ABSTRACT
The bacteriophage ϕ29 infects Gram-positive Bacillus subtilis with a short noncontractile tail. Recent studies showed that the ϕ29 tail protein gp9 forms a hexameric tube with six long loops of membrane-active peptides blocking in the tube at the distal end of the tail. The long loops exit on genome release and form a membrane pore for passage of the genome. The membrane penetration mechanism of the ϕ29 tail might be common among tailed bacteriophages.
KEYWORDS: membrane penetration, bacteriophages, cryoEM, crystal structure, gp9, membrane-active peptide, ϕ29, short noncontractile tail, tail knob
INTRODUCTION
The goal of viral infection is the delivery of the viral genome into the host cell and the subsequent replication. Cells are well protected by the cell membrane, a major barrier that all viruses must conquer to accomplish genome delivery. Eukaryotic viruses must penetrate only the cell membrane, whereas prokaryotic viruses must penetrate a complex cell wall. Although viruses deploy various strategies to infect host cells, they may have evolved to breach common membrane barriers by using similar mechanisms.
MEMBRANE PENETRATION BY EUKARYOTIC VIRUSES
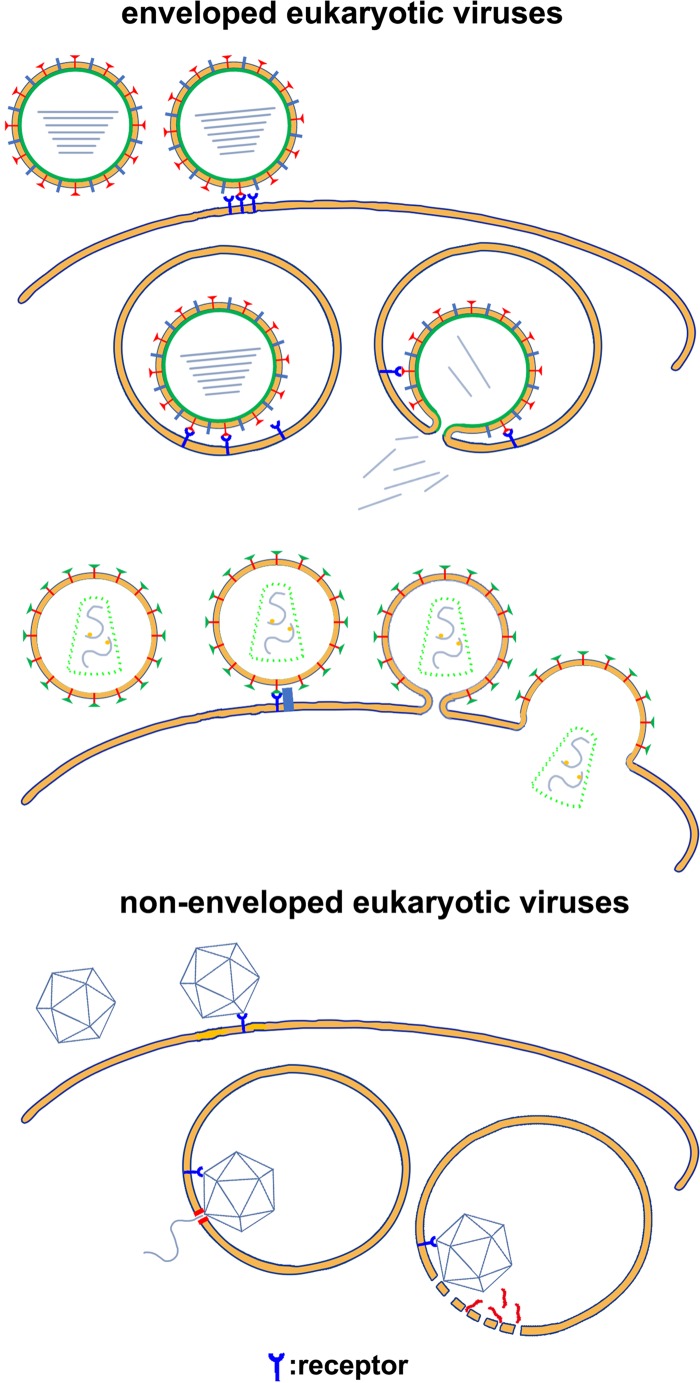
Although the membrane penetration mechanisms of most eukaryotic viruses have been roughly established, the finer details of the molecular processes involved remain largely unknown. The genetic material of enveloped eukaryotic viruses is enclosed by a lipid membrane, which is decorated or covered with envelope proteins (1). Eukaryotic viral envelope proteins contain a hydrophobic peptide called the fusion peptide, which is often part of a polypeptide that is anchored to the viral membrane by its distal end (1, 2). Enveloped eukaryotic viruses recognize host cells through specific interactions between viral envelope proteins and host cell surface receptors. For some viruses, envelope protein-receptor interactions trigger conformational changes in the envelope proteins that directly expose the fusion peptide and lead to fusion of the viral and cellular membranes (3, 4) (Fig. 1). For many other enveloped eukaryotic viruses, the interactions trigger endocytosis and the engulfment of virions. Typically, a change in the pH of late endosomes triggers conformational changes in the viral envelope protein that expose the fusion peptide, and subsequent conformational changes of the envelope proteins lead to membrane fusion (5) (Fig. 1). Proteolysis of the envelope proteins is frequently required in both pH-dependent and pH-independent entry of enveloped eukaryotic viruses (2, 5, 6). On exposure to a hydrophilic environment, the hydrophobic fusion peptide inserts into the nearby cellular membrane and anchors the viral envelope protein onto the cellular membrane. Further conformational changes of the envelope protein shift the distal end that is anchored to the viral membrane close to the fusion peptide, which brings the cellular membrane and viral membrane