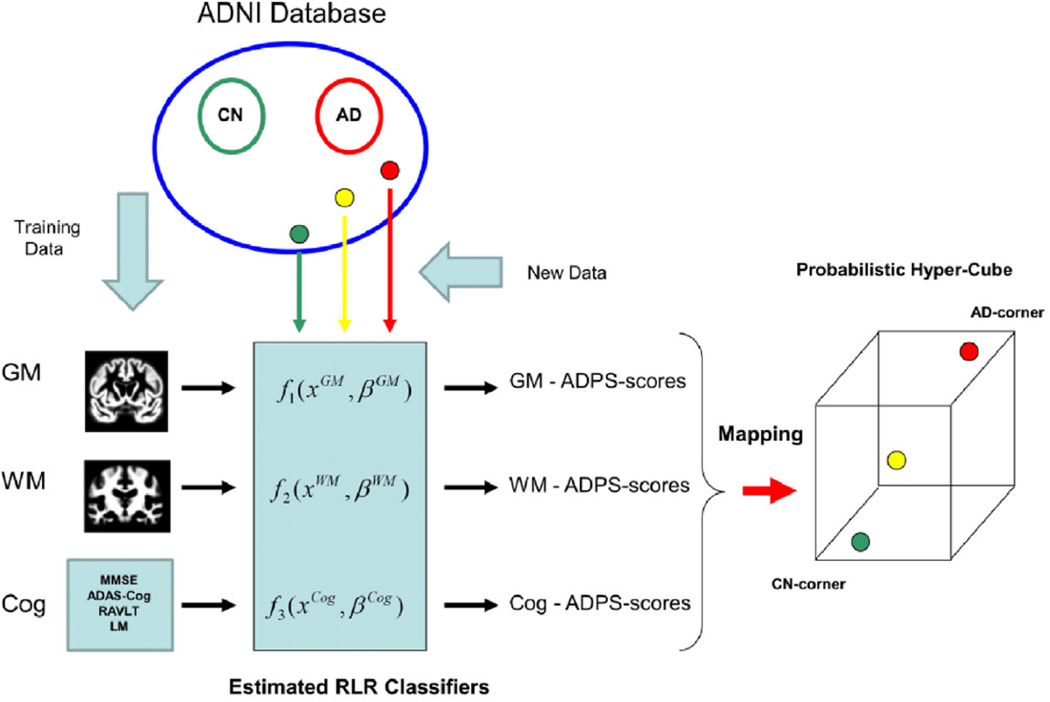
Abstract
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) is an ongoing, longitudinal, multicenter study designed to develop clinical, imaging, genetic, and biochemical biomarkers for the early detection and tracking of Alzheimer’s disease (AD). The initial study, ADNI-1, enrolled 400 subjects with early mild cognitive impairment (MCI), 200 with early AD, and 200 cognitively normal elderly controls. ADNI-1 was extended by a 2-year Grand Opportunities grant in 2009 and by a competitive renewal, ADNI-2, which enrolled an additional 550 participants and will run until 2015. This article reviews all papers published since the inception of the initiative and summarizes the results to the end of 2013. The major accomplishments of ADNI have been as follows: (1) the development of standardized methods for clinical tests, magnetic resonance imaging (MRI), positron emission tomography (PET), and cerebrospinal fluid (CSF) biomarkers in a multicenter setting; (2) elucidation of the patterns and rates of change of imaging and CSF biomarker measurements in control subjects, MCI patients, and AD patients. CSF biomarkers are largely consistent with disease trajectories predicted by β-amyloid cascade (Hardy, J Alzheimer’s Dis 2006;9(Suppl 3):151–3) and tau-mediated neurodegeneration hypotheses for AD, whereas brain atrophy and hypometabolism levels show predicted patterns but exhibit differing rates of change depending on region and disease severity; (3) the assessment of alternative methods of diagnostic categorization. Currently, the best classifiers select and combine optimum features from multiple modalities, including MRI, [18F]-fluorodeoxyglucose-PET, amyloid PET, CSF biomarkers, and clinical tests; (4) the development of blood biomarkers for AD as potentially noninvasive and low-cost alternatives to CSF biomarkers for AD diagnosis and the assessment of α-syn as an additional biomarker; (5) the development of methods for the early detection of AD. CSF biomarkers, β-amyloid 42 and tau, as well as amyloid PET may reflect the earliest steps in AD pathology in mildly symptomatic or even nonsymptomatic subjects and are leading candidates for the detection of AD in its preclinical stages; (6) the improvement of clinical trial efficiency through the identification of subjects most likely to undergo imminent future clinical decline and the use of more sensitive outcome measures to reduce sample sizes. Multimodal methods incorporating APOE status and longitudinal MRI proved most highly predictive of future decline. Refinements of clinical tests used as outcome measures such as clinical dementia rating-sum of boxes further reduced sample sizes; (7) the pioneering of genome-wide association studies that leverage quantitative imaging and biomarker phenotypes, including longitudinal data, to confirm recently identified loci, CR1, CLU, and PICALM and to identify novel AD risk loci; (8) worldwide impact through the establishment of ADNI-like programs in Japan, Australia, Argentina, Taiwan, China, Korea, Europe, and Italy; (9) understanding the biology and pathobiology of normal aging, MCI, and AD through integration of ADNI biomarker and clinical data to stimulate research that will resolve controversies about competing hypotheses on the etiopathogenesis of AD, thereby advancing efforts to find disease-modifying drugs for AD; and (10) the establishment of infrastructure to allow sharing of all raw and processed data without embargo to interested scientific investigators throughout the world.
Keywords: Alzheimer’s disease, Mild cognitive impairment, Amyloid, Tau, Biomarker
1. Introduction to Alzheimer’s Disease Neuroimaging Initiative: Goals, history, and organization
1.1. Background
Alzheimer’s disease (AD), the most common form of dementia, is a complex disease characterized by an accumulation of β-amyloid (Aβ) plaques and neurofibrillary tangles composed of tau amyloid fibrils [1] associated with synapse loss and neurodegeneration leading to memory impairment and other cognitive problems. There is currently no known treatment that slows the progression of this disorder. According to the 2014 World Alzheimer report, there are an estimated 44 million people worldwide living with dementia at a total cost of more than US$600 billion in 2010, and the incidence of AD throughout the world is expected to triple by 2050. There is a pressing need to find and validate biomarkers to both predict future clinical decline and for use as outcome measures in clinical trials of disease-modifying agents to facilitate phase II–III studies and foster the development of innovative drugs [2]. To this end, Alzheimer’s Disease Neuroimaging Initiative (ADNI) was conceived at the beginning of the millennium and began as a North American multicenter collaborative effort funded by public and private interests in October 2004. Although special issues focused on North American ADNI have been published in Alzheimer’s and Dementia [3] and Neurobiology of Aging [4] in addition to a number of other review articles [5–12], the purpose of this review is to provide a detailed and comprehensive overview of the approximately 500 papers that have been published as a direct result of ADNI to the end of 2013. The original review [350] covered approximately 200 papers to the end of 2010. The first update [351] detailed an additional 150 papers published from 2011 to mid-2012, and this material is highlighted in yellow. The current iteration adds around 200 more publications from mid-2012 to the end of 2013, and these are highlighted in green. To mid-2014, an additional 70 publications indicate the continuing impact of ADNI.
1.2. Disease model and progression
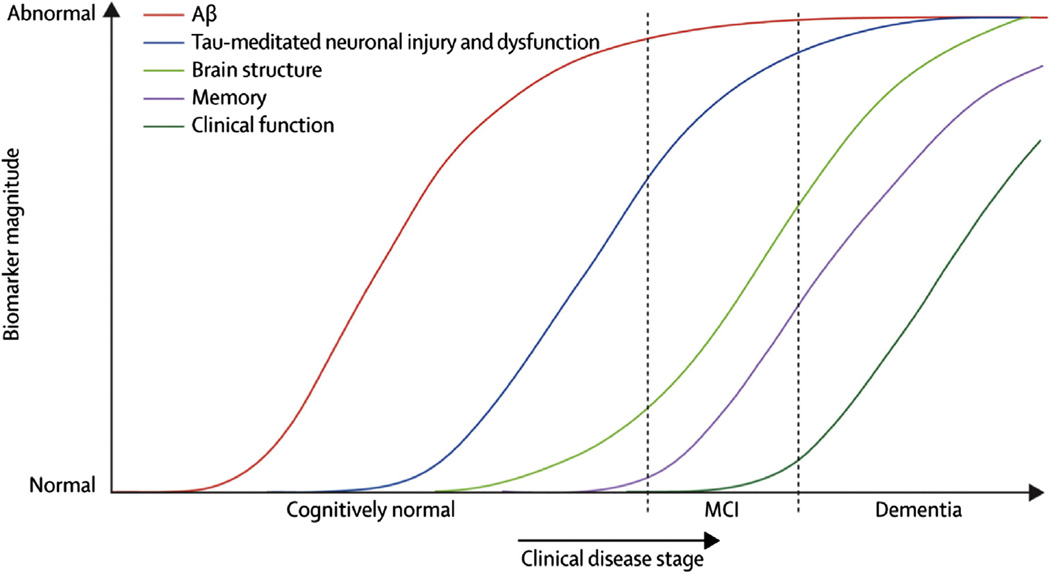
One approach toward a greater understanding of the events that occur in AD is the formulation of a disease model [3,12– 16]. According to the Aβ hypothesis, AD begins with the abnormal processing of the transmembrane Aβ precursor protein. Proteolysis of extracellular domains by sequential β and γ secretases result in a family of peptides that form predominantly β-sheets, the β-amyloids (Aβ) (Fig. 1). The more insoluble of these peptides, mostly Aβ42, have a propensity for self-aggregation into fibrils that form the senile plaques characteristic of AD pathology. Subsequently, it is thought that the microtubule-associated tau protein in neurons becomes abnormally hyperphosphorylated and forms neurofibrillary tangles that disrupt neurons. However, although ADNI and other biomarker data support this sequence of events, by direct examination of postmortem human brains, Braak and Del Tredici have shown that tau pathology in the medial temporal limbic isocortex precedes the development of Aβ deposits with advancing age in the human brain [17]. Downstream processes such as oxidative and inflammatory stress contribute to loss of synaptic and neuronal integrity, and eventually, neuron loss results in brain atrophy. Jack et al [14,16] presented a hypothetical model for biomarker dynamics in AD pathogenesis. The model begins with the abnormal deposition of Aβ fibrils, as evidenced by a corresponding drop in the levels of soluble Aβ42 in cerebrospinal fluid (CSF) and increased retention of the positron emission tomography (PET) radioactive tracer [11C]-labeled Pittsburgh compound B (11C-PiB) in the cortex. Sometime later, neuronal damage begins to occur, as evidenced by increased levels of CSF tau protein. Synaptic dysfunction follows, resulting in decreased [18F]-fluorodeoxyglucose (FDG) uptake measured by PET. As neuronal degeneration progresses, atrophy in certain areas typical of AD becomes detectable by magnetic resonance imaging (MRI). The model provided by Jack et al [14] is highly relevant to many papers reviewed in section 4 (Studies of the ADNI cohort), which often provide empirical evidence to support it. An example of a model that proposes a series of pathological events leading to cognitive impairment and dementia is summarized in Fig. 2.
Fig. 1.
Generation of soluble β-amyloid (Aβ) fragments from amyloid precursor protein. Reproduced with permission from Ref [7].
Fig. 2.
Model for Alzheimer’s disease (AD) progression. Reproduced with permission from Ref [14].
1.3. Mild cognitive impairment
Similar to many disease processes that originate in microscopic environments and are asymptomatic until the start of organ failure, the course of AD pathology is likely to be 20 to 30 years. It is now generally accepted that the initial AD pathology develops in situ while the patient is cognitively normal, sometimes termed the “preclinical stage” [18,19]. At some point in time, sufficient brain damage accumulates to result in cognitive symptoms and impairment. Originally defined in 1999, this has been classified in a number of ways, including as predementia AD or as mild cognitive impairment (MCI), a condition in which subjects are usually only mildly impaired in memory with relative preservation of other cognitive domains and functional activities and do not meet the criteria for dementia [5], or as the prodromal state AD [18]. Epidemiological studies of participants aged 70 to 89 years who were nondemented found the prevalence of MCI in this population to be approximately 15%, with an approximate 2:1 ratio of two identified phenotypes, amnestic and nonamnestic [20,21]. Studies showed that MCI patients progressed to AD at a yearly rate of 10% to 15%, and that predictors of this conversion included whether the patient was a carrier of the ε4 allele of the apolipoprotein E (APOE) gene, clinical severity, brain atrophy, certain patterns of CSF biomarkers and of cerebral glucose metabolism, and Aβ deposition [5]. The National Institute on Aging-Alzheimer’s Association (NIA-AA) has developed criteria for MCI due to AD that use biomarkers to determine the likelihood of AD pathology and classify patients accordingly [350]. The application of these criteria to ADNI is described later in this review [352–354].
1.4. History of biomarker development
Although the etiology of AD was not known, there was sufficient knowledge of the mechanisms of AD pathology at the beginning of the past decade to allow the development of new drugs. Once transgenic mice expressing Aβ in their brains were available [22], development of treatments to slow the progression of AD began in earnest. Although considerable work had been done to develop quantitative measurements of cognitive function and activities of daily living for clinical trials of symptomatic treatments such as acetylcholinesterase inhibitors, it was recognized that changes in cognition did not necessarily signify “disease modification.” Therefore, investigators from academia and the pharmaceutical industry became interested in how “disease modification” of AD could be detected using a variety of biomarkers, including brain MRI scanning, and blood and CSF analytes. This led to a decision by the National Institute on Aging (NIA) to fund the ADNI and to structure it as a public–private partnership.
The development of AD biomarkers for clinical trials, both for use in subject selection and as outcome measures, is paramount to the success of ADNI. During the genesis of the initiative, Frank et al [23] described the importance of biomarkers to ADNI and to clinical trials. In the first paper to come out of ADNI, Trojanowski [24] reviewed candidate AD biofluid biomarkers thought to be most promising at the time, homocysteine, isoprostanes, sulfatide, tau, and Aβ, and described how ADNI was poised, as a large public–private collaboration, to identify and validate the best candidate AD biomarkers. Mueller et al [25] reported on the scientific background at the beginning of ADNI and the limitations of the clinical and neuropsychological tests available for monitoring disease progression at that time. Principally, a definitive diagnosis of AD required severe cognitive deficits and autopsy confirmation, whereas the clinical criteria for the detection of the MCI transitional phase were much less certain. Accordingly, outcome measures for assessing the efficacy of new drugs relied primarily on neurocognitive tests such as ADAS-cog (cognitive subscale of the Alzheimer’s Disease Assessment Scale), the efficacy of which was limited by substantial ceiling effects and variability in subject performance over time. There was a clear need to develop biomarkers, biological tools that “mark” the presence of pathology, for the early diagnosis of AD and for measuring clinical drug trial outcomes [8].
Relatively early in the initiative, a major concern was developing an AD biomarker that distinguished AD from other dementias, such as Lewy body dementia, frontotemporal degeneration, and Parkinson disease with dementia [10]. Based on a model of AD pathogenesis fundamentally similar to that described in the paper by Jack et al [14], Shaw et al [10] reviewed a number of potential biomarkers, including some, such as isoprostanes and total plasma homocysteine, that did not subsequently prove to be of use. Others, such as levels of soluble Aβ42 or tau protein in CSF, reflected the increase in deposition of Aβ in fibrillar plaques or the later release of tau protein as a result of neuronal damage. Neuronal metabolism and neuronal degeneration could be measured using FDG-PET and by examining the concentrations of total tau protein (t-tau) and tau phosphorylated at serine 181 (p-tau181p) in CSF, respectively. Volumetric changes to brain structure could be assessed by MRI of specific regions such as the hippocampus, entorhinal cortex, temporal and parietal lobes, and ventricles. Additional potential risk biomarkers included genetic susceptibility factors, such as the APOE genotype, plasma homocysteine levels, and isoprostanes as non-AD-specific indicators of oxidative stress. By the following year, the wide range of potential biomarkers had been substantially narrowed to include CSFAβ42, t-tau and p-tau181p hippocampal volume, voxel-based volumetry, deformation-based morphometry (DBM), functional MRI, and FDG-PET [26]. In tandem with the development of these biomarkers, a new imaging technology using 11C-PiB in PET scans was being developed [27,28], and the possibility of a diagnostic approach predicated on the concept of certain combinations of biomarkers providing complementary information was raised [8,26].
In 2008, twin reviews were published in Neurosignals [8,15] by members of the ADNI Biomarker Core at the University of Pennsylvania. The first paper reviewed potential biomarkers for the early detection of AD. In addition to the potential biomarkers described previously, these included MRI T1ρ relaxation times to image neuritic plaques and single-photon emission computed tomography (SPECT) using a 125I-labeled imidazole derivative (6-iodo-2-(4’-dimethylamino-)phenyl-imidazo[1, 2]pyridine) as an alternative approach to amyloid PET imaging [29]. The second paper distinguished between diagnostic biomarkers and risk biomarkers, such as the APOE ε4 allele and plasma total homocysteine levels, suggesting that although they were not sufficiently sensitive for diagnostic purposes, they were indicative of increased risk for AD and were predictive of disease progression. Finally, in 2010, Hampel et al [7] presented a review that updated our current understanding of tau and Aβ biomarkers, including levels of Aβ42 and activity of BACE1 (the major amyloid precursor protein-cleaving β-secretase in the brain) in CSF, blood plasma levels of Aβ40 and Aβ42, and human antibodies against Aβ-related proteins. Thus, the search for biomarkers to fulfill a variety of niches is an ongoing quest and is without doubt set to evolve even further as research progresses.
1.5. Goals of ADNI
A comprehensive description of the goals of ADNI is given in papers by Mueller et al [2] and Weiner et al [3]. At initiation, ADNI had the overall objective of characterizing clinical, genetic, imaging, and biochemical biomarkers of AD and identifying the relationships between them over the course of disease progression from normal cognition to MCI to dementia. Specific goals of ADNI included the development of optimized and standardized methods for use across multiple centers, the enrollment of a large cohort (>800) of healthy elderly subjects, MCI patients, and AD patients for baseline characterization and longitudinal studies, and the establishment of repositories of data and biological samples, both of which were to be accessible to the wider scientific community without embargo. A specific pre-specified goal was to identify those imaging (MRI and PET) and image analysis techniques and blood/CSF biomarkers that had the highest statistical power to measure change (defined as the sample size required to detect a 25% reduction of rate of change in 1 year) and thus, it was hoped, detect effects of treatments that would slow the progression of AD. With these goals, ADNI hoped to identify a combination of biomarkers that could act as a signature for a more accurate and earlier diagnosis of AD, and that could be used to monitor the effects of AD treatment [2,3].
When originally conceived, ADNI had not included aims around genetic or proteomic analysis. Additional add-on studies supported the evolution of the Genetics Core (see later in the text) and the study of protein changes in plasma and CSF. Plasma proteomic data from a 190-analyte multiplex panel have been posted to the ADNI Web site and are available for additional data mining.
1.6. The evolution of an idea: ADNI-1, ADNI Grand Opportunities, and ADNI-2
Drs. Neil Buckholz and William Potter had discussed the overall concept of a large biomarker project to study AD for many years. Dr. Buckholz convened an NIA meeting focused on AD biomarkers in 2000. In 2001, Drs. Michael Weiner and Leon Thal (since deceased) proposed a longitudinal MRI study of AD, MCI, and control subjects. Subsequently, Dr. Buckholz brought together a number of investigators from the field of AD as well as industry leaders, all of whom strongly supported the overall concept. The NIA published a Request for Applications, and ADNI was funded in 2004. The initial ADNI was projected to run for 5 years and to collect serial information, every 6 months, on cognitive performance; brain structural and metabolic changes; and biochemical changes in blood, CSF, and urine in a cohort of 200 elderly control subjects, 200 MCI patients, and 400 AD patients [2–4]. It was funded as a public–private partnership, with $40 million from the NIA and $27 million from 20 companies in the pharmaceutical industry and 2 foundations for a total of $67 million, with the funds from private partners provided through the Foundation for the National Institutes of Health. An interesting perspective of the process by which potential competitors in the race to develop new drugs for AD were brought together in a consortium under the auspices of the Foundation for the National Institutes of Health is given in the paper by Schmidt et al [30], who emphasize the importance of the cooperative, precompetitive nature of ADNI. When the ADNI grant was first submitted and funded, the significance and impact of 11C-PiB [27,28] studies were not fully appreciated, and there was no infrastructure to conduct multisite clinical trials with 11C-PiB. Therefore, Aβ imaging with 11C-PiB was not included in the application. However, after the first year of funding, Chet Mathis proposed adding an 11C-PiB substudy to ADNI, which was funded by the Alzheimer’s Association and General Electric. In addition, further industry and foundation funding was secured to allow supplemental or “add-on” genomewide association studies (GWAS), and for additional lumbar punctures to obtain CSF, as new technologies emerged to make these studies feasible in a large-scale initiative such as ADNI.
In 2009, toward the end of the ADNI study, a Grand Opportunities grant, ADNI-GO, was secured to extend the original ADNI-1 studies with both longitudinal studies of the existing cohort and the enrollment of a new cohort of early MCI patients to investigate the relationship between biomarkers at an earlier stage of disease progression. Technical advances made it possible to add analyses of the new cohorts using AV45 (Florbetapir; Eli Lilly, Indianapolis, IN) amyloid imaging. Additional experimental MRI sequences included for evaluation ofADNI-GOandADNI-2 are arterial spin labeling perfusion imaging and diffusion tensor imaging. The development of the [18F]-labeled AV45 amyloid imaging agent with a substantially longer radioactive half-life than the 11C form made it practicable to extend amyloid imaging studies to additional sites beyond those undertaken in ADNI-1 [7].
A competitive renewal of the ADNI-1 grant, ADNI-2, was awarded with total funding of $69 million on October 1, 2010, together with funding from the pharmaceutical industry in a cooperative agreement similar to the original initiative, to further extend these studies with additional cohorts [3,4,31]. It is anticipated that the study of very mild MCI patients in ADNI-GO and ADNI-2 will help identify subjects at risk who are candidates for preventative therapy when they are mildly symptomatic or asymptomatic [30]. Table 1 summarizes details of the three initiatives.
Table 1.
Comparison of ADNI-1, ADNI-GO, and ADNI-2
| Study characteristics | ADNI-1 | ADNI-GO | ADNI-2 |
|---|---|---|---|
| Primary goal | Develop CSF/blood and imaging biomarkers as outcome measures |
Act as bridging grant between ADNI-1 and ADNI-2, examine biomarkers in earlier stage of disease progression |
Develop CSF/blood and imaging biomarkers as predictors of cognitive decline, and as outcome measures |
| Funding | $40 million federal (NIA), $20 million industry and foundation, $7 million industry for supplemental studies |
$24 million American Recovery Act funds (stimulus finds) |
$40 million federal (NIA), $27 million expected industry and foundation |
| Duration/start date | 5 years/October 2004 | 2 years/September 2009 | 5 years/September 2011 |
| Cohort | 200 elderly control subjects200 MCI400 AD |
Existing ADNI-1 cohort plus: 200 EMCI |
Existing ADNI-1 and ADNI-GO cohort plus:150 elderly control subjects100 EMCI150 MCI150 AD |
| Study techniques | |||
| MRI | X | X | X |
| fMRI | X | X | |
| FLAIR (microhemorrhage detection) | X | X | |
| T2* GRE (microhemorrhage detection) | X | X | |
| Vendor-specific protocols (1) resting state (task-free) fMRI to Phillips systems, (2) perfusion imaging (ASL) to Siemens, and (3) DTI to General Electric |
X | X | |
| FDG-PET | X | X | X |
| AV45 | X | X | |
| Biosamples | X | X | X |
| “Add-on” studies | GWAS, PiB-PET, lumbar puncture |
Abbreviations: ADNI, Alzheimer’s Disease Neuroimaging Initiative; ADNI-GO, Grand Opportunities grant; CSF, cerebrospinal fluid; NIA, National Institute on Aging; MCI, mild cognitive impairment; AD, Alzheimer’s disease; EMCI, early mild cognitive impairment; MRI, magnetic resonance imaging; fMRI, functional magnetic resonance imaging; FLAIR, fluid attentuated inversion recovery; T2* GRE, T2* gradient echo; ASL, arterial spin labeling; DTI, diffusion tensor imaging; FDG-PET, [18F]-fluorodeoxyglucose-positron emission tomography; GWAS, genomewide association studies; PiB-PET, Pittsburgh compound B-positron emission tomography.
1.7. Structure and organization of ADNI
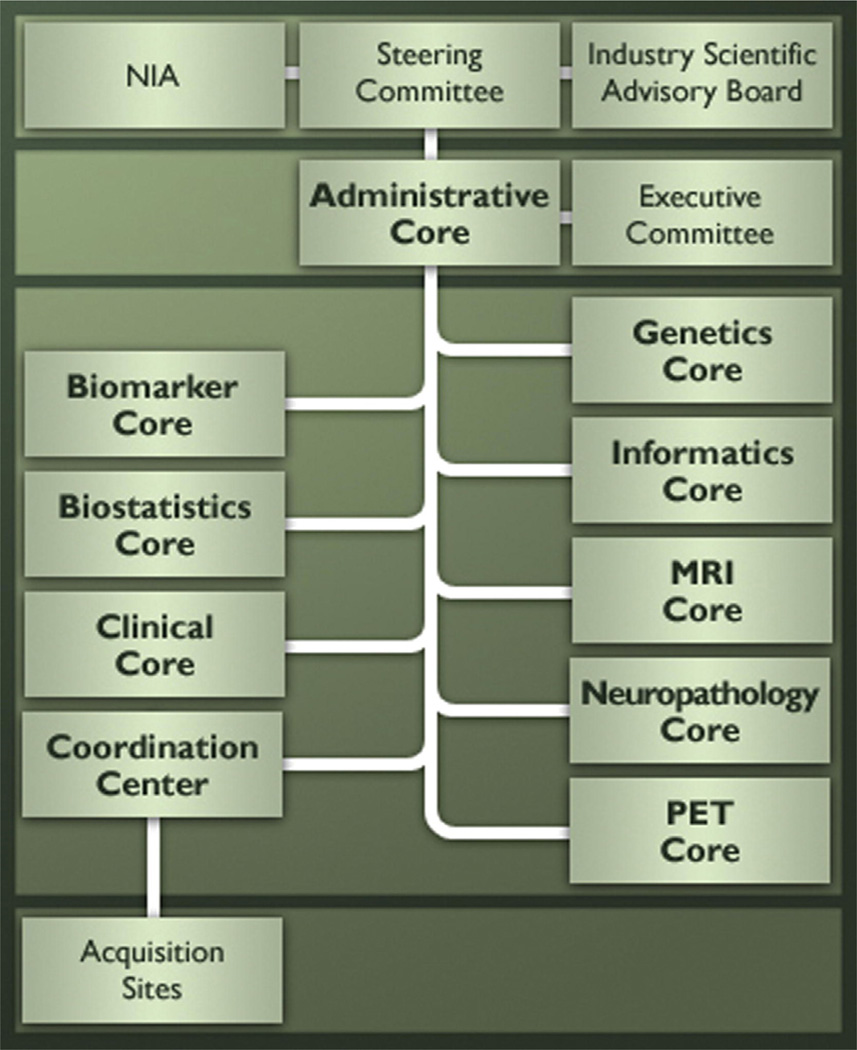
A full description of ADNI structure is given in the paper by Weiner et al [3]. Briefly, ADNI is governed by a Steering Committee that includes representatives from all funding sources as well as principal investigators of the ADNI sites and is organized as eight cores, each with different responsibilities, under the direction of an Administrative Core, led by Dr. Weiner, as well as a Data and Publications Committee (DPC), led by Dr. Green (Fig. 3). The eight cores comprise (1) the Clinical Core, led by Drs. Aisen and Petersen, responsible for subject recruitment, collection and quality control of clinical and neuropsychological data, testing clinical hypotheses, and maintaining databases; (2) the MRI and (3) PET Cores, led by Drs. Jack and Jagust, respectively, responsible for developing imaging methods, ensuring quality control between neuroimaging centers, and testing imaging hypotheses; (4) the Biomarker Core, led by Drs. Shaw and Trojanowski, responsible for the receipt, storage, and analysis of biological samples; (5) the Genetics Core, led by Dr. Saykin, responsible for genetic characterization and analysis of participants as well as banking DNA, RNA, and immortalized cell lines at the National Cell Repository for Alzheimer’s Disease; (6) the Neuropathology Core, led by Drs. Morris and Cairn, responsible for analyzing brain pathology obtained at autopsies of ADNI participants; (7) the Biostatistics Core, led by Dr. Beckett, responsible for statistical analyses of ADNI data; and (8) the Informatics Core, led by Dr. Toga, responsible for managing data sharing functions [2,3]. Additionally, Dr. Robert Green directs a recently funded project involving whole genome sequencing of ADNI DNA. The Private Partner Scientific Board (PPSB), convened by the Foundation for the National Institutes of Health and chaired in 2013 by Dr. Adam Schwarz, provides an independent, precompetitive forum for study-related scientific exchange among industry partners. Representatives of the PPSB liaise with the Steering and Executive Committee and with the ADNI Cores. A schematic of ADNI structure is given in Fig. 3. In addition to the Core leaders, the NIA established a completely independent committee, chaired by Tom Montine (U. Washington), to review and make recommendations concerning requests for ADNI blood, CSF, or DNA samples. Instructions concerning the preparation of requests for samples can be found at www.ADNI-info.org. Since the founding of ADNI in 2004, 11 batches of samples have been provided to requestors. The results of all sample analyses can be found in the ADNI data base at UCLA/LONI/ADNI.
Fig. 3.
Alzheimer’s Disease Neuroimaging Initiative (ADNI) structure and organization.
1.8. Data sharing and informatics
An objective of ADNI, in addition to its scientific goals outlined in section 1.5, was to make data available to the scientific community, without embargo. To this end, the Informatics Core of ADNI at the Laboratory of Neuro Imaging (LONI) at the University of Southern California in conjunction with the DPC, has developed policies and procedures for immediate, open-access data sharing on a previously unprecedented scale. The principles for this data sharing were developed in the initial months of the ADNI project in consultation with the Executive Committee and presented to the Steering Committee for adoption in the first year. The infrastructure for implementing this policy is through the LONI data archive (LDA), enabling the widespread sharing of imaging, clinical, genetics, and proteomic ADNI results, while overcoming fundamental hurdles such as the question of ownership of the disseminated scientific data, and the collection of data from multiple sites in a form that supports data analysis [32]. Briefly, LONI has developed automated systems that deidentify and upload data from the 57 ADNI sites, ensure quality control of images before removing them from quarantine status and make them available for download, manage preprocessing and postprocessing of images and their linkage to associated metadata, support search functions, and manage user access and approval. Clinical data are collected by the Alzheimer’s Disease Cooperative Study through their online data capture system and transferred to the ADNI repository at LONI through nightly data transfers. After these data are received at LONI, portions of the clinical data are used to update data in the ADNI repository to ensure consistency of demographic and examination data and to update the status of image data based on quality assessment results. Additional nightly processes integrate other clinical data elements, so they may be used in querying the data in the repository. Any researcher who has been granted access to ADNI data is able to analyze any part of the available data and can post results to LONI. In addition to ADNI data, LDA also contains data from the parallel Australian Imaging Biomarkers and Lifestyle (AIBL) Flagship Study of Ageing, which were collected using protocols comparable with those of ADNI. To date, from 35 countries worldwide, more than 1300 investigators from academic and governmental institutions, the pharmaceutical and biotechnology industries, and the scanner manufacturing sector have accessed ADNI data through the LDA [32]. The number of downloads of ADNI data has increased yearly since 2006, and in 2014, more than 5.5 million images, 322,000 sets of clinical data (including cognitive tests and levels of CSF biomarkers), and sets of genetic data were downloaded by more than 3000 distinct downloaders.
Recently, ADNI has been held up as an example of comprehensive and elegant data sharing in the clinical research community. It has been estimated that the costs of data sharing, which include infrastructure costs and administration, amount to approximately 10%to 15%of the monetary cost of the initiative and that providing data in a standardized form with documentation takes approximately 15% of investigators’ time [355]. A considerable number of NIH grants have been funded to investigators not directly funded by ADNI for analysis of ADNI data. A query of the NIH database indicates that a total of 23 separate NIH grants of this type have been funded. In 2014, there were 20 active grants of this sort, indicating the continued impact of ADNI-generated data throughout the research community.
To further enhance the utility of ADNI T1-weighted screening and baseline MR images to the scientific community, Heckemann et al[226] automatically segmented images of 816 healthy elderly, MCI, and AD patients in the ADNI database. They used the MAPER approach to generate WM, GM and CSF labels in 83 regions from the raw ADNI data with the aim of reducing future computation times. The automatic segmentations were in strong agreement with independent atlas-subset based segmentations of the target images, making this work a highly significant contribution to the repository.
Although LONI acts as the ADNI data repository, the DPC is responsible for developing policy around publication, granting access to the data to investigators around the world, and reviewing publications that result from this data use. Briefly, members of the scientific community can apply for access to ADNI data for either research or teaching purposes and must submit a data use agreement (available at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Data_Use_Agreement.pdf) for approval. Several thousand data applications from across the world had been approved, predominantly from academia, but also from the biotechnology, pharmaceutical, and other industries. Part of the data use agreement requires applicants to include certain language in manuscripts prepared from ADNI data, including citing “for the Alzheimer’s Disease Neuroimaging Initiative” as an ADNI group acknowledgment, and the recognition of ADNI’s role in data gathering in the Methods section and of ADNI’s funding in the Acknowledgments. Manuscripts must be submitted for approval to the DPC before publication. The full publication policy can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_DSP_Policy.pdf. The role of the DPC in this step is primarily to check that manuscripts are compliant with ADNI publication policy, and not to provide a scientific peer review. Papers found to be noncompliant are returned to the authors for editing and can subsequently be resubmitted for approval. This process is primarily designed to track, tabulate, and standardize the publication of manuscripts using ADNI data.
The approval by the Food and Drug Administration (FDA) of Florbetapir for clinical use brought into question the ADNI policy of nondisclosure of research results to study participants. Shulman et al [356] surveyed ADNI investigators about their willingness to disclose amyloid imaging results to different clinical groups and found that a majority were in favor of releasing results to MCI patients, whereas fewer supported data release to cognitively normal controls. The study also reported a desire for guidance on this issue from investigators and may reflect a need for ADNI to respond to a changing research/clinical environment with a review of disclosure policy.
1.9. The ADNI special issue of Alzheimer’s and Dementia
Weiner et al [3] introduced the special ADNI issue of Alzheimer’s and Dementia in 2010 with an overview of ADNI’s background, rationale, goals, structure, methods, impact, and future directions. A set of papers followed highlighting the achievements of individual ADNI cores and perspectives of the Industry Scientific Advisory Board (or ISAB), which is now referred to as the Private Partner Scientific Board (or PPSB). Jack et al [33] described the achievements of the MRI Core of ADNI in areas ranging from the development of MRI technology to the elucidation of AD biology, and concluded that this Core had succeeded in demonstrating the feasibility of multicenter MRI studies in ADNI and validity of this method as a biomarker in clinical trials. The progress of the PET Core of ADNI in developing FDG-PET and 11C-PiB PET protocols, ensuring quality control, and acquiring and analyzing longitudinal data was reviewed by Jagust et al [34], who similarly concluded that the Core had successfully demonstrated both the feasibility of this technology in a multicenter setting and the potential of FDG-PET to reduce sample sizes in clinical trials. Trojanowski et al [12] reviewed progress by the Biomarker Core of ADNI in developing profiles of CSF or plasma biomarkers that would act as a “signature” of mild AD or predict future MCI to AD conversion. Moreover, the review described studies in support of a temporal sequence of changes in individual biomarkers that reflected proposed trajectories of Aβ deposition and the formation of neurofibrillary tangles in AD progression [14]. The accomplishments of the Clinical Core of ADNI were reviewed by Aisen et al [35], who reported that the Core had successfully recruited a cohort of >800 subjects, characterizing them both clinically and cognitively at baseline and following them longitudinally over the course of the study. As the Clinical Core provided data management support to ADNI, this review also reported on the contribution of ADNI biomarker and MRI findings to improving clinical trial design by determining the most powerful outcome measures and reducing sample size using subject selection strategies. The contribution of the Genetics Core of ADNI to untangling the apparently complex genetic contributions to AD was reviewed by Saykin et al [6], who reported considerable progress in the identification of novel AD susceptibility loci and of candidate loci worthy of further investigation, often using AD biomarkers as quantitative traits (QTs) in imaging genetics and GWAS. The role of the Neuropathology Core in developing procedures to improve the autopsy rate of ADNI patients and to standardize neuropathological assessment was reviewed by Cairns et al [36]. Finally, Schmidt et al [30] discussed the contributions of the Industry Scientific Advisory Board, including acting as a conduit of information to and from sponsoring companies and foundations, supporting add-on studies, and contributing to the scientific review of protocols and procedures.
2. Development and assessment of treatments for AD: Perspectives of academia and the pharmaceutical industry
Given that the ultimate goal of ADNI is to develop biomarkers to facilitate clinical trials of AD therapeutics, it is germane to consider the perspective of investigators from academia and the pharmaceutical industry on the development of these biomarkers. The aim of this section is to review those papers that focus on this issue.
Although ADNI is a natural history study, and it is not known whether its biomarkers can measure the effect of candidate treatments in drug trials, the primary focus of ADNI has been the development of diagnostic biomarkers for the early detection of AD and development of prognostic biomarkers that would be used to monitor disease progression [37]. Mueller et al [38] and Weiner et al [3] reaffirmed the definition of an ideal biomarker formulated at the first meeting of the NIA working group on AD biomarkers, which proposed that an ideal AD biomarker should detect a fundamental feature of AD pathology; be minimally invasive, simple to analyze, and inexpensive; and meet criteria with regard to specificity and sensitivity outlined in Table 2. Prognostic biomarkers should be representative of a stage of AD at which the treatment has maximal effect, and also be representative of the proposed mechanism of action of the treatment [3,38].
Table 2.
Characteristics of an ideal biomarker
| Characteristic | Ideal |
|---|---|
| Sensitivity: % of patients correctly identified as having AD | >80%–85% |
| Specificity: % of patients correctly identified as not having AD. |
>80% |
| Positive predictive value: % of patients who are positive for biomarker and have definite AD pathology at autopsy |
>80% |
| Negative predictive value: % of patients who, at autopsy, prove not to have the disease |
>80% |
Both diagnostic and prognostic biomarkers are required for clinical trials. To date, such clinical trials have been frustratingly unsuccessful. It was thought that the failures of phase III clinical trials of high-profile putative antiamyloid therapies, flurizan and Alzhemed, were in part due to methodological difficulties, such as the initial subject selection, and the statistical comparison of results from multiple centers [7,9,39]. In the case of the first generation of clinical trials focusing on patients with MCI, there was a lack of consistency in numbers of patients progressing to AD over a certain period, likely due to the heterogeneous nature of MCI; it is possible that one-half of study participants did not have underlying AD pathology [7,11,40]. Correctly distinguishing patients with AD pathology is critical, especially considering the overlap that exists between various late-life neurodegenerative pathologies. For example, the Lewy bodies that characterize Parkinson’s disease are found in >50% of patients with AD, in addition to neuritic plaques and tangles. Therefore, there is a real need for biomarkers that reliably distinguish between different types of dementias [8,10].
Diagnostic biomarkers that meet the criteria outlined previously are urgently needed for subject selection, thereby allowing the stratification and enrichment of clinical trials. There is a need to select subjects at an early stage of the Alzheimer’s continuum who are likely to progress through MCI to dementia, and also to eliminate subjects with other pathologies. In phase I, II, and III trials, biomarkers that detect the earliest indications of AD pathology, Aβ deposition, such as CSF Aβ42, and 11C-PiB PET are most likely to be useful. FDG-PET as a measure of metabolism could also have potential [41].
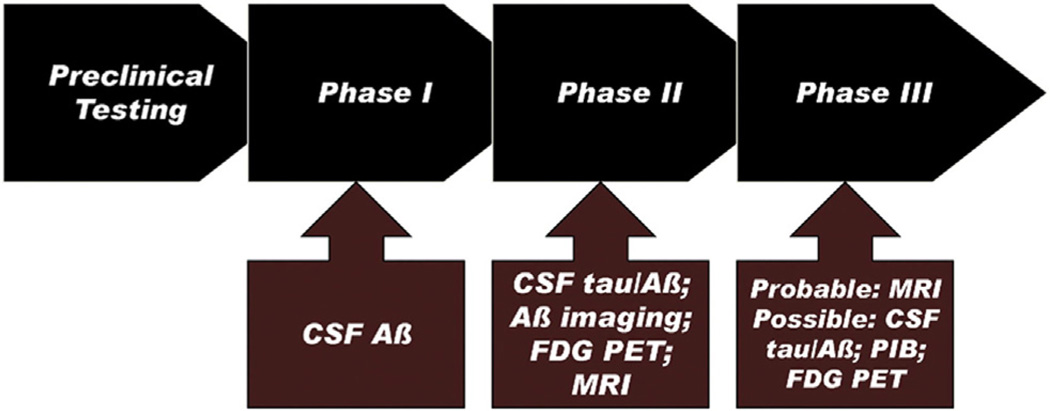
The biomarkers used in a clinical trial will differ depending on the mechanism of action of the therapeutic, the goals of the trial, and questions at hand. In small, short phase I trials, CSF and plasma measures can be used to monitor Aβ turnover in healthy subjects. In phase II proof-of-principle or proof-of-concept trials, Aβ biomarkers in brain can be used to confirm the mechanism of action of a new treatment and “target engagement.” For phase II and III trials, CSF tau and phosphorylated tau, MRI, and Aβ PET can be used to determine whether there is evidence of an effect of treatment on disease progression. Clinical MRI is used routinely for subject selection, to exclude confounding medical conditions, and for detection of vasogenic edema as a safety end point of “immune”-based treatments [41]. Finally, Aβ PET imaging, MRI, CSF and plasma biomarkers, and FDG-PET are candidates as prognostic biomarkers in phase II trials for selection of nondemented subjects at risk for developing AD to test whether treatments have the potential of preventing or delaying the onset of AD. The predictive power of these biomarkers in isolation or in combination varies and will need to be factored into consideration. None of the current generation of treatments proposed to modify the progression of AD is free of safety concerns. Estimation of the probability of developing AD will be required for assessing the risk versus possible benefit of participating in research trials [41]. Figure 4 shows ADNI biomarkers that could be used at different stages of the drug development process.
Fig. 4.
AD drug development. Black arrows show the phases of drug development; the brick-colored arrows show the ADNI biomarkers that could be used in that stage. Reproduced with permission from Ref [37].
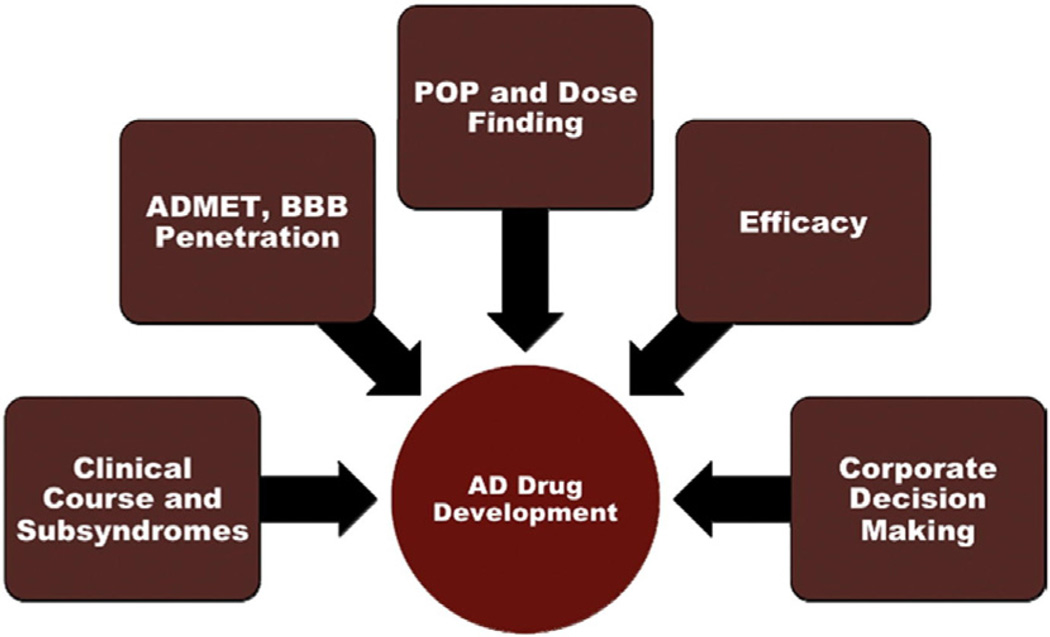
Looking at drug development as a whole, Cummings [37] saw a wide variety of roles for biomarkers, from identifying disease pathology and tracking disease progression, to demonstrating pharmacokinetic effects of the body on the drug, to facilitating proof-of-principle and determining doses for subsequent trials, to determining drug efficacy, and, finally, to contributing to corporate decision making, such as whether to proceed with riskier and more expensive later-phase trials (Fig. 5). Fleisher et al [9] reviewed progress in developing neuroimaging biomarkers, either alone or in conjunction with CSF biomarkers, for subject selection, and in developing biomarkers functioning at later stages in disease, such as MRI measures of brain atrophy or changes in cerebral glucose metabolism detected by FDG-PET as outcome measures. This review also highlighted the need for biomarkers in drug development and discussed the use of imaging biomarkers in replacing cognitive end points in clinical trials.
Fig. 5.
Roles of biomarkers in AD drug development. Abbreviations: AD-MET, absorption, distribution, metabolism, excretion, toxicity; BBB, blood–brain barrier; POP, proof of principle. Reproduced with permission from Ref [37].
Both common sense and regulatory policies of the Food and Drug Administration (FDA) and regulators in other countries require that treatment trials need to demonstrate a significant effect on cognition and function. Although effects on biomarkers would provide additional evidence of treatment effect and evidence of disease modification, there are no validated surrogates for AD trials, and such surrogates will take many years to develop. Different biomarkers are likely to be effective over different phases of the disease [11,41]. To be used as surrogates for clinical measures, biomarkers would need to be validated as reflecting clinical and/or pathological disease processes with a high degree of specificity and sensitivity. To qualify for validation as an outcome measure, the biomarker must be shown to predict clinical outcome over several trials and several classes of relevant agents by following subjects through disease progression and even possibly to autopsy [3,9,37]. This validation process is likely to be aided by the contribution of ADNI to standardizing procedures, particularly for imaging techniques, to reduce measurement errors in clinical trials [42]. A review by Petersen and Jack [11] discussed neuroimaging and chemical biomarkers, either alone or in combination, for the prediction of the development of dementia in MCI patients. These authors provided an excellent and succinct summary of the issues facing clinical trials for AD-modifying drugs and the role of both U.S. and worldwide ADNI in developing biomarkers to facilitate these trials.
A detailed discussion of the position of the FDA on biomarker validation is given by Carrillo et al [31], and it is likely that the process will require a wider population of well-characterized subjects than is available through ADNI. To this end, and for the further study of therapeutic interventions for AD, Petersen [40] proposed the establishment of a national registry of aging. In their editorial in the Journal of the American Medical Association, Petersen and Trojanowski [39] introduced a paper that reports on the evaluation of CSF biomarkers in a large multicenter study. Placing this in the context of other work in the same area, and in research undertaken as part of ADNI, they concluded that as biomarkers become more sophisticated, they will play even greater roles in AD clinical trials, and may one day be of use in clinical practice in a diagnostic capacity. Hill [41] concluded in his perspective on neuroimaging and its role in assessing safety and efficacy of disease-modifying therapies for AD: “…there is now sufficient experience of imaging for Alzheimer’s disease in both natural history and therapeutic trials for a clear recipe for success to be emerging.” Weiner [43] concluded that the use of biomarkers to select cognitively normal subjects who have AD-like pathology and as validated outcome measures in clinical trials “is the path to the prevention of AD.”
ADNI has proven to be a rich data set for industry-sponsored research, including an assessment of disease progression in the AD population [44]. Results from ADNI data have been combined with additional placebo data from clinical trials conducted in AD and are publicly available on the Coalition Against Major Diseases (CAMD) Web site for additional data mining [227]. Modeling efforts have highlighted the importance of age, baseline cognitive status, and APOE status on disease progression rates; a model is currently under qualification review through newly developed European Medicines Agency (EMA) and FDA qualification procedures. These types of models will inform clinical trial design and streamline analysis for drug studies conducted in mild-to-moderate AD.
ADNI has also enabled clinical studies in predementia, and many have been posted to www.clinicaltrials.gov, highlighting the use of CSF and amyloid PET biomarkers in cognitively impaired subjects to enrich for predementia clinical trials. Application to registration-level, phase III studies remains a challenge, as the biomarkers in ADNI have not yet been qualified for use or received regulatory approval. To address some of the remaining challenges, precompetitive and industry-sponsored initiatives were recently conducted to qualify CSFAβ-42 and t-tau as biomarkers for enrichment in predementia study with the EMA, and a positive qualification opinion was posted on the EMA site for these particular biomarkers. Additional efforts are ongoing with the FDA. For the most part, industry has been using the biomarkers as enrichment tools in predementia and mild-to-moderate AD studies, and as secondary or exploratory efficacy measures to assess impact of exploratory drugs on biomarker measures of disease progression.
In 2012–2013, ADNI has continued to provide an ever richer data set and important venue for precompetitive public-private interaction around biomarkers and clinical trial methodologies for AD, greatly facilitating the application of biomarkers and new methods in clinical trials. CSF and hippocampal volume biomarkers remain the focus of ongoing qualification efforts with the FDA. Amyloid biomarkers are actively used for subject selection in clinical trials of candidate therapeutics.
Amyloid biomarker substudies in the recent solanezumab and bapineuzumab phase III programs revealed that even in AD dementia populations, more than 20% of enrolled mild and moderate AD subjects were amyloid negative by CSF Aβ or amyloid PET. Subsequent trials of antiamyloid therapeutic candidates are requiring amyloid biomarkers at screening and amyloid positivity as an inclusion criterion. Longitudinal measures of amyloid are also being increasingly used later in the drug development process to assess potential disease-modifying effects.
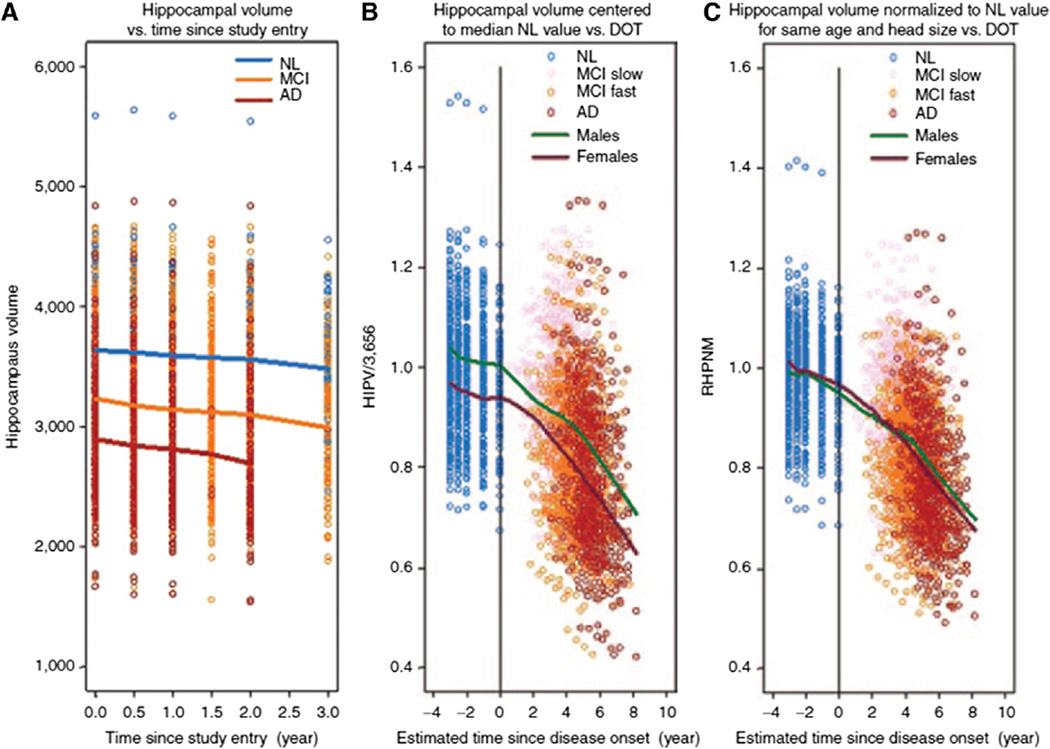
Hippocampal volume, as measured from structural MRI scans, decreases rapidly in the MCI phase preceding transition to AD and is strongly associated with imminent clinical decline. While not pathologically specific, screening for reduced baseline hippocampal volume selects a more homogeneous population of rapidly declining subjects, decreasing variability in longitudinal clinical outcome measures. Hippocampal volume was also recently qualified by the EMA for enrichment of amnestic MCI clinical trial populations, based in part on de novo analyses of ADNI data and coordinated in a precompetitive fashion by CAMD [357].
The widespread application of biomarkers in large, global trials owes much to the methods and data generated by ADNI.
3. Methods papers
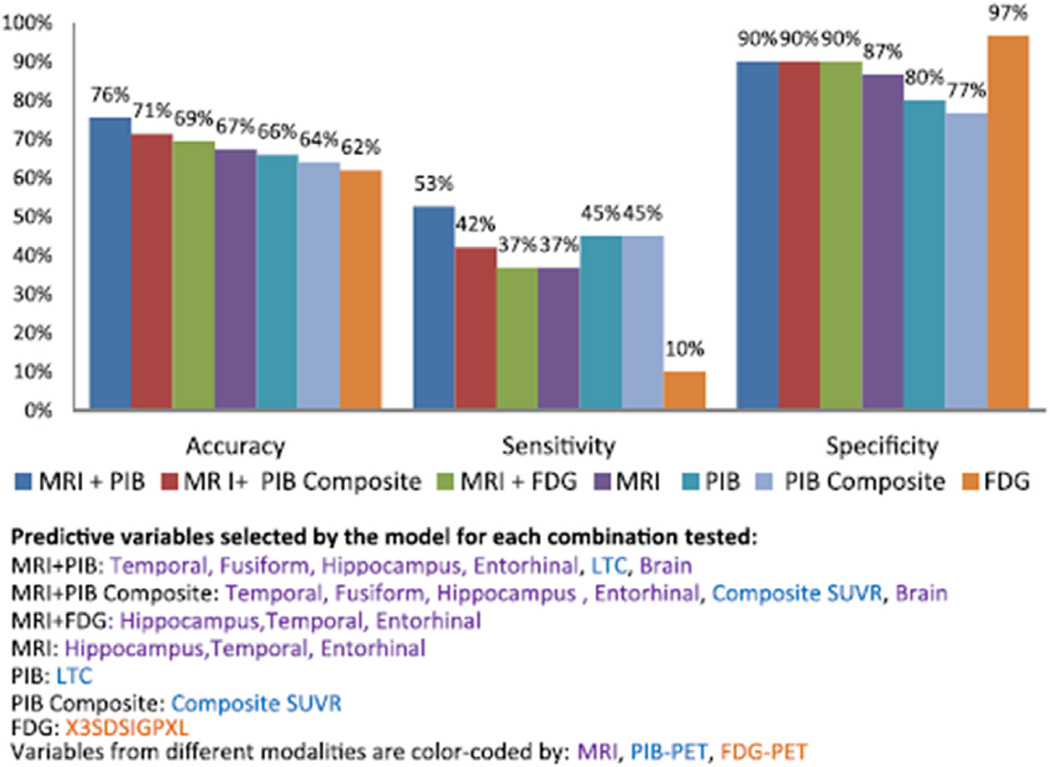
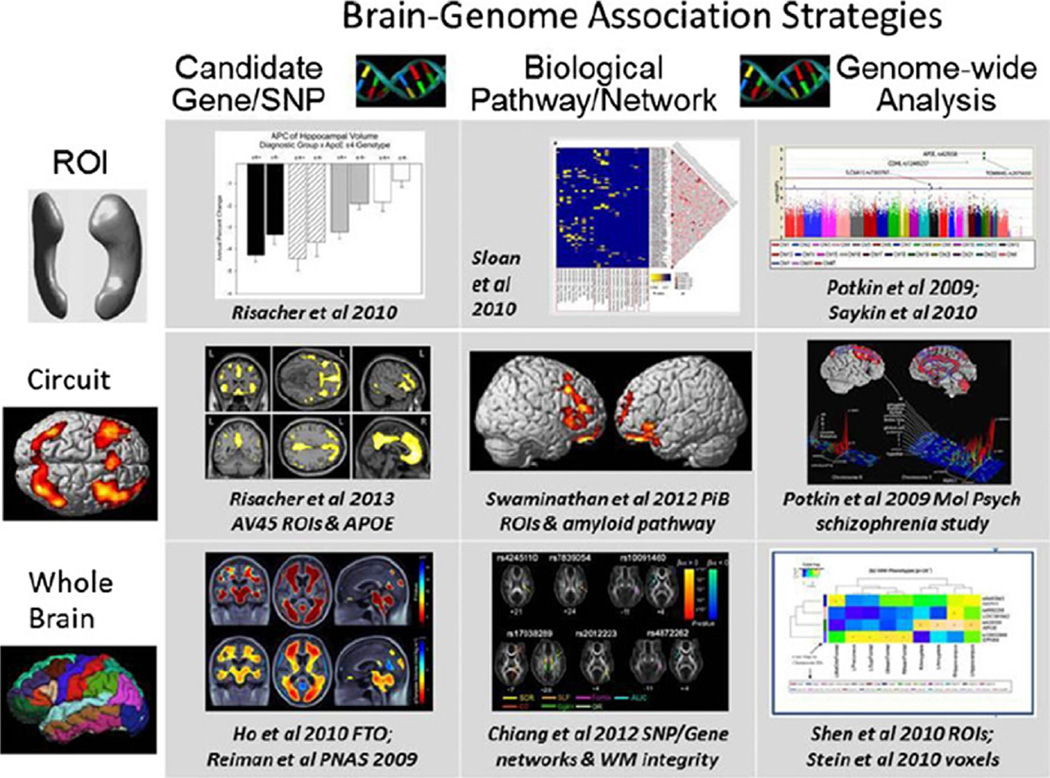
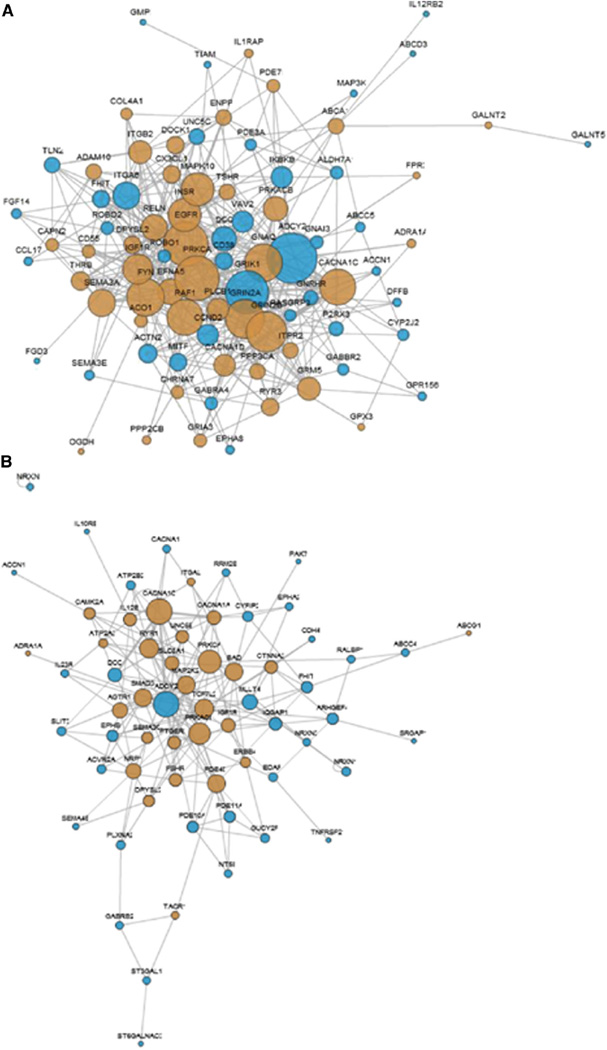
A considerable proportion of papers published as a result of ADNI concerns the development and testing of methods for use in ADNI, in the cohorts of other studies, or in clinical trials. These run the gamut from papers examining the best way to reduce differences between scanners in multicenter studies to those describing a new way to discriminate between AD, MCI, and control subjects, to methods for enriching clinical trials to reduce required sample sizes and therefore the associated cost, to new methods for examining genotype–phenotype relationships in neuroimaging GWAS. This section presents an overview of these papers.
3.1. Standardization of ADNI procedures
3.1.1. Magnetic resonance imaging
3.1.1.1. Assessment of scanner reliability
A key feature of assessing the reliability of scanner hardware over longitudinal scans is the use of a high-resolution geometric “phantom” that can detect linear and nonlinear spatial distortion, signal-to-noise ratio, and image contrast, allowing these artifactual problems to be identified and subsequently eliminated. Although these are commonly used for periodic adjustments to quality control, they are scanned after every patient in the ADNI MRI protocol. Gunter et al [45] estimated that these artifactual problems would contribute to >25% imprecision in the metric used, and found that phantom analysis helped correct scanner scaling errors and/or miscalibration, thereby increasing the potential statistical power of structural MRI for measuring rates of change in brain structure in clinical trials of AD-modifying agents. The utility of a scanner phantom was once again underscored by Kruggel et al [46], who examined the influence of scanner hardware and imaging protocol on the variability of morphometric measures longitudinally and also across scanners in the absence of a phantom in a large data set from the ADNI cohort. Using different acquisition conditions on the same subject, the variance in volumetric measures was up to 10 times higher than under the sample acquisition conditions, which were found to be sufficient to track changes. Their results suggested that the use of a phantom could reduce between-scanner imaging artifacts in longitudinal studies. Accurate ventricular segmentation also has an important role in estimating disease progression. Khan et al. [358] described the construction and testing of a physical brain ventricle phantom constructed to accurately simulate brain tissue T1 relaxation times. The phantom proved suitable for the validation of ventricular segmentation algorithms as it was life size, easy to fabricate, inexpensive, and accurately mimicked brain tissue.
Kruggel et al [46] also investigated the effect of scanner strength and the type of coil used on image quality and found that a 3.0-T array coil system was optimal in terms of image quality and contrast between white matter (WM) and gray matter (GM). Ho et al [47] similarly tested the ability of 3.0-Tand 1.5-T scanners to track longitudinal atrophy in AD and MCI patients using tensor-based morphometry (TBM). They saw no significant difference on the ability of either scanner type to detect neurodegenerative changes over a year, and found that TBM used at both field strengths gave excellent power to detect temporal lobe atrophy longitudinally. Marchewka et al [359] used voxel-based morphometry (VBM) with diffeomorphic registration to examine the effect of the different magnetic field strengths on the estimation of AD-related atrophy. They found slight grey matter volume differences related to field strength in the cerebellum, precentral cortex, and thalamus. The authors concluded that diffeomorphic registration was most accurate when scans were pooled across the study to create a common anatomic template regardless of scanner type or disease status.
While the scanning of a geometric phantom helps eliminate artifacts introduced by the machine, Mortamet et al [48] described an automated method for accounting for patient artifacts that can affect image quality, such as edge, flow, and aliasing artifacts. They developed two quality indices and tested their ability to differentiate between high- and low-quality scans, as assigned by an expert reader at the ADNI MRI center. Both indices accurately predicted the “gold standard” quality ratings (sensitivity and specificity >85%), and the authors proposed that this method could be integrated into a real-time or online MRI scanning protocol to eliminate the need to rescan at a later date due to a poor-quality scan, in keeping with the goal of placing as minimal burden on the patient as possible. Clarkson et al [49] examined within-scanner geometric scaling drift over serial MRI scans, as assessed by geometric phantoms, and developed a nine degrees-of-freedom registration algorithm to correct these scaling errors in longitudinal brain scans of patients. They found that the nine degrees-of-freedom registration was comparable with geometric phantom correction, allowing atrophy to be measured accurately, and the authors suggest that this registration-based scaling correction was the preferred method to correct for linear changes in gradient scaling over time on a given scanner. This in turn could obviate the need for scanning a phantom with every patient. Bauer et al [50] assessed the utility of collecting whole brain quantitative T2 MRI from multiple scanners using fast spin echo (FSE)/dual spin echo sequences, which have been shown to be useful in the early detection of AD pathology in MCI patients. Although FSE–T2 relaxation properties were related to the global dementia status, the authors concluded that the utility of the method was affected by the variability between scanners. Several papers were aimed at reducing between-scanner effects, including those by Gunter et al [45] and Clarkson et al [49]. Leung et al [51] presented a method aimed at overcoming variability in serial MRI scans for the detection of longitudinal atrophy by modifying the boundary shift integral (BSI) method of image analysis. Two improvements to the BSI method were made: (1) tissue-specific normalization was introduced to improve consistency over time, and (2) automated selection of BSI parameters was based on image-specific brain boundary contrast. The modified method, termed KN-BSI, had enhanced robustness and reproducibility and resulted in a reduction in the estimated sample sizes, required to see a 25% reduction in atrophy in clinical trials of AD-modifying drugs, from 120 to 81 AD patients (80% power, 5% significance).
3.1.1.2. Development of protocols
Jack et al [52] described the development of standardized MRI procedures for use in the multiple ADNI centers, a process guided by the principle of maximizing the scientific benefit of a scan while minimizing the burden on the patient. Using technology widely available in 2004 to 2005, and limiting scanner platforms to three vendors, they succeeded in developing a protocol that could be run in <30 minutes and that included the use of a phantom scan to monitor scanner performance over time and across different centers, back-to-back T1-weighted magnetization-prepared rapid gradient echo scans to capture structural information while minimizing the need to rescan patients due to technical difficulties, and T2-weighted dual-contrast FSE sequences for the detection of pathologies. Postacquisition corrections were instituted to remove certain image artifacts. Serial MRI scans, such as those used in ADNI, often suffer from problems associated with the uniformity of signal intensity that introduce artifacts into the results. Boyes et al [53] tested the ability of nonparametric nonuniform intensity normalization (N3) to eliminate these artifacts on higher-field 3-T scanners, which had a newer generation of receiver coils, in serial 2-week scans of healthy elderly control subjects. They found that the robustness and reliability of the N3 correction were highly dependent on the selection of the correct mask to identify the region of the scan over which the N3 worked, and on the smoothing parameter used for head scans at different pulse sequences. Leow et al [54] also used serial scans, 2 weeks apart, of healthy elderly control subjects to investigate the stability of different pulse sequences. They used TBM to generate maps of computed changes that could be statistically analyzed and to give information on MRI reliability, reproducibility, and variability. This optimization of pulse sequences contributed to the design of the ADNI MRI protocol, and authors concluded that TBM is a useful tool for the study of longitudinal changes in brain structure.
Vuong et al. [360] compared the accuracy or T1- versus T2-weighed MRI sequences in determining intracranial volume and found that the T2-based measurement improved the strength of associations between cognitive function and brain volume in MCI and AD patients compared with T1-weighted sequences in the standard ADNI protocol.
Although many methods have focused on the development of automated hippocampal segmentation, manual hippocampal segmentation remains the gold standard. Boccardi et al. [361] determined the most reliable orientation for manual segmentation to be anterior-posterior permissive and defined four segmentation units able to account for inter-protocol differences (the minimum hippocampus, the alveolus/fimbria, the tail, and the subiculum). The results were presented to a Delphi panel to develop a harmonized manual hippocampal segmentation protocol. Nestor et al. [362] directly compared the performance of five hippocampal-labeling protocols for multiatlas-based segmentation selected by the Hippocampal Harmonization Initiative. Using ADNI-1 baseline and 24-month scans and a fully automated multiatlas segmentation technique, the Sunnybrook Hippocampal Volumetry (SBHV) tool, they found that protocols differed in voxel overlap accuracies between automatic and manual labels, the ability to distinguish between MCIc and MCInc patients, and sample size estimates for clinical trials, suggesting that the choice of protocol for automatic multitemplate-based segmentation can influence segmentation accuracy.
3.1.1.3. Standardization of ADNI data sets
The lack of standardized ADNI data sets has hampered the direct comparison of results and the replication and evaluation of techniques published using ADNI data. The MRI core proposed offering a series of standardized data sets that include scans that have both passed quality control measures and have been performed on the same scanner to reduce interscanner variability. They proposed that deviation from the use of these prespecified data sets be reported in the published papers to facilitate comparison and replication of results. With the increasing role of multimodal studies, other ADNI cores are also working toward making available standardized data sets to achieve the same goals across the breadth of the initiative. Ongoing efforts by the ADNI Biomarker Core have also been directed toward the standardization of CSF biomarker assay methods to minimize the sources of analytical variability and to develop standard reference methods.
3.1.2. Aβ- and FDG-PET
Variability across scanners is also a major factor in ADNI PET studies, which are spread over 50 different centers and involve 15 different scanner/software combinations. Joshi et al [55] tackled the problem of reducing between-scanner variability in PET images that has been observed despite the use of standardized protocols. Major sources of between-scanner variability are high-frequency differences, mostly related to image resolution, and low-frequency differences, mostly related to image uniformity and also to corrections for scatter and attenuation. Joshi et al [55] scanned a Hoffmann phantom at each participating center, and by comparing the scans to the Hoffman “gold standard” digital phantom, they developed corrections for both types of variability, which were tested on scans from the ADNI cohort. They found that the high-frequency correction, by smoothing all images to a common resolution, reduced interscanner variability by 20% to 50%, but that the low-frequency correction was ineffective, perhaps due to differences in geometry between the Hoffman phantom and the human brain. Jagust et al [34] reported the development of a standardized protocol for the acquisition of FDG-PET and 11C-PiB PET data that first granted approval to participating sites based on the results from a pair of phantom scans on the three-dimensional (3-D) Hoffman brain phantom using defined acquisition and reconstruction parameters. These were assessed for image resolution and uniformity using a quality control process that used the digital gold standard phantom for comparison. In this way, corrections were made for differences in PET images across sites. A stable reference region is also critical to detecting metabolic changes with optimum sensitivity across different sites and scanners. Rasmussen et al [363] used an approach based on differences in coefficients of variation of FDG ratios over time across selected anatomic regions. They found the superior portion of the cerebellum to be the optimum reference region because of its stability over time compared with AD patients.
Amyloid PET using the longer half-life ligand Florbetapir was introduced into ADNI-2. Landau et al [364] compared amyloid imaging results in 32 individuals from ADNI cohort with both Florbetapir and PiB scans acquired on a variety of scanner types and processed using a variety of methods. They found that the numeric scale of cortical retention ratios was affected predominantly by the type of radioligand and the reference region used for scanning, but not by image preprocessing or analysis methods. By estimating a conversion factor in a study population scanned by both ligands, they were able to account for differences in scales and found that both ligands were in excellent agreement in the categorization of patients as amyloid positive or negative.
3.1.3. Biomarkers
The measurement of CSF concentrations of Aβ-42, t-tau, and p-tau is recognized to reflect early AD pathology. Within ADNI, levels of these analytes are measured by flow cytometry using monoclonal antibodies provided in the INNO-BIA Alz Bio3 immunoassay kit (Innogenetics, Ghent, Belgium) with xMAP technology (Luminex, Austin, TX) [56,57]. The Biomarker Core of ADNI has worked to make this a standardized procedure across multiple ADNI sites, and Shaw et al [56] presented an analysis of withinsite and intersite assay reliability across seven centers using aliquots of CSF from normal control subjects and AD patients. Five CSF pools were tested, each pool made up of either AD patients (n = 2) or controls (n = 3). Each center performed three analytical runs using separate fresh aliquots of each CSF sample and data were analyzed using mixed-effects modeling to determine assay precision. The coefficient of variation was 5.3% for Aβ-42, 6.7% for t-tau, and 10.8% for p-tau within center, and 17.9% for Aβ-42, 13.1% for t-tau, and 14.6% for p-tau between centers. The authors concluded that although they found good within-laboratory assay precision, the reason for the reduced inter-laboratory precision is not fully understood and may be caused by many sources of variability. As for any test method, strict attention to the laboratory standard operating procedures, inclusion of CSF quality control specimens in each analytical run, and following the manufacturer’s guidance for test performance are essential to assure best performance of this immunoassay test system [228].
