Abstract
High-molecular-weight proteins of Haemophilus influenzae mediate attachment to epithelial cells. Previous reports describe several allelic versions of hmwA genes that have different adherence properties. Here we report three new alleles of hmwA (hmwA from strain AAr96, hmwA from strain AAr105, and hmwA from strain G822), demonstrating the high degree of DNA variation of these genes among different strains.
Nontypeable Haemophilus influenzae (NTHi), a gram-negative, nonencapsulated, human-specific microorganism, commonly inhabits the upper respiratory tract and causes otitis media, conjunctivitis, sinusitis, pneumonia, and acute exacerbation of chronic bronchitis. Occasionally, NTHi causes severe invasive diseases, such as meningitis, endocarditis, and bacteremia (14).
NTHi high-molecular-weight (HMW) proteins mediate bacterial attachment to epithelial cells (7, 11, 17, 18) and have been implicated as possible virulence factors for otitis media (12, 20) or chronic obstructive pulmonary disease (21). This adhesin is produced by the action of three genes (hmwA, hmwB, and hmwC) located in the hmw locus (1, 2). Many H. influenzae strains contain two distinct hmw loci, hmw-1 and hmw-2 (3, 4). The hmwA genes of these loci encode the HMWA adhesive proteins, which are 52 to 62% identical at the amino acid level (4) among several NTHi strains. hmwB genes encode outer membrane translocator proteins, which are 99% identical in NTHi strain 12. HMWB proteins are located in the outer membrane and serve to translocate HMWA across the outer membrane and prevent degradation by periplasmic proteases. hmwC genes encode cytoplasmic proteins, which are 97% identical in NTHi strain 12, and appear to stabilize HMWA (2, 17) and to influence glycosylation of HMWA1 (10). HMWA1 of strain 12 mediates binding to α-2,3-linked sialylated glycoproteins, and the epithelial cell receptor structure for HMWA2 of this strain is unknown (17).
While initial studies of hmwA described the alleles hmwA1 and hmwA2, one in each hmw locus of strain 12 (3), Van Schilfgaarde et al. (21) described a third hmwA allele from the chronic obstructive pulmonary disease H. influenzae strain A950006, whose 4,671-bp gene encodes a predicted protein with 70% homology to HMWA1 and 68% homology to HMWA2 of strain 12. More recently, Buscher et al. (4) have identified four additional hmwA alleles with either HMWA1- or HMWA2-like binding characteristics from two NTHi strains and showed that the genes encoding these differential binding characteristics were variably located downstream of either HI01679 or HI01598 in the Rd genome.
Although type b strains lack hmw loci, 55 to 80% of NTHi strains have these genes (1, 8, 20). Among the other encapsulated H. influenzae types, hmw loci were detected in 26% of type a, 8% of type e, and 5% of type f strains (16). In a previous study by our group using dot blot hybridization, 51% of NTHi isolates hybridized with strain 12 hmwA1-specific probes, 23% hybridized with hmwA2-specific probes, and 48% hybridized with hmwC-specific probes (8). While 18% hybridized with all three probes, 23% hybridized with only hmwA1 and hmwC probes, 1% hybridized with only hmwA2 and hmwC probes, and 6% (10 isolates) hybridized with only the hmwC probe, suggesting that many strains may contain hmwA genes that do not hybridize the strain 12-specific probes. Specifically, NTHi strains AAr96, AAr105, and G822 failed to hybridize with gene probes targeting the unique regions hmwA1 and hmwA2 of strain 12 but hybridized with a probe targeting the conserved hmwC genes. In the present study, we investigate the possibility that these three strains contain allelic versions of hmwA that failed to hybridize with the strain 12-specific probes.
Table 1 lists the primers synthesized by the University of Michigan Biomedical Research Core Facility that were used to amplify regions of the hmwA genes from NTHi strains AAr96, AAr105, and G822. For all PCRs, NTHi strain 12 served as the positive control and the hmw-deficient NTHi strain 11 served as the negative control (3). All PCRs were carried out as previously described (8). The resulting PCR products were cloned into the plasmid vector pCR4-TOPO (Invitrogen, Carlsbad, Calif.), and the resulting recombinant plasmids were transformed into TOP10 Escherichia coli host cells (Invitrogen). Insert regions were sequenced at the University of Michigan Sequencing Core Facility, using Applied Biosystems model 3700 and 3730 automated sequencers. DNA and protein sequences were aligned and compared by using Lasergene Biocomputing software (DNASTAR, Inc., Madison, Wis.). Multiple alignments were performed with the CLUSTAL W program. The Lipman-Pearson algorithm (Ktuple = 2; gap penalty = 4; gap length penalty = 12) was used for pairwise alignment.
TABLE 1.
