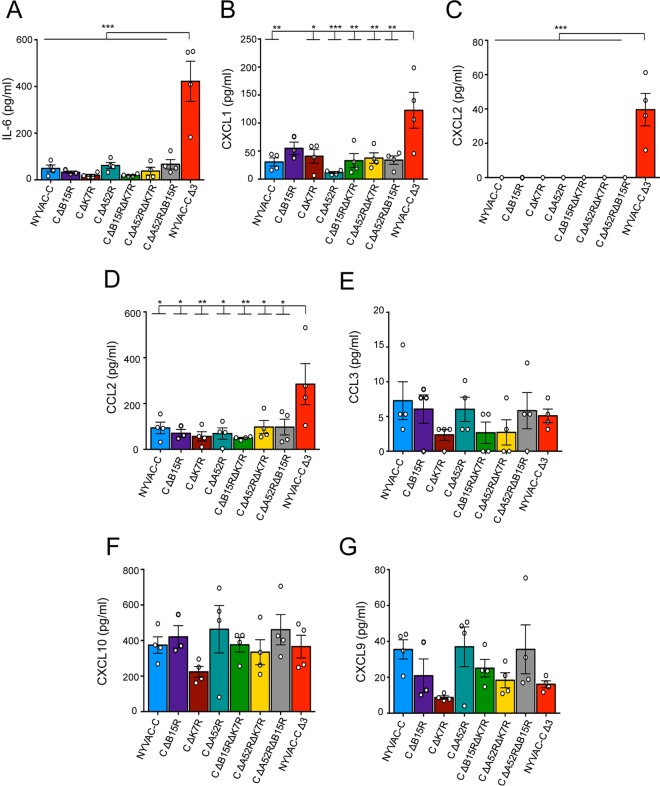
To determine whether this altered innate immune response correlates with a change in cytokine/chemokine secretion at the infection site, we analyzed peritoneal exudates (PEs) of 6-h NYVAC-C- and NYVAC-C deletion mutant-infected mice. IL-6, CXCL1 (KC), CXCL2 (MIP-2), CXCL9 (MIG), CXCL10 (IP-10), CCL2 (MCP-1), CCL3/4 (MIP-1α/β), and CCL5 (RANTES) cytokines/chemokines are essential for recruitment of several cell types (20, 26). The proinflammatory cytokine IL-6 and the chemokines CXCL1 and CXCL2, which mainly induce neutrophil migration (27, 28), were significantly overexpressed in PEs of NYVAC-C Δ3-injected mice compared to those of mice injected with the other deletion mutants (Fig. 3A to C). CCL3 is involved in DC migration (29), whereas CXCL9, CXCL10, and to some extent CCL2 induce NK cell migration (30, 31). While CCL2 increased significantly after i.p. injection of NYVAC-C Δ3 compared to the other viruses (Fig. 3D), there was no notable difference in expression of CCL3, CXCL10, and CXCL9 in these groups (Fig. 3E to G).
FIG 3.
Deletion of the A52R, B15R, and K7R genes affects cytokine/chemokine levels. Shown are concentrations of cytokine IL-6 (A) and chemokines CXCL1 (B), CXCL2 (C), CCL2 (D), CCL3 (E), CXCL10 (F), and CXCL9 (G) at 6 h postinfection in the peritoneal cavity of NYVAC-C- or NYVAC-C deletion mutant-injected mice. Graphs show the mean ± SEM; each point represents an individual mouse. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The data suggest that infection by the triple-deletion mutant induced a chemokine pattern that influences mainly neutrophil migration and partially affects DC and NK cell migration. The significant cytokine/chemokine increase in NYVAC-C Δ3-injected mice correlated with enhanced neutrophil migration in these mice.
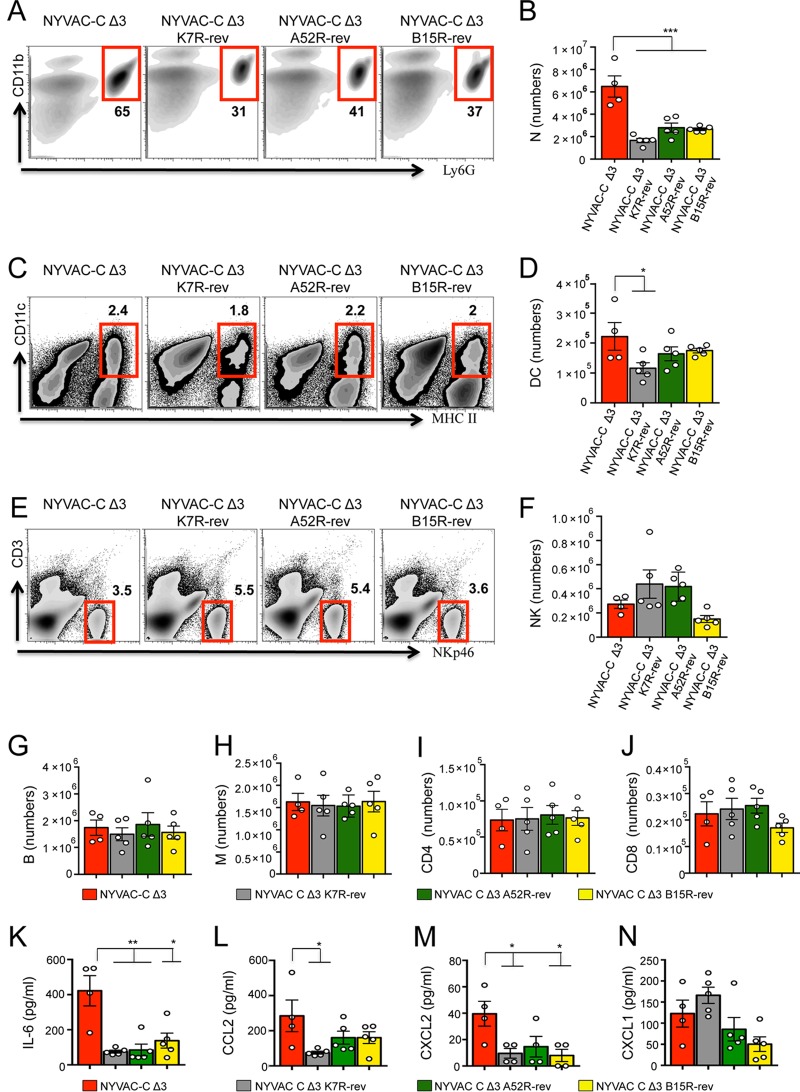
To define the distinct roles of these immunomodulatory genes in inducing innate immune responses, we used NYVAC-C Δ3 K7R-rev virus (18) and generated revertant NYVAC-C Δ3 A52R-rev and NYVAC-C Δ3 B15R-rev viruses, in which the B15R and A52R genes were reinserted at the hemagglutinin locus (HA); expression was confirmed by RT-PCR (Fig. 4A and B). The percentage of neutrophils was lower in the peritoneal cavities of all revertant virus-injected mice compared to triple-deletion mutant-infected mice (Fig. 5A). Absolute neutrophil numbers in revertant virus-infected mice were significantly lower than those in mice injected with NYVAC-C Δ3 (Fig. 5B). DC percentages were similar for all virus-injected mice (Fig. 5C), and absolute DC numbers were only significantly lower between NYVAC-C Δ3 K7R-rev- and triple-deletion mutant-infected mice (Fig. 5D). The percentages (Fig. 5E) and absolute numbers (Fig. 5F) of NK cells showed no differences between triple-deletion mutant- and revertant virus-treated mice. Again, no differences were found in absolute numbers of B cells (Fig. 5G), monocytes (Fig. 5H), or CD4 (Fig. 5I) or CD8 T cells (Fig. 5J) among the virus-injected mice. Peritoneal concentrations of IL-6, CCL2, and CXCL2 (Fig. 5K to M) were significantly lower after infection with revertant viruses compared to NYVAC-C Δ3. We found no marked differences in CXCL1 levels between NYVAC-C Δ3- and revertant virus-injected mice (Fig. 5N). These data show a correlation between chemokine levels and neutrophil migration and indicate that these VACV proteins have a central role in inhibiting innate immune responses.
