Abstract
The hallmark of enteropathogenic Escherichia coli (EPEC) infection is the ability of EPEC to cause attaching and effacing (A/E) lesions on intestinal epithelium. This event is reproducible in in vitro tissue culture models of infection. We used real-time PCR to measure transcription from several locus of enterocyte effacement (LEE) operons (LEE1 to LEE5) and from bfp during a 5-h infection of HEp-2 cells with EPEC. We found that after the initial formation of A/E lesions, which occurs as early as 5 min postinfection, EPEC continues to increase transcription from LEE3 to LEE5 as well as from bfp. These levels are maximized by 3 h postinfection and remain constant throughout the course of infection. This increase in transcription from LEE3 to LEE5 occurs when LEE1 (ler) transcription is decreasing. EspA, EspB, intimin, Tir, and bundle-forming pilus expression is detectable during the entire 5-h infection. These results indicate that the EPEC genes involved in localized and intimate adherence are continually expressed after the initial stages of A/E lesion formation on host cells.
Enteropathogenic Escherichia coli (EPEC) is a leading bacterial cause of infant diarrhea in developing countries (46). Additionally, EPEC has been implicated in sporadic outbreaks in industrialized countries, such as the United States and the United Kingdom (3, 30, 45). The hallmark of EPEC infection is the ability of EPEC to cause attaching and effacing (A/E) lesions on intestinal epithelium (44, 46). All of the genes necessary for the A/E phenotype in EPEC are located on a 35.6-kb pathogenicity island containing 41 predicted open reading frames and termed the locus of enterocyte effacement (LEE) (39, 46). The LEE is organized into five major operons, LEE1 to LEE5. LEE1 to LEE3 encode portions of a type III secretion apparatus and the transcriptional regulator Ler (15, 16, 42). LEE4 encodes the secreted proteins EspA, EspB, and EspD and the translocated effector protein EspF (33). The LEE5 operon (also known as the tir operon) encodes an intimate adhesin (intimin), its associated translocated receptor (Tir), and the Tir chaperone CesT (14, 42, 49).
The central regulator of the LEE pathogenicity island is Ler, the LEE-encoded regulator (42). Ler increases the expression of downstream polycistronic operons of the LEE and was shown to directly bind upstream of LEE2 and LEE5 (15, 24, 42, 51). Ler is an H-NS homologue which is able to relieve H-NS-mediated repression of the LEE2, LEE3, and LEE5 operons (4, 24, 49). Per, a regulator encoded on the EPEC adherence factor (EAF) plasmid, has been associated with activation of the LEE, specifically through the activation of LEE1 (4, 21, 42). In addition to EPEC-specific genes, the E. coli proteins IHF, Fis, and BipA have also been shown to activate Ler expression (17, 20, 23, 42). In addition, GrlA and GrlR have been shown to act as an activator and as a repressor of LEE gene expression, respectively, in Citrobacter rodentium (10).
Several candidate adhesins have been proposed to mediate the nonintimate adhesion of EPEC to host cells; these include the plasmid-encoded bundle-forming pilus (BFP) (5, 36, 53), flagella (19), and EspA (5). The BFP is responsible for the formation of three-dimensional microcolonies on the surface of epithelial cells, a process also known as localized adherence (LA) (18, 26). During A/E lesion formation, Tir is translocated into the host cell, where it undergoes phosphorylation and insertion into the host cell membrane (32). There it is able to act as a receptor for intimin in the bacterial membrane, mediating a more intimate association between the bacteria and the host cell (9, 25, 32). Tir, along with specific host proteins, mediates the rearrangement of the host cell cytoskeleton, which results in the formation of actin-rich pedestals beneath the intimately adherent bacteria (22, 32). Other LEE-encoded translocated effector proteins—EspB, EspD, EspF, EspG, EspH, and Map—affect additional processes in epithelial cells. EspB, along with EspD, forms a pore in the host cell membrane and is essential for protein translocation (28). EspF disrupts tight junctions and is associated with the induction of apoptosis (7, 41), and Map induces the formation of transient filopodia on the host cell and interrupts mitochondrial membrane potential (31). EspH modulates the host actin cytoskeleton structure and represses the formation of filopodia (55). The role of EspG is not yet known (13).
It has been proposed that A/E lesion formation occurs in three distinct stages (12). First, initial interactions with the host cell are mediated through the BFP and/or other factors. Second, the type III secretion apparatus is formed, and effector proteins are translocated into the host cell cytoplasm. Finally, Tir-induced cytoskeletal rearrangement forms pedestals beneath the intimately adherent bacteria. Additionally, after successful A/E lesion formation, EPEC undergoes dispersal of microcolonies, and host cell apoptosis is induced (1, 6, 36, 53).
The above model suggests that the genes involved in the progression of this infection process would be expressed in a timely manner so as to properly coordinate the above events. We sought to examine the temporal expression of the LEE operons and bfp in order to determine whether they were expressed in a cascade fashion during the course of infection in a HEp-2 tissue culture model of EPEC infection. Interestingly, we found that transcription from the LEE1 operon was consistently decreased by approximately 50% 3 h postinfection and that this reduced level of LEE1 transcription remained constant throughout the course of infection. In contrast, we found that transcription from LEE3 to LEE5 as well as from bfp was increased over a 5-h infection of HEp-2 cells with EPEC. The transcription data correlate with the continued production of the Tir, intimin, EspA, and BfpA proteins during the later stages of infection as well.
MATERIALS AND METHODS
Bacterial and tissue cultures.
The strains used in this study are shown in Table 1. For real-time PCR experiments involving comparisons of media in the absence of epithelial cells, bacteria were grown overnight in Luria-Bertani (LB) medium, concentrated by centrifugation, and resuspended in Dulbecco modified Eagle medium (DMEM) (Gibco BRL/Life Technologies 11995-065) at a dilution of 1:25. Alternatively, bacteria were concentrated and resuspended in fresh LB medium at a dilution of 1:25. After dilution, cultures were grown to mid-logarithmic phase at 37°C.
TABLE 1.