PCR primers used in this study
| Target region amplifieda | Primerb | bp positions in corresponding gene | Nucleotide sequence (5′-3′) |
|---|---|---|---|
| hmwA gene | hmwAF | 34-57 (hmwA1/A2) | CGCCTGAATGCTTTGGTTGCTGTG |
| hmwAR | 1055-1078 (hmwA1/A2) | CGCCGCGCTCGTCACCGCCAAGGT | |
| Variable internal fragment of hmwA genes | hmwMF | 1212-1235 (hmwA1/A2) | CGCTCAAGGTAGTGGTGATATCGC |
| hmwMR | 3150-3173 (hmwA1) | CCATCTTTAGCTGTAATCTCTGCT | |
| hmwA1 probe | hmw1AF | 1610-1633 | CCACCGGTGATGATACCAGAGGTG |
| hmw1AR | 2507-2530 | CGGCTTTCCTGGAGCCAAAGGTGA | |
| hmwA2 probe | hmw2AF | 1771-1794 | GTCGCCCAGGGCACTGTAACCATT |
| hmw2AR | 2478-2501 | CCGCCCAGAATGGATATGTTGTAG | |
| 5′ fragment from AAr96 | HI01598 AAr96R | 142-163 | GTTTAGCAAGAAAATGATCGGG |
| 1309-1327 | CCGGGTCTAACAACCACTC | ||
| AAr105 | HI01679 | 455-479 | CCGATATGATTTTACAGGCACAGGG |
| AAr105R | 1341-1358 | CGTCCAGCAGGAGCATCA | |
| G822 | HI01679 G822R | CCGATATGATTTTACAGGCACAGGG | |
| 1829-1849 | TCGCGCCACTAATGTTGTGTG | ||
| 3′ fragment from AAr96 | hmwR | 82-104/79-101 (3′ of hmwA1/ A2, respectively) | CAAGATGGGTAAAGCCCGTACTG |
| AAr96F | 3042-3062 | GGGTATTGATGTAGAGAGCTC | |
| AAr105 | hmwR AAr105F | CAAGATGGGTAAAGCCCCGTACTG | |
| 3049-3069 | GATGGAGAGAGCTCTGTTCCA | ||
| G822 | hmwR G822F | CAAGATGGGTAAAGCCCGTACTG | |
| 2989-3009 | GGCGTTGATGGGGAGAGTTCT |
Strain names are given to indicate those regions of hmwA that are strain specific.
F, forward primer; R, reverse primer.
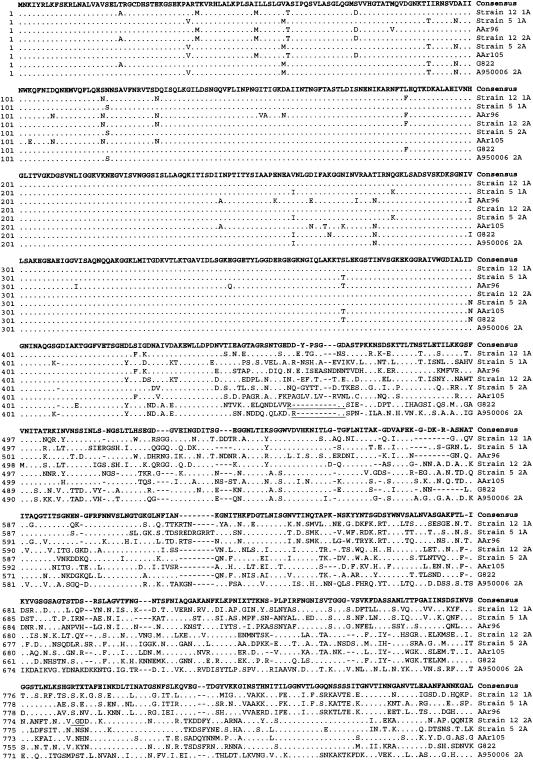
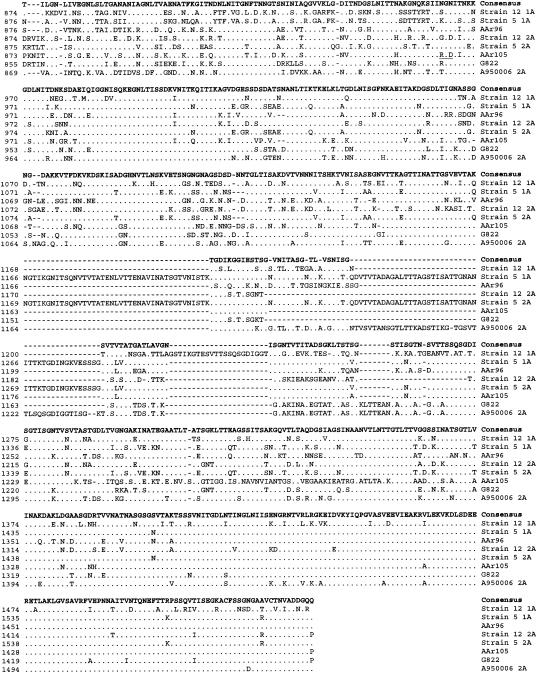
hmwA genes from strains AAr96, AAr105, and G822 were 4,428, 4,476, and 4,449 bp in length, respectively. Figure 1 compares the predicted amino acid sequences of these three hmwA genes to the five previously described and available in GenBank. In all eight proteins, the N-terminal 440 amino acids of HMWA are highly conserved and the C-terminal 100 amino acids are moderately conserved, but the region between, which corresponds to the epithelial cell binding domain (7), is extremely variable. In pairwise comparisons, using the Lipman-Pearson algorithm, amino acid identities between the eight alleles varied from 66 to 77% (Table 2). DNA sequence analysis 5′ of each hmwA gene in this study revealed that the genes from strains AAr105 and G822 were adjacent to the homologous Rd gene HI01679, while hmwA gene in strain AAr96 was adjacent to the HI01598 homolog (Table 2).