FIG 4.
Characterization of the NYVAC-C-Δ3 revertant viruses. RT-PCR analysis of RNA from BSC-40 cells infected with the NYVAC-C-Δ3 B15R-rev and NYVAC-C-Δ3 A52R-rev revertants (Rev) (1 PFU per cell, 24 h) confirms the correct B15R (A) and A52R (B) gene reinsertion and transcription.
FIG 5.
Revertant viruses reduce cell migration and cytokine/chemokine levels. Shown are the percentages and absolute numbers of Ly6G+ CD11b+ neutrophils (N) (A and B), CD11c+ MHCII+ DCs (C and D), NKp46+ CD3− NK cells (E and F), CD19+ B cells (B) (G), F4/80low CD11blow monocytes (M) (H), CD4+ CD3+ T cells (CD4) (I) and CD8+ CD3+ T cells (CD8) (J) in the peritoneal cavity of BALB/c mice at 6 h postinjection of 107 PFU of NYVAC-C triple-deletion mutants and revertant viruses. (K to N) Concentrations of IL-6 (K), CCL2 (L), CXCL2 (M), and CXCL1 (N) in the peritoneal cavity of infected mice. Data are representative of two independent experiments. Graphs show the mean ± SEM; each point represents an individual mouse. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Deletion of NF-κB inhibitory proteins enhances HIV-specific T cell and humoral responses.
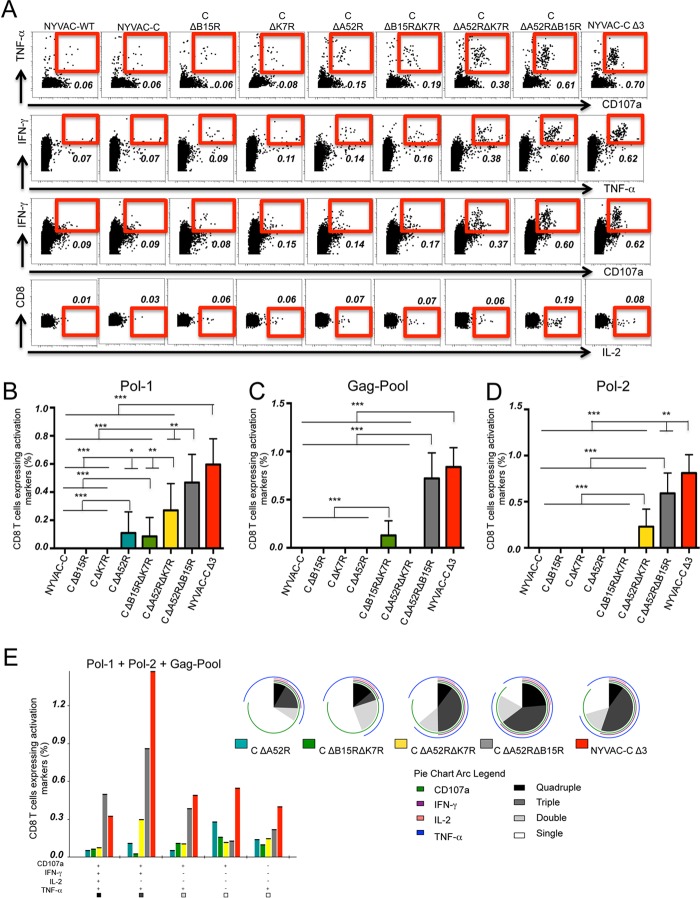
To study the ability of NYVAC-C deletion mutants to induce HIV-specific adaptive cellular responses, we tested a heterologous DNA prime/poxvirus boost immunization regimen in BALB/c mice, a protocol widely used to enhance immune responses to HIV antigens (18, 25). At 11 days after the last immunization, splenocytes from NYVAC-C- or NYVAC-C deletion mutant-infected mice were stimulated with HIV-1 Pol-1, Pol-2, or Gag-Pool (representing 60 peptides of Gag-1 plus 61 peptides of Gag-2) to study HIV-1-specific CD8 T cell responses (32). The magnitude of CD8 T cell responses was measured as the expression of IL-2 and/or IFN-γ and/or TNF-α and/or CD107a activation markers. NYVAC-C Δ3 and NYVAC-C ΔA52R ΔB15R induced the largest number of TNF-α+ CD107a+, IFN-γ+ TNF-α+, or IFN-γ+ CD107a+ double-positive cells (Fig. 6A). Only a small percentage of CD8 T cells expressed IL-2 (Fig. 6A). Of the single-deletion mutants, only NYVAC-C ΔA52R significantly enhanced adaptive Pol-1 CD8 T cell responses compared to the parental NYVAC-C virus (Fig. 6B). Compared to the single-deletion mutants, NYVAC-C ΔB15R ΔK7R and NYVAC-C ΔA52R ΔK7R significantly increased Gag-Pool and Pol-2 responses, respectively (Fig. 6C and D). NYVAC-C Δ3 and NYVAC-C ΔA52R ΔB15R elicited the strongest CD8 T cell responses to the three HIV antigens (Fig. 6B to D). Only in the case of Pol-2, did the triple-deletion mutant significantly increase CD8 T cell responses compared to NYVAC-C ΔA52R ΔB15R (Fig. 6D). The quality of the Gag and Pol responses, defined as cytokine production and cytotoxic potential, showed that the NYVAC-C ΔA52R ΔB15R and NYVAC-C ΔA52R ΔK7R double-deletion mutants and NYVAC-C Δ3 triple-deletion mutant induced a marked increase in the cytotoxic T lymphocyte (CTL) polyfunctional profile compared to the single-deletion mutant NYVAC-C ΔA52R (Fig. 6E; pie charts). The CD8 T cell subset that produced IFN-γ, TNF-α, and CD107a was the most representative population induced with HIV peptides (Fig. 6E; bar graph). This CD8 T cell subset marked the difference in the CTL polyfunctional profiles among the virus-infected mice. These results indicate that the double- and triple-deletion mutants enhanced the magnitude and polyfunctional profile of specific CD8 T cells to HIV-1 Gag and Pol intracellular antigens.