The ADNI Biomarker Core has continued to focus on improving chemical biomarker assays. Korecka et al [365] compared the diagnostic utility of ultraperformance liquid chromatography–tandem mass spectroscopy (UPLC-MS-MS) with the AlzBio3 immunoassay for quantifying Aβ42. They demonstrated that a surrogate matrix consisting of artificial CSF containing 4 mg/mL of bovine serum albumin provided linear and reproducible calibration comparable to human pooled CSF. Analyses of CSF Aβ42 showed that UPLC-MS-MS distinguished neuropathologically diagnosed AD subjects from healthy controls with diagnostic utility at least equivalent to AlzBio3 indicating that this technique provides selective, reproducible, and accurate results and should be considered as a candidate reference method.
Kang et al [366] reviewed the clinical performance and reliability of immunoassays used in clinical studies of CSF Aβ42, t-tau, and p-tau181 as biochemical markers of the presence of AD neuropathology. They found that measurements of these biomarkers using the most widely used immunoassay platforms reliably reflected the AD neuropathology in patients with MCI or even in presymptomatic patients. These CSF biomarker tests were therefore deemed useful for early diagnosis of AD, prediction of disease progression, and efficient design of drug intervention clinical trials.
3.2. Methods for MRI image preparation and processing
A large portion of ADNI research relies on the extraction of information from MRI images; therefore, the development of automated methods to reliably and robustly process thousands of scans from multiple centers is vital to the project. Processing steps include whole brain extraction, image registration, intensity normalization, tissue classification (segmentation), cortical thickness estimation, and brain atrophy estimation [58].
3.2.1. Whole brain extraction
The separation of brain from nonbrain voxels in neuroimaging data, known as whole brain extraction or “skull-stripping,” is an important initial step in image analysis. Inaccuracies at this step can lead to the introduction of artifacts adversely affecting further analysis; therefore, a robust and accurate automated method for this step is highly desirable. To this end, Leung et al [58] compared the accuracy of a technique, multiatlas propagation and segmentation (MAPS), previously developed for hippocampal segmentation ([59]; see later section), with three other widely used automated brain extraction methods: brain extraction tool, hybrid watershed algorithm, and brain surface extractor. They found that compared with the semiautomated “gold standard” segmentation, MAPS was more accurate and reliable than the other methods and that its accuracy approached that of the gold standard, with a mean Jaccard index of 0.981 using 1.5-T scans and 0.980 using 3-T scans of control, MCI, and AD subjects.
3.2.2. Automated registration and segmentation
As manual registration and segmentation of images into WM, GM, and CSF is time-consuming, rater-dependent, and infeasible for a large study because of its often prohibitive cost, a number of studies have focused on developing automated registration and segmentation methods.
3.2.2.1. Atlas-based registration
Wolz et al [60] offered a solution in which atlases are automatically propagated to a large population of subjects using a manifold learned from a coordinate embedding system that selects similar images and reduces the potentially large deformation between dissimilar images, thereby reducing registration errors. This learning embeddings for atlas propagation method resulted in a more accurate segmentation of the hippocampus compared with other multiatlas methods [60].
The use of more than one atlas on which to register brain images has been recognized as a powerful way to increase accuracy of the automatic segmentation of T1-weighted MRI images, as it addresses the problem of brain variability. The steps of the process have been described by Lotjonen et al [61] and are presented in Fig. 6. Initially, multiple atlases are nonrigidly registered to the patient image, after which majority voting is applied to produce class labels for all voxels. Then, postprocessing by a variety of algorithms takes into account intensity distributions of different structures.
Fig. 6.
Steps of multiatlas segmentation. (I) nonrigid registration used to register all atlases to patient data, (II) classifier fusion using majority voting for producing class labels for all voxels, and (III) postprocessing of multiatlas segmentation result by various algorithms, taking into account intensity distributions of different structures. Reproduced with permission from Ref [61].
The addition of atlases has been found to increase segmentation accuracy in a logarithmic manner, that is, rapidly at first, but eventually slowing toward a maximum. This increased accuracy must be balanced by the increased computation time required for each additional atlas [61]. Lotjonen et al [61] obtained the best segmentation accuracy with relatively few [8–15] atlases, and, additionally, found that postprocessing using either the graph cuts or expectation maximization algorithms contributed to an optimized multiatlas segmentation method that balanced accuracy and computation times. They also found that the use of normalized intensity differences in the nonrigid registration step produced segmentation accuracy similar to that found using the more computationally intensive normalized mutual information method.
The selection of the atlases is a critical step. Heckeman et al [62] described the case in which the use of atlases based on the brains of young people resulted in occasional gross segmentation failures due to ventricular expansion in the older AD subjects. To overcome this problem, they modified a hierarchical registration approach by changing the first three levels to a tissue classification algorithm, instead of using native magnetic resonance (MR) intensity data. This multiatlas propagation with enhanced registration approach was found to create accurate atlas-based segmentations and was more robust in the presence of pathology than previous approaches. Li et al [229] presented another approach to account for ventricular expansion and other variations in tissue composition that occur in older subjects, such as WM hyper- and hypo- intensities, and changes in subcortical shape and cortical thickness. They employed a deformable registration algorithm that embeds 3D images in surfaces in a 4D Reimannian space to topological changes caused by false deformation. The method compared favorably with other registration methods employing diffeomorphic demons when tested on MR images with lesions from the ADNI data set.
Leung et al [58] generated multiple segmentations using nonlinear registration to best-matched manually segmented library templates and combined them using a simultaneous truth and performance level estimation algorithm. MAPS was then used to measure volume change over 12 months by applying the BSI. The accuracy of MAPS was found to compare favorably to manual segmentation, with a mean difference between automated and manual volumes of approximately 1% and a Dice score of 0.89 compared with other methods developed by ADNI (0.86: Morra et al [63]; 0.85: Wolz et al [64]; and 0.89: Lotjonen et al [61]).
The efficacy of three established manifold learning techniques (Isomap, Laplacian Eigenmaps, and Locally Linear Embeddings [LLE]) for the selection of atlases for multiatlas automatic hippocampal segmentation was compared by Hoang Duc et al [367] using the ADNI data set. LLE was found to result in the most accurate segmentations with a mean Dice similarity index of 0.883 on unseen data.
In heterogeneous populations, improvements in registration and segmentation may be gained by selecting atlases based on clusters of homogeneous morphologic features. The data-driven and unsupervised framework of Ribbens et al [368], termed SPARC, automatically identifies such subgroups that largely correspond to clinical status and constructs probabilistic atlases for each cluster to guide segmentation.
3.2.2.2. Other registration methods
In addition to registration of images to one or more atlases, segmentation of images may use image statistics to assign labels for each tissue or use geometric information such as deformable models or active contours [65]. A method that combines elements of these two approaches was described by Huang et al [65], who used an edge-based geodesic active contour. They found that this method segmented a range of images more accurately and robustly than those using individual statistical or geometric features only.
Calvini et al [66] developed software for the automatic analysis of the hippocampus and surrounding medial temporal lobe (MTL) and the calculation of a novel statistical indicator, the Δ-box, computed on intensities of the automatically extracted regions Their method did not directly segment the hippocampus, relying instead on the use of the Δ-box to assess intensities after a manual extraction step. A refinement and generalization of this method that standardizes the intensity scale of MR images, automatically extracts hippocampal volume, and generates templates using a clusterization method was subsequently described by Cataldo et al [369]. The authors propose that this simple and easily reproducible method could be easily applied to other regions of the brain.
A computational processing application to measure subtle longitudinal changes using nonlinear registration to the baseline image was described by Holland and Dale [67]. This method, called quantitative anatomical regional change (QUARC), used nonrigid 12-parameter affine registration, image smoothing minimization, normalization of local intensity nonuniformity, direct calculation of the displacement field of the region of interest (ROI) rather than the Jacobian field, and bias correction. When QUARC was compared with four other common registration methods used on ADNI data, it produced significantly larger Cohen d effect sizes in several ROIs than FreeSurfer v4.3 (Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA; http://surfer.nmr.mgh.harvard.edu/), voxel-based morphometry, and TBM, and a similar whole brain effect size to the standard KN-BSI method. Although, unlike the other methods, the signal-to-noise ratio of the raw images obtained using QUARC was enhanced by back-to-back repeat scans, the authors concluded that QUARC is a powerful method for detecting longitudinal brain morphometric changes in levels varying from the whole brain to cortical areas to subcortical ROIs [230]. Lorenzi et al [370] described a novel nonlinear registration method, LCC (local correlation coefficient)–Demons, designed for both intersubject and intrasubject studies. Based on the log-Demons diffeomorphic registration algorithm, it retained the computational efficiency of the log-Demons algorithm while using stationary velocity fields to parameterize the transformation. The algorithm was flexible, robust, and efficient when tested on ADNI cross-sectional and longitudinal data.
Robitaille et al [371] presented an alternative approach to tissue-based standardization called STandardization of Intensities (STI), which uses a spatial intensity correspondence between an input image and a standard determined using joint intensity histograms. The method was tested on the ADNI data set and was found to be superior to histogram-matching techniques.
3.2.3. Automated temporal lobe and hippocampal segmentation
3.2.3.1. Temporal lobe and hippocampus
In AD, atrophy in MTL and, in particular, the hippocampus is associated with declining cognitive function. It is not surprising, then, that a substantial body of work has been published on the subject of analyzing structural MRI T1-weighted measurements of this region. Chupin et al [68] developed a fully automated method for hippocampal segmentation based on probabilistic information derived from an atlas built from the manually segmented hippocampi of 16 young subjects and anatomical information derived from stable anatomical patterns. Wolz et al [64] used a fully automated four-dimensional (4-D) graph-cut approach to hippocampal segmentation that segmented serial scans of the same patient. Power analysis of the method revealed that a clinical trial for an AD-modifying drug would require 67 AD or 206 MCI patients to detect a 25% change in volume loss (80% power and 5% significance). Morra et al [69] developed the auto context model (ACM), a fully automated method to segment the hippocampus, based on the machine learning approach, AdaBoost. After training the classifier on a training set, ACM was able to discriminate between AD, MCI, and control groups, suggesting that the automatic segmentation is sufficiently sensitive to detect changes in hippocampal volume over the course of disease progression. This method was compared with manual and other automated methods for hippocampal segmentation, and also with TBM, which was used to assess whole brain atrophy in an earlier paper by the same group [63]. These authors found that ACM compared well with hand-labeled segmentation and that the volume atrophy over clinical groups and correlation with clinical measures with ACM were comparable with that found with other automated methods and better than TBM, suggesting that the latter method may not be optimal for assessing hippocampal atrophy.
Lotjonen et al [231] developed an automatic hippocampal segmentation method using an intermediate template space between unseen data and atlas spaces to increase processing speed and partial volume modeling to increase classification accuracy. The authors reported that this method more than halved the processing time on a standard laptop computer and resulted in a Dice overlap compared to manual segmentation of 0.869, in the range of previously reported accuracies (0.85 [63] and 0.93 [59]), supporting the feasibility of the method for clinical use. Tong et al [372] combined an alternative strategy, Fixed Discriminative Dictionary and Learning for Segmentation (F-DDLS) with fast coding techniques, for image reconstruction and reported computation burden. When the method was tested on hippocampal segmentation using ADNI cohort data, a Dice overlap of 0.879 was obtained. A similar Dice overlap (0.903) was obtained using a method, STEPS (Similarity and Truth Estimation for Propagation Segmentation), based on the STAPLE algorithm, which used a local ranking strategy to estimate the voxel-by-voxel classifier performance [373]. Using STEPS, statistically significant differences were found between subject groups in both baseline hippocampal volume and hippocampal atrophy rates.
Automated hippocampal segmentation may gain accuracy by considering not only volume but surface geometric variations. Shi et al [374] used a surface fluid registration method based on the use of holomorphic one-forms to compute a global conformal parameterization that can map a service to a rectangular plane, followed by multivariate TBM to compare in morphometry between study groups. The method successfully detected differences in hippocampal shape between APOE ε 4 allele carriers and noncarriers in both control and MCI groups.
Automatic image segmentation is prone to systematic errors, which are introduced when these mostly knowledge-based protocols mistranslate manual segmentation protocols into the automatic format. Wang et al [70] presented a wrapper algorithm that can be used in conjunction with automatic segmentation methods to correct such consistent bias. The algorithm uses machine learning methods to first learn the pattern of consistent segmentation errors and then applies a bias correction to the mislabeled voxels detected in the initial step. When the algorithm was applied to four different segmentation methods, it decreased the number of mislabeled voxels by 14% (multiatlas hippocampal segmentation) to 72% (FreeSurfer hippocampal segmentation) and resulted in a higher Dice overlap than other hippocampal segmentation methods, including some of those by Leung et al, Chupin et al, and Morra et al, described in this review [59,68,69].
3.2.3.2. Cortical thickness segmentation and estimation
Cortical thickness, which is correlated with disease progression, offers an alternative approach to ROIs to assessing disease progression. Consequently, the development of automated methods to analyze this region is an important step in monitoring disease progression [232]. Cardoso et al [233] presented a new method of post-processing for accurate segmentation of cortical thickness that reduced bias towards anatomical priors, explicitly models partial volume effects and improved the modeling of sulci and gyri using a locally varying Markov Random Field model. When the algorithm was tested on ADNI data, the authors reported an improvement in accuracy over established methods.
With the progression from ADNI-1 to ADNI-GO to ADNI-2, analysis of longitudinal ROI data has become of increasing importance. Several papers have focused on the accurate comparison of time points using 4D segmentation to determine rates of atrophy. Wang et al [375] proposed a method for longitudinal cortical thickness estimation in which all scans at every time point are included in the segmentation process, cortical thickness is maintained within a reasonable range using a spatial cortical thickness constraint, and artificial variations between time points are suppressed by a temporal constraint. When the method was compared in the ADNI data set to FreeSurfer (4D segmentation that lacks the temporal constraint), it produced more consistent and smoother thickness measurements in a more computationally efficient manner. Another approach, also involving temporal constraint, was described by Li et al [376]. First, they used a deformable parametric surface method to reconstruct the cortical surfaces of a group-mean image of all longitudinal images. They then used their mean surfaces to simultaneously reconstruct all longitudinal cortical surfaces. The method was tested on the ADNI cohort and found to be sufficiently sensitive to detect subtle changes in cortical surfaces such as a thinning of 0.02 mm over 24 months in normal aging.
Specific regions of the cortex may be of particular interest to researchers. For example, the perirhinal cortex may play a critical role in memory and has been shown to accumulate neurofibrillary tangles very early in disease progression. Augustinack et al [377] used probabilistic mapping on ADNI images to predict allocation for the perirhinal cortex and confirmed this with histologic staining. Using this method, they confirmed the utility of cortical thickness in this region as a specific metric for disease progression.
Segmentation bias can also be introduced when one baseline image is used as a reference in the comparison of multi-time point longitudinal images to estimate brain atrophy from changes in cortical thickness. Leung et al [230] developed a method based on BSI that utilized affine registration, differential bias correction and symmetrical global registration for multiple time points through the concept of a geometric mean to overcome this asymmetry. They found that this locally adaptive cortical segmentation algorithm (LoAD) consistently reduced bias and increased cortical thickness estimation accuracy compared to established methods when tested on the ADNI data set. Another challenge in cortical segmentation is filtering geometric topological outliers without causing unintended shrinkage to other parts of cortical surface. Shi et al [378] used a unified Reeb analysis that offered an integrated regularization of these outliers and also enhanced tissue classification at the subvoxel level. When tested on images from ADNI, the method was shown to be robust, accurate, and computationally efficient, requiring roughly a third of the computing time of FreeSurfer.
3.2.4. TBM and DBM
Bossa et al [72] used the method of TBM, which examines the deformation fields generated when an image is registered to a template. Previous work used large deformation algorithms for the nonrigid registration step, as they have the flexibility to characterize anatomical variability in cross-sectional studies. These algorithms are, however, computationally intensive, and the authors proposed a simplified version of the large deformation algorithms, stationary velocity field diffeomorphic registration. When the method was evaluated using ADNI subjects, it provided brain atrophy maps at high spatial resolution with lower computational requirements. Hua et al [73] examined two methods of image registration in TBM and found that the method in which each image is aligned to a single template was a more effective measure of brain deterioration. They also found TBM to be better suited to analyzing morphometric changes over larger areas, such as the entire temporal lobe, rather than specific ROIs, such as the hippocampus, and that atrophic changes detected by their method correlated well with clinical measures of brain deterioration (Mini-Mental State Examination [MMSE] and clinical dementia rating [CDR] scores). The use of multi-template registration was investigated by Koikkalainen et al [234], who developed and compared the classification accuracy of four methods with a single template registration method. They found that all four multi-template methods improved classification accuracy and resulted in smaller sample size estimates.
Yushkevich et al [74] examined the use of DBM, a technique closely related to TBM in estimating longitudinal hippocampal atrophy in the ADNI cohort. They found that without a correction for asymmetry that arises during longitudinal image registration, substantial bias can result in the overestimation of the rate of change of hippocampal atrophy. Park and Seo [75] tackled the problem of accurate registration algorithms required in DBM to compute the displacement field. They proposed a method that uses multidimensional scaling to improve the robustness of the registration step, and found that this method improves the ability of DBM to detect shape differences between patients. The same group reported a further advance using a manifold learning method, ISOMAP, embedding that represents high-dimensional imaging data in the low dimensional manner [379].
3.2.5. Quantification of brain morphometric changes
Several papers have focused on the development of methods for quantifying structural changes across the whole brain from structural MRI scans. Chen et al [76] developed a semiquantitative brain and lesion index based on T1- and T2-weighted imaging. They found that both the T1-based and T2-based scores correlated with age and cognitive performance and differentiated between control, MCI, and AD subjects. Acosta et al [77] presented a new accurate and computationally efficient voxel-based method for 3-D cortical measurement. The method, which uses an initial Lagrangian step to initialize boundaries using partial volume information and a subsequent Eulerian step to compute the final cortical thickness, offered higher statistical power to detect differences between clinical groups with a slight increase in computational time compared with methods using only the Eulerian step. The authors proposed that the increased accuracy and precision are attributable to the Lagrangian step, which effectively achieves subvoxel accuracy.
The reliability of two common algorithms, Siena and Siena X, used for measuring changes in whole brain volume cross-sectionally and longitudinally in MRI studies, was assessed by Cover et al [235] using ADNI data. They found that Siena was more reproducible than Siena X, that both algorithms gave estimates of atrophy rates in the ADNI cohort in line with atrophy rates reported in other cohorts, and that the distribution of atrophy in the ADNI cohort appeared to have a non-Gaussian distribution. The study demonstrated the utility of ADNI data as a benchmark for assessing the reliability of future algorithms for measuring brain atrophy.
3.2.6. Fractal analysis
A different approach for detecting atrophy in disease progression based on fractal analysis has been described by King et al [78]. Recognizing that the cerebral cortex has fractal properties, such as being statistically self-similar, this group investigated the effect of AD on gyrification using fractal analysis. They found that fractal analysis of cortical ribbons was able to discriminate between AD and control subjects in all of the seven regions tested, apart from the hippocampus, and suggested that this method may play a complementary role to ROI approaches, especially at earlier stages of disease progression. In a subsequent work, King et al [79] presented a new method for fractal dimension analysis of the cortical ribbon that also measured cortical thickness. When this method was compared with gray/white and pial surface cortical models, they found that it was the only measurement to have a significant correlation with cortical thickness and ADAS-cog scores, and that it best discriminated between control subjects and AD patients. The authors concluded that the fractal dimension of the cortical ribbon has strong potential as a quantitative marker of cerebral cortex atrophy in AD. Li et al [80] presented a method to reliably measure cortical thickness for longitudinal studies by incorporating 4-D information from successive scans directly into processing steps. In the absence of a gold standard against which to test their method, they used power analysis of the correlation between cortical thickness and the MMSE to show that this method improved longitudinal stability compared with 3-D methods that do not take the temporal factor into account.
3.2.7. Other MRI methods
Risser et al [81] presented a new method to compare imaged shapes, either longitudinally or against an atlas, on several different scales simultaneously, and to quantify the deformations on a single scale using large-scale deformation diffeomorphic mapping. When the method was applied to examine hippocampal atrophy in ADNI patients using baseline and 24-month scans, it was found to be able to extract information at the desired scale among all the scales.
A modification of the voxel-based analysis and statistical parametric mapping method for the detailed spatial analysis of image data without a priori defined ROIs was proposed by Zhang and Davatzikos [82]. Their method, optimally discriminative voxel-based analysis, uses non-negative discriminative projection applied to the spatial neighborhood around each voxel to find the optimally discriminative direction between two groups, determines a statistic for each group, and obtains a statistical parametric map of group differences. Optimally discriminative voxel-based analysis was found to perform well compared with traditional statistical parametric mapping using an ADNI data set.
Beyond volumetric analysis of ROIs, recent research has focused on extracting more meaningful information from the shape of brain structures, but most studies have not considered the pose, or location and orientation, of the structure. Bossa et al [71] presented a method for the statistical analysis of the relative pose of subcortical nuclei. The framework of the analysis was a variety of approaches based on similarity transformations with Reimannian metrics. Significant group differences were found between control subjects, MCI patients who did or did not subsequently convert to AD (MCI-c and MCI-nc, respectively) and AD patients, and the authors suggested that the method may be particularly useful as an AD biomarker in conjunction with shape analysis, as both approaches leverage complementary information. Two new approaches for analyzing longitudinal MRI data sets were reported by Skup et al [380] and Bernal-Rusiel et al [381]. The multiscale adaptive generalized method of moments [380] tackles the problem of analysis of longitudinal MRI data sets that have multiple response images per subject. The spatiotemporal linear mixed-effects modeling approach [381] uses the large number of spatial locations in a mass univariate setting and offers large gains in statistical power over other methods especially in a small sample setting.
3.3. Methods for AD classification from imaging data
The development of automatic methods for the accurate classification of patients into clinical groups from imaging data has been the aim of multiple ADNI studies. Many of these classification methods are based on support vector machines (SVMs), a set of algorithms that uses supervised learning of pattern recognition in a training set to build a classifier to predict the category to which a new example belongs. Some methods condense imaging data into one score that is reflective of brain abnormalities associated with AD to allow the direct comparison of patients, thereby facilitating their classification into patient group [83–85], whereas others examine which combination of imaging, CSF biomarkers, genetics, and other factors results in the most accurate classifiers [86,87], or formulate novel approaches for identifying AD-like patterns [87–90]. Other methods leverage the changes in spatial connectivity between different areas of the brain that most likely occur, as functional connectivity becomes affected during disease progression [65,83]. Finally, some methods [91,92] use an alternative approach to machine learning, a relevance vector machine (RVM), which, unlike the binary SVM, is a probabilistic machine learning algorithm. A brief description of these methods is given later in the text, and their results are presented and compared with existing methods of classification in section 5.4.1.
3.3.1. Magnetic resonance imaging
Fan et al [83] used an SVM to construct a classifier based on patterns of spatial distribution of brain tissue from T1-weighted MRI scans of control subjects and AD patients and applied this classifier to scans of MCI patients. The classifier, which acts as an indicator of how the structural profile of an individual fits that of AD or control subjects, also produced a structural phenotypic score (SPS) that allowed direct comparison of patients. This approach differs from ROI or voxel-based analyses, as it examines spatial patterns of atrophy rather than individual brain regions, and is also able to examine functional connectivity. Shen et al [89] also developed a method that integrated feature selection into the learning process, but used sparse Bayesian learning methods instead of an SVM. They reported that their automatic relevance determination and predictive automatic relevance determination, in general, outperformed the SVM used for comparison and classified patients more accurately than the method of Hinrichs et al [88]. Stonnington et al [91] used regression analysis based on an RVM to analyze T1-weighted MRI data and predict clinical scores, whereas Franke et al [92] used an RVM combined with an automatic preprocessing step and dimension reduction using principal component analysis to estimate the age of healthy subjects from T1-weighted MRI data, and found the method to be reliable, efficient, and scanner independent. In contrast to the supervised SVMs used in the aforementioned studies, Filipovych and Davatzikos [93] used a semisupervised SVM to classify MCI-c and MCI-nc patients. In the supervised approach, there is an assumption that patterns in a heterogeneous construct like MCI are known, but in a semisupervised approach, only some of the data, in this instance, baseline MRIs from AD patients and control subjects, are labeled, whereas scans of MCI patients are left unlabeled. Using a leave-one-out approach, scans were then classified as having a degree of AD-like or normal-like anatomic features, as defined by Fan et al [83]. Likewise, Spulber et al [382] derived a severity index reflective of the degree of AD-like neurodegeneration based on a priori–defined MRI regions from baseline MRI scans of NC and AD patients. They then used an orthogonal projection to latent structure algorithm as an alternative to SVM for analyzing high-dimensional data.
A more data-driven approach for patient classification that circumvents the need for a priori defined ROIs by using an initial independent component analysis (ICA) step was proposed by Yang et al [94]. Their preliminary study combined the ICA step to extract defining neuroimaging features with a subsequent SVM for classification of scans into AD, MCI, and control subjects, and the resulting method was tested on two cohorts, including ADNI. Pelaez-Coca et al [95] compared ability of anatomical versus statistically defined ROIs to discriminate between control and AD subjects. Using a variety of classifiers, they sought to restrict the number of features using principal component analysis and found that a higher number of features did not necessarily correspond with higher classification accuracy. When generalizability of the algorithm was tested by analyzing classification performance of 20 different experiments in which different subsets of the cohort were used as training and testing sets, they found that the resulting variability was larger than within the different classifiers used. Finally, they found that statistically defined ROIs representing voxels with the largest significance difference in a group comparison with an unbiased atlas (belonging to voxels in the hippocampi and amygdalae) resulted in better classification accuracy than anatomically predefined ROIs in the hippocampi, lateral ventricles, and amygdalae.
Shen et al [236] described a method to leverage differences in hippocampal shape for the discrimination of AD from control patients. The approach selected a subset of landmarks by using shape descriptors from statistical shape models that were further selected by statistical means for direct involvement in AD-specific neurodegeneration. Selected landmarks, including the CA1 subfield and the subiculum, were then used in a principal component analysis with SVMs for classification and their targeted use resulted in an increase in the discriminatory power of statistical shape models.
An alternative to a priori defined ROIs in AD classification is the use of cortical thickness estimates. Pachauri et al [232] used a topology-based kernel construction algorithm to measure cortical thickness. They suggested that this automated method can leverage discriminative information found on cortical surfaces that can be included in multimodal or multi-variate models to boost the signal of interest. Cho et al [237] employed an incremental learning method that represented cortical thickness data using the manifold harmonic transform to overcome problems of noise sensitivity in vertex based methods and the lack of detailed spatial variation of cortical thickness of region-wise methods. They found that this method was more robust than traditional methods and resulted in high classification accuracy. Park et al [383] reduced the dimensionality of selected ROIs of cortical thickness and sulcal depth using principal component analysis and trained an SVM on a small sample of ADNI patients. A novel approach for leveraging more information from cortical thickness measures by taking into account the relationship between morphological features was proposed by Wee et al [384]. By constructing a similarity map of correlations between pairs of ROIs, they augmented classification accuracy over ROIs alone. Beyond cortical thickness, differences exist in cortical folding patterns between patient groups in ADNI. Cash et al [385] described an alternative method to the fractal analysis of King et al [78] that uses a gyrification index as a summary statistic representing the degree of cortical folding in addition to indices describing cortical curvature and shape.
Increasingly, the ability to discriminate between MCI patients who will either remain stable or convert to AD is paramount to the selection of clinical trial populations. To this end, Eskildsen et al [386] defined differential patterns of cortical thinning in MCI patients depending on their time to conversion. They identified disease stage–specific neurodegenerative changes: initial thinning in the parahippocampal gyrus followed by the hippocampus and then the amygdala and occipital areas closer to conversion. Selected regions were applied to a linear discriminant analysis classifier. They also tested the effects of “double dipping”—the practice of reusing the training set in the test set—and found this artificially inflated classification accuracy.
ADNI acquires MRI data across multiple centers and scanner types. Abdulkadir et al. [238] investigated the effects of hardware heterogeneity on the classification accuracy of fully automated machine learning methods using an SVM classifier. They found that the negative effects of differences in scanner strength (1.5 T versus 3.0 T) on accuracy were offset by the gain made from the larger data sets available from multiple sites. A maximum accuracy of 87% in the classification of AD patients from controls was reported using data acquired with heterogeneous scanner settings.
Selection of MR features representative of change to a more AD-like morphometry has allowed the development of models that predicts future clinical decline from MR data. Zhang et al [239] targeted the use of both baseline and longitudinal data in a method that uses a longitudinal feature selection approach developed from a sparse linear regression model of each time point and which finally extracts a set of most relevant features longitudinally for input into a multi-kernel SVM. They found that the addition of longitudinal data substantially increased prediction accuracy. Aksu et al[240] used an approach intermediate between supervised and unsupervised machine learning to construct an automatic prognosticator of MCI to AD conversion and to define a conversion point between the two disease states. When an MCI patient showed any region of the brain as being “AD-like,” they were classified as converters which resulted in a higher prognostic accuracy than a CDR-based method.
Disease classification using MRI is plagued by the curse of dimensionality in which the number of voxels for whole brain analysis is so large that a direct SVM approach becomes prohibitively computationally expensive and does not necessarily lead to the best classification as not all features are relevant to disease pathology. Feature selection is one approach frequently used to address this challenge. In 2012 and 2013, there was a trend to considering higher level imaging features such as relationships between ROIs rather than low-level features such as voxelwise GM. The method of Suk et al [387] used as a stacked autoencoder to incorporate latent high-level information residing in patterns between low-level features in combination with the lower level features, themselves. Liu et al [388] likewise took into account structural variability in pathologic degeneration both at coarse and fine levels using a tree-based method of feature selection. Cuingnet et al [389] accounted for the structural and functional connectivity of imaging data in their alternative method that included a regularization step to add spatial and anatomical priors. Using ADNI imaging data, all methods increased classification accuracy over methods with low-level feature selection alone. Interestingly, when the effects of feature selection and sample size on classification accuracy were systematically examined, larger sample size was found to generally have a greater effect in classification accuracy than feature selection [390]. One exception to this was when ROIs were used as a feature selection; in this case, a large sample size was less advantageous.
Liu et al [391] presented a modification of the Lasso algorithm for dimensionality reduction in which hierarchical tree–guided regularization is added to identify relationships between the imaging voxels. Testing this tree-guided sparse coding method on ADNI data, they found that inclusion of this step was able to achieve better classification to L1-regularized Lasso alone using fewer features that were concentrated in areas known to be most relevant to AD such as the hippocampus, entorhinal cortex, parahippocampal gyrus, and amygdala. The use of Lasso regularization for dimensionality reduction in the multimodal setting has also been reported [392–394] and described in Section 3.3.4.
Most classification methods are based on SVM and kernel approaches which, in the process of dimension reduction, may discard useful information contained in the images. An alternative approach that operates directly in the voxel space was proposed by Casanova et al [241] who used penalized logistic regression and coordinate-wise descent optimization to overcome these problems of large scale classification. A subsequent paper by the same group [395] examined classification methods from structural MRI from the perspective of linear ill-posed problems and in the absence of dimensionality reduction techniques. They found that logistic regression, linear regression, and SVM classifiers were robust to increased dimensionality. Conversely, Liu et al [396] found that LLE as a method of dimensionality reduction improved classification accuracy universally using regularized logistic regression, SVM, and linear discriminant analysis. Their algorithm had the additional advantage of selecting the most discriminatory MRI features using unsupervised learning.
3.3.2. [18F]-fluorodeoxyglucose-positron emission tomography
Haense et al [84] also used a discrimination procedure, developed by the European Network for Standardization of Dementia Diagnosis, which generates a measure reflective of scan abnormality from FDG-PET data. This measure, AD t-sum, is calculated from the sum of abnormal t-values in voxels known to be affected by AD, and was used for discrimination of clinical groups. A similar approach was used by Chen et al [85], who developed an automatically generated hypometabolic convergence index (HCI) reflective of the degree to which the patient’s pattern and magnitude of cerebral hypometabolism corresponded to that of probable AD patients. Arbizu et al [397] combined the HCI with age and gender in a multivariate model to produce an AD score in an automated analysis method. To reflect the developing idea of AD as a continuum of disease rather than a progression of discrete states, they categorized patients into sixtile groups that had a progressive monotonic increase in AD scores. They also developed a similar index, the AD-conv-score, generated from FDG-PET data from the posterior cingulate index in combination with MMSE score and APOE ε 4 genotype, gender, and age.
Huang et al [65] identified changes in spatial connectivity patterns based on sparse inverse covariance estimation using FDG-PET data. Salas-Gonsalez et al [90] developed an automated procedure to classify AD patients from FDG-PET data using a t test to select voxels of interest and factor analysis to reduce feature dimension. The resulting factor loadings were tested on three different classifiers, two Gaussian mixture models with either linear or quadratic discriminant functions and an SVM. Lemoine et al [87] used a combination of feature selection and data fusion to construct SVMs from both FDG-PET and clinical data. To extract the most meaningful features from FDG-PET scans, they used an evolutionary algorithm in which each feature corresponded to one gene, the number of features was arbitrarily selected to be 30, and which was complete when an area under the curve (AUC) of 0.98 was achieved on the training data set. SVMs were also constructed for a range of clinical features, and the results of these and the FDG-PET classifiers were weighted and data finally fused to create a final classifier. Martinez-Murcia et al [398] selected maximally discriminative voxels using significance measures and ICA for dimensionality reduction and finally used an SVM or Bayesian classifier. The method was robust and accurate when applied to ADNI FDG-PET data. Toussaint et al [399] used a combination of univariate voxel-based analysis using two sample t tests and multivariate (ICA) techniques to discern patterns of glucose hypometabolism. These patterns in conjunction with a SVM were used to discriminate between patient groups. Derado et al [400] used a Gaussian process based on Bayesian theory as an alternative to an SVM to construct a hierarchical framework that used spatial correlations in the data. The method predicted glucose uptake patterns at 6 months for good accuracy and compared favorably with other methodologies.
Inspiration for improving classification accuracy increasingly comes from fields outside of neuroimaging. An alternative method for scoring brain images based on the principles of information retrieval, a computer science technique often used in Internet search engines, was described by Clark et al [242]. In this method, PET scores were arranges in a vector space with one dimension per voxel and orthogonal vectors were subtracted to refine queries. Cosine similarity between vectors was used between residual vectors to score the PET scan relevance to a diagnostic query. The resulting cosine similarity scores were used to construct classifiers. Keator et al [401] applied a biophysically inspired hierarchical filtering model for image recognition developed in the computer vision community to ADNI FDG-PET data. The hierarchical filtering pipeline determined which steps were most important for classification accuracy and thus produced outputs for training on neural networks or logistic regression classifiers. This method performed well compared with others (e.g., [402]) and outperformed a human rater in classification accuracy.
The large, heterogeneous ADNI data set proved an ideal testing ground for assessing the efficacy of a previously described classification method [243] which included principal component analysis and Fisher discriminant analysis. Markiewicz et al [244] successfully verified their multivariate approach and found that the highest accuracy for the whole sample verification was achieved using 4 principal components.
One of the issues with the use of FDG-PET data the selection of an appropriate reference region for either longitudinal or group comparison studies that measure changes in brain metabolism that can be leveraged for classification purposes. The method described by Rasmussen et al [245] sought to improve this critical step by the selection of candidate reference regions based on heat maps of coefficients of variation of FDG ratios over time. They found that intensity normalization systematically isolated the superior portion of the cerebellum as the test reference region for detecting rates of decline and baseline deficits in AD patients.
3.3.3. Cognitive methods
Llano et al [96] developed a cognitive test based on ADAS-cog as an alternative to imaging or CSF biomarkers for use as an outcome measure or for subject enrichment in clinical trials. The ADAS.Tree composite was derived by weighting test components of ADAS-cog based on their ability to discriminate between control, MCI, and AD subjects of the ADNI cohort using a Random Forests tree-based algorithm. ADAS.Tree discriminated between patient groups as well as, or better than, the best imaging or CSF biomarkers or cognitive tests. Optimal sets of markers for the prediction of 12-month decline were then determined using machine learning algorithms, and performance of the derived cognitive marker was found to be comparable with, or better than, other individual or composite baseline CSF or neuroimaging biomarkers. The authors suggest that the ADAS.Tree might prove more widely applicable than expensive and/or invasive imaging or CSF biomarkers.
Tractenberg et al [246] presented an alternative method for quantitating neuropsychological decline using inter-individual variability in cognitive testing. They found that this approach resulted in similar effect sizes to the total scores of MMSE and Clock Drawing test for discriminating between both controls and AD, and MCI and AD patients. The authors suggested that this may be a useful addition for measuring neuropsychological performance that is reflective of underlying neurobiology. Measuring cognitive decline in longitudinal studies often requires using different versions of the same test that are assumed to be equivalent. Gross et al [403] compared three equating methods—mean, linear, and equipercentile—for testing equivalence of alternative versions of the AVLT used in ADNI. They found that the equipercentile method performed best as it accommodated tests more difficult than the reference test at different percentiles of performance and adjusted for retest effects in models of within-person change.
3.3.4. Combined modalities
The new machine learning algorithm of Hinrichs et al [88], which uses data from both MR and FDG-PET images, integrates a spatial discrimination step to identify AD-related patterns in different brain regions, rather than assessing these relationships at the pre- or postprocessing steps.
The development of a panoply of multimodal classifiers that leverage information from imaging, biological and neuropsychological sources has been a major focus of ADNI papers published in 2011–2012. Likewise, the selection of features that are most ‘AD-like’ across multiple modalities is a critical step in constructing an accurate classifier and new approaches to this step have been reported in a number of papers. Hinrichs et al. [247] developed a method based on the Multi-Kernel Learning framework to produce a classifier that, in addition to classifying control and AD patients, also produced a Multi-Modality Disease Marker (MMDM) that could be used for the prediction of MCI to AD conversion. The method leveraged information from FDG-PET and MR scans and the authors reported that this method consistently outperformed a similarly trained SVM using the ADNI data set. An alternative method for AD classification that uses a non-negative matrix factorization for feature selection in combination with SVMs with bounds of confidence for classification was reported by Padilla et al[248]. The authors found that this method was an accurate tool for classifying AD patients from a combination of SPECT and PET data. Zhang et al [249] reported the first work to combine not only imaging but also biological data in the form of levels of CSF biomarkers into multi-modal classifier. They used a linear SVM with an intrinsic feature selection mechanism to rank top features of 93 ROIs (MR or FDG-PET) and CSF biomarkers were added directly as features. This method achieved high classification accuracy.
The next step in utilizing these classifiers is to determine their effectiveness in the prediction of future cognitive decline in addition to classification problems. Combining MR, FDG-PET and CSF data is again the focus of a later paper by Zhang et al[250] who presented a method, multi-modal multi-task (M3T), that uses this disparate data to estimate both continuous variables, such as scores on neuropsychological tests (MMSE, ADAS-cog), by regression and a categorical variable (classification class). M3T combines a multi-task feature selection with a multi-modal SVM that fuses selected features for regression and classification. They found that M3T was more effective than a concatenation method of combining features in both classification and prediction of future clinical scores and comparable to other reported prediction methods such as that described by Misra et al [118]. To account for the fact that brain structures in imaging data are interconnected, Wang et al [251] proposed the Sparse Multi-task Regression and Feature Selection (SMART) method that jointly analyzed all imaging and clinical data using a single regression model with sparse multi-task learning, and found that this method was an improvement on multi-variate regression when used to predict decline in AVLT scores.
In 2012 and 2013, there has been substantial improvement in the accuracy and computational efficiency of methods that use multimodal data. A modification by Liu et al [404] of M3T learning [25] tried to preserve complementary information between modalities by introducing a new constraint after treating the selection of features from each modality as a task. A further paper by the same group [392] describes a novel multiple kernel learning framework that improves on by the method of Zhang et al using Fourier transform approaches to approximate Gaussian kernels and reduce learning complexity to a linear scale. They used group Lasso regularization to enforce sparsity in the different modalities. The tree-based random forests algorithm was used by Gray et al [405] to derive consistent pairwise similarity measures from multiple modalities. The resulting embedding simultaneously encoded information on all features for multimodal classification. To combine data from both high- and low-dimensional modalities, Singh et al [406] used a partial least squares approach to allow the weighted fusion of these data and then achieved optimum classification accuracy using a quadratic discriminant analysis.
The prediction of continuous variables such as clinical scores has also received additional attention. Cheng et al [407] used a semisupervised multimodal relevance vector regression to predict MMSE and ADAS-cog scores of MCI patients. In a similar manner, Zhou et al [407] attempted to predict the scores over 4 years from baseline MRI data. The prediction of each time point (6, 12, 18, 24 months) was considered as a separate task, and the authors used two novel Lasso-based multitask regression formulations and longitudinal stability selection to identify patterns of biomarker change through disease progression. A similar method, temporally constrained group Lasso (tgLasso) used longitudinal data to predict clinical scores [394]. TgLasso trains a linear regression model and uses group regularization to group together weights corresponding to the same brain regions at different time points. When the model was used to predict clinical scores from longitudinal ADNI data, it outperformed other Lasso-based methods.
A number of studies have focused on using multimodal data to predict MCI to AD conversion. Young et al [408] made this prediction using a Gaussian process classification that integrates multimodal data in a probabilistic manner and reported that their method integrated multimodal data more efficiently than SVMs. Cheng et al [409] leveraged imaging and biomarker data from patient groups to discriminate between MCI converters and nonconverters. Their method, domain transfer SVM classification, uses auxiliary domain data combined with a modified SVM to help infuse domain knowledge. Yu et al [410] used a Bayesian classifier to determine which combinations of structural MRI, FDG-PET, and CSF biomarker measures, along with APOE genotype and cognitive scores, most accurately predicted progression from amnestic MCI to AD within 2 years. Overall, structural MRI measures were found to be the most predictive (78% accuracy), and a combination of MRI, genotype, and cognitive scores was found to provide the best trade-off between trial cost and time, in the context of clinical trial enrichment.
A multimodal approach reported by Casanova et al [411] distills information from multiple indices into a single index representative of the degree of AD-like features, the AD Pattern and Similarity (AD-PS) score. This method differs from scores such as STAND and SPARE-AD, which use an SVM, in that it uses logistic regression with sparsity regularization combined with feature selection and dimensionality reduction techniques. The probabilistic model creates a hypercube in which each dimension represents a range from normal to AD characteristics of a component such as GM, WM, CSF, and cognitive score. The AD-PS score of an individual is determined by their position in the hypercube (Fig. 32). The AD-PS score can then be analyzed directly using an index-based receiver operational characteristic (ROC). An alternative to this is described by Wu et al [412] who constructed a multivariate ROC that directly incorporated information from multiple indices.
Fig. 32.
A probabilistic hypercube. This can be interpreted as a geometrical representation of the output of a seat of classifiers, each one estimated with different types of data. The set of AD-PS scores corresponding to a given individual define a position inside the hypercube. The position of three individuals is illustrated. From Casanova et al [411]. Abbreviations: ADNI, Alzheimer’s Disease Neuroimaging Initiative; AD, Alzheimer’s Disease; CN, cognitively normal; GM, gray matter; WM, white matter.
With the development of multimodal classification and prediction, it has become increasingly recognized that the incompleteness of data can be problematic in these approaches. For example, in the ADNI data set, there are half as many controls as AD cases with proteomic measurements, but 40% more control cases than AD cases with MRI measurements [413]. An assumption of most longitudinal studies is that missing data is random. Lo et al [414] used univariate and multivariate approaches to examine associations between baseline demographic and clinical features and loss of data at follow-ups for CSF biomarkers and found that the missingness of data was nonrandom but tended to be blockwise and predictable. Yuan et al [415] described an incomplete Multi-Source Feature (iMSF) learning model, which initially partitioned patients into disjoint groups possessing the same data source combinations. They then applied independent feature learning for each group before combining results from all groups and found that this method was an improvement over other missing value estimation methods and over single modality classification alone. However, this model was not able to discern the most relevant data sources or provide a consistent prediction model for a specific data source across groups. Xiang et al [416] proposed a new bi-level multisource learning framework (incomplete source feature selection [iSFS]) that unifies feature- and source-level information and avoids the direct imputation of missing data, instead presenting efficient algorithms for calculating the missing data.
One impact of missing or imbalanced data on classification performance is that they achieve a much lower sensitivity and specificity. Dubey et al [413] examined this problem by testing a variety of sampling approaches, six feature reduction techniques, and two classifiers and by determining the optimum ensemble of these techniques for classification accuracy and balance of sensitivity and specificity. They demonstrated that the K-Medoid under sampling approach was superior to other data resampling techniques. In combination with sparse logistic regression with stability selection, it yielded competitive results with both an SVM classifier and a tree-based random forest algorithm using both MRI data and proteomic data separately. This comprehensive study outlines promising methodology to aid other researchers in improving classification performance using ADNI data.
For methods based on combined modalities to ultimately be useful in a clinical setting, they must present patient data clearly to aid in the physician’s diagnosis and ideally help reduce diagnostic errors. Mattila et al [252] and Soininen [253] created a diagnostic decision support system by representing cognitive, imaging, biological and genetic data in a graphical form termed a Disease State Fingerprint (DSF), as well as statistically distilling a score, the Disease State Index (DSI), that reflects the likelihood of a patient having AD (Fig. 26). The DSI can be used for both diagnostic classification and prediction of future decline. The PredictAD tool developed by this group has been further evaluated. This software increased classification accuracy over clinicians provided with test results directly in paper form and improved inter-rater agreement and the rater’s confidence in their decision [417]. Liu et al [418] examined the efficacy of PredictAD software in predicting AD conversion within 3 years in comparison with currently recommended criteria for prodromal AD: episodic memory impairment, visual assessment of medial temporal atrophy, and abnormal CSF biomarkers. They found that the software significantly outperformed these criteria and were equally effective with the aid of clinician. Both papers provide further support for the utility of software that can integrate heterogeneous data and provide objective, evidence-based information on the state of the patient that is not limited to binary classification. Escudero et al [419] created a similar metric to the Disease State Index using an unsupervised K-means machine learning technique. The bioprofile was characteristic of the disease and predictive of MCI to AD progression, whereas individual bioindices measured the closeness of an individual’s data pattern to the profile.
Fig. 26.
Disease State Index values of a patient with subtle indication of AD (total DSI value = 0.56). The name of the test and DSI value is shown next to each node. Larger nodes discriminate better between healthy and diseased patients (visualization of relevance). ‘Hot,’ i.e., red, nodes highlights patient data that fits AD profile (visualization of DSI). Here, ADAS and MRI contribute most to the AD DSI, indicated by the largest node size. MRI variables, especially hippocampal volume, whose computation is depicted on the right hand side, push the total DSI value towards AD population. Reproduced with permission from Ref [252].
An overarching consideration in the clinical setting is cost as many of the current biomarker tests are expensive to perform. Today’s health care environment requires biomarker tests that are maximally informative but minimally expensive. Escudero et al [420] proposed an approach to minimize the number of biomarkers required for diagnosis. This personalized and iterative approach initially attempts to classify the patient by comparison of available variables with data from a pool of local diagnosed patients. If this classification is below a threshold value of confidence, the method then selects a biomarker test that either minimizes the number of biomarkers required or maximizes cost-effectiveness.
3.5.5. Blood-based biomarkers
The identification of a blood-based biomarker for AD has been the goal of researchers for many years [254] and AD-NI’s extensive collection of biological specimens provides an ideal testing ground for new methods developed to this end [255]. The improved precision performance of a robotized version of the multiplex xMAP INNO-BIA plasma Aβ immunoassay for measurement of Aβ1–40 and Aβ1–42 in a longitudinal study of ADNI study subjects was described by Figurski et al [256]. Using this method in a longitudinal study of complementary measures of Aβ pathology (PiB, CSF and plasma Aβ) and other biomarkers in the ADNI cohort, Toledo et al [257] correlated baseline Aβ1–40 and Aβ1–42 plasma measurements in 205 cognitively normal subjects (CN), 348 patients with MCI and 162 with AD with PiB PET, MRI, and CSF tau and Aβ1–42 measures. Plasma Aβ1–42 levels were mildly correlated with other biomarkers of Aβ pathology and were associated with infarctions in MRI. They were also related to baseline and longitudinal diagnoses in addition to a number of health conditions. Longitudinal measurement of Aβ1–40 and Aβ1–42 plasma levels showed modest value as a prognostic factor for clinical progression, suggesting that plasma Aβ measurements have limited value for disease classification and prediction over the three year follow-up. However, with longer follow-up, within subject plasma Aβ measurements could be used as a simple and minimally invasive screen to identify those at increased risk for AD. This study and a recent review [258] emphasized the need for a better understanding of the biology and dynamics of plasma Aβ as well as for longer term studies to determine the clinical utility of measuring plasma Aβ. Finally, Soares et al [259] recently reported a study conducted in collaboration with Biomarkers Consortium Alzheimer’s Disease Plasma Proteomics Project that sought to develop a blood-based test as a screen for AD for early intervention. A multiplex immunoassay panel was used to identify plasma biomarkers of AD using ADNI plasma samples at baseline and at 1 year. These were analyzed from 396 (345 at 1 year) patients with MCI, 112 (97 at 1 year) patients with AD, and 58 (54 at 1 year) healthy control subjects. Multivariate and univariate statistical analyses across diagnostic groups and relative to the APOE genotype revealed increased levels of eotaxin 3, pancreatic polypeptide, and N-terminal protein B-type brain natriuretic peptide in MCI and AD patients, paralleling changes reported in CSF samples. Increases in tenascin C levels and decreases in IgM and ApoE levels were also observed. All participants with APOE ε 3/ ε 4 or ε 4/ ε 4 alleles showed a distinct biochemical profile characterized by low C-reactive protein and ApoE levels and by high cortisol, interleukin 13, apolipoprotein B, and gamma interferon levels. The use of plasma biomarkers improved specificity in differentiating patients with AD from controls, supporting the potential usefulness of these analytes as a screening tool. These studies have been extended by comparing the ADNI dataset with similar data obtained from ADNI independent cohorts followed at the University of Pennsylvania and Washington University as described by Hu et al [260]. This study used the same targeted proteomic approach described above and measured levels of 190 plasma proteins and peptides in 600 participants from two independent. 17 analytes were identified as being associated with the diagnosis of very mild dementia/MCI or AD. Four analytes (ApoE, B-type natriuretic peptide, C-reactive protein, pancreatic polypeptide) were also found to be altered in clinical MCI/AD in the ADNI cohort (n = 566). Regression analysis showed CSF Aβ42 levels and t-tau/Aβ42 ratios to correlate with the number of APOE ε /4 alleles and plasma levels of B-type natriuretic peptide and pancreatic polypeptide. Notably, 4 plasma analytes were consistently associated with the diagnosis of very mild dementia/MCI/AD in these 3 independent clinical cohorts, but further studies are need to determine if these plasma biomarkers may predict underlying AD through their association with CSF AD biomarkers.
Most studies have examined either serum or plasma for potential biomarkers, but not both. O’Bryant et al [255] sought to identify blood-based markers that were highly correlated across both plasma and serum and to construct a classifier using them. They found 11 suitable proteins, including C-reactive protein, factor VIII, fatty acid binding protein and adiponectin, and tested the classifier using ADNI biological samples.
In 2012 and 2013, the investigation of blood- and plasma-based biomarkers as a less invasive and therefore more clinically useful tool for AD diagnosis has continued to produce promising results. A blood-based panel of analytes identified from the AIBL study and validated in the ADNI cohort [421] identified biomarkers that overlapped biomarkers selected in studies by O’Bryant et al [255] and Soares et al [259]. Llano et al [422] used multivariate analysis to identify from a panel of 146 plasma analytes four proteomic signatures able to discriminate between AD and control patients. Associations between a panel of plasma analytes and amyloid burden as assessed by PiB-PET were investigated by Kiddle et al [423] who found a set of 13 analytes that along with the covariates age, APOE status, gender and education accounted for more than 30% of amyloid burden. This was more than double that of covariates alone, suggesting that these analytes reflect amyloid burden. Similarly, Burnham et al [424] selected five analytes in addition to APOE status, age, and CDR-SB scores to construct a blood-based biomarker signature able to predict amyloid burden. The most frequently reported analytes from these studies that are associated with diagnostic status are pancreatic polypeptide, brain natriuretic peptide, C-reactive protein, vascular cell adhesion molecule-1, α2-microglobulin, tenascin-C, and lymphocyte secreted protein B, and those most commonly associated with increased amyloid load are cortisol and interleukin 15. Panels of these analytes have been shown to classify AD patients with similar accuracy to other more established methods (Section 4.5.5) and therefore have exciting clinical potential.
3.4. Other imaging methods
Rousseau [97] presented a method for generating a high-resolution image from a low-resolution input, using jointly one low-resolution image and intermodality priors from another high-resolution image to create a super-resolution framework, for instance, a high-resolution T1-weighted image and a low-resolution T2-weighted image from the same patient. The method, when tested on clinical images from ADNI data, automatically generated high-resolution images from low-resolution input, and the authors suggest that this method may permit the investigation of multimodal imaging at high resolution.
The problem of representing a high dimensionality of brain images amassed in common neuroimaging applications was tackled by Gerber et al [98], who proposed that these images can be approximated by a low-dimensional, nonlinear manifold representative of variability in brain anatomy. They constructed a generative manifold model through kernel regression and tested this using ADNI data, and their finding was that important clinical trends were captured by this manifold when learned manifold coordinates and clinical parameters were subjected to analysis by linear regression.
3.5. Statistical methods
Interpretation of imaging data is a key facet in the process of extracting meaningful information from these scans. As the volume of neuroimaging data generated by ADNI studies burgeons, there is an obvious need for more sophisticated analysis techniques. Habeck and Stern [99] reviewed advances in multivariate analysis techniques that are being developed to supersede the more commonly used univariate, voxel-by-voxel analysis of imaging data. By evaluating the correlation or covariance of activation across brain regions, these multivariate techniques produce results that can be interpreted as neural networks, thereby addressing brain functional connectivity. Habeck and Stern [99] directed this review specifically at neuroscientists to explain the “bewildering variety of (multivariate) approaches …presented…typically by people with mathematics backgrounds.” In an effort to further spread the word to neuroscientists about this technique, a video article is also available [100].
Wu et al [101] presented a method to assess the reliability of hypometabolic voxels during the statistical inference stage of analysis. The aim of this method was to incorporate the differential involvement of each voxel into the multiple comparison correction, as opposed to current methods in which each location is treated equally. They used statistical parametric mapping and bootstrap resampling to create a bootstrap-based reliability index and compared this approach with the commonly used type I error approach, and found a strong, but nonlinear, association between the two methods. The authors suggest that this approach could have utility in both cross-sectional and longitudinal studies, in the early detection of AD, and in tracking disease progression in clinical trials.
A method to control for the effects of confounding variables was described by Dukart et al [261] and applied to the problem of controlling for the effects of age in group comparisons. Using a linear detrending model in terms of the general linear model, the method is able to control for the effects of age between groups of subjects. The application of this correction to either SVM classification or to the detection of disease-related GM using VBM in AD patients who differed in age from control subjects resulted in substantial gains in accuracy.
Singh et al [102] presented a new method to relate complex anatomical changes observed in AD patients with changes in cognition based on a statistical analysis of large deformation diffeomorphic metric mapping. In this method, the diffeomorphic transformations were analyzed using a multivariate and partial least squares approach without segmentation or the use of a priori defined ROIs. They found that this approach associated ventricular expansion, cortical thinning, and hippocampal atrophy with worsening scores on neuropsychological variables such as ADAS-cog, Rey Auditory Verbal Learning Test (AVLT), and clinical dementia rating-sum of boxes (CDR-SB), confirming that this data-driven approach was able to reach similar conclusions as other studies based on predefined ROIs [261,262].
The selection of MCI patients likely to progress for clinical trials is made more challenging by well-recognized heterogeneity of the MCI construct. Tatsuoka et al [425] proposed an alternative approach for analyzing the neuropsychiatric tests given to MCI patients based on the statistical method of partially ordered set (poset) models. Poset models in this instance allow direct links to be established between a specific cognitive function and the risk of conversion to AD. When poset models were applied to baseline data of ADNI MCI patients, specific cognitive domains were correlated with conversion within 2 years.
Ziegler et al [426] present an overview of current statistical approaches for modeling age-related structural brain decline and interindividual variations within this process. All analytical approaches are applicable to voxel- and surface-based whole brain MRI and include both parametric and nonparametric models. The focus of the overview is on estimating age trajectories of structural change and accounting for both nonlinear trajectories and differences in rates of change in specific regions.
3.6. Genetics methods
Genetic contributions to AD are being revealed by GWAS that search for associations between QTs in the form of imaging or biomarker data and genetic loci. The standard approach (mass univariate linear modeling), which compares each phenotype–genetic loci pair individually and then ranks the association in terms of significance, is extremely computing-intensive and can miss information from areas surrounding a particular association. Methods that boost power to detect genetic associations and therefore reduce sample sizes are considered necessary for replicable genetics results. Vounou et al [103] proposed a new method, sparse reduced rank regression, which overcomes these problems by enforcing sparsity of regression. They found sparse reduced rank regression to be less computing-intensive and to have better power to detect deleterious genetic variants than mass univariate linear modeling. Kohannim et al [427] reported a similar approach that used Lasso regression for dimensionality reduction of a multivariate GWAS investigating gene effects on temporal lobe volume. This gene-centric approach jointly considers groups of genetic variants that are correlated through a sparsity-driven LI penalty and associates them jointly in partial F tests with the MRI-derived temporal lobe volume measure. It identified more genes and at higher significance than traditional univariate methods. An alternative approach to reducing computational requirements, while retaining a high degree of significance to AD, has been presented by Chen et al [104], who used each of 142 preselected imaging ROIs as QTs in a GWAS. Heat maps and hierarchical mapping were then used to organize and visualize results and to select target SNPs, QTs, or associations for further analysis.
Meda et al [263] presented a method for multivariate analysis of GWAS data based on the premise that genetic determinants are not randomly distributed throughout the genome, but tend to cluster in specific biological processes related to AD. Their method used a parallel ICA and a hypothesis-free, data-driven statistical technique to simultaneously examine multiple modalities. They found that the parallel ICA was effective on the large sample, sizes in ADNI and that it identified clusters of SNPs potentially related in different metabolic pathways associated with AD. Similarly, to address the issue of underlying interactions between SNPs and QTs such as imaging data, Wang et al [264] developed a novel method, Group-Sparse Multi-task Regression and Feature selection (G-SMuRFS) that is built on multivariate regression analysis with a new form of regularization. Application of the method using the ADNI data-set demonstrated its ability to predict continuous responses of brain imaging measures and to select relevant SNPs in a more efficient manner than conventional multivariate linear regression. Univariate and multivariate genetic analysis techniques and sparse regression methods aimed at reducing the dimensionality of imaging and genomics domains are reviewed by Shen et al [428].
In addition to computational challenges, imaging genetics studies with multiple testing are also prone to false-positive results, and both familywise error and false discovery rate corrections are used to adjust significance thresholds across multiple voxels. Silver et al [105] measured false-positive rates using VBM to investigate the effect of 700 null SNPs on GM volume in the ADNI cohort. They found that although false-positive rates were generally found to be well controlled, under certain conditions, such as under low cluster-forming thresholds, the false-positive rates were substantially elevated. Consequently, they proposed the use of parametric random field theory cluster size inference and alternative nonparametric methods under different circumstances.
3.7. Methods for Clinical Trials
ADNI data has recently been utilized to test methods for improving clinical trials of compounds with the potential to attenuate the progression of AD. These are commonly designed as long-term, randomized, placebo-controlled trials (RPCTs), which present the problem of whether the exposure of pre-symptomatic AD patients to placebos long term is ethical. Spiegel et al [265] proposed a method to overcome this ethical dilemma with a placebo group simulation approach (PGSA) which involved construction of univariate and multivariate models based on baseline data of MCI patients in ADNI. Gender, obesity, Functional Activities Questionnaire (FAQ), MMSE, ADAS-cog and Neuropsychological Battery scores were used to predict ADAS-cog scores after 24 months and models corresponded closely to real observed values (R2 = .63, residual S.D = 0.67). These results suggest that the PGSA approach has the potential to complement future RCPTs for AD drugs. Another issue with RCPTs is the selection of a primary end-point, which is often either time-to-event (for example, progression to dementia) or a continuous measure of disease severity such as ADAS-cog to assess the effect of the treatment. Donohue et al[266] compared the power to detect an effect of these two methods by using Cox proportional hazard models to estimate time-to endpoint, and linear mixed models to estimate continuous variables and found that linear models consistently demonstrated greater power than Cox proportional hazard models when tested on the ADNI data-set (Fig. 27). The authors concluded that linear models may be more robust and appropriate for the detection of MCI to AD progression in clinical trials of MCI patients.
Fig. 27.
Simulated power for studies in MCI and MCI with amyloid dysregulation (MCI-Aβ) versus total sample size, n. Lines represent LOESS smooths. Abbreviation: PH, proportional hazard. Reproduced with permission from Ref [266].
3.8. Methods papers: Summary and conclusions
Papers focused on method development have been instrumental in facilitating ADNI research thus far and promise to deliver improvements in reliability, efficiency, and effectiveness in ADNI-GO and ADNI-2. The establishment of standardized protocols that account for problems of variability, both across the multicenter setting of ADNI and longitudinally, has been a primary accomplishment. Likewise, the development of methods for automatic tissue registration and segmentation that avoid the necessity of time-consuming and costly manual segmentation is critical for the analysis of ADNI data. The majority of these approaches are atlas-based, although statistically based registration has also been proposed. Automatic segmentation of the hippocampus, a prominent AD biomarker, poses particular challenges because of its size and location, and several studies have made contributions to the analysis of its volume, shape, and pose. TBM and DBM methods and fractal approaches offer an alternative to volumetric ROI analysis. Methods to allow the classification of patients according to disease status have primarily been based on SVMs and the related RVMs, which are used to build classifiers that can include MRI, FDG-PET, biomarker, APOE ε4, and cognitive data. Finally, statistical methods have been developed to deal with the complexities of the volume and diverse types of data generated by ADNI studies.
In the 2011–2012 year, focus has shifted from the establishment of standardized protocols and methods for automatic segmentation and registration of the hippocampus to other areas including extracting information from cortical thickness data and developing increasingly sophisticated and powerful classification methods that select and combine AD-like features from multiple modalities. Methods to predict future clinical decline have appeared, sometimes in conjunction with classifiers – ‘multi-tasking’ is a recent area of interest in methods development. Another trend has been the use of the ADNI data set, either images or biological materials, as a test set for approaches beyond the bounds of the original ADNI objectives, such as the development of blood-based biomarkers for AD. The importance of GWAS studies in unraveling the genetic contribution to AD is reflected in the publication of methods that capitalize on the underlying interconnectedness of genes with quantitative traits. Finally, finding solutions to ethical problems associated with RCPTs and increasing clinical trial efficiency have been the focus of two reports.
In 2012–2013, significant methodological advances have been made in the areas of MRI segmentation and Florbetapir imaging, multimodal classification, blood-based biomarkers, and genetics. In imaging, major advances include methods that address the four-dimensional segmentation challenges of longitudinal MRI studies and that leverage information contained within relationships between regions and in shape, rather than simple volumetric changes, and studies describing the use of Florbetapir radiotracer introduced into ADNI for amyloid imaging. Multimodal classification has largely evolved beyond identifying optimal combinations of modalities to finding the best way to combine them using feature selection methods and by maintaining data dimensionality. This research has also shifted away from binary disease classification toward probabilistic approaches that view AD as a continuum of disease and the prediction of MCI to AD conversion. The problem of missingness of data in the ADNI data set and its effect on classification accuracies has also been recognized. Some ADNI sections have proposed standardized data sets to circumvent these challenges. Blood- and plasma-based biomarker development has continued at an increasing rate, and this approach shows promise as a less-invasive first-screen clinical option. Software to aid clinicians in diagnosis and to optimize cost-effective use of biomarkers in a clinical environment has been developed and tested. Finally, genetics methods have progressed toward gene-centric approaches to reduce dimensionality.
4. Studies of the ADNI cohort
4.1. Clinical characterization
Central to achieving the goals of ADNI was the recruitment of a study population that mirrors cohorts used in MCI and mild AD trials. Petersen et al [106] presented a baseline and 12-month longitudinal clinical characterization of the ADNI cohort, comprising 229 normal control subjects, 398 subjects with MCI, and 192 subjects with mild AD, and provided clear support for the success of ADNI in this regard. The demographic characteristics of the participant groups, given in Table 3, indicate that the cohort was mostly white and well educated, and that there were a high proportion of APOE ε4 carriers, consistent with populations recruited for clinical trials. At baseline, each study group differed significantly in a range of cognitive measures, with the MCI group intermediate between the control and AD groups in measures of memory impairment and in levels of CSF biomarkers (Table 4). In contrast to AD subjects who were impaired in virtually all cognitive measures, MCI subjects were only mildly impaired in nonmemory cognitive measures. After 12 months, 16.5% of MCI subjects had converted to AD, and a greater increase in the ADAS-cog was seen in the AD group compared with the MCI group. Little change was observed in control subjects. The study also found that baseline Aβ-42 levels were predictive of the progression of clinical measures over 12 months.
Table 3.
Demographic characteristics of ADNI participant groups
| Characteristic | Control subjects (n = 229) | MCI group (n = 398) | AD (CDR: 1.0) group (n = 192) | P value | P < .05* |
|---|---|---|---|---|---|
| Age, mean ± SD, years | 75.8 ± 5.0 | 74.7 ± 7.4 | 75.3 ± 7.5 | .137 | |
| Education, mean ± SD, years | 16.0 ± 2.9 | 15.7 ± 3.0 | 14.7 ± 3.1 | <.001 | b, c |
| Years from symptom onset | Not available | Not available | 3.9 ± 2.5 | NA | |
| % Female | 48.0 | 35.4 | 47.4 | .002 | a, c |
| Marital status, % | .002 | a | |||
| Married | 80.2 | 81.2 | |||
| Widowed | 17.5 | 12.1 | 10.4 | ||
| Divorced | 7.4 | 6.3 | 4.7 | ||
| Never married | 6.6 | 1.5 | 3.6 | ||
| Unknown | 0.4 | 0 | 0 | ||
| APOE ε4, % | <.001 | a, b, c | |||
| Carriers | 26.6 | 53.3 | 66.1 | ||
| Noncarriers | 73.4 | 46.7 | 33.9 | ||
| Ethnicity | |||||
| American Indian | 0 | 0.3 | 0 | ||
| Asian American | 1.3 | 2.3 | 1.0 | ||
| African American | 7.0 | 3.5 | 4.2 | ||
| Hispanic | 0.9 | 3.5 | 2.1 | ||
| White | 90.8 | 90.5 | 92.2 | ||
| Other | 0 | 0 | 0.5 |
NOTE. Reproduced with permission from Ref [106].
Multiple comparisons abbreviated as:
control subjects differ from subjects with AD,
subjects with MCI differ from subjects with AD,
control subjects differ from subjects with MCI.
Table 4.
Baseline assessments of the ADNI cohort
| Control subjects | MCI | AD | |||||
|---|---|---|---|---|---|---|---|
| Assortment variable | Mean SD | Mean SD | Z score MCI–control |
Mean SD | Z score AD–MCI | P value | P < .05* |
| MMSE score | 29.1 ± 1.0 | 27.0 ± 1.8 | −18.8 | 23.3 ± 2.1 | −21.3 | <.001 | a, b, c |
| CDR global score | 0.0 ± 0.0 | 0.5 ± 0.0 | 397 | 0.7 ± 0.3 | 13.4 | <.001 | a, b, c |
| CDR-SB | 0.0 ± 0.1 | 1.6 ± 0.9 | 34.9 | 4.3 ± 1.6 | 21.3 | <.001 | a, b, c |
| Memory | 0.0 ± 0.0 | 0.6 ± 0.2 | 61.3 | 1.0 ± 0.3 | 16.5 | <.001 | a, b, c |
| Orientation | 0.0 ± 0.0 | 0.2 ± 0.3 | 17.5 | 0.8 ± 0.4 | 17.7 | <.001 | a, b, c |
| Judgment | 0.0 ± 0.1 | 0.4 ± 0.3 | 21.8 | 0.8 ± 0.4 | 14.6 | <.001 | a, b, c |
| Community affairs | 0.0 ± 0.0 | 0.2 ± 0.2 | 13.1 | 0.7 ± 0.4 | 16.9 | <.001 | a, b, c |
| Hobbies | 0.0 ± 0.0 | 0.2 ± 0.3 | 15.1 | 0.8 ± 0.5 | 15.9 | <.001 | a, b, c |
| Personal care | 0.0 ± 0.0 | 0.1 ± 0.2 | 4.4 | 0.2 ± 0.4 | 4.3 | <.001 | a, b, c |
| Hachinski score | 0.6 ± 0.7 | 0.6 ± 0.7 | 0.8 | 0.7 ± 0.7 | 0.7 | .418 | NA |
| GDS score | 0.8 ± 1.1 | 1.6 ± 1.4 | 7.3 | 1.7 ± 1.4 | 0.6 | <.001 | a, b |
| FAQ | 0.1 ± 0.6 | 3.9 ± 4.5 | 16.2 | 13.0 ± 6.9 | 16.8 | <.001 | a, b, c |
| ADAS-cog total | 6.2 ± 2.9 | 11.5 ± 4.4 | 18.1 | 18.6 ± 6.3 | 14.0 | <.001 | a, b, c |
| ADAS word list immediate recall | 2.9 ± 1.1 | 4.6 ± 1.4 | 16.8 | 6.1 ± 1.5 | 12.2 | <.001 | a, b, c |
| ADAS word list recognition | 2.6 ± 2.3 | 4.6 ± 2.7 | 10.1 | 6.6 ± 2.8 | 8.2 | <.001 | a, b, c |
| ADAS-cog without word list | 0.8 ± 0.9 | 2.3 ± 2.0 | 12.9 | 5.9 ± 4.1 | 11.4 | <.001 | a, b, c |
| ADAS word list delayed recall | 2.9 ± 1.7 | 6.2 ± 2.3 | 20.8 | 8.6 ± 1.6 | 15.0 | <.001 | a, b, c |
| AVLT trials 1–5 | 43.3 ± 9.1 | 30.7 ± 9.0 | −16.7 | 23.2 ± 7.7 | −10.4 | <.001 | a, b, c |
| AVLT delayed recall | 7.4 ± 3.7 | 2.87 ± 3.3 | −15.6 | 0.7 ± 1.6 | −10.3 | <.001 | a, b, c |
| AVLT DR/trial, 5% | 65.8 ± 27.6 | 32.1 ± 33.1 | −13.9 | 11.2 ± 22.0 | −9.3 | <.001 | a, b, c |
| Trails A | 36.5 ± 13.2 | 44.9 ± 22.8 | 5.9 | 68.0 ± 36.9 | 8.0 | <.001 | a, b, c |
| Trails B | 89.2 ± 44.3 | 130.7 ± 73.5 | 8.8 | 198.9 ± 87.2 | 9.2 | <.001 | a, b, c |
| Category fluency (animal) | 19.9 ± 5.6 | 15.9 ± 4.9 | −9.1 | 12.4 ± 4.9 | −8.1 | <.001 | a, b, c |
| Category fluency (vegetable) | 14.7 ± 3.9 | 10.7 ± 3.5 | −12.7 | 7.8 ± 3.3 | −9.8 | <.001 | a, b, c |
| Number cancellation | 0.4 ± 0.7 | 1.0 ± 0.9 | 8.0 | 1.8 ± 1.3 | 7.6 | <.001 | a, b, c |
| Boston Naming Test | 27.9 ± 2.3 | 25.5 ± 4.1 | −9.4 | 22.4 ± 6.2 | −6.2 | <.001 | a, b, c |
| Digit backward | 7.2 ± 2.2 | 6.2 ± 2.0 | −6.0 | 5.0 ± 1.8 | −7.2 | <.001 | a, b, c |
| Clock drawing | 4.7 ± 0.7 | 4.2 ± 1.0 | −7.6 | 3.4 ± 1.3 | −7.5 | <.001 | a, b, c |
| CSF biomarkers (pg/mL) | (n = 114) | (n = 199) | (n = 102) | ||||
| Tau | 69.7 ± 30.4 | 101.4 ± 62.2 | 6.0 | 119.1 ± 59.6 | 2.4 | <.001 | a, b |
| Aβ–42 | 205.6 ± 55.1 | 162.8 ± 56.0 | −6.6 | 143.0 ± 40.8 | −3.5 | <.001 | a, b, c |
| p-tau181P | 24.9 ± 14.6 | 35.5 ± 18.0 | 5.7 | 41.6 ± 19.8 | 2.6 | <.001 | a, b, c |
Abbreviations: GDS, Geriatric Depression Score; FAQ, Functional Activities Questionnaire; ADAS-cog, cognitive subscale of the Alzheimer’s Disease Assessment Scale; AVLT, Rey Auditory Verbal Learning Test; CDR-SB, clinical dementia rating-sum of boxes.