FIG. 1.
Comparison of the predicted amino acid sequences of HMWA from the strains AAr96, AAr105, and G822 with the deduced amino acid sequences of HMW-1 and HMW-2 from nontypeable Hi strains 12, 5, and A950006 previously reported. The HMWA protein sequences were aligned by using the CLUSTAL algorithm in the DNAStar software. Identical amino acids in all HMWA sequences are indicated with dots, and gaps introduced to maximize alignments are indicated by dashed lines. At the top in bold is a consensus amino acid sequence in which all letters except X represent an amino acid present in at least three of the six predicted proteins, and X means there is no consensus amino acid at that position. The RGD sequences are underlined.
TABLE 2.
Comparison of predicted amino acid sequences of HMWA from strains AAr96, AAr105, and G822 with HMWA protein sequences from strains 12, 5, and A950006
| HMW proteina | % Identity with sequence from straina:
|
||
|---|---|---|---|
| AAr96 (HI01598) | AAr105 (HI01679) | G822 (HI01679) | |
| Strain 12 A1 (HI01679) | 72 | 69 | 66 |
| Strain 12 A2 (HI01598) | 67 | 74 | 76 |
| Strain 5 A1-like (HI1598) | 73 | 66 | 68 |
| Strain 5 A2-like (HI1679) | 67 | 75 | 76 |
| A950006 A2-like (HI1598) | 69 | 68 | 73 |
Parentheses contain 5′ flanking gene based on the Rd genome sequence.
HMW proteins are structural and functional analogs of the filamentous hemagglutinin of Bordetella pertussis (1), which mediates binding by an arginine-glycine-aspartic acid (RGD) motif. Because this motif is also present in the derived amino acid sequence of HMWA2 of strain 12, RGD-mediated adherence of HMWA2 to the integrin CR3 has been suggested (15, 21). The RGD tripeptide motif of HMWA2 from strain 12 (amino acids 785 to 787) (1) differs in location from the motif in strain AAr105 (amino acids 963 to 965), in strain G822 (amino acids 459 to 461), and in strain A950006 (amino acids 460 to 462). Examination of the previously reported HMW amino acid sequences of strain AAr96 and strain 5 (4) reveals no RGD tripeptide motifs (Fig. 1).
HMWA1 and HMWA2 of strain 12 localize to the surface of NTHi by a two-step process involving first cleavage between amino acids 68 and 69 and then cleavage of a 441-amino-acid N-terminal fragment (1, 9, 19). All eight HMWA proteins show strong N-terminal amino acid homology (Fig. 1) in this region, suggesting that the immature proteins are cleaved by the same mechanisms in all strains.
Previous studies reveal that the binding domains of HMWA1 and HMWA2 of strain 12 are located in the approximately 360 amino acids near the N termini of the mature proteins (7). This region (Fig. 1) shows significant sequence diversity among the eight different alleles, which may result both in functional differences in adherence to human cells and in antigenic variation (21, 4).
The 5′ regions immediately upstream of the initiation codons of hmwA1 and hmwA2 in strain 12 and of hmwA of strain A950006 all contain 16 to 22 copies of a 7-bp tandem direct repeat sequence (ATCTTTC) whose variation in number can result in phase variation of expression of the hmwA genes (1, 6, 21). Strains AAr96, AAr105, and G822 in this study all had 16 copies of the repeat sequence.
In conclusion, we identified three additional hmwA alleles of NTHi, which is reminiscent of the allelic diversity of genes encoding both the structural protein (hifA) and the adhesin (hifE) of H. influenzae hemagglutinating pili (5, 13). The most conserved domain of the HMW-like proteins is the N terminus region of the immature protein, which traffics the proteins to the cell surface and is then cleaved from the mature proteins, while the most variable domain is the receptor binding region (6). While the antigenic domains corresponding to protective antibodies of HMW remain unknown, the extreme sequence diversity of the binding domain suggests that these proteins may vary antigenically as well as functionally.