FIG 6.
Deletion of the A52R, B15R, and K7R genes influences adaptive HIV-specific CD8 T cell immune responses. Shown are the vaccine-induced HIV-specific CD8 T cell response in mice (n = 4/group) immunized following a heterologous DNA prime/virus boost regimen (107 PFU of NYVAC-C or NYVAC-C deletion mutants). (A) Percentages of TNF-α+ CD107a+, IFN-γ+ TNF-α+, IFN-γ+ CD107a+ double-positive and IL-2 single-positive T cells. The total value (magnitude) is the sum of percentages of CD8 T cells that express IFN-γ and/or TNF-α and/or IL-2 and/or CD107a, measured by intracellular cytokine staining (ICS). Nonspecific responses of mice infected with control NYVAC-WT were subtracted from the total magnitude. (B to D) Magnitude of Pol-1 (B)-, Gag-Pool (C)-, or Pol-2 (D)-specific CD8 T cell responses. Graphs show the mean ± confidence interval (CI). (E) Functional profile of adaptive Gag-Pol-specific CD8 T cells. Combinations of responses (x axis) and percentages of functionally distinct cell subsets (y axis) are shown in the bar graph. Responses are grouped and color coded based on the number of functions. Pie chart colors indicate the percentage of cytokine-producing cells based on the number of functions (inside) and the different activation markers (outside). Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine whether the altered innate immune response correlates with changes in HIV-specific adaptive humoral responses, we measured Env-specific IgG antibodies in the sera of mice immunized as described above. Compared to the parental virus, NYVAC-C ΔA52R was the only single-deletion mutant that significantly increased Env-specific total IgG and IgG1 antibody titers (Fig. 7A and B). Double-deletion mutant-infected mice showed significantly higher Env-specific total IgG, IgG1, and IgG3 antibody levels compared to those infected with NYVAC-C and NYVAC-C single-deletion mutants (Fig. 7A to C). NYVAC-C ΔA52R ΔB15R significantly increased Env-specific total IgG, IgG1, and IgG3 responses compared to the other double-deletion mutants (Fig. 7A to C). Mice infected with NYVAC-C ΔA52R ΔB15R and NYVAC-C Δ3 showed similar Env-specific IgG1 and IgG3 antibody titers (Fig. 7B and C). Compared to NYVAC-C ΔA52R ΔB15R, the triple-deletion mutant only significantly increased the total IgG response (Fig. 7A).
FIG 7.
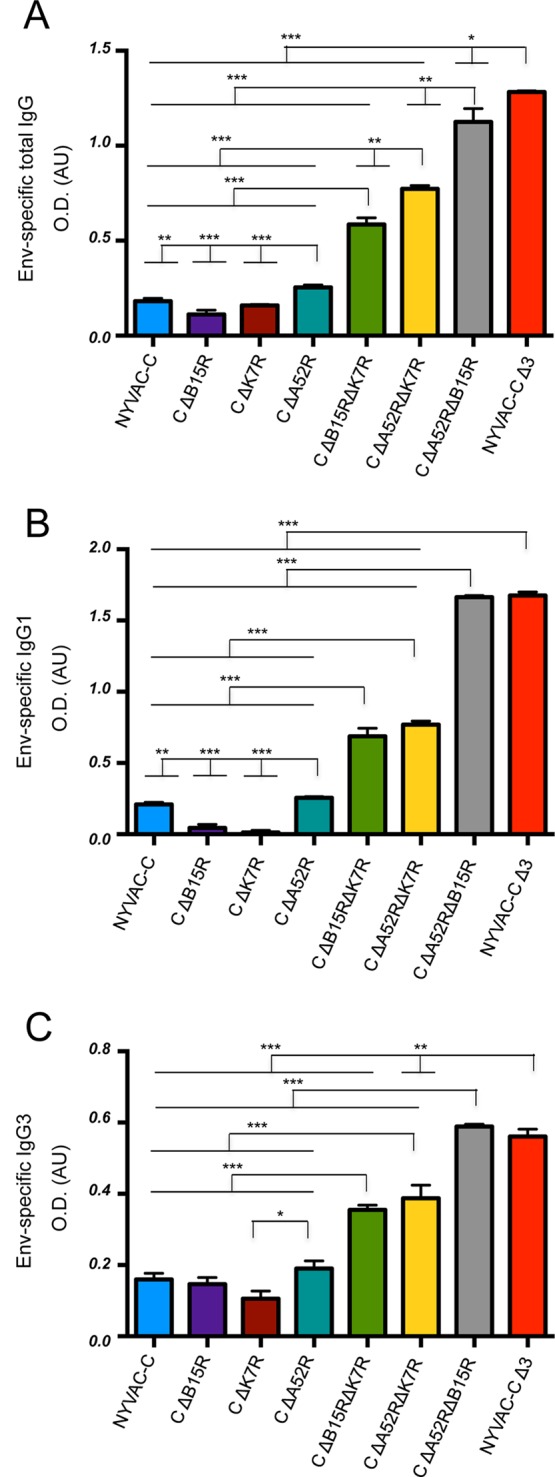
Deletion of the A52R, B15R, and K7R genes affects humoral Env-specific IgG immune responses. Shown are vaccine-induced HIV-specific IgG responses in mice (n = 4/group) immunized following a heterologous DNA prime/virus boost regimen (107 PFU of NYVAC-C or NYVAC-C deletion mutants). (A to C) Levels of Env-specific total IgG (A), IgG1 (B), and IgG3 (C) antibodies in serum of mice (n = 4/group) 11 days postinfection with NYVAC-C or NYVAC-C deletion mutants. Data are represented as optical density (OD) in arbitrary units (AU). Nonspecific responses of mice infected with control NYVAC-WT were subtracted from total values. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
These data show that A52R single-gene deletion partially influences HIV-specific cell-mediated and humoral immune responses, represented by Pol-1 CD8 T cell responses and by Env-specific total IgG and IgG1 levels. Of the double-deletion mutants, NYVAC-C ΔA52R ΔB15R induced the highest HIV-specific CD8 T cell and IgG responses. Additional deletion of K7R in NYVAC-C ΔA52R ΔB15R had a further positive effect on Pol-2- and Env-specific total IgG immune responses.