Strains used in this study
For real-time PCR studies involving cultured epithelial cells, HEp-2 cells were grown in DMEM-10% fetal bovine serum in six-well plates (Falcon 353046) to 90% confluence. Prior to infection, HEp-2 cells were washed twice with phosphate-buffered saline (PBS), and fresh DMEM without supplements was added. Cells were infected with preactivated bacteria at a multiplicity of infection (MOI) of 100. Bacteria were preactivated by subculturing from an overnight LB medium culture into DMEM; they were allowed to grow for 2 h at 37°C in 5% CO2. To synchronize infections, bacteria were centrifuged onto HEp-2 cells (500 × g for 3 min). Infections were allowed to proceed at 37°C in 5% CO2 for 10 min, 3 h, or 5 h.
For mock infection experiments (in the absence of HEp-2 cells), 150-cm2 tissue culture flasks (Falcon 355001) were incubated overnight with DMEM-10% fetal bovine serum. Prior to the addition of preactivated bacteria, the flasks were washed twice with PBS, and fresh DMEM without supplements was added. Bacteria were added at the same concentration (CFU per milliliter of medium) as that used to achieve an MOI of 100 in the HEp-2 cell-containing experiments.
Immunofluorescence microscopy and confocal microscopy.
HEp-2 cells were grown on 18-mm coverslips in six-well plates to 75% confluence. Prior to infection, HEp-2 cells were washed twice with PBS, and fresh DMEM without supplements was added. Cells were infected with either 30 μl of an overnight bacterial culture grown in LB medium or preactivated bacteria at an MOI of 100. Infections were allowed to proceed for 3 or 5 h. Following infection, the coverslips were washed twice with PBS and fixed with 4% formaldehyde-PBS. The coverslips were washed three times with PBS, and the cells were permeabilized with 0.1% Triton X-100-PBS. Cells were washed three times with PBS and blocked with 10% normal goat serum-PBS-0.1% Triton X-100. The coverslips were probed with rabbit anti-Tir, anti-intimin, anti-BfpA, or anti-EspA antibodies, washed three times with PBS, and stained with Alexa-Fluor 568-goat anti-rabbit antibody (Molecular Probes). After the coverslips were washed with PBS, actin was stained with Alexa-Fluor 488-phalloidin (Molecular Probes), and 4′,6′-diamidino-2-phenylindole (DAPI) (Molecular Probes) was used to stain bacterial and host cell DNA. After the coverslips were washed with PBS, they were mounted by use of a ProLong antifade kit (Molecular Probes) and viewed with a Zeiss LSM510 Meta confocal microscope. The anti-Tir, anti-intimin, anti-BfpA, and anti-EspA antibodies were used at a 1:200 dilution. Alexa-Fluor 568-goat anti-rabbit antibody and Alexa-Fluor 488-phalloidin were used at a 1:100 dilution, and DAPI was used at 2 μg/ml.
RNA extraction and cDNA synthesis.
At the end of the incubation period, total RNA was extracted by use of Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA was quantified by use of the A260 determined by spectrophotometry. The quality of RNA was assessed by use of the A260/A280 ratio and by visualization of 16S and 23S rRNAs by 2% agarose gel electrophoresis. Ten micrograms of RNA was treated with DNase I (Invitrogen) at a concentration of 1 U/μg of RNA for 30 min at room temperature. DNase I was inactivated by adding 25 mM EDTA and heating the samples at 65°C for 10 min. First-strand cDNA synthesis was performed on 3.5 μg of DNase I-treated total RNA with gene-specific reverse primers (Table 2) by use of a SuperScript first-strand synthesis system for reverse transcription-PCR (Invitrogen) as described by the manufacturer.
TABLE 2.
Primers used in this study
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| ler | TGGGATATACTAATGTGCCTGATGA | ACCAGGTCTGCCCTTGTTGA |
| escV | AGTGCTCGTTTTTCCCTTGA | AGCGAAGAACTTTTGCCTCA |
| tir | GCAGAAGACGCTTCTCTGAATA | CCCAACTTCAGCATATGGATTA |
| eae | GGCGATTACGCGAAAGATAC | GATTAACCTATGCCGTTCCA |
| espA | GCTGCAATTCTCATGTTTGC | GGGCAGTGGTTGACTCCTTA |
| bfpA | TGATTGAATCTGCAATGGTG | AGCATTCTGCGACTTATTGG |
| rrsB | TGCAAGTCGAACGGTAACAG | AGTTATCCCCCTCCATCAGG |
Real-time PCR studies.
Real-time PCR was performed by use of a QuantiTect SYBR green PCR kit (Qiagen) and an ABI Prism 7900HT sequence detection system (Applied Biosystems). For each reaction, 1 μl of reverse-transcribed cDNA was subjected to PCR amplification in a 25-μl final volume containing 900 nM each primer and 12.5 μl of 2× SYBR green master mix. The following conditions were used for amplification: 1 cycle at 95°C for 10 min and 40 cycles at 95°C for 15 s and 60°C for 1 min. To ensure the specificity of the PCR products, melting curve analysis was performed by heating products to 95°C for 15 s, followed by cooling to 60°C and slowly heating to 95°C while monitoring fluorescence (data not shown).
Detection, quantitation, and statistical analysis.
Data collection was carried out by use of ABI Prism SDS 2.0 software (Applied Biosystems). Data were normalized to levels of rrsB (16S rRNA) and analyzed by use of the comparative critical threshold (CT) method as described previously (2). The relative comparison method was used to compare the levels of expression of target genes at different time points (2). No contamination by genomic DNA was detectable by PCR for tir after DNase I treatment of RNA (data not shown). For real-time data presentation, error bars represent standard deviations of the mean. All results are expressed as the mean plus or minus 1 standard deviation of the mean for three independent experiments. Statistical significance was calculated with Student's t test. A P value of <0.05 was considered significant.
Triton X-100 fractionation and Western blotting.