NOTE. Reproduced with permission from Ref [106].
Multiple comparisons abbreviated as:
control subjects differ from subjects with MCI,
control subjects differ from subjects with AD,
subjects with MCI differ from subjects with AD.
4.2. Medication use
Medication use among the ADNI cohort was investigated by Epstein et al [107]. They found a high rate of polypharmacy, with 85% of participants taking more than four medications, the average being eight (SD = 4). Moreover, 22% of participants reported taking one or more Beers list medications deemed to be potentially dangerous in the elderly population. The most common medications for symptomatic treatment of AD or MCI were the cholinesterase inhibitor donepezil and the N-methyl-d-aspartate partial receptor agonist memantine, which were frequently taken as a combination therapy. Despite the lack of FDA approval for use of these drugs to treat MCI, donepezil, memantine, and other cholinesterases were commonly used by MCI patients. Women, less educated, and more elderly participants were less likely to receive treatment. Schneider et al [108] focused on the use of cholinesterase inhibitors and memantine in the ADNI cohort. They found that 44% of MCI patients and 85% of mild AD patients were treated with cholinesterase inhibitors, and that 11% of MCI patients and 46% of mild AD patients were treated with memantine. In both patient groups, use of these medications was associated with increased cognitive impairment at baseline, a higher rate of clinical decline over 2 years, and a more rapid progression to dementia in MCI patients. Cholinesterase inhibitors and memantine appeared to be more frequently prescribed to patients diagnosed as having MCI due to AD, despite a lack of evidence from clinical trials and lack of FDA approval for this treatment. The authors suggested that use of these medications may affect the interpretation of clinical trial outcomes. Medication has been implicated in an increased risk of falls in the elderly. Epstein et al [429] investigated the association between medications in mild MCI (memantine and cholinesterase inhibitors) and falls in the ADNI cohort. They found that the use of these medications increased the hazard of fall by approximately 63%.
4.3. Baseline and longitudinal studies of biomarker changes during disease progression
ADNI has afforded a unique opportunity to examine biomarker changes that occur during disease progression in a large, well-defined cohort. Using MRI, CSF, 11C-PiB PET, and FDG-PET data, cross-sectional and longitudinal studies focused either on evaluating spatial pattern and regional rates of atrophy or on characterizing biomarkers for varying disease stages have together resulted in a more detailed and coherent picture of this complex process.
4.3.1. Magnetic resonance imaging
A cross-sectional study by Fennema-Notestine et al [109] examined the feasibility of high-throughput image analysis to detect subtle brain structural changes in the early stages of AD. They further divided the MCI group, based on neuropsychological performance, into single-domain and multidomain groups, which they proposed represented earlier and later stages in disease progression, respectively. Using comparisons of cortical thickness, they found a pattern of progressive atrophy from normal control subjects to single-domain MCI subjects, to multidomain MCI subjects, and finally to subjects with AD (Fig. 7). When ROIs were examined, they found that the regions that differed between the control group and the single-domain MCI group included not only the hippocampus and entorhinal cortex, which had the largest effect sizes, but also other temporal regions, the temporal horn of the lateral ventricle, rostral posterior cingulate, and several parietal and frontal regions. Relative to control subjects, multidomain MCI patients had greater differences in the same regions as well as in the lateral inferior, middle, and superior temporal gyri and fusiform cortices. Additional atrophy was seen in AD patients relative to control subjects in the inferior parietal, banks of the superior temporal sulcus, retrosplenial, and some frontal regions. Similar results were reported in a cross-sectional study by Karow et al [110], who found a pattern of atrophy spreading from the mesial temporal lobe in MCI patients to widespread areas in AD patients patients and by Asku et al [240] who differentiated between MCI converters and non-converters and found the most active areas of degeneration in converters tended to lie in the parietal and temporal cortex, whereas those areas discriminating between AD and control patients included occipital and frontal regions.
Fig. 7.
Group differences in average thickness (mm) for left hemisphere. Top row: NC vs. SMCI; middle row: normal controls (NC) vs. MMCI; bottom row: NC vs. AD. Left mesial views, right lateral views. The scale ranges from < −0.3 (yellow) to > +0.3 (cyan) mm thickness. Areas on the red-yellow spectrum indicate regions of thinning with disease: approximate color scale in mm is −0.05 to −0.15 dark red, −0.20 bright red, −0.25 orange, and < −0.30 yellow. For thicker regions: +0.05 to +0.15 blue. Any differences smaller than ± 0.05 mm are gray. Reproduced with permission from Ref [109].
An emerging view of disease progression is that spatial patterns of structural changes are coordinated with distributed cognitive networks. Carmichael et al [430] identified data-driven groupings of cortical regions that exhibited highly correlated rates of atrophy in amnestic MCI patients over 2 years. Conversion to AD was associated with groupings that included sections of the default-mode network and the prefrontal and medial-temporal structures, suggesting that multiple coherent modes of longitudinal brain atrophy may act simultaneously in the early stages of the AD pathologic process, with each grouping corresponding to a distinct biological substrate.
Fennema-Notestine et al [109] also explored the trajectories of change of ROIs over the course of the disease and found that although some regions, such as mesial temporal regions, exhibited a linear rate of atrophy through both MCI stages to AD, other regions, such as the lateral temporal middle gyrus, retrosplenial cortex, inferior parietal cortex, and rostral middle frontal cortex, exhibited accelerated atrophy later in the disease.
The idea that rates of change of atrophy are not uniform but vary by disease stage is supported by several studies. When MCI groups were classified according to subsequent clinical outcome, Leung et al [59] found higher rates of hippocampal atrophy in MCI-c than MCI-nc patients. McDonald et al [111] examined regional rates of neocortical atrophy in the ADNI cohort, dividing MCI subjects into two groups by their CDR-SB scores. The less impaired MCI group had CDR-SB scores of between 0.5 and 1.0, whereas the more impaired group had CDR-SB scores of between 1.5 and 2.5 (AD subjects had CDR-SB scores of >2.5). They found that over the course of disease progression, atrophy changed from the medial and inferior lateral temporal, inferior parietal, and posterior cingulate cortices initially, to the superior parietal, prefrontal, and lateral occipital cortices, and finally to the anterior cingulate cortex (Fig. 8). Moreover, the rates of change differed among the three groups. The least impaired MCI patients showed the greatest rates of atrophy in the medial temporal cortex, whereas later in disease progression, rates of atrophy were higher in the prefrontal, parietal, and anterior regions. Similar patterns were found by several other groups using a range of MRI methods. Hua et al [112] and Leow et al [113] both used TBM to create 3-D maps of structural changes over 12 months. Risacher et al [114,115] examined a variety of structural MRI markers for their sensitivity to longitudinal change and clinical status using multiple methods, including VBM and ROIs, whereas Schuff et al [116] focused on changes in hippocampal volume, and McEvoy et al [117] calculated an atrophy score based on ROIs most associated with AD atrophy. A more recent comprehensive analysis of longitudinal ADNI MRI data by Leung et al [431] focused on global cerebral atrophy, hippocampal atrophy, and ventricular expansion over 6-month intervals to 3 years. They found expected between-group differences in atrophy rates and an acceleration of rates of ventricular enlargement in both MCI and AD patients of 0.27 and 0.88 mL/y, respectively. An acceleration of hippocampal atrophy rates in MCI patients of 0.22% was also observed, which appeared to be primarily driven by MCI patients who later converted to AD. This rate of acceleration is small compared with the actual rate of atrophy in MCI patients (around 3.5%), suggesting that the transition to pathologic losses observed in AD occurs slowly. Collectively, these studies showed atrophy spreading from the MTL to the parietal, occipital, and frontal lobes over the course of the disease, with MCI patients, in general, having a more anatomically restricted AD-like pattern of change. MCI subjects who converted to AD within the time frame of the study (MCI-c) had a more AD-like pattern of atrophy, and nonconverters (MCI-nc) had a pattern more intermediate between control and AD subjects (Fig. 9). Several studies [114,115,118,119] divided the MCI group into those patients who converted to AD within a year and those who remained stable. Each group had distinct profiles when assessed using a score derived from patterns of structural abnormality, the future converters having mostly positive scores that reflected a largely AD-like pattern of brain atrophy. Conversely, the distribution of abnormality scores in the MCI-nc group was bimodal, reflecting the heterogeneity of this group that appears to contain some members who, with abnormality scores close to those of AD patients, are likely to convert in the near future.
Fig. 8.
Annual atrophy rates as a function of degree of clinical impairment. Clinical impairment measured using baseline clinical dementia rating-sum of boxes (CDR-SB) scores. Mean atrophy rates are represented as a percent change in neocortical volume and mapped onto the lateral (left), ventral (middle), and medial (right) pial surface of the left hemisphere. These data demonstrate that atrophy rates are most prominent in posterior brain regions early in the course of disease, spreading to anterior regions as the level of impairment increases, with relative sparing of sensorimotor regions. Reproduced with permission from Ref [111].
Fig. 9.
Distribution of atrophy scores used to classify subjects with MCI. MCI atrophy score was derived from LONI data archive trained on data from all control subjects and subjects with AD. Discriminant model assumed equal prior group probabilities. Individuals were classified as having control phenotype if their scores were above −0.33. Cutoff score was chosen to maximize overall accuracy of classifying control subjects and subjects with AD on whom this model was trained. Average atrophy score for subjects with MCI was −0.50. Atrophy score is not normally distributed (Kolmogorov–Smirnov test = 0.73, df = 175, P =.025) but shows evidence of bimodal distribution. Reproduced with permission from Ref [117].
The highest rates of change occurred in AD subjects and MCI-c patients in measures of hippocampal volume and entorhinal cortex thickness [115,120]. Schuff et al [121] found that atrophy was detectable at 6 months and accelerated with time to 12 months in MCI and AD subjects, with the highest rates of atrophy seen in AD patients (Fig. 10). Hua et al [120] used TBM to examine the effects of age and sex on atrophic rates and found that the atrophic rates of women were 1% to 1.5% higher than for men. They also observed a 1% increase in atrophic rate and a 2% increase in ventricular expansion for every 10-year decrease in age, with correlations strongest in the temporal lobe.
Fig. 10.
Individual trajectories of hippocampal volume change. Thick black lines indicate the mean trajectory change of each group. Reproduced with permission from Ref [121].
A different data-driven approach to determining the time course of brain volume changes in healthy elderly, MCI, and AD subjects without using a priori models was taken by Schuff et al [116]. Using generalized additive models to analyze serial MRI scans over 30 months, they found that atrophy rates varied nonlinearly with age and cognitive status, most noticeably in temporal regions, and that atrophy tended to level off in control and MCI-nc subjects, but decline further in MCI-c and AD patients. The authors suggest that these differences are a reflection of the different processes involved in healthy versus disease-related neurodegeneration. The regions with the greatest effect sizes between young control and AD subjects were the entorhinal cortex, the hippocampus, and the lateral ventricles, suggesting that rates of change in these regions have potential as biomarkers for the early detection of AD.
Beyond simple volumetric analysis, one approach to analyzing brain morphometric changes in greater detail has been to assess changes in shape of ROIs. Qiu et al [122] used large deformation diffeomorphic metric mapping to reveal that the anterior of the hippocampus and the basolateral complex of the amygdala had the most surface inward deformation in MCI and AD patients, whereas the most surface outward deformation was found in the lateral ventricles (Fig. 11). These results are in agreement with the volumetric findings of Apostolova et al [123] and also with many findings documenting the enlargement of the lateral ventricles with disease progression. Greene et al [267] examined atrophy of sub-regions of the hippocampus over time and found that the head appears to be initially affected followed by the body and tail of the structure. Cash et al [385] examined changes in cortical folding patterns throughout disease progression, using several metrics to represent the degree of cortical folding, curvature, and shape. Stable MCI, MCI-c, and AD patient groups had a progressively lower degree of cortical folding and a progressive reduction in the depth and area of sulcal folds, suggesting a widening and flattening of these areas due to neurodegeneration.
Fig. 11.
Group differences in regional shape deformations. Abbreviations: Am, amygdala; Hp, hippocampus; V, ventricles; iLV, inferior lateral ventricles; Cd, caudate; Pu, putamen; Pa, globus pallidus; Th, thalamus. Reproduced with permission from Ref [122].
Disease progression appears to be influenced by other factors such as genotype, gender and age differences. The influence of the APOE ε 4 allele on GM loss in MCI patients was investigated by Spampinato et al [268] who found greater atrophy in a variety of regions including the hippocampus, temporal and parietal lobes and insulae in MCI converters who were carriers compared to non-converter carriers. GM loss was greatest in the first 12 months, supporting the idea of non-linearity of atrophy throughout disease progression. Furthermore, they found no difference in cognitive decline between carriers and non-carriers of the APOE ε 4 allele, suggesting that accelerated hippocampal and neocortical atrophy did not completely account for the cognitive deterioration in this study. Skup et al [269] examined longitudinal atrophy in selected ROIs to look for sex-specific patterns of atrophy. They found that female MCI and AD patients differed from controls in right caudate nucleus atrophy, that between MCI and AD patients, there were female-specific differences in insula and amygdala atrophy and male specific differences in the atrophy of the left precuneus, and that sex differences tended to be bilateral in MCI patients and side-specific in those with AD. These results suggest that disease progression has gender differences that may be more widespread during the MCI stage. The question of whether disease progression is a set process over a range of ages was examined by Stricker et al [270] who compared changes in cognition and brain morphometry in the young old (ages 60–75) and the very old (ages > 80 years) compared to age-matched controls. In the very old group, there was comparatively less atrophy in a number of regions and less impairment in a number of cognitive domains than in the young-old group, likely a reflection of normal age-related changes in the control group. Conversely, atrophy of the hippocampus and MTL substantially eclipsed these age-related changes and remained salient markers of AD, regardless of age. Rates of clinical decline and brain atrophy decreased with age in controls, MCI patients, and AD patients but increased with age in the normal elderly [432]. Consequently, rates of change of many measures converged at a point close to 85 years. Baseline clinical measures did not differ by age, implying that the association between neuropathologic markers and AD attenuates with age and that in the oldest old, AD can be difficult to distinguish from normal aging.
While aging is associated with cognitive decline, improvements in cognition are well documented in MCI and even mild-AD patients, suggesting that cognition in aging is a dynamic process. Song et al [433] studied changes in cognition (MMSE and ADAS-cog) and structural brain changes (brain and lesion index [BALI] and Medial Temporal Atrophy Score [MTAS]) over 2 years. Although cognition and brain structure declined overall, increases in both the BALI score and cognitive improvements were observed in some patients. Two-thirds of patients who showed structural improvement on BALI also showed cognitive improvement, and no MCI patient with structural improvement converted to AD. These results suggest that brain aging is a dynamic process in which improvements in both structural and cognitive aspects may be observed despite the fact that decline dominates overall.
4.3.2 CSF biomarkers
In the first longitudinal study of CSF biomarkers in ADNI cohort, Toledo et al [434] investigated biomarker dynamics and trajectories in controls, MCI patients, and AD patients for up to 4 years. The longitudinal stability of Aβ42, p-tau181, and t-tau varied in patients with normal baseline levels of biomarkers: one group had stable biomarker levels, whereas another had decreasing Aβ42 and increasing p-tau181 levels over time. When the stable population was excluded from analysis, the time taken to reach cut point levels of biomarkers was significantly shortened, with changes in p-tau181 occurring before changes in Aβ42. Furthermore, whereas low baseline Aβ42 predicted greater increases in p-tau181 levels, the converse was not true, implying that changes in Aβ42 levels precede those in p-tau181 levels (Fig. 33).
Fig. 33.
Estimated trajectories of Aβ-42 (A) and p-tau181 (B) based on ADNI longitudinal data. The estimated time to reach the AD cut point threshold is indicated based on a model that includes all subjects (blue) or subjects with abnormal baseline values or changes during follow-up (red). From Toledo et al [434].
4.3.3. PET
Toussaint et al [399] identified hypometabolic patterns in the areas constituting the default mode network known to be involved in memory processing and found that these patterns evolve in a specific manner with disease progression. Leveraging newly differentiated EMCI and LMCI cohorts from ADNI-GO and ADNI-2, Wu et al [435] studied the dynamics of amyloid deposition and its relationship to glucose hypometabolism in MCI patients. AV45 retention increased from control to EMCI to LMCI groups in diffuse areas, but no further amyloid burden was detected in AD patients. In contrast, FDG-PET indicated that hypometabolism was essentially unchanged in EMCI patients compared with controls but then increased sequentially in LMCI and AD patients in a variety of regions. These observed dynamics are in accordance with the model by Jack et al [15].
4.3.4. Cognitive
Johnson et al [436] examined baseline and longitudinal change over 36 months of five cognitive factors (memory, executive function/processing speed, language, attention, and visuospatial awareness) in control and MCI patients. In the cognitively normal elderly, memory was the sole measure observed to decline over the study period, whereas all measures declined in MCI patients. Interestingly, executive function declined more rapidly than memory in these patients suggesting that different patterns of cognitive change exist in MCI compared with normal cognition. Executive function may therefore play an important role in distinguishing MCI from cognitively normal patients.
4.4. Associations between characteristics of the ADNI cohort
A major area of focus in research using ADNI data has been the elucidation, both at baseline and longitudinally, of associations between various imaging, CSF, genetic, and clinical correlates in different clinical groups to gain a better understanding of the interplay of biomarkers throughout disease progression.
4.4.1. Magnetic resonance imaging
4.4.1.1. Temporal lobe
Structures within the temporal lobe have long been associated with AD decline because of their critical role in the formation of long-term memory, one of the first functions to be affected in disease progression. Leow et al [113] found temporal lobe atrophy to be associated with increased cognitive impairment in MCI patients, as indicated by changes in CDR, MMSE scores, and the AVLT (Fig. 12). Among the structures of the temporal lobe, hippocampal atrophy is the best studied structural biomarker, as it is one of the earliest structures to degenerate in AD. In a small initial study, Morra et al [63] found that bilateral hippocampal atrophy at baseline was strongly correlated with both MMSE and CDR-SB (Table 5). A further larger study by the same group [124] examined rates of hippocampal atrophy over 12 months and found that these correlated with both baseline cognitive scores on MMSE and global and sum of boxes CDR and with longitudinal change in these measures (Table 5). Wolz et al [64] also revealed significant correlations between rates of hippocampal atrophy and both baseline MMSE and CDR, and changes in these measures over 12 months (Table 5). Additionally, a study by Schuff et al [121] found that rates of change of MMSE and ADAS-cog were associated with rates of hippocampal atrophy (Table 5). Using TBM, Hua et al [73] found that baseline temporal lobe atrophy was associated with both baseline and change in the CDR-SB in MCI and AD patients, but with change in the MMSE only in the AD group, providing further evidence for the acceleration of atrophic change with disease progression.
Fig. 12.
Cumulative distribution function (CDF) plots for voxelwise correlation of progressive temporal lobe tissue loss in MCI, AD, and pooled groups. (A) Correlations with various biomarker indices, including Aβ-42 (AB142), tau protein (TAU), phosphorylated-tau 181 (PTAU), tau/Aβ-42 ratio (TAUAB), and p-tau/Aβ-42 ratio (PTAUAB), and (B) correlations with various clinical measures. Reproduced with permission from Ref [113].
Table 5.
Associations between biomarker and clinical measures in ADNI cohort (correlation coefficients)
| Clinical correlates | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biomarker | N | Clinical group | MMSE | ΔMMSE | CDR-SB | ΔCDR-SB | ADAS-cog | ΔADAS-cog | LM-II-DR | AVLT | TMT A and B | Reference | |
| Hippocampal volume (L/R) |
21 | Pooled sample | 0.423*/0.529† | −0.369*/−0.705† | [63] | ||||||||
| 12-month hippocampal atrophy rate (L/R) |
490 | Pooled sample | −0.191†/−0.168† | 0.117†/0.136† | 0.173†/0.181† | −0.174†/−0.171† | [124] | ||||||
| 12-month hippocampal atrophy rate (L/R) |
555 | Pooled sample | −0.52‡/−0.43‡ | 0.36‡/0.30‡ | 0.47‡/0.38‡ | −0.27‡/−0.21‡ | [64] | ||||||
| Hippocampal radial distance (L/R) |
245 | MCI | −0.20†/−0.17† | 0.24§/0.31§ | 0.27§/0.25§ | [123] | |||||||
| 98 | AD | −0.21*/NS | NS/NS | 0.21*/NS | |||||||||
| Hippocampal atrophy rate |
498 | Pooled sample | 0.18* | [121] | |||||||||
| 607 | MCI | Learning | Retention | [140] | |||||||||
| 0.36‡ | 0.37‡ | ||||||||||||
| Cortical thickness | |||||||||||||
| Entorhinal | 0.33‡ | 0.33‡ | |||||||||||
| Parahippocampal | 0.22‡ | 0.23‡ | |||||||||||
| Frontal caudal middle |
0.16‡ | NS | |||||||||||
| Rostral middle | 0.23‡ | 0.16c | |||||||||||
| Lateral orbitofrontal | 0.16‡ | NS | |||||||||||
| Inferior parietal | 0.24‡ | 0.17‡ | |||||||||||
| Precuneus | 0.25‡ | 0.16‡ | |||||||||||
| Cortical thickness (L/ R) |
536 | Pooled sample | |||||||||||
| Posterior cingulate | 0.14‡/0.13† | 0.22†/0.19† | [138] | ||||||||||
| Caudal middle | NS/NS | 0.17†/0.15† | High EF: TM A + B Low EF: AVLT |
||||||||||
| Rostral middle | 0.30†/0.13‡ | 0.18†/0.21† | |||||||||||
| Superior frontal | NS/0.13† | 0.16†/0.17† | |||||||||||
| Operculum | NS/NS | 0.14†/0.16† | |||||||||||
| Lateral bifrontal | NS/NS | 0.15†/NS | |||||||||||
| Frontal polar | NS/NS | 0.17†/NS | |||||||||||
| STAND score | 399 | Pooled sample | −0.50‡ | 0.59‡ | [132] | ||||||||
| 192 | MCI | −0.19† | 0.26‡ | ||||||||||
| 98 | AD | −0.29† | 0.34‡ | ||||||||||
| Aβ–42 | 399 | Pooled sample | 0.31‡ | −0.37‡ | |||||||||
| 192 | MCI | NS | NS | ||||||||||
| 98 | AD | NS | NS | ||||||||||
| Caudate volume | 400 | Pooled sample | 0.175* | −0.209* | [130] | ||||||||
| Hippocampal volume [130] |
0.349* | −0.365* | |||||||||||
| Ventricular volume | −0.205* | 0.225* | |||||||||||
| Predicted scores from whole brain gray matter volumes |
586 | Pooled sample | [91] | ||||||||||
| MMSE | 0.47‡ | ||||||||||||
| ADAS-cog | 0.49‡ | ||||||||||||
| AVLT | NS | ||||||||||||
| Biomarker | N | Clinical group | Clinical correlates | ||||||||||
| MMSE | ΔMMSE | CDR-SB | ΔCDR-SB | FAQ | ADAS-cog | LM-II-DR | AVLT | TMT A and B | |||||
| FDG 12 months ROI decline in CMRgl |
154 | MCI | 0.22† | −0.19* | NS | [104] | |||||||
| 69 | AD | NS | −0.25* | NS | |||||||||
| FDG-PET regional- to-whole brain CMRgl (L/R) |
[134] | ||||||||||||
| Posterior cingulate | 298 | Pooled sample | NS | −0.47§/NS | |||||||||
| Precuneus | 0.36§/0.37§ | −0.46§/−0.49§ | |||||||||||
| Parietal | 0.26§/0.36§ | −0.42§/−0.47§ | |||||||||||
| Temporal | 0.43§/0.32§ | −0.41§/−0.41§ | |||||||||||
| Frontal | 0.23§/0.22§ | −0.24§/−0.26§ | |||||||||||
| Medial temporal | NS | −0.36§/−0.41§ | |||||||||||
| Occipital | 0.31§/0.22§ | −0.37§/−0.26§ | |||||||||||
| ΔPiB uptake | 61¶ | Pooled sample | −0.22 (P = .09) | NS | [16] | ||||||||
| Ventricular expansion |
−0.52† | −0.42† | |||||||||||
| 161 | Pooled sample | Learning | Recognition | [136] | |||||||||
| Hippocampal volume |
0.35§,** | 0.34§,** | |||||||||||
| Parahippocampal complex volume |
NS | 0.17*,** | |||||||||||
| Precuneus cortical thickness |
0.22†,** | NS | |||||||||||
| Inferior parietal lobe metabolism |
0.23†,** | NS | |||||||||||
| Hippocampal metabolism |
0.15*,** | 0.25§,** | |||||||||||
| APOE genotype | NS | 0.14*,** | |||||||||||
| FDG-ROIs baseline | 95 | AD | NS | −1.95†,** | [135] | ||||||||
| ΔFDG-ROIs | −1.21†,** | −3.25‡,** | |||||||||||
| FDG-ROIs baseline | 208 | MCI | −0.88‡,** | −0.66†,** | |||||||||
| ΔFDG-ROIs | NS | −1.08‡,** | |||||||||||
| Hypometabolic convergence index |
188 | Pooled sample | −0.48 | 0.54 | 0.53 | −0.43 | 0.45 | [85] | |||||
| 50 | MCI | [155] | |||||||||||
| Hippocampal volume |
NS | NS | 0.41* | ||||||||||
| Retrosplenial volume | −0.42* | −0.43* | NS | ||||||||||
| Retrosplenial metabolism |
0.47* | NS | NS | ||||||||||
| Entorhinal metabolism |
0.38* | NS | NS | ||||||||||
| Δ LM-II | Boston Naming |
Category fluency |
TMT B | [142] | |||||||||
| Right medial lobe atrophy rate |
−0.41‡,†† | ||||||||||||
| Left entorhinal cortex |
0.47‡,†† | ||||||||||||
| Left lateral lobe thinning |
0.31‡,†† | ||||||||||||
| Left temporal lobe atrophy rate |
0.38‡,†† | ||||||||||||
| Left frontal lobe —pars orbitalis |
0.33‡,†† | ||||||||||||
| Learning | Delayed recall | [110] | |||||||||||
| Hippocampal volume |
156 | MCI | −0.388 | 0.279 | 0.459 | ||||||||
| Entorhinal metabolism |
−0.406 | 0.175 | 0.318 | ||||||||||
| Hippocampal subregions |
120 | CN, MCI, AD (40 per group) |
Right entorhinal cortex |
Left entorhinal cortex |
Clock command | Boston naming | AVLT | ||||||
| Head | 0.614‡ | 0.628‡ | 0.316† | 0.424‡ | 0.460‡ | ||||||||
| Body | 0.547‡ | 0.494‡ | 0.281† | 0.374‡ | |||||||||
| Tail | 0.317‡ | 0.366‡ | 0.269† | 0.211* | |||||||||
Abbreviations: ROI, region of interest; CMR-gl, cerebral metabolic rate for glucose; TMT, Trail Making Test; NS, not significant.
P < .05.
P < .01.
P < .001.
P < .0001.
Includes 23 subjects from the Mayo Clinic Study of Aging.
β values from regression model.
Figures from a mixed effect model that examined baseline level and longitudinal change as independent variables as predictors of change.
The relationships between hippocampal volume and memory retention were examined by Apostolova et al [123], who found that MCI patients had bilateral associations between hippocampal volume and radial distance and three tests of delayed recall (DR): ADAS-cog-DR, AVLT-DR, and the Wechsler Logical Memory Test II-DR, whereas associations between these tests in AD patients were stronger in the left hippocampus both at baseline and at the 12-month follow-up (Table 5). In addition, they found highly significant regional associations for memory performance, especially in the CA-1 subregion and the subiculum on the anterior hippocampal surface. Greene et al [267] examined the relationship between subregions of the hippocampus and neuropsychological measures and atrophy in other regions. Most cognitive decline measures were correlated most strongly with the hippocampal head, a subregion that includes the histologically defined CA1 and CA3 subfields (Table 5). With both cognitive and volumetric measures, the strength of association diminished from the head to the body to the tail of the hippocampus (Table 5). In cognitively normal participants, Carmichael et al [437] reported that localized atrophy in several hippocampal subregions, and not whole hippocampal atrophy, was significantly associated with CSF measures of Aβ and tau. In contrast, both total hippocampal volume and various subregional measures were significantly associated with a wide range of neurocognitive measures included in the ADNI neuropsychological battery. These results suggest that amyloid and tau abnormalities may be associated with atrophy in specific regions of the hippocampus in asymptomatic patients, whereas changes in cognition occur later in disease and are related to overall volumetric changes in the hippocampus.
Associations between temporal lobe degeneration and memory performance (Wechsler Memory Scale-Revised—Logical Memory, immediate recall and DR) were also found by Hua et al [73].
Along with hippocampal atrophy, ventricular expansion is a hallmark of brain morphometric changes that occur during AD progression and has great potential as a structural biomarker, as the lateral ventricles are comparatively easy to measure, because of their high contrast under MRI, and are highly sensitive to disease progression. Evans et al [125] found that ventricular expansion differentiated between patient groups was associated with ADAS-cog scores in AD patients, and that MCI-c patients had higher rates of ventricular expansion than MCI-nc patients. Chou et al [126] automatically mapped ventricular geometry and examined correlations between surface morphology, clinical decline, and CSF biomarkers. They found that ventricular enlargement at baseline correlated with diagnostic group, depression severity, both baseline and rates of change of cognitive function (MMSE and CDR-SB), and lower CSF Aβ-42. In a subsequent study by the same group [127] using automated radial mapping to generate statistical maps, ventricular enlargement was found to correlate with a large number of measures of clinical decline as well as with lower levels of CSF Aβ-42 and the APOE ε4 allele (Fig. 13). Chou et al [126] also noted expansion of the posterior regions of the ventricles in MCI patients and in the frontal regions of the superior horns in AD patients compared with control subjects, suggesting a topographic sequence of morphometric change throughout disease progression.
Fig. 13.
Apolipoprotein E (APOE) gene effects on regional brain volumes. Maps show the mean percent differences in regional brain volumes for four different group comparisons. Percent differences are displayed on models of the regions implicated: (A) ventricular cerebrospinal fluid (CSF), (B) sulcal CSF, (C) hippocampi, and (D) temporal lobes; dotted lines show the boundary of the hippocampus. Reproduced with permission from Ref [112].
The relationship between hippocampal atrophy and regional neocortical thinning was investigated by Desikan et al [163] who sought to determine whether disruptions to the medial temporal lobe and heteromodal association areas, shown to preferentially accumulate amyloid plaques and neurofibrillary tangles, contribute to a functional isolation of the hippocampus. In all patients, the strongest associations with hippocampal atrophy were with thinning of regions in the temporal lobe. However, additional associations were identified in control and MCI, but not AD patients within the occipital, frontal and parietal cortices.
The studies of Morra et al [124], Wolz et al [64], Hua et al [112], and Risacher et al [115] all found that carriers of the APOE ε4 allele had higher rates of hippocampal atrophy than noncarriers. In contrast, Schuff et al [121] found that increased rates of hippocampal atrophy were associated with APOE ε4 in the AD, but not MCI or control, group. Using Structural Abnormality Index (STAND) scores to reflect the overall level of AD-like anatomic features, Vemuri et al [128] also found that the APOE ε4 allele contributed to MRI atrophy. Hua et al [112] found that the APOE ε4 allele had a dose-dependent detrimental risk with greater atrophy in the hippocampus and temporal lobe in homozygotes than heterozygotes in MCI and AD groups (Fig. 13). Hostage et al [438] quantified the effect of the APOE ε 4 and APOE ε 2 alleles on hippocampal volume across the disease spectrum, reporting that the APOE ε 4 allele had a dose-dependent effect on MCI and AD patients equating to an approximate loss of 4% volume per allele below mean hippocampal volume. Cognitively normal participants were unaffected and APOE ε 2 appeared to confer a modest protective effect.
The recently identified AD risk allele GRIN2b was associated with higher rates of temporal lobe atrophy in the pooled group, but more weakly than APOE ε4 [120]. Other thus far unidentified genetic risk factors likely contribute to AD, with epidemiological studies suggesting maternal history of the disease increases the risk of developing AD. Andrawis et al [129] examined the influence of maternal history of dementia on hippocampal atrophy and found smaller baseline and 12-month follow-up hippocampal volumes in MCI patients with maternal, but not paternal, history. APOE ε4-positive patients also had decreased hippocampal volumes, regardless of parental history. These results suggest the involvement of maternally inherited genetic material, encoded on either the X chromosome or mitochondrial genome. The latter may be more likely, given that decline in mitochondrial function has been found to lead to increased generation of reactive oxygen species, enhanced apoptosis, cell loss, and brain atrophy [129].
4.4.1.2. Other ROIs
Although the caudate has not been the subject of intensive AD research, it plays a crucial role in the formation of new associations required for the acquisition of explicit memories. Madsen et al [130] found that baseline caudate atrophy was associated with a number of clinical and biochemical measures, including, most strongly, body mass index (BMI), in the AD group alone and in the pooled sample, and CDR-SB and MMSE scores at baseline (Table 5). There appeared to be preferential right caudate atrophy in AD patients, and the authors proposed that caudate atrophy might function as a complementary biomarker to other structural measures. The inferior parietal lobe (IPL) is involved in sensory and motor association and possibly comprises part of the memory circuitry. Greene and Killiany [131] examined the associations between subregions of the IPL (gyrus, banks, and fundus) and cognitive measures in control, MCI, and AD subjects. They found that compared with control subjects, MCI patients differed only in the thickness of the banks of the left IPL, a change that correlated with decreased scores in the AVLT-DR, whereas AD patients had significant morphometric changes in all subregions of the right IPL. These results suggest a temporal sequence of changes during disease progression, with atrophy beginning in the left IPL and spreading to the right.
Like the IPL and caudate, the role of the amygdala in AD has received comparatively little attention despite postmortem evidence to suggest that atrophy is similar to that observed in the hippocampus. Poulin et al [271] found a similar degree of atrophy in both structures in patients with early AD. They also found that amydalal atrophy had a comparable association with decline in the MMSE but a weaker association with decline in the CDR-SB than hippocampal atrophy (Table 5), suggesting that cognitive changes in mild AD may be caused by atrophy of both these MTL structures.
4.4.1.3. Multiple ROIs and whole brain studies
Other MRI studies have used approaches based on the whole brain or multiple ROIs, rather than specific ROIs. Evans et al [125] examined brain atrophy rates using the brain BSI technique and found atrophy to be associated with MMSE and ADAS-cog scores in MCI and AD patients. Within the MCI group, they found greater rates of change, in a range similar to that observed in the AD group, in subjects who converted to AD within the time frame of the study. Stonnington et al [91] found that whole brain GM at baseline predicted baseline scores on the ADAS-cog and, MMSE, but not on the AVLT (Table 5). Similarly, Zhang et al [272] used the Brain and Lexion Index (BALI), a score summarizing brain structural changes in aging, to assess changes in cognition throughout disease progression and found that it correlated significantly with baseline MMSE (β = −0.310, P = .008) and 2 year follow-up MMSE (β = −0.725, P = .0010 and ADAS-cog scores at baseline (β = 0.612, P = .013) and at follow-up (β = 0.126, P = .003). The latter is a more specific test of memory, and the authors suggest that whole brain methods may be preferentially more highly sensitive to tests, unlike the AVLT, that involve diverse brain regions. Vemuri et al [132] used STAND scores as a measure of the degree of AD-like anatomic features to assess correlations between brain morphometric changes and cognitive scores, and found that STAND scores were highly correlated with CDR-SB and MMSE scores in individual groups and the pooled sample (Table 5). These studies lend support for atrophy of the whole brain or multiple ROIs as biomarkers, based on their ability to differentiate between patient groups and healthy control subjects, and to track disease progression and clinical decline.
In addition to memory loss, AD is commonly associated with other neuropsychiatric symptoms such as depression, apathy, agitation, and aggression. Trzepacz et al [439] reported that frontolimbic ROIs involved in the salience network were associated with greater severity of agitation and aggression in MCI and AD patients, suggesting that AD may disrupt behavioral control via modulation of this neural network. Zahodne et al [440] examined the associations between both apathy and depression and longitudinal regional cortical atrophy in MCI patients. Apathy was not predictive of longitudinal cortical atrophy, but depression was associated with both baseline entorhinal cortex thickness and increased longitudinal atrophy in the anterior cingulate cortex. These results are consistent with hypotheses regarding the relationship between depression and AD: either depression is indicative of increased neurodegeneration or depression lowers cognitive reserve, which in turn allows more rapid progression of AD neuropathology.
A measure derived from a multidimensional scaling method for quantifying shape differences using DBM [75] had a strong inverse correlation with the MMSE (r = −0.53), although the findings were limited by small sample size. Using the related method of TBM, Ho et al [133] created regional maps of changes in brain tissue and used the resulting Jacobian values to represent brain tissue excess or deficit relative to a template. They found that lower brain volume in the frontal, parietal, occipital, and temporal lobes was associated with higher BMI in MCI and AD patients, and that ventricular expansion correlated with higher BMI in AD, but not MCI, patients (Fig. 14). Every unit increase in BMI was associated with a 0.5% to 1.5% decrease in brain volume in patients of the ADNI cohort.
Fig. 14.
Association of regional brain tissue volumes with body mass index. These represent the estimated degree of tissue excess or deficit at each voxel, as a percentage, for every unit increase in body mass index, after statistically controlling for the effects of age, sex, and education on brain structure. Images are in radiological convention (left side of the brain shown on the right) and are displayed on a specially constructed average brain template created from the subjects within each cohort (mean deformation template). Reproduced with permission from Ref [133].
Elevated levels of homocysteine, a risk factor for AD, are associated with cortical and sub-cortical atrophy and may promote the magnitude of atrophy in the brain. Rajagopalan et al [273] found that elevated homocysteine levels (> 14µM) was significantly associated with atrophy in frontal, parietal and occipital WM irrespective of disease status and in the MCI group alone, suggesting that Vitamin B supplements such as folate that reduce homocysteine concentrations may help prevent AD.
4.4.1.4. White matter changes
In addition to the well-recognized Aβ plaque and tau fibril deposition considered to be emblematic of AD, there is increasing evidence that white matter abnormalities play a role in exacerbating cognitive problems. Although patients with high risk for cerebrovascular disease are excluded from study in ADNI, cardiovascular risk factors are consistently associated with cognitive decline and AD. Provenzano et al [441] found that higher white matter hyperintensity (WMH) volume, an indicator of cerebrovascular disease, was associated with AD diagnosis independent of amyloid deposition, providing support for the idea that white matter damage may provide a “second hit” required for clinical manifestation of AD symptoms. Guzman et al [442] reported that temporal lobe WMH volume was independently associated with entorhinal cortex volume to a greater extent than Aβ42 or p-tau181. Lo et al [443] also investigated whether the presence of WMHs was associated with established AD biomarkers. Baseline WMHs were not predictive of changes in CSF biomarkers, glucose metabolism, or hippocampal atrophy but were significantly associated with cognitive decline, particularly executive function. These results suggest that vascular disease may target different cognitive domains than AD-type pathology and thus may be an independent additive factor contributing to the disease. However diagnostic groups within ADNI share most vascular factors and rates of longitudinal cardiovascular events, suggesting that vascular factors may already have contributed to cognitive decline [444].
Changes in the white matter architecture have been modeled as structural brain networks, and changes in the functional connectivity over the course of the disease can be studied using diffusion tensor imaging (DTI) included in ADNI-2. A preliminary study found that network measures of baseline connectivity were predictive of future white matter changes [445], suggesting that less optimal baseline small-world architecture may be a useful biomarker in predicting WM changes in the prodromal phase of the disease. Widespread group differences between MCI and AD patients were also detected using DTI, with the greatest effect sizes in the left hippocampal cingulate and throughout the temporal lobe and posterior brain regions. White matter disruptions were associated with MMSE, ADAS-cog, and CDR-SB scores [446]. Rowley et al [447] measured both mean diffusivity and fractional anisotropy in patients with amnestic MCI, nonamnestic MCI, and AD. No difference in either measure was detected between nonamnestic MCI patients and cognitively normal controls. However, patients classified as amnestic MCI had around 28% white matter abnormality compared with the control group, and AD patients had reduced structural connectivity between the hippocampus and the temporal, inferior parietal, posterior cingulate, and frontal regions suggesting that white matter abnormalities in the advanced patients have a significant influence on connectivity.
4.4.2. Glucose metabolism
4.4.2.1. [18F]-fluorodeoxyglucose-positron emission tomography
FDG-PET has been used by several groups to investigate relationships between cerebral glucose hypometabolism and other factors, including cognitive measures and CSF biomarkers. Several papers confirmed that there is a characteristic regional pattern of hypometabolism in MCI and AD patients. Wu et al [101] found that hypometabolic voxels were associated with the posterior cingulate/precuneus and parietotemporal regions. Lower bilateral cerebral metabolic rate for glucose (CMRgl) at baseline in these regions and in the frontal cortex was associated with higher CDR-SB and lower MMSE scores in MCI and AD groups [134] (Table 5). Although the pattern of hypometabolism was similar in the two groups, the magnitude and spatial extent were greater with increasing disease severity. In the AD group alone, however, lower MMSE correlated with lower left frontal and temporal CMRgl, suggesting that the characteristic pattern of baseline reductions in glucose metabolism shifts to the frontal cortex after the onset of dementia. Chen et al [104] investigated declines in CMRgl in statistically predefined ROIs associated with AD over 12 months in the ADNI cohort and found significant changes in MCI and AD groups compared with control subjects bilaterally in the posterior cingulate, medial and lateral parietal, medial and lateral temporal, frontal, and occipital cortices. These changes correlated with CDR-SB, but not ADAS-cog, scores in both groups, and with MMSE scores in the MCI group (Table 5). Habeck et al [448] used the recently developed psychometric composite score of ADNI-mem [449] to represent memory. Impaired memory in MCI patients was associated with reduced metabolism in regions of the parietal and temporal lobes, whereas memory deficits in AD patients were associated with hypometabolism in the frontal and orbitofrontal regions. The differential location of hypometabolism in relation to memory in MCI and AD groups suggests that frontal regions may play a role in compensating for memory as the disease progresses. Landau et al [135] found a greater decline in CMRgl in all a priori defined ROIs in AD patients and in a composite score of ROIs in MCI patients compared with control subjects. Longitudinal glucose decline was associated with concurrent ADAS-cog scores and decline on the Functional Activities Questionnaire (FAQ), validating the relevance of longitudinal measures of glucose metabolism to both cognitive and functional decline. The annual decline in the ADAS-cog and FAQ was greatest in AD patients, followed by the MCI and control groups, in accordance with an acceleration of the disease process over time (Table 5). FDG-PET and hippocampal volume, but not whole brain volume, were independently related to ADAS-cog as a measure of disease progression [450]. The independence of glucose metabolism from other biomarkers in its relationship with cognitive decline suggests that cognitive decline in MCI patients with reduced hippocampal volume but higher glucose metabolism may be delayed or reduced. The hypometabolism index reported by Chen et al [85] correlated with cognitive measures of disease severity, hippocampal volume, and CSF biomarkers (Table 5). These papers support the use of glucose metabolism as a sensitive measure of cognition in AD.
4.4.2.2. Arterial spin labeling
Arterial spin labeled (ASL) MRI is an imaging technique that reveals alterations in cerebral blood flow (CBF) and that has been shown to overlap with FDG-PET measures of hypometabolism. ASL can be performed at the same time as structural MRI, obviating the need for a separate FDG-PET scan with its attendant costs and exposure to radioactive tracer. ADNI-2 has added an ASL protocol to a portion of the cohort, presenting an opportunity to assess the utility of this method as a biomarker in a multicenter study. Wang et al [451] examined associations between ASL measures in ROIs previously determined to be most emblematic of AD in FDG-PET studies and hippocampal volume with disease severity as measured by CDR-SB. They found that mean meta-ROI CBF was associated with group status in a manner comparable to hippocampal volume. However, whereas hippocampal volume differences only reached significance between control and AD groups, differences in CBF reached significance between control and LMCI groups. Regression analysis indicated that CBF and hippocampal volume provide complementary information regarding disease severity. The results suggest that CBF may be an effective biomarker for an equivalent point in disease progression to FDG-PET.
4.4.3. Cognitive
4.4.3.1. Association with imaging or CSF biomarkers
A number of studies have focused on the relationship between cognitive function and imaging or CSF biomarkers. Atrophic changes in the episodic memory network (Fig. 15), which is composed of MTL structures, medial and lateral parietal cortical areas, and prefrontal cortical areas and is involved in the formation of new episodic memories, are presumed to underlie ongoing memory loss in AD. Walhovd et al [136] studied how baseline brain morphometry and metabolism within the episodic memory network and APOE genotype predicted memory, as assessed by the AVLT. They found that in the total sample of the ADNI cohort, hippocampal volume and metabolism, parahippocampal thickness, and APOE genotype predicted recognition, whereas hippocampal volume and metabolism, cortical thickness of the precuneus, and inferior parietal metabolism predicted learning, suggesting that MTL structures are related to learning, recall, and recognition, whereas parietal structures are involved solely in learning (Table 5). The authors concluded that MRI and FDG-PET imaging have differential sensitivity to memory in AD and thus provide complementary information. Episodic memory likely involves a number of different cognitive processes, such as initial encoding, learning on repeated exposure, and DR, which may be subserved by disparate components of the episodic memory network. Wolk and Dickerson [137] investigated whether verbal episodic memory could be fractionated into dissociable anatomic regions in mild AD patients, using cortical thickness of predefined “AD signature” ROIs and hippocampal volume as structural measures and different stages of the AVLT as a verbal memory measure. They found that initial immediate recall trials were most significantly associated with the temporal pole region, but that regions in the MTL became more significantly associated in later trials. In tests of DR, only the hippocampus correlated with performance, whereas the perirhinal/entorhinal cortex was most strongly associated with delayed recognition discrimination. The authors concluded that their results lend support to models hypothesizing that dissociable brain regions are involved in differential episodic memory processes. Associations between memory learning and brain morphometry in the MTL were found in a study by Chang et al [138]. MCI patients were differentiated into learning-deficit and retention-deficit subgroups using the AVLT. Low memory retention was associated with changes in the medial temporal regions, particularly the hippocampus and entorhinal cortex, whereas low memory learning correlated with a more widespread pattern of morphometric changes beyond the temporal lobe, including areas of the frontal and parietal lobes (Table 5). While memory loss is a hallmark of AD, a subset of MCI patients is impaired primarily in their executive function. Dickerson and Wolk [139] identified dysexecutive and amnestic phenotypes in patients with MCI or very mild AD based on performance on the Trail Making Test and ADAS-cog subscale: Word recognition. They found that the memory-impaired group had a more frequent occurrence of the APOE ε4 allele status than the dysexecutive group, and that patients with low executive function had thinner frontoparietal cortical regions and were more impaired in daily life than those with predominantly memory impairment. A further study by Chang et al [140] found that MCI patients with high executive function performed better on tests of verbal memory than those with low executive function, and that morphometric measures of the two groups differed primarily in the dorsolateral prefrontal and posterior cingulate cortices, where more thinning was evident in low executive function patients (Table 5). Results from both studies suggest that the dysexecutive phenotype may reflect differences in underlying pathology in brain regions beyond the MTL.
Fig. 15.
The episodic memory network. Along with the hippocampal formation, the cortical areas shown here are part of the episodic memory network. Shown here are pial cortical representations of selected parcellations in the left hemisphere. From left to right: medial, ventral, and lateral views. Reproduced with permission from Ref [136].
4.4.3.2. Neuroanatomic regions and cognition
The ideas that different brains regions subserve different cognitive functions and that MCI is a heterogeneous construct led Wolk et al [141] to examine the influence of APOE genotype on memory and executive function in AD. When cortical thickness in predefined ROIs was examined in carriers and noncarriers of the APOE ε4 allele who had a CSF biomarker profile consistent with AD, carriers were more impaired in measures of memory retention and had greater atrophy in medial temporal regions, whereas noncarriers were more impaired in tests of executive function, working memory, and lexical access and had greater frontoparietal atrophy. The finding that neuroanatomic regions thought to subserve different cognitive processes are differentially affected by APOE ε4 allele status supports the hypothesis that this allele exerts its effect on AD by influencing different large-scale brain networks.
The question of whether domain-specific cognitive deficits in MCI are caused by global atrophy or progressive atrophy within specific regions was studied by McDonald et al [142], who examined 2-year regional atrophy rates in MCI patients. Stepwise regression models revealed that left entorhinal atrophy, left lateral lobe thinning, left temporal lobe atrophy, left frontal lobe atrophy rate, and the right MTL atrophy rate were associated with memory decline (Logical Memory II), naming decline (Boston Naming Test), semantic fluency decline (Category Fluency Test), executive function (Trail Making Test B; TMT-B), and clinical decline (CDR-SB), respectively (Table 5). Semantic memory impairment (Boston Naming Test) in mild AD cases was associated with anterior temporal lobe atrophy, with the strongest correlation being with the dorsal temporal pole [452]. These studies afford a glimpse into the specific structure– function relationships that occur early in disease progression and enhance our understanding of the neural basis of cognitive impairments.
An alternative method to individual cognitive tests and ROI approaches for studying brain structure-cognition relationships was reported by Nho et al [453]. They used recently reported psychometric composite scores of memory (ADNI-Mem [449]) and executive function (ADNI-Exec [454]) in concert with voxelwise and cortical thickness whole brain measures of brain atrophy. Across the ADNI cohort, they found strong positive associations between memory and temporal lobe atrophy, particularly hippocampal atrophy in the MCI and AD groups. Executive function was strongly associated with cortical thickness and GM density across large areas of the brain, particularly bilateral parietal and temporal lobes. After adjusting for memory and GM density/cortical thickness in the pooled sample, executive function was additionally related to frontal lobe GM density and cortical thickness. This study provides insight into brain changes associated with different cognitive processes involved.
4.4.3.3. Functional decline
Although studies, such as those described previously, have focused on the relationship between brain atrophy, APOE ε4 status, and cognitive decline, relatively little is known about the biomarkers of functional decline, a hallmark of AD. Impairment of instrumental activities of daily living (IADL) such as driving, handling finances or preparing meals, leads to a greater burden on care-givers and institutionalization as the patient loses independence. A number of papers have focused on the prevalence of functional impairment, its rate of decline and its association with cerebral atrophy, other cognitive measures and biological biomarkers. Brown et al [274] examined IADL in MCI and AD patients and found that, despite a definition that does not include substantial impairment of daily function, MCI was associated with a high prevalence of IADLs; nearly three-quarters of MCI patients reported deficits in some items of the FAQ, a measure of the ability of patients to maintain daily function, compared to 97.4% of patients with AD and 7.9% of cognitively normal controls. Functional impairment was also associated with deficits in memory, processing speed and atrophy of the hippocampus and entorhinal cortex. The rate of decline in the FAQ, a measure of the ability of patients to maintain daily function, and how it is affected by cerebral atrophy and APOE ε4 allele status, was studied by Okonkwo et al [143]. They found that AD patients had a higher rate of functional decline than control subjects, with the rate of MCI patients intermediate between the two. Moreover, MCI patients who subsequently progressed to dementia had higher rates of decline on the FAQ than stable MCI patients. Increasing ventricle-to-brain ratio, the measure of neurodegeneration chosen for the study, correlated with increased functional impairment in MCI patients. Those patients who were both APOE ε4-positive and had elevated ventricle-to-brain ratio were the most functionally impaired. These results have shown neurodegeneration and APOE ε4 status to be associated with cognitive decline. Whereas the APOE ε 4 allele is detrimental to disease progression, the APOE ε 4 allele may have a protective effect. Bonner-Jackson et al [275] found that at 24 months, carriers of the APOE ε 4 allele showed significantly less functional decline that non-carriers in the pooled ADNI cohort and that individual groups showed the same trend. This allele was also associated with better scores in composite measures of memory and executive function in the pooled sample, suggesting that the APOE ε 4 allele may slow the rate of functional decline as well as positively influence neurocognition.
Okonkwo et al [144] investigated the relationships between CSF biomarkers and everyday function, as assessed by the FAQ. They found that biomarkers were more sensitive to functional decline in control subjects and MCI patients than in AD patients, and that in the latter group, scores on the ADAS-cog were more highly correlated with functional activity. Combinations of tau and Aβ-42 abnormalities had the steepest rates of functional decline across clinical groups. The authors suggested that the effect of CSF abnormalities on functional decline is partially mediated by their effect on cognitive status. The relationship between functional impairment and amyloid burden as assessed by 11C-PiB PET imaging was subsequently investigated by Marshall et al [276] in control and MCI patients of the ADNI cohort. They found that increased cortical PiB retention was associated with greater IADL impairment in the pooled sample (r2 = 0.40, P = .0002) and in the MCI group (r2 = 0.28, P = .003) and that poorer performance on FAQ was also associated with poorer performance in the AVLT and MMSE in all subjects. A companion paper by the same group [277] examined the relationship between functional impairment and executive function in a longitudinal study of the ADNI cohort. Executive dysfunction was strongly correlated with IADL impairment across all subjects (r2 = 0.60, P < .0001). MCI patients with impaired executive function also had greater impairment of IADL than patients with no executive dysfunction, possibly representing a portion of the heterogeneous MCI construct more likely to progress to AD.
Depression may also influence functional decline. A study of the effect of subsyndromal symptoms of depression (SSDs) on functional ability of MCI patients [455] reported that these symptoms, present in 77% of this group, increased the risk of having poorer FAQ scores by 1.77-fold. However, although SSDs were associated with higher disability at baseline, they were not associated with longitudinal decline in FAQ scores or faster conversion to AD. The authors posited that treatment of SSD may therefore reduce the burden of disability in MCI patients but is unlikely to slow AD progression.
4.4.3.4. Other neuropsychiatric symptoms
A number of neuropsychiatric symptoms and apathy, anxiety, hallucinations, depression, and psychosis are associated with AD and are highly prevalent in the MCI population. Despite this, their association with functional impairment has not been fully evaluated. Wadsworth et al [456] found that baseline apathy, anxiety, and hallucinations were all significantly associated with functional impairment, as measured by CDR-SB scores. Hallucinations and apathy at baseline were also associated with functional impairment over time and increased the risk of MCI to AD conversion, suggesting that these symptoms may be predictive of current and future disease progression. Similarly, Ramakers et al [457] reported that anxiety, but not depression or apathy, was associated with abnormal levels of Aβ42, t-tau, and t-tau/Aβ42, supporting the idea that anxiety may be a result of underlying disease pathology in these MCI patients. Richard et al [458] focused on the association between apathy and disease progression, finding that symptoms of apathy alone, but not symptoms of depression alone or symptoms of apathy and depression together, increased the risk of conversion to AD within the study period. Therefore, apathy and depression may differ in their effects on cognitive decline. One distinct variant of AD is AD with psychosis, which is associated with more rapid functional decline related to frontal lobe impairments. Koppel et al [459] investigated associations between psychosis and glucose metabolism and found both a decline in orbitofrontal brain metabolism and an accelerated functional decline in patients with active psychosis.
4.4.3.5. Association of cognition with body mass index
In elderly populations, in addition to brain atrophy or genetic studies, BMI has been associated with cognitive decline. Cronk et al [145] examined the relationship between BMI and cognition in MCI patients and found that lower BMI at baseline was associated with a decline in the MMSE, ADAS-cog, and a global composite of the ADNI neuropsychological battery, but not with CDR-SB scores or conversion to AD. The causal relationships between BMI and cognitive decline in MCI remain to be elucidated, but the authors suggest either that low BMI is a result of factors associated with MCI or that MCI patients with low BMI are predisposed to more rapid disease progression.
4.4.3.6. Cognitive reserve
The concept of the cognitive reserve describes the mind’s resilience to neuropathological changes in the brain and may account for the observed dissociation between AD pathology and cognition. Vemuri et al [278] investigated whether a measure of cognitive reserve, the American National Adult Reading Test (AMNART) modified the relationship between biomarkers of pathology and cognition in AD. In cognitively normal patients, they observed that the AMNART, but not CSF biomarkers or STAND scores correlated with cognitive measures (MMSE, ADAS-cog, AVLT-memory, Trails B and Boston Naming tests) whereas in MCI patients, all three were associated with cognitive performance in an additive manner. The authors propose a model (Fig. 28) in which cognitive reserve acts to shift curves of cognitive decline relative to biomarker trajectories over time; high cognitive reserve delays cognitive decline whereas low cognitive reserve results in an earlier cognitive decline. This evidence is consistent with the early increase in Aβ levels and subsequent later increase in cerebral atrophy in the disease progression model of Jack et al [14]. Further evidence that cognitive reserve buffers the effects of the disease comes from studies by Ewers et al [460] and Pillai et al [461]. In cognitively normal patients identified as having preclinical AD by the abnormal levels of Aβ42, higher education was associated with lower glucose metabolism in the posterior cingulate and angular gyrus [460]. Thus, education appeared to buffer cognitive function in the presence of AD pathology that impairs glucose metabolism. This supports the idea that cognitive reserve has a compensatory function that acts to preserve a clinical state despite more advanced pathology. Higher education was associated with lower, not higher, cortical thickness in selected areas [461]. In addition, Guo et al [462] reported that larger initial brain volumes, as measured by intracranial volume (ICV), were associated with reduced clinical deterioration in amnestic MCI patients and also with a lesser impact of APOE status. ICV did not impact the effect of APOE on atrophy progression or the rate of atrophy itself. These studies support the idea that the nature of this compensatory function is an improvement in connectivity rather than a passive reserve of increased neural substrate. Guo et al [462] proposed that the brain reserve confers protective effects earlier in the disease, but beyond a certain threshold of neurodegenerative burden, it is no longer advantageous.
Fig. 28.
Model illustrating the independent effect of cognitive reserve on the relationship between biomarkers of pathology and cognition in subjects with (A) low, (B), average and (C) high cognitive reserve. In (A) and (C), the levels of Aβ are indicated by a square and the levels of atrophy are indicated by a circle at the point where cognitively normal subjects progress to MCI. This illustrates that at an equivalent clinical diagnostic threshold, subjects with high cognitive reserve have greater biomarker abnormalities than those with low cognitive reserve. Reproduced with permission from Ref [278].
4.4.3.7. The ADNI special issue of Brain Imaging and Behavior, 2012
A special issue of Brain Imaging and Behavior was published in 2012 focusing on cognition research within ADNI. Mungas et al [463] introduced papers that had been presented at the Advanced Psychometric Methods in Cognitive Aging Conference, in June 2011 at Friday Harbor, WA. These papers will be discussed individually in this review and fall into four primary groups. The first set of articles focused on the measurement of cognition and on improving reliability, sensitivity, and validity of measurements of different cognitive domains [449, 454, 464, 465]. A second set examined neuroimaging-cognition relationships [448, 453, 466], and a third set examined sequencing of biomarker and cognitive changes in relation to the model by Jack et al [14, 467, 468]. A final area examined genetic contributions to cognition using high-dimensional genetic data [469–471].
4.4.3.8. Psychometric analysis of cognitive tests
As cognitive testing plays such a critical role in ADNI, confidence that the tests used accurately reflect subject cognition is paramount. To this end, psychometric analysis has been applied to a number of cognitive tests to assess the validity of test components and their combined performance. The ADAS-cog plays a critical role in detecting clinical change in ADNI. However, as the test was originally developed for AD, its sensitivity to cognitive decline at the increasingly targeted earlier stages of the disease has not been fully explored. Posner et al [472] reviewed two critical papers by Hobart et al that use different analytical approaches to evaluate the performance of ADAS-cog in ADNI data. Using traditional psychometric methods, Hobart et al [473] found that 8 of 11 components of the ADAS-cog had a limited response distribution that may underestimate performance differences and therefore limit the usefulness of this test in detecting clinical change in mild MCI populations. Other weaknesses of the test were identified using Rasch measurement theory methods [474]: the range of cognitive performance in every cohort was not well matched to the range of scale (Fig. 34), and 6 of 11 components of scale were bunched instead of evenly spread out across the continuum. In addition, the validity of the way in which the 11 components were combined to create a simple variable was questioned given that the scale was not constructed on a specific definition of cognition. These analyses may provide a starting point for specific improvements to make the ADAS-cog better suited to its role in the upcoming early intervention clinical trials.
Fig. 34.
Suboptimal targeting of ADAS-cog. The distribution of person measurements (upper pink histogram) and the distribution of item locations of the 11 ADAS-cog components (lower blue histogram) are presented. From Hobart et al [474].
Crane et al [449] derived a single composite memory score from elements of the RAVLT, MMSE, and Logical Memory, and ADAS-cog tests firstly to address questions of equality of different versions of word lists in ADAS-cog and RAVLT administered in ADNI study, and secondly to facilitate a statistically more simple analysis of relationships between memory and other factors such as imaging data, biomarkers, and clinical diagnoses. The memory composite score, ADNI-Mem, performed comparably to other memory measures in the prediction of clinical change over time and was able to differentiate changes over time in participants with or without the AD CSF biomarker signature. It was strongly associated with neuroimaging parameters previously associated with memory performance, suggesting this improvement could simplify ADNI protocol by allowing a single test for memory to be administered.
Trzepacz et al [475] developed three subscales of the Neuropsychiatric Inventory Questionnaire (NPI-Q) to measure agitation/aggression, mood, and frontal syndromes. They used principal component analysis to determine how the subscales relate to each other and evaluated them in two cohorts including ADNI. They concluded that the subscales were valid based on the similarity of results across the two cohorts.
4.4.4. CSF biomarkers
4.4.4.1. β-amyloid and tau
The relationship between CSF biomarkers and neuronal degeneration has been investigated by a number of groups within and outside ADNI following the seminal publication by Shaw et al [57], which defined cut points for CSF tau and Aβ-42 based on an ADNI-independent cohort of autopsy-confirmed AD patients as well as normal control subjects and then applied these cut points successfully to the ADNI cohort. Follow-up studies went on to test the hypothesis that changes in levels of biomarkers occur early in disease and thus are likely predictive of future brain atrophy, if not directly associated with all parts of the degenerative process. For example, Tosun et al [146] examined how rates of regional brain trophy were related to levels of CSF biomarkers in MCI patients and healthy elderly control subjects. They found that lower CSF Aβ-42 levels and higher tau levels were associated with increased atrophy in numerous brain regions, beginning primarily in the temporal and parietal cortices in MCI patients and extending to regions not normally associated with amyloid pathology, such as the caudate and accumbens areas, in AD patients. Schuff et al [121] also found that increased rates of hippocampal atrophy were associated with lower levels of Aβ-42 in the MCI, but not AD or control, group. Leow et al [113] used TBM to examine rates of atrophy and found that lower CSF Aβ-42 levels, higher tau levels, and a higher p-tau/Aβ-42 ratio were significantly associated with temporal lobe atrophy in the pooled group, and, additionally, that within the AD group, levels of CSF p-tau and the p-tau/Aβ-42 ratio were also significantly associated. Fjell et al [147] investigated whether baseline levels of CSF biomarkers were associated with baseline brain morphometric differences between control, MCI, and AD subjects, as measured by cortical thickness in a number of ROIs. They found that although CSF biomarkers levels could not account for baseline differences, they were moderately associated with longitudinal change in multiple areas, including medial temporal regions and beyond. Stricker et al [467] examined associations between baseline levels of Aβ-42 and p-tau and rates of atrophy of the precuneus, the first area to be affected by amyloid deposition, and hippocampus, the first area to be affected by neurofibrillary tangles. Neither of the baseline CSF measures was associated with the thinning of the precuneus, but both were associated with the rate of hippocampal atrophy. Furthermore, lower baseline Aβ-42 levels in cognitively normal controls and higher baseline p-tau in the MCI and AD groups were associated with increased rates of hippocampal atrophy, suggesting that Aβ-42 exerts its effect earlier and that tau exerts its effect later in disease progression.
A second focus of research into CSF biomarkers has been how they are modulated by APOE genotype and their association with cognitive measures. Shaw et al [57] reported that Aβ-42 concentrations were dose dependent on the number of APOE ε4 alleles, with the highest concentrations found in homozygotes. Vemuri et al [128] found that Aβ-42 is more closely associated with APOE genotype than cognitive function (MMSE, CDR-SB), but that APOE genotype had no significant effect on levels of t-tau (Fig. 16). An earlier study by the same group [132] investigated the relationship between CSF biomarkers and cognitive function (MMSE and CDR-SB), and found that the CSF biomarkers Aβ-42, t-tau, and p-tau181p were only significantly correlated with cognitive function in the pooled sample (Table 5). Ott et al [148] studied the relationship between CSF biomarkers and ventricular expansion with the hypothesis that ventricular dilation may reflect faulty CSF clearance mechanisms resulting in reduced levels of Aβ. They found that ventricular expansion was associated with reduced CSF Aβ levels in normal elderly carriers of APOE ε4, but that in APOE ε4-positive AD patients, ventricular expansion was associated with increased levels of tau and not Aβ. The authors suggested that the APOE ε4 allele may exert its effect through modulation of CSF–blood–brain barrier function.
Fig. 16.
Correlations between biomarker levels, structural abnormalities, and cognitive performance in APOE ε4 carriers and noncarriers. Smoothed biomarker (A and B) or STAND (C) z score curves plotted as a function of cognitive performance (Mini-Mental State Examination, MMSE). Abbreviation: STAND, Structural Abnormality Index. Reproduced with permission from Ref [128].
Toledo et al [476] examined the relationship between CSF and plasma apoE protein levels and APOE genotype with cognition and AD biomarker changes. At baseline, higher CSF apoE levels were associated with higher total and phosphorylated CSF tau levels. CSF apoE levels were also associated with longitudinal cognitive decline, MCI to AD conversion, and GM atrophy rate but not changes in CSF Aβ or tau levels. Plasma apoE levels showed a mild correlation with CSF apoE levels but were not associated with longitudinal cognitive and MRI changes. The authors proposed that increased CSF apoE2 or apoE3 levels might represent a protective response to injury in AD and may have neuroprotective effects by decreasing neuronal damage independent of tau and amyloid deposition in addition to their effects on amyloid clearance.
The results from these studies support a model in which changes in the levels of CSF biomarkers are an early step in the course of the disease that reflects the degree of AD pathology, and in which Aβ-42 is modulated by the APOE ε4 allele, which functions in the early stages of pathology by reducing the efficiency of Aβ-42 clearance. As described in the Genetics section 5.3, Kim et al [149] performed a genomewide search for markers associated with CSF analyte levels in the ADNI cohort. Overall, CSF Aβ-42 and tau, in conjunction with imaging measures of atrophy, are promising biomarkers for early detection of AD.
Two recent studies by Ewers et al [279] and Vidoni et al [280] investigated the relationship between markers of early AD and BMI, which appears to have a paradoxical association with the disease; high BMI in mid-life increases the risk of the disease whereas it appears to be protective in later life [279, 280]. Vidoni et al found that the association between amyloid burden (measured by both CSF Aβ levels and global PiB uptake) and low BMI was strongest in MCI patients and cognitively normal controls (Table 6). Ewers et al found that BMI was significantly lower in patents with levels of CSF Aβ and t-tau above a pre-defined cut-point (F = 27.7, df = 746, P < .001), regardless of diagnosis. These results provide further evidence that AD pathology is present before the disease becomes a clinically evident and suggest that low BMI may either be a systemic response due to the presence of this pathology, or constitute a trait that predisposes an individual to its development. If the latter scenario is correct, then therapies that address BMI issues are of interest in the treatment of AD. Rajagopalan et al [477] investigated associations between BMI, regional brain volumes, and leptin, a hormone produced by adipose tissue which regulates appetite and energy expenditure. Higher levels of leptin were associated with lower volumes of the frontal, parietal, temporal, and occipital lobes, and the brainstem and cerebellum, and these associations persisted after controlling for BMI. As leptin levels can be manipulated, it may represent a promising therapeutic target for the treatment of AD.
Table 6.
Associations between imaging, clinical, and CSF biomarkers (correlation coefficients)
| CSF biomarker correlates | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Imaging or clinical biomarker |
N | Clinical group |
Aβ–42 | t-tau | p-tau181p | t-tau/Aβ–42 | FDG-PET composite ROI |
Mean cortical PiB SUVR |
Reference |
| Hippocampal volume (L/R) |
388 | Pooled sample |
0.11*/ 0.17† |
−0.17†/0.21‡ | −0.17†/ −0.23‡ |
−0.17†/ −0.21‡ |
−0.24*/ −0.23* |
[151] | |
| Mean cortical PiB SUVR |
55 | Pooled sample |
−0.73‡ | −0.42§ | 0.49‡ | 0.28* | [34] | ||
| FDG-PET composite ROI |
0.33† | 0.24 (P = .08) | 0.34† | ||||||
| MMSE | NS | 0.26 (P = .055) | 0.28* | 0.63‡ | NS | ||||
| Aβ–42 | 0.38† | ||||||||
| APOE ε4 | 77 | CN | −0.50‡ | NS | NS | [146] | |||
| 119 | MCI | −0.49‡ | 0.39‡ | 0.34† | |||||
| 54 | AD | −0.53‡ | NS | NS | |||||
| BMI | 112 | CN | 0.19‡ | −0.20‡ | −0.43† | [280] | |||
| 193 | MCI | NS | −0.17* | −0.32‡ | |||||
| 100 | AD | NS | NS | NS | |||||
Abbreviations: NS, not significant; SUVR, standard uptake value ratio.
P < .05.
P < .01.
P < .001.
P < .0001.
4.4.4.2. α-Synuclein and dementias with Lewy bodies
AD is frequently comorbid with Lewy body diseases such as Parkinson’s disease, with α-synuclein (α-syn)–positive Lewy bodies found in 40% to 50% of AD patients. Faster cognitive decline has been noted in these patients, and this neuronal damage may be accelerated by the α-syn–mediated aggregation of Aβ and tau in the ADNI cohort. Levels of α-syn and p-tau181 were strongly correlated in AD, MCI, and Parkinson’s disease patients [478]. There was an inverse correlation between levels of α-syn and p-tau181, suggesting that this inverse CSF signature indicates concomitant AD and Lewy body disease pathology [478]. Korff et al [479] reported that CSF α-syn differentiated between patient groups and that increased α-syn was associated with decreased MMSE scores but not with APOE status. Clearly, further investigation of α-syn as a biomarker for AD is warranted.
At autopsy, a high percentage of coincident pathologies are reported in demented patients. In a consecutive series of the first 22 ADNI autopsies, Toledo et al [480] found that only four patients had pure AD pathology. Coincident dementia with Lewy bodies (DLB), medial temporal lobe pathology (TDP-43 proteinopathy, argyrophilic grain disease, and hippocampal sclerosis), and vascular pathology occurred in 45.5%, 40.0%, and 22.7% of patients, respectively. DLB was predicted by hallucinations and a severe dysexecutive profile, and coincident DLB was classified by occipital FDG-PET hypometabolism. These results suggest that biomarkers can be used to independently predict coincident AD and DLB pathology.