DISCUSSION
A52, K7, and B15 vaccinia virus proteins, members of the B cell lymphoma-2 family (Bcl-2) (33, 34), have an important NF-κB inhibitory role (18) and thus induce suppression and evasion of host immune responses (14). During infection, A52 and K7 interact with TRAF6 and inhibit its kinase activity (15, 16), whereas B15 binds the IKK complex to inhibit IκBα phosphorylation and degradation (17). Using single-, double- and triple-deletion mutants, we showed that combined deletion of the A52R, K7R, and B15R genes efficiently triggers innate immune responses. With the generation of revertants NYVAC-C Δ3 K7R-rev, NYVAC-C Δ3 A52R-rev, and NYVAC-C Δ3 B15R-rev, we also determined that one of these inhibitory molecules is enough to reduce chemokine release and cell migration to infection sites. A46 and C49 are other NYVAC NF-κB inhibitors (35, 36) whose contribution to induction of innate immune responses might be better determined by removing these genes from the backbone of the NYVAC triple-deletion mutant, followed by testing under conditions of strong NF-κB activation and cell migration.
Effective antigen presentation by DCs is NF-κB dependent (37), and direct DC stimulation of CD8/CD4 T and B cells induces cytokine and antibody secretion (38, 39). We show that A52 is partially involved in DC evasion and suggest that DCs recruited after infection are involved in HIV-specific CD8 T cell and IgG responses. Neutrophils cross-prime naive T cells and induce adaptive immune responses (40, 41). A52R, B15R, and K7R gene deletion enhances chemokines that can directly induce neutrophil migration, and thus HIV-specific CD8 T cell and IgG responses, which supports neutrophil mediation of HIV immune responses (18). NK cells also mediate adaptive immune responses (42), and A52, B15, and K7 together are needed to reduce NK cell migration after NYVAC infection. Ablation of their genes is sufficient to increase HIV-specific CD8 T cells and IgG responses, suggesting that NK cells also mediate these antigen-specific immune responses. Our results indicate that the A52, B15, and K7 VACV proteins influence immune responses differently (Table 1), which suggests new approaches to more efficient poxvirus vaccine design.
TABLE 1.
Immune response levels induced by A52R, B15R, and K7R deletion mutants
| Virus mutation(s) | Immune responsea |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| DCs | NK cells | Neutrophils | CD8 cells |
Env-specific IgG |
|||||
| Pol-1 | Pol-2 | Gag | Total IgG | IgG1 | IgG3 | ||||
| ΔB15R | |||||||||
| ΔK7R | |||||||||
| ΔA52R | ++ | +++ | ++ | ++ | |||||
| ΔB15R ΔK7R | ++ | +++ | +++ | +++ | +++ | +++ | |||
| ΔA52R ΔK7R | ++ | ++++ | +++ | ++++ | +++ | +++ | |||
| ΔA52R ΔB15R | ++ | ++ | +++++ | ++++ | +++++ | +++++ | +++++ | ++++ | |
| ΔA52R ΔB15R ΔK7R | +++ | ++ | +++ | +++++ | +++++ | +++++ | ++++++ | +++++ | ++++ |
+ indicates a significant increase in innate, adaptive cell-mediated, or humoral immune responses of NYVAC-C deletion mutants compared to NYVAC-C.
Attenuated VACV vectors (NYVAC and MVA) are used as vaccine candidates for emerging infectious diseases and cancer in humans (43). Whereas distinct VACV vaccine approaches for HIV have yielded promising results in primates (44, 45), their effectiveness was limited in a human clinical trial (2), which prompted improvement of poxvirus vectors as HIV vaccine candidates. Immunomodulatory viral gene deletion is a strategy for VACV vectors to improve immunogenicity to HIV (46). Combined or single deletion of the A52R, B15R, and K7R genes might thus be relevant for the generation of new HIV-VACV vaccines that specifically modify innate immune responses and thus cellular or humoral adaptive immune responses.
The abilities of double-deletion and NYVAC-C Δ3 triple-deletion mutants to generate robust polyfunctional Gag- and Pol-specific CD8 T cell responses and Env-specific IgG antibodies are important immune properties for future vaccine applications. In the RV144 phase III clinical trial, effective vaccination correlated with humoral responses to Env, specifically human antibodies to the V1/V2 loops of HIV-1 gp120 (47). Here we determined that the combined deletion of the A52R, B15R, and K7R genes increased Env-specific total IgG, IgG1, and IgG3 responses compared to single deletions, which suggests that VACV vectors with NF-κB inhibitory protein ablation are encouraging therapeutic strategies.