HEp-2 cells were grown in T-75 flasks (Corning 430725) to 95% confluence. Prior to infection, HEp-2 cells were washed twice with PBS, and fresh DMEM without supplements was added. Cells were infected at an MOI of 100 with bacteria preactivated in DMEM. Infections were allowed to proceed for 3 or 5 h. Following infection, the cells were washed twice with ice-cold PBS and scraped into a final volume of 1.5 ml of ice-cold PBS. Cells were collected by centrifugation, and the pellet was lysed for 30 min at room temperature with 75 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 1% Triton X-100, 1.0 mM Na2VO4, 2× complete protease inhibitor cocktail [Roche]). Ten microliters of the lysate was diluted and plated to determine the CFU per milliliter for each sample to standardize sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis loading volumes. The lysate was centrifuged at 4°C for 30 min. Laemmli sample buffer (25 μl) was added to the supernatant to become the Triton X-100-soluble fraction. The pellet was washed twice with PBS, resuspended in 75 μl of lysis buffer-25 μl of 4× Laemmli sample buffer, and used as the Triton X-100-insoluble fraction. Samples were boiled, cleared by centrifugation, and standardized to equivalent CFU per lane prior to loading on SDS-14% polyacrylamide gels. Uninfected samples were loaded to the same volumes as 5-h infection samples. Western transfer blots were probed with rabbit polyclonal anti-Tir, anti-intimin, anti-BfpA, and anti-EspB antisera in PBS-0.5% bovine serum albumin-0.5% Tween 20. The primary antibody was detected by use of a horseradish peroxidase-conjugated anti-rabbit secondary antibody in conjunction with enhanced chemiluminescence Western blotting detection reagents (Amersham Biosciences).
RESULTS
Genes chosen for real-time PCR analysis.
It has been shown that the LEE of EPEC E2348/69 is sufficient to cause A/E lesion formation on tissue culture cells when it is placed in a nonpathogenic E. coli strain (40). The LEE encodes factors involved in the production of this phenotype, such as the Ler regulator, a type III secretion apparatus, and effector proteins. We sought to examine whether the operons encoding these genes were regulated in a distinct temporal pattern during the course of infection. We used real-time PCR to monitor the patterns of transcription of representative genes from LEE1 (ler), LEE2 (escJ), LEE3 (escV), LEE5 (tir and eae), and LEE4 (espA). Additionally, we examined the transcriptional profile of bfpA from the bfp operon carried on the EAF plasmid. During preliminary studies, we were unable to obtain consistent and reproducible results for LEE2 (escJ) transcription with our real-time PCR assay system for unknown reasons (data not shown). LEE2 and LEE3 contain divergent overlapping promoters, and Ler is able to alleviate H-NS-mediated repression of these operons by binding to the LEE2 regulatory region (4, 51). These divergent promoters may contain other undescribed regulatory regions which may be responsible for the widely variable transcription that we detected from LEE2 but not from LEE3. Due to the highly variable results obtained, LEE2 (escJ) was removed from this study. 16S rRNA (rrsB) levels were used to normalize real-time data and to adjust for various bacterial numbers present at each of the time points. The genes chosen and the primers used for real-time PCR are shown in Table 2.
EPEC virulence gene expression is increased in DMEM.
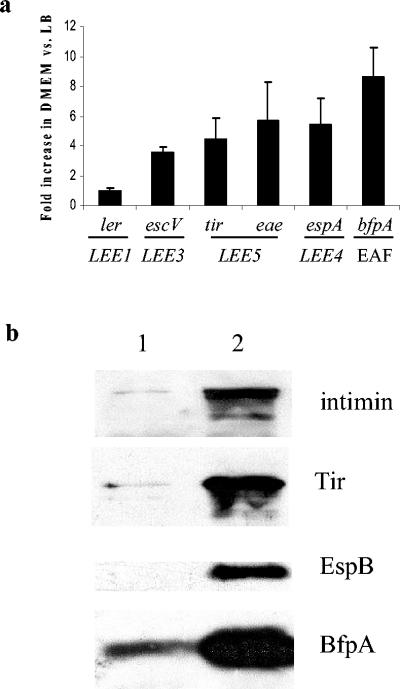
Previous work showed that the expression of EPEC virulence factors is increased when the bacteria are grown in DMEM verses rich medium, such as LB medium (47, 48). We sought to reproduce these findings with our real-time PCR assay system. We performed initial real-time PCR experiments measuring the transcription of rrsB under these two culture conditions and found that the transcription levels were not statistically different (data not shown), thus validating our use of 16S rRNA levels as a normalization control for our experiments. When EPEC was grown to mid-log phase in DMEM, we found an approximate fivefold increase in transcription from LEE5 (eae and tir) and LEE4 (espA) compared to transcription seen with growth in LB medium (Fig. 1a). Our results showed that tir and eae levels were increased to the same degrees; therefore, under these growth conditions, we did not see evidence of transcrption from an eae-specific promoter. Transcription from LEE3 (escV) showed a fourfold increase in DMEM, while a more dramatic eightfold increase was seen in the transcription of bfp. These findings correlate with reports by other investigators (47, 48).
FIG. 1.
EPEC virulence gene expression is increased by growth in DMEM. (a) Fold change in gene transcription in E2348/69 grown to mid-log phase in DMEM versus LB medium, as measured by real-time PCR. Primers used for each gene are shown in Table 2. Results shown are means and standard deviations from triplicate experiments. CT values were normalized to levels of 16S rRNA to correct for variations in bacterial numbers. Fold differences were calculated by using the relative comparison method. (b) Western blot showing the expression of intimin, Tir, EspB, and BfpA in E2348/69 grown to mid-log phase in LB medium (lane 1) and DMEM (lane 2). Bacterial cultures were adjusted to equivalent optical densities at 600 nm prior to loading.
In contrast to the increase in transcription from LEE3 to LEE5, LEE1, encoding a transcriptional activator of the downstream LEE operons, showed no change in transcription in the two media tested. These results suggest that Ler may not be responsible for the increased transcription from LEE3 to LEE5 seen when EPEC was grown in DMEM versus LB medium. Previous studies showed that H-NS is involved in repression of the LEE operons and that Ler alleviates that repression (4, 24, 49). There may be additional Ler-independent activating factors acting on those promoter regions as well. We correlated these transcription findings with protein levels for intimin and Tir (LEE5), EspB (LEE4), and BfpA by Western blot analysis. All proteins showed an increase in expression with growth in DMEM versus LB medium (Fig. 1b).
EPEC increases expression from LEE3 to LEE5 and bfp during HEp-2 cell infection.