4.4.5. Amyloid imaging
A complementary method for assessing amyloid deposition is 11C-PiB PET imaging. Jack et al [16] investigated the relationship between amyloid deposition and ventricular expansion in the ADNI cohort by examining serial 11C-PiB PET and MRI scans. They found no difference in the rate of global PiB retention between clinical groups, and changes in global PiB retention only weakly correlated with concurrent decline on MMSE and CDR-SB. In contrast, ventricular expansion increased from control subjects to MCI to AD groups and correlated strongly with concurrent cognitive decline (Table 5). The relationship between PET and CSF biomarkers and cognitive measures in the ADNI cohort at baseline was investigated by Jagust et al [150]. CSF Aβ-42 and 11C-PiB PET were found to be in substantial agreement as measures of amyloid deposition, and neither measure correlated with MMSE scores. In contrast, FDG-PET, as a measure of cerebral glucose metabolism, was strongly correlated with MMSE scores, but much less so with CSF biomarkers (Table 6). Apostolova et al [151] also examined associations between hippocampal atrophy, CSF biomarkers, and average cortical, precuneal, and parietal uptake of 11C-PiB. They found that although all CSF biomarkers were associated with hippocampal atrophy, the strongest correlations were with p-tau181p and the weakest with Aβ-42. Precuneal 11C-PiB uptake was most strongly associated with hippocampal atrophy. Jack et al [152] examined the relationship between log relative hazard of progressing from MCI to AD and both hippocampal atrophy and amyloid load, measured as a composite of 11C-PiB PET and CSF Aβ-42 data. They found that although the risk profile was linear throughout the range of hippocampal atrophy, amyloid load reached a ceiling at a certain concentration earlier in disease progression. These papers support a disease model in which initial amyloid deposition occurs in the early stages and does not correlate with cognitive decline, but stabilizes later in disease, and in which neurodegeneration accelerates with disease progression with concomitant cognitive decline. A protocol using the longer half-life Florbetapir amyloid ligand was added to ADNI-GO and ADNI-2. An initial study compared Florbetapir binding with FDG uptake as differential indicators of metabolism [481]. The percentage of patients classified as Florbetapir positive increased from 29% of cognitively normal controls to 43% of EMCI patients to 62% of LMCI patients to 77% of AD patients. Additionally, cognitive decline in cognitively normal patients was more closely linked to Florbetapir binding than to FDG abnormalities, but the reverse was found in MCI and AD patients. Murphy et al [482] investigated associations between amyloid deposition, age, clinical status, and APOE status and found that while age was not associated with Florbetapir signal, APOE status had a strong influence on the uptake of this radiotracer. In all patient categories, APOE ε 4 + patients had higher densities of amyloid plaques across all cortical regions than patients lacking this risk allele. APOE ε 4 status had a far larger effect on Florbetapir uptake than clinical status (Fig. 35). These results suggest that APOE status is a better predictor of amyloid deposition than age or clinical status and that high amyloid deposition precedes the manifestation of clinical symptoms, whereas glucose hypometabolism occurs later in progression in accordance with the Jack model of disease progression [14]. A second study by Landau et al [483] found that Florbetapir binding and levels of CSF Aβ were inversely correlated in 86% of subjects both longitudinally and cross-sectionally. The number of subjects who were discordant for these measurements dropped threefold when a 5% confidence interval was applied to the cutoff point.
Fig. 35.
The effect of APOE ε 4 status on the β-amyloid PET standard uptake value ratio (SUVR) in four cortical regions across diagnostic categories. (−) APOE ε 4 negative; (+) APOE ε 4 positive. From Murphy et al [482]. Abbreviations: EMCI, early mild cognitive impairment; LMCI, late mild cognitive impairment; AD, Alzheimer’s disease.
4.4.6. Combined modalities
The dynamics of CSF, MRI, and FDG-PET biomarkers in the ADNI cohort were studied by Caroli and Frisoni [153] in an effort to understand how they change over the course of the disease. Each biomarker differed between clinical groups after post hoc analysis, and the authors found that these measures of disease progression fit better in sigmoidal, rather than linear, models, suggesting that individual biomarkers vary in their rate of change during disease progression. Aβ-42 imaging signals increased early in disease progression and then plateaued, whereas CSF Aβ-42 declined early and then plateaued, and hippocampal volume followed a similar trajectory, with volumes increasing later in disease progression. In contrast, FDG-PET measures of glucose metabolism and CSF tau began to increase early in disease progression and only stabilized at later stages of disease, suggesting that there is an ongoing reduction in glucose metabolism and tau-mediated neurodegeneration throughout the early stages of AD (blue line in Figs. 2 and 17). Carriers of the APOE ε4 allele had earlier hippocampal atrophy. A similar study by Beckett et al [154] also found that measures associated with early disease, such as Aβ-42, had greater changes in MCI patients than in AD patients, and that those associated with later changes, such as those in FDG-PET ROIs, were more evident in AD patients (Table 7). The authors hypothesized that changes in biomarkers may not be linear and that for each biomarker, there may be steeper rates of change in some stages of disease progression than others. An extension of this study examined trajectories of CSF Aβ42, FDG uptake and hippocampal volume loss and the influence of the APOE ε 4 allele study up to 36 months from the original 12 months [281]. Aβ42 levels declined most rapidly in cognitively normal participants, glucose metabolism declined most rapidly in AD patients and hippocampal atrophy accelerated with disease progression. Presence of the APOE ε 4 allele acted primarily to accelerate hippocampal atrophy in MCI and AD patients. These results are in keeping with the model of Jack et al [14], which was subsequently empirically tested in a further paper by the same group [282]. Using cut-points demarcating normal from abnormal levels of CSF Aβ42 and t-tau, and of hippocampal volume, Jack et al examined the distribution of these biomarkers in control, MCI and AD patients at baseline and 12 months using ADNI cohort data. They found support for the model in that the percentage of abnormal biomarker findings increased with disease severity as assessed by clinical status and MMSE score, and in the temporal progression of the appearance of biomarker abnormalities: Aβ42 first followed by t-tau and lastly by hippocampal volume (Fig. 29). Biomarker dynamics continue to be the subject of intense interest. With substantial longitudinal data now available through ADNI, ADNIGO, and ADNI-2 and with the addition of an E-MCI cohort in ADNI-2, a convincing body of evidence has accumulated for the temporal ordering of biomarkers and the shape of their trajectories. Jedynak et al [484] computed a disease progression score for each patient in the ADNI cohort and produced a data-driven plot of biomarker dynamics that was in substantial agreement with the model by Jack et al [14]. One exception was the AVLT-30 test added to ADNI-GO and ADNI-2 protocols, which appeared to be the first biomarker to become abnormal (Fig. 36). The sequential nature of these models predicts that in order to affect cognition, Aβ must be dependent on tau and neuroimaging variables, whereas tau must only be dependent on neuroimaging variables. The longitudinal study of Han et al [468] largely supported the prediction with two notable exceptions: Aβ was found to have effect on brain structure and function independent of tau, and tau was found to have effect on baseline cognition independent of neuroimaging measures. All study variables had significant overlap between clinical groups, suggesting that these groups differ quantitatively but not qualitatively. Risacher et al [485] characterized cognition and biomarker levels in the EMCI cohort. The status was associated with impaired cognition, abnormally high levels of tau, and brain atrophy, but not with increased amyloid deposition. In contrast, APOE ε 4 status was significantly associated with increased amyloid deposition and abnormal levels of tau in not only EMCI patients but also the cognitively normal controls. These results suggest that amyloid deposition occurs at an extremely early stage of the disease and that this event is substantially modulated by the APOE ε 4 allele in agreement with the results obtained in the Florbetapir study of Murphy et al [482]. Mouiha et al [486] examined biomarker trajectories, predicted in the model by Jack et al [14] to be sigmoidal in shape. Fitting the data to one of six possible models, they reported that the FDG-PET trajectory appeared to be linear in contrast to all other biomarkers, which best fit either a quadratic model (Aβ42) or a penalized B-spline model (p-tau, t-tau, and hippocampal volume). Some facets of the Jack model [14] were not supported by a study by Yang et al [487] who found that t-tau, p-tau181, and Aβ42 discriminated between controls and both MCI and AD groups, but only Aβ42 discriminated between MCI and AD groups, suggesting that Aβ levels may plateau after tau in the pathologic cascade.
Fig. 17.
Biomarker trajectories through disease progression. For each biomarker, individual z scores are plotted against ADAS-cog (cognitive subscale of the Alzheimer’s Disease Assessment Scale) scores, and the fitted sigmoid curve is displayed. Full circles denote healthy control subjects, full squares MCI patients converted to AD, empty circles early AD, and full triangles late AD patients. Sigmoid fitting was better than linear fitting for tau, Aβ-42, and hippocampus (for the latter: sigmoid nonsignificantly better than linear); linear fitting was better for [18F]-fluorodeoxyglucose-positron emission tomography (FDG-PET). Reproduced with permission from Ref [153].
Table 7.
Mean (standard deviation) of annualized change for selected ADNI variables
| Annualized mean change by diagnosis | ||||||
|---|---|---|---|---|---|---|
| Normal control | MCI | AD | ||||
| Variable name | Mean (SD) | CV | Mean (SD) | CV | Mean (SD) | CV |
| CSF Aβ−42 | −0.94 (18) | 1914 | −1.4 (17) | 1214 | −0.1 (14) | 14,000 |
| CSF tau | 3.45 (13) | 377 | 2.34 (21) | 897 | 1.24 (24) | 1935 |
| PiB | 0.098 (0.18) | 184 | −0.008 (0.18) | 2250 | −0.004 (0.25) | 6250 |
| FDG-PET: ROI-avg | −0.006 (0.06) | 1000 | −0.015 (0.064) | 426 | −0.081 (0.047) | 58 |
| Hippocampus | −40 (84) | 210 | −80 (91) | 114 | −116 (93) | 80 |
| Ventricles | 848 (973) | 115 | 1551 (1520) | 98 | 2540 (1861) | 73 |
| ADAS-cog total | −0.54 (3.05) | 565 | 1.05 (4.40) | 384 | 4.37 (6.60) | 151 |
| MMSE | 0.0095 (1.14) | 12,000 | −0.64 (2.5) | 390 | −2.4 (4.1) | 171 |
| CDR-SB | 0.07 (0.33) | 471 | 0.63 (1.16) | 184 | 1.62 (2.20) | 135 |
| AVLT 5-trial total | 0.29 (7.8) | 2690 | 21.37 (6.6) | 31 | 23.62 (5.6) | 24 |
NOTE. Reproduced with permission from Ref [154].
Fig. 29.
Box plots and superimposed data points showing the distribution of AD biomarkers by baseline diagnosis and visit. The dotted horizontal line extending across all box plots represents the cut point delineating normal from abnormal for each biomarker. Reproduced with permission from Ref [282].
Fig. 36.
(A) Estimated biomarker dynamics as a function of the ADPS score. (B) 90% Confidence intervals for the inflection point of each biomarker. From Jedynak et al [484]. Abbreviations: MMSE, Mini-Mental State Examination; CDR-SB, clinical dementia rating-sum of boxes; MCI, mild cognitive impairment; AD, Alzheimer’s disease.
In seeking an optimum combination of imaging and CSF biomarkers to predict normal control/AD classification, Walhovd et al [155] examined the relationships between the best predictive biomarkers and changes in cognitive scores in the MCI group. They found that changes in MMSE scores correlated with retrosplenial volume and metabolism as well as entorhinal volume, but that only hippocampal volume was associated with the Logical Memory II-DR, and only retrosplenial volume was associated with changes in CDR-SB. No CSF biomarkers were significantly associated with cognitive scores in this clinical group (Table 5). Once again, these results are consistent with the disease progression model in that earlier changes that are reflected in CSF biomarkers do not correlate with clinical measures, whereas changes in brain metabolism and morphometry occur at later stages of the disease and therefore correlate better with cognitive measures. Further support for this model comes from the study of the annual change in MRI and CSF biomarkers and how these are influenced by APOE genotype in control, MCI, and AD subjects [156]. Levels of neither Aβ-42 nor t-tau changed significantly over 12 months in any clinical group, but annual changes in ventricular volume increased with disease severity and were correlated with worsening cognitive and functional indices. APOE ε4 carriers had higher rates of change in ventricular volume, but not in levels of CSF biomarkers, consistent with the model in which levels of Aβ and tau plateau as neurodegeneration becomes detectable by MR measures.
The question of whether structural or metabolic measures are the most sensitive biomarkers of changes associated with early stages of AD was investigated by Karow et al [110]. Directly comparing the ability of MR and FDG-PET measures in prespecified ROIs to detect such changes by quantifying and comparing their effect sizes (Cohen d), they found that largest morphometric effect size (hippocampal volume: 1.92) was significantly greater than the largest metabolic effect size (entorhinal metabolism: 1.43). Both measures were significantly associated with ADAS-cog and AVLT scores in AD patients, but in MCI patients, the relationship was only maintained with hippocampal volume (Table 5). The authors concluded that for the detection of early AD, MRI may be preferable to FDG-PET, as it is more sensitive, more widely available, less invasive, and less costly.
4.4.7. Genetic associations
Following the identification of novel AD genetic risk factors (Section 5), several studies have focused on associations between these risk variants and biomarkers such as amyloid burden and brain atrophy. Nondemented elderly carriers of the AD risk variant rS3818361 SNP in CR1 (A/A or A/G) had a lower brain amyloid burden relative to noncarriers [488]. In addition, noncarriers of the allele (G/G) had a greater variability of amyloid burden, which could be partially accounted for by APOE genotype as the APOE ε 4 allele was associated with higher amyloid burden in noncarriers. However, in carriers of the risk allele, there was no significant difference between the amyloid burden of carriers and noncarriers of APOE ε 4 allele. The finding that carriers of this risk allele have a reduced amyloid burden and that the presence of APOE does not influence this suggests that the CR1 × APOE interaction might modify early changes in AD pathogenesis, influencing brain amyloid levels in nondemented older individuals. Sabuncu et al [489] generated a polygenic score from the aggregate of many genetic markers for disease susceptibility and examined its associations with clinical status, cortical thickness in defined ROIs, and CSF biomarkers. Their polygenic score was significantly associated with clinical status and cortical thickness measurements at predefined ROIs in cognitively normal individuals. Aβ, but not tau, was also significantly associated with a polygenic score and remained associated in individuals with subthreshold levels of CSF Aβ42. These results suggest that AD susceptibility genes may modulate neurodegeneration even in individuals who are cognitively normal and lack amyloid burden.
Recently, maternal history of AD has been linked to risk of LOAD. Honea et al [490] reported a confirmatory study in a larger ADNI data set that examined relationships between family AD history and biomarkers of AD pathophysiology. Patients with maternal but not paternal family history of the disease had significantly higher global PiB uptake and greater PiB uptake in parietal cortex, precuneus, and sensimotor cortex than those with no family history group. In MCI patients, only a maternal family history was associated with significantly increased CSF Aβ42 and higher t-tau/Aβ42. Results are consistent with earlier findings that suggest that early changes in the disease process are associated with a maternally inherited genetic factor.
4.4.8. Summary and conclusions of papers concerning associations of the ADNI cohort
ADNI has succeeded in recruiting a cohort of MCI and mild AD patients that mirrors populations used for clinical trials of AD therapies. A number of cross-sectional and longitudinal studies have lent support to a model of disease progression in which the earliest indications of neurodegeneration occur within the MTL, particularly the hippocampus, and atrophy becomes more widespread in later stages, ultimately encompassing areas of the parietal, occipital, and frontal lobes. Rates of atrophy are initially fastest in the temporal lobe, but accelerate in other regions as the disease progresses. Cortical atrophy and that of specific regions identified in the model of disease progression as well as ventricular enlargement have been correlated with measures of clinical severity. Structure–function relationships within the brain are being elucidated with findings that atrophy in dissociable anatomic regions, especially within the episodic memory network, is associated with different cognitive functions. Patterns of glucose hypometabolism associated with AD have been identified, with the precuneus and posterior cingulate typically displaying the most reduced CMRgl and with reduced metabolism in these key areas being associated with lower scores on cognitive tests. The differential effects of an SNP in brain-derived neurotrophic factor suggest that genetics may modulate glucose metabolism. Levels of CSF biomarkers, particularly Aβ and tau, have been associated with earlier stages of neurodegeneration. 11C-PiB PET Aβ imaging has largely confirmed that decreased levels of CSF Aβ and increasing 11C-PiB PET represent an early event in disease progression, and neither amyloid imaging nor studies of CSF biomarkers have found that levels of these biochemicals are strongly associated with cognitive decline. Levels of CSF biomarkers have been found to be abnormal (i.e., decreased CSF Aβ and increased CSF tau) early in disease and then plateau with little detectable change, whereas glucose metabolism remains relatively stable until the latest stages of disease progression. Presence of the APOE ε4 allele has been shown to enhance neurodegeneration and to modulate levels of CSF biomarkers, but the exact mechanism by which it exerts its effect remains unclear. Likewise, the role of BMI has been the subject of contradictory reports, and it is unknown whether changes in BMI influence disease development or occur as a result of the disease.
In 2011–2012, evidence accumulated supporting the disease model of Jack et al [14], and detailing how hippocampal atrophy is associated with neocortical atrophy or neuropsychological measurements. The relationship between amygdalal atrophy and cognitive decline revealed parallels with the hippocampal atrophy – cognitive decline relationship, suggesting that this structure warrants further investigation. There was further development of the use of summary scores based on MRI data reflecting the degree of AD-like neuroanatomical changes as an indicator of disease status. The importance of functional decline, in addition to decline in the traditional cognitive domains was reflected in a group of studies highlighting the associations between difficulties in performing daily living activities and various biomarkers. The association of the cognitive reserve with cognition and biomarkers was reported and provided evidence for a model in which the degree of cognitive reserve affects curves of CSF biomarkers throughout disease progression. Finally, studies provided insight into possible mechanisms by which the known AD risk factors, BMI and high homocysteine levels may act by investigating their associations with AD biomarkers.
The model by Jack et al [14] was again well supported in numerous studies published in 2012 and 2013, including a data-driven observational study across all ADNI participants. Florbetapir binding was shown to correlate with clinical status and CSF Aβ levels and to be modulated by APOE ε 4 allele, indicating this radiotracer to be an accurate biomarker of amyloid deposition. The deleterious effects of the APOE ε 4 allele were documented in repeated works, and studies of genetic associations extended to include novel risk variants identified and confirmed in GWAS studies. Glucose hypometabolism in specific regions was associated with measures of cognitive decline. The newly introduced ASL modality revealed that the changes in CBF in regions identified by FDG-PET as typically hypometabolic in AD occur later in disease progression. This technique warrants further investigation as a potential biomarker. Increasing recognition that AD patients may also harbor white matter changes leads to the inclusion of DTI in the ADNI-2 protocol. WMHs detected by this method were correlated with clinical status and may significantly disrupt structural connectivity in later stages of the disease. The common comorbidity of AD with Lewy body dementias suggests that α-syn, which was associated with clinical groups, CSF biomarkers, and memory measures, may also have potential as a biomarker. Finally, studies have suggested that cognitive reserve is the result of improved functional connectivity and not increased neural substrate.
4.5. Diagnostic classification of study participants
The ability to accurately diagnose to which clinical group a subject belongs is a crucial one in the clinical trial design. To this end, some researchers have investigated the ability of individual MRI, FDG-PET, and CSF biomarkers to discriminate between ADNI AD participants and ADNI control subjects, and between MCI-c and MCI-nc subjects. Others have tried to determine the optimum combination of these biomarkers for ADNI participant classification, with many studies leveraging knowledge of associations between various structural and fluid biomarkers and the sequence of brain morphometric change over the course of disease to guide development of marker combinations. Discrimination between the clinically distinct ADNI participant groups offers an important first step in identifying biomarker diagnostic tools that can be validated in representative population-based studies before clinical use. More recently, a view has emerged of AD as a continuum of increasing pathology and clinical manifestation of symptoms. Accordingly, more studies have focused on predicting continuous variables instead of binary classification.
4.5.1. Magnetic resonance imaging
4.5.1.1. Temporal lobe structures
Atrophy of the hippocampus, the best studied structure affected by AD, has been used in patient classification by a number of groups. Chupin et al [68] correctly distinguished AD patients from control subjects 76% of the time, and MCI patients who would convert within 18 months from control subjects 71% of the time (Table 8). Karow et al [110] found that hippocampal volume discriminated between control subjects and AD patients with an AUC of 0.90, and between control subjects and MCI patients with an AUC of 0.75 (Table 8). The discriminative ability of the rate of hippocampal atrophy was investigated by Wolz et al [64], who found that their method correctly classified 75% to 82% of AD patients and 70% of MCI patients who converted to AD over 12 months. Their method was also able to discriminate between MCI-c and MCI-nc patients at a rate of 64% (Table 8). Lotjonen et al [231] compared Wolz’s method to their own automatic hippocampal extraction method using the same data and found that it resulted in significantly more accurate classification of both AD patients from controls and MCI converters from non-converters (Table 8). The division of the hippocampus into head, body and tail subregions extracted further information from this structure for use in diagnostic classification. Greene et al [267] found that the combined left and right hippocampal head produced the most accurate classifications of any hippocampal subregions, but that the best accuracy was attained by a combination of left hippocampal body, right hippocampal tail, AVLT and Digit Symbol which classified controls, MCI and AD patients with accuracies of 95.5%, 82.4% and 78.9%, respectively. Calvini et al [66] derived a statistical indicator from the hippocampus and other MTL structures and were able to discriminate between AD and control groups, and between MCI and control groups, with AUCs of 0.863 and 0.746, respectively (Table 8). The classification index of Chincarini et al [283] used seven maximally discriminative small volumes in the medial temporal lobe to distinguish between AD and control groups, MCI and control groups and MCI converters from non-converters with AUCs of 0.97, 0.92 and 0.74, respectively (Table 8).
Table 8.
Methods for the classification of MCI and AD patients
| Control vs AD | Control vs MCI | Control vs MCI-c | MCI-c vs MCI-nc | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | SEN | SPE | ACC | AUC | SEN | SPE | ACC | AUC | SEN | SPE | ACC | AUC | SEN | SPE | ACC | AUC | Cross-validated? | Reference |
| Hippocampal volume | 75 | 77 | 76 | 61 | 71 | 72 | 72 | 67 | 66 | 60 | [68] | |||||||
| Hippocampal volume | 82 | 89 | 49 | 89 | 62 | 67 | [68]** | |||||||||||
| 12-month hippocampal atrophy rates | 81 | 83 | 82 | 0.88 | 59 | 71 | 63 | 0.71 | 73 | 78 | 76 | 62 | 68 | 66 | [60] | |||
| Hippocampal volume | 79.7 | 63 | [283] | |||||||||||||||
| Compared to 12 month hippocampal atrophy rates using same data |
71.6 | 60 | ||||||||||||||||
| MTL structural atrophy | 74 | 85 | 0.86 | 45 | 85 | 0.75 | 85 | 83 | 0.88 | [66] | ||||||||
| SPS score | 82 | 0.97 | 76 | 0.85 | Yes | [83] | ||||||||||||
| SPS score | 71 | 77 | 70 | 73 | 62 | 69 | [83]** | |||||||||||
| ROI atrophy score | 83 | 93 | 89 | 0.92 | Yes | [117] | ||||||||||||
| DBM-multidimensional scaling | 86.3 | [75] | ||||||||||||||||
| Cortical thickness | 82 | 93 | 66 | 76 | 63 | 76 | [107] | |||||||||||
| Single template TBM | 74 | 84 | 80 | 68 | 65 | 66 | Yes | [81] | ||||||||||
| Multiple template TBM | ||||||||||||||||||
| Mean deformation | 78 | 88 | 84 | 64 | 69 | 68 | ||||||||||||
| Mean of Jacobians | 79 | 91 | 86 | 74 | 68 | 70 | ||||||||||||
| Mean of features | 81 | 91 | 86 | 64 | 63 | 63 | ||||||||||||
| Combination of classifiers | 79 | 90 | 85 | 65 | 63 | 64 | ||||||||||||
| Classification index | 89 | 94 | 0.97 | 89 | 80 | 0.92 | 72 | 65 | 0.74 | Yes | [215] | |||||||
| ROI atrophy score | 81.5 | 0.70 | Yes | [118] | ||||||||||||||
| Semisupervised SVM, SPS score | 79.6 | 85.7 | 82.9 | 0.69 | Yes | [93] | ||||||||||||
| ICA and SVM–gray matter | 81.9 | 79.5 | 80.7 | 73.2 | 68.6 | 71.1 | [94] | |||||||||||
| Combined MR features | Yes | [102] | ||||||||||||||||
| Linear Discriminant analysis | 85 | 93 | 82 | 86 | 69 | 67 | ||||||||||||
| SVM | 75 | 94 | 67 | 93 | 14 | 92 | ||||||||||||
| Age correction MR data | Yes | [261] | ||||||||||||||||
| No age correction | 83 | 85 | ||||||||||||||||
| Age correction applied | ||||||||||||||||||
| FDG-PET abnormality index | 83 | 78 | 0.90 | [84] | ||||||||||||||
| FDG-PET functional connectivity | 88 | 88 | Yes | [65] | ||||||||||||||
| FDG-PET factor analysis feature selection | 98.1 | 92.5 | 95.2 | 91.2 | 80.8 | 88.0 | Yes | [90] | ||||||||||
| FDG-PET information retrieval/residual vectors plus FAQ scores |
94.4 | 84.8 | 93.6 | 89.7 | 62.9 | 76.5 | [242] | |||||||||||
| FDG-PET baseline and 12 month ROIs combined |
83.2 | 93.6 | 88.4 | 79.8 | 82.9 | 81.3 | 52.2 | 73.2 | 63.1 | [285] | ||||||||
| FDG-PET non-negative matrix factorization | 87.5 | 85.4 | 86.6 | [249] | ||||||||||||||
| AD-like brain regions | Yes | [88] | ||||||||||||||||
| MRI | 85 | 80 | 82 | 0.88 | ||||||||||||||
| FDG-PET | 84 | 82 | 84 | 0.87 | ||||||||||||||
| Hippocampal volume | [110] | |||||||||||||||||
| MRI | 0.90 | 0.75 | ||||||||||||||||
| Entorhinal metabolism | ||||||||||||||||||
| FDG-PET | 0.71 | 0.63 | ||||||||||||||||
| AD-like brain regions–Bayesian approach | 87.6 | [89] | ||||||||||||||||
| Hippocampal volume, ventricular expansion, APOE, age |
82 | 0.95 | Yes | [86] | ||||||||||||||
| Hippocampal volume, ventricular expansion, age |
71 | 0.86 | ||||||||||||||||
| T-tau | 69.3 | 92.3 | 80.6 | 0.83 | [57] | |||||||||||||
| Aβ–42 | 96.4 | 76.9 | 87.0 | 0.91 | ||||||||||||||
| p-tau181p | 67.9 | 73.1 | 70.4 | 0.75 | ||||||||||||||
| t-tau/Aβ–42 | 85.7 | 84.6 | 85.2 | 0.92 | ||||||||||||||
| p-tau181p/Aβ–42 | 91.1 | 71.2 | 81.5 | 0.86 | ||||||||||||||
| LRTAA model | 98.2 | 79.5 | 89.9 | 0.94 | ||||||||||||||
| Cortical normalized thickness index (NTI) | 0.76 | Yes | [165] | |||||||||||||||
| AVLT | 0.67 | |||||||||||||||||
| ADAS-cog–DR | 0.67 | |||||||||||||||||
| MMSE | 0.64 | |||||||||||||||||
| Longitudinal cortical thickness: static, dynamic, and network features |
96.1 | 81.7 | 0.88 | [225] | ||||||||||||||
| MRI: Hippocampal volume, entorhinal thickness, retrosplenial thickness |
85.0 | [155] | ||||||||||||||||
| FDG-PET: Entorhinal, retrosplenial, lateral orbitofrontal metabolism |
82.5 | |||||||||||||||||
| CSF: t-tau/Aβ–42 | 81.2 | |||||||||||||||||
| Combination: Hippocampal volume, retrosplenial thickness; entorhinal, retrosplenial, orbitofrontal metabolism, t- tau/Aβ–42 |
88.8 | |||||||||||||||||
| t-tau/Aβ–42, left entorhinal cortex, hippocampal volume |
82.5 | 90.1 | 86.7 | Yes | [161] | |||||||||||||
| t-tau/ Aβ–42, RAVLT immediate and delayed recall, TMT–B* |
93.8 | 95.6 | 94.8 | |||||||||||||||
| LTRAA, left entorhinal cortex, hippocampal volume |
90.1 | 92.1 | 91.1 | |||||||||||||||
| LTRAA, left entorhinal cortex, hippocampal volume, RAVLT immediate and delayed recall, TMT-B |
92.2 | 97.5 | 95.2 | |||||||||||||||
| ADAS-cog | 0.93 | Yes | [87] | |||||||||||||||
| FDG-PET–30 best features | 0.94 | |||||||||||||||||
| Combined classifier | 0.97 | |||||||||||||||||
| Multiple Kernel Learning | Yes | [247] | ||||||||||||||||
| Imaging modalities | 78.9 | 93.8 | 87.6 | 0.94 | ||||||||||||||
| Biological markers | 58.1 | 79.4 | 70.4 | 0.77 | ||||||||||||||
| Cognitive scores | 89.2 | 92.6 | 91.2 | 0.98 | ||||||||||||||
| All modalities | 86.7 | 96.6 | 92.4 | 0.98 | ||||||||||||||
| Multi-modal multi-task learning | 93.3 | 83.2 | Yes | [250] | ||||||||||||||
| Multi-modal: kernel combination | 93 | 93.3 | 93.2 | 81.8 | 66.0 | 76.4 | Yes | [249] | ||||||||||
| Multi-modal automatic data-driven | 96.4 | 48.3 | 67.1 | 0.80 | Yes | [287] | ||||||||||||
| Blood-based biomarkers | ||||||||||||||||||
| 11 serum/plasma proteins | 54 | 78 | 0.70 | |||||||||||||||
| 11 serum/plasma proteins + age, sex, education and APOE |
79 | 87 | 0.88 | |||||||||||||||
| Blood-based biomarkers | [286] | |||||||||||||||||
| 11 item signature + APOE | 64.8 | 85.9 | ||||||||||||||||
| 11 item signature − APOE | 71.1 | 77.0 | ||||||||||||||||
| Metafeature signature + APOE | 86.7 | 85.6 | ||||||||||||||||
| Metafeature signature − APOE | 83.0 | 90.7 | ||||||||||||||||
| Age stratification | CN vs MCI and AD | [288] | ||||||||||||||||
| ACC | AUC | |||||||||||||||||
| <75 | >75 | All | <75 | >75 | all | |||||||||||||
| Neuropsychological measures | 84 | 85 | 84 | 0.92 | 0.94 | 0.93 | ||||||||||||
| CSF biomarkers (t-tau/Aβ42) | 77 | 70 | 73 | 0.83 | 0.71 | 0.77 | ||||||||||||
| MRI ROIs | 84 | 83 | 82 | 0.88 | 0.88 | 0.88 | ||||||||||||
| FDG-PET ROIs | 79 | 75 | 76 | 0.86 | 0.69 | 0.77 | ||||||||||||
| Combined modalities | 92 | 88 | 85 | 0.96 | 0.95 | 0.93 | ||||||||||||
Abbreviations: t-tau, total tau; SEN, sensitivity; SPE, specificity; ACC, accuracy; AUC, area under the curve; SPS, structural phenotypic score; MTL, medial temporal lobe; DBM, deformation-based morphometry; SVM, support vector machine; ICA, independent component analysis.
MRI measures no longer significant in this model
in [158].
4.5.1.2. Multiple ROIs and whole brain
Other methods have focused on many ROIs across the brain, using the degree of association with AD to construct a score reflective of the anatomic profile of AD. These include temporal, cingulate, and orbitofrontal regions. The classifier developed by Fan et al [83] produced an SPS that allowed direct comparison of patients and was able to discriminate between AD and control subjects, between MCI and control subjects, and between AD and MCI subjects with AUCs of 0.965, 0.846, and 0.750, respectively (Table 8). Similarly, Misra et al [118] extracted an abnormality score that discriminated MCI-c patients from MCI-nc patients with a classification accuracy of 81.5 and an AUC of 0.77 (Table 8). Using a semisupervised SVM, Filipovych and Davatzikos [93] discriminated between MCI-c and MCI-nc patients with an AUC of 0.69, comparing favorably with fully supervised SVM methods (Table 8). They also found that 79.4% of all converters were classified as AD-like (the remainder being classified as normal-like). In addition, 51.7% of nonconverters were classified as normal-like and the remainder as AD-like, perhaps representing a proportion of MCI patients who would convert to AD further in the future. The authors also found that semisupervised SVM performed better than a fully supervised SVM in instances when there were a small number of labeled images. The classifier developed by Yang et al [94], which relied on image features defined by ICA, discriminated between control and AD subjects with an accuracy of 80.7%, a sensitivity of 81.9%, and a specificity of 79.5%, and between control and MCI subjects with an accuracy of 71.1%, a sensitivity of 73.2%, and a specificity of 68.6%, based on GM images and a training set-to-test set ratio of 90%:10% (Table 8).
McEvoy et al [117] presented data from their fully cross-validated linear discriminant model compared with partially cross-validated models, and found that the fully cross-validated model discriminated between AD and control subjects with an accuracy of 89%, a sensitivity of 83%, a specificity of 93%, and an AUC of 0.915 (Table 8). They noted that these numbers were lower than those obtained using the partially cross-validated model, suggesting that numbers presented by other studies using partially cross-validated models may be artificially high. Hinrichs et al [88] used a classifier based on GM probability maps and found that it discriminated between AD and control subjects with a sensitivity of 85% and a specificity of 80%. Park and Seo [75] tested their method of multidimensional scaling (MDS) of DBM and compared it with the ability of hippocampal volume to discriminate between AD and control subjects. They found that their MDS method outperformed hippocampal volume, yielding accuracies of 86.3% and 75.0%, respectively (Table 8). Further details of classifier construction using SVMs are given in the Methods section 3.3.
Longitudinal measurements of cortical thickness were the focus of a classifier constructed by Li et al [157]. They found that although the pattern of cortical thinning was similar in all patient groups, the rate of thinning and ratio of follow-up to baseline measures provided a better tool for distinguishing between MCI-c and MCI-nc patients. An additional complementary component in the form of a brain network feature computed from the correlations of cortical thickness changes with ROIs further improved classification accuracy. The final classifier, comprising static, dynamic, and network measures, discriminated between normal control subjects and AD patients with an accuracy of 96.1%, and between MCI-c and MCI-nc patients with an accuracy of 81.7% (Table 8). The classifier of Park et al [383] based on cortical shape and sulcal depths discriminated between control and MCI patients with an accuracy of 73%, a sensitivity of 73%, and a specificity of 73%. Noise sensitivity and spatial variation problems of other cortical thickness estimation methods were overcome by more robust method of Cho et al [237] which discriminated successfully between control and AD patients or MCI converters, or between MCI converters and non-converters (Table 8). An automated method developed by Pachauri et al [232] to leverage information found in cortical surface topology boosted the classification accuracy of hippocampal volume in discriminating between AD and control patients by 4% and of other ROIs by around 3%.
The penalized logistic regression approach of Casanova et al [241] to the high dimensional classification of patients from MRI data discriminated between AD patients and controls with accuracies, specificities and sensitivities of 85.7%, 90% and 82.9%, respectively, using GM and 81.1%, 82.5% and 80.6%, respectively, using WM. The effect of registration to multiple templates on classification accuracy of TBM was investigated by Koikkalainen et al [234] who found that all 4 multi-template methods investigated resulted in better discrimination of both AD from controls patients and MCI converters from non-converters (Table 8).
Applying a correction to account for age-related atrophy in controls was shown by Franke et al [92] to increase the accuracy of classification of AD patients from controls from 83% to 85%, indicating that controlling for the effects of such confounding variables as age is critical to achieving clinically useful classification accuracies with MR data.
Leveraging information contained within structural and functional connectivities in imaging data has increasingly been a focus for improving classification. Cuingnet et al [389] included a regularization step to take these relationships into account and found that the addition of the step to an SVM increased classification accuracy over an SVM alone. Using cortical thickness measures, AD patients were discriminated from normal controls with an accuracy of 87%, a sensitivity of 83%, and a specificity of 90% compared with the SVM alone, which achieved an accuracy of 83%, a sensitivity of 74%, and a specificity of 90% in the same classification challenge. Likewise, a method [384] that takes into account the relationships between morphologic features of ROIs augmented classification accuracy over ROIs alone, and an approach that integrated the correlative data via multikernel SVMs further improved classification accuracy. The integrated approach discriminated between AD and controls with an accuracy of 92.4% and between MCIc and MCInc patients with an accuracy of 75%. The OLPS score of Spulber et al [382], reflective of the degree of AD-like neurodegeneration, achieved a sensitivity of 86.1%, a specificity of 90.4%, an accuracy of 88.4%, and an AUC of 0.948 for the same classification challenge. An improvement to the feature selection step proposed by Liu et al [388], which takes into account pathologic degeneration, achieved classification accuracies of 90.2%, 87.2%, and 70.7% for the classification of AD patients versus controls, MCIc versus controls, and MCIc versus MCInc patients, respectively.
Combining automatically estimated features from different structural MRI analysis techniques augmented classification accuracy in a study by Wolz et al [284]. When TBM, hippocampal volume, cortical thickness and a manifold-based learning framework were combined, they improved classification accuracy over single features using both a SVM and linear discriminant analysis (Table 8). A novel dimensionality reduction approach [379] improved classification accuracies over a DBM–based technique and discriminated between AD and control patients and between MCI and control patients with accuracies of 84% and 76%, respectively. Likewise, the tree-guided sparse coding method of Liu et al [391] was able to achieve better classification to L1-regularized Lasso alone using fewer features that were concentrated in areas known to be most representative of AD such as the hippocampus, entorhinal cortex, parahippocampal gyrus, and amygdala. The addition of LLE as a method of dimensionality reduction improved the efficacy of three different classifiers [396]. For example, MRI features alone using a SVM discriminated between control and AD patients with an accuracy of 50%, a sensitivity of 48%, and a specificity of 51%. The results improved to an accuracy of 90%, a sensitivity of 87%, and a specificity of 92% with the addition of LLE.
Normalization approaches in automated structural MRI processing pipelines can affect classification accuracy. Using non-normalized cortical thickness measures and volumetric measures normalized to intracranial volume, Westman et al [491] achieved a classification accuracy of 91.5% between control and AD patients and of 75.9% between MCI converters and nonconverters.
4.5.1.3. White matter hyperintensities
Provenzano et al [441] used the level of WMHs in patients who were PiB positive for diagnostic classification. At a cutoff of 1.25 cm3 for “high” WMH, they were able to discriminate between AD and control patients with a sensitivity of 83% and a specificity of 64%.
4.5.1.4. Comparison of MRI methods
Cuingnet et al [158] directly compared 10 methods for the automatic classification of AD patients from anatomical MR data using the ADNI database. Five voxel-based approaches, three cortical approaches, and two methods based on hippocampal shape and volume were tested for their ability to discriminate between control, MCI-c, MCI-nc, and AD subjects. They found that voxel- or cortical thickness-based whole brain methods yielded highest sensitivities for AD versus control subjects (maximum of 81%), but that sensitivities were substantially lower for discriminating between MCI-c and MCI-nc subjects (maximum of 70%). Casanova et al [411] compared classification data from a number of studies and illustrated the difficulty in directly comparing studies that differ in many aspects beyond simple methodology. Eskildsen et al [386] directly tested the effects of “double dipping” (the practice of reusing the training set as the test set) by using a dependent test set. They found that the practice artificially inflated classification accuracies, in one case to 87.4% from 74.6%. Young et al [408] noted the difficulty in comparing studies of the prediction of conversion because of differences in the length of time to conversion or because of the use of longitudinal data versus baseline data. These studies again emphasize the difficulty in comparing the results from different studies using different methodologies. For this reason, the 2012–2013 update has not included additional classification data in Table 8. As ADNI progresses, it is increasingly clear that standardized systematic studies of the cohort such as those reported by Cuingnet et al [158] are required to make sound conclusions regarding the efficacy of each technique.
Combining automatically estimated features from different structural MRI analysis techniques augmented classification accuracy in a study by Wolz et al [284]. When TBM, hippocampal volume, cortical thickness and a manifold-based learning framework were combined, they improved classification accuracy over single features using both a SVM and linear discriminant analysis (Table 8).
4.5.2. [18F]-fluorodeoxyglucose-positron emission tomography
As AD affects not only morphology but also metabolism in the brain, Haense et al [84] used the AD t-sum measure of scan abnormality from FDG-PET data to discriminate between AD and control subjects with a sensitivity of 83% and a specificity of 78% (Table 8). The HCI of Chen et al [85], which also capitalized on hypometabolism data across the entire brain, was significantly different in control, MCI-nc, MCI-c, and AD subject groups. The method of Hinrichs et al [88], described in the MRI section, was also used with FDG-PET data and was able to discriminate between AD and control subjects with a sensitivity of 78% and a specificity of 78% (Table 8). Huang et al [65] used FDG-PET data to examine functional connectivity between brain regions and then leveraged the patterns they found to be typical of AD for classification purposes. They found that compared with control subjects, AD patients had decreased temporal lobe inter-regional connectivity, especially in the hippocampus, and weaker between-lobe and between-hemisphere connectivity. In contrast, MCI patients had increased connectivity between occipital and frontal lobes compared with control subjects, illustrating the uniqueness of this condition. This method discriminated between AD and control subjects with a specificity of 88% and a sensitivity of 88% (Table 8). Using their method based on feature selection using factor analysis and an SVM, Salas-Gonzalez et al [90] discriminated between AD and control subjects with sensitivity, specificity, and accuracy of 98.1%, 92.5%, and 95.2%, respectively, and between MCI and control subjects with sensitivity, specificity, and accuracy of 92.1%, 80.8%, and 88.0%, respectively (Table 8). The classifier constructed by Clark et al [242] based on information retrieval techniques was able to discriminate between control and AD patients with a sensitivity, specificity and accuracy of 94.4%, 92.5% and 93.6%, respectively and between MCI converters and non-converters with a sensitivity, specificity and accuracy of 89.7%, 62.9% and 76.5%, respectively when the model include FAQ scores (Table 8). An alternative approach using non-negative matrix factorization was described by Padilla et al [248] and achieved an accuracy of 86.6%, a sensitivity of 87.5% and a specificity of 85.4% in the classification of AD patients from controls (Table 8). Having identified entorhinal metabolism as the FDG-PET measure with the largest effect size for the detection of early AD, Karow et al [110] found that this measure discriminated between control and AD subjects with an AUC of 0.71, and between control and MCI subjects with an AUC of 0.63 (Table 8). Mormino et al used 11C-PiB PET imaging to deduce a cutoff point to optimally separate PiB-positive from PiB-negative MCI patients, and found that PiB-positive MCI patients had lower hippocampal volumes and greater episodic memory loss compared with MCI patients with 11C-PiB levels below the cutoff point of 1.465. The selection of maximally discriminative voxels using significance measures was used in a classifier described by Martinez-Murcia [398] and applied to ADNI FDG-PET data. It discriminated between AD and control patients with an accuracy of 91%, a sensitivity of 92%, and a specificity of 89%. The addition of longitudinal data to baseline data to improve classification accuracy from anatomically selected features of FDG-PET scans was the approach taken by Gray et al [285]. Across all categories, improved classification accuracies were reported, ranging from 65% in the MCI converter versus non-converter classification to 88% in discriminating between control and AD patients (Table 8).
Arbizu et al [397] used their AD-conv score, based primarily on hypometabolism in the posterior cingulate area in addition to clinical and cognitive variables to divide MCI patients into groups with different probabilities of progressing to AD. These ranged from 8% in the first sixtile to 38% in the fourth sixtile to 100% in the top sixtile. This approach differs from conventional binary approaches to classification and emphasizes the concept of AD as a continuum of disease. The classification of MCIc versus MCInc patients by this method was improved significantly with the addition of clinical variables: the combined modalities reached an accuracy of 82%, a sensitivity of 85%, and a specificity of 80%.
A direct comparison of the diagnostic ability of three methods summarizing FDG PET data into a single score was made by Caroli et al [492] in three data sets including ADNI. In the ADNI cohort, the hyperbolic convergence index (HCI) [85] significantly outperformed the PMOD Alzheimer discrimination analysis tool [84] and a set of meta-analytically derived regions of interest (metROI) [135]. However, other methods were superior in other cohorts. Classification using all indices improved with increasing disease severity with AUCs ranging from 0.800 to 0.949 (PALZ), 0.774 to 0.967 (metaROI), and 0.801 to 0.983 (HCI), for the classification of MCIc and moderate AD compared with controls, respectively. The authors concluded that all three indices are differentially sensitive to disease severity and are therefore of utility in the detection of disease in both research and clinical settings.
4.5.3. CSF biomarkers
Shaw et al [57] examined CSF biomarkers in the ADNI cohort as well as in a cohort of non-ADNI autopsy-confirmed AD patients, with the goal of developing a “biomarker signature” best able to predict AD and to classify patients correctly. Like many smaller studies, they found that t-tau and p-tau181p, as well as the t-tau/Aβ-42 and p-tau181p/Aβ-42 ratios, all increased in MCI patients compared with control subjects, whereas CSF Aβ-42 decreased. The best single measure for discriminating between AD and control subjects was CSF Aβ-42, which had an AUC of 0.913, a sensitivity of 96.4%, a specificity of 76.0%, and an accuracy of 87% (Table 8). Linear regression analyses determined which variables, including APOE genotype, contributed most to the discrimination, and a final linear regression model, which included Aβ-42, APOE ε4 carriers, and t-tau (LRTAA model), resulted in enhanced discrimination over individual factors (Table 8). De Meyer et al [159] used an unsupervised learning method that did not presuppose clinical diagnosis to identify biomarkers of AD. A mixture modeling approach derived a signature, consisting of both Aβ-42 and t-tau concentrations, which had a sensitivity of 94% in autopsy-confirmed AD patients from an independent cohort and was present in 90%, 72%, and 36% of patients with AD, MCI, and no cognitive impairment, respectively (Fig. 18). APOE ε4 carriers were over-represented in those patients with the AD biomarker signature by a factor of 6.88:1. Interestingly, when modeling single biomarkers, the cutoff concentration of Aβ-42 that optimally delineated AD patients from healthy elderly subjects was found to be 188 pg/mL, close to that found by Shaw et al [57] and Schott et al [160]. Moreover, the proportion of healthy elderly subjects with an identifying AD CSF biomarker signature was similar to that found by Schott et al [160], and likely reflects a proportion of cognitively normal elderly subjects who will progress to MCI and AD in the future. Further, De Meyer et al [159] examined another data set with MCI patients (n = 57) followed up for 5 years, and they showed that their model had a sensitivity of 100% in patients progressing to AD. The finding that AD pathology is detectable in significant numbers of healthy elderly control subjects has important implications for future clinical trials and suggests the possibility of presymptomatic treatment studies of potential AD-preventive compounds. Initial studies have reported that given the often shared pathology between AD and Lewy body dementias such as Parkinson’s disease, CSF levels of α-syn may have potential as a biomarker of AD. Korff et al [479] found that the marker alone offered only modest sensitivity (65%) and specificity (74%) and an AUC of 0.719 for the classification of AD from control patients. However, the addition of either α-syn levels alone or the ratio of α-syn to p-tau181 to established CSF biomarkers improved classification accuracy between both controls and AD patients and between MCI converters and nonconverters [478], suggesting that in combination of other markers, α-syn may represent a useful additional tool for AD diagnostics.
Fig. 18.
Separation of control, MCI, and AD subjects using a CSFAβ-42/t-tau mixed model signature. A combined CSFAβ-42/t-tau mixed model was applied to the subject groups. Densities of each signature are represented with confidence ellipses, and signature membership of the subject based on the mixture is indicated with the corresponding color (signature 1 is the AD signature [red]; signature 2 is the healthy signature [green]). Reproduced with permission from Ref [159].
4.5.4. Clinical
Llano et al [96] compared the ADAS-cog and MMSE tests with a new form of ADAS-cog in which the subscores were given weights using a Random Forests tree algorithm, thereby resulting in a new metric, the composite ADAS.-Tree. Therefore, ADAS.Tree represents a multivariate model in which subscales have been weighted according to their importance in discriminating between AD and control subjects. When the ability of ADAS.Tree to classify control, MCI, and AD subjects was compared with that of ADAS-cog and MMSE, the composite model generated a numerically highest test statistic. The authors suggest that this derivative of an internationally recognized and easily administered test may offer a more widely useful and less expensive approach to other imaging and CSF biomarkers that can be invasive and/or expensive.
Another cognitive test that may have utility in diagnosing MCI is the FAQ. Brown et al [274] identified two items of the FAQ, the ability to keep business papers organized and the ability to remember important dates and occasions, that effectively differentiated between control and MCI patients. Ito et al [493] proposed a bounded model which takes into consideration the nonnormal distribution of data at the boundaries of the FAQ scores. The censored approach greatly improved predictability of disease progression in FAQ scores over a standard approach.
4.5.5. Blood based biomarkers
A new direction of research in 2011–2012 has been the development of blood-based biomarkers for diagnostic classification as a potentially more clinically feasible alternative to more costly or invasive modalities as a first line screening method for the disease. O’Bryant et al [255] constructed a classifier from blood based markers that were highly correlated across both serum and plasma. These 11 proteins were comparable to CSF biomarkers in their ability to discriminate AD patients from cognitively normal controls, but the addition of demographic data (age, sex, education, APOE status) resulted in a model with similar classification accuracies to the best CSF-based models (Table 8). A study of potential plasma based markers by Johnstone et al [286] identified 11 analytes that were maximally discriminative between controls and MCI converters. Once again, APOE status increased classification accuracy (Table 8). The refinement of the model by the addition of ‘metafeatures,’ able to identify and leverage information from potentially biologically linked features, further enhanced accuracy (Table 8). A proteomic signature identified by Llano et al [422] from a bank of plasma analytes using a multivariate analysis contained 14 analytes and discriminated between control and AD patients with a sensitivity of 86.5%, a specificity of 84.2%, and an AUC of 85.3%. The blood-based panel of analytes developed by Doecke et al [421], which overlapped with biomarkers selected by both O’Bryant et al [494], was able to discriminate AD from control patients with an accuracy of 83% and an AUC of 85%. They found that the inclusion of APOE status, education, BMI, age, and sex also strengthened the model. Dubey et al [413] compared the diagnostic ability of proteomic versus MRI data and found that proteomic data were on a par with MRI data in discriminating between controls and AD patients and had increased accuracy, sensitivity, and specificity in diagnosing MCI patients.
4.5.6. Combined modalities
4.5.6.1. Early approaches
The approach of Kohannim et al [86] combined multiple factors, including MRI and FDG-PET measures, CSF biomarkers, APOE genotype, age, sex, and BMI, to enhance machine learning methods for AD diagnosis. They found that the optimum combination of factors to discriminate between AD and control subjects—hippocampal volume, ventricular expansion, APOE genotype, and age—yielded an AUC of 0.945 with an accuracy of 82%, whereas to detect MCI patients, the optimum combination of hippocampal volume, ventricular expansion, and age yielded an AUC of 0.860 and an accuracy of 71% (Table 8). Walhovd et al [155] likewise sought the optimum discriminatory combination of biomarkers. They found that the best MRI combination to discriminate between AD and control subjects consisted of hippocampal volume, entorhinal thickness, and retrosplenial thickness (85% accuracy); the best FDG-PET combination was entorhinal, retrosplenial, and orbitofrontal metabolism (82.5% accuracy); and the best CSF combination was t-tau/Aβ-42 (81.2% accuracy). Using stepwise linear regression, they developed a final model that included retrosplenial thickness and the t-tau/Aβ-42 ratio as predictors and which achieved 88.8% accuracy in the classification of AD versus control subjects. For the discrimination of MCI from control subjects, the optimum combination of factors was found to be hippocampal volume and the t-tau/Aβ-42 ratio, with an accuracy of 79.1 % (Table 8). Ewers et al [161] tested a variety of cross-validated models of single or multiple predictors for their ability to discriminate between control and AD subjects. They found that the addition of neuropsychological tests, specifically the AVLT immediate free recall and DR and the TMT-B, to models that included only CSF and/or genetic biomarkers and imaging measures resulted in increased overall classification accuracy. The best model, which included CSF t-tau/Aβ-42, the number of APOE ε4 alleles (the previously described LRTAA model [57]), left entorhinal volume, and hippocampal volume, in addition to the aforementioned neuropsychological tests, resulted in an accuracy of 95.2%, a sensitivity of 92.2%, and a specificity of 97.5% (Table 8). Van Gils et al [162] also demonstrated that cognitive tests such as the CDR, MMSE, and the neuropsychological battery comprised the most important feature category of all classifiers designed to discriminate between different patient groups. The classifier constructed by Lemoine et al [87] from data fusion of both FDG-PET and clinical data discriminated between control and AD subjects with an AUC of 0.97, an improvement over the best single FDG-PET classifier (AUC = 0.94) or the best clinical classifier (derived from ADAS-cog data: AUC = 0.93) (Table 8). Vemuri et al [132] compared STAND score measures from MRI with CSF and concluded that CSF and MRI biomarkers independently contribute to intergroup diagnostic discrimination, and the combination of CSF and MRI provides better prediction than either source of data alone.
4.5.6.2. Multimodal classification
A significant technical advance in classification was the development of classifiers that combine information from multiple modalities. The multi-kernel learning framework developed by Hinrichs et al [247] combined multiple modalities for classification of AD patients. They found that while the classifier based on all modalities performed best overall, cognitive scores alone separated AD patients from controls to almost the same level of accuracy (Table 8). Similarly, two studies by Zhang et al [249] focused on combining MR, FDG-PET and CSF biomarker data using different methods (multi-modal multi-task [250], and kernel combination [249]). They found that combination of different modalities outperformed single modalities in classification accuracy [250] (Table 8) and that the kernel combination method correctly identified 91.5% of MCI converters and 73.4% of MCI non-converters. Likewise, both Westman et al [495] and Yang et al [487] found that combinations of MRI measures representative of AD brain morphology changes and CSF biomarkers were most effective at discriminating MCI from control patients with accuracies of 77.6% and 72%, respectively. Yang et al [487] also reported improved classification accuracies using hippocampal and ventricular volumes rather than shapes.
With the optimum combination of modalities for classification largely established (usually a combination of MRI and CSF data, with APOE status, gender, and age), new methodologies have been developed that have realized further gains in accuracy by optimizing the ways in which features are selected and modalities combined. The multimodal classifier of Gray et al [405], based on the random forests algorithm, enhanced classification accuracy for AD versus controls, and MCI versus controls over all single-modality classifications. However, it was no more accurate in discriminating between MCI converters and nonconverters than MRI data alone. The novel multiple kernel classifier of Liu et al [392] combined MRI and CSF data to improve on the classification accuracies attained by the SVM of Zhang et al [249]. The authors attributed this improvement to the use of group Lasso regularization to enforce sparsity in the different modalities. Instead of focusing on binary classification, the multimodal approach of Cheng et al [393] estimated clinical scores from MRI, FDG-PET, and CSF data using relevance vector regression. Their approach attained correlation coefficients of 0.80 and 0.78 for the estimation of MMSE and ADAS-cog scores, respectively. Not all multimodal approaches have proven optimal. Casanova et al [411], who condensed multimodal information to a single score, AD-PS, found that although this index improved MCIc versus MCInc classification accuracy beyond other summary indices such as STAND and SPARE-AD, it did not perform as well as individual structural or cognitive measures in other classification problems.
Can multimodal techniques improve diagnosis in the clinical setting? Simonsen et al [417] compared the accuracy of their PredictAD software with that of clinical raters asked to classify patients into a range of categories depending on their likelihood of developing the disease. The software increased classification accuracy over paper information alone from 63.2% to 70% and additionally improved interrater agreement and increased the raters’ confidence in their decision. The PredictAD appears capable of beneficially combining data from different modalities and therefore may prove a useful adjunct for decision support in a clinical environment. The study of Escudero et al [420] addressed cost-effectiveness of biomarker-based diagnosis. They found that when the number of biomarkers was minimized, classification accuracies were comparable to methods that include all biomarkers. However, the selection of biomarkers solely to minimize costs lowered accuracies. This proof-of-concept study supports the feasibility of a personalized clinical diagnostic aid for AD that can be optimized for cost and time efficiency.
4.5.6.3. Feature selection
Given the high dimensionality of imaging techniques, the selection of a minimum set of features that optimally preserve complementary information between modalities is a critical step. A number of feature selection techniques have been borrowed from computer graphics fields, and the incorporation of these into AD classification represents a significant advance. Using an automatic data-driven method for the selection of multi-modal features and SVM trained on AD and control patients, Cui et al [287] also found that combined optimal MR, CSF and neuropsychological features outperformed any single modality in the classification of MCI converters versus non-converters. From baseline features, they predicted conversion of MCI to AD within 24 months with an accuracy of 67.1%, a sensitivity of 96.4%, a specificity of 48.3% and an AUC of 0.796 (Table 8). Liu et al [404] reported an accuracy of 67.8%, a sensitivity of 64.9%, and a specificity of 70% in the classification of MCIc versus MCInc groups. Dukart et al [496] used meta-analyses to combine MRI and FDG PET data for improved classification. The combination discriminated between control and AD patients with an accuracy of 85.7%, a sensitivity of 89.3%, and a specificity of 82.1%. Additionally, when ADNI data were used for training the SVM, classification in another cohort reached similar accuracies, showing the applicability of ADNI data to the wider community. Suk et al [387] applied deep learning techniques developed for vision processing to neuroimaging to improve feature selection. The incorporation of high-level information such as relationships between features improved classification accuracy over the use of only low-level features, such as gray matter volumes.
4.5.6.4. The effect of age on classification
The possibility that different modalities may not be equally useful in classification of patients of different ages was explored by Schmand et al [288] who analyzed the efficacy of neuropyschological measures, CSF biomarkers and FDG-PET and MRI measures in classifying two age-stratified groups (younger and older than 75 years) within the ADNI cohort. They found that, regardless of age, neuropsychological and MRI measures resulted in the most accurate classification. Classifications based on CSF biomarkers were more accurate in those aged younger than 75 but neither FDG-PET nor CSF data augmented accuracy in older individuals (Table 8). Once again, combined features resulted in the most accurate discrimination of the cognitively impaired (AD and MCI) from the cognitively normal.
4.5.6.5. Missing data
In any large study involving multiple tests, it is inevitable that some data will be missing from the overall data set. Two approaches for addressing the issue of missing data in multimodal classification represented by Yuan et al [415] and Xiang et al [416] who tested their methods (iMSF and iSFS, respectively) on the ADNI cohort and found both improved classification over other missing value estimation methods and over single-modality classification. A comparison of their results is presented in Fig. 37.
Fig. 37.
The effect of accounting missing data on classification. In addition to the feature selection of an incomplete Multi-Source Feature (iMSF) learning method, the incomplete Source Feature Section (iSFS) model accounts for missing data. From Xiang et al [416]. Abbreviations: AUC, area under the curve; AD, Alzheimer’s disease; NC, normal control; MCI, mild cognitive impairment.
4.5.7. Diagnosis of MCI using National Institute on Aging-Alzheimer’s Association criteria
The NIA-AA criteria for MCI due to AD recently incorporated the use of biomarkers for both amyloid deposition (CSF Aβ42, 11C-PiB-PET) and neuronal injury (CSF tau, hippocampal and whole brain atrophy on MRI, FDG-PET) in addition to clinical information [497]. According to these criteria, MCI patients are classified into four categories: MCI high (a high likelihood of AD—both Aβ and neuronal biomarkers positive), MCI intermediate (an intermediate likelihood of AD—one or other of the biomarker types positive), MCI unlikely (unlikely to progress to AD—neither biomarker type positive), and MCI core (conflicting/missing biomarkers, no probability assigned). Petersen et al [353] compared these criteria in MCI patients in two cohorts including ADNI. Similar trends were observed in both cohorts regarding progression to AD, namely, a similar percentage of subjects progressed within 12 to 15 months, and of these, the highest proportion were positive for both neurodegeneration (MRI, FDG-PET) and amyloid (CSF Aβ42 and PiB-PET) biomarkers. Progressors also included a smaller proportion of patients who tested positive for neurodegeneration biomarkers only, but none who were amyloid positive and neurodegeneration negative. The MCI group displayed expected heterogeneity: 55% had evidence for both neurodegeneration and amyloid deposition, 12% were amyloid positive but neurodegeneration negative, 17% were neurodegeneration positive but amyloid negative, and 16% were biomarker negative. This last group may include patients with cognitive impairment unrelated to AD. The results suggest that neurodegeneration is of primary importance in progression to AD and support the use of biomarkers as an adjunct to the clinical diagnosis of AD. In the ADNI cohort alone, Guo et al [352] examined disease progression over 6 to 60 months in MCI patients classified by these criteria. The MCI-high subgroup was 2.3 times more likely to progress than MCI core (95% CI: 1.36–3.89, P = .002), and no patients in MCI-low group progressed within the time frame. The authors concluded that the NIA-AA criteria are a useful set of diagnostic guidelines for stratification of MCI patients in terms of risk of progression to AD but that binary nature of biomarker cutoff points and also division of biomarkers into only two categories may miss opportunity for a more nuanced interpretation of the data. As only 48% of ADNI MCI patients were found to have consistent AD-like patterns across all biomarkers and could be classified as high probability for AD using the NIA-AA criteria, Lowe et al [354] proposed modifications to these guidelines (Fig. 38). Using these adjustments, when abnormal amyloid markers were prioritized over neuronal markers, only a positive amyloid biomarker and a single neuronal injury marker were required for diagnosis of an AD pattern. Eighty seven percent of subjects were then classified as having a high probability for AD, and 1% of subjects lacking abnormal amyloid and neuronal markers were categorized as having a “high probability of non-AD,” avoiding some of the NIA-AA categorizations of patients as “undefined” and “uninformative.” Clearly, operationalizing these criteria is a current priority to ensure the optimal stratification of patients across the AD spectrum.
Fig. 38.
Flow charts showing the categorization of ADNI subjects with AD using the strict NIA-AA criteria (A) and Mayo-modified NIA-AA criteria (B). Abbreviations: AD, Alzheimer’s disease; ADNI, Alzheimer’s Disease Neuroimaging Initiative; FDG, fluorodeoxyglucose; MRI, magnetic resonance imaging; NIA-AA-C, National Institute on Aging-Alzheimer’s Association clinical diagnostic guidelines. From Lowe et al [354].
4.5.8. Summary and conclusions of diagnostic classification papers
A variety of approaches have been used to diagnose MCI and AD, some based on single measures, others on composite scores of a single modality, and still others on a combination of factors from different modalities. It should be emphasized that ADNI was not designed as a diagnostic classification study; none of the imaging methods used in ADNI is as accurate as a clinical diagnosis, and the enrolled cohort represents typical cases rather than the types of difficult diagnostic problems that clinicians often confront. However, a number of conclusions can be drawn from the results of these studies. Single features, such as hippocampal volume, are not as accurate as multiple features, such as whole brain or cortical thickness measurements. The best classifiers combine optimum features from different modalities, including CSF biomarkers, MRI, FDG-PET, and cognitive measures, as well as factors such as age and APOE ε4 allele status. The most discriminative measures include hippocampal volume, entorhinal cortical thickness, entorhinal metabolism, the t-tau/Aβ-42 ratio, and ADAS-cog scores. In some of these models, FDG-PET measures appear to lose significance to cognitive and MRI measures; however, glucose hypometabolism alone has been shown to have high classification accuracy. ADAS-cog scores, either used directly or in a model using weighted components, appear to be an excellent diagnostic tool, although the highest accuracies were found with the addition of MRI measures. Although most classifiers used baseline measurements, there is some evidence to suggest that longitudinal data may provide even more accurate diagnoses, but it remains to be seen whether this approach is more generally applicable to other modalities. Currently, the best classifiers are able to discriminate between control and AD subjects with accuracies in the mid-90% range, but have considerably lower accuracies when discriminating between control and MCI subjects or between MCI-nc and MCI-c subjects, although data for the latter diagnoses, arguably the more important distinction to make, are far less reported. It is as yet unknown whether the application of some of the promising classifiers to these problems will result in increased diagnostic accuracy. Another key question is how methods that perform well in ADNI, with its sharply delineated diagnostic groups and exclusion of mixed dementias and borderline cases, will translate to the community or general clinic setting for wider diagnostic use. Validation studies in population-based samples will be required to address this issue.
Studies published in 2011–2012 continued to seek ways to improve classification accuracy within the ADNI cohort. Some works detailed approaches to leveraging more information from the hippocampus, from cortical thickness and topology and from maximally discriminative volumes by deriving statistical indicators. Others dealt with improving classification through methodological improvements such as registration to multiple templates and by accounting for age-related cognitive decline in control groups. While 2011–2012 studies did not improve on the best classification accuracies of previous studies, they became consistently more accurate, and were able to discriminate between controls and MCI or AD patients with accuracies in the mid-90s and mid-80s, respectively. Classification of MCI converters and non-converters reached accuracies in the low 80s. The most accurate classification methods were generally longitudinal and combined multiple modalities and multiple features within each modality. The first reports of blood based biomarkers appeared and, despite being exploratory and preliminary, showed great promise for future clinical diagnosis. Classification methods developed in ADNI still remain to be validated in independent, population based cohorts. Burgeoning research in numerous fields led to many exciting developments in classification during 2012 and 2013. The widespread use of dimensionality reduction and feature selection techniques improved classifier performance over many SVMs. Shapes of subcortical structures and changes in structural and functional connectivities during disease progression were used to improve classification. New potential biomarkers such as WMHs and α-syn showed promise in distinguishing between patient groups, and the continued development of blood-based biomarkers led to their incorporation into classifiers for the first time. The nature of the classification problems targeted by research groups transitioned from primarily distinguishing AD patients from controls to the far more challenging problem of distinguishing MCI converters from nonconverters. However, there was increasing recognition of the difficulties in comparing results from studies using nonstandardized data sets and/or methodologies. Finally, introduction of new NIA-AA criteria for classifying MCI patients underscored the importance of assessing these criteria and operationalizing them for future ADNI studies.
4.6. Improvement of clinical trial efficiency
One of the primary goals of ADNI is to improve the efficiency of clinical trials of AD-modifying treatments. Selection of the study population and development of more sensitive outcome measures are two approaches to increasing the power of clinical trials and therefore reducing the number of participants required, the length of time required before a disease-modifying effect is observed, and therefore the overall cost. With the advent of early intervention trials, the prediction challenge has shifted toward detecting persons with normal cognition who are likely to progress to MCI or even AD within the time frame of the clinical trial. This involves the selection of a subset of those subjects who harbor AD pathology but to present no clinical manifestation of the disease. This section details the results of studies examining the use of structural, fluid, and genetic biomarkers in the improvement of clinical trial efficiency.
4.6.1. Prediction of cognitive decline
Beyond the simple classification of clinical trial participants, an important strategy for increasing clinical trial efficiency is the enrichment of clinical trial populations, normally MCI patients, with participants who are likely to progress to AD within a short time frame. In particular, the early and reliable detection of MCI subjects who convert early to AD could support clinical decisions for or against therapy with disease-modifying drugs. Many studies have therefore focused on identifying baseline predictors of future decline, with “future decline” meaning both decline in clinical measures such as the MMSE, ADAS-cog, and CDR-SB, and conversion of MCI to AD status. However it is measured, it is desirable for appreciable decline to occur over a relatively short time frame, typically 12 months. Imaging measures, CSF biomarkers, and APOE ε4 allele status, in combination or alone, have been identified as baseline future predictors, and several studies have focused on determining the optimum combination of all modalities that results in the most power for clinical trials.
4.6.1.1. Magnetic resonance imaging
4.6.1.1.1. Temporal lobe
Hua et al [112] used TBM to create Jacobian maps of temporal lobe atrophy at baseline and examined the relationship between the maps and cognitive decline over the following year, as assessed by both the CDR-SB and the MMSE. They found that baseline temporal lobe atrophy predicted decline in the MMSE in AD patients and also predicted the conversion of MCI to AD over 12 months (Fig. 19; Table 9). Baseline atrophy of MTL structures was also found to best predict the progression of MCI patients to AD in a study by Desikan et al [163]. These measures, including the volumes of the hippocampus and amygdala and the thickness of the entorhinal cortex, temporal lobe, and parahippocampal gyrus, were found to be better predictors of clinical decline than levels of CSF Aβ-42 or FDG-PET ROIs. The combination of CSF biomarkers and FDG-PET ROIs predicted time to progression of MCI to AD with an AUC of 0.70, a sensitivity of 93%, and a specificity of 48% compared with MRI temporal lobe factors, which had an AUC of 0.83, a sensitivity of 87%, and a specificity of 66%. The addition of CSF or FDG-PET measures to the combined Cox proportional hazards model did not significantly increase prediction accuracy, with the combined model predicting conversion with an AUC of 0.83, a sensitivity of 90%, and a specificity of 69% (Table 9). Similar structures were found to predict future decline in cognitive status by Kovacevic et al [164], who used high-throughput volumetry to segment ROIs in control, MCI, and AD subjects. They found that after adjusting for age, education, and APOE genotype, smaller baseline volumes of the hippocampus and the amygdala and larger temporal horn volume predicted 6-month decline in both the MMSE (β [P] = 0.14 [.04], 0.18 [.004], and −0.2 [.003], respectively) and CDR-SB (β [P] = −0.19 [.005], −0.12 [.06], and 0.2 [.005, respectively) in all groups (Table 9). Risacher et al [114] also found atrophy of structures within the MTL to be the best antecedent of imminent conversion of MCI to AD. The largest effect sizes were for hippocampal and amygdalar volume and cortical thickness of the entorhinal cortex and inferior, middle, and superior gyri (Fig. 20; Table 9).
Fig. 19.
Association between temporal lobe atrophy and conversion to AD. Subjects who converted from MCI to AD over a period of 1 year after their first scan were coded as “1”; nonconverters were coded as “0.” A negative correlation suggests that temporal lobe degeneration predicts future conversion to AD. Reproduced with permission from Ref [112].
Table 9.