Human HIV nonprogressor individuals preferentially maintain highly functional HIV-specific CD8 T cells (48). Gag and Pol, the most conserved HIV-1 proteins (49), can shift the CD8 T cell response from the variable Env epitope in the first years of HIV-1 infection (50). In chronically HIV-1-infected persons, a Gag CD8 T cell response correlates with lower HIV titers (51) and with decreased viremia in HIV-1-infected patients with suspension of antiretroviral therapy (50). A prophylactic vaccine that induces a Gag T cell response controls simian immunodeficiency virus (SIV) infection (52). Since the Gag/Pol-specific CD8 T cell response in double- and triple-deletion mutant-infected mice is robust and mainly polyfunctional compared to that in single-deletion mutants, these vectors could show promise for prophylactic and therapeutic treatment.
In summary, here we define the immune biological functions of three NF-κB inhibitors encoded by NYVAC and show that the vaccinia virus NYVAC deletion mutant lacking the A52R, K7R, and B15R genes is the most effective vector to trigger immunologically relevant markers of innate and adaptive immune responses. Based on these immunomodulatory properties, these vectors can be used for new HIV vaccine designs.
MATERIALS AND METHODS
Mice and injections.
BALB/c mice (6 to 8 weeks old) were purchased from Harlan. In the DNA prime immunization protocol, mice received 100 μg DNA-C (50 μg pcDNA-CN54GP120 plus 50 μg pcDNA-CN54GPN) or 100 μg sham DNA-ϕ (100 μg pcDNA) by the intramuscular route. We purified plasmids using the Endofree plasmid megakit (Qiagen). After 2 weeks, mice were immunized intraperitoneally (i.p.) with 107 PFU of wild-type NYVAC (NYVAC-WT), NYVAC-C, and deletion mutants. Animal studies were approved by the Ethical Committee of Animal Experimentation (CEEA) of the Centro Nacional de Biotecnologia (CNB, Madrid, Spain) in accordance with national and international guidelines and the Royal Decree (RD 1201/2005) (permit no. 13013).
Cells.
African green monkey kidney cells (BSC-40; American Type Culture Collection) and primary chicken embryo fibroblasts (CEFs [Intervet, Salamanca, Spain]) in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) were used to grow the viruses.
Viruses.
NYVAC-WT and NYVAC-C (Sanofi Pasteur), the NYVAC-C deletion mutants, and NYVAC-C Δ3 K7R-rev have been described previously (18, 25). NYVAC-C Δ3 A52R-rev and NYVAC-C Δ3 B15R-rev were constructed using the A52R and B15R VACV genes and the β-glucuronidase gene as a reporter gene. For the generation of revertant viruses, we used the virus hemagglutinin (HA) flanking regions to insert K7R, B15R, and A52R genes. BSC-40 cells were infected with 0.01 PFU/cell of NYVAC-C Δ3 (1 h) and transfected with VACV insertional plasmid vector pCAR-2/A52R or pCAR-2/B15R to insert the genes at the HA locus. Recombinant viruses were selected from progeny virus by consecutive rounds of plaque purification, as reported previously (53). All viruses were purified through two 36% (wt/vol) sucrose cushions, and their titers were determined by immunostaining plaque assay in BSC-40 cells (54).
Plasmids.
The plasmid transfer vectors pCAR-2/A52R and pCAR-2/B15R were obtained by cloning the A52R and B15R sequence into the plasmid pCAR-2 (patent no. WO2007132052 A1 [55]). The NYVAC genome was used as the template to amplify the left flank, the repeated left flank, and the right flank of the genes. The following oligonucleotides were used: for A52R, forward, CGCGGATCCATGGACATAAAGATAG, and reverse, AAGGAAAAAAGCGGCCGCCTATGACATTTCCAC; and for B15R, forward, CTAGGCCTGGTACCCGGGGATGACAGCCAACTTTAGTACCC, and reverse, GGCCGCTCTAGAACTAGTGTCAATTCATACGCCGGAATATG. The resulting plasmids were confirmed by DNA sequence analysis.
PCR and RT-PCR.
Viral DNA was extracted by the SDS-proteinase K-phenol method. We extracted total RNA using the RNeasy minikit (Qiagen) with RNase-free DNase I recombinant. First-strand cDNA was synthesized with oligo(dT)12–18 primers using SuperScript III reverse transcriptase (both from Invitrogen), and cDNA was used for PCR amplification.
Cytokine/chemokine analysis.
Peritoneal washes from phosphate-buffered saline (PBS)- or virus-injected mice were used to detect cytokines and chemokines in multiplex analysis using LuminexXMAP technology.
Peptides.
The HIV-1 peptides Pol-1 (LVGPTPVNI) and Pol-2 (YYDPSKDLI) (32) were provided by the CNB-CSIC Proteomics Service. The HIV-1 Gag-Pool peptides (representing 60 peptides of Gag-1 plus 61 peptides of Gag-2) (25) were provided by the EuroVacc Foundation.
Flow cytometry.
For intracellular cytokine staining (ICS), splenocytes were resuspended in RPMI 1640 with 10% FCS and 1 μg/ml Golgiplug (BD), monensin (eBioscience), and anti-CD107a (1D4B; BD), restimulated with peptides (6 h, 37°C, 5% CO2), stained for surface markers with anti-CD3 (145-2C11), anti-CD4 (GK1.5), and anti-CD8 (53-6.7) (all from BD), fixed, permeabilized (Cytofix/Cytoperm kit; BD), and stained intracellularly with anti-IL-2 (JES6-5H4), anti-IFN-γ (XMG 1.2), and anti-TNF-α (MP6-X722) (all from BD). Peritoneal exudate cells (PECs) were stained with anti-Ly6G (1A8), anti-CD3 (145-2C11), anti-CD11b (M1/70), anti-CD19 (1D3), anti-MHC class II (1-A/1-E; 2G9), anti-CD4 (GK1.5), and anti-CD8 (53-6.7) (all from BD), anti-CD45 (30-F11; Biolegend), anti-CD11c (N418) and anti-F4/80 (BM8; both from eBioscience), and anti-NKp46 (29A1.4; BioLegend). Dead cells were stained using the violet LIVE/DEAD stain kit (Invitrogen). Cells were acquired using a Gallios (Beckman Coulter) flow cytometer, and analyses were performed with FlowJo software v.8.5.3 (Tree Star). Boolean combinations of single functional gates were used to determine the frequency of each response based on all combinations of activation markers.