We examined the transcriptional profiles for the LEE operons and the bfp operon during a typical in vitro tissue culture model of infection known to produce LA and A/E lesions. Real-time PCR was used to measure gene transcription periodically during a 5-h infection of HEp-2 tissue culture cells with EPEC strain E2348/69. HEp-2 cells were infected with E2348/69 for 10 min, 3 h, or 5 h. Rosenshine et al. (48) demonstrated that EPEC that had been preactivated in DMEM was able to induce A/E lesion formation as early as 5 min after infection of tissue culture cells (48). We found the same to be true with HEp-2 cells; therefore, we used 10 min as our first time point for transcriptional measurements of EPEC gene expression. Additionally, when the infections were allowed to continue to 6 h, marked host cell detachment and cell rounding were observed. Therefore, all infections were stopped at 5 h to provide a more accurate view of infections involving mostly viable HEp-2 cells.
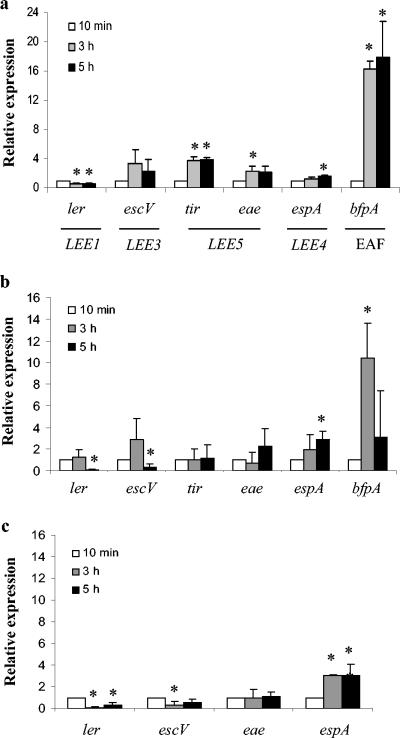
Figure 2a shows the results of a representative triplicate set of experiments. Transcription from LEE1 (ler) was consistently decreased 50% at 3 and 5 h versus 10 min (P values of <0.01 for both) Transcription from LEE5 (tir and eae) and bfp was statistically significantly increased by 3 h postinfection (P values of <0.01, <0.05, and <0.0001, respectively). Initially, we expected that these genes would be down-regulated during the course of infection, since they encode proteins involved in the initial stages of attachment and intimate adherence. However, we consistently observed an increase in the expression of bfp, eae, and tir over time even after the cells had produced a fluorescent actin staining (FAS)-positive phenotype (indicative of A/E lesions) and large microcolonies had already formed (data not shown). Transcription from LEE3 (escV) was also consistently up-regulated over time; however, the actual fold differences seen were not statistically significant at 3 and 5 h versus 10 min (P > 0.05). Although the data for LEE3 (escV) were not statistically significant, the overall trend of increased transcription was seen in multiple experiments.
FIG. 2.
Transcriptional profiles of LEE expression and bfp expression during a 5-h infection for EPEC on HEp-2 cells (a), for EPEC in the absence of HEp-2 cells (b), and for JAC719 (tir mutant) on HEp-2 cells (c). Relative expression represents the fold change in E2348/69 gene transcription from 10 min to 3 and 5 h postinfection, as measured by real-time PCR. Primers used for each gene are indicated in Table 2. Results shown are means and standard deviations from triplicate infections. CT values were normalized to levels of 16S rRNA to correct for variations in bacterial numbers. Fold differences from the 10-min time point were calculated by using the relative comparison method. Values that were significantly different from values at 10 min are indicated by an asterisk (P < 0.05).
Mock infection experiments were carried out in the absence of HEp-2 cells to determine whether the transcriptional patterns that we observed were due to dilution of the preactivated bacteria into tissue culture vessels. Figure 2b shows the results for a triplicate set of experiments. In the absence of HEp-2 cells, ler transcription remained constant through the 3-h time point. At 5 h, we see saw a decrease in ler transcription similar to that shown in Fig. 2a, but this effect was delayed in the mock infection experiment, indicating that the presence of HEp-2 cells was affecting the transcriptional pattern for LEE1. tir and eae transcription remained fairly constant during the mock infection compared to the two- to fourfold increase seen at 3 h when HEp-2 cells were present. bfp and LEE3 also showed altered transcriptional patterns in the absence of HEp-2 cells. espA transcription remained consistent in the presence or absence of HEp-2 cells, indicating that the transcription of LEE4 may not be directly affected by the presence of HEp-2 cells.
To further examine whether the effects that we observed were due to actual intimate contact with host cells or merely the presence of host cells, we examined the transcriptional profile of JAC719, an EPEC tir mutant unable to form A/E lesions (Fig. 2c). LEE1 transcription decreased significantly at both 3 h and 5 h (P values of <0.02 for both), suggesting that intimate adherence was not required for the transcriptional modulation of ler during the infection. LEE3 (escV) transcription and LEE5 (eae) transcription remained fairly constant during a 5-h infection. espA transcription increased over time in a manner similar to that seen in wild-type EPEC in the presence or absence of HEp-2 cells.
EPEC protein production and translocation occur at 5 h postinfection.
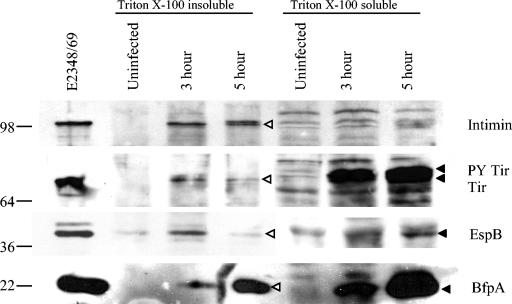
In order to correlate our transcription results with protein expression, we performed Triton X-100 lysate fractionations of HEp-2 cells infected with E2348/69. After either 3 or 5 h of infection of HEp-2 cells, bacteria and host cells were fractionated with Triton X-100, which separates the lysate into a soluble fraction (containing eukaryotic host cell membrane proteins and translocated bacterial proteins) and an insoluble fraction (containing eukaryotic host cell cytoskeletal components and bacteria). In order to accurately represent the relative amounts of bacterial proteins present in the samples, bacterial CFU per milliliter of lysate were determined prior to fractionation, and equivalent bacterial CFU were loaded for the samples. Uninfected samples were loaded to the same volumes as 5-h infection samples. The fractions then were subjected to Western blotting with rabbit polyclonal antisera to intimin, Tir, EspB, and the BFP.