within close proximity for fusion. After fusion, a fusion pore is formed, allowing the release of the viral genetic material into the cytoplasm of the host cell (2, 5, 6). Most nonenveloped eukaryotic viruses enter host cells through receptor-mediated endocytosis (7, 8). Unlike entry of enveloped eukaryotic viruses, membrane fusion is not involved in the entry of nonenveloped eukaryotic viruses. Studies on the entry of several nonenveloped eukaryotic viruses show that viral capsids contain membrane-active peptides that are either exposed or cleaved from the capsid protein in late endosomes (7, 9, 10). To release the viral genome, which is encapsulated by the viral capsid during capsid assembly, membrane-active peptides disrupt the cellular membrane locally or form a pore in the cellular membrane (Fig. 1).
FIG 1.
Schematic diagrams showing host cell entry by eukaryotic viruses. The membrane bilayers are represented by thick yellow lines. Membrane-active peptides and proteins are red.
MEMBRANE PENETRATION BY PROKARYOTIC VIRUSES
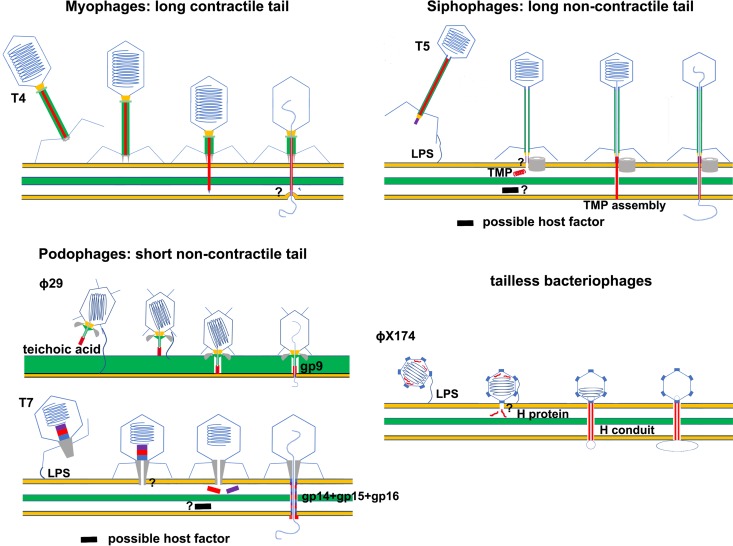
The mechanisms of membrane penetration by prokaryotic viruses are less clear than those of penetration by eukaryotic viruses. Most prokaryotic viruses are tailed viruses that consist of a capsid head attached to a tail, with the viral genome encapsulated in the head. Compared with eukaryotic viruses, prokaryotic viruses (bacteriophages) use quite different mechanisms for genome delivery. Bacteriophages inject their genetic materials into the cytoplasm of the host cell, leaving an empty viral capsid shell on the host cell surface. In contrast to eukaryotic viruses, bacteriophage genomes are not encapsulated during capsid assembly. Instead, an empty capsid is assembled first, and then the viral genome is packaged into the empty capsid via a genome-packaging motor. Viral genome packaging is accompanied by accumulating pressure in the capsid, which can be up to 60 atmospheres for some bacteriophages and is the major force driving the injection of the phage genome into host cells during infection (11–13). Bacterial cells are protected by a peptidoglycan cell wall and one or two membranes. Gram-negative bacteria are surrounded by an outer membrane and an inner membrane. Located between the two membranes are the periplasmic space and a thin peptidoglycan layer. Gram-positive bacteria do not have an outer membrane, but they do have a thick peptidoglycan layer that can reach a thickness of ∼50 nm. Macromolecules with a molecular weight of >50 kDa cannot pass through the highly cross-linked peptidoglycan layer of bacterial cells (14). Bacteriophage tails are responsible for host cell recognition; host cell wall and membrane penetration; signal transmission, which triggers genome injection; and channel formation, which facilitates genome injection (15). Bacteriophage tails are complex molecular machines. Without considering other aspects of the bacteriophage life cycle, bacteriophages can be classified by their tail morphology into three groups: myophages with long contractile tails, siphophages with long noncontractile tails, and podophages with short noncontractile tails (16) (Fig. 2). Phage tails use various strategies for infection.