Nucleotide sequence accession numbers.
Sequences determined in this work have been submitted to GenBank with the following accession numbers: for hmwA from strain AAr96, AY601284; for hmwA from strain AAr105, AY601283; and for hmwA from strain G822, AY601282.
Acknowledgments
We thank May Patel for her expert technical assistance.
This study was supported by awards AI 25630 and DC05840 to J.R.G. from the National Institutes of Health.
Editor: J. N. Weiser
REFERENCES
- 1.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of surface-exposed B-cell epitopes on high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae. Infect. Immun. 64:3032-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by nontypable Haemophilus influenzae. Mol. Microbiol. 19:1215-1223. [DOI] [PubMed] [Google Scholar]
- 4.Buscher, A., Z. K. Burmeister, S. J. Barenkamp, and J. W. St. Geme III. 2004. Evolutionary and functional relationships among the nontypeable Haemophilus influenzae HMW family of adhesins. J. Bacteriol. 186:4209-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemans, D. L., C. F. Marrs, M. Patel, M. Duncan, and J. R. Gilsdorf. 1998. Comparative analysis of Haemophilus influenzae hifA (pilin) genes. Infect. Immun. 66:656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawid, S., S. J. Barenkamp, and J. W. St. Geme III. 1999. Variation in expression of the Haemophilus influenzae HMW adhesins: a prokaryotic system reminiscent of eukaryotes. Proc. Natl. Acad. Sci. USA 96:1077-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawid, S., S. Grass, and J. W. St. Geme III. 2001. Mapping of binding domains of nontypeable Haemophilus influenzae HMW1 and HMW2 adhesins. Infect. Immun. 69:307-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ecevit, I. Z., K. W. McCrea, M. M. Pettigrew, A. Sen, C. F. Marrs, and J. R. Gilsdorf. 2004. Prevalence of hifBC, hmw1A, hmw2A, hmwC, and hia genes in Haemophilus influenzae isolates. J. Clin. Microbiol. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grass, S., and J. W. St. Geme III. 2000. Maturation and secretion of the non-typable Haemophilus influenzae HMW1 adhesin: roles of the N-terminal and C-terminal domains. Mol. Microbiol. 36:55-67. [DOI] [PubMed] [Google Scholar]
- 10.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St. Geme III. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 11.Hultgren, S. J., S. Abraham, M. Caparon, P. Falk, J. W. St. Geme III, and S. Normark. 1993. Pilus and nonpilus bacterial adhesions: assembly and function in cell recognition. Cell 73:887-901. [DOI] [PubMed] [Google Scholar]
- 12.Krasan, G. P., D. Cutter, S. L. Block, and J. W. St. Geme III. 1999. Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect. Immun. 67:449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCrea, K. W., J. L. St. Sauver, C. F. Marrs, D. Clemans, and J. R. Gilsdorf. 1998. Immunologic and structural relationship of the minor pilus subunits among Haemophilus influenzae isolates. Infect. Immun. 66:4788-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy, T. F., and M. A. Apicella. 1987. Nontypeable Haemophilus influenzae: a review of clinical aspects, surface antigens, and the human immune response to infection. Rev. Infect. Dis. 9:1-15. [DOI] [PubMed] [Google Scholar]
- 15.Relman, D., E. Tuomanen, S. Falkow, D. T. Golenbock, K. Saukkonen, and S. D. Wright. 1990. Recognition of a bacterial adhesion by an integrin: macrophage CR3 (alpha M beta 2, CD11b/CD18) binds filamentous hemagglutinin of Bordetella pertussis. Cell 61:1375-1382. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez, C. A., V. Avadhanula, A. Buscher, A. L. Smith, J. W. St. Geme III, and E. E. Adderson. 2003. Prevalence and distribution of adhesins in invasive non-type b encapsulated Haemophilus influenzae. Infect. Immun. 71:1635-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.St. Geme, J. W., III. 1994. The HMW1 adhesin of nontypeable Haemophilus influenzae recognizes sialylated glycoprotein receptors on cultured human epithelial cells. Infect. Immun. 62:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.St. Geme, J. W., III, S. Falkow, and S. J. Barenkamp. 1993. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 90:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.St. Geme, J. W., III, and S. Grass. 1998. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 27:617-630. [DOI] [PubMed] [Google Scholar]
- 20.St. Geme, J. W., III, V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66:364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Schilfgaarde, M., P. van Ulsen, P. Eijk, M. Brand, M. Stam, J. Kouame, L. van Alphen, and J. Dankert. 2000. Characterization of adherence of nontypeable Haemophilus influenzae to human epithelial cells. Infect. Immun. 68:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]