Figure 3 indicates that intimin was produced at 3 and 5 h postinfection. Intimin was not found in the Triton X-100-soluble fraction, indicating that the soluble fraction was not contaminated with nontranslocated bacterial proteins. Additionally, Tir appeared to be translocated and phosphorylated at both 3 h and 5 h, as shown by the presence of two Tir bands in the Triton X-100-soluble fraction. Tir was also seen in the insoluble bacterial fraction. However, the bacterial Tir level appeared to be slightly reduced at 5 h. This reduction in the bacterial Tir level did not result in a reduction in the amount of translocated Tir present at 5 h. EspB was used to demonstrate expression from LEE4. Figure 3 shows the EspB protein levels at 3 and 5 h postinfection in the insoluble bacterial fraction; however, the level was lower at 5 h. EspB was still abundant in the insoluble host cell fraction at 5 h postinfection. Finally, we examined BFP production and found an increase in BFP production in both the insoluble bacterial fraction and the soluble host cell fraction from 3 to 5 h. The BFP found in the soluble host cell fraction most likely represented extracellular BFP which had been dissociated from the bacterial membrane during the fractionation procedure.
FIG. 3.
EPEC protein production and translocation are evident 5 h postinfection. A Western blot of Triton X-100 fractions shows E2348/69 protein expression during 3-h and 5-h infections of HEp-2 cells. Triton X-100 total lysates (soluble and insoluble) were plated prior to fractionation to determine total CFU. Lanes of SDS-polyacrylamide gels were loaded with equivalent CFU. Uninfected lanes were loaded with the same volume as that used for the 5-h lysate. Transfer membranes were probed with anti-intimin, anti-Tir, anti-EspB, or anti-BfpA antisera. Sea-Blue 2 molecular size markers (in kilodaltons) are indicated at the left. The E2348/69 whole-cell lysate was prepared from bacteria grown to mid-log phase in DMEM. Protein bands in the Triton X-100-soluble fraction are indicated by solid arrowheads; protein bands in the Triton X-100-insoluble fraction are indicated by open arrowheads. PY, tyrosine phosphorylated.
Intimin and Tir are expressed at 3 and 5 h after infection with EPEC.
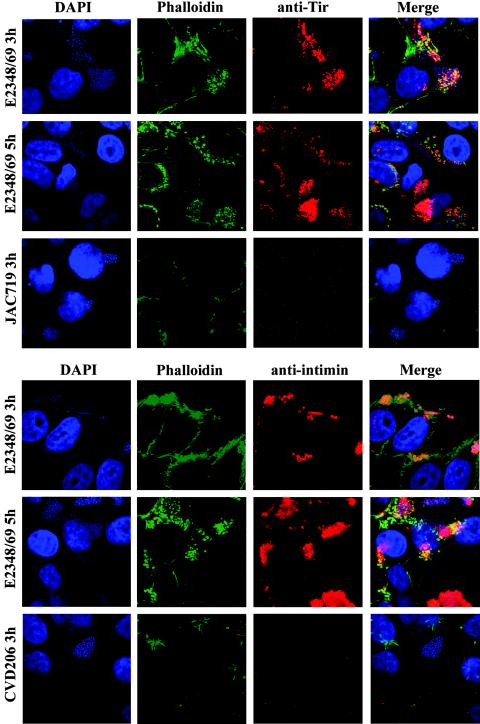
Our transcription data for LEE5 showed an increase in transcription occurring by 3 h postinfection. Additionally, using Triton X-100 fractionation, we showed that intimin and Tir were being produced even in the later stages of the infection process. We wished to confirm these results by using immunofluorescence combined with confocal microscopy at both 3 h and 5 h postinfection. The FAS assay reveals actin condensation beneath bacteria and indicates that these cells are A/E positive (35). Specific proteins tested were visualized with rabbit polyclonal anti-Tir or anti-intimin antisera. Actin condensation was visualized with Alexa-Fluor 488-phalloidin, and bacterial and host cell DNA was stained with DAPI.
Tir was expressed in the majority of cells at 3 and 5 h, with no apparent or evident reduction in expression between the two time points (Fig. 4). Intimin was expressed by nearly all bacteria at 3 h postinfection. However, by 5 h a small percentage of cells no longer appeared to express intimin compared to what was seen at 3 h. Knutton et al. (34) previously demonstrated that intimin expression was nearly abolished by 6 h postinfection compared to what was seen at 3 h. While we did not see such dramatic results at 5 h, we did see a small reduction in the number of bacteria expressing intimin on their surface. JAC719 (tir mutant) and CVD206 (eae mutant) showed LA patterns, were FAS negative, and failed to show the expression of Tir and of intimin, respectively. Similar results were seen when bacterial cultures grown overnight in LB medium were compared to those preactivated in DMEM (data not shown).
FIG. 4.
Intimin and Tir are coexpressed at 3 and 5 h after HEp-2 cell infection with E2348/69. Intimin expression and Tir expression were demonstrated at 3 and 5 h postinfection by immunofluorescence microscopy and confocal microscopy. Intimin and Tir were not visualized in isogenic mutant strains at either time point (CVD206 and JAC719, respectively). Cells were triply labeled with either anti-Tir or anti-intimin antisera, for actin with Alexa-Fluor 488-phalloidin (green), and for bacterial and HEp-2 cell DNA with DAPI (blue). The primary antibody was visualized by using Alexa-Fluor 568-goat anti-rabbit secondary antibody (red). Bacteria were preactivated in DMEM prior to infection of HEp-2 cells at an approximate MOI of 100.
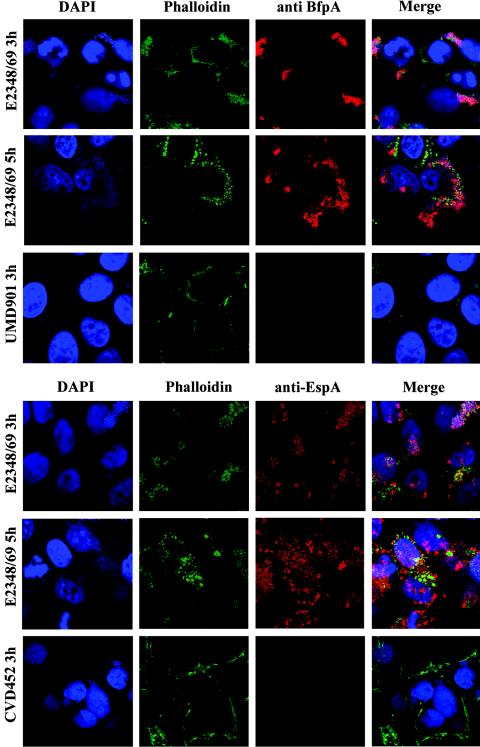
EPEC EspA filaments and the BFP are expressed at 3 and 5 h postinfection.