FIG 2.
Schematic diagrams showing host cell entry by prokaryotic viruses. The membrane bilayers are represented by thick yellow lines. The peptidoglycan layers of the bacterial cells are represented by thick green lines or green layers. Membrane-active peptides and proteins are red. The question marks indicate unknown host factors or unclear membrane penetration mechanisms. TMP, tape measure protein; LPS, lipopolysaccharide.
PROKARYOTIC VIRUSES WITH LONG CONTRACTILE TAILS
The bacteriophage T4, which infects Gram-negative Escherichia coli cells with its long contractile tail, is a representative myophage. A layer of sheath proteins surrounds the long T4 tail tube through which the genome is ejected. The sheath proteins are arranged in helical arrays with one end attached to the terminator complex and the other end attached to the baseplate at the distal end of the tail (17). On binding to the cell surface receptors, the baseplate undergoes a hexagon-to-star shape transition, and a contraction signal is transferred from the tail baseplate to the sheath, triggering contraction of the sheath (18, 19). The sheath contracts to approximately half of the original length, and the mechanical force generated by this sheath contraction drives the inner tail tube to penetrate the outer membrane of the host cell (19). It has been proposed that the tail tip spike complex falls off the tail and supports penetration of the tail tube through the peptidoglycan layer by digesting peptidoglycans with the lysozyme-like tip spike complex component gp5 (20). Further contraction of the sheath propels the tube to pierce the inner membrane. However, recent cryoelectron tomography (cryoET) studies of T4-infected E. coli minicells showed that the tail tube does not directly penetrate the cytoplasmic membrane and that the distal end of the tail tube is fused with the inner membrane (21). These cryoET results suggest alternative mechanisms for breaching the inner membrane, most likely through a membrane pore structure (Fig. 2). There is no clear evidence to indicate which T4 or host protein is involved in membrane pore formation. The adaptor protein gp27, which is located at the distal end of the T4 tail tube, and the ejected tape measure protein (TMP) gp29 were hypothesized to be the pore-forming proteins (21). It is also possible that a host membrane pore protein is involved in inner membrane penetration. Further investigation is required to reveal these mechanistic details. Infection of Gram-positive bacteria by phages with long contractile tails has not been well characterized. It would be interesting to determine whether sheath contraction can directly drive tail tube penetration of the inner membrane of Gram-positive host cells.