Antibody measurement.
Env-binding antibodies were measured by enzyme-linked immunosorbent assay (ELISA) (25). Serum samples from mice were reacted in plates coated with 2 μg/ml recombinant CN54 gp120 purified protein (ARP683, HIV-1 CN54 gp120 clade C; EU Programme EVA).
Statistical analysis.
For statistical analysis of CD8 T cell responses to HIV-1 antigens, we used an approach to correct measurements for medium response and allow calculation of confidence intervals and P values of hypothesis tests (53). Only antigen response values significantly larger than the corresponding RPMI values are shown. Background levels (splenocytes in RPMI) were subtracted from all values used. For distribution analysis and presentation, we used SPICE version 5.1 (http://exon.niaid.nih.gov). For statistical analysis of cell migration and cytokine/chemokine expression, one-way analysis of variance (ANOVA) with Tukey's honestly significant difference (HSD) post hoc tests was applied. Student's t test was used for antibody analysis to establish the differences between two groups.
ACKNOWLEDGMENTS
We are grateful to Victoria Jiménez and Cristina Sánchez Corzo for tissue culture and Catherine Mark for editorial assistance.
Mauro Di Pilato received a Formación en Investigación en Salud PhD fellowship and Ernesto Mejías-Pérez a Formación del Profesorado Universitario PhD fellowship, from the Spanish Ministries of Health and Education, respectively. This research was supported by Spanish SAF-2013-45232R-Mineco-FEDER and AIDS Research Network RD16/0025/0014-ISCIII-FEDER, and by the PTVDC Program with support from the Bill & Melinda Gates Foundation. The authors declare they have no conflicting financial interests.
REFERENCES
- 1.Garcia-Arriaza J, Esteban M. 2014. Enhancing poxvirus vectors vaccine immunogenicity. Hum Vaccin Immunother 10:2235–2244. doi: 10.4161/hv.28974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 3.Harari A, Rozot V, Cavassini M, Bellutti Enders F, Vigano S, Tapia G, Castro E, Burnet S, Lange J, Moog C, Garin D, Costagliola D, Autran B, Pantaleo G, Bart PA. 2012. NYVAC immunization induces polyfunctional HIV-specific T-cell responses in chronically-infected, ART-treated HIV patients. Eur J Immunol 42:3038–3048. doi: 10.1002/eji.201242696. [DOI] [PubMed] [Google Scholar]
- 4.Bart PA, Huang Y, Karuna ST, Chappuis S, Gaillard J, Kochar N, Shen X, Allen MA, Ding S, Hural J, Liao HX, Haynes BF, Graham BS, Gilbert PB, McElrath MJ, Montefiori DC, Tomaras GD, Pantaleo G, Frahm N. 2014. HIV-specific humoral responses benefit from stronger prime in phase Ib clinical trial. J Clin Invest 124:4843–4856. doi: 10.1172/JCI75894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mooij P, Koopman G, Drijfhout JW, Nieuwenhuis IG, Beenhakker N, Koestler J, Bogers WM, Wagner R, Esteban M, Pantaleo G, Heeney JL, Jacobs BL, Melief CJ. 2015. Synthetic long peptide booster immunization in rhesus macaques primed with replication competent NYVAC-C-KC induces a balanced CD4/CD8 T-cell and antibody response against the conserved regions of HIV-1. J Gen Virol 96:1478–1483. doi: 10.1099/vir.0.000074. [DOI] [PubMed] [Google Scholar]
- 6.Flynn BJ, Kastenmuller K, Wille-Reece U, Tomaras GD, Alam M, Lindsay RW, Salazar AM, Perdiguero B, Gomez CE, Wagner R, Esteban M, Park CG, Trumpfheller C, Keler T, Pantaleo G, Steinman RM, Seder R. 2011. Immunization with HIV Gag targeted to dendritic cells followed by recombinant New York vaccinia virus induces robust T-cell immunity in nonhuman primates. Proc Natl Acad Sci U S A 108:7131–7136. doi: 10.1073/pnas.1103869108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harari A, Bart PA, Stohr W, Tapia G, Garcia M, Medjitna-Rais E, Burnet S, Cellerai C, Erlwein O, Barber T, Moog C, Liljestrom P, Wagner R, Wolf H, Kraehenbuhl JP, Esteban M, Heeney J, Frachette MJ, Tartaglia J, McCormack S, Babiker A, Weber J, Pantaleo G. 2008. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J Exp Med 205:63–77. doi: 10.1084/jem.20071331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Arriaza J, Gomez CE, Sorzano CO, Esteban M. 2014. Deletion of the vaccinia virus N2L gene encoding an inhibitor of IRF3 improves the immunogenicity of modified vaccinia virus Ankara expressing HIV-1 antigens. J Virol 88:3392–3410. doi: 10.1128/JVI.02723-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Arriaza J, Najera JL, Gomez CE, Tewabe N, Sorzano CO, Calandra T, Roger T, Esteban M. 2011. A candidate HIV/AIDS vaccine (MVA-B) lacking vaccinia virus gene C6L enhances memory HIV-1-specific T-cell responses. PLoS One 6:e24244. doi: 10.1371/journal.pone.0024244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perdiguero B, Gomez CE, Di Pilato M, Sorzano CO, Delaloye J, Roger T, Calandra T, Pantaleo G, Esteban M. 2013. Deletion of the vaccinia virus gene A46R, encoding for an inhibitor of TLR signalling, is an effective approach to enhance the immunogenicity in mice of the HIV/AIDS vaccine candidate NYVAC-C. PLoS One 8:e74831. doi: 10.1371/journal.pone.0074831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaloye J, Roger T, Steiner-Tardivel QG, Le Roy D, Knaup Reymond M, Akira S, Petrilli V, Gomez CE, Perdiguero B, Tschopp J, Pantaleo G, Esteban M, Calandra T. 2009. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog 5:e1000480. doi: 10.1371/journal.ppat.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Hutchens MA, Luker KE, Sonstein J, Nunez G, Curtis JL, Luker GD. 2008. Protective effect of Toll-like receptor 4 in pulmonary vaccinia infection. PLoS Pathog 4:e1000153. doi: 10.1371/journal.ppat.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araki H, Katayama N, Yamashita Y, Mano H, Fujieda A, Usui E, Mitani H, Ohishi K, Nishii K, Masuya M, Minami N, Nobori T, Shiku H. 2004. Reprogramming of human postmitotic neutrophils into macrophages by growth factors. Blood 103:2973–2980. doi: 10.1182/blood-2003-08-2742. [DOI] [PubMed] [Google Scholar]