Using immunofluorescence, we were able to demonstrate BFP and EspA filament expression during the course of HEp-2 cell infection with EPEC (Fig. 5). Similar results were seen when overnight cultures of bacteria grown in LB medium were used instead of preactivated bacteria for infecting HEp-2 cells (data not shown). CVD452 (escN mutant) was LA positive, FAS negative, and lacked surface expression of EspA. UMD901 (bfpA mutant) was FAS positive and did not produce the BFP. We also observed the elongation of BFP fibrils at 5 h compared to the tightly formed BFP aggregates visible at 3 h (Fig. 5), similar to the data reported by Knutton et al. (36).
FIG. 5.
EspA filaments and the BFP are expressed at 3 and 5 h after HEp-2 cell infection with E2348/69. EspA expression and BFP expression were demonstrated by immunofluorescence microscopy and confocal microscopy. Isogenic mutants CVD452 and UMD901 did not express surface EspA and BFP, respectively. Cells were labeled with either anti-EspA or anti-BfpA antisera, for actin with Alexa-Fluor 488-phalloidin (green), and for bacterial and HEp-2 cell DNA with DAPI (blue). The primary antibody was visualized by using Alexa-Fluor 568-goat anti-rabbit secondary antibody (red). Bacteria were preactivated in DMEM prior to infection of HEp-2 cells at an approximate MOI of 100.
In Fig. 5 there are areas where bacteria appeared not to be expressing the BFP or EspA at 5 h. However, this phenomenon was due to discrepancies in the focal planes of the three-dimensional microcolony and did not represent an actual absence of staining. When the focal planes of this sample were adjusted during confocal microscopic examination, actin pedestals and BFP (or EspA filament) staining became visible. We did not have as much difficulty with discrepancies in focal planes at the 3-h time point because of the smaller numbers of bacteria adhering to host cells. However, focal plane discrepancies do not account for the decreased intimin staining observed at 5 h (Fig. 4). When the focal planes were altered during microscopic examination, we could see that a few of the bacteria remained intimin negative (data not shown). Together, these micrograph experiments showed that most, if not all, of the bacteria were efficiently expressing intimin, Tir, the BFP, and EspA on their surface at both 3 h and 5 h postinfection. These findings correlate with our transcription data indicating that the genes encoding these proteins continue to be expressed up to 5 h after infection with EPEC.
DISCUSSION
The hallmark of EPEC infection is the formation of A/E lesions on the intestinal epithelium (44). This phenotype is reproducible in in vitro experiments with tissue culture cell lines such as HEp-2, Caco-2, HeLa, and T84 (46). We sought to examine the transcriptional profile of EPEC virulence gene expression during a typical tissue culture experiment under conditions which are commonly used to demonstrate the FAS-positive and LA-positive phenotypes characteristic of EPEC infection. We chose to examine EPEC gene transcription during a 5-h infection of HEp-2 cells by real-time PCR. We compared the levels of transcription of several LEE operons as well as the bfp operon at 10 min, 3 h, and 5 h postinfection.
Our results showed that transcription from LEE3 to LEE5 as well as bfp was increased at 3 and 5 h, while transcription from LEE1 was decreased at these time points (Fig. 2a). Ler, encoded by the first gene of the LEE1 operon, is known to activate the downstream LEE operons (42). Therefore, we had initially hypothesized that transcription from LEE1 and the downstream LEE operons would occur in a cascade fashion. Instead, we observed that ler transcription levels did not appear to correlate with the levels of transcription from LEE3 to LEE5 during the course of infection. ler transcription levels decreased approximately 50% by 3 h postinfection and remained constant thereafter. Using mock infection experiments and a tir mutant, we were able to determine that this effect was dependent on the presence of host cells but not on intimate adherence of EPEC to the host cells (Fig. 2). In contrast, the transcriptional profile of LEE4 (espA) appeared to be independent of host cells. Cleary et al. (5) showed that EspA can be a sufficient adherence factor for EPEC independent of intimin and the BFP. espA transcription may respond to other environmental factors or even bacterial cell numbers in order to “prime” the bacteria for potential adherence opportunities. Additionally, we saw an increase in LEE3 to LEE5 and bfp transcription when the bacteria were grown in DMEM versus LB medium. In contrast, ler RNA levels remained the same regardless of the medium in which the bacteria were grown (Fig. 1). It is possible that Ler activity is actually increased under some conditions (such as growth in DMEM) and therefore increased transcription of ler is unnecessary. It also remains possible that other factors are able to affect transcription from the LEE in a Ler-independent fashion. In either scenario, an increase in ler transcription is not necessary for the increase in LEE3 to LEE5 transcription observed during the course of infection.
We correlated our transcription findings with protein expression levels at 3 and 5 h postinfection. Triton X-100 lysate fractionation and immunofluorescence on EPEC-infected HEp-2 cells showed that intimin was produced up to 5 h postinfection. Data reported previously by Knutton et al. (34) demonstrated that intimin production was drastically down-regulated at 6 h postinfection. Although we did not observe such a dramatic reduction in intimin production at 5 h, we did see a slight decrease in the number of bacteria expressing intimin (Fig. 4). Tir production appeared to be slightly decreased in the bacterial cytoplasm at 5 h; however, no change in the amount of translocated Tir was shown by Western blot analysis (Fig. 3) or confocal microscopy between the two time points that we used (Fig. 4). A previous report from our laboratory with CyaA fusions to Tir showed that Tir is translocated most efficiently at 40 min postinfection (8). Therefore, it is possible that the decrease in the amount of bacterial Tir present in the insoluble fraction at 5 h would not correlate with an immediate decrease in the amount of translocated Tir present in the host cell soluble fraction at that same time point.