PROKARYOTIC VIRUSES WITH LONG NONCONTRACTILE TAILS
Long noncontractile tails are characterized by a long noncontractile tail tube that is blocked at the distal end by a tail tip complex (TTC). The TTC is the essential element for receptor recognition and genome release. Typically, interactions between the TTC and a membrane protein receptor directly trigger the release of siphophage genomes (22–24). For siphophages that infect Gram-negative hosts, such as the bacteriophages λ, T5, and HK97, an injectable tail TMP is required to penetrate the membrane and bridge membrane bilayers (25–29) (Fig. 2). Studies on bacteriophage λ indicate that the TMP (gpH) becomes protease resistant after it irreversibly binds to a liposome (30). Similarly, the TMP (pb2) of bacteriophage T5 was shown to be able to associate with liposome membranes (31, 32). It was hypothesized that TMPs assemble to form a conduit that bridges the outer and inner membranes, providing a channel for genome release (33). Host cell factors might be involved in assembling such a conduit and in mediating membrane penetration, as shown in studies of bacteriophage HK97 (28). The TMP contains a glycine-rich hydrophobic peptide fragment that has been suggested to form a pore in the inner membrane for genome transport. However, it was also proposed that the TMP may bridge the two membranes by interacting with host membrane channel proteins (28). Similarly, it has been suggested that siphophages that infect Gram-positive hosts use host membrane channels for genome transport over the membrane. However, only a few such membrane channels have been identified and characterized, and their role in siphophage genome delivery remains controversial (34).
PROKARYOTIC VIRUSES WITH SHORT NONCONTRACTILE TAILS AND PROKARYOTIC VIRUSES WITH NO TAIL
Based on recent crystal and cryoET structural studies, mechanisms similar to those used by siphophages have been proposed for the tailless phage ϕχ174 and the podophage T7 that infect Gram-negative hosts (Fig. 2) (35, 36). The encapsulated viral H protein of the bacteriophage ϕχ174 forms a 170-Å-long conduit. The length of the conduit is sufficient to bridge the periplasmic space, and the inner diameter of the conduit is wide enough to permit the circular single-stranded DNA genome transport (36). The N terminus of the H protein contains a glycine-rich hydrophobic sequence, which is not visible in the structure but has been suggested to form a membrane pore. The podophage T7 has a short tail that is not long enough to span the cell wall and membranes. CryoET structural studies of the bacteriophage T7 suggested that a conduit structure is assembled in the periplasmic space by the encapsulated viral core proteins gp14, gp15, and gp16, which are ejected from the head after breaching the outer membrane (35). The mechanisms of outer membrane penetration for these viruses remain elusive. Mechanisms have been proposed that involve viral tail pore-forming proteins or host membrane proteins. However, firm experimental data supporting either are still lacking.