Predictors of future decline
| Predictor | Measurement of decline | Statistical measurement | Patient group | Cross-validated? | Reference |
|---|---|---|---|---|---|
| Baseline temporal lobe measures | MMSE | P <.05 | MCI | [112] | |
| MCI to AD conversion | P < .05 | MCI | |||
| CDR-SB | P < .05 | CN, MCI, AD | |||
| Baseline temporal lobe measures | CDR-SB | AUC = 0.83, SEN = 87%, SPE = 66% | MCI | Yes | [163] |
| CSF biomarkers + FDG-PET ROIs | AUC = 0.70, SEN = 93%, SPE = 48% | ||||
| TL measures + CSF + FDG-PET ROIs | AUC = 0.83, SEN = 90%, SPE = 69% | ||||
| Baseline hippocampal, amygdala, temporal horn volume |
MMSE | β (P) = 0.14 (.04), 0.18 (.004), −0.2 (.003) | Pooled sample | [164] | |
| CDR-SB | β (P) = −0.19 (.005), −0.12 (.06), 0.2 (.005) | ||||
| Baseline hippocampal volume | MCI to AD conversion | Cohen d = 0.603 | MCI-nc vs MCI-c | [114] | |
| Baseline inferior temporal gyrus volume | Cohen d = 0.535 | ||||
| Baseline middle temporal gyrus volume | Cohen d = 0.529 | ||||
| Baseline entorhinal cortical volume | Cohen d = 0.493 | ||||
| Baseline ventricular expansion | MMSE, global CDR, CDR-SB | P <.05 | Pooled sample | [126] | |
| Baseline ventricular expansion | MMSE, global CDR, CDR-SB | P <.05 | Pooled sample | [127] | |
| Baseline right caudate volume | MMSE | P < .05 | Pooled sample | [130] | |
| MCI to AD conversion | P <.05 | ||||
| Baseline cortical thickness in ROIs | MCI to AD conversion | Accuracy = 76% | MCI | Yes | [165] |
| Baseline cortical thickness in ROIs | [147] | ||||
| Longitudinal cortical thickness | MCI to AD conversion | Accuracy = 81.7% | MCI | Yes | [157] |
| Baseline white matter hyperintensity volume | ADAS-cog | β (P) = 0.34 (.05) | Pooled sample | [166] | |
| MMSE | β (P) = −0.096 (<.001) | ||||
| Multiple ROI atrophy score | MMSE | r (P) = 0.39 (<.001) | MCI | [117] | |
| Structural phenotypic score | MCI to AD conversion | AUC = 0.77 | MCI | Yes | [118] |
| STAND score | CDR-SB | MCI, AD | [167] | ||
| MCI to AD conversion | Cox proportional hazards ratio = 2.6 | MCI | |||
| Log (t-tau/Aβ–42) | MCI to AD conversion | Cox proportional hazards ratio = 2.0 | |||
| SPARE-AD score | MCI to AD conversion | AUC = 0.734, SEN = 94.7%, SPE = 37.8% | MCI | Yes | [119] |
| MMSE | P <.05 | ||||
| FDG-PET hypermetabolic convergence index | MCI to AD conversion | Cox proportional hazards ratio = 7.38 | MCI | [85] | |
| FDG-PET HCI + hippocampal volume | Cox proportional hazards ratio = 36.72 | ||||
| FDG-PET sco | MCI to AD conversion | AUC = 0.75, sens – 57%, spe = 67% | MCI | [291] | |
| Aβ load | MCI to AD conversion | 75th vs 25th percentile Cox HR = 2.6 (P < .001) | MCI | [152] | |
| Baseline hippocampal volume | 25th vs 75th percentile Cox HR = 2.6 (P<.001) | ||||
| Baseline ADAS-cog (from meta-analysis) | ADAS-cog | Slope of disease progression = 5.49 points/yr, baseline five point increase in ADAS-cog effect on slope = 0.669/yr |
MCI, AD | [171] | |
| Baseline ADAS. Tree | MCI to AD conversion | P = 6.23E-10, AUC = 0.746 | MCI | Yes | [96] |
| Baseline MMSE | P = .0188, AUC = 0.589 | ||||
| Baseline hippocampal volume | CDR-SB, MMSE, LM delayed change |
r = −0.29, 0.29, 0.41 | MCI | [155] | |
| Baseline entorhinal volume | r = −0.17, 0.23, 0.34 | ||||
| Baseline retrosplenial volume | r = −0.43, 0.42, 0.35 | ||||
| Baseline entorhinal metabolism | r = −0.30, 0.38, 0.28 | ||||
| Baseline retrosplenial volume | r = −0.22, 0.47, 0.11 | ||||
| t-tau/Aβ−42 | r = 0.02, 0.08, −0.23 | ||||
| APOE ε4+ | Hippocampal volume change (P<.05). Multivariate model |
Coefficient of effect on annual change = −0.36 | MCI | [154] | |
| FDG-PET ROI-avg | Coefficient of effect on annual change = 9.3 | ||||
| CSF tau | Coefficient of effect on annual change = −8.7 | AD | |||
| FDG-PET ROIs | MCI to AD conversion | β (SE) = 1.00 (0.51), Cox HR = 2.72 | MCI | [173] | |
| AVLT | β (SE) = 1.46 (0.64), Cox HR = 4.30 | ||||
| FDG-PET ROIs | ADAS-cog | β (SE) = 1.26 (0.43) | |||
| p-tau181/Aβ–42 | β (SE) = 1.10 (0.53) | ||||
| Right entorhinal cortical volume | MCI to AD conversion | Prediction accuracy (95% CI) = 68.5% (59.5, 77.4) | MCI | Yes | [161] |
| TMT-B test | Prediction accuracy = 64.6% (55.5, 73.4) | ||||
| p-tau181/Aβ−42, hippocampal volume, TMT–B, age | Prediction accuracy = 76.3 (68.4, 84.2) | ||||
| AVLT-delayed, LM-delayed, left middle temporal lobe thickness |
MCI to AD conversion | AUC = 0.80 | MCI | Yes | [294] |
| Baseline multi-modal multi-task learning: MR, FDG- PET CSF |
MMSE | r = 0.511 | MCI | Yes | [239] |
| ADAS-cog | r = 0.531 | MCI | |||
| Multi-modality disease marker | MCI to AD conversion | MCI | Yes | [247] | |
| Biological (baseline) | AUC = 0.5292 | ||||
| Imaging (baseline) | AUC = 0.7378 | ||||
| Imaging (longitudinal) | AUC = 0.7911 | ||||
| Neuropsych (baseline) | AUC = 0.6693 | ||||
| Neuropsych (longitudinal) | AUC = 0.7385 | ||||
| Combined modalities | AUC = 0.7667 | ||||
| Disease state index | MCI to AD conversion | AUC = 0.752 | MCI | [252] | |
| Disease state index | MCI to AD conversion | Prediction accuracy = 68.6% | All MCI | [253] | |
| Prediction accuracy = 84.4% | MCI – strong evidence of AD pathology |
||||
| Prediction accuracy = 93.7% | MCI – very strong evidence of AD pathology |
||||
| Baseline and longitudinal multimodal data | MCI to AD conversion | AUC = 0.768, ACC = 78.4%, SEN = 79%, SPE = 78% |
MCI | Yes | [239] |
Fig. 20.
Effect size of imaging biomarkers for MCI converters versus MCI nonconverters. Effect sizes (Cohen d) of the comparison between MCI stable (MCI nonconverter) and MCI converter groups evaluated for selected imaging biomarkers. Reproduced with permission from Ref [114].
A major challenge for the selection of clinical trial populations is the heterogeneity of the amnestic MCI population as only 60% to 70% of these patients harbor AD pathology. Hippocampal volume has been shown to be effective in identifying subjects more likely to progress to dementia, but its use as a practical tool in subject selection has not been addressed. A systematic study by Yu et al [498] characterized hippocampal volume as an enrichment biomarker, investigating the selection of an appropriate cut point, the effect of outcome measures, and the performance of different hippocampal volume algorithms that generate different absolute hippocampal volumes. The study developed cut points based on percentiles of distribution of hippocampal volumes from a normal control cohort. At more stringent cut points of less than 25% of normal, effect sizes were maximized and sample sizes minimized, but trial costs were increased because of higher screen failure rates. Investigator costs were minimized with cut points in the range of 10% to 25% of normal, which corresponded to an approximate 30% to 40% saving. In general, the estimated trial time was unchanged. All four hippocampal volume algorithms tested had similar sample sizes, trial costs and trial duration, and effects of cut points on effect sizes. The authors hoped that the study would allow informed choice of hippocampal volume cut point, leading to the operationalization of hippocampal volume in clinical trials.
If hippocampal atrophy is predictive of future cognitive decline, what biomarkers are then predictive of hippocampal atrophy itself? Answering this question has clear implications for powering early intervention CTs for AD in which the ability to predict cognitive decline from an even earlier time point in the disease is crucial. Desikan et al [163] examined whether factors such as CSF biomarkers and measures of cortical thinning were able to predict hippocampal atrophy. They found that hippocampal atrophy was significantly predicted by decreased levels of Aβ and increased levels of tau in MCI and AD patients and by the baseline thickness of the entorhinal cortex and inferior temporal gyrus Aβ and tau positive individuals.
4.6.1.1.2. Ventricles
Baseline ventricular morphology has been shown to predict future clinical decline in studies of the ADNI cohort. Chou et al [126] found that this measure predicted decline in MMSE, global CDR, and CDR-SB over 12 months (Fig. 21; Table 9). These findings were confirmed in a subsequent larger study by the same group [127], and further extended by examining additional cognitive criteria. Only right ventricular baseline anatomy was correlated with future decline in DR memory scores, but there was no correlation between ventricular anatomy and changes in depression scores, despite a baseline association between these measures (Table 9). Yang et al [487] reported that hippocampal and ventricular shapes outperformed corresponding volumetric measures, predicting MCI conversion within 2 years with an accuracy of 66.7%, a sensitivity of 82%, and a specificity of 51.4%.
Fig. 21.
Significance maps of correlation between ventricular shape and cognitive decline. Significance maps correlate baseline ventricular shape with subsequent decline, over the following year, in three commonly used clinical scores. Reproduced with permission from Ref [126].
4.6.1.1.3. Other regions
Targeting the caudate, a region not traditionally associated with AD, Madsen et al [130] found that baseline atrophy in the right caudate predicted both the conversion of MCI patients to AD and cognitive decline of this group, as assessed by the MMSE (Fig. 22; Table 9). The predictive ability of atrophy in the posterior regions (posterior cingulate gyrus, precuneus, and parietal lobe) compared with MTL regions was investigated by Lehman et al [499]. MTL atrophy was strongly associated with MCI to AD conversion and with decreased Aβ, whereas posterior atrophy was more weakly associated with conversion but more strongly associated with levels of tau. This differential association with biomarkers may be indicative of widespread neuronal loss in posterior regions at latter stages of disease. These results suggest that in late-onset MCI patients without MTL atrophy, posterior atrophy may predict conversion.
Fig. 22.
Maps of associations with MMSE scores at baseline and 1 year later, MCI-to-AD conversion, and CSF concentrations of tau. Three-dimensional maps show areas of significant associations between local volumetric atrophy in the caudate and MMSE scores at baseline and after a 1-year follow-up interval, with P values color-coded at each surface voxel. Reproduced with permission from Ref [130].
Querbes et al [165] created a normalized thickness index, which was derived from the cortical thicknesses of regions most likely to show atrophy in AD and to distinguish between MCI-c and MCI-nc patients, primarily the left lateral temporal, right medial temporal, and right posterior cingulate. They found that the normalized thickness index predicted conversion of MCI patients to AD with 76% accuracy compared with accuracies ranging from 63% to 72% by cognitive scores (Table 9). The additional dimension of time increased the ability of cortical thickness measurements to predict the conversion of MCI to AD in a study by Li et al [157]. By incorporating both static baseline and follow-up measures, dynamic measures of thinning speed, the ratio of follow-up to baseline thicknesses in ROIs, and a network feature that examined correlations between longitudinal thickness change in different ROIs, Li et al constructed a classifier that correctly identified 81.7% of MCI-c patients 6 months ahead of their conversion (Table 9).
White matter hyperintensities (WMH) may represent an accrual of nonspecific neuronal injury over a lifetime. Carmichael et al [166] investigated the relationship between WM disease and cognition over a year, and found that both baseline and longitudinal change in WMH were associated with worsening of ADAS-cog and MMSE scores over 12 months (Table 5), raising the possibility of the use of WMH as a biomarker and highlighting its ability to predict future clinical decline (Table 9). When the MCI group was divided according to the level of baseline WMH and amyloid burden (PiB uptake), Provenzano et al [441] found a monotonic increase in proportion of individuals converting to AD within the follow-up visit time frame from amyloid negative/low WMH to amyloid negative/high WMH to amyloid positive/low WMH to amyloid positive/high WMH (Fig. 39). The amyloid positive/high WMH group was strongly associated with future conversion to AD, providing further support for involvement of vascular factors in the pathogenesis of AD.
Fig. 39.
Contribution of white matter hyperintensities (WMHs) to AD. Proportion of subjects with MCI who converted to Alzheimer’s disease (AD) during the follow-up period as a function of β-amyloid deposition (PiB) and WMHs. From Provenzano et al [441].
A number of studies have leveraged information on atrophy from multiple brain regions to distill a number or a score that is more predictive of future clinical decline than single regions alone. McEvoy et al [117] found that an atrophy score derived from mesial and lateral temporal, isthmus cingulate, and orbitofrontal areas was predictive of 1-year decline in MMSE scores and progression of MCI patients to AD. They found that the atrophy score was a better predictor than right or left hippocampal volume or the thickness of the left or right entorhinal cortex (Table 9). Similarly, a structural abnormality score extracted from baseline MRI data by Misra et al [118] was higher in MCI patients who converted to AD over the following year than stable MCI patients, and an SPS derived by Fan et al [83] from a complex pattern of spatial atrophy predicted decline in MMSE scores within a year from baseline (Table 9). Vemuri et al [167] found that STAND scores that reflected greater baseline atrophy in regions associated with AD predicted greater subsequent decline on the CDR-SB and also a shorter time to conversion for MCI patients than CSF analytes (Table 9). Davatzikos et al [119] focused on structural changes occurring at the early stages of AD and derived SPARE-AD scores (Spatial Pattern of Abnormalities for Recognition of Early AD) largely from changes in the temporal regions, posterior cingulate cortex, precuneus, and orbitofrontal cortex. They found that higher SPARE-AD scores predicted conversion of MCI to AD (Table 9). In a follow-up study, Da et al [500] found that the inclusion of neither APOE status nor CSF biomarkers significantly improved the prediction, although ADAS-cog was equally effective as SPARE-AD alone in predicting time to conversion. Intriguingly, when the MCI patients were stratified by amyloid status, SPARE-AD remained highly predictive in the amyloid negative group, contrary to the model of disease progression put forward by the amyloid hypothesis. Zhang et al [501] compared the ability of two further summary scores of medial temporal lobe and whole brain atrophy, Medial Temporal Lobe Atrophy Score (MTAS) and Brain and Lesion Index (BALI), to predict MCI to AD conversion over 2 years. MTAS was a stronger predictor than common changes in the aging brain represented by BALI. However, BALI scores increased prediction accuracy when combined with MTAS (Table 9), suggesting that structural brain changes outside and within the MTL have an additive effect. The OLPS score of Spulber et al [382] distinguished between MCIc and MCInc patients, with an accuracy of 67.6%, a sensitivity of 69.6%, a specificity of 66.8%, and an AUC of 0.675. Moreover, the distribution of scores in MCI patients who did not convert during the course of the 3-year study was broad, and higher scores were associated with greater age and a higher likelihood of an APOE ε 4 allele. These results support the idea of the MCI nonconverter group being highly heterogeneous and consisting of a subgroup displaying structural hallmarks of AD likely to progress in the near future.
Longitudinal data are increasingly being leveraged for the prediction of future decline. Macdonald et al [502] compared the ability of hippocampal and temporal horn baseline volumes and 12-month volumetric rates of change to predict conversion in MCI patients over the subsequent 12 months. All measures were predictive of future decline, but rates of volumetric change were better predictors than baseline volumetric measures. In addition, they found some evidence that temporal horn expansion is more predictive than hippocampal atrophy. The longitudinal stability selection technique of Zhou et al [407] revealed MRI regions predictive of decline in ADAS-cog and MMSE scores at different stages of disease progression. Strikingly different patterns were observed for ADAS-cog and MMSE. No regions were strongly predictive of MMSE scores for more than 2 years, whereas a number of medial temporal regions were predictive of ADAS-cog decline for as long as 3 years (Fig. 40).
Fig. 40.
The stability of different MRI features in predicting (A) ADAS-cog and (B) MMSE scores. MRI regions aligning with red in the stability vector most accurately predict cognitive scores. Several regions (e.g., cortical thickness averages of the left and right entorhinal, and left middle temporal, and hippocampal volume) predict ADAS-cog over 3 years, whereas most regions predict MMSE scores only over 6 to 12 months. From Zhou et al [407]. Abbreviations: MRI, magnetic resonance imaging; MMSE, Mini-Mental State Examination.
Similarly, disease state–specific neurodegenerative changes were found by Eskildsen et al [386], who divided the ADNI cohort into groups determined by their time to conversion: 6, 12, 24, and 36 months. In each group, ROIs representing differential patterns of cortical thinning were identified. MTL regions predominated. Initially, the parahippocampal gyrus was selected in the 36-month group followed by the hippocampus in the 24-month group. The amygdala and occipital areas were selected in groups closer to conversion, implying a progression of the disease through these areas. Selected regions were applied to a linear discriminant analysis classifier, and the authors found that prediction of conversion improved in groups closer to conversion, with an accuracy of 74.6% obtained in the 6-month group compared with 67.8% in the whole MCI cohort (Table 9).
Prediction of future cognitive decline in cognitively normal individuals has been an increasingly important focus in ADNI studies in 2011–2012. Dickerson et al [289] used a predefined cortical thickness measure as an MRI biomarker suggestive of early AD neurodegeneration to examine this group over 3 years. They found that cognitively normal individuals with the low cortical thickness signature were at increased risk of cognitive decline (CDR-SB, AVLT and TMT) and were more likely to have Aβ42 levels below the 192 pg/ml cut-point designating AD-like higher risk (Fig. 30). Chiang et al [290] identified 8 baseline MRI ROIs from predominantly the temporal lobe that predicted 12 month cognitive decline of greater than one standard deviation from the mean with an accuracy of 79% in cognitively normal individuals. These results suggest that these MRI biomarkers may have utility in identifying individuals harboring AD pathology with a greater likelihood of imminent cognitive decline emblematic of AD.
Fig. 30.
(A) Expression of cortical signature of Alzheimer’s disease is associated with future cogntive decline. (B) Expression of cortical signature of Alzheimer’s disease is associated with AD-like spinal fluid. Reproduced with permission from Ref [139].
McEvoy et al [168] also investigated enrichment strategies for constraining recruitment into clinical trials by selecting MCI patients most likely to progress. Their first strategy, which selected MCI patients with an APOE ε4 allele, reduced sample sizes by an estimated 10% to 40%, but this was discounted because of the possibility that restricting patient genotype may invalidate trial findings. Their second strategy, based on baseline MRI atrophy in regions previously shown to be predictive of disease progression, resulted in an estimated sample size reduction of 43% to 60% (Table 11).
Table 11.
Comparison of outcome measure methods in clinical trials: sample size estimates per arm required to detect a 25% reduction in atrophy with 80% power, 5% significance
| Outcome measure | Method tested | Sample size AD | Sample size MCI | Reference |
|---|---|---|---|---|
| Hippocampus | Two scans, 0–6 months | 462 | 949 | [121] |
| Three scans, 0–6–12 months | 255 | 673 | ||
| Three scans + Markov Chain + APOE ε4 | 86 | 341 | ||
| Clinical | ADAS-cog two tests, 0–6 months | 745 | 4663 | |
| ADAS-cog three tests, 0–6–12 months | 569 | 8354 | ||
| MMSE two tests, 0–6 months | 1280 | 6300 | ||
| MMSE three tests, 0–6–12 months | 780 | 3353 | ||
| Hippocampal atrophy | 12-(24)-month | 67 (46) | 206 (121) | [64] |
| Hippocampal atrophy | 12-month | 78 | 285 | [59] |
| Ventricular expansion | 6-month change | 342 | 1180 | [175] |
| Clinical | MMSE | 7056 | 7712 | |
| ADAS-cog | 1607 | >20,000 | ||
| MRI (Model T/Model D) | Entorhinal | 45/65 | 135/241 | [176] |
| Inferior temporal | 79/117 | 199/449 | ||
| Fusiform | 72/114 | 185/485 | ||
| Mid temporal | 83/122 | 229/501 | ||
| Hippocampus | 67/118 | 179/510 | ||
| Inferior lateral ventricle | 76/157 | 160/550 | ||
| Whole brain | 101/189 | 158/541 | ||
| Ventricles | 86/240 | 189/1141 | ||
| Clinical (Model T/Model D) | CDR-SB | 226/236 | 490/551 | |
| ADAS-cog | 324/283 | 1232/804 | ||
| MMSE | 482/494 | 1214/1304 | ||
| Whole brain atrophy | KN-BSI | 81 | NA | [51] |
| Classic-BSI | 120 | NA | ||
| TBM | 1.5-T MRI/3.0-T MRI | 37/48 | 107/159 | [47] |
| SIENA* | 1.5-T MRI/3.0-T MRI | 116/92 | 207/265 | |
| TBM | sKL-MI S6L8† | 48 | 88 | [177] |
| Clinical | ADAS-cog | 619 | 6797 | |
| MMSE | 1078 | 3275 | ||
| CDR-SB | 408 | 796 | ||
| TBM | Gray matter atrophy | 43 | 86 | [120] |
| Temporal lobe atrophy | 43 | 82 | ||
| CSF biomarkers | Aβ–42 | 5,721,531 | 75,816 | |
| t-tau | 81,292 | 19,098 | ||
| t-tau/Aβ–42 | 66,293 | 533,091 | ||
| PET | ROI-avg‡ | 4605 | [154] | |
| logSumZ2PNS§ | 2176 | |||
| logSumZ2PR§ | 1629 | |||
| DD-fROI¶ | 249 | |||
| MRI | VBSI** | 284 | ||
| Ventricles†† | 277 | |||
| Hippocampus†† | 202 | |||
| BSI‡‡ | 177 | |||
| DD-ROI† | 73 |
Structural Image Evaluation, using Normalization, of Atrophy (SIENA). See text for more details.
A nonlinear registration algorithm driven by mutual information cost function and with a regularizing term based on the symmetric Kullback–Leibler (sKL) distance.
Jagust laboratory method.
Foster laboratory method, measures of glucose hypometabolism, log transformed.
Reiman laboratory method, data-driven summaries applied to independent test set.
Fox laboratory method, ventricular boundary shift interval as a percentage of baseline brain volume.
Schuff laboratory method (FreeSurfer).
Fox laboratory method, brain shift interval.
4.6.1.2. [18F]-fluorodeoxyglucose-positron emission tomography
Chen et al [85] reported that their HCI outperformed other measures such as hippocampal volume, cognitive scores, APOE genotype, and CSF biomarkers in the prediction of conversion of MCI patients to AD. In a univariate model, patients with an HCI above a predefined cutoff had an average Cox proportional hazards ratio for the estimated risk of conversion to probable AD within 18 months of 7.38 compared with 6.34 for hippocampal volume, 4.94 for p-tau181p, and 3.91 for ADAS-cog, the most significant of the other measures tested. Moreover, patients with a combination of both high HCI score and hippocampal volume below a similarly defined threshold value had a Cox proportional hazards ratio of 36.72 (Table 9). This study suggests that data from FDG-PET analyses represent a powerful tool for the prediction of future decline in AD that is complementary to MRI data. Herholz et al [291] assessed the utility of an alternative composite score in predicting MCI to AD conversion within 24 months from baseline data and found that their PET score predicted disease progression with a sensitivity of 57%, a specificity of 67% and a AUC of 0.75, compared to AUCs of 0.68 and 0.66 for ADAS-cog and MMSE scores, respectively (Table 9). The PET score appeared to be reflective of AD pathology and highlighted the heterogeneous nature of both MCI and control groups, especially evident after 24 months (Fig. 31). When an alternative composite, the AD-conv score of Arbizu et al [397] was used to divide MCI patients into groups with different probabilities of converting to AD within 18 months, the overall AUC for conversion within this time frame was 0.804. While the overall conversion rate was 29.7% for the pooled group, it ranged from 75% in the high-probability group to 7.5% in the very low probability group (Fig. 41). This approach differs from conventional approaches that interpret a biomarker as being positive or negative by using the AD-conv score to render probabilities of conversion that may more accurately reflect the slow progressive nature of the disease.
Fig. 31.
Box plot of baseline PET AD scores for diagnostic groups. AD patients and MCI patients progressing to AD have significantly higher scores than stable subjects (arrows in top insert, P < .05 in Tukey multiple comparisons). Abbreviation: C, control. Reproduced with permission from Ref [291].
Fig. 41.
Application of the AD-conv score in the ADNI population. (A) Box plot of MCI converters and nonconverters; (B) ROC curves for MCIc versus MCInc; (C) distribution of probabilities among the stratified groups according to the AD-conv score. From Arbizu et al [397].
The ability of FDG-PET and Florbetapir imaging to predict future cognitive decline and conversion from normal cognition to LMCI and from LMCI to AD was compared by Landau et al [481]. In normal subjects, Florbetapir status but not FDG uptake was associated with ADAS-cog change. However, LMCI patients who were positive for both FDG uptake and Florbetapir had greater change in ADAS-cog scores. FDG positive status was associated with an OR of 10.9 of conversion from LMCI to AD compared with an OR of 3.0 for Florbetapir positive status, underscoring the differential temporal involvement of amyloid deposition and glucose metabolism in disease progression.
4.6.1.3. CSF and blood biomarkers
Vemuri et al [167] examined the ability of CSF biomarkers to predict decline in CDR-SB and MMSE scores over 2 years and the time to conversion from MCI to AD. Although all CSF biomarkers were predictive of future decline, the best predictor was log (t-tau/Aβ-42), which was comparable with the MRI-derived STAND scores. In contrast, Aβ-42 alone was only weakly predictive of conversion to AD, reflecting its status as a marker of early AD pathology. Used in combination with STAND scores, only log (t-tau/Aβ-42) improved the predictive ability of the MRI measure (Table 9). Jack et al [152] compared the ability of amyloid load, measured either by levels of CSF Aβ-42 or by 11C-PiB PET imaging, and hippocampal volume to predict MCI to AD progression. Using a new method to pool CSF and 11C-PiB PET data [169] and to extract a score representative of Aβ load from the pooled information, they found that the group of MCI patients classified as being Aβ positive had higher frequencies of the APOE ε4 allele and smaller baseline hippocampal volumes and a threefold higher chance of progressing to AD within 3 years than the Aβ-negative group (Fig. 23; Table 9). Thus, both baseline hippocampal atrophy and Aβ load were significant predictors of future decline. Interestingly, when risk profiles were constructed from the log relative hazard of progressing and degree of hippocampal atrophy or Aβ load, the relationship was linear for hippocampal atrophy, but plateaued at higher Aβ loads, consistent with a model in which Aβ deposition is an early event in AD progression, whereas neurodegeneration, as evidenced by hippocampal atrophy, occurs later and is thus a better indicator of progression toward dementia.
Fig. 23.
Pittsburgh compound B-positron emission tomography (PiB-PET) and magnetic resonance imaging (MRI) comparisons of MCI converters versus MCI nonconverters. Left: MCI progressor. Top: positive PiB-PET. Bottom: MRI illustrating atrophic hippocampi and ventricular enlargement. Right: MCI nonprogressor. Top: negative PiB-PET with nonspecific white matter retention but no cortical retention. Bottom: MRI illustrating normal hippocampi and no ventricular enlargement. Reproduced with permission from Ref [152].
Using the ADNI database, Schneider et al [170] empirically tested the recommendation that low Aβ-42 and a high t-tau/Aβ-42 ratio can help select those MCI patients most likely to progress to AD throughout the course of a clinical trial. After statistically simulating a number of different clinical trial scenarios with MCI patients with or without biomarker enrichment, they found that selection with either of the biomarker criteria resulted in only minor increases in power for the trial, and concluded that the use of these criteria would likely not result in more efficient clinical trials. In contrast, Beckett et al [154] calculated that restricting a trial population to MCI subjects with CSF Aβ-42 levels of <192 pg/mL would reduce the sample size required from 375 to 226 subjects per arm to detect a 25% change using ADAS-cog as an outcome measure, demonstrating a clear beneficial use of CSF biomarkers in clinical trial population selection (Table 10). Schott et al [160] tested the use of the same cutoff point of CSF Aβ-42 levels in cognitively normal elderly subjects as a selection tool for presymptomatic treatment studies in AD. Those participants with CSF Aβ-42 levels of <192 pg/mL had higher levels of t-tau and p-tau and higher ratios of tau/CSF Aβ-42 and p-tau/CSF Aβ-42, were more likely to be carriers of the APOE ε4 allele, and had significantly higher whole brain atrophy, ventricular expansion, and hippocampal atrophy over 1 year than participants with higher CSF Aβ-42 levels. Of the six participants who later converted to MCI or AD, five had low or borderline baseline CSF Aβ-42 levels, suggesting that the roughly one-third of healthy elderly subjects with a CSF profile consistent with AD were at greater risk for development of the disease. When sample sizes for clinical trials were calculated for both CSF Aβ-42 levels and APOE ε4 genotype as selection criteria and using whole brain atrophy, ventricular expansion, or hippocampal atrophy as the outcome measure, the smallest size per arm [140] was calculated using selection by CSF Aβ-42 levels and whole brain atrophy as an outcome measure (Table 10). Samtani et al [503] studied the relationships between CSF biomarkers and disease progression measured by ADAS-cog scores in MCI patients. Baseline Aβ42, p-tau181, and ptau181/Aβ42, but not t-tau levels, had a bimodal distribution, supporting the idea of the state of prodromal disease being a heterogeneous population of those likely and unlikely to progress. Baseline levels of these biomarkers had good negative predictive value. Eighty-four percent of subjects below the cutoff did not convert to AD within 3 years. In contrast, the positive predictive value of this measure was lower as only 67% of those above the cutoff converted within this time frame. The study suggests that in clinical trials, CSF biomarkers may play a more important role in the exclusion of MCI candidates unlikely to progress than in the selection of candidates likely to progress.
Table 10.
Comparison of methods for increasing power in clinical trials: sample sizes per arm required to detect a 25% reduction in atrophy with 80% power, 5% significance
| Outcome measure: MCI (AD) | |||||||
|---|---|---|---|---|---|---|---|
| Strategy | ADAS-cog | CDR- SB |
Whole brain |
Ventricular expansion |
Hippocampal volume |
Entorhinal complex |
Reference |
| Subject selection by multiple biomarker classifier |
<40 (<40) |
[86] | |||||
| No baseline adjustments, no aging | 149 (81) | 234 (118) | 201 (88) | [174] | |||
| Best baseline adjustments, no aging | 122 (68) | 167 (84) | 178 (74) | ||||
| No baseline adjustments, with aging | 739 (235) | 944 (254) | 648 (179) | ||||
| Best baseline adjustments, with aging | 605 (197) | 675 (181) | 573 (150) | ||||
| Aβ–42 <192 pg/mL | 141 | 225 | 467 | [160] | |||
| Normal elderly | |||||||
| APOE ε4 carrier | 224 | 222 | 703 | ||||
| Normal elderly | |||||||
| All MCI | 834 | 674 | [172] | ||||
| Screening in, best enrichment | 260* | 191† | |||||
| Screening out, best enrichment | 517* | 351‡ | |||||
| All MCI | 978 | 437 | 181 | 161 | 186 | 140 | [168] |
| APOE ε4 enrichment | 774 | 397 | 135 | 129 | 133 | 100 | |
| Atrophy enrichment | 458 | 191 | 141 | 121 | 107 | 67 | |
| All MCI | 375 | [154] | |||||
| Enrichment with Aβ–42 | 225 | ||||||
| QUARC entorhinal volume vs baseline | 131 (44) | [297] | |||||
| QUARC entorhinal volume vs Aβ negative controls |
293 (74) | ||||||
FDG-PET.
Hippocampal volume.
11C-PiB-PET.
Llano et al [422] reported that they were unable to derive a proteomic signature derived from plasma analytes that was able to predict MCI to AD conversion over 12 to 24 months with an accuracy greater than 55% to 60%. However, more promising results were reported for the prediction of amyloid burden rather than disease progression. A panel of 13 plasma analytes developed by Kiddle et al [423] predicted PiB positivity with a sensitivity of 0.918 and a specificity of 0.545. The combination of five analytes, APOE status, age, and CDR-SB scores predicted abnormal neocortical amyloid burden in all AD patients, 69% of MCI patients, and 34% of cognitively normal controls with a sensitivity of 79% and a specificity of 76%. These studies suggest that while blood-based biomarkers do not yet predict disease progression sufficiently accurately to be clinically relevant, they represent a promising approach that warrants further development.
4.6.1.4. Cognitive
Ito et al [171] evaluated disease progression in clinical studies and drug trials performed between 1990 and 2008 by using a model to assess the effect of cholinesterase inhibitors and placebos on longitudinal ADAS-cog scores in mild-to-moderate AD patients. They found no significant differences in the rate of disease progression between patients taking the placebo versus patients receiving cholinesterase treatment. The only significant covariate in disease progression was baseline ADAS-cog score, suggesting that those patients with a higher (worse) ADAS-cog score at baseline had a significantly worse prognosis and higher rates of cognitive deterioration than those with lower (better) baseline scores (Table 9). In a further work by the same group [44], longitudinal ADAS-cog data from ADNI was used to construct a model that included baseline severity, APOE status, age and gender identified as covariates to predict a curvilinear rate of disease progression. Samtani et al [262] also used longitudinal changes in ADAS-cog scores and developed a non-linear mixed effects model for disease progression in AD. They found that years since disease onset and hippocampal and ventricular volume were the primary covariates affecting baseline disease status, whereas age, total cholesterol, APOE status, and cognitive scores (TMT-B and ADAS-cog) most influenced the rate of disease progression in the model. The time of entry into the study may not be equivalent to disease onset time. Delor et al [504] developed a natural history population disease progression model based on CDR-SB scores that allows biomarker profiles to be synchronized at disease onset rather than at study entry. This approach virtually expanded the observation period of the population from 3 to 8 years (Fig. 42).
Fig. 42.
The effect of modeling disease onset time on hippocampal volume-time profiles. (A) Evolution of hippocampal volumes for each disease status. Disease onset times are shown with hippocampal volume (B) centered to the median of the normal population and (C) normalized for age and head size. From Delor et al [504]. Abbreviations: MCI, mild cognitive impairment; AD, Alzheimer’s disease.
Llano et al [96] used a new Random Forests tree-based multivariate model of ADAS-cog in which the subscores had been weighted according to their contribution to patient discrimination. This model, ADAS. Tree, predicted conversion of MCI to AD more accurately than baseline MMSE or ADAS-cog and, in addition, was a better predictor of conversion than the best single imaging (left inferior temporal cortex), metabolism (left precuneus), or CSF (p-tau181p/Aβ-42) biomarkers. The significance of association varied by several orders of magnitude, with the ADAS. Tree four orders of magnitude higher than the next MRI marker, and FDG-PET and CSF biomarkers several orders of magnitude lower than the MRI marker. Moreover, the addition of these markers to the ADAS. Tree model did not result in substantial improvement, providing support for this modified form of ADAS-cog as a useful and effective predictor of future decline (Table 9).
The analysis of neuropsychiatric tests using poset models by Tatsuoka et al [425] allowed correlations to be made between specific cognitive domains and MCI to AD conversion within 2 years. While the overall conversion rate for all MCI subjects was 37.7%, the rate for subjects with a high level of episodic memory impairment at baseline was 61.2%. Those subjects who additionally possessed an APOE ε 4 allele had a conversion rate of 84.2%. In contrast, subjects who were APOE ε 4 negative and whose episodic memory functioned relatively higher had less than a 10% chance of conversion in 2 years. Lower scores in cognitive flexibility and perceptual motor speed were also associated with greater rates of conversion. This preliminary study suggests that poset modeling may be an extremely useful tool in the selection of clinical trial populations.
As depression is a recognized risk factor for AD, there has been some interest in depression as a symptom of prodromal AD and therefore as a surrogate clinical marker. Mackin et al [292] investigated whether subsyndromal symptoms of depression (SSD), with a prevalence of up to 70% in MCI patients may be associated with conversion to AD and thus may predict future cognitive decline. They found that increased endorsement of only one symptom – memory problems – longitudinally, predicted MCI to AD conversion. Lee et al [293] used TBM to compare patterns of brain atrophy over 2 years in MCI patients with or without depressive symptoms. They detected greater frontal (P = .024), parietal (P = .030) and temporal (P = .038) WM atrophy, and larger cognitive deficits in a range of neuropsychological tests in subjects with depression and found that 62% of those with stable depressive symptoms converted to AD within the time of the study compared to 27% of asymptomatic individuals. These studies suggest that depression and its related syndromes may have potential as clinical markers for the identification of patients likely to progress.
4.6.1.5. Combined modalities
As in diagnostic classification, combinations of different modalities are proving to be powerful tools in the prediction of future cognitive decline [239,247,251,252]. Lorenzi et al [172] tested two strategies for the enrichment of MCI patients in clinical trials using changes in brain structure or metabolism, or changes in CSF biomarkers well known to herald future disease progression. They used hippocampal atrophy (MRI); temporoparietal hypometabolism (FDG-PET); CSF Aβ-42, t-tau, and p-tau; and cortical amyloid deposition (11C-PiB PET) as biomarkers to either screen in MCI-c or screen out MCI-nc. Although both strategies substantially reduced the estimated sample sizes required, the authors found that there was a trade-off between the high proportion of converters screened out in the first strategy and the decreased power and increased estimated sample sizes using the second strategy (Table 10). Kohannim et al [86] investigated the utility of their machine learning classifier, based on MRI hippocampal and ventricular summaries, APOE genotype, and age as features, in subject stratification and found that it reduced the numbers of AD and MCI patients required to detect a 25% slowing of temporal lobe atrophy with 80% power to fewer than 40, a substantial reduction over other methods (Table 10). Walhovd et al [155] examined baseline MRI, FDG-PET, and CSF biomarker data to determine the optimum combination of these biomarkers for the prediction of decline over 2 years. They found that in MCI patients, retrosplenial and cortical thickness predicted decline on the CDR-SB, retrosplenial and entorhinal metabolism predicted decline on the MMSE, and hippocampal volume predicted decline in delayed logical memory. The tau/Aβ-42 ratio also predicted decline in the CDR-SB and MMSE, but less significantly than the MRI and FDG-PET measures (Table 9). Beckett et al [154] found that in MCI and AD patients, baseline glucose metabolism in a range of ROIs predicted cognitive decline, as measured by ADAS-cog in a multivariate model. In univariate models, hippocampal and ventricular volume, Aβ-42, and tau also predicted cognitive decline in MCI patients (Table 9). Both papers support the idea that reduced metabolism and greater brain atrophy at baseline are associated with more rapid cognitive decline, and that CSF biomarkers are less useful indicators of future change. Shaffer et al [505] reported that while the addition of MRI or FDG PET ROIs, or CSF biomarkers to clinical covariates (age, APOE status, ADAS-cog, education) improved both prediction accuracy and reduced the percentage of misclassification, the effect of MRI and CSF biomarkers was minimal and FDG PET data added the greatest gains (Table 9). This may be due to a shared variance between the APOE ε 4 allele and Aβ and also between the APOE ε 4 allele and hippocampal volume; FDG-PET is a modality that provides truly novel data that are not related to other covariates. A degree of agreement with these results was found by Landau et al [173], who studied a range of predictors of conversion to AD and cognitive decline, including FDG-PET measures, CSF biomarkers, APOE ε 4 status, and hippocampal atrophy, that were defined dichotomously according to their ability to separate AD and control subjects. Although all biomarkers were predictive of decline in univariate models, only reduced glucose metabolism and episodic memory (measured by the AVLT) predicted conversion to AD and, in contrast to the studies by Beckett et al [154] and Walhovd et al [155], only p-tau181p/Aβ-42 predicted decline in ADAS-cog scores in multivariate models (Table 9). Ewers et al [161] compared the effectiveness of single variables and multiple variables in predicting the conversion of MCI to AD. They found that these best single predictors (right entorhinal cortex and the TMT-B) were comparable in accuracy with the best multiple predictor models, which included right hippocampal volume, CSF p-tau181p/Aβ-42, TMT-B, and age (Table 9). In ADNI and an additional cohort, Prestia et al [506] reported that the optimal combination of biomarkers for identifying prodromal AD was hippocampal volume and CSF Aβ. Examining MR, CSF, cognitive and demographic data, Gomar et al [294] found that their most predictive model included 2 measures of episodic memory (AVLT-delayed memory and Logical memory delayed total) and one MR measure (left middle temporal lobe thickness) (Table 9).
In 2011–2012, the emphasis of these studies has shifted toward using methods that automatically combine and leverage the most pertinent information from a range of modalities and away from the construction and comparison of individual linear regression models. The multi-modal multi-task learning method of Zhang et al [239] was able to combine most predictive features from MRI, FDG-PET and CSF data and predict 2 year changes in both MMSE (r = 0.511) and ADAS-cog (r = 0.511) scores in MCI patients (Table 9). A subsequent paper by Zhang et al [239] used both baseline and longitudinal data to achieve even higher prediction accuracies. Their best predictions of 2 year changes in both MMSE (r = 0.786) and ADAS-cog (r = 0.777) scores used baseline, 6, 12 and 18 month data. The conversion to AD from MCI within the same time frame was predicted with accuracy of 78.4%, a sensitivity of 79%, a specificity of 78% and an AUC of 0.768 (Table 9). Similarly, the Multi-Modality Disease marker developed by Hinrichs et al [118] used longitudinal data to predict conversion more accurately than baseline data alone and found that combined biological, imaging and neuropsychological data outperformed single modalities (Table 9). Another method recently developed by Wang et al [251], SMART, which takes into account the interconnectedness of brain structures and other measures, consistently resulted in better prediction of AVLT scores in control, MCI and AD patients. An approach using the weighted fusion of data from both high- and low-dimensional modalities [406] found that MCI to AD conversion was optimally predicted with an accuracy of 68% by a combination of FDG PET, MRI shape information (ventricular expansion and cortical thinning in specific regions), and CSF biomarkers, although CSF biomarkers added only minor improvement. Integration of multimodal data in a probabilistic manner using a Gaussian process classification approach [408] predicted conversion of MCI to AD over 3 years with an accuracy of 72.2% compared with 69.4% using the multimodal method of Zhang et al [249]. The disease state index of Mattila et al [252], which included demographic and genetic information as well as imaging data and cognitive scores, was able to predict the conversion of MCI to AD with an AUC of 0.752 (Table 9). Soininen et al [253] used the same tool and found that it could discriminate between MCI converters and stable MCI patients with an accuracy of 68.6%, but that when patients were assigned to categories of risk for AD based on threshold values, the prediction accuracy increased to 84.4% for those having strong evidence and to 93.7% for those with very strong evidence of AD pathology (Table 9). The Predict AD software developed by this group significantly outperformed currently recommended criteria for prodromal AD in predicting conversion within 3 years. Liu et al [418] found that, with the aid of a clinician, this tool had an accuracy of 72%, a sensitivity of 75%, and a specificity of 68%, supporting the utility of software that can integrate heterogeneous data and provide objective and evidence-based information about the state of the patient that is not limited to yes/no answers and that places patients on a disease spectrum on the basis of this information.
While the above papers developed a range of automatic multi-modal methods of the prediction of disease progression, Heister et al [295] asked whether MCI to AD conversion can be predicted using clinically available biomarker systems (commercially available software for fully automated volumetric MRI and commercial CSF analysis). They stratified the MCI cohort by degree of MR atrophy, CSF biomarker levels or the degree of learning impairment on AVLT. Cox proportional hazard models were used to assess the contribution of each factor to MCI to AD conversion. They found that a single risk factor resulted in a 1.8 to 4.1 fold risk of converting to AD within 3 years and that more than one risk factor was associated with a greater risk of conversion. Patients with both learning impairment and increased MR atrophy were at the highest risk with a HR of 29.0 for conversion. This study supports the use of commercially available CSF and MRI biomarkers in combination with neuropsychological tests in predicting the risk of MCI to AD conversion.
The degree to which prediction models developed in one cohort are generalizable to different settings was investigated by Devanand et al [296]. They developed a variety of models that included different combinations of imaging, cognitive and demographic data in the Questionable Dementia study and tested these in the ADNI cohort. Prediction accuracy of the MCI to AD conversion was consistently lower by a similar degree in the ADNI setting, suggesting that these models are portable and robust in clinical settings. Gross et al [466] combined cortical thickness and cognitive data to predict MCI to AD conversion. They used regression analysis to develop cortical signatures of cognition specific to memory, executive function, language, and visuospatial processing domains, empirically defined by their correlation with domain-specific cognitive factor scores derived from new ADNI neuropsychological battery scores [112]. The combination of the baseline thickness of these cortical signatures of cognition for memory and language and the corresponding neuropsychological factor scores predicted future clinical decline. One standard deviation decrease in the combined memory cortical signature and memory factor score corresponded to a 2.3-fold increase in hazard of conversion. Casanova et al [411] developed AD-PS scores that, when combined structural MRI and cognitive scores, were able to detect AD-like patterns of atrophy and cognitive decline across clinical groups and were strongly associated with MCI to AD conversion times.
What role does age play in the prediction of future decline in MCI patients? The logistic regression analyses conducted by Schmand et al [507] revealed that some biomarkers had differential predictive abilities depending on the age of the participants. Whereas MRI and neuropsychological tests were relatively stable predictors regardless of age, CSF biomarkers were only effective in patients younger than 75 years and FDG-PET did not significantly currently predict conversion at any age. None of the measures predicted MCI to AD conversion with an AUC of greater than 0.73 at any age, although combined markers had improved predictive abilities (AUCs of 0.79 and 0.74 in participants younger and older than 75 years, respectively). The study supports the idea that age is an important characteristic of the observed heterogeneity in MCI patients.
Does the predictive ability of biomarkers change over different lengths of time? Dickerson et al [508] examined how a structural MRI measure of neurodegeneration, the AD signature, and hippocampal volume compared with a measure of amyloid deposition (CSF Aβ) in predicting conversion of MCI patients over both 1- and 3-year periods. The AD signature biomarker outperformed CSF Aβ at predicting dementia within 1 year, whereas both biomarkers had similar prognostic abilities over 3 years. In addition, 1-year conversion to AD in MCI subjects with normal CSF Aβ levels was best predicted by the AD signature biomarker, suggesting that evidence of neurodegeneration is prognostically useful in these individuals. These results are consistent with the model of development of AD pathophysiology by Jack et al [14] in that the deposition of Aβ is an earlier event and therefore should predict conversion over a longer period than the later-occurring neurodegeneration.
The need to select clinical trial participants who are cognitively normal but who are likely to progress to MCI or AD has driven research into predicting clinical progression in the elderly healthy subjects. Ewers et al [509] examined 54 healthy control subjects from ADNI and tested the ability of MRI regional GM volume, FDG-PET a priori–defined ROIs, and executive function measures to predict clinical change over 3 to 4 years. The combination of FDG-PET measures in the medial temporal and parietal regions combined with TMT-B scores proved to be the most accurate predictor of clinical progression with an accuracy of 93.4%, a sensitivity of 82%, and a specificity of 93%. Of the 54 subjects, 11 converted to MCI or AD over the course of study.
4.6.1.6. Comparison of modalities
The trend toward a multimodal approach to prediction is fueled by studies reporting improved accuracy with the use of more than one modality. Most studies report that optimum combinations of modalities for prediction include MRI measures of the temporal lobe, hippocampus and entorhinal cortex, and another imaging modality, perhaps with the addition of CSF biomarkers, genetic, or clinical information. However, for the first time, the effectiveness of three different imaging modalities (MRI, PiB-PET, and FDG-PET) in the prediction of conversion of MCI patients to AD was directly compared by Trzepacz et al [510] in ADNI MCI patients who converted within 2 years. Eliminating genetic or clinical data from their models, they found that the best single predictors were MRI measures of temporal lobe, hippocampal and entorhinal cortex volume, along with amyloid deposition in the temporal cortex measured by PiB-PET. MRI was the single modality with the highest predictive accuracy (67%) followed by PiB-PET (66%) and FDG-PET (62%). In combination, MRI increased the accuracy and sensitivity, but not specificity of both PiB-PET and FDG-PET measures (Fig. 43). MRI in combination with PiB-PET increased accuracy to 76% with a sensitivity of 53% and a specificity of 90%. This systematic review confirms results from a variety of previous studies.
Fig. 43.
Prediction accuracies, sensitivities, and specificities for individual and combined neuroimaging modalities. From Trzepacz et al [510]. Abbreviations: MRI, magnetic resonance imaging; FDG, fluorodeoxyglucose; SUVR, standard uptake value ratio; PET, positron emission tomography.
4.6.2. Adjustments for normal aging and baseline characteristics
McEvoy et al [168] also examined the effect of normal aging on the detection of longitudinal change and found that although this did not affect clinical outcome measures such as ADAS-cog and CDR-SB, neuroimaging outcome measures were far more sensitive to atrophy associated with normal aging. They suggested that larger sample sizes are required in clinical trials to account for this effect, and that clinical trials run the risk of being severely underpowered if normal aging is not taken into account. Schott et al [174] proposed an alternative method for increasing the statistical power of clinical trials without resorting to subject selection procedures that can potentially limit the applicability of studies. They found that by statistically adjusting for a range of baseline characteristics that might account for interindividual differences, and also for normal aging, sample sizes were reduced by 15% to 30% in AD subjects and by 10% to 30% in MCI subjects (Table 10). The importance of appropriate controls in AD disease-modifying clinical trials was studied by Holland et al [297] who estimated required sample sizes using either absolute change relative to baseline, change relative to controls or change relative to healthy controls who tested negative for Aβ. While their calculations suggested that larger sample sizes were required for measures relative to Aβ negative controls, the authors felt that this approach would most accurately reflect the actual effect of a drug on AD pathology. The study compared 5 publically available methododologies to measure structural changes in neuroanatomical subregions and smallest sample sizes were calculated using the QUARC approach to quantify the entorhinal cortex (Table 10).
4.6.3. Development of outcome measures
A number of studies have focused on determining the effectiveness of different biomarkers as outcomes in clinical trials by calculating sample size estimates for a hypothetical clinical trial, per arm at either 90% (N90) or 80% (N80) power to detect a 25% improvement in annual rate of decline. Schuff et al [121] used hippocampal volume loss over time, assessed by MRI, as an outcome measure and found that the greatest reductions in sample size were achieved when three serial scans (0, 6, and 12 months) were combined with APOE ε4 data using Markov chain analysis to exploit correlations between observations (Table 11). The inclusion of Aβ-42 level data did not further reduce sample size. All MRI hippocampal measures were substantially better than cognitive measures (ADAS-cog and MMSE) as outcome measures. Wolz et al [64] used a 4-D graph cut method to segment the hippocampus and subsequently calculated N80s in the same range as the best combinations of Schuff et al [121] (Table 11). Nestor et al [175] investigated the use of ventricular expansion as an outcome measure and found that ventricular expansion over 6 months was sufficiently sensitive to produce N80s for a hypothetical trial at least an order of magnitude lower than clinical scores (MMSE and ADAS-cog). Moreover, sample sizes were further reduced when the trial population of AD subjects was restricted to carriers of the APOE ε4 allele (Table 11). Holland et al [176] examined the utility of longitudinal volumetric change in a variety of ROIs as an outcome measure with which to measure putative disease-modifying medications for AD and MCI. ROIs, including temporal lobe structures and ventricles, and whole brain atrophy were compared with clinical measures in two separate models, one in which the putative drug was presumed to affect both disease and aging-related changes (model T for “total”), and one in which the drug putatively affected only disease-specific changes (model D for “disease-specific”). They found that although imaging measures generally resulted in smaller sample sizes than cognitive measures in both models, model T was the more conservative model for cognitive measures, whereas model D was more conservative for imaging measures. The authors emphasized the importance of comparing both models when comparing across imaging and cognitive outcome measures (Table 11).
Hua et al [177] compared a variety of nonlinear registration methods used in TBM with standard clinical outcome measures and found that a substantial reduction in sample size at 80% power (N80s) was achieved over clinical measures using all TBM methods, with the best TBM measure presenting an eightfold improvement over the best clinical measure (CDR-SB) (Table 11). The same group [120] subsequently compared the use of TBM to measure GM of the entire brain and WM atrophy in the temporal lobe with 1-year changes in CSF biomarkers as outcome measures in a hypothetical clinical trial. The N80s for CSF biomarkers were much larger than those from neuroimaging measures, reflecting their poorer reproducibility, especially in later stages of the disease process (Table 11). Ho et al [47] compared 3.0-T and 1.5-T MRI for tracking disease progression using TBM and an alternative method for measuring the overall percentage brain volume change, Structural Image Evaluation, using Normalization, of Atrophy. The lowest calculated N80 resulted from using TBM on a 1.5-T MRI scanner to detect changes in brain atrophy as an outcome measure (Table 11). Leung et al [51] estimated N80s for both the classic brain BSI MRI technique and their improvement on this, the KN-BSI method, and found that the improved method resulted in lower N80s (Table 11). More recently, using a newly revised TBM method that enforces inverse consistency, Hua et al [178] reported that to demonstrate a 25% slowing of atrophic rates with 80% power, 62 AD and 129 MCI subjects would be required for a 2-year trial and 91 AD and 192 MCI subjects for a 1-year trial. A longitudinally unbiased method that uses machine learning techniques to maximize the ability to track three-dimensional change in lateral ventricles over time was described by Gutman et al [511]. Using linear discriminant analysis on the generated data, they estimated N80s over 1 year to be 104 for MCI patients and 75 for AD patients, compared with estimates of 165 and 94 for MCI and AD patients, respectively, using the statistical ROI method. Pardoe et al [512] derived a model using a genetic programming approach that takes into account study-specific parameters such as the type I error rate, the level of smoothing applied, and the thickness difference to be detected in clinical trials measuring cortical thickness. Sample size estimates were heterogeneous over the cortical surface, with lobe-specific N80s for the detection of a 0.25-mm thickness change ranging from approximately 25 to 30 to 50 in the occipital lobe, frontal and parietal lobes, and the temporal lobe, respectively. Thus, accounting for cortical thickness spatial variability may be critical when designing clinical trials.
Beckett et al [154] compared a number of promising MRI and FDG-PET outcome measures. They calculated the sample size that would be required in a two-arm, 1-year clinical trial with 80% power to detect a 25% effect, and found that MRI measures of overall brain change, using either ROIs or BSI techniques, or hippocampal volume required fewest subjects. Brain metabolism measures were generally less effective, requiring substantially larger sample sizes, although the best FDG-PET measure, a data-driven functional ROI, was comparable with many of the MRI measures (Table 11). In contrast, Herholz et al [291] found their composite PET score, based on FDG-PET data, to be a better outcome measure than ADAS-cog scores due mostly to its higher test-retest reliability which resulted in smaller required sample sizes. Relative to a sample size of 100 required at 12 months with ADAS-cog as an outcome measure, the PET score outcome measure required a sample size of 28. At 6 and 24 months, the PET and ADAS-cog sample sizes were 120 and 397, and 13 and 35, respectively. The PET score was linearly associated with ADAS-cog scores, emphasizing its validity as a measure of cognitive impairment.
The accepted standard outcome measure in AD disease modifying clinical trials is the ADAS-cog. Schrag et al [298] empirically determined the minimum clinically relevant change in ADAS-cog and compared it to the current standard of expert consensus. Using MMSE, CDR and FAQ scores, they ascertained that a 3 point decline in ADAS-cog over 6 months was clinically relevant, a point less than the consensus FDA recommendation, suggesting that the FDA standard may be too stringent and may consequently miss an important drug effect.
4.6.4. Genetic risk factors in subject selection
The addition of APOE status has consistently been shown to improve the accuracy of predictive models. Kohannim et al [513] examines the utility of the recently identified AD genetic risk alleles, CR1, CLU, and PICALM, in selecting cohorts for clinical trials. Control and MCI subjects from ADNI were ranked depending on the relative risk from these four genes, and N80s were obtained using an MRI-derived 2-year atrophy rate as an outcome measure. They found a statistically significant reduction in sample size of approximately 50% in the combined control and MCI group beyond the effect of the APOE ε 4 allele alone when these three risk alleles were included in the model. In a mixture of MCI and control APOE ε 4 carriers, the N80 was reduced from 94 to 69 using selection with these three genetic risk factors. The results suggest that genetic profiling with additional risk genes may be an effective strategy in reducing sample sizes in clinical trials of early intervention therapies.
4.6.5. Combining enrichment and stratification strategies
Increasing attention has been given to optimizing enrichment and stratification strategies to select the small cohorts of cognitively asymptomatic and MCI subjects required for viable clinical trials. Using the full standardized ADNI data set [514], Hua et al [515] calculated sample size estimates using temporal lobe atrophy measured by TBM as an outcome measure combined with stratification based on APOE ε 4 status. For a 2-year trial, N80s of 73 (95% CI: 57–94) and 122 (95% CI: 80–229) were estimated for MCI and cognitively normal patients, respectively. In MCI patients, Holland et al [516] found that stratification with p-tau181 combined with the use of entorhinal cortex atrophy as an outcome measure produced the smallest estimated sample sizes. All MRI atrophy outcome measures outperformed the best clinical measure, CDR-SB (Fig. 44). However, as the FDA has not yet accepted biomarkers as outcome measures, Grill et al [517] tested combination of enrichment strategies and clinical outcome measures. In MCI subjects, the CDR-SB as an outcome measure consistently produced lowest sample sizes over virtually all enrichment strategies in agreement with Cedarbaum et al [518]. Estimates for N80s with this outcome measure ranged from 258 with enrichment using the t-tau/Aβ to 458 with no enrichment. In contrast, the lowest sample size in the cognitively normal group (499) was obtained using enrichment with APOE ε 4 status and AVLT-total as an outcome measure. The studies support the use of biomarker inclusion criteria for predementia and early dementia clinical trials and suggest that the CDR-SB may be a more sensitive clinical outcome measure of these trials. However, both studies using cognitive tests as outcome measures reported that in contrast to biomarker outcome measures, sample size estimates for cognitively normal populations had prohibitively high upper bounds. This suggests that in order for clinical trials in presymptomatic cohorts to be feasible, a biomarker-based outcome measure should be considered.
Fig. 44.
Enrichment strategies for the selection of MCI participants for clinical trials. Estimated N80s are indicated assuming a 24-month trial with scans every 6 months. From Holland et al [516]. Abbreviations: MCI, mild cognitive impairment; MRI, magnetic resonance imaging; CDR-SB, clinical dementia rating-sum of boxes.
4.6.6. Other improvements to clinical trials
Clinical trials for AD modifying treatments require special considerations due to the advanced age of the participants and their high rates of medical co-morbidities. Hendley et al [299] studied patients taking placebo in recent AD clinical trials and ADNI control participants to determine the rates of adverse events, serious adverse events, discontinuation from trials and frequencies of death. The authors hoped that the accumulated reference data would aid in the design of future long term AD studies. Thompson et al [300] reviewed methodologies for characterizing disease trajectories over a lifespan using ADNI as an illustration of a longitudinal unstructured multi-cohort study. They reported that, while this study design is superior to a cross-sectional design in terms of eliminating a number of confounding factors, it is still susceptible to age cohort effects due to the randomness of participant ages (ranging from 55 to 90). They suggest an improved structured longitudinal model in which age cohorts would be tiered but overlapping. As ADNI is a convenience sample and not population based, the ADNI cohort may select people more predisposed to cognitive decline. Therefore, sample sizes based on hippocampal atrophy may result in trials underpowered to detect treatment effects in the general population [519]. Hua et al [515] tested the effect of selective data exclusion, that is, the removal of apparent outliers, and found that this practice resulted in the underestimation of N80s and therefore constitutes a major source of bias in ADNI experiments. The use of the standardized ADNI data set [514] will hopefully circumvent this issue.
The clearance of Aβ by immunotherapy approaches under study in some clinical trials may increase the risk of microhemorrhage and siderosis. Kantarci et al [520] investigated the prevalence of these conditions in the ADNI cohort and their association with amyloid load. Focal hemosiderin deposits indicating microhemorrhages occurred in 25% of the cohort. Their prevalence increased with age and Aβ deposition, and the risk of subsequent microhemorrhage increased with increasing baseline occurrence. The study suggests that microhemorrhages are a common imaging finding that should be taken into consideration in the planning of clinical trials of amyloid-modifying agents for disease prevention and treatment.
4.6.7. Summary and conclusions of papers focused on the improvement of clinical trial efficiency
Strategies for the reduction of sample sizes in clinical trials by the selection of subjects with a significantly worse prognosis and through the use of more effective outcome measures have been developed over the course of ADNI. Studies have found that baseline MRI measures, particularly of hippocampal volume and of whole brain atrophy, outperform measures of glucose hypometabolism or CSF biomarkers in the prediction of future decline. In one instance, a score derived from AD-like patterns of hypometabolism outperformed other single MRI, cognitive, or CSF biomarker measures, but this too was enhanced by the addition of MRI measures. Of the CSF biomarkers, the t-tau/Aβ-42 ratio and the use of a cutoff value of approximately 192 pg/mL Aβ-42 have been shown to best predict future decline. In a manner similar to classification of AD subjects, the use of multiple modalities appears to enhance the prediction of future decline. Interestingly, a weighted version of the ADAS-cog [96] has been shown to outperform any single MRI measure tested as a predictor of future change and was not improved by the addition of any MRI measure tested. In contrast, MRI and FDG-PET, which have strikingly better signal-to-noise ratios, clearly outperformed cognitive tests as outcome measures of rates of change. Calculated sample sizes for clinical trials required to see a 25% effect at 80% power were lowest for MRI measures of overall morphometric change or of hippocampal volume, followed by those for hypometabolism ROIs and cognitive scores. CSF biomarkers were the least effective outcome measures by several orders of magnitude. Finally, it also will be necessary to study the comparative effectiveness and cost-effectiveness of the AD biomarkers studied in ADNI to determine the optimal way to make use of these biomarkers in the diverse applications needed in AD research. For example, based on the recent studies of Wiegand et al [169], it is possible to impute Aβ measures determined by Aβ imaging using far less expensive measures of CSF Aβ-42 levels. Additional similar studies as well as others focused on the economics of the use of biomarkers in clinical trials and clinical practice are needed.
A major emphasis in papers published in 2011–2012 is the prediction of future decline at an even earlier stage, with some works focusing on identifying cognitively normal individuals at high risk of disease development. Both levels of CSF biomarkers and volumetric MRI were successfully used in this application, in agreement with the Jack model for disease progression [14]. As in classification, the prediction of MCI to AD conversion was most accurate when longitudinal data and/or combined modalities were used, and a number of papers focused on the use of automated methods to select the most pertinent information from multiple modalities. Depression was identified as a novel predictor. From a methodological standpoint, the use of an Aβ negative control group in clinical trials was recommended to reflect the largest drug effect, and the minimum significant change in ADAS-cog scores was calculated to be lower than the FDA-recommended change. Both methods reduced sample sizes.
Approaches for the prediction of future decline continued to be developed in 2012 and 2013. In particular, studies of longitudinal data found that the predictive ability of biomarkers is dynamic in relation to the time to conversion, with modalities reflecting amyloid deposition having a greater predictive ability at time points further from conversion and glucose metabolism being more predictive closer to conversion. The predictive ability of brain atrophy also progressed from temporal to frontal lobes with time to conversion. There was a focus on distilling a single composite score of brain atrophy to predict decline, as structural changes appeared more useful over a longer period of time, reflecting the ongoing nature of atrophy in disease progression. Blood-based biomarkers were not reported to be effective predictors, but their development is ongoing, and they do hold some promise as a cost-effective alternative to imaging techniques. For the first time, the genetic risk alleles other than APOE ε 4 were shown to enhance prediction accuracy, and these may be a useful addition to the clinical trial subject selection process. Other studies suggested that hippocampal volume was an effective enrichment biomarker but that CSF biomarkers may function better to exclude participants. Although MRI measures were shown to be superior as outcome measures, a lack of FDA approval turned the focus to evaluating and improving existing cognitive tests. CDR-SB was shown to outperform ADAS-cog in more than one study. Together, these studies suggest that sample sizes of MCI patients will soon be sufficiently small and have a high enough probability of progressing to make clinical trials of disease-preventing or altering therapies viable.
5. Identification of genetic risk factors for AD
The influence of genetics on the dynamic trajectory of brain development and aging is well established, if not well understood. Studies of twins have estimated the heritability of AD to be between approximately 60% and 80% [179], and until recently the only established genetic risk factor for AD was the APOE ε4 allele, which accounts for approximately 50% of AD heritability [180]. The question of accounting for the up to 30% of heritability remaining has only begun to be addressed, and although there have been a number of candidate genes proposed, the majority of them await independent confirmation. ADNI is in the unique position of providing a large cohort with genotype information in addition to imaging and biochemical data that can be leveraged as QTs in uncovering new genetic associations, and as such plays an increasingly important role in the discovery and confirmation of novel genetic risk alleles. As of the end of 2014, there were 2065 distinct APOE genotype data results available, and GWAS data were available for 1252 participants within ADNI.
Three main approaches have been taken to investigating the genetic basis of AD. Case–control studies that search for loci with differential frequency between patient groups have identified a number of candidate genes. Typically, markers are used to tag susceptibility loci, usually in 10-kb to 20-kb regions in the genome, that are rarely found to be causal. Using this method, the association of APOE ε4 allele with AD has been confirmed, and three new risk loci, CLU, PICALM, and CR1, have been identified and confirmed [181–183]. Further studies have focused on examining relationships between SNPs in a limited number of genes of interest and quantifiable phenotypic characteristics or QTs, such as imaging data or levels of CSF biomarkers. GWAS evaluate a large and dense set of SNP markers distributed throughout the genome, providing an unbiased search for the discovery of new candidate genes. With more than 500,000 markers typically included in a GWAS, a stringent correction for multiple testing is required with typical thresholds of P < 10−8 used to reduce false detections. These stringent corrections also greatly reduce power and require extremely large sample sizes to achieve significance in case–control designs. However, the use of quantitative phenotypes such as cognitive, imaging, and fluid biomarker measures can greatly increase the power to detect associations. Where a binary case–control design might require many thousands of samples to detect a gene effect, samples on the scale of ADNI are sufficient for detecting associations with quantitative phenotypes [184]. Structural neuroimaging data have been most commonly used as a quantitative phenotype as the whole brain, brain circuits involving multiple regions of interest, or individual regions of interest [521]. The emerging field of imaging genetics, which uses imaging data as QTs in GWAS, promises the power to reveal patterns of genetic associations throughout the brain, but is hampered by the computational load required for such high-dimensional studies. Further development of this field, including improvement of existing GWAS methods, is a major goal of the Genetics Core of ADNI [6].
The rich multimodal ADNI data set also offers the possibility of discovering novel susceptibility loci associated with risk for AD or for the disease itself [521]. By the end of 2013, APOE and nine other risk genes had been identified and confirmed. However, together they account for only about 50% of the estimated 80% of AD heritability, indicating that other genes must be involved. The hunt for the missing heritability continues, even as newly discovered susceptibility loci are examined for their potential as targets for the development of disease-modifying agents.
5.1. Case–control studies
Jun et al [185] conducted a meta-analysis case–control study of AD patients and healthy elderly control subjects from 12 different studies, including ADNI, to examine the association of APOE ε4, CLU, PICALM, and CR1 with AD. They found that CLU, PICALM, and CR1 were significantly associated with AD only in Caucasian populations. In contrast, APOE ε4 was significantly associated with AD in all ethnic groups and with PICALM in white populations, suggesting that APOE ε4 and PICALM act synergistically and may participate in a common pathological pathway (Table 12). Two of the largest case–control GWAS studies of AD were recently published as companion reports in Nature Genetics [186,187]. Both reports included the ADNI-1 data in their analyses (Table 12). These multistage meta-analytic reports included discovery and replication data sets and confirmed each other. These new results bring the total set of confirmed and replicated candidate genes to 10 (APOE/TOMM40, ABCA7, BIN1, CD2AP, CD33, CLU, CR1, EPHA1, MS4A4/MS4A6A, PICALM).
Table 12.
AD susceptibility and quantitative trait loci identified by genetic studies of ADNI cohort
| Confirmed AD risk loci identified using ADNI data | |||
|---|---|---|---|
| Gene | Protein | Putative protein function | Reference |
| TOMM40 | Translocase of outer mitochondrial membrane | Protein transport across mitochondrial membrane | [184] [199] [194] [149] [152] [313] |
| CLU | Clusterin | Clearance of A? | [185] |
| CR1 | Complement component[3b/4b] receptor | Clearance of A? | [185] [193] |
| PICALM | Phosphatidylinositol-binding clathrin assembly protein |
Synaptic vesicle cycling and/or affects APP processing via endocytic pathways |
[185] [193] [197] |
| BIN1 | Myc box-dependent-interacting protein 1 | Synaptic vesicle endocytosis | [193] |
| CD2AP | CD2-associated protein | Regulation of receptor-mediated endocytosis | [186] |
| CD33 | Siglec-3 | Clathrin-independent endocytosis | [186] |
| MSA4 | Membrane Spanning 4 Domains Subfamily A gene cluster |
Cell surface protein – receptor? | [187] [186] |
| ABCA7 | ATP-binding cassette sub-family A member 7 | Membrane transporter highly expressed in brain | [187] |
| EFHA1 | EF-hand domain family member A1 | Regulation of cell morphology and motility in epithelial tissues |
[181] |
| Candidate AD risk loci identified using ADNI data | |||
| ARSB | Arylsulfatase b | Oxidative necrosis, dementia | [184] |
| ATXN1 | Ataxin-1 | Upregulates A? | |
| CADPS2 | Calcium-dependent secretion activator 2 | Synaptic vesicle priming | [195] |
| CAND1 | Cullin-associated and neddylation-associated 1 | Ubiquination, apoptosis | [184] |
| CDH8 | cadherin 8, type 2 | calcium-dependent cell adhesion protein implicated in synaptic adhesion; interacts with presenilin |
[6] |
| CHRFAM7A | Cholinergic receptor, nicotinic, alpha7/FAM 7A | unknown | [317] |
| CNTN5 | Contactin-5 | Neurite growth | [301] |
| CSMD1 | CUB and sushi domain-containing protein 1 | Central nervous system regulator | [314] |
| CSMD2 | CUB and sushi domain-containing protein 2 | Oligodendroglioma suppressor ? | [195] |
| CYP19A1 | Cytochrome P450, family 19, subunit a, polypeptide 1 | Conversion of androgens to estrogens | [199] |
| DOPEY | Dopey family member 2 | Down syndrome candidate gene | |
| EFNA5 | Ephrin-A5 | Hippocampal development | [184] |
| EPC2 | Enhancer of polycomb homolog 2 | Formation of heterochromatin | [149] |
| EPHA4 | EPH receptor A4 | Synapse morphology | [194] |
| ERBB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 |
Brain tyrosine kinase | |
| GRINB | N-methyl-D-aspartate glutamate receptor | Learning, memory, excitotoxic cell death | [196] |
| GSTT1 | Glutathione S-synthetase | Oxidative stress | |
| HFE | Hemochromatosis | Increases redox-active iron and oxidative stress | [189] |
| HLA-DPB1 | Major histocompatibility complex, class II | Immune system | |
| LOC10012 | Unknown function, overlaps with APOE | Unknown | [149] |
| IMMPL2 | Inner mitochondrial protein peptidase-like | Mitochondrial function − oxidative stress | |
| MAGI2 | Membrane associated guanylate kinase | Ubiquination, dementia | [184] |
| NCAM2 | Neural cell adhesion molecule 2 | Neural adhesion, fasciculation of neurons | [199] |
| NRXN1 | Neurexin 1 | Synaptic contacts | |
| NXPH1 | Neurexophilin 1 | Dendrite-axon adhesion | [194] |
| PPP3CA | Protein phosphatase B | Affects tau phosphorylation | [190] |
| PPP3R1 | Protein phosphatase B | Affects tau phosphorylation | [190] |
| PPP3R1 | Protein phosphatase B | Affects tau phosphorylation | [191] |
| PRUNE2 | Prune homolog 2 | Apoptosis | [184] |
| RELN | Reelin | Neuronal migration | |
| TF | Transferrin | Increased redox-active iron + oxidative stress | [189] |
| TP63 | Tumor protein 63 | Unknown | [194] |
| ZNF292 | zinc finger protein 292 | Expressed in brain | [197] |
Mitochondrial genes are also of great interest in AD, and Lakatos et al [188] studied the incidence of AD in patients belonging to different subgroups (HV, JT, UK, and IWX) of mitochondrial haplogroup N in the ADNI cohort. They found that haplogroup UK had the strongest association with AD, and that this relationship remained significant after adjusting for APOE ε4 allele dose. Additionally, they identified five mitochondrial SNPs that were associated with increased risk of AD and suggested that, given the vital role of mitochondria in maintaining cellular energy balance, dysfunctional mitochondria may contribute to AD by causing neuronal oxidative damage. In another case–control design, Kauwe et al. [189] attempted to replicate a study that found that epistatic linkage between two SNPs in the transferrin and hemochromatosis genes was associated with AD risk, suggesting a role for iron in AD pathology. Using synergy factor analysis, they found significant association between bicarriers of the minor alleles of both SNPs and risk for AD in several U.S. and European study populations, including ADNI, providing support for the iron hypothesis (Table 12). Erten-Lyons et al [301] investigated the association between microencephaly genes, responsible for regulating brain growth in utero, and AD in two cohorts including ADNI, but were unable to detect any increase risk associated with common variants of these genes.
5.2. Studies of limited loci using quantitative phenotypes
Several studies have used knowledge of the model for AD progression by testing the associations between genes potentially involved in AD pathology and CSF biomarkers. Cruchaga et al [190] examined associations between SNPs in 35 genes putatively involved in tau posttranslational modification and CSF levels of p-tau181p. They found that SNPs in the gene for protein phosphatase B were associated with higher levels of p-tau181p, and that an SNP in the regulatory subunit of protein phosphatase B was more highly expressed in AD patients compared with control subjects (Table 12). These results suggest that genetic variants that alter the activity of protein phosphatase B could contribute to AD pathology by affecting tau phosphorylation. A further study by the same group [191] found that the SNP in the regulatory subunit of protein phosphatase B was associated with the rate of disease progression, and not with the age of onset or risk of AD. In contrast, APOE ε4 was associated with lower levels of CSF Aβ-42, increased disease risk, and lower age of onset, providing support for a model in which amyloid deposition is an early event in disease progression and accumulation of hyperphosphorylated tau occurs at a later stage (Table 12). Kauwe et al [192] also used levels of CSF biomarkers as a QT to investigate the predicted biological effects of SNPs in three genes associated with AD. They found that a nonsynonymous coding substitution in the gene for calcium homeostasis modulator 1 (CAHLM1), proposed to affect levels of Aβ by modulating intracellular calcium levels, was associated with increased CSF levels of Aβ-42 (Table 12). Associations between levels of CSF biomarkers and SNPs in the two other genes for growth factor receptor-bound protein-associated binding protein 2 (GAB2; proposed to influence tau phosphorylation) and sortilin-related receptor (SORL1; an apoE receptor proposed to bind Aβ) were not found, perhaps because of power limitations of the study.