The findings presented here clearly show expression, both transcriptional and translational, from LEE3 to LEE5 at time points at which we observed a significant decrease in ler expression. Although we cannot show with our system that Ler protein expression decreases over time, we do show that the levels of transcription and translation of the other genes tested are correlated. Therefore, it is possible that a Ler-independent mode of activation is responsible for the increase in transcription (and subsequent translation) of the downstream LEE operons. Sperandio et al. demonstrated that quorum sensing is involved in the activation of LEE1 in EPEC (52) and that this regulation occurs though QseA (51). It is possible that this quorum-sensing system activates the LEE in a Ler-independent manner as well or that there is an as-yet-undescribed regulator of the downstream LEE operons. Alternatively, Ler may be extremely stable when bound to DNA, and the initial surge of ler transcription (and subsequent translation) that we observed at 10 min postinfection may be able to produce enough Ler to allow the continued activation of the downstream LEE operons to occur efficiently. It is possible that the transcripts that we measured are extremely stable, so that the transcript levels that we detected after the initial increase may not have been due to new transcription. The average E. coli RNA half-life has been shown to be less than 10 min (43, 50). Since we measured transcript levels at 10, 180, and 300 min, it is unlikely that RNA stability affected our measurements at all of the time points. However, studies of the stability of LEE transcripts would help to address this possibility. Additional investigations are also necessary to determine whether another factor or other factors in EPEC are responsible for the potentially novel regulation pattern that we have demonstrated.
Additionally, we showed that EspA and the BFP are expressed on the bacterial cell surface at 3 and 5 h postinfection and that there is no down-regulation in protein expression. Of all the genes tested, bfp showed the most dramatic increase in transcription. We also showed that the BFP is clearly present on the bacterial surface 5 h after infection with EPEC. The BFP is essential for the apoptosis seen during the later stages of tissue culture infection with EPEC (1). Additionally, Tir and EspF have been shown to play a role in the induction of apoptosis in HeLa and COS cells as well (7, 38). Since a strain lacking the BFP was unable to induce apoptosis (1), we would expect the BFP to still be expressed during the later stages of infection. Additionally, we observed the initial stages of microcolony dissociation into blankets of bacteria coating host cells (Fig. 5), as previously described (36, 53). These morphological changes may be involved in the initial stages of BFP-mediated apoptosis. PerA, encoded on the EAF plasmid, is the transcriptional activator of the bfp operon (27, 54) and therefore is most likely responsible for the increase in BFP expression that we observed in our in vitro tissue culture infection. It would be interesting to expand this study in the future to examine the transcriptional profile of perA over time and to correlate that with BFP expression levels and with the morphological changes in the BFP which occur over the course of infection.
The data presented here help shed light on the manner in which EPEC virulence genes are expressed during the course of infection. The real-time PCR assay system has an advantage over a more traditional transcriptional reporter system (such as β-galactosidase) in that we are able to examine “snapshots” of the transcription of specific genes without the use of multicopy reporter plasmids and/or indirect assay systems. Our analysis of LEE gene expression during the infection process suggests that Ler, which is known to alleviate H-NS-mediated repression of the LEE, may actually be most necessary at the early stages of infection. The decrease in ler transcription to 50% that at the early stages suggests that ler activity is not as essential after the infection process has been well established. The connection between H-NS activity on the LEE and the need for Ler antirepression of H-NS to achieve LEE gene expression also bring into question the role of H-NS during the later stages of infection. Recently, two new regulators of the C. rodentium LEE, GrlA and GrlR, which act as an activator and as a repressor, respectively, were described (10). Both of these regulators were shown to act upstream of Ler in C. rodentium (10). It is possible that these proteins play a similar role in EPEC LEE regulation and may modulate ler gene expression during the course of infection. EPEC LEE gene regulation is a complex system with many components affecting the final outcome. Our study shows that LEE gene expression patterns vary at different stages of infection and suggest that this tightly controlled transcriptional modulation may also prove to be necessary in producing a productive infection in vivo.
Acknowledgments
We thank Alfredo Torres, Jane Michalski, and Kristen Kanack for helpful discussions and critical review of the manuscript. We also thank S. Vogel for the use of the ABI Prism 7900HT system.
This research was supported by NIH grant A121657 (to J.B.K.) and NIH training grant 2T32AI007540-06 (to L.Q.L.).
Editor: V. J. DiRita
REFERENCES
- 1.Abul-Milh, M., Y. Wu, B. Lau, C. A. Lingwood, and D. B. Foster. 2001. Induction of epithelial cell death including apoptosis by enteropathogenic Escherichia coli expressing bundle-forming pili. Infect. Immun. 69:7356-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 1997. Applied Biosystems user bulletin 2. The Perkin-Elmer Corp., Norwalk, Conn.
- 3.Bokete, T. N., T. S. Whittam, R. A. Wilson, C. R. Clausen, C. M. O'Callahan, S. L. Moseley, T. R. Fritsche, and P. I. Tarr. 1997. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J. Infect. Dis. 175:1382-1389. [DOI] [PubMed] [Google Scholar]
- 4.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 5.Cleary, J., L. C. Lai, R. K. Shaw, A. Straatman-Iwanowska, M. S. Donnenberg, G. Frankel, and S. Knutton. 2004. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 150:527-538. [DOI] [PubMed] [Google Scholar]
- 6.Crane, J. K., S. Majumdar, and D. F. Pickhardt III. 1999. Host cell death due to enteropathogenic Escherichia coli has features of apoptosis. Infect. Immun. 67:2575-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crane, J. K., B. P. McNamara, and M. S. Donnenberg. 2001. Role of EspF in host cell death induced by enteropathogenic Escherichia coli. Cell. Microbiol. 3:197-211. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, J. A., and J. B. Kaper. 2002. The N-terminus of enteropathogenic Escherichia coli (EPEC) Tir mediates transport across bacterial and eukaryotic cell membranes. Mol. Microbiol. 46:855-868. [DOI] [PubMed] [Google Scholar]
- 9.de Grado, M., A. Abe, A. Gauthier, O. Steele-Mortimer, R. DeVinney, and B. B. Finlay. 1999. Identification of the intimin-binding domain of Tir of enteropathogenic Escherichia coli. Cell. Microbiol. 1:7-17. [DOI] [PubMed] [Google Scholar]