Bacteriophage ϕ29 infects Gram-positive Bacillus subtilis cells and is one of the most-studied podophages. The short noncontractile tail of bacteriophage ϕ29 is ∼300 Å long, which is just long enough to penetrate the host cell wall and membrane. The ϕ29 tail comprises a thin tail tube with an enlarged distal end called a tail knob. Twelve appendages (or tail spikes) extend from the head-tail junction. The function of the tail appendages is to recognize the host cell and anchor the viral particle to the surface of the host cell (37, 38). The tail knob is a protein assembly of gp9 and the peptidoglycan-degrading enzyme gp13 (39). Our recent study on tail knob protein gp9 showed that six gp9 molecules form a tube structure that is blocked by a long loop of gp9 (40). Because in vitro genome-release studies have shown that the tail knob is part of the genome-release channel, the long loops must exit from the tube on genome release. Sequence analysis of the loop found that it contains a glycine-rich hydrophobic peptide that is similar to that of the HIV fusion peptide. Additional cryoEM structural studies of genome-emptied virions showed that the long loops exit upon genome release and form a cone-shaped structure at the distal end of the tail. Taken together, these data strongly suggest that the long loop of gp9 may be involved in membrane penetration. To test this hypothesis, an in vitro system was established using liposomes and ϕ29 virions. In this system, genome release can be triggered by using a low pH and ammonium sulfate, and membrane penetration can be directly observed to determine whether the hydrophobic long loops assemble into a cone-shaped structure that functions as a membrane pore. As expected, the ϕ29 tail penetrated the liposome membrane and injected the viral genome into the liposome. CryoEM studies of the ϕ29 virions on the liposomes further showed that the cone-shaped structure, which was assembled from the released long loops, inserts into the lipid bilayer and functions as a membrane pore for genome release (Fig. 2). The reconstituted in vitro system mimics in vivo membrane penetration by the ϕ29 tail. However, artificial conditions, such as a low pH and ammonium sulfate, were used to trigger genome release in the reconstituted system. Several aspects of the genome injection process still need to be defined, especially those regarding the location and functional mechanism of gp13 and the in vivo signaling that triggers genome release. These studies on the bacteriophage ϕ29 tail, perhaps for the first time, establish a nearly complete mechanism for the penetration of a host membrane by a tailed phage at the molecular level.
PERSPECTIVES AND CHALLENGES
Membrane-active peptides are generally used by eukaryotic viruses. As shown in the accumulated data, they likely exist in prokaryotic viral proteins as well. The basic function of these peptides is membrane insertion. It is not a complete surprise that these membrane-active peptides share common features, such as being rich in hydrophobic and glycine residues (41). These common features imply the possible convergent evolution of eukaryotic and prokaryotic viruses in response to a similar barrier, the cell membrane. Although the bacteriophage C1 contains a tail protein, gp12, that assembles to form a similar hexameric tube structure, the sequence similarity between ϕ29 gp9 and C1 gp12 is low (42). Based on structure and sequence information, a similar long loop can also be mapped on the C1 gp12 sequence. However, the sequence of the C1 long loop is quite different from that of the ϕ29 gp9. It may be coincidental that the ϕ29 tail knob loop is similar to that of the HIV fusion loop. Because of the sequence diversity of these membrane-active peptides, predicting their presence in viral protein sequences is difficult. The pore-forming mechanism for membrane penetration is likely more widely used by prokaryotic viruses than expected. However, the accurate prediction of these membrane-active peptides and verification of their function in vitro and in vivo remain a challenge.
ACKNOWLEDGMENTS
We apologize to all those colleagues whose excellent work could not be discussed due to space limitations.
This work was supported by funds from the Ministry of Science and Technology of China (grant numbers 2016YFA0501100 and 2015CB910102), the National Natural Science Foundation of China (31470721), the Junior Thousand Talents Program of China, and the Beijing Advanced Innovation Center for Structure Biology (to Y.X.).