- 14.Smith GL, Benfield CT, Maluquer de Motes C, Mazzon M, Ember SW, Ferguson BJ, Sumner RP. 2013. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol 94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 15.Schroder M, Baran M, Bowie AG. 2008. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J 27:2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harte MT, Haga IR, Maloney G, Gray P, Reading PC, Bartlett NW, Smith GL, Bowie A, O'Neill LA. 2003. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J Exp Med 197:343–351. doi: 10.1084/jem.20021652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen RA, Ryzhakov G, Cooray S, Randow F, Smith GL. 2008. Inhibition of IkappaB kinase by vaccinia virus virulence factor B14. PLoS Pathog 4:e22. doi: 10.1371/journal.ppat.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Pilato M, Mejias-Perez E, Zonca M, Perdiguero B, Gomez CE, Trakala M, Nieto J, Najera JL, Sorzano CO, Combadiere C, Pantaleo G, Planelles L, Esteban M. 2015. NFkappaB activation by modified vaccinia virus as a novel strategy to enhance neutrophil migration and HIV-specific T-cell responses. Proc Natl Acad Sci U S A 112:E1333–E1342. doi: 10.1073/pnas.1424341112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medzhitov R. 2007. Recognition of microorganisms and activation of the immune response. Nature 449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 20.Luster AD. 2002. The role of chemokines in linking innate and adaptive immunity. Curr Opin Immunol 14:129–135. doi: 10.1016/S0952-7915(01)00308-9. [DOI] [PubMed] [Google Scholar]
- 21.Teigler JE, Phogat S, Franchini G, Hirsch VM, Michael NL, Barouch DH. 2014. The canarypox virus vector ALVAC induces distinct cytokine responses compared to the vaccinia virus-based vectors MVA and NYVAC in rhesus monkeys. J Virol 88:1809–1814. doi: 10.1128/JVI.02386-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beauchamp NM, Yammani RD, Alexander-Miller MA. 2012. CD8 marks a subpopulation of lung-derived dendritic cells with differential responsiveness to viral infection and Toll-like receptor stimulation. J Virol 86:10640–10650. doi: 10.1128/JVI.01413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duffy D, Perrin H, Abadie V, Benhabiles N, Boissonnas A, Liard C, Descours B, Reboulleau D, Bonduelle O, Verrier B, Van Rooijen N, Combadiere C, Combadiere B. 2012. Neutrophils transport antigen from the dermis to the bone marrow, initiating a source of memory CD8+ T cells. Immunity 37:917–929. doi: 10.1016/j.immuni.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 24.Lee SC, Srivastava RM, Lopez-Albaitero A, Ferrone S, Ferris RL. 2011. Natural killer (NK): dendritic cell (DC) cross talk induced by therapeutic monoclonal antibody triggers tumor antigen-specific T cell immunity. Immunol Res 50:248–254. doi: 10.1007/s12026-011-8231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez CE, Najera JL, Jimenez V, Bieler K, Wild J, Kostic L, Heidari S, Chen M, Frachette MJ, Pantaleo G, Wolf H, Liljestrom P, Wagner R, Esteban M. 2007. Generation and immunogenicity of novel HIV/AIDS vaccine candidates targeting HIV-1 Env/Gag-Pol-Nef antigens of clade C. Vaccine 25:1969–1992. doi: 10.1016/j.vaccine.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 26.De Filippo K, Henderson RB, Laschinger M, Hogg N. 2008. Neutrophil chemokines KC and macrophage-inflammatory protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J Immunol 180:4308–4315. doi: 10.4049/jimmunol.180.6.4308. [DOI] [PubMed] [Google Scholar]
- 27.Ritzman AM, Hughes-Hanks JM, Blaho VA, Wax LE, Mitchell WJ, Brown CR. 2010. The chemokine receptor CXCR2 ligand KC (CXCL1) mediates neutrophil recruitment and is critical for development of experimental Lyme arthritis and carditis. Infect Immun 78:4593–4600. doi: 10.1128/IAI.00798-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramos CD, Fernandes KS, Canetti C, Teixeira MM, Silva JS, Cunha FQ. 2006. Neutrophil recruitment in immunized mice depends on MIP-2 inducing the sequential release of MIP-1alpha, TNF-alpha and LTB(4). Eur J Immunol 36:2025–2034. doi: 10.1002/eji.200636057. [DOI] [PubMed] [Google Scholar]
- 29.Caux C, Ait-Yahia S, Chemin K, de Bouteiller O, Dieu-Nosjean MC, Homey B, Massacrier C, Vanbervliet B, Zlotnik A, Vicari A. 2000. Dendritic cell biology and regulation of dendritic cell trafficking by chemokines. Springer Semin Immunopathol 22:345–369. doi: 10.1007/s002810000053. [DOI] [PubMed] [Google Scholar]
- 30.Thapa M, Welner RS, Pelayo R, Carr DJ. 2008. CXCL9 and CXCL10 expression are critical for control of genital herpes simplex virus type 2 infection through mobilization of HSV-specific CTL and NK cells to the nervous system. J Immunol 180:1098–1106. doi: 10.4049/jimmunol.180.2.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allavena P, Bianchi G, Zhou D, van Damme J, Jilek P, Sozzani S, Mantovani A. 1994. Induction of natural killer cell migration by monocyte chemotactic protein-1, -2 and -3. Eur J Immunol 24:3233–3236. doi: 10.1002/eji.1830241249. [DOI] [PubMed] [Google Scholar]
- 32.Wild J, Bieler K, Kostler J, Frachette MJ, Jeffs S, Vieira S, Esteban M, Liljestrom P, Pantaleo G, Wolf H, Wagner R. 2009. Preclinical evaluation of the immunogenicity of C-type HIV-1-based DNA and NYVAC vaccines in the Balb/C mouse model. Viral Immunol 22:309–319. doi: 10.1089/vim.2009.0038. [DOI] [PubMed] [Google Scholar]