Using six imaging measures reflective of AD pathology as QTs, Biffi et al [193] searched for associations between these and SNPs in a range of established and candidate genes for AD risk. They first sought to confirm associations of APOE, PICALM, CLU, and CR1 with AD, and found that although APOE had a strong association with diagnosis, of the remaining identified risk alleles, only CR1 was associated with AD in the ADNI cohort, possibly reflecting sample size limitations for case–control studies. Two novel loci, CNTN5 and BIN1, were also found to have significant association with AD (Table 12). When the relationship of APOE ε4, CR1, CNTN5, and BIN1 with imaging measures was examined, it appeared that APOE ε4 was associated with virtually all brain regions, whereas the other loci had a more limited pattern of association, consistent with APOE ε4 being the primary AD genetic risk factor and other loci making more modest contributions to the disease.
While the APOE ε 4 allele remains the major risk allele for AD, the question of its influence on other alleles remains to be clarified. Murphy et al [302] investigated the effect of APOE status on 2 alleles of the cholesteryl ester transfer protein (CETP) and their relationship with brain atrophy in the ADNI control, MCI and AD patients. Using atrophy of the hippocampus, entorhinal cortex and parahippocampal gyrus as a QT, they found that the V and A alleles of I405V and C629A, which decrease CETP activity and therefore increase high density lipoproteins, had differential effects depending on APOE status. In carriers of the APOE ε 4 allele, the V and A alleles were associated with less atrophy whereas results were reversed in non-carriers, suggesting that CETP polymorphisms may influence neurodegenerative disease susceptibility in an APOE-dependent manner.
Given that glucose metabolism reflects cognition, the effect of genetic risk factors for AD that influence brain atrophy and subsequently cognition may be reflected in altered cerebral metabolism. Xu et al [303] investigated the influence of one genetic factor, the V66M polymorphism of brain-derived neurotrophic factor (BDNF), essential for neuron survival, on brain glucose metabolism and identified patterns of changed metabolism in carriers of the V66M polymorphism compared to non-carriers in the ADNI cohort. The regions affected by this polymorphism changed with disease severity, with MCI carriers exhibiting alterations in regions affected in both cognitively normal carriers (parahippocampal gyrus and temporal cortex) and those with AD (bilateral insula), providing further support for polymorphisms in BDNF as a genetic risk factor for AD. In a further study, Honea et al [522] tested associations between SNPs in the BNDFI gene and established cognitive and imaging AD phenotypes. In a pooled sample of 645 ADNI participants, no SNPs in this gene were associated with AD diagnosis. However, additional SNPs, other than the SNP containing the V66M polymorphism, were associated with baseline ADAS-cog scores and with hippocampal atrophy over 2 years. Additional SNPs were also associated with cognitive decline and whole brain atrophy over 2 years in cognitively normal patients. These results suggest that while BDNF genetic variation is not specifically associated with AD, it does play a role in memory-related performance and brain morphometry in aging individuals.
The fat mass and obesity associated (FTO) gene, an obesity genetic risk factor, has been associated with AD. Reitz et al [523] identified SNPs within introns 1 and 2 and within exon 2 of the FTO gene that were significantly associated with AD in several data sets including ADNI. Moreover, the expression of FTO was lower in AD cases compared with that in controls.
Delta opioid receptors have been implicated in neurodegeneration. Roussotte et al [524] investigated associations between a common variant of the opioid receptor gene (OPRD1), rs 678849, and regional brain volumes in the ADNI cohort. The minor C allele at this locus (which alters Aβ processing by changing receptor structure) was associated with smaller volumes in frontal, temporal, and occipital brain regions and had a trend toward association with CSF biomarkers in the healthy elderly.
5.3. GWAS of quantitative phenotypes
In the first ADNI GWAS using the ADNI AD cases and control subjects, Potkin et al [184] confirmed the association of APOE with AD and identified a novel AD risk gene, TOMM40, encoding a regulatory subunit of a protein translocase in the outer mitochondrial membrane, as being significantly associated with AD. A further GWAS using VBM-derived estimates of hippocampal volume as a QT identified 21 loci with significant association with hippocampal volume including, in addition to APOE ε4, genes involved in hippocampal development (EFNA5), ubiquination (MAGI2, CAND1), apoptosis (PRUNE2, CAND1), necrosis (ARSB), and dementia (MAGI2, ARBS) (Table 12). The involvement of TOMM40 in numerous brain regions of AD patients was confirmed by Shen et al [194]. This study used a novel whole brain set of ROIs from both VBM and FreeSurfer parcellation as QTs in a GWAS. Of the three SNPs additionally identified as significantly associated with brain volumetric changes, only one, proximal to the NXPH1 gene encoding neurexophilin (known to promote adhesion between dendrites and axons), had a bilateral pattern of association and was chosen for further study (Table 12). AD patients homozygous for the T allele at this locus displayed reduced GM most significantly in hallmark regions of AD atrophy, such as the hippocampus. This study illustrates the potential power of imaging genetics to identify novel candidate genes that warrant further investigation as AD candidates.
While Shen et al [194] used ROIs covering the brain, Stein et al [195] further extended the dimensionality of imaging genetics studies by carrying out a voxelwise GWAS, which explored associations between hundreds of thousands of SNPs and each of the nearly 32,000 voxels of the entire brain. Although no SNP was found significant at the stringent criteria used in the study, a number of SNPs of interest were identified in or near genes known to have functions relating to brain structure, such as monoamine uptake in neurons (CAPDS2), psychiatric illness (CSMD2 and CAPDS2), and neurite growth (SHB and ARP1) (Table 12). In a second GWAS of a targeted region of TBM-derived structural brain degeneration on MRI, Stein et al [196] identified an SNP located in the gene encoding N-methyl-d-aspartate receptor NR2B subunit (GRIN2B) that was significantly associated with lower volumes in the temporal lobe bilaterally. Risk alleles at this locus were more prevalent in AD patients of the AD cohort than in healthy elderly control subjects and were additionally associated with decreased MMSE scores (Table 12).
Furney et al [197] also used targeted imaging measures (entorhinal cortex thickness and volume, hippocampal volume, whole brain volume, and ventricular volume) as QTs in a large GWAS involving two cohorts (AddNeuroMed and ADNI). In addition to confirming a role of PICALM as a susceptibility gene for AD and as related to entorhinal thickness, they identified two other loci, ZNF292 and ARPP-21, as potential candidate genes based on associations of flanking SNPs with entorhinal cortex thickness and volume (Table 12). Kohannim et al [427] used the Lasso method of reducing dimensionality of multivariate GWAS to identify 22 genes associated with temporal lobe volumes in ADNI cohort. These included the previously identified GRIN2B and NRXN3 and a number of novel candidate genes. Of these, the greatest effect size was reported with MAC-ROD2 (macro-domain-containing 2), a gene expressed in the brain, which has been previously associated with schizophrenia and MRI-defined brain infarcts.
Most imaging GWAS reports have addressed baseline ADNI data; however, genetic variants predicting rate of progression are of great interest. Saykin et al [6] reported an initial longitudinal analysis of hippocampal volume and GM density using baseline and 12-month scans. In a candidate gene analysis [198], five AD genes from the AlzGene database (alzgene.org) were found to have significant SNPs associated with hippocampal volume or GM density changes, after accounting for APOE, baseline diagnosis, and other factors (NEDD9, SORL1, DAPK1, IL1B, and SORCS1). Next, a longitudinal GWAS was performed on hippocampal volume and GM density, using the MRI measures reported in the paper by Risacher et al [115]. A number of interesting potential candidate genes were identified by this GWAS. In addition to APOE and TOMM40, an SNP (rs12449237) located at 16q22.1 between CDH8 (cadherin 8, type II) and LOC390735 was strongly associated with change in hippocampal volume. CDH8 codes for a calcium-dependent cell adhesion protein related to synaptic integrity (neuronal adhesion and axonal growth and guidance). Although the cadherin protein has been implicated in AD and is known to interact with presenilin, this was the first indication that genetic variation in CDH8 may be associated with rate of neurodegenerative changes in the hippocampus. Several other markers did not reach genomewide significance but also showed association signals worthy of follow-up (for volume change: SLC6A13; for GM density change: MAD2L2, LOC728574, QPCT, and GRB2).
In a QT GWAS of CSF biomarker levels instead of imaging variables, Kim et al [149] examined levels of Aβ-42, t-tau, and p-tau181p and the ratios of p-tau181p/Aβ-42 and t-tau/Aβ-42 in the ADNI cohort. They found five SNPs that reached genomewide significance for associations with one or more biomarkers, including the known candidates (APOE and TOMM40) as well as one hypothetical gene (LOC10012950) that partially overlaps APOE. Most interestingly, several SNPs in the vicinity of the novel gene EPC2 (enhancer of polycomb homolog 2) were associated with t-tau levels. EPC2 is involved in chromatin remodeling and has not been previously associated with AD, yet this gene may be causally associated with mental retardation in a microdeletion syndrome. Along with EPC2, SNPs near CCDC134, ABCG2, SREBF2, and NFATC4 approached significance (P < 105) in their association with CSF biomarkers and can be considered potential candidate genes for future studies (Table 12). Han et al [199] also used levels of CSF biomarkers as QTs in a GWAS of the ADNI cohort. They found that increasing APOE ε4 allele dose was associated with lowered Aβ-42 and elevated t-tau and p-tau181p levels. After adjusting for age and APOE genotype, several SNPs were found to be significantly associated with increased Aβ-42 levels in normal subjects, the most strongly associated being within or proximal to the TOMM40, NCAM2, and CYP19A1 genes (Table 12). NCAM2 encodes neural adhesion molecule 2, a poorly characterized protein implicated in neuronal adhesion and fasciculation of neurons, whereas CYP19A1 encodes cytochrome P450 aromatase, an enzyme that catalyzes the conversion of androgens to estrogens.
Cruchaga et al [525] used t-tau and p-tau181 as quantitative phenotypes in a large GWAS (n = 1269 from four cohorts including ADNI) and identified three novel loci, one between GEMC1 and OSTN, one within GLIS4, and one within the TREM gene cluster. In an independent data set, the SNP between GEMC1 and OSTN was associated with CSF tau/ptau181 levels as well as tangle pathology and the rate of cognitive decline. The introduction of Florbetapir amyloid imaging in ADNI-GO and ADNI-2 has provided another quantitative phenotype for GWAS studies. Using these data, Ramanan et al [526] conducted a GWAS that identified, in addition to APOE, an SNP upstream of BCHE that was independently associated with Aβ levels. Together, the two loci accounted for 15% of the variance in cortical Aβ levels (APOE, 10.7% and BCHE, 4.3%). BCHE (butyrylcholinesterase) is an AD risk gene that has been found in senile plaques. Genetic variation at this locus may increase enzymatic activity, decreasing acetylcholine levels and disrupting synaptic functioning. This result is of particular interest as acetylcholinesterase inhibitors are currently first-line symptomatic therapies for AD.
In addition to risk for AD itself, age at onset (AAO) of the disease has an estimated heritability of 42%, some of it accounted for by APOE. Kamboh et al [307] conducted a GWAS of AAO data from 3 cohorts including ADNI to identify additional loci involved in AAO. They confirmed the involvement of APOE and neighboring loci (TOMM40 and APOC1) but no other SNPs reached significance. However, SNPs in 11 loci approached significance and as they lie in or near genes expressed in the brain, the authors suggested that they may be worthy candidates for further investigation.
GWAS appear to be a powerful tool for detecting associations between genes and phenotypes, but they are limited by the large sample sizes (typically thousands) required to gain sufficient statistical power to find these links and may also fail to detect connectivities between genetic loci by considering all SNPs separately. Imaging GWAS are also particularly extremely computationally intensive. In 2011–2012, various studies have focused on approaches to overcome these difficulties [305]. Schott et al [306] reported a method to increase the power of GWAS by defining cases and controls more accurately to reflect the presence of AD pathology rather than on the basis of clinical diagnosis. To this end, they divided the ADNI cohort into CSF positive and CSF negative groups based on previously established cut-points for Aβ42 and ptau181 and examined minor allele frequencies for 7 SNPs in previously identified AD risk genes. They found significant associations between the CSF negative group and SNPs in CR1, PICALM, TOMM40 and APOE using only slightly more than 300 subjects, an order of magnitude fewer than generally required to detect associations in GWAS. An alternative approach to reducing sample sizes and to leveraging information from potentially linked genes, was taken by Swaminathan et al [307] who used SNPs in 15 amyloid pathway associated genes and PiB uptake in 4 regions affected by AD to study genetic associations in 103 ADNI AD patients. This approach identified a minor allele (A) of an SNP in the DHCR24 gene that confers a protective effect and in a subsequent whole brain analysis, they found they found a higher mean PiB uptake for the major allele in frontal regions. Hu et al [308] also used a pathway approach, investigating multiple SNPs in canonical AD pathways, and identified SNPs in the Gleevec pathway, a cancer drug shown to modulate APP cleavage by γ-secretase, as being involved in AD. This targeted pathway-based approach may be more effective in identifying genes involved in AD pathology than traditional GWAS. The issue of reducing dimensionality was tackled by Hibar et al [305], who proposed that condensing the number of SNPs (around 400,000) to genes (slightly over 18,000) would avoid having to restrict phenotypes to a priori defined ROIs to enable a practical computational burden. They used principal components regression to test for gene association at each voxel and identify the most significant gene on a voxel basis. Although no genes identified remained significant after correction for multiple comparisons, many top genes, including GAB2, an established AD risk gene, had been previously identified as being associated with brain diseases, suggesting that this multivariate gene-based approach holds promise for future investigations.
The vGeneWAS approach of Hibar et al [527] refined the “brute force” approach of voxelwise GWAS by operating at the gene level. In 2012–2013, the GWAS SNP approach has been further refined to reduce computational burden and the high rate of false positives in the search for the full complement of genetic susceptibility genes for AD. Pathway and network analyses have been used to narrow the selection of variants to be tested to a biologically relevant pathway. Thus, a biochemical understanding of pathologic mechanisms in combination with genetic approaches may together discover functional relationships that cannot be seen at the level of individual SNPs or genes. Similarly, pathways of interest have been selected that constrain the search space for such computationally demanding approaches as gene-gene or SNP-SNP interactions. A summary of brain-genome association strategies is shown in Fig. 45. Interaction analyses have been used to help identify epistatic relationships that may explain some of the “missing heritability” of AD. Next generation sequencing (whole genome or whole exome sequencing) and the combined use of proteomics data and genetics data represent further significant developments.
Fig. 45.
Genetic approaches used with ADNI data. From Shen et al [521]. Abbreviation: ROI, region of interest.
Several studies have used different quantitative traits to target a particular set of biochemical pathways putatively involved in AD pathogenesis. Koran et al [528] examined interactions of SNPs within genes identified as being on the AD pathway by the Kyoto Encyclopedia of Genes and Genomes (KEGG). Using a discovery set and two independent validation data sets from the ADNI cohort and amyloid deposition measured by Florbetapir uptake as a variable phenotype, they identified an interaction between the RYR3 and CACNA1C genes in which a minor allele in genes corresponded to a higher amyloid load than one or no minor alleles. Both genes are involved in calcium homeostasis, a process that is recognized as important in amyloid formation and deposition. RYR3 encodes ryanodine receptor-3, which regulates intracellular calcium homeostasis, while CACNA1C encodes a calcium channel subunit. Together, they accounted for between 4% and 9% of variance in amyloid load in the three data sets. In a similar approach, Meda et al [529] used 12-month hippocampal and entorhinal cortex atrophy rates as quantitative traits and used a priori knowledge to target biological pathways known to be associated with atrophy in these regions. They identified 109 SNP-SNP interactions in 78 genes that were significantly associated with right hippocampal atrophy and 125 SNP-SNP interactions in 102 genes significantly associated with right entorhinal cortex atrophy (Fig. 46). These were located in three KEGG pathways for hippocampal interactions including calcium signaling, axon guidance, and ErbB signaling, and 14 pathways for entorhinal cortex atrophy including calcium signaling, axon guidance, long-term depression and potentiation, and neuroactive ligand-receptor interaction. The analysis confirmed the involvement of some genes as well as identifying novel candidate genes for further investigation and suggested that different biogenetic mechanisms may mediate atrophy in different brain regions. A psychometrically determined memory score was used as a phenotype to enrich pathways involved in memory impairment in a study by Ramanan et al [428]. Using GWA data from ADNI participants, they enriched 27 pathways that included not only processes well understood to be involved in memory such as long-term potentiation and neurotransmitter receptor-mediated calcium signaling but also pathways involved in cell adhesion and differentiation. Expression in some of these identified pathways was coordinated; a large gene set was regulated by the SP1 transcription factor (Fig. 47). The newly identified enriched pathways may provide targets future studies of memory impairment. These studies demonstrate that a pathway-based approach to analyzing GWA data has great promise in untangling the relationships between genes in these complex phenotypes.
Fig. 46.
Gene-gene interaction networks for (A) entorhinal atrophy and (B) hippocampal atrophy. Each circle is a gene that participated in a significant SNP-SNP interaction model. Circles colored orange are genes previously identified as a possible AD risk gene. From Meda et al [529].
Fig. 47.
Involvement of genes of interest identified from pathways enriched in memory impairment in a transcriptional regulation network centered on SP1. From Ramanan et al [471].
Recent technological advances in proteomics have made it feasible to use protein analytes as diagnostic, prognostic, or treatment biomarkers for disease. The identification of important functional genetic variants that affect levels of protein analytes known to be associated with disease is an important step in the development of these biomarkers. Thus, Kim et al [530] integrated ADNI GWAS array data with baseline multiplex panel proteomics data to investigate the effects of SNPs within genes on the corresponding plasma protein level for 140 gene-protein association pairs. They detected 112 significant associations within this cohort, of which 50 were replicated in an independent cohort. The top replicated associations included two SNPs in CHFH1 (complement factor H-related protein 1), along with gene-protein associations for interleukin-6 receptor, pulmonary and activation-regulated chemokine, chemokine CC-4, and apolipoprotein A4. Each SNP accounted for between 14% and 16% of the total variation in plasma protein levels, emphasizing the large role of genetic variation in proteomics.
An alternative to a pathway approach was reported by Benitez et al [531] who restricted their search for new risk loci to the known risk genes, APP, PSEN1, PSEN2, GRN, APOE, and MAPT. Using extreme levels of CSF Aβ, t-tau, and p-tau as quantitative traits, they confirmed one known pathogenic mutation in PSEN1 (p.A426P) and found a number of high-risk and novel variants in these genes. A new PSEN1 variant, p.E318G, was associated with high levels of CSF t-tau and p-tau and carriers of this variant who were also carriers of APOE ε 4 had a twofold increased risk of AD over APOE ε 4 carriers alone. Moreover, in APOE ε 4 carriers, the p.E318G variant was associated with more Aβ plaques and faster cognitive decline. These results suggest that p.E314G in PSEN1 interacts with the APOE ε 4 allele to raise the risk of AD via increased amyloid deposition.
Although GWAS have been successful in identifying novel risk variants for AD, the observed effects of these are relatively small and substantial heritability has yet to be accounted for. One explanation for this is that the genetic makeup of AD is complex and involves epistatic relationships that go beyond single genes. Hohman et al [532] examined epistatic relationships between four top candidates SNPs: PICALM, BIN1, CR1, and CLU, using amyloid burden measured by Florbetapir uptake as a quantitative endophenotype. They reported a novel interaction between PICALM and BIN1 in that the minor allele of BIN1 was associated with higher levels of amyloid deposition but only in noncarriers of the protective minor allele of PICALM. The interaction suggests that variation in the genes may together modulate amyloid deposition by as yet unknown mechanisms. Glycogen synthase kinase 3 (GSK-3) has been posited to regulate both tau-phosphorylation and Aβ production, and overactivity of this kinase influences cognitive impairment, neuroinflammatory response, and the pathologic cascade of AD. Hohman et al [533] again searched for epistatic relationships, this time between GSK-3 and kinases involved in Aβ pathology using Florbetapir binding as a quantitative trait. They found three interactions involving a GSK-3β SNP and SNPs within APP and APBB2, each accounting for over 1% of the variance in amyloid deposition. GSK3β may therefore modify risk for amyloid deposition and increase amyloid burden together with APP-related genes. Clearly, the examination of epistatic relationships between carefully selected gene pairs is a promising approach that leverages both genetic and biochemical knowledge to fit more pieces into the complex puzzles of AD pathology and genetics.
Brain phenotypic measures are only partially explained by genetic variation as represented by SNPs identified in GWAS studies. Bryant et al [534] mapped the proportion of phenotypic variance of multiple regions throughout the brain that are caused by genetic variation using genome-wide complex trait analysis. They reported that 85% variability in intracranial volume and 57% of the variability in cerebrospinal fluid volume was explained by considering the complete set of SNPs (512,905). In contrast, genetic variability for white and gray matter was close to zero. Within regional volumes, high genetic variability was found in several medial cortical regions, subcortical nuclei, and perceptual cortical pathways including the entorhinal cortex, caudate, and insula.
Next generation sequencing of either the whole genome or, more feasibly, the whole exome, is a logical next step in genetic analysis. Nho et al [535] reported the first application of whole exome sequencing to identification of risk alleles for LOAD. They selected MCI participants who were also APOE ε 3/ ε 3 homozygotes with an extreme change in hippocampal volume over 2 years. Two single nucleotide variants in CARD10 and PARP1 accounted for the greatest group difference. Further quantitative trait analysis in the remaining ADNI ε 3/ ε 3 group revealed that genetic variation in both CARD10 and PARP1 was associated with greater hippocampal atrophy. PARP1 was further found to be associated with baseline hippocampal volume in a meta-analysis of ε 3/ ε 3 subjects from five studies. CARD10 (caspase recruitment domain family, member 10) had not been previously associated with AD but is known to activate NFκB which is activated in the disease. PARP1 (poly (ADP-ribose) polymerase-1) is involved in many cellular genetic processes, expressed in many regions of the brain, and has important role in long-term memory formation.
Shen et al (2012) [521] analyzed ADNI genetic associations using pathway and network enrichment and identified a number of pathways involved in cell adhesion, neurophysiological processes, immune response, and development in addition to neurogenesis, synaptic contact, angiogenesis, cell adhesion, and synaptogenesis process networks. This work highlighted pathways already identified in AD pathogenesis and also identified novel targets for future investigation.
5.4. Replication studies and meta-analyses
In 2011–2012, genetic data from the ADNI cohort have been used in a number of studies both confirming candidate AD risk genes, by attempting to replicate results in different cohorts, or by conducting meta-analyses of previously published work, and providing more detailed mapping of candidate genes. An independent confirmation of the involvement of CR1 in AD was reported by Antunez et al [309], who found a trend supporting association in a Spanish cohort of approximately 3500 and a stronger association in a meta-analysis of over 30,000 individuals. Further confirmation for CR1 as an AD risk gene came from Hu et al [308] who conducted a GWAS on combined cohorts including ADNI. They also replicated the BIN locus by testing top SNPs from the GWAS in an independent cohort, and used haplotype conditional analysis to show that multiple variants at the BIN locus had conditionally independent associations with AD. PICALM variants were also replicated, but their association with AD was attenuated by APOE status. Cruchaga et al [310] focused on replicating the association between APOE3-TOMM40 haplotypes and AD as well as age of onset of the disease. They found it difficult to identify the genetic variant driving the association of the genes because of extensive linkage disequilibrium around TOMM40 and APOE and possibly an insufficient sample size. Consequently, they were not able to replicate results, identifying instead a polymorphism of TOMM40 associated with decreased risk of AD. An additional study by Antunez et al [311] independently identified the MSA4A gene cluster as being associated with AD after a meta-analysis of 4 public GWAS sets including ADNI, and a new Spanish cohort. This gene cluster was previously identified by Naj et al [186], and the use of a combined total of over 10,000 cases and over 14,000 controls in this study underscores the importance of combining cohorts to increase power to detect genetic associations that may have small effect sizes. Kauwe et al [312] investigated whether common variants of BIN1, CLU, CR1 and PICALM were associated with Aβ42 and p-tau181. No associations between these SNPs and CSF biomarkers were found in two cohorts including ADNI, suggesting that these candidate genes may affect risk for AD via other mechanisms than a direct effect on AD pathology. CSF biomarkers were also used as a QT in a study by Alexopoulos et al [313], who investigated the association between SORL1 (neuronal sortilin-related receptor with A-type repeats), likely involved in sorting of APP in the Golgi, and levels of Aβ42, ptau181 and t-tau. They found that Aβ42 was significantly associated with the A allele for SORL1 SNP233 in the AD group and marginally associated with Tallele of SNP24. Levels of some SNPs in SORL1 were modulated by the APOE ε 4 allele.
ADNI data continue to play a vital role as a subset of meta-analyses of GWAS results which have been necessary to gain sufficient statistical power to identify risk variants. For example, Rhinn et al [536] investigated regulatory mechanisms affecting AD and AD risk using differential co-expression analysis and identified candidate genes predicted to mediate transcriptional changes in APOE ε 4 carriers including modifiers of APP processing and endocytic trafficking. A meta-analysis of GWAS including ADNI data was then used to confirm that common genetic variants in two genes of interest, FYN and RNF219, affected amyloid deposition and age of disease onset in APOE ε 4 carriers. Likewise, five of nine previously identified AD risk loci (PICALM, BIN1, ABAC7, MS4A4/MS4A6E, and EPHA1) were confirmed in a GWAS by Kamboh et al [537] using a University of Pittsburgh cohort. When the top 1% most significant SNPs from this GWAS were then analyzed in a meta-analysis including the ADNI cohort, the authors found that the most significant SNP, located in PPP1R3B (protein phosphatase 1, regulatory subunit 1B), carried an OR for AD risk of 2.43. A replication study by Peterson et al [538] confirmed the association of another protein phosphatase gene, PPP3R1, as well as MAPT with CSF tau levels. The high-risk allele in MAPT was also associated with a 30% faster change in CDR-SB scores, and patients with the high-risk alleles at both loci progressed to AD six times faster than those with the low-risk alleles. A meta-analysis of five GWAS studies and subsequent replication of results in an independent sample by Martinez-Murcia et al [398] identified a marker in the NRXN3 gene with a consistent protective effect in men. These results support a role for neurexins, synaptic cell adhesion molecules processed by presenilin, in LOAD.
5.5. Genomic copy number analysis
One method of genetic analysis not extensively used in the field of AD research is that of copy number variation analysis. Copy number variants (CNVs) are sequence alterations involving differences in gene copy numbers usually cause by deletions or duplications of genomic sequences. Swaminathan et al [314] used this technique to analyze the ADNI cohort and compared CNV calls generated in AD and MCI cases to those in controls using whole genome and candidate gene association approaches. While no excess CNV burden was observed in cases versus controls, a number of genes already implicated in AD were identified (CHRFAM7A, NRXN1), in addition to some novel loci (CSMD1, HNRNPCL1, SLC35F2, ERBB4) (Table 12). Of these candidate genes, three (CHRFAM7A, NRXN1, ERBB4) were replicated in an analysis of a different cohort by the same group [315]. This analysis also identified other loci previously identified as possible AD candidate genes (ATXN1, HLA-DPB1, RELN, DOPEY2, GSTT1) in addition to a novel candidate gene, IMMP2L which codes for a mitochondrial enzyme and may play a role in AD susceptibility through influencing oxidative damage (Table 12). A follow-up paper by the same group [539] confirmed in a separate cohort that the CHRFAM7A, RELN, and DOPEY2 genes were associated with AD and identified a novel gene, HLA-DRA (major histocompatibility complex, class II DR alpha). A subsequent meta-analysis that included the ADNI cohort found that the CHRFAM7A gene was significantly associated with MCI/AD risk (OR = 3.986; 95% CI: 1.490–10.667). The gene has a putative function in synaptic transmission and cholinergic anti-inflammatory response.
Guffanti et al [540] used intensity variation in SNP microarrays to study differences in CNVs between control and AD/MCI patients and identified a number of CNV regions that included heterozygous deletions over-represented in MCI and AD patients. Genome resequencing identified genes putatively affected by these deletions, and functional pathway analysis revealed that these genes were involved in processes such as cell-cell adhesion, axon guidance, differentiation, and neuronal morphogenesis. The authors hypothesized that, although rare, these CNV regions may confer an increased susceptibility to cognitive decline by acting in combination with additional genetic or epigenetic mechanisms.
5.6. Other genetic studies using ADNI data
Like other fields discussed in this review, studies have recently emerged that utilize ADNI genetic and/or imaging data for uses not directly related to AD research. Stein et al [316] conducted a GWAS investigating genetic influences in caudate volume, a structure involved in many disorders including depression and schizophrenia as well as in AD. While no SNPs reached genome-wide significance, loci involved in dopaminergic neuron development and with links to schizophrenia were identified suggesting that MRI phenotypes may be powerful phenotypes when searching for genetic associations. The ADNI cohort was also used in 2 GWAS, one identifying SNPs associated with variability in the surface of the visual cortex [317] and the other determining that circadian clock SNPs are not associated with the breakdown of sleep-wake consolidation observed in AD [318]. Hibar et al [541] used ADNI genetic data to perform a GWAS to discover common genetic variants associated with lentiform nucleus volume, implicated in disorders such as Parkinson’s disease, ADHD, and schizophrenia. They identified variants within the flavin-containing monooxygenase gene cluster.
5.7. Summary and conclusions of genetic risk factor studies
Genetic studies of the ADNI cohort have confirmed that the APOE ε4 allele is the major genetic risk factor for late-onset AD and that it is associated with atrophy in widespread areas of the brain. Case–control GWAS that have included ADNI data have also confirmed CLU, CRI, and PICALM as AD risk loci and identified a number of other candidate genes. QT GWAS using ADNI phenotypes such as Aβ-42 and tau or imaging measures of brain atrophy have detected genes implicated in the modification or modulation of Aβ or tau proteins, mitochondrial oxidative pathways, iron metabolism, neural adhesion and growth, synaptic plasticity, epigenetic processes, and memory function. A particular contribution of ADNI imaging genetic studies has been to develop methods to expand the dimensionality of GWAS studies to include all regions or voxels of an imaging scan, significantly expanding the potential of the field of imaging genetics to pinpoint specific brain regions influenced by different loci. Although candidate genes await confirmation by independent studies, they promise to unveil biological mechanisms underlying AD pathology.
Publications of genetic findings using ADNI data have continued to increase in 2011 and 2012. From only 1 paper published in 2009 [184] and 19 in 2010 [6,103,128,141,146,184,188,189,191–193,303,194–196,199,208–210], 2011 saw 20 new publications [105,139,149,186,187,194,197,305,308–311,314,316,318–323] and the first three-quarters of 2012 saw 32 new publications [262–264,302,304,306,307,315,317,324–346]. This significant expansion in number has been matched by an equally impressive expansion in scope. While new candidate risk loci continue to be reported, the focus of many studies has been to replicate previous work, sometimes using meta-analysis of combined cohorts, to independently confirm candidate genes. These studies have demonstrated that the increased power resulting from the larger sample sizes is critical to success in this endeavor. Other approaches to increasing power to identify candidate genetic loci have been reported, such as targeting SNPs in selected pathways rather than using a genome-wide approach or using genes instead of SNPs in a genome wide search, and defining controls and cases on the basis of pathological rather than clinical criteria. The analysis of copy number variations in AD has been reported and appears to be an important additional tool for untangling the contributions of AD susceptibility loci to the disease. Finally, ADNI genetics data have been used in fields outside of Alzheimer’s research, demonstrating a pleasing contribution of the project to the greater scientific community. The ultimate goal of genetics research in AD is to identify novel candidates as targets for the development of disease-modifying agents. ADNI genetics data have now been used in the identification of 13 novel AD susceptibility genes in addition to APOE, 10 of which are not in linkage disequilibrium with APOE: BIN1, CD2AP, CLU, CR1, EPHA1, FTO, GRIN2B, MAGI2, MS4A4A, and PICALM. In 2012–2013, rapid progress toward this goal has been made notably by the use of more targeted approaches that reflect an increasing understanding of the biochemistry of AD. The selection of suspected pathologic pathways or a particular set of genes has helped to narrow the search for risk alleles. Interaction studies of epistatic relationships have revealed additional heritability factors, and the first studies using ADNI whole exome sequencing and proteomics data have been reported. Through these and other approaches, ADNI has continued to make substantial contributions to untangling the genetic basis of AD and has thereby deepened our understanding of the biological pathways involved in pathogenesis.
6. Studies of normal control subjects
With the realization that AD pathology most likely begins to accumulate years in advance of any detectable cognitive effect, a major issue has been determining the proportion of apparently normal control subjects who harbor preclinical AD. As more sensitive biomarkers have been developed, studies have emerged with the goals of ascertaining the utility of these biomarkers in healthy elderly subjects and determining the earliest stage at which incipient AD pathology can be detected. This clearly has implications for development of AD therapies: if AD pathology can be reliably detected at such an early stage, then would existing or novel AD-modifying treatments be more effective when used before clinical symptoms become evident? In tandem with these studies, ADNI’s cohort of well-characterized normal control subjects has been used to investigate processes occurring in the brain during healthy aging when there are no clinically detectable underlying pathologies. These two thrusts are often interwoven within the same study, as it becomes more obvious that healthy elderly subjects, although cognitively normal, are in fact a heterogeneous group when examined by other means.
6.1. MRI studies
The effect of developmental brain changes on neurocognitive late-life functions was investigated by Tamnes et al [542] who compared longitudinal volume changes in a developing cohort (8–22 years) with cognitively normal ADNI participants. Developmental reductions in GM volume proceeded in a generally posterior to anterior gradient and were generally greater than the cortex then subcortical structures. Late developing cortices were more vulnerable to atrophy in aging, with the exception of the medial temporal lobe. This study provides new insights into the relationships between brain changes during development and in normal aging. Insights into normal aging and into strategies for preventing age- or disease-related cognitive decline were gained by Harrison et al [543] who studied a group of “superagers”: people over 80 years who perform at a level equivalent to 20 to 30 years younger in tests of episodic memory and at age level for other cognitive tests. Cortical volume and thickness in super-agers with superior memory function were greater than those of their age-matched peers and equivalent to those of middle-aged controls. This is contrary to the dictum of normal cognitive aging involving slow global atrophy, and future studies may identify other factors that may contribute to unusually successful cognitive aging.
The question of whether atrophy observed in normal aging is due primarily to normal aging processes or to the development of underlying pathologies is the subject of much debate. Fjell et al [200] presented the first detailed longitudinal study of brain atrophy in healthy elderly subjects aimed at understanding age-related changes in cognitive function. When volume changes in multiple ROIs and across the entire cortex were compared in healthy elderly subjects and AD patients, these authors found that the healthy elderly subjects had an atrophy rate of about 0.5% per year and that volume loss was widely distributed across the brain and included both regions typical of AD-associated atrophy and areas not typically associated with AD, such as the inferior, superior, and middle frontal cortices. The rate of change accelerated with age, especially in those regions associated with AD, possibly because of the existence of preclinical AD pathology superimposed on normal aging processes. The authors believe, however, that the majority of volumetric changes observed in healthy aging are not related to those caused by degenerative diseases. Davatzikos et al [119] used the SPARE-AD index (see section 4.4.2.1. for further description) to examine the degree of AD pathology in healthy elderly subjects and its association with cognitive decline in ADNI and another cohort with longitudinal data available. They found that SPARE-AD scores increased with age, as did the rate of change of the SPARE-AD score. When healthy elderly subjects were divided into groups of high versus low SPARE-AD score, the majority had negative scores. However, a small group with positive scores had significantly lower MMSE scores at baseline, suggesting that a subset of cognitively normal elderly subjects harbored underlying AD preclinical pathology.
In response to a paper by Burgmans et al [201] suggesting that underlying preclinical disorders may lead to the overestimation of GM atrophy in normal aging studies, Fjell et al [202] conducted a meta-analysis of a number of cross-sectional studies. They found that atrophy correlated with age in virtually all ROIs studied, even at younger ages, suggesting a linear trajectory of brain atrophy over time. When 2-year follow-up cognitive data of healthy elderly subjects from the ADNI cohort were used to exclude participants with any indication of cognitive decline, significant atrophy in all ROIs was still found in the remaining “super-stable” cohort. These results support the view that brain atrophy is part of normal aging and not necessarily caused by underlying neuropathological processes. To detect unusually fast atrophy in cognitively normal healthy elderly subjects, Franke et al [92] developed a model of healthy aging by estimating age from MRI scans of normal brain anatomy. Their method (described in more detail in section 3.7) accurately estimated the age of healthy subjects (r = 0.92 between real and calculated ages). Using the same method, they also estimated ages of patients with early AD and found that the predicted ages were an average of 10 years higher than the actual ages, implying that the pattern of AD atrophy does accelerate relative to healthy elderly control subjects.
Murphy et al [203] used an automated method to examine volume changes in 14 cortical and subcortical regions over 6 months in an effort to determine whether atrophy was detectable over the short period in healthy elderly subjects and whether this atrophy was related to 2-year declines in memory-specific neuropsychological tests. They found that volume changes in these regions could be measured and that they were predictive of future clinical decline. The most significant associations were found in the MTL, suggesting that this atrophy could represent the earliest stages of AD and that MRI may be a useful tool in complementing neuropsychological tests in the early detection of those at risk for subsequent cognitive decline.
Furthermore, cognitively normal individuals who were amyloid positive had greater thinning of the medial portion of the orbital frontal cortex than amyloid negative patients, and those who were tau-positive were distinguished from tau-negative individuals by greater thinning of the entorhinal cortex. These results suggest that in asymptomatic individuals, Aβ and tau pathology affects GM thinning in select neocortical regions that potentially influence hippocampal atrophy at a later stage of the disease [163].
6.2. Studies of CSF biomarkers and amyloid deposition (11C-PiB PET)
In a manner similar to the examination of MRI markers of AD pathology, there has been interest in assessing the utility of CSF biomarkers in healthy elderly subjects on the basis that an “earlier biomarker horizon” [204] would have great clinical significance. Nettiksimmons et al [204] examined healthy elderly subjects in the ADNI cohort and found three clusters of participants when 11 biomarker and imaging measures were subjected to unsupervised cluster analysis. The first, compact cluster had the most “normal” CSF and MRI measures, whereas the measures of the third, more dispersed group more closely resembled those of MCI patients included in the study for comparison (the second cluster was placed in an intermediate position). The third cluster had a significantly higher proportion of APOE ε4 carriers and scored worse on tests of cognition (ADAS-cog, AVLT), suggesting that this group may harbor the earliest manifestations of AD symptoms. These results provide support for the notion that cognitively normal elderly subjects are in fact a heterogeneous group, a portion of which may progress to MCI in the future. The second cluster of normal controls found by Nettiksimmons et al [204] lacked the CSF biomarker signature for AD but had atrophy in multiple brain regions, approaching levels observed in MCI patients. A further study by the same group [544] examined whether vascular damage could account for these differences. They found that this subgroup had a higher mean frequency of WMHs in the periventricular area and higher BMI, triglycerides, blood glucose, and Haschinki scores. There was no difference in the APOE ε 4 allele frequency of this cluster compared with the first “normal” cluster, but it had a worse trajectory in longitudinal cognitive tests (AVLT, FAQ). The results support the involvement of a vascular component in cerebral atrophy observed in a subset of normal controls.
In a study of the relationship between levels of CSF biomarkers and 1-year atrophy in 15 subcortical and 33 cortical ROIs in healthy elderly subjects, Fjell et al [205] reached similar conclusions. They found that levels of CSF biomarkers, especially Aβ-42, correlated with atrophy in many of the regions tested and that atrophy was not restricted to regions most typically associated with AD. When Aβ-42 concentration was plotted against the percentage of annual change in ROIs, there was an inflection point at approximately 175 pg/mL, below which participants had larger brain volume changes over a year, suggesting that Aβ-42 may play a role in changes in brain volume observed in healthy elderly subjects below a certain threshold level. De Meyer et al [159] found that when a biomarker “signature” for AD using levels of Aβ-42, t-tau, and p-tau181p was tested in healthy elderly subjects, there was a bimodal distribution of Aβ-42 levels with a separation point at 188 pg/mL. Although it was unknown whether those participants with low levels of Aβ-42 in these two studies would develop AD pathology, they once again highlighted the heterogeneity of the cognitively normal healthy elderly group.
In the current model of AD pathogenesis, it is well established that deposition of amyloid plaques is an early event that, in conjunction with subsequent tau pathology, causes neuronal damage typically beginning in the hippocampus and resulting in the first clinical manifestations of the disease in the form of episodic memory deficits. Mormino et al [206] investigated the relationship between Aβ deposition, as measured by 11C-PiB PET uptake, hippocampal atrophy, and episodic memory loss in cognitively normal healthy elderly subjects. They found an inverse relationship between 11C-PiB uptake and hippocampal volume and that episodic memory loss was predicted by hippocampal volume, but not by 11C-PiB uptake. The results suggest that low levels of CSF Aβ-42 (high levels of brain accumulation) in healthy elderly subjects may reflect early stages of AD pathogenesis and may subsequently mediate dementia through an effect on hippocampal volume and the resulting declines in episodic memory. These findings warrant further investigation. Aβ deposition may require abnormal p-tau181 to induce neuronal and synaptic damage. Desikan et al [545] found that CSF Aβ-42 was significantly associated with longitudinal change in cognition (CDR-SB, ADAS-cog) only in the presence of elevated p-tau181 in cognitively normal subjects. There was no significant association between abnormal levels of Aβ-42 alone and cognitive decline over 3 years. Results suggest that early intervention trials should take into account both increased p-tau181 and decreased Aβ-42 as individuals with this profile are likely to have a different rate of clinical progression from that of individuals with decreased Aβ-42 alone.
6.3. Genetic studies of normal control subjects
Although the APOE ε4 allele has been clearly identified as an AD risk allele, the question of whether a second variant in the APOE gene, the ε2 allele, confers a protective effect has been less well studied. Evidence for the protective effect of the APOE ε2 allele came from a study by Hua et al [120], who found reduced CSF volume in the ventricular system of healthy elderly subjects who had the highest frequency of this allele compared with MCI and AD patients. Chiang et al [207] sought to determine the effect of APOE ε2 allele on hippocampal volume and levels of CSF biomarkers in healthy elderly subjects. They found that carriers of the APOE ε2 genotype, constituting approximately 5%of the population, had lower rates of hippocampal atrophy and higher Aβ-42 and lower t-tau and p-tau181p levels compared with the more common (~70% of population) APOE ε3/ε3 homozygotes, suggesting that lower rates of atrophy could be related to decreased underlying AD pathology and may explain the lower rates of AD among carriers of this allele. A similar finding was reported by Fan et al [208], who examined the relationship between cortical thickness at multiple regions across the brain and APOE genotype in healthy elderly subjects who were grouped as ε2 carriers, ε3 homozygotes, and ε4 carriers. After adjusting for multiple comparisons, they found greater thickness in the superior temporal cortex in ε2 carriers compared with ε3 homozygotes, and in the dorsolateral prefrontal cortex in ε2 compared with ε4 carriers. Moreover, CSF concentrations of Aβ-42, t-tau, and p-tau181p were significantly different in all groups (Fig. 24), although no differences were found in the MMSE between groups. The results of these two studies provided support for the differential effect of APOE alleles on brain structure and on CSF biomarkers.
Fig. 24.
Mean biomarker levels (t-tau, p-tau, and Aβ-42) for the APOE genotype groups. The APOE ε2 carriers are represented in black, the ε3 homozygotes in gray, and the ε4 carriers in white. The CSF Aβ-42 levels show a significant stepwise trend downward, from APOE ε2 carriers to ε3 homozygotes to ε4 carriers, whereas the t-tau and the p-tau levels show the opposite trend. Reproduced with permission from Ref [208].
In addition to risk factors like age and APOE genotype, increased BMI has been associated with frontal, temporal, and subcortical atrophy and may increase susceptibility to AD. Recent studies identified a novel obesity genetic risk factor, a variant of the fat mass and obesity associated (FTO) gene, carried by almost one-half of Western Europeans. Ho et al [209] examined the effect of the FTO risk allele on brain volumes in healthy elderly subjects and compared its effects on brain structure with that of increased BMI. They found that carriers of the FTO risk allele had an 8% to 12% deficit in a subset of areas affected by BMI, predominantly in the frontal and occipital lobes, compared with noncarriers, suggesting that the FTO risk allele contributes to, but does not fully account for, the effect of increasing BMI on brain atrophy. Bertam and Heekeren [198] discussed the findings of the study and the need for corroborating the results to determine the influence of genetics on normal brain structure and function.
The idea that common variance in brain structure may be primarily controlled not by polymorphisms resulting in altered protein structure, but by changes in regulatory elements found support in a study by Rimol et al [210]. Using the ADNI cohort, they found that two SNPs located in nonexonic regions of genes for primary microencephaly were correlated with reduced cortical surface in males only, regardless of disease status, and suggested that these polymorphisms may affect gene regulation and result in gross abnormalities in brain structure observed in this disease. More data on the role of common genetic sequence variations in accounting for commonly occurring brain structure variations came from a study by the same group [211] on associations between a common haplotype of the MECP2 gene and brain structure. Mutations in MECP2, encoding methyl-CpG binding protein 2, cause microencephalopathy and are associated with other severe neurodevelopmental disorders, but Joyner et al [211] found that common sequence variations in this region correlated with reduced cortical surface area in males only of the ADNI cohort. As MECP2 is thought to transcriptionally activate or repress thousands of genes, studies of the influence of such common sequence variations may reveal profound insights into brain structure and development.
Hypothesizing that multiple brain pathologies may share common pathways such as inflammation, protein misfolding and mitochondrial dynamics, De Jager et al [326] searched for genetic variants affected the rate of age-related cognitive decline. In addition to identifying the APOE locus, they found an SNP close to PDE7A and MTFR1, genes potentially involved in inflammation and oxidative injury, respectively.
6.4. Summary and conclusions of papers focusing on normal control subjects
Heterogeneity of cognitively normal healthy elderly subjects seems to be well supported by these studies, with a number suggesting the existence of a subset of cognitively normal elderly subjects that bears the hallmarks of early AD pathogenesis in terms of changes in brain volume and levels of CSF biomarkers. The extent to which these changes are separate from those of normal aging remains to be fully elucidated. Fjell et al [202] concluded, “We need more knowledge about which factors mediate brain atrophy in healthy elderly and what consequences the changes have for cognitive function.” Likewise, several intriguing studies have pointed to the role of genetics in healthy aging, and suggest a protective effect of the APOE ε2 allele and increased susceptibility to brain atrophy and perhaps AD conferred by a risk allele at the novel FTO locus. Clearly, studies of the healthy elderly control subjects are revealing information not only about the processes of healthy aging but also the initial development of preclinical AD pathology. In 2011–2012 there has been a further shift toward considering cognitively normal elders as a heterogeneous population, some of whom harbor the earliest pathological manifestations of AD and are therefore part of the disease continuum. Many papers studying this group have therefore been included in other more relevant sections of this review.
7. Worldwide ADNI
Since the inception of ADNI in North America in 2004, there has been worldwide interest in creating programs that are at least partially modeled on the ADNI platform, and that use protocols developed by ADNI for at least part of their studies. Combined, the initiatives represent a concerted effort toward globalization of this concept. Society may well reap the rewards of having not just a well-characterized North American cohort for the development of AD biomarkers but also similarly characterized cohorts globally that may represent diverse ethnic groups, important for determining the applicability of ADNI findings to the world population. Like ADNI, these initiatives from Europe, Japan, and Australia are predicated on the sharing of data, and infrastructure is beginning to be developed to allow full transparency of global results. Future ADNIs are expected to begin in Argentina and China and have recently begun in Korea and Taiwan. All worldwide ADNIs share common goals of increasing understanding of AD onset and progression, both cognitively and physically, establishing globally recognized standards for diagnosis, and ultimately developing methods to allow more efficient clinical trials.
7.1. European ADNI
Frisoni [212] provides an overview of all programs, either completed or underway, in Europe that are in some way related to ADNI. The ADNI platform was first introduced into Europe in the form of a small cross-sectional pilot study, E-ADNI, which aimed to assess the feasibility of importing ADNI procedures to a European multicenter multicountry setting [213]. E-ADNI was initiated under the auspices of the Alzheimer’s Association through the generosity of the HEDCO Foundation and enrolled 49 control, MCI, and AD participants over seven sites in seven countries. The pilot study used all ADNI protocols, with the exception of PET imaging, the feasibility of which had been previously demonstrated, and MRI sequences for the detection of cerebral small vessel damage, a slightly different emphasis of the study. Buerger et al [214] conducted a multicenter feasibility study within E-ADNI and found that the use of fresh, rather than frozen, biological samples increased diagnostic accuracy. Overall, the study demonstrated that apart from age and education, the enrolled cohort was similar to the ADNI cohort in MRI and CSF measures and that implementation of the ADNI platform in Europe was feasible [213].
Other data collection programs in Europe include (1) AddNeuroMed, a public–private initiative with a cohort of 700 control, MCI, and AD subjects across Europe that used ADNI protocols for structural MRI; (2) Pharma-cog, which overlaps to the greatest extent with ADNI and which aims to predict cognitive properties of new drug candidates for neurodegenerative diseases; (3) Swedish ADNI, a small-scale initiative funded by the Alzheimer’s Association that used ADNI protocol and which has merged into the larger Swedish BrainPower initiative; and (4) Italian ADNI, a larger project with 480 patients enrolled. These initiatives vary in the size and composition of enrolled cohorts, the length of study, and the frequency and type of data collection. However, they all have the use of standardized ADNI protocols in common for at least some of their data collection [212].
Two additional European programs funded by the Alzheimer’s Association focused on harmonization of measurements of both CSF biomarkers [215] and hippocampal volume [216], aiming to create worldwide protocols for standardized hippocampal segmentation and measurement of CSF biomarker concentrations to allow the direct comparison of results generated globally. Westman et al [347] investigated whether, based on shared MRI data acquisition methodologies, it was possible to combine data from Add-NeuroNet and ADNI to produce a combined cohort more representative of the general public that could be analyzed for classification and disease prediction purposes. They demonstrated that the 2 cohorts showed similar patterns of atrophy and that data from the 2 programs produced similar classification accuracies and concluded that the combination of large data sets such as these was feasible and could improve overall knowledge of the disease.
Finally, initiatives inspired by ADNI to build infrastructure including a central repository of all data, like that developed at LONI, have been implemented in Europe. NeuGRID is being developed at the European equivalent of LONI, and outGRID aims to synergize neuGRID, LONI, and the Canadian repository CBRAIN and to develop full interoperability. CATI (Centre pour l’Acquisition et le Traitement de l’Image) is the French repository for data sets within that country.
ADNI-related programs and initiatives in Europe are summarized in Table 13.
Table 13.
European initiatives related to ADNI
| Purpose | Program name | Funding agency | Time frame | Countries |
|---|---|---|---|---|
| Data collection | Pilot E-ADNI | Alzheimer’s Association | 2006–2007 | IT, FR, GE, NL, SW, DE |
| AddNeuroMed | EC | Ongoing, 40 months | FI, PL, UK, IT, GR, FR | |
| Pharma-Cog WorkPackage 5 (E-ADNI) |
EC IMI | Ongoing 5 years | SP, IT, GE, FR | |
| Swedish ADNI | Alzheimer’s Association | 2007–2009 | SW | |
| Italian ADNI | NHS | 2009–2011 | IT | |
| SOP development | International harmonization of CSF Aβ42, t-tau, and p-tau |
Alzheimer’s Association | 2009–2013 | 40 laboratories (EU, US, Japan, Australia, Brazil) |
| EADC-ADNI harmonization of hippocampal volume |
Alzheimer’s Association Lily-Wyeth |
2010–2012 | 24 centers in EU, US, Canada, Australia |
|
| Infrastructure development | NeuGRID | FP7 | 2008–2011 | IT, FR, SP, CH, UK, SW |
| OutGRID | FP7 | 2009–2011 | IT, FR, UK, US, CD | |
| Centre pour l’Acquisition et le Traitement de l’Image (CATI) |
French National Foundation on AD and RD |
2010–2013 | FR |
Abbreviations: ADNI, Alzheimer’s Disease Neuroimaging Initiative; EC, European Commission; IMI, Innovative Medicines Initiatives; NHS, National Health System; EADC, European Alzheimer’s Disease Consortium; FP7, 7th Framework Programme; AD and RD, Alzheimer’s disease and related diseases; DE, Denmark; CD, Canada; CH, Switzerland; FI, Finland; FR, France; GE, Germany; GR, Greece; IT, Italy; NL, Netherlands; PL, Poland; SP, Spain; SW, Sweden; UK, United Kingdom; US, United States.NOTE. Reproduced with permission from Ref [212].
7.2. AIBL study: The Australian ADNI
Often termed the “Australian ADNI,” the AIBL has similar goals to ADNI, namely, to better understand disease pathogenesis and to develop tests for an earlier diagnosis of AD, and, to this end, uses ADNI protocols for its imaging studies [217]. Some methodological differences between the two studies include the omission of FDG-PET metabolic investigations and the comparison of amyloid pathology using 11C-PiB PET and Aβ-42 levels in blood plasma instead of from CSF on the basis that obtaining blood plasma is both less expensive and less invasive than lumbar punctures. Perhaps the greatest difference between AIBL and ADNI lies in the approach AIBL is taking to investigating lifestyle factors involved in AD. By collecting extensive neuropsychological and lifestyle data, the study aims to understand which health and lifestyle factors protect or contribute to AD. Like ADNI, however, all data are made available through LONI and are funded by the Alzheimer’s Association. Ellis et al [217] reported that one recent finding from the study found that hippocampal atrophy was regionally associated with 11C-PiB retention only in the inferior lobe, leading to a new hypothesis of how Aβ accumulation could disrupt connections between the hippocampus through accumulation in this area (Bourgeat et al., Beta-amyloid burden in the temporal neocortex is related to elderly subjects without dementia. Neurology 2010:74:121–7; see Appendix).
Rowe et al [218] reported on the progress of the neuroimaging arm of the AIBL in characterizing a cohort of 177 healthy elderly subjects, 57 MCI patients, and 53 AD patients. The patient groups had increasing numbers of APOE ε4 carriers, increased hippocampal atrophy, and increased cognitive impairment with disease progression. The distribution of 11C-PiB binding in control subjects did not follow a normal distribution, and cluster analysis determined a separation point between low and high 11C-PiB binding groups at a neocortical standardized uptake value threshold of 1.5. This bimodal distribution in normal healthy elderly subjects again echoes the idea of heterogeneity within this group and the existence of a subset of patients with the first manifestations of AD pathogenesis well in advance of any effects on cognition. 11C-PiB binding may therefore play a role in populating and monitoring clinical trials of antiamyloid therapies. Rowe et al [218] also used 11C-PiB PET imaging for diagnosis and found that 11C-PiB scans discriminated between AD and control subjects with an accuracy of 73%, a sensitivity of 98%, and a specificity of 63%, comparable with results obtained using hippocampal volume (accuracy = 73%, specificity = 80%, sensitivity = 78%).
7.3. Japanese ADNI
The need for a Japanese ADNI (J-ADNI) was realized in 2006 when ADNI was beginning in North America and at the end of the Japanese study J-COSMIC (Japan Cooperative SPECT Study on Assessment of Mild Impairment of Cognitive Function) [219,221]. Iwatsubo [220] reported that J-ADNI was needed not only to meet requirements for global clinical trials of AD drugs about to begin in Japan and to develop the necessary infrastructure for these trials, but was also motivated by the desire of Japanese researchers to improve their clinical science through international collaboration. A special issue of Rinsho Shinkeigaku near the inception of J-ADNI in 2007 reported on ADNI and the need for the establishment of a Japanese version [221], the goals of early detection of AD and biomarker development [222], the methods used by ADNI and adopted by J-ADNI for achieving these goals [219], and the use of ADNI approaches for detecting MCI in neuropathological studies [223]. Funding for J-ADNI was sought and received from both the public and private sector, including Japanese and international companies, to a total of approximately Ұ300 million per year [220]. The study began in 2008 and aimed to recruit 300 amnestic MCI patients, 150 patients with early AD, and 150 healthy elderly control subjects from 30 centers across Japan by the end of 2010; participants would then be followed until 2013 using a research protocol designed to maximize compatibility with ADNI [220,224]. Compatibility with ADNI protocols was designed to allow sharing and direct comparison of data and as a way to contribute to global standardization of protocols. Arai et al [224] reported that initial results from ADNI supporting the use of biomarkers in clinical trials contributed to a paradigm shift in Japanese geriatric medicine from defining AD solely by cognitive measures to considering the information available from biomarkers.
7.4. Worldwide ADNI future directions
The establishment of Worldwide ADNI, an umbrella organization of global ADNI efforts, is coordinated by the Alzheimer’s Association and is a direct result of ADNI. Information on the countries that have established or plan to establish ADNI sties in their countries can be found at http://www.alz.org/research/funding/partnerships/WW-ADNI_overview.asp. (Fig. 25). Information on the countries that have established or plan to establish ADNI sties in their countries can be found at http://www.alz.org/research/funding/partnerships/WW-ADNI_overview.asp. Using standardized protocols developed by ADNI, these programs collectively aim to help define the rate of progression of MCI and AD, and to develop improved methods for identifying the appropriate patient populations to participate in clinical trials. It is anticipated that data generated by these global initiatives will ultimately be shared through a common infrastructure with international researchers. It is clear that ADNI has had and will continue to have a profound and far-reaching impact on the development of methods for the prediction and monitoring of the onset and progression of AD and in gaining a worldwide picture of the physical changes that lead to AD.
Fig. 25.
Worldwide ADNI sites. Abbreviations: NA-ADNI, North American ADNI; Arg-ADNI, Argentinean ADNI; E-ADNI, European ADNI; C-ADNI, Chinese ADNI; K-ADNI, Korean ADNI; J-ADNI, Japanese ADNI; T-ADNI, Taiwanese ADNI; A-ADNI, Australian ADNI.
8. Other papers using ADNI data
In addition to generating numerous papers related to its primary goals, ADNI is becoming a source of data for other fields of study in which a well-characterized cohort is desirable. Papers published from these studies may have some connection to AD, or may be completely unrelated.
Cuingnet et al [348] presented an improved method for the detection of regional changes in apparent diffusion coefficients (ADCs) that are indicative of irreversible ischemic damage in stroke victims. MR images of ADNI participants were used to test the method, based on a SVM in which spatial consistency is enforced by Laplacian regularization and then followed by statistical analysis to detect group differences in brain images, they found that the method was able to detect ADC changes that were not detected by standard univariate approaches.
Hypertension is a risk factor for AD and is associated with brain atrophy. Jennings et al [349] used longitudinal MR scans of ADNI controls as a normotensive control group in an investigation of whether hypertensive medication remediated the reduction in grey matter volume observed in hypertensive individuals over a year. They found that successful treatment of hypertension did not prevent brain atrophy in regions especially vulnerable to negative modification by hypertension. Another risk factor for AD is depression. Arnold et al [546] used the plasma sample collection of ADNI to identify a number of biochemical markers from a multianalyte biochemical panel that were associated with the number of depressive symptoms endorsed by participants.
Bakken et al [319] used ADNI MRI and genetic data to investigate the relationship between skull and brain morphology and European geography. They found a significant gradient of skull shape, predominantly in the frontotemporal cortical areas that extends across Europe in a NW-SE direction, supporting previous studies of European gene flow. This represents an intriguing contribution of ADNI to unlocking the mysteries of historical population movements.
Acknowledgments
Michael W. Weiner has served on the scientific advisory boards for Lilly, Araclon and Institut Catala de Neurociencies Aplicades, Gulf War Veterans Illnesses Advisory Committee, VACO, Biogen Idec, and Pfizer; has served as a consultant for Astra Zeneca, Araclon, Medivation/Pfizer, Ipsen, TauRx Therapeutics LTD, Bayer Healthcare, Biogen Idec, Exonhit Therapeutics, SA, Servier, Synarc, Pfizer, and Janssen; has received funding for travel from NeuroVigil, Inc., CHRU-Hopital Roger Salengro, Siemens, AstraZeneca, Geneva University Hospitals, Lilly, University of California, San Diego – ADNI, Paris University, Institut Catala de Neurociencies Aplicades, University of New Mexico School of Medicine, Ipsen, CTAD (Clinical Trials on Alzheimer’s Disease), Pfizer, AD PD meeting, Paul Sabatier University, Novartis, Tohoku University; has served on the editorial advisory boards for Alzheimer’s & Dementia and MRI; has received honoraria from NeuroVigil, Inc., Insitut Catala de Neurociencies Aplicades, PMDA /Japanese Ministry of Health, Labour, and Welfare, and Tohoku University; has received commercial research support from Merck and Avid; has received government research support from DOD and VA; has stock options in Synarc and Elan; and declares the following organizations as contributors to the Foundation for NIH and thus to the NIA funded Alzheimer’s Disease Neuroimaging Initiative: Abbott, Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, Anonymous Foundation, AstraZeneca, Bayer Healthcare, BioClinica, Inc. (ADNI 2), Bristol-Myers Squibb, Cure Alzheimer’s Fund, Eisai, Elan, Gene Network Sciences, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly & Company, Medpace, Merck, Novartis, Pfizer Inc., Roche, Schering Plough, Synarc, and Wyeth.
Paul S. Aisen serves on a scientific advisory board for NeuroPhage; serves as a consultant to Elan Corporation, Wyeth, Eisai Inc., Bristol-Myers Squibb, Eli Lilly and Company, NeuroPhage, Merck & Co., Roche, Amgen, Abbott, Pfizer Inc, Novartis, Bayer, Astellas, Dainippon, Biomarin, Solvay, Otsuka, Daiichi, AstraZeneca, Janssen, Medivation, Inc., Theravance, Cardeus, and Anavex; receives research support from Pfizer Inc, and Baxter International Inc., and the NIH [NIA U01-AG10483 (PI), NIA U01-AG024904 (Coordinating Center Director), NIA R01-AG030048 (PI), and R01-AG16381 (Co-I)]; and has received stock options from Medivation, Inc.
Laurel A. Beckett receives funding from the following NIH grants: 2P30CA093373-09 (deVere White, Ralph), 2P30AG010129-21 (deCarli, Charles), 5U01AG024904-07 (Weiner), 5RC2AG036535-02 (Weiner), 3UL1RR024146-06S2 (Berglund), 5R01GM088336-03 (Villablanca), 5R01AG012975-14 (Haan), 5R25RR026008-03 (Molinaro). In addition, she has received funding from the following California Breast Cancer Research Program grant: CBCRP # 16BB-1600 (von Friederichs-Fitzwater). She also has received funding from the nonprofit Critical Path Institute (Arizona) for consultation on analysis of potential biomarkers for Alzheimer’s disease clinical trials. She has served as a speaker and consultant on biomarkers for Roche.
Nigel J. Cairns has been supported by grants P50-AG05681 and P01-AG03991 from the National Institute on Aging and, National Institutes of Health, Bethesda, MD, and by the Charles and Joanne Knight Alzheimer’s Research Initiative, Washington, and the ADNI (National Institute of Health Grant U01 AG024904) which is funded by the National Institute on Aging.
Clifford R. Jack serves as a consultant for Janssen, Bristol-Meyer-Squibb, General Electric, Johnson and Johnson, and Lilly, and is involved in clinical trials sponsored by Allon and Baxter, Inc.
William Jagust is a consultant for Genentech, Bayer Healthcare, GE Healthcare, Synarc, Janssen Alzheimer Immunotherapy, Elan, and TauRx.
Enchi Liu is an employee of Janssen Alzheimer Immunotherapy, R&D, and was the 2011 Chair for the Private Partner Scientific Board for ADNI.
John C. Morris has participated in clinical trials of antidementia drugs sponsored by Janssen Immunotherapy, Eli Lilly and Company, and Pfizer, and has served as a consultant or has received speaking honoraria for Eisai, Janssen Alzheimer Immunotherapy Program/Elan, Glaxo-Smith-Kline, Novartis, Otsuka Pharmaceuticals, and Pfizer/Wyeth. Neither Dr. Morris nor his family owns stock or has equity interest (outside of mutual funds or other externally directed accounts) in any pharmaceutical or biotechnology company.
Ronald C. Petersen is a Chair, Data Monitoring Committee, for Pfizer, Inc. and Janssen Alzheimer Immunotherapy; is a consultant for Elan Pharmaceuticals; and has a CME presentation for Novartis.
Andrew J. Saykin has received support from NIA R01 AG19771 and P30 AG10133, as well as investigator initiated research support from Welch Allyn and Siemens Healthcare.
Mark E. Schmidt is a full-time employee of Janssen Pharmaceutica, NV.
Leslie Shaw has received grant support from ADNI 1, ADNI GO, ADNI 2, NIH/NIA, and Pfizer/UPenn rbm studies, and is a consultant for Innogenetics/Fujirebio, Janssen Research & Development, Bristol-Meyers Squibb, and Saladax Biomedical, Inc.
Holly Soares is a full-time employee of Bristol-Myers Squibb and a BMS shareholder.
John Q. Trojanowski has received funding for travel and honoraria from Takeda Pharmaceutical Company Ltd.; has received speaker honoraria from Pfizer Inc.; serves as an associate Editor of Alzheimer’s & Dementia; may accrue revenue on patents re: Modified avidin-biotin technique, Method of stabilizing microtubules to treat Alzheimer’s disease, Method of detecting abnormally phosphorylated tau, Method of screening for Alzheimer’s disease or disease associated with the accumulation of paired helical filaments, Compositions and methods for producing and using homogeneous neuronal cell transplants, Rat comprising straight filaments in its brain, Compositions and methods for producing and using homogeneous neuronal cell transplants to treat neurodegenerative disorders and brain and spinal cord injuries, Diagnostic methods for Alzheimer’s disease by detection of multiple MRNAs, Methods and compositions for determining lipid peroxidation levels in oxidant stress syndromes and diseases, Compositions and methods for producing and using homogenous neuronal cell transplants, Method of identifying, diagnosing and treating alpha-synuclein positive neurodegenerative disorders, Mutation-specific functional impairments in distinct tau isoforms of hereditary frontotemporal dementia and parkinsonism linked to chromosome-17: genotype predicts phenotype, Microtubule stabilizing therapies for neurodegenerative disorders, and Treatment of Alzheimer’s and related diseases with an antibody; and receives research support from the NIH (NIA P01 AG 09215-20 [PI], NIA P30 AG 10124-18 [PI], NIA PO1 AG 17586-10 [Project 4 Leader], NIA 1PO1 AG-19724-07 [Core C Leader], NIA 1 U01 AG 024904-05 [Co-PI Biomarker Core Laboratory], NINDS P50 NS053488-02 [PI], NIA UO1 AG029213-01 [Co-I]; RC2NS069368 [PI], RC1AG035427 [PI], and NIA P30AG036468 [PI]), and from the Marian S. Ware Alzheimer Program.
Appendix
1 Publications arising from AIBL
1.1 AIBL Publication list, 2009–present
1.1.1 2009
- 09.03.Bourgeat P, Chetelat G, Villemagne VL, Fripp J, Raniga P, Acosta O, et al. B-amyloid burden in the temporal neocortex is related to hippocampal atrophy in elderly subjects without dementia. Neurology. 2010;74:121–127. doi: 10.1212/WNL.0b013e3181c918b5. [DOI] [PubMed] [Google Scholar]
- 09.04.Ellis KA, Bush AI, Darby D, De Fazio D, Foster J, Hudson P, et al. The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. Int Psychogeriatr. 2009;21:672–687. doi: 10.1017/S1041610209009405. [DOI] [PubMed] [Google Scholar]
- 09.05.Fodero-Tavoletti MT, Cappai R, McLean CA, Pike KE, Adlard PA, Cowie T, et al. Amyloid imaging in Alzheimer’s disease and other dementias. Brain Imaging Behav. 2009;3:246–261. doi: 10.1007/s11682-009-9067-2. [DOI] [PubMed] [Google Scholar]
- 09.06.Fodero-Tavoletti MT, Rowe CC, McLean CA, Leone L, Li QX, Masters CL, Cappai R, Villemagne VL. Characterization of PiB binding to white matter in AD and other dementias. J Nucl Med. 2009;50:198–204. doi: 10.2967/jnumed.108.057984. [DOI] [PubMed] [Google Scholar]
- 09.07.Fodero-Tavoletti MT, Mulligan RS, Okamura N, Furumoto S, Rowe CC, Kudo Y, et al. In vitro characterisation of BF227 binding to α-synuclein/Lewy Bodies. Eur J Pharmacol. 2009;617:54–58. doi: 10.1016/j.ejphar.2009.06.042. [DOI] [PubMed] [Google Scholar]
- 09.08.Villemagne VL, McLean CA, Reardon K, Boyd A, Lewis V, Klug G, et al. 11C-PiB PET studies in typical sporadic Creutzfeldt-Jakob disease. J Neurol Neurosurg Psychiatr. 2009;80:998–1001. doi: 10.1136/jnnp.2008.171496. [DOI] [PubMed] [Google Scholar]
- 09.09.Villemagne VL, Ataka S, Mizuno T, Brooks WS, Wada Y, Kondo M, et al. High striatal amyloid β-peptide deposition across different autosomal Alzheimer’s disease mutation types. Arch Neurol. 2009;66:1537–1544. doi: 10.1001/archneurol.2009.285. [DOI] [PubMed] [Google Scholar]
- 09.10.Okamura N, Fodero-Tavoletti MT, Kudo Y, Rowe CC, Furumoto S, Arai H, Masters CL, Yanai K, Villemagne VL. Advances in molecular imaging for the diagnosis of dementia. Expert Opin Med Diagn. 2009;3:705–716. doi: 10.1517/17530050903133790. [DOI] [PubMed] [Google Scholar]
- 09.11.Acosta O, Bourgeat P, Fripp J, Zuluaga MA, Fripp J, Salvado O, Ourselin S. Automated voxel-based 3D cortical thickness measurement in a combined Lagrangian-Eulerian PDE approach using partial volume maps. Med Image Anal. 2009;13:730–743. doi: 10.1016/j.media.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
1.1.2 2010
- 10.01.Bourgeat P, Chételat G, Villemagne VL, Fripp J, Raniga P, Pike K, et al. β-amyloid burden in the temporal neocortex is related to hippocampal atrophy in elderly subjects without dementia. Neurology. 2010;74:121–127. doi: 10.1212/WNL.0b013e3181c918b5. [DOI] [PubMed] [Google Scholar]
- 10.02.Ellis KA, Rowe CC, Villemagne VL, Martins RN, Masters CL, Salvado O, Szoeke C, Ames D the AIBL Research Group. Addressing population aging and Alzheimer’s disease through the Australian imaging biomarkers and lifestyle study: collaboration with the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement. 2010;6:291–296. doi: 10.1016/j.jalz.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 10.03.Villemagne VL, Perez KA, Pike KE, Kok WM, Rowe CC, White AR, et al. Blood borne amyloid-β dimer correlates with clinical markers of Alzheimer’s disease. J Neurosci. 2010;30:6315–6322. doi: 10.1523/JNEUROSCI.5180-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.04.Chételat G, Villemagne VL, Bourgeat P, Pike KE, Jones G, Ames D, et al. Relationship between atrophy and β-amyloid deposition in Alzheimer disease. Ann Neurol. 2010;67:317–324. doi: 10.1002/ana.21955. [See also editorial: Rabinovici GD, Roberson ED. Beyond diagnosis: what biomarkers are teaching us about the “bio”logy of Alzheimer disease. Ann Neurol 2010;67:283–5.] [DOI] [PubMed] [Google Scholar]
- 10.05.Villemagne VL, Pike K, Pejoska S, Boyd A, Power M, Jones G, Masters CL, Rowe CC. 11C-PiB PET ABri imaging in Worster-Drought syndrome (Familial British Dementia): a case report. J Alzheimers Dis. 2010;19:423–428. doi: 10.3233/JAD-2010-1241. [DOI] [PubMed] [Google Scholar]
- 10.06.Lui JK, Laws SM, Li QX, Villemagne VL, Ames D, Brown B, et al. Plasma amyloid-β as a biomarker in Alzheimer’s disease: the AIBL Study of Aging. J Alzheimers Dis. 2010;20:1233–1242. doi: 10.3233/JAD-2010-090249. [DOI] [PubMed] [Google Scholar]
- 10.07.Rowe CC, Ellis KA, Rimajova M, Bourgeat P, Pike KE, Jones G, et al. Amyloid imaging results from the Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging. Neurobiol Aging. 2010;31:1275–1283. doi: 10.1016/j.neurobiolaging.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 10.08.Chételat G, Villemagne VL, Pike KE, Baron JC, Bourgeat P, Jones G, et al. Larger temporal volume in elderly with high versus low beta-amyloid deposition. Brain. 2010;133:3349–3358. doi: 10.1093/brain/awq187. [DOI] [PubMed] [Google Scholar]
- 10.09.Rueda A, Acosta O, Couprie M, Bourgeat P, Fripp J, Dowson N, Romero E, Salvado O. Topology-corrected segmentation and local intensity estimates for improved partial volume classification of brain cortex in MRI. J Neurosci Methods. 2010;188:305–315. doi: 10.1016/j.jneumeth.2010.02.020. [DOI] [PubMed] [Google Scholar]
1.1.3 2011
1.1.3.1 Published
- 11.01.Villemagne VL, Pike KE, Chételat G, Ellis KA, Mulligan R, Bourgeat P, et al. Longitudinal assessment of Aβ burden and cognition in aging and Alzheimer’s disease. Ann Neurol. 2011;69:181–192. doi: 10.1002/ana.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.02.Ellis KA, Rowe CC, Szoeke C, Villemagne VL, Ames D, Chételat G, et al. Advances in structural and molecular neuroimaging in Alzheimer’s disease. Med J Aust. 2011;194:S20–S23. doi: 10.5694/j.1326-5377.2011.tb02938.x. [DOI] [PubMed] [Google Scholar]
- 11.03.Bahar-Fuchs A, Moss S, Pike KE, Villemagne VL, Masters CL, Rowe C, Savage G. Olfactory deficits and Aβ burden in AD, MCI and healthy ageing: a PiB PET Study. J Alzheimers Dis. 2010;22:1081–1087. doi: 10.3233/JAD-2010-100696. [DOI] [PubMed] [Google Scholar]
- 11.04.Gupta VB, Laws SM, Villemagne VL, Ames D, Bush AI, Ellis KA, et al. Plasma Apolipoprotein E and Alzheimer’s disease risk: the AIBL study of ageing. Neurology. 2011;76:1091–1098. doi: 10.1212/WNL.0b013e318211c352. [DOI] [PubMed] [Google Scholar]
- 11.05.Sittironnait G, Ames D, Bush AI, Faux N, Flicker L, Foster J, et al. Effects of anticholinergic drugs on cognitive function in older Australians: results from the AIBL Study. Dement Geriatr Cogn Disord (Special ASIA issue) 2011;31:173–178. doi: 10.1159/000325171. [DOI] [PubMed] [Google Scholar]
- 11.06.McBride S, Good N, Szoeke C, Ames D, Martins R, Masters C, et al. A web-based normative data tool for assessing cognitive performance in healthy older Australians. Med J Aust. 2011;194:S12–S14. doi: 10.5694/j.1326-5377.2011.tb02936.x. [DOI] [PubMed] [Google Scholar]
- 11.07.Chételat G, Villemagne VL, Pike KE, Ellis KA, Bourgeat P, Jones G, et al. Research Group. Independent contribution of temporal Aβ deposition to memory decline in the predementia phase of Alzheimer’s disease. Brain. 2011;134(Pt 3):798–807. doi: 10.1093/brain/awq383. [DOI] [PubMed] [Google Scholar]
- 11.08.Watt AD, Perez KA, Faux NG, Pike KE, Rowe CC, Bourgeat P, et al. Increasing the predictive accuracy of beta-amyloid blood-borne biomarkers in Alzheimer’s disease. J Alzheimers Dis. 2011;24:47–59. doi: 10.3233/JAD-2010-101722. [DOI] [PubMed] [Google Scholar]
- 11.09.Villemagne VL, Okamura N, Pejoska S, Drago J, Mulligan RS, Chételat G, et al. In vivo assessment of vesicular monoamine transporter type 2 in dementia with Lewy bodies and Alzheimer’s disease. Arch Neurol. 2011;68:905–912. doi: 10.1001/archneurol.2011.142. [DOI] [PubMed] [Google Scholar]
Footnotes
Disclosures
Dallas P. Veitch has no conflicts to report.