REFERENCES
- 1.Podbilewicz B. 2014. Virus and cell fusion mechanisms. Annu Rev Cell Dev Biol 30:111–139. doi: 10.1146/annurev-cellbio-101512-122422. [DOI] [PubMed] [Google Scholar]
- 2.Harrison SC. 2015. Viral membrane fusion. Virology 479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melikyan GB. 2014. HIV entry: a game of hide-and-fuse? Curr Opin Virol 4:1–7. doi: 10.1016/j.coviro.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilen CB, Tilton JC, Doms RW. 2012. Molecular mechanisms of HIV entry. Adv Exp Med Biol 726:223–242. doi: 10.1007/978-1-4614-0980-9_10. [DOI] [PubMed] [Google Scholar]
- 5.White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol 43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morizono K, Chen IS. 2011. Receptors and tropisms of envelope viruses. Curr Opin Virol 1:13–18. doi: 10.1016/j.coviro.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JE, Vogt PK. 2010. Cell entry by non-enveloped viruses. Curr Top Microbiol Immunol 343:v–vii. [PubMed] [Google Scholar]
- 8.Wiethoff CM, Nemerow GR. 2015. Adenovirus membrane penetration: tickling the tail of a sleeping dragon. Virology 479–480:591–599. doi: 10.1016/j.virol.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee M, Johnson JE. 2008. Activation, exposure and penetration of virally encoded, membrane-active polypeptides during non-enveloped virus entry. Curr Protein Pept Sci 9:16–27. doi: 10.2174/138920308783565732. [DOI] [PubMed] [Google Scholar]
- 10.Odegard A, Banerjee M, Johnson JE. 2010. Flock house virus: a model system for understanding non-enveloped virus entry and membrane penetration. Curr Top Microbiol Immunol 343:1–22. doi: 10.1007/82_2010_35. [DOI] [PubMed] [Google Scholar]
- 11.Molineux IJ, Panja D. 2013. Popping the cork: mechanisms of phage genome ejection. Nat Rev Microbiol 11:194–204. doi: 10.1038/nrmicro2988. [DOI] [PubMed] [Google Scholar]
- 12.Rickgauer JP, Fuller DN, Grimes S, Jardine PJ, Anderson DL, Smith DE. 2008. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage phi29. Biophys J 94:159–167. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. 2001. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 14.Demchick P, Koch AL. 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J Bacteriol 178:768–773. doi: 10.1128/jb.178.3.768-773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leiman PG, Shneider MM. 2012. Contractile tail machines of bacteriophages. Adv Exp Med Biol 726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 16.Ackermann HW. 2003. Bacteriophage observations and evolution. Res Microbiol 154:245–251. doi: 10.1016/S0923-2508(03)00067-6. [DOI] [PubMed] [Google Scholar]
- 17.Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. 2010. Morphogenesis of the T4 tail and tail fibers. Virol J 7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crowther RA, Lenk EV, Kikuchi Y, King J. 1977. Molecular reorganization in the hexagon to star transition of the baseplate of bacteriophage T4. J Mol Biol 116:489–523. doi: 10.1016/0022-2836(77)90081-X. [DOI] [PubMed] [Google Scholar]
- 19.Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. 2004. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 20.Kao SH, McClain WH. 1980. Baseplate protein of bacteriophage-T4 with both structural and lytic functions. J Virol 34:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu B, Margolin W, Molineux IJ, Liu J. 2015. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci U S A 112:E4919–E4928. doi: 10.1073/pnas.1501064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sao-Jose C, Lhuillier S, Lurz R, Melki R, Lepault J, Santos MA, Tavares P. 2006. The ectodomain of the viral receptor YueB forms a fiber that triggers ejection of bacteriophage SPP1 DNA. J Biol Chem 281:11464–11470. doi: 10.1074/jbc.M513625200. [DOI] [PubMed] [Google Scholar]
- 23.Plisson C, White HE, Auzat I, Zafarani A, Sao-Jose C, Lhuillier S, Tavares P, Orlova EV. 2007. Structure of bacteriophage SPP1 tail reveals trigger for DNA ejection. EMBO J 26:3720–3728. doi: 10.1038/sj.emboj.7601786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zivanovic Y, Confalonieri F, Ponchon L, Lurz R, Chami M, Flayhan A, Renouard M, Huet A, Decottignies P, Davidson AR, Breyton C, Boulanger P. 2014. Insights into bacteriophage T5 structure from analysis of its morphogenesis genes and protein components. J Virol 88:1162–1174. doi: 10.1128/JVI.02262-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreiseikelmann B. 1994. Translocation of DNA across bacterial membranes. Microbiol Rev 58:293–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scandella D, Arber W. 1976. Phage lambda DNA injection into Escherichia coli pel− mutants is restored by mutations in phage gene V or gene H. Virology 69:206–215. doi: 10.1016/0042-6822(76)90207-5. [DOI] [PubMed] [Google Scholar]