- 33.Graham SC, Bahar MW, Cooray S, Chen RA, Whalen DM, Abrescia NG, Alderton D, Owens RJ, Stuart DI, Smith GL, Grimes JM. 2008. Vaccinia virus proteins A52 and B14 share a Bcl-2-like fold but have evolved to inhibit NF-kappaB rather than apoptosis. PLoS Pathog 4:e1000128. doi: 10.1371/journal.ppat.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalverda AP, Thompson GS, Vogel A, Schroder M, Bowie AG, Khan AR, Homans SW. 2009. Poxvirus K7 protein adopts a Bcl-2 fold: biochemical mapping of its interactions with human DEAD box RNA helicase DDX3. J Mol Biol 385:843–853. doi: 10.1016/j.jmb.2008.09.048. [DOI] [PubMed] [Google Scholar]
- 35.Stack J, Haga IR, Schroder M, Bartlett NW, Maloney G, Reading PC, Fitzgerald KA, Smith GL, Bowie AG. 2005. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J Exp Med 201:1007–1018. doi: 10.1084/jem.20041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansur DS, Maluquer de Motes C, Unterholzner L, Sumner RP, Ferguson BJ, Ren H, Strnadova P, Bowie AG, Smith GL. 2013. Poxvirus targeting of E3 ligase beta-TrCP by molecular mimicry: a mechanism to inhibit NF-kappaB activation and promote immune evasion and virulence. PLoS Pathog 9:e1003183. doi: 10.1371/journal.ppat.1003183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimura S, Bondeson J, Foxwell BM, Brennan FM, Feldmann M. 2001. Effective antigen presentation by dendritic cells is NF-kappaB dependent: coordinate regulation of MHC, co-stimulatory molecules and cytokines. Int Immunol 13:675–683. doi: 10.1093/intimm/13.5.675. [DOI] [PubMed] [Google Scholar]
- 38.Hasegawa K, Noguchi Y, Koizumi F, Uenaka A, Tanaka M, Shimono M, Nakamura H, Shiku H, Gnjatic S, Murphy R, Hiramatsu Y, Old LJ, Nakayama E. 2006. In vitro stimulation of CD8 and CD4 T cells by dendritic cells loaded with a complex of cholesterol-bearing hydrophobized pullulan and NY-ESO-1 protein: identification of a new HLA-DR15-binding CD4 T-cell epitope. Clin Cancer Res 12:1921–1927. doi: 10.1158/1078-0432.CCR-05-1900. [DOI] [PubMed] [Google Scholar]
- 39.Wan S, Zhou Z, Duan B, Morel L. 2008. Direct B cell stimulation by dendritic cells in a mouse model of lupus. Arthritis Rheum 58:1741–1750. doi: 10.1002/art.23515. [DOI] [PubMed] [Google Scholar]
- 40.Chtanova T, Schaeffer M, Han SJ, van Dooren GG, Nollmann M, Herzmark P, Chan SW, Satija H, Camfield K, Aaron H, Striepen B, Robey EA. 2008. Dynamics of neutrophil migration in lymph nodes during infection. Immunity 29:487–496. doi: 10.1016/j.immuni.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beauvillain C, Delneste Y, Scotet M, Peres A, Gascan H, Guermonprez P, Barnaba V, Jeannin P. 2007. Neutrophils efficiently cross-prime naive T cells in vivo. Blood 110:2965–2973. doi: 10.1182/blood-2006-12-063826. [DOI] [PubMed] [Google Scholar]
- 42.Paust S, Senman B, von Andrian UH. 2010. Adaptive immune responses mediated by natural killer cells. Immunol Rev 235:286–296. doi: 10.1111/j.0105-2896.2010.00906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez CE, Najera JL, Krupa M, Perdiguero B, Esteban M. 2011. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr Gene Ther 11:189–217. doi: 10.2174/156652311795684731. [DOI] [PubMed] [Google Scholar]
- 44.Mooij P, Balla-Jhagjhoorsingh SS, Koopman G, Beenhakker N, van Haaften P, Baak I, Nieuwenhuis IG, Kondova I, Wagner R, Wolf H, Gomez CE, Najera JL, Jimenez V, Esteban M, Heeney JL. 2008. Differential CD4+ versus CD8+ T-cell responses elicited by different poxvirus-based human immunodeficiency virus type 1 vaccine candidates provide comparable efficacies in primates. J Virol 82:2975–2988. doi: 10.1128/JVI.02216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, Liu J, Abbink P, Maxfield LF, Seaman MS, Dugast AS, Alter G, Ferguson M, Li W, Earl PL, Moss B, Giorgi EE, Szinger JJ, Eller LA, Billings EA, Rao M, Tovanabutra S, Sanders-Buell E, Weijtens M, Pau MG, Schuitemaker H, Robb ML, Kim JH, Korber BT, Michael NL. 2013. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell 155:531–539. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. 2012. Poxvirus vectors as HIV/AIDS vaccines in humans. Hum Vaccin Immunother 8:1192–1207. doi: 10.4161/hv.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goulder PJ, Watkins DI. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol 8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang OO, Daar ES, Ng HL, Shih R, Jamieson BD. 2011. Increasing CTL targeting of conserved sequences during early HIV-1 infection is correlated to decreasing viremia. AIDS Res Hum Retroviruses 27:391–398. doi: 10.1089/aid.2010.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 52.Iwamoto N, Takahashi N, Seki S, Nomura T, Yamamoto H, Inoue M, Shu T, Naruse TK, Kimura A, Matano T. 2014. Control of simian immunodeficiency virus replication by vaccine-induced Gag- and Vif-specific CD8+ T cells. J Virol 88:425–433. doi: 10.1128/JVI.02634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Najera JL, Gomez CE, Garcia-Arriaza J, Sorzano CO, Esteban M. 2010. Insertion of vaccinia virus C7L host range gene into NYVAC-B genome potentiates immune responses against HIV-1 antigens. PLoS One 5:e11406. doi: 10.1371/journal.pone.0011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramirez JC, Gherardi MM, Esteban M. 2000. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J Virol 74:923–933. doi: 10.1128/JVI.74.2.923-933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Najera GJL, Gomez RCE, Rodriguez ME. November 2007. Vectors into which the c71 gene is inserted and use of said vectors in the production of vaccines and of compositions for gene therapy.European patent WO2007132052 A1.