Robert C. Green has no conflicts of interest to report.
Danielle Harvey has no conflicts of interest to report.
Judith A. Siuciak has no conflicts of interest to report.
Arthur W. Toga has no conflicts of interest to report.
References
- 1.Hardy J. Alzheimer’s disease: the amyloid cascade hypothesis: an update and reappraisal. J Alzheimers Dis. 2006;9(Suppl 3):151–153. doi: 10.3233/jad-2006-9s317. [DOI] [PubMed] [Google Scholar]
- 2.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, et al. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15:869–877. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiner MW, Aisen PS, Jack CR, Jr, Jagust WJ, Trojanowski JQ, Shaw L, et al. The Alzheimer’s Disease Neuroimaging Initiative: progress report and future plans. Alzheimers Dement. 2010;6:202.e7–211.e7. doi: 10.1016/j.jalz.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frisoni GB, Weiner MW. Alzheimer’s disease neuroimaging initiative special issue. Neurobiol Aging. 2010;31:1259–1262. doi: 10.1016/j.neurobiolaging.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Petersen RC, Roberts RO, Knopman DS, Boeve BF, Geda YE, Ivnik RJ, et al. Mild cognitive impairment: ten years later. Arch Neurol. 2009;66:1447–1455. doi: 10.1001/archneurol.2009.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimers Dement. 2010;6:265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hampel H, Shen Y, Walsh DM, Aisen P, Shaw LM, Zetterberg H, et al. Biological markers of amyloid beta-related mechanisms in Alzheimer’s disease. Exp Neurol. 2010;223:334–346. doi: 10.1016/j.expneurol.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark CM, Davatzikos C, Borthakur A, Newberg A, Leight S, Lee VM, et al. Biomarkers for early detection of Alzheimer pathology. Neurosignals. 2008;16:11–18. doi: 10.1159/000109754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleisher AS, Donohue M, Chen K, Brewer JB, Aisen PS. Applications of neuroimaging to disease-modification trials in Alzheimer’s disease. Behav Neurol. 2009;21:129–136. doi: 10.3233/BEN-2009-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw LM, Korecka M, Clark CM, Lee VM, Trojanowski JQ. Biomarkers of neurodegeneration for diagnosis and monitoring therapeutics. Nat Rev Drug Discov. 2007;6:295–303. doi: 10.1038/nrd2176. [DOI] [PubMed] [Google Scholar]
- 11.Petersen RC, Jack CR., Jr Imaging and biomarkers in early Alzheimer’s disease and mild cognitive impairment. Clin Pharmacol Ther. 2009;86:438–441. doi: 10.1038/clpt.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trojanowski JQ, Vandeerstichele H, Korecka M, Clark CM, Aisen PS, Petersen RC, et al. Update on the biomarker core of the Alzheimer’s Disease Neuroimaging Initiative subjects. Alzheimers Dement. 2010;6:230–238. doi: 10.1016/j.jalz.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 14.Jack CR, Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaw LM. PENN biomarker core of the Alzheimer’s disease Neuroimaging Initiative. Neurosignals. 2008;16:19–23. doi: 10.1159/000109755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jack CR, Jr, Lowe VJ, Weigand SD, Wiste HJ, Senjem ML, Knopman DS, et al. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer’s disease: implications for sequence of pathological events in Alzheimer’s disease. Brain. 2009;132(Pt 5):1355–1365. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braak H, Del Tredici K. The pathological process underlying Alzheimer’s disease in individuals under thirty. Acta Neuropathol. 2011;121:171–181. doi: 10.1007/s00401-010-0789-4. [DOI] [PubMed] [Google Scholar]
- 18.Dubois B, Feldman HH, Jacova C, Cummings JL, Dekosky ST, Barberger-Gateau P, et al. Revising the definition of Alzheimer’s disease: a new lexicon. Lancet Neurol. 2010;9:1118–1127. doi: 10.1016/S1474-4422(10)70223-4. [DOI] [PubMed] [Google Scholar]
- 19.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts RO, Geda YE, Knopman DS, Cha RH, Pankratz VS, Boeve BF, et al. The Mayo Clinic Study of Aging: design and sampling, participation, baseline measures and sample characteristics. Neuroepidemiology. 2008;30:58–69. doi: 10.1159/000115751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen RC, Roberts RO, Knopman DS, Geda YE, Cha RH, Pankratz VS, et al. Prevalence of mild cognitive impairment is higher in men. The Mayo Clinic Study of Aging. Neurology. 2010;75:889–897. doi: 10.1212/WNL.0b013e3181f11d85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Games D, Adams D, Alessandrini R, Barbour R, Berthelette P, Blackwell C, et al. Alzheimer-type neuropathology in transgenic mice overexpressing V717F beta-amyloid precursor protein. Nature. 1995;373:523–527. doi: 10.1038/373523a0. [DOI] [PubMed] [Google Scholar]
- 23.Frank RA, Galasko D, Hampel H, Hardy J, de Leon MJ, Mehta PD, et al. Biological markers for therapeutic trials in Alzheimer’s disease. Proceedings of the biological markers working group; NIA initiative on neuroimaging in Alzheimer’s disease. Neurobiol Aging. 2003;24:521–536. doi: 10.1016/s0197-4580(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 24.Trojanowski J. Searching for the biomarkers of Alzheimer’s. Pract Neurol. 2004;3:30–34. [Google Scholar]
- 25.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, et al. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative. Cogn Dement. 2006;5:56–62. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hampel H, Burger K, Teipel SJ, Bokde AL, Zetterberg H, Blennow K. Core candidate neurochemical and imaging biomarkers of Alzheimer’s disease. Alzheimers Dement. 2008;4:38–48. doi: 10.1016/j.jalz.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, et al. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 28.Klunk WE, Mathis CA. The future of amyloid-beta imaging: a tale of radionuclides and tracer proliferation. Curr Opin Neurol. 2008;21:683–687. doi: 10.1097/WCO.0b013e3283168e1a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kung MP, Hou C, Zhuang ZP, Skovronsky D, Kung HF. Binding of two potential imaging agents targeting amyloid plaques in postmortem brain tissues of patients with Alzheimer’s disease. Brain Res. 2004;1025:98–105. doi: 10.1016/j.brainres.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt ME, Siemers E, Snyder PJ, Potter WZ, Cole P, Soares H. The Alzheimer’s Disease Neuroimaging Initiative: perspectives of the Industry Scientific Advisory Board. Alzheimers Dement. 2010;6:286–290. doi: 10.1016/j.jalz.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Carrillo MC, Sanders CA, Katz RG. Maximizing the Alzheimer’s Disease Neuroimaging Initiative II. Alzheimers Dement. 2009;5:271–275. doi: 10.1016/j.jalz.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Toga AW, Crawford KL. The informatics core of the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement. 2010;6:247–256. doi: 10.1016/j.jalz.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jack CR, Jr, Bernstein MA, Borowski BJ, Gunter JL, Fox NC, Thompson PM, et al. Update on the magnetic resonance imaging core of the Alzheimer’s disease neuroimaging initiative. Alzheimers Dement. 2010;6:212–220. doi: 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA, et al. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimers Dement. 2010;6:221–229. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aisen PS, Petersen RC, Donohue MC, Gamst A, Raman R, Thomas RG, et al. Clinical core of the Alzheimer’s Disease Neuroimaging Initiative: progress and plans. Alzheimers Dement. 2010;6:239–246. doi: 10.1016/j.jalz.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cairns NJ, Taylor-Reinwald L, Morris JC. Autopsy consent, brain collection, and standardized neuropathologic assessment of ADNI participants: the essential role of the neuropathology core. Alzheimers Dement. 2010;6:274–279. doi: 10.1016/j.jalz.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cummings JL. Integrating ADNI results into Alzheimer’s disease drug development programs. Neurobiol Aging. 2010;31:1481–1492. doi: 10.1016/j.neurobiolaging.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, et al. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen RC, Trojanowski JQ. Use of Alzheimer disease biomarkers: potentially yes for clinical trials but not yet for clinical practice. JAMA. 2009;302:436–437. doi: 10.1001/jama.2009.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen RC. Early diagnosis of Alzheimer’s disease: is MCI too late? Curr Alzheimer Res. 2009;6:324–330. doi: 10.2174/156720509788929237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill D. Neuroimaging to assess safety and efficacy of AD therapies. Expert Opin Investig Drugs. 2010;19:23–26. doi: 10.1517/13543780903381320. [DOI] [PubMed] [Google Scholar]
- 42.Becker RE, Greig NH. Alzheimer’s disease drug development: old problems require new priorities. CNS Neurol Disord Drug Targets. 2008;7:499–511. doi: 10.2174/187152708787122950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiner MW. Editorial: imaging and biomarkers will be used for detection and monitoring progression of early Alzheimer’s disease. J Nutr Health Aging. 2009;13:332. doi: 10.1007/s12603-009-0032-y. [DOI] [PubMed] [Google Scholar]
- 44.Ito K, Corrigan B, Zhao Q, French J, Miller R, Soares H, et al. Disease progression model for cognitive deterioration from Alzheimer’s Disease Neuroimaging Initiative database. Alzheimers Dement. 2011;7:151–160. doi: 10.1016/j.jalz.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 45.Gunter JL, Bernstein MA, Borowski BJ, Ward CP, Britson PJ, Felmlee JP, et al. Measurement of MRI scanner performance with the ADNI phantom. Med Phys. 2009;36:2193–2205. doi: 10.1118/1.3116776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kruggel F, Turner J, Muftuler LT. Impact of scanner hardware and imaging protocol on image quality and compartment volume precision in the ADNI cohort. Neuroimage. 2010;49:2123–2133. doi: 10.1016/j.neuroimage.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho AJ, Hua X, Lee S, Leow AD, Yanovsky I, Gutman B, et al. Comparing 3 T and 1.5 T MRI for tracking Alzheimer’s disease progression with tensor-based morphometry. Hum Brain Mapp. 2010;31:499–514. doi: 10.1002/hbm.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mortamet B, Bernstein MA, Jack CR, Jr, Gunter JL, Ward C, Britson PJ, et al. Automatic quality assessment in structural brain magnetic resonance imaging. Magn Reson Med. 2009;62:365–372. doi: 10.1002/mrm.21992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarkson MJ, Ourselin S, Nielsen C, Leung KK, Barnes J, Whitwell JL, et al. Comparison of phantom and registration scaling corrections using the ADNI cohort. Neuroimage. 2009;47:1506–1513. doi: 10.1016/j.neuroimage.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bauer CM, Jara H, Killiany R. Whole brain quantitative T2 MRI across multiple scanners with dual echo FSE: applications to AD, MCI, and normal aging. Neuroimage. 2010;52:508–514. doi: 10.1016/j.neuroimage.2010.04.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leung KK, Clarkson MJ, Bartlett JW, Clegg S, Jack CR, Jr, Weiner MW, et al. Robust atrophy rate measurement in Alzheimer’s disease using multi-site serial MRI: tissue-specific intensity normalization and parameter selection. Neuroimage. 2010;50:516–523. doi: 10.1016/j.neuroimage.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyes RG, Gunter JL, Frost C, Janke AL, Yeatman T, Hill DL, et al. Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils. Neuroimage. 2008;39:1752–1762. doi: 10.1016/j.neuroimage.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leow AD, Klunder AD, Jack CR, Jr, Toga AW, Dale AM, Bernstein MA, et al. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joshi A, Koeppe RA, Fessler JA. Reducing between scanner differences in multi-center PET studies. Neuroimage. 2009;46:154–159. doi: 10.1016/j.neuroimage.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw LM, Vanderstichele H, Knapik-Czajka M, Figurski M, Coart E, Blennow K, et al. Qualification of the analytical and clinical performance of CSF biomarker analyses in ADNI. Acta Neuropathol. 2011;121:597–609. doi: 10.1007/s00401-011-0808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leung KK, Barnes J, Modat M, Ridgway GR, Bartlett JW, Fox NC, et al. Brain MAPS: an automated, accurate and robust brain extraction technique using a template library. Neuroimage. 2011;55:1091–1108. doi: 10.1016/j.neuroimage.2010.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leung KK, Barnes J, Ridgway GR, Bartlett JW, Clarkson MJ, Macdonald K, et al. Automated cross-sectional and longitudinal hippocampal volume measurement in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2010;51:1345–1359. doi: 10.1016/j.neuroimage.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolz R, Aljabar P, Hajnal JV, Hammers A, Rueckert D. LEAP: learning embeddings for atlas propagation. Neuroimage. 2010;49:1316–1325. doi: 10.1016/j.neuroimage.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lotjonen JM, Wolz R, Koikkalainen JR, Thurfjell L, Waldemar G, Soininen H, et al. Fast and robust multi-atlas segmentation of brain magnetic resonance images. Neuroimage. 2010;49:2352–2365. doi: 10.1016/j.neuroimage.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 62.Heckemann RA, Keihaninejad S, Aljabar P, Rueckert D, Hajnal JV, Hammers A. Improving intersubject image registration using tissue-class information benefits robustness and accuracy of multi-atlas based anatomical segmentation. Neuroimage. 2010;51:221–227. doi: 10.1016/j.neuroimage.2010.01.072. [DOI] [PubMed] [Google Scholar]
- 63.Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, et al. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer’s disease mild cognitive impairment, and elderly controls. Neuroimage. 2008;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolz R, Heckemann RA, Aljabar P, Hajnal JV, Hammers A, Lotjonen J, et al. Measurement of hippocampal atrophy using 4D graph-cut segmentation: application to ADNI. Neuroimage. 2010;52:109–118. doi: 10.1016/j.neuroimage.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 65.Huang S, Li J, Sun L, Ye J, Fleisher A, Wu T, et al. Learning brain connectivity of Alzheimer’s disease by sparse inverse covariance estimation. Neuroimage. 2010;50:935–949. doi: 10.1016/j.neuroimage.2009.12.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calvini P, Chincarini A, Gemme G, Penco MA, Squarcia S, Nobili F, et al. Automatic analysis of medial temporal lobe atrophy from structural MRIs for the early assessment of Alzheimer disease. Med Phys. 2009;36:3737–3747. doi: 10.1118/1.3171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holland D, Dale AM. Nonlinear registration of longitudinal images and measurement of change in regions of interest. Med Image Anal. 2011;15:489–497. doi: 10.1016/j.media.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chupin M, Gerardin E, Cuingnet R, Boutet C, Lemieux L, Lehericy S, et al. Fully automatic hippocampus segmentation and classification in Alzheimer’s disease and mild cognitive impairment applied on data from ADNI. Hippocampus. 2009;19:579–587. doi: 10.1002/hipo.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, et al. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp. 2009;30:2766–2788. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H, Das S, Pluta J, Craige C, Altinay M, Avants B, et al. Standing on the shoulders of giants: improving medical image segmentation via bias correction. Med Image Comput Comput Assist Interv. 2010;13(Pt 3):105–112. doi: 10.1007/978-3-642-15711-0_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bossa M, Zacur E, Olmos S. Statistical analysis of relative pose information of subcortical nuclei: application on ADNI data. Neuroimage. 2011;55:999–1008. doi: 10.1016/j.neuroimage.2010.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bossa M, Zacur E, Olmos S. Tensor-based morphometry with stationary velocity field diffeomorphic registration: application to ADNI. Neuroimage. 2010;51:956–969. doi: 10.1016/j.neuroimage.2010.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hua X, Leow AD, Lee S, Klunder AD, Toga AW, Lepore N, et al. 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. Neuroimage. 2008;41:19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yushkevich PA, Avants BB, Das SR, Pluta J, Altinay M, Craige C. Bias in estimation of hippocampal atrophy using deformation-based morphometry arises from asymmetric global normalization: an illustration in ADNI 3 T MRI data. Neuroimage. 2010;50:434–445. doi: 10.1016/j.neuroimage.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park H, Seo J. Application of multidimensional scaling to quantify shape in Alzheimer’s disease and its correlation with Mini Mental State Examination: a feasibility study. J Neurosci Methods. 2011;194:380–385. doi: 10.1016/j.jneumeth.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 76.Chen W, Song X, Zhang Y, Darvesh S, Zhang N, D’Arcy RC, et al. An MRI-based semiquantitative index for the evaluation of brain atrophy and lesions in Alzheimer’s disease, mild cognitive impairment and normal aging. Dement Geriatr Cogn Disord. 2010;30:121–130. doi: 10.1159/000319537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Acosta O, Bourgeat P, Zuluaga MA, Fripp J, Salvado O, Ourselin S. Automated voxel-based 3D cortical thickness measurement in a combined Lagrangian-Eulerian PDE approach using partial volume maps. Med Image Anal. 2009;13:730–743. doi: 10.1016/j.media.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.King RD, George AT, Jeon T, Hynan LS, Youn TS, Kennedy DN, et al. Characterization of atrophic changes in the cerebral cortex using fractal dimensional analysis. Brain Imaging Behav. 2009;3:154–166. doi: 10.1007/s11682-008-9057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.King RD, Brown B, Hwang M, Jeon T, George AT. Fractal dimension analysis of the cortical ribbon in mild Alzheimer’s disease. Neuroimage. 2010;53:471–479. doi: 10.1016/j.neuroimage.2010.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y, Wang Y, Xue Z, Shi F, Lin W, Shen D. Consistent 4D cortical thickness measurement for longitudinal neuroimaging study. Med Image Comput Comput Assist Interv. 2010;13(Pt 2):133–142. doi: 10.1007/978-3-642-15745-5_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Risser L, Vialard FX, Wolz R, Holm DD, Rueckert D. Simultaneous fine and coarse diffeomorphic registration: application to atrophy measurement in Alzheimer’s disease. Med Image Comput Comput Assist Interv. 2010;13(Pt 2):610–617. doi: 10.1007/978-3-642-15745-5_75. [DOI] [PubMed] [Google Scholar]
- 82.Zhang T, Davatzikos C. ODVBA: Optimally-Discriminative Voxel-Based Analysis. IEEE Trans Med Imaging. 2011;30:1441–1454. doi: 10.1109/TMI.2011.2114362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan Y, Batmanghelich N, Clark CM, Davatzikos C. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. Neuroimage. 2008;39:1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haense C, Herholz K, Jagust WJ, Heiss WD. Performance of FDG PET for detection of Alzheimer’s disease in two independent multicentre samples (NEST-DD and ADNI) Dement Geriatr Cogn Disord. 2009;28:259–266. doi: 10.1159/000241879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen K, Ayutyanont N, Langbaum JB, Fleisher AS, Reschke C, Lee W, et al. Characterizing Alzheimer’s disease using a hypometabolic convergence index. Neuroimage. 2011;56:52–60. doi: 10.1016/j.neuroimage.2011.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kohannim O, Hua X, Hibar DP, Lee S, Chou YY, Toga AW, et al. Boosting power for clinical trials using classifiers based on multiple biomarkers. Neurobiol Aging. 2010;31:1429–1442. doi: 10.1016/j.neurobiolaging.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lemoine B, Rayburn S, Benton R. Data fusion and feature selection for Alzheimer’s disease. Lect Notes Comput Sci. 2010;6334:320–327. [Google Scholar]
- 88.Hinrichs C, Singh V, Mukherjee L, Xu G, Chung MK, Johnson SC. Spatially augmented LPboosting for AD classification with evaluations on the ADNI dataset. Neuroimage. 2009;48:138–149. doi: 10.1016/j.neuroimage.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shen L, Qi Y, Kim S, Nho K, Wan J, Risacher SL, et al. Sparse bayesian learning for identifying imaging biomarkers in AD prediction. Med Image Comput Comput Assist Interv. 2010;13(Pt 3):611–618. doi: 10.1007/978-3-642-15711-0_76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salas-Gonzalez D, Gorriz JM, Ramirez J, Illan IA, Lopez M, Segovia F, et al. Feature selection using factor analysis for Alzheimer’s diagnosis using 18F-FDG PET images. Med Phys. 2010;37:6084–6095. doi: 10.1118/1.3488894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stonnington CM, Chu C, Kloppel S, Jack CR, Jr, Ashburner J, Frackowiak RS. Predicting clinical scores from magnetic resonance scans in Alzheimer’s disease. Neuroimage. 2010;51:511–517. doi: 10.1016/j.neuroimage.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Franke K, Ziegler G, Kloppel S, Gaser C. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage. 2010;50:883–892. doi: 10.1016/j.neuroimage.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 93.Filipovych R, Davatzikos C. Semi-supervised pattern classification of medical images: application to mild cognitive impairment (MCI) Neuroimage. 2011;55:1109–1119. doi: 10.1016/j.neuroimage.2010.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang W, Hong CX, Xie H, Huang X. ICA-based automatic classification of magnetic resonance images from ADNI data. Lect Notes Comput Sci. 2010;6330:340–347. [Google Scholar]
- 95.Pelaez-Coca M, Bossa M, Olmos S. Discrimination of AD and normal subjects from MRI: anatomical versus statistical regions. Neurosci Lett. 2011;487:113–117. doi: 10.1016/j.neulet.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 96.Llano DA, Laforet G, Devanarayan V. Derivation of a new ADAS-cog composite using tree-based multivariate analysis: prediction of conversion from mild cognitive impairment to Alzheimer disease. Alzheimer Dis Assoc Disord. 2010;25:73–84. doi: 10.1097/WAD.0b013e3181f5b8d8. [DOI] [PubMed] [Google Scholar]
- 97.Rousseau F. A non-local approach for image super-resolution using intermodality priors. Med Image Anal. 2010;14:594–605. doi: 10.1016/j.media.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gerber S, Tasdizen T, Thomas Fletcher P, Joshi S, Whitaker R. Manifold modeling for brain population analysis. Med Image Anal. 2010;14:643–653. doi: 10.1016/j.media.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Habeck C, Stern Y. Multivariate data analysis for neuroimaging data: overview and application to Alzheimer’s disease. Cell Biochem Biophys. 2010;58:53–67. doi: 10.1007/s12013-010-9093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Habeck CG. Basics of multivariate analysis in neuroimaging data. J Vis Exp. 2010;41 doi: 10.3791/1988. pii: 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu X, Chen K, Yao L, Ayutyanont N, Langbaum JB, Fleisher A, et al. Assessing the reliability to detect cerebral hypometabolism in probable Alzheimer’s disease and amnestic mild cognitive impairment. J Neurosci Methods. 2010;192:277–285. doi: 10.1016/j.jneumeth.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Singh N, Fletcher PT, Preston JS, Ha L, King R, Marron JS, et al. Multivariate statistical analysis of deformation momenta relating anatomical shape to neuropsychological measures. Med Image Comput Comput Assist Interv. 2010;13(Pt 3):529–537. doi: 10.1007/978-3-642-15711-0_66. [DOI] [PubMed] [Google Scholar]
- 103.Vounou M, Nichols TE, Montana G. Discovering genetic associations with high-dimensional neuroimaging phenotypes: a sparse reduced-rank regression approach. Neuroimage. 2010;53:1147–1159. doi: 10.1016/j.neuroimage.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen K, Langbaum JB, Fleisher AS, Ayutyanont N, Reschke C, Lee W, et al. Twelve-month metabolic declines in probable Alzheimer’s disease and amnestic mild cognitive impairment assessed using an empirically pre-defined statistical region-of-interest: findings from the Alzheimer’s Disease Neuroimaging Initiative. Neuroimage. 2010;51:654–664. doi: 10.1016/j.neuroimage.2010.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Silver M, Montana G, Nichols TE. False positives in neuroimaging genetics using voxel-based morphometry data. Neuroimage. 2011;54:992–1000. doi: 10.1016/j.neuroimage.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74:201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Epstein NU, Saykin AJ, Risacher SL, Gao S, Farlow MR. Differences in medication use in the Alzheimer’s disease neuroimaging initiative: analysis of baseline characteristics. Drugs Aging. 2010;27:677–686. doi: 10.2165/11538260-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schneider LS, Insel PS, Weiner MW. Treatment with cholinesterase inhibitors and memantine of patients in the Alzheimer’s Disease Neuroimaging Initiative. Arch Neurol. 2011;68:58–66. doi: 10.1001/archneurol.2010.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fennema-Notestine C, Hagler DJ, Jr, McEvoy LK, Fleisher AS, Wu EH, Karow DS, et al. Structural MRI biomarkers for preclinical and mild Alzheimer’s disease. Hum Brain Mapp. 2009;30:3238–3253. doi: 10.1002/hbm.20744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karow DS, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jr, Jennings RG, Brewer JB, et al. Relative capability of MR imaging and FDG PET to depict changes associated with prodromal and early Alzheimer disease. Radiology. 2010;256:932–942. doi: 10.1148/radiol.10091402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McDonald CR, McEvoy LK, Gharapetian L, Fennema-Notestine C, Hagler DJ, Jr, Holland D, et al. Regional rates of neocortical atrophy from normal aging to early Alzheimer disease. Neurology. 2009;73:457–465. doi: 10.1212/WNL.0b013e3181b16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, et al. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008;43:458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leow AD, Yanovsky I, Parikshak N, Hua X, Lee S, Toga AW, et al. Alzheimer’s disease neuroimaging initiative: a one-year follow up study using tensor-based morphometry correlating degenerative rates, biomarkers and cognition. Neuroimage. 2009;45:645–655. doi: 10.1016/j.neuroimage.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res. 2009;6:347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Risacher SL, Shen L, West JD, Kim S, McDonald BC, Beckett LA, et al. Longitudinal MRI atrophy biomarkers: relationship to conversion in the ADNI cohort. Neurobiol Aging. 2010;31:1401–1418. doi: 10.1016/j.neurobiolaging.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schuff N, Tosun D, Insel PS, Chiang GC, Truran D, Aisen PS, et al. Nonlinear time course of brain volume loss in cognitively normal and impaired elders. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2010.07.012. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McEvoy LK, Fennema-Notestine C, Roddey JC, Hagler DJ, Jr, Holland D, Karow DS, et al. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251:195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neuroimage. 2009;44:1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2010.05.023. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hua X, Hibar DP, Lee S, Toga AW, Jack CR, Jr, Weiner MW, et al. Sex and age differences in atrophic rates: an ADNI study with n=1368 MRI scans. Neurobiol Aging. 2010;31:1463–1480. doi: 10.1016/j.neurobiolaging.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, et al. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009;132(Pt 4):1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Qiu A, Fennema-Notestine C, Dale AM, Miller MI. Regional shape abnormalities in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2009;45:656–661. doi: 10.1016/j.neuroimage.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Apostolova LG, Morra JH, Green AE, Hwang KS, Avedissian C, Woo E, et al. Automated 3D mapping of baseline and 12-month associations between three verbal memory measures and hippocampal atrophy in 490 ADNI subjects. Neuroimage. 2010;51:488–499. doi: 10.1016/j.neuroimage.2009.12.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, et al. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Neuroimage. 2009;45(Suppl 1):S3–S15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Evans MC, Barnes J, Nielsen C, Kim LG, Clegg SL, Blair M, et al. Volume changes in Alzheimer’s disease and mild cognitive impairment: cognitive associations. Eur Radiol. 2010;20:674–682. doi: 10.1007/s00330-009-1581-5. [DOI] [PubMed] [Google Scholar]
- 126.Chou YY, Lepore N, Avedissian C, Madsen SK, Parikshak N, Hua X, et al. Mapping correlations between ventricular expansion and CSF amyloid and tau biomarkers in 240 subjects with Alzheimer’s disease, mild cognitive impairment and elderly controls. Neuroimage. 2009;46:394–410. doi: 10.1016/j.neuroimage.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chou YY, Lepore N, Saharan P, Madsen SK, Hua X, Jack CR, et al. Ventricular maps in 804 ADNI subjects: correlations with CSF biomarkers and clinical decline. Neurobiol Aging. 2010;31:1386–1400. doi: 10.1016/j.neurobiolaging.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Shaw LM, Trojanowski JQ, et al. Effect of apolipoprotein E on biomarkers of amyloid load and neuronal pathology in Alzheimer disease. Ann Neurol. 2010;67:308–316. doi: 10.1002/ana.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Andrawis JP, Hwang KS, Green AE, Kotlerman J, Elashoff D, Morra JH, et al. Effects of ApoE4 and maternal history of dementia on hippocampal atrophy. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2010.07.020. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Madsen SK, Ho J, Hua X, Saharan PS, Toga AW, Jack CR, Jr, et al. 3D maps localize caudate nucleus atrophy in 400 Alzheimer’s disease, mild cognitive impairment, and healthy elderly subjects. Neurobiol Aging. 2010;31:1312–1325. doi: 10.1016/j.neurobiolaging.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Greene SJ, Killiany RJ. Subregions of the inferior parietal lobule are affected in the progression to Alzheimer’s disease. Neurobiol Aging. 2010;31:1304–1311. doi: 10.1016/j.neurobiolaging.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: diagnostic discrimination and cognitive correlations. Neurology. 2009;73:287–293. doi: 10.1212/WNL.0b013e3181af79e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ho AJ, Raji CA, Becker JT, Lopez OL, Kuller LH, Hua X, et al. Obesity is linked with lower brain volume in 700 AD and MCI patients. Neurobiol Aging. 2010;31:1326–1339. doi: 10.1016/j.neurobiolaging.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Langbaum JB, Chen K, Lee W, Reschke C, Bandy D, Fleisher AS, et al. Categorical and correlational analyses of baseline fluorodeoxyglucose positron emission tomography images from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Neuroimage. 2009;45:1107–1116. doi: 10.1016/j.neuroimage.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Landau SM, Harvey D, Madison CM, Koeppe RA, Reiman EM, Foster NL, et al. Associations between cognitive, functional, and FDG-PET measures of decline in AD and MCI. Neuro0biol Aging. 2011;32:1207–1218. doi: 10.1016/j.neurobiolaging.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Walhovd KB, Fjell AM, Dale AM, McEvoy LK, Brewer J, Karow DS, et al. Multi-modal imaging predicts memory performance in normal aging and cognitive decline. Neurobiol Aging. 2010;31:1107–1121. doi: 10.1016/j.neurobiolaging.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wolk DA, Dickerson BC. Fractionating verbal episodic memory in Alzheimer’s disease. Neuroimage. 2011;54:1530–1539. doi: 10.1016/j.neuroimage.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chang YL, Bondi MW, Fennema-Notestine C, McEvoy LK, Hagler DJ, Jr, Jacobson MW, et al. Brain substrates of learning and retention in mild cognitive impairment diagnosis and progression to Alzheimer’s disease. Neuropsychologia. 2010;48:1237–1247. doi: 10.1016/j.neuropsychologia.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dickerson BC, Wolk DA. Dysexecutive versus amnesic phenotypes of very mild Alzheimer’s disease are associated with distinct clinical, genetic and cortical thinning characteristics. J Neurol Neurosurg Psychiatry. 2011;82:45–51. doi: 10.1136/jnnp.2009.199505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chang YL, Jacobson MW, Fennema-Notestine C, Hagler DJ, Jr, Jennings RG, Dale AM, et al. Level of executive function influences verbal memory in amnestic mild cognitive impairment and predicts prefrontal and posterior cingulate thickness. Cereb Cortex. 2010;20:1305–1313. doi: 10.1093/cercor/bhp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wolk DA, Dickerson BC, Weiner M, Aiello M, Aisen P, Albert MS, et al. Apolipoprotein E (APOE) genotype has dissociable effects on memory and attentional-executive network function in Alzheimer’s disease. Proc Natl Acad Sci USA. 2010;107:10256–10261. doi: 10.1073/pnas.1001412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McDonald CR, Gharapetian L, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jr, Holland D, et al. Relationship between regional atrophy rates and cognitive decline in mild cognitive impairment. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2010.03.015. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Okonkwo OC, Alosco ML, Jerskey BA, Sweet LH, Ott BR, Tremont G. Cerebral atrophy, apolipoprotein E varepsilon4, and rate of decline in everyday function among patients with amnestic mild cognitive impairment. Alzheimers Dement. 2010;6:404–411. doi: 10.1016/j.jalz.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Okonkwo OC, Alosco ML, Griffith HR, Mielke MM, Shaw LM, Trojanowski JQ, et al. Cerebrospinal fluid abnormalities and rate of decline in everyday function across the dementia spectrum: normal aging, mild cognitive impairment, and Alzheimer disease. Arch Neurol. 2010;67:688–696. doi: 10.1001/archneurol.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cronk BB, Johnson DK, Burns JM. Body mass index and cognitive decline in mild cognitive impairment. Alzheimer Dis Assoc Disord. 2010;24:126–130. doi: 10.1097/WAD.0b013e3181a6bf3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tosun D, Schuff N, Truran-Sacrey D, Shaw LM, Trojanowski JQ, Aisen P, et al. Relations between brain tissue loss, CSF biomarkers, and the ApoE genetic profile: a longitudinal MRI study. Neurobiol Aging. 2010;31:1340–1354. doi: 10.1016/j.neurobiolaging.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, et al. CSF biomarkers in prediction of cerebral and clinical change in mild cognitive impairment and Alzheimer’s disease. J Neurosci. 2010;30:2088–2101. doi: 10.1523/JNEUROSCI.3785-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ott BR, Cohen RA, Gongvatana A, Okonkwo OC, Johanson CE, Stopa EG, et al. Brain ventricular volume and cerebrospinal fluid biomarkers of Alzheimer’s disease. J Alzheimers Dis. 2010;20:647–657. doi: 10.3233/JAD-2010-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim S, Swaminathan S, Shen L, Risacher SL, Nho K, Foroud T, et al. Genome-wide association study of CSF biomarkers Abeta1–42, t-tau, and p-tau181p in the ADNI cohort. Neurology. 2011;76:69–79. doi: 10.1212/WNL.0b013e318204a397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jagust WJ, Landau SM, Shaw LM, Trojanowski JQ, Koeppe RA, Reiman EM, et al. Relationships between biomarkers in aging and dementia. Neurology. 2009;73:1193–1199. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Apostolova LG, Hwang KS, Andrawis JP, Green AE, Babakchanian S, Morra JH, et al. 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiol Aging. 2010;31:1284–1303. doi: 10.1016/j.neurobiolaging.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jack CR, Jr, Wiste HJ, Vemuri P, Weigand SD, Senjem ML, Zeng G, et al. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer’s disease. Brain. 2010;133:3336–3348. doi: 10.1093/brain/awq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Caroli A, Frisoni GB. The dynamics of Alzheimer’s disease biomarkers in the Alzheimer’s Disease Neuroimaging Initiative cohort. Neurobiol Aging. 2010;31:1263–1274. doi: 10.1016/j.neurobiolaging.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Beckett LA, Harvey DJ, Gamst A, Donohue M, Kornak J, Zhang H, et al. The Alzheimer’s Disease Neuroimaging Initiative: annual change in biomarkers and clinical outcomes. Alzheimers Dement. 2010;6:257–264. doi: 10.1016/j.jalz.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Walhovd KB, Fjell AM, Brewer J, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jr, et al. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. Am J Neuroradiol. 2010;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Trojanowski JQ, Shaw LM, et al. Serial MRI and CSF biomarkers in normal aging, MCI, and AD. Neurology. 2010;75:143–151. doi: 10.1212/WNL.0b013e3181e7ca82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li Y, Wang Y, Wu G, Shi F, Zhou L, Lin W, et al. Discriminant analysis of longitudinal cortical thickness changes in Alzheimer’s disease using dynamic and network features. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2010.11.008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehericy S, Habert MO, et al. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage. 2010;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 159.De Meyer G, Shapiro F, Vanderstichele H, Vanmechelen E, Engelborghs S, De Deyn PP, et al. Diagnosis-independent alzheimer disease biomarker signature in cognitively normal elderly people. Arch Neurol. 2010;67:949–956. doi: 10.1001/archneurol.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schott JM, Bartlett JW, Fox NC, Barnes J. Increased brain atrophy rates in cognitively normal older adults with low cerebrospinal fluid Abeta1–42. Ann Neurol. 2010;68:825–834. doi: 10.1002/ana.22315. [DOI] [PubMed] [Google Scholar]
- 161.Ewers M, Walsh C, Trojanowski JQ, Shaw LM, Petersen RC, Jack CR, Jr, et al. Prediction of conversion from mild cognitive impairment to Alzheimer’s disease dementia based upon biomarkers and neuropsychological test performance. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2010.10.019. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.van Gils M, Koikkalainen J, Mattila J, Herukka S, Lotjonen J, Soininen H. Discovery and use of efficient biomarkers for objective disease state assessment in Alzheimer’s disease. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:2886–2889. doi: 10.1109/IEMBS.2010.5626311. [DOI] [PubMed] [Google Scholar]
- 163.Desikan RS, Cabral HJ, Settecase F, Hess CP, Dillon WP, Glastonbury CM, et al. Automated MRI measures predict progression to Alzheimer’s disease. Neurobiol Aging. 2010;31:1364–1374. doi: 10.1016/j.neurobiolaging.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kovacevic S, Rafii MS, Brewer JB. High-throughput, fully automated volumetry for prediction of MMSE and CDR decline in mild cognitive impairment. Alzheimer Dis Assoc Disord. 2009;23:139–145. doi: 10.1097/WAD.0b013e318192e745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Querbes O, Aubry F, Pariente J, Lotterie JA, Demonet JF, Duret V, et al. Early diagnosis of Alzheimer’s disease using cortical thickness: impact of cognitive reserve. Brain. 2009;132:2036–2047. doi: 10.1093/brain/awp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Carmichael O, Schwarz C, Drucker D, Fletcher E, Harvey D, Beckett L, et al. Longitudinal changes in white matter disease and cognition in the first year of the Alzheimer disease neuroimaging initiative. Arch Neurol. 2010;67:1370–1378. doi: 10.1001/archneurol.2010.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology. 2009;73:294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McEvoy LK, Edland SD, Holland D, Hagler DJ, Roddey JC, Fennema-Notestine C, et al. Neuroimaging enrichment strategy for secondary prevention trials in Alzheimer disease. Alzheimer Dis Assoc Disord. 2010;24:269–277. doi: 10.1097/WAD.0b013e3181d1b814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Weigand SD, Vemuri P, Wiste HJ, Senjem ML, Pankratz VS, Aisen PS, et al. Transforming cerebrospinal fluid Abeta42 measures into calculated Pittsburgh Compound B units of brain A beta amyloid. Alzheimers Dement. 2011;7:133–141. doi: 10.1016/j.jalz.2010.08.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schneider LS, Kennedy RE, Cutter GR. Requiring an amyloid-beta1–42 biomarker for prodromal Alzheimer’s disease or mild cognitive impairment does not lead to more efficient clinical trials. Alzheimers Dement. 2010;6:367–377. doi: 10.1016/j.jalz.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ito K, Ahadieh S, Corrigan B, French J, Fullerton T, Tensfeldt T. Disease progression meta-analysis model in Alzheimer’s disease. Alzheimers Dement. 2010;6:39–53. doi: 10.1016/j.jalz.2009.05.665. [DOI] [PubMed] [Google Scholar]
- 172.Lorenzi M, Donohue M, Paternico D, Scarpazza C, Ostrowitzki S, Blin O, et al. Enrichment through biomarkers in clinical trials of Alzheimer’s drugs in patients with mild cognitive impairment. Neurobiol Aging. 2010;13:1443.e1–1451.e1. doi: 10.1016/j.neurobiolaging.2010.04.036. [DOI] [PubMed] [Google Scholar]
- 173.Landau SM, Harvey D, Madison CM, Reiman EM, Foster NL, Aisen PS, et al. Comparing predictors of conversion and decline in mild cognitive impairment. Neurology. 2010;75:230–238. doi: 10.1212/WNL.0b013e3181e8e8b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schott JM, Bartlett JW, Barnes J, Leung KK, Ourselin S, Fox NC. Reduced sample sizes for atrophy outcomes in Alzheimer’s disease trials: baseline adjustment. Neurobiol Aging. 2010;31:1452.e2–1462.e2. doi: 10.1016/j.neurobiolaging.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nestor SM, Rupsingh R, Borrie M, Smith M, Accomazzi V, Wells JL, et al. Ventricular enlargement as a possible measure of Alzheimer’s disease progression validated using the Alzheimer’s disease neuroimaging initiative database. Brain. 2008;131(Pt 9):2443–2454. doi: 10.1093/brain/awn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Holland D, Brewer JB, Hagler DJ, Fenema-Notestine C, Dale AM, Weiner M, et al. Subregional neuroanatomical change as a biomarker for Alzheimer’s disease. Proc Natl Acad Sci USA. 2009;106:20954–20959. doi: 10.1073/pnas.0906053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hua X, Lee S, Yanovsky I, Leow AD, Chou YY, Ho AJ, et al. Optimizing power to track brain degeneration in Alzheimer’s disease and mild cognitive impairment with tensor-based morphometry: an ADNI study of 515 subjects. Neuroimage. 2009;48:668–681. doi: 10.1016/j.neuroimage.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hua X, Gutman B, Boyle CP, Rajagopalan P, Leow AD, Yanovsky I, et al. Accurate measurement of brain changes in longitudinal MRI scans using tensor-based morphometry. Neuroimage. 2011;57:5–14. doi: 10.1016/j.neuroimage.2011.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 180.Ashford JW, Mortimer JA. Non-familial Alzheimer’s disease is mainly due to genetic factors. J Alzheimers Dis. 2002;4:169–177. doi: 10.3233/jad-2002-4307. [DOI] [PubMed] [Google Scholar]
- 181.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 182.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, et al. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Arch Neurol. 2010;67:961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS One. 2009;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, et al. Meta-analysis confirms CR1, CLU, and PICALM as Alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lakatos A, Derbeneva O, Younes D, Keator D, Bakken T, Lvova M, et al. Association between mitochondrial DNA variations and Alzheimer’s disease in the ADNI cohort. Neurobiol Aging. 2010;31:1355–1363. doi: 10.1016/j.neurobiolaging.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kauwe JS, Bertelsen S, Mayo K, Cruchaga C, Abraham R, Hollingworth P, et al. Suggestive synergy between genetic variants in TF and HFE as risk factors for Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:955–959. doi: 10.1002/ajmg.b.31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cruchaga C, Bertelsen S, Kauwe JS, Nowotny P, Shah AR, et al. SNPs in the regulatory subunit of calcineurin are associated with CSF tau protein levels, brain mRNA levels. Alzheimers Dement. 2009;5(Suppl 4):P471–P472. [Google Scholar]
- 191.Cruchaga C, Kauwe JS, Mayo K, Spiegel N, Bertelsen S, Nowotny P, et al. SNPs associated with cerebrospinal fluid phospho-tau levels influence rate of decline in Alzheimer’s disease. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001101. pii: e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kauwe JS, Cruchaga C, Bertelsen S, Mayo K, Latu W, Nowotny P, et al. Validating predicted biological effects of Alzheimer’s disease associated SNPs using CSF biomarker levels. J Alzheimers Dis. 2010;21:833–842. doi: 10.3233/JAD-2010-091711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, et al. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: a study of the ADNI cohort. Neuroimage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, et al. Voxelwise genome-wide association study (vGWAS) Neuroimage. 2010;53:1160–1174. doi: 10.1016/j.neuroimage.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stein JL, Hua X, Morra JH, Lee S, Hibar DP, Ho AJ, et al. Genome-wide analysis reveals novel genes influencing temporal lobe structure with relevance to neurodegeneration in Alzheimer’s disease. Neuroimage. 2010;51:542–554. doi: 10.1016/j.neuroimage.2010.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, et al. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol Psychiatry. doi: 10.1038/mp.2010.123. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bertram L, Heekeren H. Obesity and the brain: a possible genetic link. Alzheimers Res Ther. 2010;2:27. doi: 10.1186/alzrt51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Han MR, Schellenberg GD, Wang LS. Genome-wide association reveals genetic effects on human Abeta42 and tau protein levels in cerebrospinal fluids: a case control study. BMC Neurol. 2010;10:90. doi: 10.1186/1471-2377-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, et al. One-year brain atrophy evident in healthy aging. J Neurosci. 2009;29:15223–15231. doi: 10.1523/JNEUROSCI.3252-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Burgmans S, van Boxtel MP, Vuurman EF, Smeets F, Gronenschild EH, Uylings HB, et al. The prevalence of cortical gray matter atrophy may be overestimated in the healthy aging brain. Neuropsychology. 2009;23:541–550. doi: 10.1037/a0016161. [DOI] [PubMed] [Google Scholar]
- 202.Fjell AM, Westlye LT, Espeseth T, Reinvang I, Dale AM, Holland D, et al. Cortical gray matter atrophy in healthy aging cannot be explained by undetected incipient cognitive disorders: a comment on Burgmans, et al. (2009) Neuropsychology. 2010;24:258–263. doi: 10.1037/a0018827. discussion 264–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Murphy EA, Holland D, Donohue M, McEvoy LK, Hagler DJ, Jr, Dale AM, et al. Six-month atrophy in MTL structures is associated with subsequent memory decline in elderly controls. Neuroimage. 2010;53:1310–1317. doi: 10.1016/j.neuroimage.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Nettiksimmons J, Harvey D, Brewer J, Carmichael O, Decarli C, Jack CR, Jr, et al. Subtypes based on cerebrospinal fluid and magnetic resonance imaging markers in normal elderly predict cognitive decline. Neurobiol Aging. 2010;31:1419–1428. doi: 10.1016/j.neurobiolaging.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, et al. Brain Atrophy in Healthy Aging Is Related to CSF Levels of A{beta}1–42. Cereb Cortex. 2010;20:2069–2079. doi: 10.1093/cercor/bhp279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mormino EC, Kluth JT, Madison CM, Rabinovici GD, Baker SL, Miller BL, et al. Episodic memory loss is related to hippocampal-mediated beta-amyloid deposition in elderly subjects. Brain. 2009;132(Pt 5):1310–1323. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chiang GC, Insel PS, Tosun D, Schuff N, Truran-Sacrey D, Raptentsetsang ST, et al. Hippocampal atrophy rates and CSF biomarkers in elderly APOE2 normal subjects. Neurology. 2010;75:1976–1981. doi: 10.1212/WNL.0b013e3181ffe4d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fan M, Liu B, Zhou Y, Zhen X, Xu C, Jiang T. Cortical thickness is associated with different apolipoprotein E genotypes in healthy elderly adults. Neurosci Lett. 2010;479:332–336. doi: 10.1016/j.neulet.2010.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ho AJ, Stein JL, Hua X, Lee S, Hibar DP, Leow AD, et al. A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc Natl Acad Sci USA. 2010;107:8404–8409. doi: 10.1073/pnas.0910878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rimol LM, Agartz I, Djurovic S, Brown AA, Roddey JC, Kahler AK, et al. Sex-dependent association of common variants of microcephaly genes with brain structure. Proc Natl Acad Sci USA. 2010;107:384–388. doi: 10.1073/pnas.0908454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Joyner AH, Roddey JC, Bloss CS, Bakken TE, Rimol LM, Melle I, et al. A common MECP2 haplotype associates with reduced cortical surface area in humans in two independent populations. Proc Natl Acad Sci USA. 2009;106:15483–15488. doi: 10.1073/pnas.0901866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Frisoni GB. Alzheimer’s disease neuroimaging Initiative in Europe. Alzheimers Dement. 2010;6:280–285. doi: 10.1016/j.jalz.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 213.Frisoni GB, Henneman WJ, Weiner MW, Scheltens P, Vellas B, Reynish E, et al. The pilot European Alzheimer’s Disease Neuroimaging Initiative of the European Alzheimer’s Disease Consortium. Alzheimers Dement. 2008;4:255–264. doi: 10.1016/j.jalz.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Buerger K, Frisoni G, Uspenskaya O, Ewers M, Zetterberg H, Geroldi C, et al. Validation of Alzheimer’s disease CSF and plasma biological markers: the multicentre reliability study of the pilot European Alzheimer’s Disease Neuroimaging Initiative (E-ADNI) Exp Gerontol. 2009;44:579–585. doi: 10.1016/j.exger.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 215.Mattsson N, Andreasson U, Persson S, Arai H, Batish SD, Bernardini S, et al. The Alzheimer’s Association external quality control program for cerebrospinal fluid biomarkers. Alzheimers Dement. 2011;7:386–3956. doi: 10.1016/j.jalz.2011.05.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Frisoni GB, Jack CR. Harmonization of magnetic resonance-based manual hippocampal segmentation: a mandatory step for wide clinical use. Alzheimers Dement. 2011;7:171–174. doi: 10.1016/j.jalz.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 217.Ellis KA, Rowe CC, Villemagne VL, Martins RN, Masters CL, Salvado O, et al. Addressing population aging and Alzheimer’s disease through the Australian imaging biomarkers and lifestyle study: collaboration with the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement. 2010;6:291–296. doi: 10.1016/j.jalz.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 218.Rowe CC, Ellis KA, Rimajova M, Bourgeat P, Pike KE, Jones G, et al. Amyloid imaging results from the Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging. Neurobiol Aging. 2010;31:1275–1283. doi: 10.1016/j.neurobiolaging.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 219.Fukuyama H. Neuroimaging in mild cognitive impairment,. [in Japanese] Rinsho Shinkeigaku. 2006;46:791–794. [PubMed] [Google Scholar]
- 220.Iwatsubo T. Japanese ADNI: present status and future. Alzheimers Dement. 2010;6:297–299. doi: 10.1016/j.jalz.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 221.Arai H. Alzheimer’s disease neuroimaging initiative and mild cognitive impairment,. [in Japanese] Rinsho Shinkeigaku. 2007;47:905–907. [PubMed] [Google Scholar]
- 222.Ihara Y. Overview on Alzheimer’s disease,. [in Japanese] Rinsho Shinkeigaku. 2007;47:902–904. [PubMed] [Google Scholar]
- 223.Murayam S, Saito Y. Neuropathology of mild cognitive impairment Alzheimer’s disease,. [in Japanese] Rinsho Shinkeigaku. 2007;47:912–914. [PubMed] [Google Scholar]
- 224.Arai H, Okamura N, Furukawa K, Kudo Y. Geriatric medicine, Japanese Alzheimer’s disease neuroimaging initiative and biomarker development. Tohoku J Exp Med. 2010;221:87–95. doi: 10.1620/tjem.221.87. [DOI] [PubMed] [Google Scholar]
- 225.Li Y, Wang Y, Wu G, Shi F, Zhou L, Lin W, et al. Discriminant analysis of longitudinal cortical thickness changes in Alzheimer’s disease using dynamic and network features. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2010.11.008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Heckemann RA, et al. Automatic morphometry in Alzheimer’s disease and mild cognitive impairment. Neuroimage. 2011;56:2024–2037. doi: 10.1016/j.neuroimage.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Romero K, Corrigan B, Neville J, Kopko S, Cantillon M. Striving for an integrated drug development process for neurodegeneration: The Coalition Against Major Diseases. Neurodegen Dis Manage. 2011;1:379–385. [Google Scholar]
- 228.Kang JH, Vanderstichele H, Trojanowski JQ, Shaw LM. Simultaneous analysis of cerebrospinal fluid biomarkers using microsphere-based xMAP multiplex technology for early detection of Alzheimer’s disease. Methods. 2012;56:484–493. doi: 10.1016/j.ymeth.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 229.Li X, Long X, Laurienti P, Wyatt C. Registration of images with varying topology using embedded maps. IEEE Trans Med Imaging. 2012;31:749–765. doi: 10.1109/TMI.2011.2178609. [DOI] [PubMed] [Google Scholar]
- 230.Leung KK, Ridgway GR, Ourselin S, Fox NC. Consistent multi-time-point brain atrophy estimation from the boundary shift integral. Neuroimage. 2012;59:3995–4005. doi: 10.1016/j.neuroimage.2011.10.068. [DOI] [PubMed] [Google Scholar]
- 231.Lotjonen J, Wolz R, Koikkalainen J, Julkunen V, Thurfjell L, Lundqvist R, et al. Fast and robust extraction of hippocampus from MR images for diagnostics of Alzheimer’s disease. Neuroimage. 2011;56:185–196. doi: 10.1016/j.neuroimage.2011.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Pachauri D, Hinrichs C, Chung MK, Johnson SC, Singh V. Topology-based kernels with application to inference problems in Alzheimer’s disease. IEEE Trans Med Imaging. 2011;30:1760–1770. doi: 10.1109/TMI.2011.2147327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Cardoso MJ, Clarkson MJ, Ridgway GR, Modat M, Fox NC, Ourselin S. LoAd: a locally adaptive cortical segmentation algorithm. Neuroimage. 2011;56:1386–1397. doi: 10.1016/j.neuroimage.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Koikkalainen J, Lotjonen J, Thurfjell L, Rueckert D, Waldemar G, Soininen H. Multi-template tensor-based morphometry: application to analysis of Alzheimer’s disease. Neuroimage. 2011;56:1134–1144. doi: 10.1016/j.neuroimage.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Cover KS, van Schijndel RA, van Dijk BW, Redolfi A, Knol DL, Frisoni GB, et al. Assessing the reproducibility of the SienaX and Siena brain atrophy measures using the ADNI back-to-back MP-RAGE MRI scans. Psychiatry Res. 2011;193:182–190. doi: 10.1016/j.pscychresns.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 236.Shen KK, Fripp J, Meriaudeau F, Chetelat G, Salvado O, Bourgeat P. Detecting global and local hippocampal shape changes in Alzheimer’s disease using statistical shape models. Neuroimage. 2012;59:2155–2166. doi: 10.1016/j.neuroimage.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 237.Cho Y, Seong JK, Jeong Y, Shin SY. Individual subject classification for Alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. Neuroimage. 2012;59:2217–2230. doi: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Abdulkadir A, Mortamet B, Vemuri P, Jack CR, Jr, Krueger G, Kloppel S. Effects of hardware heterogeneity on the performance of SVM Alzheimer’s disease classifier. Neuroimage. 2011;58:785–792. doi: 10.1016/j.neuroimage.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhang D, Shen D. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS One. 2012;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Aksu Y, Miller DJ, Kesidis G, Bigler DC, Yang QX. An MRI-derived definition of MCI-to-AD conversion for long-term, automatic prognosis of MCI patients. PLoS One. 2011;6:e25074. doi: 10.1371/journal.pone.0025074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Casanova R, Whitlow CT, Wagner B, Williamson J, Shumaker SA, Maldjian JA, et al. High dimensional classification of structural MRI Alzheimer’s disease data based on large scale regularization. Front Neuroinform. 2011;5:22. doi: 10.3389/fninf.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Clark DG. Residual vectors for Alzheimer disease diagnosis and prognostication. Brain Behav. 2011;1:142–152. doi: 10.1002/brb3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Markiewicz PJ, Matthews JC, Declerck J, Herholz K. Robustness of multivariate image analysis assessed by resampling techniques and applied to FDG-PET scans of patients with Alzheimer’s disease. Neuroimage. 2009;46:472–485. doi: 10.1016/j.neuroimage.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 244.Markiewicz PJ, Matthews JC, Declerck J, Herholz K. Verification of predicted robustness and accuracy of multivariate analysis. Neuroimage. 2011;56:1382–1385. doi: 10.1016/j.neuroimage.2011.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rasmussen JM, Lakatos A, van Erp TG, Kruggel F, Keator DB, Fallon JT, et al. Empirical derivation of the reference region for computing diagnostic sensitive (1)fluorodeoxyglucose ratios in Alzheimer’s disease based on the ADNI sample. Biochim Biophys Acta. 2012;1822:457–466. doi: 10.1016/j.bbadis.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tractenberg RE, Pietrzak RH. Intra-individual variability in Alzheimer’s disease and cognitive aging: definitions, context, and effect sizes. PLoS One. 2011;6:e16973. doi: 10.1371/journal.pone.0016973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuroimage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Padilla P, Lopez M, Gorriz JM, Ramirez J, Salas-Gonzalez D, Alvarez I. NMF-SVM based CAD tool applied to functional brain images for the diagnosis of Alzheimer’s disease. IEEE Trans Med Imaging. 2012;31:207–216. doi: 10.1109/TMI.2011.2167628. [DOI] [PubMed] [Google Scholar]
- 249.Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. Neuroimage. 2012;59:895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wang H, Nie F, Huang H, Risacher S, Saykin A, Ding C, Shen L. ADNI*, Sparse Multi-Task Regression and Feature Selection to Identify Brain Imaging Predictors for Memory Performance; IEEE Conference on Computer Vision; 2011. pp. 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Mattila J, Koikkalainen J, Virkki A, Simonsen A, van Gils M, Waldemar G, et al. A disease state fingerprint for evaluation of Alzheimer’s disease. J Alzheimers Dis. 2011;27:163–176. doi: 10.3233/JAD-2011-110365. [DOI] [PubMed] [Google Scholar]
- 253.Soininen H, Mattila J, Koikkalainen J, van Gils M, Hviid Simonsen A, Waldemar G, et al. Software tool for improved prediction of Alzheimer’s disease. Neurodegener Dis. 2012;10:149–152. doi: 10.1159/000332600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Mayeux R, Schupf N. Blood-based biomarkers for Alzheimer’s disease: plasma Abeta40 and Abeta42, and genetic variants. Neurobiol Aging. 2011;32(Suppl 1):S10–S19. doi: 10.1016/j.neurobiolaging.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.O’Bryant SE, Xiao G, Barber R, Huebinger R, Wilhelmsen K, Edwards M, et al. A blood-based screening tool for Alzheimer’s disease that spans serum and plasma: findings from TARC and ADNI. PLoS One. 2011;6:e28092. doi: 10.1371/journal.pone.0028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Figurski MJ, Waligorska T, Toledo J, Vanderstichele H, Korecka M, Lee VM, et al. Improved protocol for measurement of plasma beta-amyloid in longitudinal evaluation of Alzheimer’s Disease Neuroimaging Initiative study patients. Alzheimers Dement. 2012;8:250–260. doi: 10.1016/j.jalz.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Toledo JB, Vanderstichele H, Figurski M, Aisen PS, Petersen RC, Weiner MW, et al. Factors affecting A beta plasma levels and their utility as biomarkers in ADNI. Acta Neuropathol. 2011;122:401–413. doi: 10.1007/s00401-011-0861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Rissman RA, Trojanowski JQ, Shaw LM, Aisen PS. Longitudinal plasma amyloid beta as a biomarker of Alzheimer’s disease. J Neural Transm. 2012;119:843–850. doi: 10.1007/s00702-012-0772-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Soares HD, Potter WZ, Pickering E, Kuhn M, Immermann FW, Shera DM, et al. Plasma Biomarkers Associated With the Apolipoprotein E Genotype and Alzheimer Disease. Arch Neurol. 2012:1–8. doi: 10.1001/archneurol.2012.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hu WT, Holtzman DM, Fagan AM, Shaw LM, Perrin R, Arnold SE, et al. Plasma multianalyte profiling in mild cognitive impairment and Alzheimer disease. Neurology. 2012 doi: 10.1212/WNL.0b013e318266fa70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Dukart J, Schroeter ML, Mueller K. Age correction in dementia–matching to a healthy brain. PLoS One. 2011;6:e22193. doi: 10.1371/journal.pone.0022193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Samtani MN, Farnum M, Lobanov V, Yang E, Raghavan N, Dibernardo A, et al. An improved model for disease progression in patients from the Alzheimer’s disease neuroimaging initiative. J Clin Pharmacol. 2012;52:629–644. doi: 10.1177/0091270011405497. [DOI] [PubMed] [Google Scholar]
- 263.Meda SA, Narayanan B, Liu J, Perrone-Bizzozero NI, Stevens MC, Calhoun VD, et al. A large scale multivariate parallel ICA method reveals novel imaging-genetic relationships for Alzheimer’s disease in the ADNI cohort. Neuroimage. 2012;60:1608–1621. doi: 10.1016/j.neuroimage.2011.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wang H, Nie F, Huang H, Kim S, Nho K, Risacher SL, et al. Identifying quantitative trait loci via group-sparse multitask regression and feature selection: an imaging genetics study of the ADNI cohort. Bioinformatics. 2012;28:229–237. doi: 10.1093/bioinformatics/btr649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Spiegel R, Berres M, Miserez AR, Monsch AU. For debate: substituting placebo controls in long-term Alzheimer’s prevention trials. Alzheimers Res Ther. 2011;3:9. doi: 10.1186/alzrt68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Donohue MC, Gamst AC, Thomas RG, Xu R, Beckett L, Petersen RC, et al. The relative efficiency of time-to-threshold and rate of change in longitudinal data. Contemp Clin Trials. 2011;32:685–693. doi: 10.1016/j.cct.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Greene SJ, Killiany RJ. Hippocampal subregions are differentially affected in the progression to Alzheimer’s disease. Anat Rec (Hoboken) 2012;295:132–140. doi: 10.1002/ar.21493. [DOI] [PubMed] [Google Scholar]
- 268.Spampinato MV, Rumboldt Z, Hosker RJ, Mintzer JE. Apolipoprotein E and gray matter volume loss in patients with mild cognitive impairment and Alzheimer disease. Radiology. 2011;258:843–852. doi: 10.1148/radiol.10100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Skup M, Zhu H, Wang Y, Giovanello KS, Lin JA, Shen D, et al. Sex differences in grey matter atrophy patterns among AD and aMCI patients: results from ADNI. Neuroimage. 2011;56:890–906. doi: 10.1016/j.neuroimage.2011.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Stricker NH, Chang YL, Fennema-Notestine C, Delano-Wood L, Salmon DP, Bondi MW, et al. Distinct profiles of brain and cognitive changes in the very old with Alzheimer disease. Neurology. 2011;77:713–721. doi: 10.1212/WNL.0b013e31822b0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Poulin SP, Dautoff R, Morris JC, Barrett LF, Dickerson BC. Amygdala atrophy is prominent in early Alzheimer’s disease and relates to symptom severity. Psychiatry Res. 2011;194:7–13. doi: 10.1016/j.pscychresns.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zhang N, Song X, Zhang Y, Chen W, D’Arcy RC, Darvesh S, et al. An MRI brain atrophy and lesion index to assess the progression of structural changes in Alzheimer’s disease, mild cognitive impairment, and normal aging: a follow-up study. J Alzheimers Dis. 2011;26(Suppl 3):359–367. doi: 10.3233/JAD-2011-0048. [DOI] [PubMed] [Google Scholar]
- 273.Rajagopalan P, Hua X, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Homocysteine effects on brain volumes mapped in 732 elderly individuals. Neuroreport. 2011;22:391–395. doi: 10.1097/WNR.0b013e328346bf85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Brown PJ, Devanand DP, Liu X, Caccappolo E. Functional impairment in elderly patients with mild cognitive impairment and mild Alzheimer disease. Arch Gen Psychiatry. 2011;68:617–626. doi: 10.1001/archgenpsychiatry.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Bonner-Jackson A, Okonkwo O, Tremont G. Apolipoprotein E epsilon2 and functional decline in amnestic mild cognitive impairment and Alzheimer disease. Am J Geriatr Psychiatry. 2012;20:584–593. doi: 10.1097/JGP.0b013e3182203c32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Marshall GA, Olson LE, Frey MT, Maye J, Becker JA, Rentz DM. Instrumental activities of daily living impairment is associated with increased amyloid burden. Dement Geriatr Cogn Disord. 2011;31:443–450. doi: 10.1159/000329543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Marshall GA, Rentz DM, Frey MT, Locascio JJ, Johnson KA, Sperling RA. Executive function and instrumental activities of daily living in mild cognitive impairment and Alzheimer’s disease. Alzheimers Dement. 2011;7:300–308. doi: 10.1016/j.jalz.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Vemuri P, Weigand SD, Przybelski SA, Knopman DS, Smith GE, Trojanowski JQ, et al. Cognitive reserve and Alzheimer’s disease biomarkers are independent determinants of cognition. Brain. 2011;134(Pt 5):1479–1492. doi: 10.1093/brain/awr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ewers M, Schmitz S, Hansson O, Walsh C, Fitzpatrick A, Bennett D, et al. Body mass index is associated with biological CSF markers of core brain pathology of Alzheimer’s disease. Neurobiol Aging. 2012;33:1599–1608. doi: 10.1016/j.neurobiolaging.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Vidoni ED, Townley RA, Honea RA, Burns JM. Alzheimer disease biomarkers are associated with body mass index. Neurology. 2011;77:1913–1920. doi: 10.1212/WNL.0b013e318238eec1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Lo RY, Hubbard AE, Shaw LM, Trojanowski JQ, Petersen RC, Aisen PS, et al. Longitudinal change of biomarkers in cognitive decline. Arch Neurol. 2011;68:1257–1266. doi: 10.1001/archneurol.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Jack CR, Jr, Vemuri P, Wiste HJ, Weigand SD, Aisen PS, Trojanowski JQ, et al. Evidence for ordering of Alzheimer disease biomarkers. Arch Neurol. 2011;68:1526–1535. doi: 10.1001/archneurol.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Chincarini A, Bosco P, Calvini P, Gemme G, Esposito M, Olivieri C, et al. Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer’s disease. Neuroimage. 2011;58:469–480. doi: 10.1016/j.neuroimage.2011.05.083. [DOI] [PubMed] [Google Scholar]
- 284.Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D, et al. Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. PLoS One. 2011;6:e25446. doi: 10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Gray KR, Wolz R, Heckemann RA, Aljabar P, Hammers A, Rueckert D. Multi-region analysis of longitudinal FDG-PET for the classification of Alzheimer’s disease. Neuroimage. 2012;60:221–229. doi: 10.1016/j.neuroimage.2011.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Johnstone D, Milward EA, Berretta R, Moscato P. Multivariate protein signatures of pre-clinical Alzheimer’s disease in the Alzheimer’s disease neuroimaging initiative (ADNI) plasma proteome dataset. PLoS One. 2012;7:e34341. doi: 10.1371/journal.pone.0034341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Cui Y, Liu B, Luo S, Zhen X, Fan M, Liu T, et al. Identification of conversion from mild cognitive impairment to Alzheimer’s disease using multivariate predictors. PLoS One. 2011;6:e21896. doi: 10.1371/journal.pone.0021896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Schmand B, Eikelenboom P, van Gool WA. Value of neuropsychological tests, neuroimaging, and biomarkers for diagnosing Alzheimer’s disease in younger and older age cohorts. J Am Geriatr Soc. 2011;59:1705–1710. doi: 10.1111/j.1532-5415.2011.03539.x. [DOI] [PubMed] [Google Scholar]
- 289.Dickerson BC, Wolk DA. MRI cortical thickness biomarker predicts AD-like CSF and cognitive decline in normal adults. Neurology. 2012;78:84–90. doi: 10.1212/WNL.0b013e31823efc6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Chiang GC, Insel PS, Tosun D, Schuff N, Truran-Sacrey D, Raptentsetsang S, et al. Identifying cognitively healthy elderly individuals with subsequent memory decline by using automated MR temporoparietal volumes. Radiology. 2011;259:844–851. doi: 10.1148/radiol.11101637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Herholz K, Westwood S, Haense C, Dunn G. Evaluation of a calibrated (18)F-FDG PET score as a biomarker for progression in Alzheimer disease and mild cognitive impairment. J Nucl Med. 2011;52:1218–1226. doi: 10.2967/jnumed.111.090902. [DOI] [PubMed] [Google Scholar]
- 292.Mackin RS, Insel P, Aisen PS, Geda YE, Weiner MW. Longitudinal stability of subsyndromal symptoms of depression in individuals with mild cognitive impairment: relationship to conversion to dementia after 3 years. Int J Geriatr Psychiatry. 2012;27:355–363. doi: 10.1002/gps.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Lee GJ, Lu PH, Hua X, Lee S, Wu S, Nguyen K, et al. Depressive symptoms in mild cognitive impairment predict greater atrophy in Alzheimer’s disease-related regions. Biol Psychiatry. 2012;71:814–821. doi: 10.1016/j.biopsych.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Gomar JJ, Bobes-Bascaran MT, Conejero-Goldberg C, Davies P, Goldberg TE. Utility of combinations of biomarkers, cognitive markers, and risk factors to predict conversion from mild cognitive impairment to Alzheimer disease in patients in the Alzheimer’s disease neuroimaging initiative. Arch Gen Psychiatry. 2011;68:961–969. doi: 10.1001/archgenpsychiatry.2011.96. [DOI] [PubMed] [Google Scholar]
- 295.Heister D, Brewer JB, Magda S, Blennow K, McEvoy LK. Predicting MCI outcome with clinically available MRI and CSF biomarkers. Neurology. 2011;77:1619–1628. doi: 10.1212/WNL.0b013e3182343314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Devanand DP, Liu X, Brown PJ, Huey ED, Stern Y, Pelton GH. A two-study comparison of clinical and MRI markers of transition from mild cognitive impairment to Alzheimer’s disease. Int J Alzheimers Dis. 2012;2012:483469. doi: 10.1155/2012/483469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Holland D, McEvoy LK, Dale AM. Unbiased comparison of sample size estimates from longitudinal structural measures in ADNI. Hum Brain Mapp. 2012;33:2586–2602. doi: 10.1002/hbm.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Schrag A, Schott JM. What is the clinically relevant change on the ADAS-Cog? J Neurol Neurosurg Psychiatry. 2012;83:171–173. doi: 10.1136/jnnp-2011-300881. [DOI] [PubMed] [Google Scholar]