- 27.Guihard G, Boulanger P, Letellier L. 1992. Involvement of phage T5 tail proteins and contact sites between the outer and inner membrane of Escherichia coli in phage T5 DNA injection. J Biol Chem 267:3173–3178. [PubMed] [Google Scholar]
- 28.Cumby N, Reimer K, Mengin-Lecreulx D, Davidson AR, Maxwell KL. 2015. The phage tail tape measure protein, an inner membrane protein and a periplasmic chaperone play connected roles in the genome injection process of E. coli phage HK97. Mol Microbiol 96:437–447. doi: 10.1111/mmi.12918. [DOI] [PubMed] [Google Scholar]
- 29.Davidson AR, Cardarelli L, Pell LG, Radford DR, Maxwell KL. 2012. Long noncontractile tail machines of bacteriophages. Adv Exp Med Biol 726:115–142. doi: 10.1007/978-1-4614-0980-9_6. [DOI] [PubMed] [Google Scholar]
- 30.Roessner CA, Ihler GM. 1984. Proteinase sensitivity of bacteriophage lambda tail protein gpJ and protein pH* in complexes with the lambda receptor. J Bacteriol 157:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boulanger P, Jacquot P, Plancon L, Chami M, Engel A, Parquet C, Herbeuval C, Letellier L. 2008. Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J Biol Chem 283:13556–13564. doi: 10.1074/jbc.M800052200. [DOI] [PubMed] [Google Scholar]
- 32.Feucht A, Schmid A, Benz R, Schwarz H, Heller KJ. 1990. Pore formation associated with the tail-tip protein pb2 of bacteriophage T5. J Biol Chem 265:18561–18567. [PubMed] [Google Scholar]
- 33.Bohm J, Lambert O, Frangakis AS, Letellier L, Baumeister W, Rigaud JL. 2001. FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr Biol 11:1168–1175. doi: 10.1016/S0960-9822(01)00349-9. [DOI] [PubMed] [Google Scholar]
- 34.Vinga I, Baptista C, Auzat I, Petipas I, Lurz R, Tavares P, Santos MA, Sao-Jose C. 2012. Role of bacteriophage SPP1 tail spike protein gp21 on host cell receptor binding and trigger of phage DNA ejection. Mol Microbiol 83:289–303. doi: 10.1111/j.1365-2958.2011.07931.x. [DOI] [PubMed] [Google Scholar]
- 35.Hu B, Margolin W, Molineux IJ, Liu J. 2013. The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science 339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Young LN, Zhang XZ, Boudko SP, Fokine A, Zbornik E, Roznowski AP, Molineux IJ, Rossmann MG, Fane BA. 2014. Icosahedral bacteriophage phi X174 forms a tail for DNA transport during infection. Nature 505:432–435. doi: 10.1038/nature12816. [DOI] [PubMed] [Google Scholar]
- 37.Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG. 2006. Structural changes of bacteriophage phi29 upon DNA packaging and release. EMBO J 25:5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang Y, Leiman PG, Li L, Grimes S, Anderson DL, Rossmann MG. 2009. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol Cell 34:375–386. doi: 10.1016/j.molcel.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang Y, Morais MC, Cohen DN, Bowman VD, Anderson DL, Rossmann MG. 2008. Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage phi29 tail. Proc Natl Acad Sci U S A 105:9552–9557. doi: 10.1073/pnas.0803787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J, Gui M, Wang D, Xiang Y. 2016. The bacteriophage phi29 tail possesses a pore-forming loop for cell membrane penetration. Nature 534:544–547. doi: 10.1038/nature18017. [DOI] [PubMed] [Google Scholar]
- 41.Del Angel VD, Dupuis F, Mornon JP, Callebaut I. 2002. Viral fusion peptides and identification of membrane-interacting segments. Biochem Biophys Res Commun 293:1153–1160. doi: 10.1016/S0006-291X(02)00353-4. [DOI] [PubMed] [Google Scholar]
- 42.Aksyuk AA, Bowman VD, Kaufmann B, Fields C, Klose T, Holdaway HA, Fischetti VA, Rossmann MG. 2012. Structural investigations of a Podoviridae streptococcus phage C1, implications for the mechanism of viral entry. Proc Natl Acad Sci U S A 109:14001–14006. doi: 10.1073/pnas.1207730109. [DOI] [PMC free article] [PubMed] [Google Scholar]