- 10.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg, M. S., and J. B. Kaper. 1992. Enteropathogenic Escherichia coli. Infect. Immun. 60:3953-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott, S. J., E. O. Krejany, J. L. Mellies, R. M. Robins-Browne, C. Sasakawa, and J. B. Kaper. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 69:4027-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, S. J., C. B. O'Connell, A. Koutsouris, C. Brinkley, M. S. Donnenberg, G. Hecht, and J. B. Kaper. 2002. A gene from the locus of enterocyte effacement that is required for enteropathogenic Escherichia coli to increase tight-junction permeability encodes a chaperone for EspF. Infect. Immun. 70:2271-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, S. J., V. Sperandio, J. A. Girón, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 17.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 18.Girón, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 19.Girón, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg, M. D., M. Johnson, J. C. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41:549-559. [DOI] [PubMed] [Google Scholar]
- 21.Gómez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goosney, D. L., S. Gruenheid, and B. B. Finlay. 2000. Gut feelings: enteropathogenic E. coli (EPEC) interactions with the host. Annu. Rev. Cell Dev. Biol. 16:173-189. [DOI] [PubMed] [Google Scholar]
- 23.Grant, A. J., M. Farris, P. Alefounder, P. H. Williams, M. J. Woodward, and C. D. O'Connor. 2003. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48:507-521. [DOI] [PubMed] [Google Scholar]
- 24.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowlkes, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71:384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartland, E. L., M. Batchelor, R. M. Delahay, C. Hale, S. Matthews, G. Dougan, S. Knutton, I. Connerton, and G. Frankel. 1999. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol. 32:151-158. [DOI] [PubMed] [Google Scholar]
- 26.Hicks, S., G. Frankel, J. B. Kaper, G. Dougan, and A. D. Phillips. 1998. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect. Immun. 66:1570-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ibarra, J. A., M. I. Villalba, and J. L. Puente. 2003. Identification of the DNA binding sites of PerA, the transcriptional activator of the bfp and per operons in enteropathogenic Escherichia coli. J. Bacteriol. 185:2835-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ide, T., S. Laarmann, L. Greune, H. Schillers, H. Oberleithner, and M. A. Schmidt. 2001. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell. Microbiol. 3:669-679. [DOI] [PubMed] [Google Scholar]
- 29.Jarvis, K. G., J. A. Girón, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenkins, C., A. J. Lawson, T. Cheasty, G. A. Willshaw, P. Wright, G. Dougan, G. Frankel, and H. R. Smith. 2003. Subtyping intimin genes from enteropathogenic Escherichia coli associated with outbreaks and sporadic cases in the United Kingdom and Eire. Mol. Cell Probes 17:149-156. [DOI] [PubMed] [Google Scholar]
- 31.Jepson, M. A., S. Pellegrin, L. Peto, D. N. Banbury, A. D. Leard, H. Mellor, and B. Kenny. 2003. Synergistic roles for the Map and Tir effector molecules in mediating uptake of enteropathogenic Escherichia coli (EPEC) into non-phagocytic cells. Cell. Microbiol. 5:773-783. [DOI] [PubMed] [Google Scholar]
- 32.Kenny, B. 2002. Mechanism of action of EPEC type III effector molecules. Int. J. Med. Microbiol. 291:469-477. [DOI] [PubMed] [Google Scholar]
- 33.Kenny, B., S. Ellis, A. D. Leard, J. Warawa, H. Mellor, and M. A. Jepson. 2002. Co-ordinate regulation of distinct host cell signalling pathways by multifunctional enteropathogenic Escherichia coli effector molecules. Mol. Microbiol. 44:1095-1107. [DOI] [PubMed] [Google Scholar]
- 34.Knutton, S., J. Adu-Bobie, C. Bain, A. D. Phillips, G. Dougan, and G. Frankel. 1997. Down regulation of intimin expression during attaching and effacing enteropathogenic Escherichia coli adhesion. Infect. Immun. 65:1644-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knutton, S., R. K. Shaw, R. P. Anantha, M. S. Donnenberg, and A. A. Zorgani. 1999. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol. Microbiol. 33:499-509. [DOI] [PubMed] [Google Scholar]
- 37.Levine, M. M., E. J. Bergquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, and S. Sotman. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet 1:1119-1122. [DOI] [PubMed] [Google Scholar]
- 38.Malish, H. R., N. L. Freeman, D. V. Zurawski, P. Chowrashi, J. C. Ayoob, J. W. Sanger, and J. M. Sanger. 2003. Potential role of the EPEC translocated intimin receptor (Tir) in host apoptotic events. Apoptosis 8:179-190. [DOI] [PubMed] [Google Scholar]
- 39.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 41.McNamara, B. P., A. Koutsouris, C. B. O'Connell, J. P. Nougayrede, M. S. Donnenberg, and G. Hecht. 2001. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Investig. 107:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 43.Mohanty, B. K., and S. R. Kushner. 1999. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 34:1094-1108. [DOI] [PubMed] [Google Scholar]
- 44.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moyenuddin, M., I. K. Wachsmuth, S. L. Moseley, C. A. Bopp, and P. A. Blake. 1989. Serotype, antimicrobial resistance, and adherence properties of Escherichia coli strains associated with outbreaks of diarrheal illness in children in the United States. J. Clin. Microbiol. 27:2234-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puente, J. L., D. Bieber, S. W. Ramer, W. Murray, and G. K. Schoolnik. 1996. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20:87-100. [DOI] [PubMed] [Google Scholar]
- 48.Rosenshine, I., S. Ruschkowski, and B. B. Finlay. 1996. Expression of attaching/effacing activity by enteropathogenic Escherichia coli depends on growth phase, temperature, and protein synthesis upon contact with epithelial cells. Infect. Immun. 64:966-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez-SanMartin, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selinger, D. W., R. M. Saxena, K. J. Cheung, G. M. Church, and C. Rosenow. 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sperandio, V., J. L. Mellies, R. M. Delahay, G. Frankel, J. A. Crawford, W. Nguyen, and J. B. Kaper. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781-793. [DOI] [PubMed] [Google Scholar]
- 52.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tobe, T., and C. Sasakawa. 2001. Role of bundle-forming pilus of enteropathogenic Escherichia coli in host cell adherence and in microcolony development. Cell. Microbiol. 3:579-585. [DOI] [PubMed] [Google Scholar]
- 54.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 55.Tu, X., I. Nisan, C. Yona, E. Hanski, and I. Rosenshine. 2003. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol. Microbiol. 47:595-606. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, H. Z., and M. S. Donnenberg. 1996. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol. Microbiol. 21:787-797. [DOI] [PubMed] [Google Scholar]