- 299.Henley DB, Sundell KL, Sethuraman G, Siemers ER. Safety profile of Alzheimer’s disease populations in Alzheimer’s Disease Neuroimaging Initiative and other 18-month studies. Alzheimers Dement. 2012;8:407–416. doi: 10.1016/j.jalz.2011.05.2413. [DOI] [PubMed] [Google Scholar]
- 300.Thompson WK, Hallmayer J, O’Hara R. Design considerations for characterizing psychiatric trajectories across the lifespan: application to effects of APOE-epsilon4 on cerebral cortical thickness in Alzheimer’s disease. Am J Psychiatry. 2011;168:894–903. doi: 10.1176/appi.ajp.2011.10111690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Erten-Lyons D, Wilmot B, Anur P, McWeeney S, Westaway SK, Silbert L, et al. Microcephaly genes and risk of late-onset Alzheimer disease. Alzheimer Dis Assoc Disord. 2011;25:276–282. doi: 10.1097/WAD.0b013e31820a1d32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Murphy EA, Roddey JC, McEvoy LK, Holland D, Hagler DJ, Jr, Dale AM, et al. CETP polymorphisms associate with brain structure, atrophy rate, and Alzheimer’s disease risk in an APOE-dependent manner. Brain Imaging Behav. 2012;6:16–26. doi: 10.1007/s11682-011-9137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Xu C, Wang Z, Fan M, Liu B, Song M, Zhen X, et al. Effects of BDNF Val66Met polymorphism on brain metabolism in Alzheimer’s disease. Neuroreport. 2010;21:802–807. doi: 10.1097/WNR.0b013e32833ccaf4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Kamboh MI, Barmada MM, Demirci FY, Minster RL, Carrasquillo MM, Pankratz VS, et al. Genome-wide association analysis of age-at-onset in Alzheimer’s disease. Mol Psychiatry. 2012;17:1340–1346. doi: 10.1038/mp.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Hibar DP, Stein JL, Kohannim O, Jahanshad N, Saykin AJ, Shen L, et al. Voxelwise gene-wide association study (vGeneWAS): multivariate gene-based association testing in 731 elderly subjects. Neuroimage. 2011;56:1875–1891. doi: 10.1016/j.neuroimage.2011.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Schott JM. Using CSF biomarkers to replicate genetic associations in Alzheimer’s disease. Neurobiol Aging. 2012;33(7):1486.e9–1486.e15. doi: 10.1016/j.neurobiolaging.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Swaminathan S, Shen L, Risacher SL, Yoder KK, West JD, Kim S, et al. Amyloid pathway-based candidate gene analysis of [(11)C] PiB-PET in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. Brain Imaging Behav. 2012;6:1–15. doi: 10.1007/s11682-011-9136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Hu X, Pickering E, Liu YC, Hall S, Fournier H, Katz E, et al. Meta-analysis for genome-wide association study identifies multiple variants at the BIN1 locus associated with late-onset Alzheimer’s disease. PLoS One. 2011;6:e16616. doi: 10.1371/journal.pone.0016616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Antunez C, Boada M, Gonzalez-Perez A, Gayan J, Ramirez-Lorca R, Marin J, et al. Genetic association of complement receptor 1 polymorphism rs3818361 in Alzheimer’s disease. Alzheimers Dement. 2011;7:e124–e129. doi: 10.1016/j.jalz.2011.05.2412. [DOI] [PubMed] [Google Scholar]
- 310.Cruchaga C, Nowotny P, Kauwe JS, Ridge PG, Mayo K, Bertelsen S, et al. Association and expression analyses with single-nucleotide polymorphisms in TOMM40 in Alzheimer disease. Arch Neurol. 2011;68:1013–1019. doi: 10.1001/archneurol.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Antunez C, Boada M, Gonzalez-Perez A, Gayan J, Ramirez-Lorca R, Marin J, et al. The membrane-spanning 4-domains, subfamily A (MS4A) gene cluster contains a common variant associated with Alzheimer’s disease. Genome Med. 2011;3:33. doi: 10.1186/gm249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Kauwe JS, Cruchaga C, Karch CM, Sadler B, Lee M, Mayo K, et al. Fine mapping of genetic variants in BIN1, CLU, CR1 and PICALM for association with cerebrospinal fluid biomarkers for Alzheimer’s disease. PLoS One. 2011;6:e15918. doi: 10.1371/journal.pone.0015918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Alexopoulos P, Guo LH, Kratzer M, Westerteicher C, Kurz A, Perneczky R. Impact of SORL1 single nucleotide polymorphisms on Alzheimer’s disease cerebrospinal fluid markers. Dement Geriatr Cogn Disord. 2011;32:164–170. doi: 10.1159/000332017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Swaminathan S, Kim S, Shen L, Risacher SL, Foroud T, Pankratz N, et al. Genomic Copy Number Analysis in Alzheimer’s Disease and Mild Cognitive Impairment: An ADNI Study. Int J Alzheimers Dis. 2011;2011:729478. doi: 10.4061/2011/729478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Swaminathan S, Shen L, Kim S, Inlow M, West JD, Faber KM, et al. Analysis of copy number variation in Alzheimer’s disease: the NIALOAD/ NCRAD Family Study. Curr Alzheimer Res. 2012;9:801–814. doi: 10.2174/156720512802455331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Stein JL, Hibar DP, Madsen SK, Khamis M, McMahon KL, de Zubicaray GI, et al. Discovery and replication of dopamine-related gene effects on caudate volume in young and elderly populations (N=1198) using genome-wide search. Mol Psychiatry. 2011;16:927–937. 881. doi: 10.1038/mp.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Bakken TE, Roddey JC, Djurovic S, Akshoomoff N, Amaral DG, Bloss CS, et al. Association of common genetic variants in GPCPD1 with scaling of visual cortical surface area in humans. Proc Natl Acad Sci U S A. 2012;109:3985–3990. doi: 10.1073/pnas.1105829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Yesavage JA, Noda A, Hernandez B, Friedman L, Cheng JJ, Tinklenberg JR, et al. Circadian clock gene polymorphisms and sleep-wake disturbance in Alzheimer disease. Am J Geriatr Psychiatry. 2011;19:635–643. doi: 10.1097/JGP.0b013e31820d92b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Bakken TE, Dale AM, Schork NJ. A geographic cline of skull and brain morphology among individuals of European Ancestry. Hum Hered. 2011;72:35–44. doi: 10.1159/000330168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.David R, Friedman L, Mulin E, Noda A, Le Duff F, Kennedy Q, et al. Lack of Association Between COMT Polymorphisms and Apathy in Alzheimer’s Disease. J Alzheimers Dis. 2011;27:155–161. doi: 10.3233/JAD-2011-110491. [DOI] [PubMed] [Google Scholar]
- 321.Hibar DP, Kohannim O, Stein JL, Chiang MC, Thompson PM. Multilocus genetic analysis of brain images. Front Genet. 2011;2:73. doi: 10.3389/fgene.2011.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Nho K, Shen L, Kim S, Swaminathan S, Risacher SL, Saykin AJ. The effect of reference panels and software tools on genotype imputation. AMIA Annu Symp Proc. 2011;2011:1013–1018. [PMC free article] [PubMed] [Google Scholar]
- 323.Wan J, Kim S, Inlow M, Nho K, Swaminathan S, Risacher SL, et al. Hippocampal surface mapping of genetic risk factors in AD via sparse learning models. Med Image Comput Comput Assist Interv. 2011;14(Pt 2):376–383. doi: 10.1007/978-3-642-23629-7_46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Cruchaga C, Kauwe JS, Nowotny P, Bales K, Pickering EH, Mayo K, et al. Cerebrospinal fluid APOE levels: an endophenotype for genetic studies for Alzheimer’s disease. Hum Mol Genet. 2012;21:4558–4571. doi: 10.1093/hmg/dds296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Damoiseaux JS, Seeley WW, Zhou J, Shirer WR, Coppola G, Karydas A, et al. Gender modulates the APOE epsilon4 effect in healthy older adults: convergent evidence from functional brain connectivity and spinal fluid tau levels. J Neurosci. 2012;32:8254–8262. doi: 10.1523/JNEUROSCI.0305-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.De Jager PL, Shulman JM, Chibnik LB, Keenan BT, Raj T, Wilson RS, et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol Aging. 2012;33:1017.e1–1017.e15. doi: 10.1016/j.neurobiolaging.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Ge T, Feng J, Hibar DP, Thompson PM, Nichols TE. Increasing power for voxel-wise genome-wide association studies: the random field theory, least square kernel machines and fast permutation procedures. Neuroimage. 2012;63:858–873. doi: 10.1016/j.neuroimage.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Hibar DP, Stein JL, Ryles AB, Kohannim O, Jahanshad N, Medland SE, et al. Genome-wide association identifies genetic variants associated with lentiform nucleus volume in N = 1345 young and elderly subjects. Brain Imaging Behav. 2012 doi: 10.1007/s11682-012-9199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Jahanshad N, Kohannim O, Hibar DP, Stein JL, McMahon KL, de Zubicaray GI, et al. Brain structure in healthy adults is related to serum transferrin and the H63D polymorphism in the HFE gene. Proc Natl Acad Sci U S A. 2012;109:E851–E859. doi: 10.1073/pnas.1105543109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Keenan BT, Shulman JM, Chibnik LB, Raj T, Tran D, Sabuncu MR, et al. A coding variant in CR1 interacts with APOE-epsilon4 to influence cognitive decline. Hum Mol Genet. 2012;21:2377–2388. doi: 10.1093/hmg/dds054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Kohannim O, Hibar DP, Jahanshad N, Stein JL, Hua X, Toga AW, et al. Predicting Temporal Lobe Volume on Mri from Genotypes Using L(1)–L(2) Regularized Regression. Proc IEEE Int Symp Biomed Imaging. 2012:1160–1163. doi: 10.1109/ISBI.2012.6235766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Kohannim O, Hibar DP, Stein JL, Jahanshad N, Hua X, Rajagopalan P, et al. Discovery and Replication of Gene Influences on Brain Structure Using LASSO Regression. Front Neurosci. 2012;6:115. doi: 10.3389/fnins.2012.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Lourdusamy A, Newhouse S, Lunnon K, Proitsi P, Powell J, Hodges A, et al. Identification of cis-regulatory variation influencing protein abundance levels in human plasma. Hum Mol Genet. 2012;21:3719–3726. doi: 10.1093/hmg/dds186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Mattila J, Koikkalainen J, Virkki A, van Gils M, Lotjonen J. Design and application of a generic clinical decision support system for multiscale data. IEEE Trans Biomed Eng. 2012;59:234–240. doi: 10.1109/TBME.2011.2170986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Melville SA, Buros J, Parrado AR, Vardarajan B, Logue MW, Shen L, et al. Multiple loci influencing hippocampal degeneration identified by genome scan. Ann Neurol. 2012;72:65–75. doi: 10.1002/ana.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ramanan VK, Kim S, Holohan K, Shen L, Nho K, Risacher SL, et al. Genome-wide pathway analysis of memory impairment in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain Imaging Behav. 2012;6:634–648. doi: 10.1007/s11682-012-9196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Mukherjee S, Kim S, Gibbons LE, Nho K, Risacher SL, Glymour MM, et al. Genetic architecture of resilience of executive functioning. Brain Imaging Behav. 2012;6:621–633. doi: 10.1007/s11682-012-9184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Silver M, Janousova E, Hua X, Thompson PM, Montana G. Identification of gene pathways implicated in Alzheimer’s disease using longitudinal imaging phenotypes with sparse regression. Neuroimage. 2012;63:1681–1694. doi: 10.1016/j.neuroimage.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Silver M, Montana G. Fast identification of biological pathways associated with a quantitative trait using group lasso with overlaps. Stat Appl Genet Mol Biol. 2012;11 doi: 10.2202/1544-6115.1755. Article 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Thambisetty M, An Y, Nalls M, Sojkova J, Swaminathan S, Zhou Y, et al. Effect of Complement CR1 on Brain Amyloid Burden During Aging and Its Modification by APOE Genotype. Biol Psychiatry. 2012;73:334–341. doi: 10.1016/j.biopsych.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Vounou M, Janousova E, Wolz R, Stein JL, Thompson PM, Rueckert D, et al. Sparse reduced-rank regression detects genetic associations with voxel-wise longitudinal phenotypes in Alzheimer’s disease. Neuroimage. 2012;60:700–716. doi: 10.1016/j.neuroimage.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Wang H, Nie F, Huang H, Risacher SL, Saykin AJ, Shen L. Identifying disease sensitive and quantitative trait-relevant biomarkers from multidimensional heterogeneous imaging genetics data via sparse multimodal multitask learning. Bioinformatics. 2012;28:i127–il36. doi: 10.1093/bioinformatics/bts228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Wang H, Nie F, Huang H, Yan J, Kim S, Nho K, et al. From phenotype to genotype: an association study of longitudinal phenotypic markers to Alzheimer’s disease relevant SNPs. Bioinformatics. 2012:28i619–28i625. doi: 10.1093/bioinformatics/bts411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2012;8(1 Suppl):S1–S68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Ye J, Farnum M, Yang E, Verbeeck R, Lobanov V, Raghavan N, et al. Sparse learning and stability selection for predicting MCI to AD conversion using baseline ADNI data. BMC Neurol. 2012;12:46. doi: 10.1186/1471-2377-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Westman E, Simmons A, Muehlboeck JS, Mecocci P, Vellas B, Tsolaki M, et al. AddNeuroMed and ADNI: similar patterns of Alzheimer’s atrophy and automated MRI classification accuracy in Europe and North America. Neuroimage. 2011;58:818–828. doi: 10.1016/j.neuroimage.2011.06.065. [DOI] [PubMed] [Google Scholar]
- 348.Cuingnet R, Rosso C, Chupin M, Lehericy S, Dormont D, Benali H, et al. Spatial regularization of SVM for the detection of diffusion alterations associated with stroke outcome. Med Image Anal. 2011;15:729–737. doi: 10.1016/j.media.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 349.Jennings JR, Mendelson DN, Muldoon MF, Ryan CM, Gianaros PJ, Raz N, et al. Regional grey matter shrinks in hypertensive individuals despite successful lowering of blood pressure. J Hum Hypertens. 2012;26:295–305. doi: 10.1038/jhh.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2012;8(1 Suppl):S1–S68. doi: 10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. Alzheimer’s Disease Neuroimaging. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2013;9:e111–e194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Guo LH, Alexopoulos P, Eisele T, Wagenpfeil S, Kurz A, Perneczky R. The National Institute on Aging-Alzheimer’s Association research criteria for mild cognitive impairment due to Alzheimer’s disease: predicting the outcome. Eur Arch Psychiatry Clin Neurosci. 2012;263:325–333. doi: 10.1007/s00406-012-0349-0. [DOI] [PubMed] [Google Scholar]
- 353.Petersen RC, Aisen P, Boeve BF, Geda YE, Ivnik RJ, Knopman DS, et al. Mild cognitive impairment due to Alzheimer’s disease in the community. Ann Neurol. 2013;74:199–208. doi: 10.1002/ana.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Lowe VJ, Peller PJ, Weigand SD, Montoya Quintero C, Tosakulwong N, Vemuri P, et al. Application of the National Institute on Aging-Alzheimer’s Association AD criteria to ADNI. Neurology. 2013;80:2130–2137. doi: 10.1212/WNL.0b013e318295d6cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Wilhelm EE, Oster E, Shoulson I. Approaches and costs for sharing clinical research data. JAMA. 2014;311:1201–1202. doi: 10.1001/jama.2014.850. [DOI] [PubMed] [Google Scholar]
- 356.Shulman MB, Harkins K, Green RC, Karlawish J. Using AD biomarker research results for clinical care: a survey of ADNI investigators. Neurology. 2013;81:1114–1121. doi: 10.1212/WNL.0b013e3182a55f4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Hill DL, Schwarz AJ, Isaac M, Pani L, Vamvakas S, Hemmings R, et al. Coalition Against Major Diseases/European Medicines Agency biomarker qualification of hippocampal volume for enrichment of clinical trials in predementia stages of Alzheimer’s disease. Alzheimers Dement. 2014;10:421–4293. doi: 10.1016/j.jalz.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 358.Khan AF, Drozd JJ, Moreland RK, Ta RM, Borrie MJ, Bartha R. A novel MRI-compatible brain ventricle phantom for validation of segmentation and volumetry methods. J Magn Reson Imaging. 2012;36:476–482. doi: 10.1002/jmri.23612. [DOI] [PubMed] [Google Scholar]
- 359.Marchewka A, Kherif F, Krueger G, Grabowska A, Frackowiak R, Draganski B, et al. Disease Neuroimaging, Influence of magnetic field strength and image registration strategy on voxel-based morphometry in a study of Alzheimer’s disease. Hum Brain Mapp. 2013;35:1865–1874. doi: 10.1002/hbm.22297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Vuong P, Drucker D, Schwarz C, Fletcher E, Decarli C, Carmichael O. Effects of T2-weighted MRI based cranial volume measurements on studies of the aging brain. Proc Soc Photo Opt Instrum Eng. 2013;8669 doi: 10.1117/12.2006727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Boccardi M, Bocchetta M, Apostolova LG, Preboske G, Robitaille N, Pasqualetti P, et al. Alzheimer’s Disease Neuroimaging. Establishing magnetic resonance images orientation for the EADC-ADNI manual hippocampal segmentation protocol. J Neuroimaging. 2014 doi: 10.1111/jon.12065. [DOI] [PubMed] [Google Scholar]
- 362.Nestor SM, Gibson E, Gao FQ, Kiss A, Black SE. Alzheimer’s Disease Neuroimaging. A direct morphometric comparison of five labeling protocols for multi-atlas driven automatic segmentation of the hippocampus in Alzheimer’s disease. Neuroimage. 2012;66C:50–70. doi: 10.1016/j.neuroimage.2012.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Rasmussen JM, Lakatos A, van Erp TG, Kruggel F, Keator DB, Fallon JT, et al. Alzheimer’s Disease Neuroimaging. Empirical derivation of the reference region for computing diagnostic sensitive (1)(8)fluorodeoxyglucose ratios in Alzheimer’s disease based on the ADNI sample. Biochim Biophys Acta. 2012;1822:457–466. doi: 10.1016/j.bbadis.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Landau SM, Breault C, Joshi AD, Pontecorvo M, Mathis CA, Jagust WJ, et al. Alzheimer’s Disease Neuroimaging. Amyloid-beta imaging with Pittsburgh compound B and florbetapir: comparing radiotracers and quantification methods. J Nucl Med. 2013;54:70–77. doi: 10.2967/jnumed.112.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Korecka M, Waligorska T, Figurski M, Toledo JB, Arnold SE, Grossman M, et al. Qualification of a surrogate matrix-based absolute quantification method for amyloid-beta (4)(2) in human cerebrospinal fluid using 2D UPLC-tandem mass spectrometry. J Alzheimers Dis. 2014;41:441–451. doi: 10.3233/JAD-132489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Kang JH, Korecka M, Toledo JB, Trojanowski JQ, Shaw LM. Clinical utility and analytical challenges in measurement of cerebrospinal fluid amyloid-beta(1–42) and tau proteins as Alzheimer disease biomarkers. Clin Chem. 2013;59:903–916. doi: 10.1373/clinchem.2013.202937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Hoang Duc AK, Modat M, Leung KK, Cardoso MJ, Barnes J, Kadir T, et al. Alzheimer’s Disease Neuroimaging. Using manifold learning for atlas selection in multi-atlas segmentation. PLoS One. 2013;8:e70059. doi: 10.1371/journal.pone.0070059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Ribbens A, Hermans J, Maes F, Vandermeulen D, Suetens P. Unsupervised segmentation, clustering and groupwise registration of heterogeneous populations of brain MR images. IEEE Trans Med Imaging. 2014;33:201–224. doi: 10.1109/TMI.2013.2270114. [DOI] [PubMed] [Google Scholar]
- 369.Cataldo R, Agrusti A, De Nunzio G, Carla A, De Mitri I, Favetta M, et al. Generating a minimal set of templates for the hippocampal region in MR neuroimages. J Neuroimaging. 2013;23:473–483. doi: 10.1111/j.1552-6569.2012.00713.x. [DOI] [PubMed] [Google Scholar]
- 370.Lorenzi M, Ayache N, Frisoni GB, Pennec X. Alzheimer’s Disease Neuroimaging. LCC-Demons: a robust and accurate symmetric diffeomorphic registration algorithm. Neuroimage. 2013;81:470–483. doi: 10.1016/j.neuroimage.2013.04.114. [DOI] [PubMed] [Google Scholar]
- 371.Robitaille N, Mouiha A, Crepeault B, Valdivia F, Duchesne S. Alzheimer’s Disease Neuroimaging. Tissue-based MRI intensity standardization: application to multicentric datasets. Int J Biomed Imaging. 2012;2012:347120. doi: 10.1155/2012/347120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Tong T, Wolz R, Coupe P, Hajnal JV, Rueckert D. Alzheimer’s Disease Neuroimaging. Segmentation of MR images via discriminative dictionary learning and sparse coding: application to hippocampus labeling. Neuroimage. 2013;76:11–23. doi: 10.1016/j.neuroimage.2013.02.069. [DOI] [PubMed] [Google Scholar]
- 373.Jorge Cardoso M, Leung K, Modat M, Keihaninejad S, Cash D, Barnes J, et al. Alzheimer’s Disease Neuroimaging. STEPS: Similarity and Truth Estimation for Propagated Segmentations and its application to hippocampal segmentation and brain parcelation. Med Image Anal. 2013;17:671–684. doi: 10.1016/j.media.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 374.Shi J, Thompson PM, Gutman B, Wang Y. Alzheimer’s Disease Neuroimaging. Surface fluid registration of conformal representation: application to detect disease burden and genetic influence on hippocampus. Neuroimage. 2013;78:111–134. doi: 10.1016/j.neuroimage.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Wang L, Shi F, Li G, Shen D. 4D segmentation of brain MR images with constrained cortical thickness variation. PLoS One. 2013;8:e64207. doi: 10.1371/journal.pone.0064207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Li G, Nie J, Wu G, Wang Y, Shen D. Alzheimer’s Disease Neuroimaging. Consistent reconstruction of cortical surfaces from longitudinal brain MR images. Neuroimage. 2012;59:3805–3820. doi: 10.1016/j.neuroimage.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Augustinack JC, Huber KE, Stevens AA, Roy M, Frosch MP, van der Kouwe AJ, et al. Predicting the location of human perirhinal cortex, Brodmann’s area 35, from MRI. Neuroimage. 2013;64:32–42. doi: 10.1016/j.neuroimage.2012.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Shi Y, Lai R, Toga AW. Alzheimer’s Disease Neuroimaging. Cortical surface reconstruction via unified Reeb analysis of geometric and topological outliers in magnetic resonance images. IEEE Trans Med Imaging. 2013;32(3):511–530. doi: 10.1109/TMI.2012.2224879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Park H. ISOMAP induced manifold embedding and its application to Alzheimer’s disease and mild cognitive impairment. Neurosci Lett. 2012;513:141–145. doi: 10.1016/j.neulet.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 380.Skup M, Zhu H, Zhang H. Multiscale adaptive marginal analysis of longitudinal neuroimaging data with time-varying covariates. Biometrics. 2012;68:1083–1092. doi: 10.1111/j.1541-0420.2012.01767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Bernal-Rusiel JL, Reuter M, Greve DN, Fischl B, Sabuncu MR. Alzheimer’s Disease Neuroimaging. Spatiotemporal linear mixed effects modeling for the mass-univariate analysis of longitudinal neuroimage data. Neuroimage. 2013;81:358–370. doi: 10.1016/j.neuroimage.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Spulber G, Simmons A, Muehlboeck JS, Mecocci P, Vellas B, Tsolaki M, et al. Alzheimer Disease Neuroimaging. An MRI-based index to measure the severity of Alzheimer’s disease-like structural pattern in subjects with mild cognitive impairment. J Intern Med. 2013;273:396–409. doi: 10.1111/joim.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Park H, Yang JJ, Seo J, Lee JM. Dimensionality reduced cortical features and their use in predicting longitudinal changes in Alzheimer’s disease. Neurosci Lett. 2013;550:17–22. doi: 10.1016/j.neulet.2013.06.042. [DOI] [PubMed] [Google Scholar]
- 384.Wee CY, Yap PT, Shen D. Alzheimer Disease Neuroimaging. Prediction of Alzheimer’s disease and mild cognitive impairment using cortical morphological patterns. Hum Brain Mapp. 2013;34:3411–3425. doi: 10.1002/hbm.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Cash DM, Melbourne A, Modat M, Cardoso MJ, Clarkson MJ, Fox NC, et al. Cortical folding analysis on patients with Alzheimer’s disease and mild cognitive impairment. Med Image Comput Comput Assist Interv. 2012;15(Pt 3):289–296. doi: 10.1007/978-3-642-33454-2_36. [DOI] [PubMed] [Google Scholar]
- 386.Eskildsen SF, Coupe P, Garcia-Lorenzo D, Fonov V, Pruessner JC, Collins DL. Alzheimer’s Disease Neuroimaging. Prediction of Alzheimer’s disease in subjects with mild cognitive impairment from the ADNI cohort using patterns of cortical thinning. Neuroimage. 2013;65:511–521. doi: 10.1016/j.neuroimage.2012.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Suk HI, Lee SW, Shen D. Alzheimer’s Disease Neuroimaging. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Struct Funct. 2013 doi: 10.1007/s00429-013-0687-3. [Epub ahead of print]PMID: 24363140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Liu M, Zhang D, Shen D. Alzheimer’s Disease Neuroimaging. Identifying informative imaging biomarkers via tree structured sparse learning for AD diagnosis. Neuroinformatics. 2014;12:381–394. doi: 10.1007/s12021-013-9218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Cuingnet R, Glaunes JA, Chupin M, Benali H, Colliot O. Spatial and anatomical regularization of SVM: a general framework for neuroimaging data. IEEE Trans Pattern Anal Mach Intell. 2012 doi: 10.1109/TPAMI.2012.142. [Epub ahead of print] PMID: 22732664. [DOI] [PubMed] [Google Scholar]
- 390.Chu C, Hsu AL, Chou KH, Bandettini P, Lin C. Alzheimer’s Disease Neuroimaging. Does feature selection improve classification accuracy? Impact of sample size and feature selection on classification using anatomical magnetic resonance images. Neuroimage. 2012;60:59–70. doi: 10.1016/j.neuroimage.2011.11.066. [DOI] [PubMed] [Google Scholar]
- 391.Liu M, Zhang D, Yap PT, Shen D. Tree-guided sparse coding for brain disease classification. Med Image Comput Comput Assist Interv. 2012;15(Pt 3):239–247. doi: 10.1007/978-3-642-33454-2_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Liu F, Zhou L, Shen C, Yin J. Multiple kernel learning in the primal for multi-modal Alzheimer’s disease classification. IEEE J Biomed Health Inform. 2014;18:984–990. doi: 10.1109/JBHI.2013.2285378. [DOI] [PubMed] [Google Scholar]
- 393.Cheng B, Zhang D, Chen S, Kaufer DI, Shen D. Alzheimer’s Disease Neuroimaging. Semi-supervised multimodal relevance vector regression improves cognitive performance estimation from imaging and biological biomarkers. Neuroinformatics. 2013;11:339–353. doi: 10.1007/s12021-013-9180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Zhang D, Liu J, Shen D. Temporally-constrained group sparse learning for longitudinal data analysis. Med Image Comput Comput Assist Interv. 2012;15(Pt 3):264–271. doi: 10.1007/978-3-642-33454-2_33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Casanova R, Hsu FC, Espeland MA. Alzheimer’s Disease Neuroimaging. Classification of structural MRI images in Alzheimer’s disease from the perspective of ill-posed problems. PLoS One. 2012;7:e44877. doi: 10.1371/journal.pone.0044877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Liu X, Tosun D, Weiner MW, Schuff N. Alzheimer’s Disease Neuroimaging. Locally linear embedding (LLE) for MRI based Alzheimer’s disease classification. Neuroimage. 2013;83C:148–157. doi: 10.1016/j.neuroimage.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Arbizu J, Prieto E, Martinez-Lage P, Marti-Climent JM, Garcia-Granero M, Lamet I, et al. Alzheimer Disease Neuroimaging. Automated analysis of FDG PET as a tool for single-subject probabilistic prediction and detection of Alzheimer’s disease dementia. Eur J Nucl Med Mol Imaging. 2013;40:1394–1405. doi: 10.1007/s00259-013-2458-z. [DOI] [PubMed] [Google Scholar]
- 398.Martinez-Murcia FJ, Gorriz JM, Ramirez J, Puntonet CG, Illan IA. Alzheimer’s Disease Neuroimaging. Functional activity maps based on significance measures and independent component analysis. Comput Methods Programs Biomed. 2013;111:255–268. doi: 10.1016/j.cmpb.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Toussaint PJ, Perlbarg V, Bellec P, Desarnaud S, Lacomblez L, Doyon J, et al. Resting state FDG-PET functional connectivity as an early biomarker of Alzheimer’s disease using conjoint univariate and independent component analyses. Neuroimage. 2012;63:936–946. doi: 10.1016/j.neuroimage.2012.03.091. [DOI] [PubMed] [Google Scholar]
- 400.Derado G, Bowman FD, Zhang L. Alzheimer’s Disease Neuroimaging. Initiative, Predicting brain activity using a Bayesian spatial model. Stat Methods Med Res. 2013;22:382–397. doi: 10.1177/0962280212448972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Keator DB, Fallon JH, Lakatos A, Fowlkes CC, Potkin SG, Ihler A. Feed-forward hierarchical model of the ventral visual stream applied to functional brain image classification. Hum Brain Mapp. 2014;35:38–52. doi: 10.1002/hbm.22149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Hinrichs C, Singh V, Mukherjee L, Xu G, Chung MK, Johnson SC. Spatially augmented LP boosting for AD classification with evaluations on the ADNI dataset. Neuroimage. 2009;48:138–149. doi: 10.1016/j.neuroimage.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Gross AL, Inouye SK, Rebok GW, Brandt J, Crane PK, Parisi JM, et al. Parallel but not equivalent: challenges and solutions for repeated assessment of cognition over time. J Clin Exp Neuropsychol. 2012;34:758–772. doi: 10.1080/13803395.2012.681628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Liu F, Wee CY, Chen H, Shen D. Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer’s disease and mild cognitive impairment identification. Neuroimage. 2014;84:466–475. doi: 10.1016/j.neuroimage.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Gray KR, Aljabar P, Heckemann RA, Hammers A, Rueckert D. Alzheimer’s Disease Neuroimaging. Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease. Neuroimage. 2013;65:167–175. doi: 10.1016/j.neuroimage.2012.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Singh N, Wang AY, Sankaranarayanan P, Fletcher PT, Joshi S. Alzheimer’s Disease Neuroimaging. Genetic, structural and functional imaging biomarkers for early detection of conversion from MCI to AD. Med Image Comput Comput Assist Interv. 2012;15(Pt 1):132–140. doi: 10.1007/978-3-642-33415-3_17. [DOI] [PubMed] [Google Scholar]
- 407.Zhou J, Liu J, Narayan VA, Ye J. Alzheimer’s Disease Neuroimaging. Modeling disease progression via multi-task learning. Neuroimage. 2013;78:233–248. doi: 10.1016/j.neuroimage.2013.03.073. [DOI] [PubMed] [Google Scholar]
- 408.Young J, Modat M, Cardoso MJ, Mendelson A, Cash D, Ourselin S. Alzheimer’s Disease Neuroimaging. Accurate multimodal probabilistic prediction of conversion to Alzheimer’s disease in patients with mild cognitive impairment. Neuroimage Clin. 2013;2:735–745. doi: 10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Cheng B, Zhang D, Shen D. Domain transfer learning for MCI conversion prediction. Med Image Comput Comput Assist Interv. 2012;15(Pt 1):82–90. doi: 10.1007/978-3-642-33415-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Yu P, Dean RA, Hall SD, Qi Y, Sethuraman G, Willis BA, et al. Enriching amnestic mild cognitive impairment populations for clinical trials: optimal combination of biomarkers to predict conversion to dementia. J Alzheimers Dis. 2012;32:373–385. doi: 10.3233/JAD-2012-120832. [DOI] [PubMed] [Google Scholar]
- 411.Casanova R, Hsu FC, Sink KM, Rapp SR, Williamson JD, Resnick SM, et al. Alzheimer’s Disease Neuroimaging. Alzheimer’s disease risk assessment using large-scale machine learning methods. PLoS One. 2013;8:e77949. doi: 10.1371/journal.pone.0077949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Wu X, Li J, Ayutyanont N, Protas H, Jagust W, Fleisher A, et al. The receiver operational characteristic for binary classification with multiple indices and its application to the neuroimaging study of Alzheimer’s disease. IEEE/ACM Trans Comput Biol Bioinform. 2013;10:173–180. doi: 10.1109/TCBB.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Dubey R, Zhou J, Wang Y, Thompson PM, Ye J. Alzheimer Disease Neuroimaging. Analysis of sampling techniques for imbalanced data: an n = 648 ADNI study. Neuroimage. 2013;87C:220–241. doi: 10.1016/j.neuroimage.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Lo RY, Jagust WJ. Predicting missing biomarker data in a longitudinal study of Alzheimer disease. Neurology. 2012;78:1376–1382. doi: 10.1212/WNL.0b013e318253d5b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Yuan L, Wang Y, Thompson PM, Narayan VA, Ye J. Alzheimer’s Disease Neuroimaging. Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data. Neuroimage. 2012;61:622–632. doi: 10.1016/j.neuroimage.2012.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Xiang S, Yuan L, Fan W, Wang Y, Thompson PM, Ye J. Alzheimer Disease Neuroimaging. Bi-level multi-source learning for heterogeneous block-wise missing data. Neuroimage. 2014;102P1:192–206. doi: 10.1016/j.neuroimage.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Simonsen AH, Mattila J, Hejl AM, Frederiksen KS, Herukka SK, Hallikainen M, et al. Application of the Predict AD software tool to predict progression in patients with mild cognitive impairment. Dement Geriatr Cogn Disord. 2012;34:344–350. doi: 10.1159/000345554. [DOI] [PubMed] [Google Scholar]
- 418.Liu Y, Mattila J, Ruiz MA, Paajanen T, Koikkalainen J, van Gils M. Alzheimer’s Disease Neuroimaging. Predicting AD conversion: comparison between prodromal AD guidelines and computer assisted Predict AD tool. PLoS One. 2013;8:e55246. doi: 10.1371/journal.pone.0055246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Escudero J, Ifeachor E, Zajicek JP. Alzheimer’s Disease Neuroimaging. Bioprofile analysis: a new approach for the analysis of biomedical data in Alzheimer’s disease. J Alzheimers Dis. 2012;32:997–1010. doi: 10.3233/JAD-2012-121024. [DOI] [PubMed] [Google Scholar]
- 420.Escudero J, Ifeachor E, Zajicek JP, Green C, Shearer J, Pearson S. Alzheimer’s Disease Neuroimaging. Machine learning-based method for personalized and cost-effective detection of Alzheimer’s disease. IEEE Trans Biomed Eng. 2013;60:164–168. doi: 10.1109/TBME.2012.2212278. [DOI] [PubMed] [Google Scholar]
- 421.Doecke JD, Laws SM, Faux NG, Wilson W, Burnham SC, Lam CP, et al. Blood-based protein biomarkers for diagnosis of Alzheimer disease. Arch Neurol. 2012;69:1318–1325. doi: 10.1001/archneurol.2012.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Llano DA, Devanarayan V, Simon AJ. Alzheimer’s Disease Neuroimaging. Evaluation of plasma proteomic data for Alzheimer disease state classification and for the prediction of progression from mild cognitive impairment to Alzheimer disease. Alzheimer Dis Assoc Disord. 2013;27:233–243. doi: 10.1097/WAD.0b013e31826d597a. [DOI] [PubMed] [Google Scholar]
- 423.Kiddle SJ, Thambisetty M, Simmons A, Riddoch-Contreras J, Hye A, Westman E, et al. Alzheimers Disease Neuroimaging. Plasma based markers of [11C] PiB-PET brain amyloid burden. PLoS One. 2012;7:e44260. doi: 10.1371/journal.pone.0044260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Burnham SC, Faux NG, Wilson W, Laws SM, Ames D, Bedo J, et al. Alzheimer’s Disease Neuroimaging, and Lifestyle Study Research. A blood-based predictor for neocortical Abeta burden in Alzheimer’s disease: results from the AIBL study. Mol Psychiatry. 2014;19:519–526. doi: 10.1038/mp.2013.40. [DOI] [PubMed] [Google Scholar]
- 425.Tatsuoka C, Tseng H, Jaeger J, Varadi F, Smith MA, Yamada T, et al. Alzheimer’s Disease Neuroimaging. Modeling the heterogeneity in risk of progression to Alzheimer’s disease across cognitive profiles in mild cognitive impairment. Alzheimers Res Ther. 2013;5:14. doi: 10.1186/alzrt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Ziegler G, Dahnke R, Gaser C. Alzheimer’s Disease Neuroimaging. Models of the aging brain structure and individual decline. Front Neuroinform. 2012;6:3. doi: 10.3389/fninf.2012.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Kohannim O, Hibar DP, Stein JL, Jahanshad N, Hua X, Rajagopalan P, et al. Discovery and replication of gene influences on brain structure using LASSO regression. Front Neurosci. 2012;6:115. doi: 10.3389/fnins.2012.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428. XXXXXX. [Google Scholar]
- 429.Epstein NU, Guo R, Farlow MR, Singh JP, Fisher M. Medication for Alzheimer’s disease and associated fall hazard: a retrospective cohort study from the Alzheimer’s Disease Neuroimaging Initiative. Drugs Aging. 2013 doi: 10.1007/s40266-013-0143-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Carmichael O, McLaren DG, Tommet D, Mungas D, Jones RN. Alzheimer Disease Neuroimaging. Coevolution of brain structures in amnestic mild cognitive impairment. Neuroimage. 2013;66C:449–456. doi: 10.1016/j.neuroimage.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Leung KK, Bartlett JW, Barnes J, Manning EN, Ourselin S, Fox NC. Alzheimer’s Disease Neuroimaging. Cerebral atrophy in mild cognitive impairment and Alzheimer disease: rates and acceleration. Neurology. 2013;80:648–654. doi: 10.1212/WNL.0b013e318281ccd3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Holland D, Desikan RS, Dale AM, McEvoy LK. Alzheimer’s Disease Neuroimaging. Rates of decline in Alzheimer disease decrease with age. PLoS One. 2012;7:e42325. doi: 10.1371/journal.pone.0042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Song X, Mitnitski A, Zhang N, Chen W, Rockwood K. Alzheimer’s Disease Neuroimaging. Dynamics of brain structure and cognitive function in the Alzheimer’s Disease Neuroimaging Initiative. J Neurol Neurosurg Psychiatry. 2013;84:71–78. doi: 10.1136/jnnp-2012-303579. [DOI] [PubMed] [Google Scholar]
- 434.Toledo JB, Xie SX, Trojanowski JQ, Shaw LM. Longitudinal change in CSF tau and Abeta biomarkers for up to 48 months in ADNI. Acta Neuropathol. 2013;126:659–670. doi: 10.1007/s00401-013-1151-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Wu L, Rowley J, Mohades S, Leuzy A, Dauar MT, Shin M, et al. Alzheimer’s Disease Neuroimaging. Dissociation between brain amyloid deposition and metabolism in early mild cognitive impairment. PLoS One. 2012;7:e47905. doi: 10.1371/journal.pone.0047905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Johnson JK, Gross AL, Pa J, McLaren DG, Park LQ, Manly JJ. Alzheimer’s Disease Neuroimaging. Longitudinal change in neuropsychological performance using latent growth models: a study of mild cognitive impairment. Brain Imaging Behav. 2012;6:540–550. doi: 10.1007/s11682-012-9161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Carmichael O, Xie J, Fletcher E, Singh B, DeCarli C. Localized hippocampus measures are associated with Alzheimer pathology and cognition independent of total hippocampal volume. Neurobiol Aging. 2012;33:1124.e31–1124.e41. doi: 10.1016/j.neurobiolaging.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Hostage CA, Roy Choudhury K, Doraiswamy PM, Petrella JR. Alzheimer’s Disease Neuroimaging. Dissecting the gene dose-effects of the APOE epsilon4 and epsilon2 alleles on hippocampal volumes in aging and Alzheimer’s disease. PLoS One. 2013;8:e54483. doi: 10.1371/journal.pone.0054483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Trzepacz PT, Yu P, Bhamidipati PK, Willis B, Forrester T, Tabas L, et al. Frontolimbic atrophy is associated with agitation and aggression in mild cognitive impairment and Alzheimer’s disease. Alzheimers Dement. 2013;9(5 Suppl):S95e1–S104e1. doi: 10.1016/j.jalz.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Zahodne LB, Gongvatana A, Cohen RA, Ott BR, Tremont G. Alzheimer’s Disease Neuroimaging. Are apathy and depression independently associated with longitudinal trajectories of cortical atrophy in mild cognitive impairment? Am J Geriatr Psychiatry. 2013;21:1098–1106. doi: 10.1016/j.jagp.2013.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Provenzano FA, Muraskin J, Tosto G, Narkhede A, Wasserman BT, Griffith EY, et al. White matter hyperintensities and cerebral amyloidosis: necessary and sufficient for clinical expression of Alzheimer disease? JAMA Neurol. 2013;70:455–461. doi: 10.1001/jamaneurol.2013.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Guzman VA, Carmichael OT, Schwarz C, Tosto G, Zimmerman ME, Brickman AM. Alzheimer’s Disease Neuroimaging. White matter hyperintensities and amyloid are independently associated with entorhinal cortex volume among individuals with mild cognitive impairment. Alzheimers Dement. 2013;9(5 Suppl):S124–S131. doi: 10.1016/j.jalz.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Lo RY, Jagust WJ. Alzheimer’s Disease Neuroimaging. Vascular burden and Alzheimer disease pathologic progression. Neurology. 2012;79:1349–1355. doi: 10.1212/WNL.0b013e31826c1b9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Epstein NU, Xie H, Ruland SD, Pandey DK. Vascular risk factors and cardiovascular outcomes in the Alzheimer’s disease neuroimaging initiative. Am J Alzheimers Dis Other Demen. 2012;27:275–279. doi: 10.1177/1533317512449730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Nir T, Jahanshad N, Jack CR, Weiner MW, Toga AW, Thompson PM. Alzheimer’s Disease Neuroimaging. Small world network measures predict white matter degeneration in patients with early-stage mild cognitive impairment; Proc IEEE Int Symp Biomed Imaging; 2012. pp. 1405–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Nir TM, Jahanshad N, Villalon-Reina JE, Toga AW, Jack CR, Weiner MW, et al. Alzheimer’s Disease Neuroimaging. Effectiveness of regional DTI measures in distinguishing Alzheimer’s disease, MCI, and normal aging. Neuroimage Clin. 2013;3:180–195. doi: 10.1016/j.nicl.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Rowley J, Fonov V, Wu O, Eskildsen SF, Schoemaker D, Wu L, et al. Alzheimer’s Disease Neuroimaging. White matter abnormalities and structural hippocampal disconnections in amnestic mild cognitive impairment and Alzheimer’s disease. PLoS One. 2013;8:e74776. doi: 10.1371/journal.pone.0074776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Habeck C, Risacher S, Lee GJ, Glymour MM, Mormino E, Mukherjee S, et al. Alzheimer’s Disease Neuroimaging. Relationship between baseline brain metabolism measured using [(1)(8)F]FDG PET and memory and executive function in prodromal and early Alzheimer’s disease. Brain Imaging Behav. 2012;6:568–583. doi: 10.1007/s11682-012-9208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Crane PK, Carle A, Gibbons LE, Insel P, Mackin RS, Gross A, et al. Development and assessment of a composite score for memory in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Brain Imaging Behav. 2012;6:502–516. doi: 10.1007/s11682-012-9186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Haight TJ, Jagust WJ. Alzheimer’s Disease Neuroimaging. Relative contributions of biomarkers in Alzheimer’s disease. Ann Epidemiol. 2012;22:868–875. doi: 10.1016/j.annepidem.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Wang Z, Das SR, Xie SX, Arnold SE, Detre JA, Wolk DA. Alzheimer’s Disease Neuroimaging. Arterial spin labeled MRI in prodromal Alzheimer’s disease: a multi-site study. Neuroimage Clin. 2013;2:630–636. doi: 10.1016/j.nicl.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Domoto-Reilly K, Sapolsky D, Brickhouse M, Dickerson BC. Alzheimer’s Disease Neuroimaging. Naming impairment in Alzheimer’s disease is associated with left anterior temporal lobe atrophy. Neuroimage. 2012;63:348–355. doi: 10.1016/j.neuroimage.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Nho K, Risacher SL, Crane PK, DeCarli C, Glymour MM, Habeck C, et al. Alzheimer’s Disease A. Neuroimaging Initiative, Voxel and surface-based topography of memory and executive deficits in mild cognitive impairment and Alzheimer’s disease. Brain Imaging Behav. 2012;6:551–567. doi: 10.1007/s11682-012-9203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Gibbons LE, Carle AC, Mackin RS, Harvey D, Mukherjee S, Insel P, et al. Alzheimer’s Disease Neuroimaging. A composite score for executive functioning, validated in Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging Behav. 2012;6:517–527. doi: 10.1007/s11682-012-9176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Mackin RS, Insel P, Tosun D, Mueller SG, Schuff N, Truran-Sacrey D, et al. Alzheimer’s Disease Neuroimaging. The effect of subsyndromal symptoms of depression and white matter lesions on disability for individuals with mild cognitive impairment. Am J Geriatr Psychiatry. 2013;21:906–914. doi: 10.1016/j.jagp.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Wadsworth LP, Lorius N, Donovan NJ, Locascio JJ, Rentz DM, Johnson KA, et al. Neuropsychiatric symptoms and global functional impairment along the Alzheimer’s continuum. Dement Geriatr Cogn Disord. 2012;34:96–111. doi: 10.1159/000342119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Ramakers IH, Verhey FR, Scheltens P, Hampel H, Soininen H, Aalten P, et al. Anxiety is related to Alzheimer cerebrospinal fluid markers in subjects with mild cognitive impairment. Psychol Med. 2013;43:911–920. doi: 10.1017/S0033291712001870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Richard E, Schmand B, Eikelenboom P, Yang SC, Ligthart SA, Moll van Charante EP, et al. Alzheimer’s Disease Neuroimaging. Symptoms of apathy are associated with progression from mild cognitive impairment to Alzheimer’s disease in non-depressed subjects. Dement Geriatr Cogn Disord. 2012;33:204–209. doi: 10.1159/000338239. [DOI] [PubMed] [Google Scholar]
- 459.Koppel J, Sunday S, Goldberg TE, Davies P, Christen E, Greenwald BS. Alzheimer’s Disease Neuroimaging. Psychosis in Alzheimer’s disease is associated with frontal metabolic impairment and accelerated decline in working memory: findings from the Alzheimer’s Disease Neuroimaging Initiative. Am J Geriatr Psychiatry. 2014;22(7):698–707. doi: 10.1016/j.jagp.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 460.Ewers M, Insel PS, Stern Y, Weiner MW. Alzheimer’s Disease Neuroimaging. Cognitive reserve associated with FDG-PET in preclinical Alzheimer disease. Neurology. 2013;80:1194–1201. doi: 10.1212/WNL.0b013e31828970c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Pillai JA, McEvoy LK, Hagler DJ, Jr, Holland D, Dale AM, Salmon DP, et al. Alzheimer’s Disease Neuroimaging. Higher education is not associated with greater cortical thickness in brain areas related to literacy or intelligence in normal aging or mild cognitive impairment. J Clin Exp Neuropsychol. 2012;34:925–935. doi: 10.1080/13803395.2012.702733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Guo LH, Alexopoulos P, Wagenpfeil S, Kurz A, Perneczky R. Alzheimer’s Disease Neuroimaging. Brain size and the compensation of Alzheimer’s disease symptoms: a longitudinal cohort study. Alzheimers Dement. 2013;9:580–586. doi: 10.1016/j.jalz.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 463.Mungas D, Crane PK, Gibbons LE, Manly JJ, Glymour MM, Jones RN. Advanced psychometric analysis and the Alzheimer’s Disease Neuroimaging Initiative: reports from the 2011 Friday Harbor conference. Brain Imaging Behav. 2012;6:485–488. doi: 10.1007/s11682-012-9211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Skinner J, Carvalho JO, Potter GG, Thames A, Zelinski E, Crane PK, et al. Alzheimer’s Disease Neuroimaging. The Alzheimer’s Disease Assessment Scale-Cognitive-Plus (ADAS-Cog-Plus): an expansion of the ADAS-Cog to improve responsiveness in MCI. Brain Imaging Behav. 2012;6:489–501. doi: 10.1007/s11682-012-9166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Park LQ, Gross AL, McLaren DG, Pa J, Johnson JK, Mitchell M, et al. Alzheimer’s Disease Neuroimaging. Confirmatory factor analysis of the ADNI neuropsychological battery. Brain Imaging Behav. 2012;6:528–539. doi: 10.1007/s11682-012-9190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Gross AL, Manly JJ, Pa J, Johnson JK, Park LQ, Mitchell MB, et al. Alzheimer’s Disease Neuroimaging. Cortical signatures of cognition and their relationship to Alzheimer’s disease. Brain Imaging Behav. 2012;6:584–598. doi: 10.1007/s11682-012-9180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Stricker NH, Dodge HH, Dowling NM, Han SD, Erosheva EA, Jagust WJ. Alzheimer’s Disease Neuroimaging. CSF biomarker associations with change in hippocampal volume and precuneus thickness: implications for the Alzheimer’s pathological cascade. Brain Imaging Behav. 2012;6:599–609. doi: 10.1007/s11682-012-9171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Han SD, Gruhl J, Beckett L, Dodge HH, Stricker NH, Farias S, et al. Beta amyloid, tau, neuroimaging, and cognition: sequence modeling of biomarkers for Alzheimer’s disease. Brain Imaging Behav. 2012;6:610–620. doi: 10.1007/s11682-012-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Mukherjee S, Kim S, Gibbons LE, Nho K, Risacher SL, Glymour MM, et al. Alzheimer’s Disease Neuroimaging. Genetic architecture of resilience of executive functioning. Brain Imaging Behav. 2012;6:621–633. doi: 10.1007/s11682-012-9184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Mukherjee S, Trittschuh E, Gibbons LE, Mackin RS, Saykin A, Crane PK. Azheimer’s Disease Neuroimaging. Dysexecutive and amnesic AD subtypes defined by single indicator and modern psychometric approaches: relationships with SNPs in ADNI. Brain Imaging Behav. 2012;6:649–660. doi: 10.1007/s11682-012-9207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Ramanan VK, Kim S, Holohan K, Shen L, Nho K, Risacher SL, et al. Genome-wide pathway analysis of memory impairment in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain Imaging Behav. 2012;6:634–648. doi: 10.1007/s11682-012-9196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Posner HB, Cano S, Carrillo MC, Selnes O, Stern Y, Thomas RG, et al. Establishing the psychometric underpinning of cognition measures for clinical trials of Alzheimer’s disease and its precursors: a new approach. Alzheimers Dement. 2013;9(1 Suppl):S56–S60. doi: 10.1016/j.jalz.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 473.Hobart J, Cano S, Posner H, Selnes O, Stern Y, Thomas R, et al. Alzheimer’s Disease Neuroimaging. Putting the Alzheimer’s cognitive test to the test I: traditional psychometric methods. Alzheimers Dement. 2013;9(1 Suppl):S4–S9. doi: 10.1016/j.jalz.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 474.Hobart J, Cano S, Posner H, Selnes O, Stern Y, Thomas R, et al. Alzheimer’s Disease Neuroimaging. Putting the Alzheimer’s cognitive test to the test II: Rasch measurement theory. Alzheimers Dement. 2013;9(1 Suppl):S10–S20. doi: 10.1016/j.jalz.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 475.Trzepacz PT, Saykin A, Yu P, Bhamditipati P, Sun J, Dennehy EB, et al. Alzheimer’s Disease Neuroimaging. Subscale validation of the neuropsychiatric inventory questionnaire: comparison of Alzheimer’s Disease Neuroimaging Initiative and national Alzheimer’s coordinating center cohorts. Am J Geriatr Psychiatry. 2013;21:607–622. doi: 10.1016/j.jagp.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Toledo JB, Da X, Weiner MW, Wolk DA, Xie SX, Arnold SE, et al. Alzheimer’s Disease Neuroimaging. CSF Apo-E levels associate with cognitive decline and MRI changes. Acta Neuropathol. 2014;127:621–632. doi: 10.1007/s00401-013-1236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Rajagopalan P, Toga AW, Jack CR, Weiner MW, Thompson PM. Alzheimer’s Disease Neuroimaging. Fat-mass-related hormone, plasma leptin, predicts brain volumes in the elderly. Neuroreport. 2013;24:58–62. doi: 10.1097/WNR.0b013e32835c5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Toledo JB, Korff A, Shaw LM, Trojanowski JQ, Zhang J. CSF alpha-synuclein improves diagnostic and prognostic performance of CSF tau and Abeta in Alzheimer’s disease. Acta Neuropathol. 2013;126:683–697. doi: 10.1007/s00401-013-1148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Korff A, Liu C, Ginghina C, Shi M, Zhang J. Alzheimer’s Disease Neuroimaging. Alpha-synuclein in cerebrospinal fluid of Alzheimer’s disease and mild cognitive impairment. J Alzheimers Dis. 2013;36:679–688. doi: 10.3233/JAD-130458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Toledo JB, Cairns NJ, Da X, Chen K, Carter D, Fleisher A, et al. Alzheimer’s Disease Neuroimaging. Clinical and multimodal biomarker correlates of ADNI neuropathological findings. Acta Neuropathol Commun. 2013;1:65. doi: 10.1186/2051-5960-1-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Landau SM, Mintun MA, Joshi AD, Koeppe RA, Petersen RC, Aisen PS, et al. Amyloid deposition, hypometabolism, and longitudinal cognitive decline. Ann Neurol. 2012;72:578–586. doi: 10.1002/ana.23650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Murphy KR, Landau SM, Choudhury KR, Hostage CA, Shpanskaya KS, Sair HI, et al. Alzheimer’s Disease Neuroimaging. Mapping the effects of ApoE4, age and cognitive status on 18F-florbetapir PET measured regional cortical patterns of beta-amyloid density and growth. Neuroimage. 2013;78:474–480. doi: 10.1016/j.neuroimage.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Landau SM, Lu M, Joshi AD, Pontecorvo M, Mintun MA, Trojanowski JQ, et al. Alzheimer’s Disease Neuroimaging. Comparing PET imaging and CSF measurements of and cerebrospinal fluid measurements of β-amyloid. Ass. Ann Neurol. 2013;74:826–836. doi: 10.1002/ana.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Jedynak BM, Lang A, Liu B, Katz E, Zhang Y, Wyman BT, et al. Alzheimer’s Disease Neuroimaging. A computational neurodegenerative disease progression score: method and results with the Alzheimer’s Disease Neuroimaging Initiative cohort. Neuroimage. 2012;63:1478–1486. doi: 10.1016/j.neuroimage.2012.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Risacher SL, Kim S, Shen L, Nho K, Foroud T, Green RC, et al. Alzheimer’s Disease Neuroimaging Initiative. The role of apolipoprotein E (APOE) genotype in early mild cognitive impairment (E-MCI) Front Aging Neurosci. 2013;5:11. doi: 10.3389/fnagi.2013.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Mouiha A, Duchesne S. Alzheimer’s Disease Neuroimaging. Toward a dynamic biomarker model in Alzheimer’s disease. J Alzheimers Dis. 2012;30:91–100. doi: 10.3233/JAD-2012-111367. [DOI] [PubMed] [Google Scholar]
- 487.Yang X, Tan MZ, Qiu A. CSF and brain structural imaging markers of the Alzheimer’s pathological cascade. PLoS One. 2012;7:e47406. doi: 10.1371/journal.pone.0047406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Thambisetty M, An Y, Nalls M, Sojkova J, Swaminathan S, Zhou Y, et al. Effect of complement CR1 on brain amyloid burden during aging and its modification by APOE genotype. Biol Psychiatry. 2013;73:422–428. doi: 10.1016/j.biopsych.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA. Alzheimer’s Disease Neuroimaging. The association between a polygenic Alzheimer score and cortical thickness in clinically normal subjects. Cereb Cortex. 2012;22:2653–2661. doi: 10.1093/cercor/bhr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Honea RA, Vidoni ED, Swerdlow RH, Burns JM. Maternal family history is associated with Alzheimer’s disease biomarkers. J Alzheimers Dis. 2012;31:659–668. doi: 10.3233/JAD-2012-120676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Westman E, Aguilar C, Muehlboeck JS, Simmons A. Regional magnetic resonance imaging measures for multivariate analysis in Alzheimer’s disease and mild cognitive impairment. Brain Topogr. 2013;26(1):9–23. doi: 10.1007/s10548-012-0246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Caroli A, Prestia A, Chen K, Ayutyanont N, Landau SM, Madison CM, et al. Summary metrics to assess Alzheimer disease-related hypometabolic pattern with 18F-FDG PET: head-to-head comparison. J Nucl Med. 2012;53:592–600. doi: 10.2967/jnumed.111.094946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Ito K, Hutmacher MM, Corrigan BW. Modeling of Functional Assessment Questionnaire (FAQ) as continuous bounded data from the ADNI database. J Pharmacokinet Pharmacodyn. 2012;39:601–618. doi: 10.1007/s10928-012-9271-3. [DOI] [PubMed] [Google Scholar]
- 494.O’Bryant SE. Using blood markers for Alzheimer disease in clinical practice? Neurology. 2012;79:846–847. doi: 10.1212/WNL.0b013e318266fd21. [DOI] [PubMed] [Google Scholar]
- 495.Westman E, Muehlboeck JS, Simmons A. Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. Neuroimage. 2012;62:229–238. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 496.Dukart J, Mueller K, Barthel H, Villringer A, Sabri O, Schroeter ML. Meta-analysis based SVM classification enables accurate detection of Alzheimer’s disease across different clinical centers using FDG-PET and MRI. Psychiatry Res. 2013;212:230–236. doi: 10.1016/j.pscychresns.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 497.Albert MS, Dekosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Yu P, Sun J, Wolz R, Stephenson D, Brewer J, Fox NC, et al. Coalition Against Major, and Alzheimer’s Disease Neuroimaging. Operationalizing hippocampal volume as an enrichment biomarker for amnestic mild cognitive impairment trials: effect of algorithm, test-retest variability, and cut point on trial cost, duration, and sample size. Neurobiol Aging. 2014;35:808–818. doi: 10.1016/j.neurobiolaging.2013.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Lehmann M, Koedam EL, Barnes J, Bartlett JW, Barkhof F, Wattjes MP, et al. Visual ratings of atrophy in MCI: prediction of conversion and relationship with CSF biomarkers. Neurobiol Aging. 2013;34:73–82. doi: 10.1016/j.neurobiolaging.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 500.Da X, Toledo JB, Zee J, Wolk DA, Xie SX, Ou Y, et al. Alzheimer’s Neuroimaging. Integration and relative value of biomarkers for prediction of MCI to AD progression: spatial patterns of brain atrophy, cognitive scores, APOE genotype and CSF biomarkers. Neuroimage Clin. 2013;4:164–173. doi: 10.1016/j.nicl.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Zhang N, Song X, Zhang Y. Alzheimer’s Disease Neuroimaging. Combining structural brain changes improves the prediction of Alzheimer’s disease and mild cognitive impairment. Dement Geriatr Cogn Disord. 2012;33:318–326. doi: 10.1159/000339364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Macdonald KE, Bartlett JW, Leung KK, Ourselin S, Barnes J. The value of hippocampal and temporal horn volumes and rates of change in predicting future conversion to AD. Alzheimer Dis Assoc Disord. 2013;27:168–173. doi: 10.1097/WAD.0b013e318260a79a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Samtani MN, Raghavan N, Shi Y, Novak G, Farnum M, Lobanov V, et al. Disease progression model in subjects with mild cognitive impairment from the Alzheimer’s disease neuroimaging initiative: CSF biomarkers predict population subtypes. Br J Clin Pharmacol. 2013;75:146–161. doi: 10.1111/j.1365-2125.2012.04308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Delor I, Charoin JE, Gieschke R, Retout S, Jacqmin P. Modeling Alzheimer’s disease progression using disease onset time and disease trajectory concepts applied to CDR-SOB scores from ADNI. CPT Pharmacometrics Syst Pharmacol. 2013;2:e78. doi: 10.1038/psp.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Shaffer JL, Petrella JR, Sheldon FC, Choudhury KR, Calhoun VD, Coleman RE, et al. Alzheimer’s Disease Neuroimaging. Predicting cognitive decline in subjects at risk for Alzheimer disease by using combined cerebrospinal fluid, MR imaging, and PET biomarkers. Radiology. 2013;266:583–591. doi: 10.1148/radiol.12120010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Prestia A, Caroli A, Herholz K, Reiman E, Chen K, Jagust WJ, et al. Diagnostic accuracy of markers for prodromal Alzheimer’s disease in independent clinical series. Alzheimers Dement. 2013;9:677–686. doi: 10.1016/j.jalz.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Schmand B, Eikelenboom P, van Gool WA. Alzheimer’s Disease Neuroimaging. Value of diagnostic tests to predict conversion to Alzheimer’s disease in young and old patients with amnestic mild cognitive impairment. J Alzheimers Dis. 2012;29:641–648. doi: 10.3233/JAD-2012-111703. [DOI] [PubMed] [Google Scholar]
- 508.Dickerson BC, Wolk DA. Alzheimer’s Disease Neuroimaging. Biomarker-based prediction of progression in MCI: comparison of AD signature and hippocampal volume with spinal fluid amyloid-beta and tau. Front Aging Neurosci. 2013;5:55. doi: 10.3389/fnagi.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Ewers M, Brendel M, Rizk-Jackson A, Rominger A, Bartenstein P, Schuff N, et al. Alzheimer’s Disease Neuroimaging. Reduced FDG-PET brain metabolism and executive function predict clinical progression in elderly healthy subjects. Neuroimage Clin. 2013;4:45–52. doi: 10.1016/j.nicl.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Trzepacz PT, Yu P, Sun J, Schuh K, Case M, Witte MM, et al. Alzheimer’s Disease Neuroimaging. Comparison of neuroimaging modalities for the prediction of conversion from mild cognitive impairment to Alzheimer’s dementia. Neurobiol Aging. 2014;35:143–151. doi: 10.1016/j.neurobiolaging.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 511.Gutman BA, Hua X, Rajagopalan P, Chou YY, Wang Y, Yanovsky I, et al. Alzheimer’s Disease Neuroimaging. Maximizing power to track Alzheimer’s disease and MCI progression by LDA-based weighting of longitudinal ventricular surface features. Neuroimage. 2013;70:386–401. doi: 10.1016/j.neuroimage.2012.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Pardoe HR, Abbott DF, Jackson GD. Alzheimer’s Disease Neuroimaging. Sample size estimates for well-powered cross-sectional cortical thickness studies. Hum Brain Mapp. 2013;34:3000–3009. doi: 10.1002/hbm.22120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Hibar DP, Kohannim O, Stein JL, Chiang MC, Thompson PM. Multi-locus genetic analysis of brain images. Front Genet. 2011;2:73. doi: 10.3389/fgene.2011.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Wyman BT, Harvey DJ, Crawford K, Bernstein MA, Carmichael O, Cole PE, et al. Alzheimer’s Disease Neuroimaging. Standardization of analysis sets for reporting results from ADNI MRI data. Alzheimers Dement. 2013;9:332–337. doi: 10.1016/j.jalz.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Hua X, Hibar DP, Ching CR, Boyle CP, Rajagopalan P, Gutman BA, et al. Alzheimer’s Disease Neuroimaging. Unbiased tensor-based morphometry: improved robustness and sample size estimates for Alzheimer’s disease clinical trials. Neuroimage. 2013;66:648–661. doi: 10.1016/j.neuroimage.2012.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Holland D, McEvoy LK, Desikan RS, Dale AM. Alzheimer’s Disease Neuroimaging. Enrichment and stratification for predementia Alzheimer disease clinical trials. PLoS One. 2012;7:e47739. doi: 10.1371/journal.pone.0047739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Grill JD, Di L, Lu PH, Lee C, Ringman J, Apostolova LG, et al. Estimating sample sizes for predementia Alzheimer’s trials based on the Alzheimer’s Disease Neuroimaging Initiative. Neurobiol Aging. 2013;34:62–72. doi: 10.1016/j.neurobiolaging.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Cedarbaum JM, Jaros M, Hernandez C, Coley N, Andrieu S, Grundman M, et al. Rationale for use of the Clinical Dementia Rating Sum of Boxes as a primary outcome measure for Alzheimer’s disease clinical trials. Alzheimers Dement. 2013;9(1 Suppl):S45–S55. doi: 10.1016/j.jalz.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 519.Whitwell JL, Wiste HJ, Weigand SD, Rocca WA, Knopman DS, Roberts RO, et al. Alzheimer Disease Neuroimaging. Comparison of imaging biomarkers in the Alzheimer Disease Neuroimaging Initiative and the Mayo Clinic Study of Aging. Arch Neurol. 2012;69:614–622. doi: 10.1001/archneurol.2011.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Kantarci K, Gunter JL, Tosakulwong N, Weigand SD, Senjem MS, Petersen RC, et al. Alzheimer’s Disease Neuroimaging. Focal hemosiderin deposits and beta-amyloid load in the ADNI cohort. Alzheimers Dement. 2013;9(5 Suppl):S116–S123. doi: 10.1016/j.jalz.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, Foroud TM, et al. Alzheimer Disease Neuroimaging. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging Behav. 2014;8:183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Honea RA, Cruchaga C, Perea RD, Saykin AJ, Burns JM, Weinberger DR, et al. Alzheimer’s Disease Neuroimaging. Characterizing the role of brain derived neurotrophic factor genetic variation in Alzheimer’s disease neurodegeneration. PLoS One. 2013;8:e76001. doi: 10.1371/journal.pone.0076001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Reitz C, Tosto G, Mayeux R, Luchsinger JA N.-L.N.F.S. Group, and Alzheimer’s Disease Neuroimaging. Genetic variants in the fat and obesity associated (FTO) gene and risk of Alzheimer’s disease. PLoS One. 2012;7:e50354. doi: 10.1371/journal.pone.0050354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Roussotte FF, Jahanshad N, Hibar DP, Sowell ER, Kohannim O, Barysheva M, et al. A commonly carried genetic variant in the delta opioid receptor gene, OPRD1, is associated with smaller regional brain volumes: replication in elderly and young populations. Hum Brain Mapp. 2014;35:1226–1236. doi: 10.1002/hbm.22247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, et al. G. Consortium, Alzheimer’s Disease Neuroimaging and Alzheimer Disease Genetic. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry. 2014;19:351–357. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Hibar DP, Stein JL, Kohannim O, Jahanshad N, Saykin AJ, Shen L, et al. Voxelwise gene-wide association study (vGeneWAS): multivariate gene-based association testing in 731 elderly subjects. Neuroimage. 2011;56:1875–1891. doi: 10.1016/j.neuroimage.2011.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Koran ME, Hohman TJ, Thornton-Wells TA. Genetic interactions found between calcium channel genes modulate amyloid load measured by positron emission tomography. Hum Genet. 2014;133:85–93. doi: 10.1007/s00439-013-1354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Meda SA, Koran ME, Pryweller JR, Vega JN, Thornton-Wells TA. Alzheimer’s Disease Neuroimaging. Genetic interactions associated with 12-month atrophy in hippocampus and entorhinal cortex in Alzheimer’s Disease Neuroimaging Initiative. Neurobiol Aging. 2013;34 doi: 10.1016/j.neurobiolaging.2012.09.020. 1518.e9–151818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Kim S, Swaminathan S, Inlow M, Risacher SL, Nho K, Shen L, et al. Alzheimer’s Disease Neuroimaging. Influence of genetic variation on plasma protein levels in older adults using a multi-analyte panel. PLoS One. 2013;8:e70269. doi: 10.1371/journal.pone.0070269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Benitez BA, Karch CM, Cai Y, Jin SC, Cooper B, Carrell D, et al. Alzheimer’s Disease Neuroimaging and Genetic and Environmental Risk for Alzheimer’s Disease. The PSEN1, p.E318G variant increases the risk of Alzheimer’s disease in APOE-epsilon4 carriers. PLoS Genet. 2013;9:e1003685. doi: 10.1371/journal.pgen.1003685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Hohman TJ, Koran ME, Thornton-Wells T. Alzheimer’s Neuroimaging. Epistatic Genetic Effects among Alzheimer’s Candidate Genes. PLoS One. 2013;8:e80839. doi: 10.1371/journal.pone.0080839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Hohman TJ, Koran ME, Thornton-Wells TA. Alzheimer’s Neuroimaging. Interactions between GSK3beta and amyloid genes explain variance in amyloid burden. Neurobiol Aging. 2014;35:460–465. doi: 10.1016/j.neurobiolaging.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Bryant C, Giovanello KS, Ibrahim JG, Chang J, Shen D, Peterson BS, et al. Alzheimer’s Disease Neuroimaging. Mapping the genetic variation of regional brain volumes as explained by all common SNPs from the ADNI study. PLoS One. 2013;8:e71723. doi: 10.1371/journal.pone.0071723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, et al. Multi-Institutional Research on Alzheimer Genetic Epidemiology, AddNeuroMed, and Alzheimer’s Disease Neuroimaging. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013;18:781–787. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Rhinn H, Fujita R, Qiang L, Cheng R, Lee JH, Abeliovich A. Integrative genomics identifies APOE epsilon4 effectors in Alzheimer’s disease. Nature. 2013;500:45–50. doi: 10.1038/nature12415. [DOI] [PubMed] [Google Scholar]
- 537.Kamboh MI, Demirci FY, Wang X, Minster RL, Carrasquillo MM, Pankratz VS, et al. Alzheimer’s Disease Neuroimaging. Genome-wide association study of Alzheimer’s disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Peterson D, Munger C, Crowley J, Corcoran C, Cruchaga C, Goate AM, et al. Alzheimer’s Disease Neuroimaging. Variants in PPP3R1 and MAPT are associated with more rapid functional decline in Alzheimer’s disease: The Cache County Dementia Progression Study. Alzheimers Dement. 2014;10:366–371. doi: 10.1016/j.jalz.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Swaminathan S, Shen L, Kim S, Inlow M, West JD, Faber KM, et al. Alzheimer’s Disease Neuroimaging, and N.-L.N.F.S. Group. Analysis of copy number variation in Alzheimer’s disease: the NIALOAD/NCRAD Family Study. Curr Alzheimer Res. 2012;9:801–814. doi: 10.2174/156720512802455331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Guffanti G, Torri F, Rasmussen J, Clark AP, Lakatos A, Turner JA, et al. Increased CNV-region deletions in mild cognitive impairment (MCI) and Alzheimer’s disease (AD) subjects in the ADNI sample. Genomics. 2013;102:112–122. doi: 10.1016/j.ygeno.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Hibar DP, Stein JL, Ryles AB, Kohannim O, Jahanshad N, Medland SE, et al. Genome-wide association identifies genetic variants associated with lentiform nucleus volume in N = 1345 young and elderly subjects. Brain Imaging Behav. 2013;7:102–115. doi: 10.1007/s11682-012-9199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Tamnes CK, Walhovd KB, Dale AM, Ostby Y, Grydeland H, Richardson G, et al. Brain development and aging: overlapping and unique patterns of change. Neuroimage. 2013;68:63–74. doi: 10.1016/j.neuroimage.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Harrison TM, Weintraub S, Mesulam MM, Rogalski E. Superior memory and higher cortical volumes in unusually successful cognitive aging. J Int Neuropsychol Soc. 2012;18:1081–1085. doi: 10.1017/S1355617712000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Nettiksimmons J, Beckett L, Schwarz C, Carmichael O, Fletcher E, Decarli C. Subgroup of ADNI normal controls characterized by atrophy and cognitive decline associated with vascular damage. Psychol Aging. 2013;28:191–201. doi: 10.1037/a0031063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Desikan RS, McEvoy LK, Thompson WK, Holland D, Brewer JB, Aisen PS, et al. Alzheimer’s Disease Neuroimaging. Amyloid-beta–associated clinical decline occurs only in the presence of elevated P-tau. Arch Neurol. 2012;69:709–713. doi: 10.1001/archneurol.2011.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Arnold SE, Xie SX, Leung YY, Wang LS, Kling MA, Han X, et al. Plasma biomarkers of depressive symptoms in older adults. Transl Psychiatry. 2012;2:e65. doi: 10.1038/tp.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]