Abstract
Cells exposed to densely ionizing radiation (high-LET) experience more severe biological damage than do cells exposed to sparsely ionizing radiation (low-LET). The prevailing hypothesis is that high-LET radiations induce DNA double strand-breaks (DSB) that are more complex and clustered, and are thereby more challenging to repair. Here, we present experimental data obtained by atomic force microscopy imaging, DNA-dependent protein kinase (DNA-PK) activity determination, DNA ligation assays, and genomic studies to suggest that short DNA fragments are important products of radiation-induced DNA lesions, and that the lengths of DNA fragments may be significant in the cellular responses to ionizing radiation. We propose the presence of a subset of short DNA fragments that may affect cell survival and genetic stability following exposure to ionizing radiation, and that the enhanced biological effects of high-LET radiation may be explained, in part, by the production of increased quantities of short DNA fragments.
Introduction
Ionizing radiation induces a large variety of damage in cellular DNA; the majority of the damage can be effectively repaired by cellular repair mechanisms. The type of damage that is most difficult to repair and frequently leads to cell death has been identified as the DNA double strand break (DSB) (1).
Established biological observations implicate DNA DSBs resulting from high-LET radiations in cell death and carcinogenesis to a greater extent than that observed following low-LET radiations (2,3). The mechanism for such observation has been proposed to be the complex distribution of the ionizing events resulting in DNA damage that is more difficult to repair (4,5,6). High-LET radiations produce DSBs that are more densely distributed and clustered, while low-LET radiations produce more sparsely distributed DSBs. Clustered DSBs are believed to be an important component of the mechanism for enhanced cell killing by high-LET radiations (7,8,9).
Ward proposed the Multiply Damaged Sites model (MDS) to suggest that multiple damages that occur within a small spatial region (a few nm) may be more challenging to cellular repair mechanisms (9,10). Based on results of Monte Carlo simulations, Goodhead and colleagues later proposed a clustered DNA damage mechanism as a possible cause for the enhanced radiation biological effect (RBE) of high-LET radiations (11). These hypotheses, while significantly improving our understanding of mechanisms of radiation damage, are largely qualitative. Furthermore, experimental data in support of these hypotheses are sparse. As has been proposed in a review by Goodhead: “It is now a major challenge to devise experimental methods to recognize and quantify by measurement radiation-induced ‘complex’ clustered DNA damages of varying complexities, and to assess their biological implications” (12).
Cellular responses to DNA DSB resulting from ionizing radiation and other damaging agents follow two principal repair mechanisms: non-homologous end joining (NHEJ) and homologous recombination (HR) (13,14,15,16). In mammalian cells NHEJ is believed to be the dominant DSB repair pathway. Core proteins known to be involved in NHEJ include Ku, DNA-PKcs, XRCC4 and DNA ligase IV (17,18,19). The appearance of free DNA ends in a cell after exposure to radiation initiates a sequence of repair processes; the earliest steps involve DNA end-binding proteins. These proteins protect DNA ends from degradation and recruit other, downstream proteins to perform their repair functions.
In this report, we investigate the molecular significance of short DNA fragments resulting from clustered DSB in cellular responses to ionizing radiation, in a model system using synthesized short DNA fragments for comparison. Binding interactions between Ku/DNA-PKcs and short DNA fragments, kinase activity of DNA-PK, and the biologically significant endpoint of ligation by the NHEJ pathway are examined with respect to DNA fragment length and end structure. Furthermore, we investigate the effects of short DNA fragments on cellular genomic stability by studying the DNA copy number changes in cells transfected with short DNA fragments.
Our results indicate that short DNA fragments formed by radiation-induced DSB clustering may play an important role in cellular responses to ionizing radiation. Cells may be unable to rejoin short DNA fragments due to impaired interactions with repair proteins, and short fragments may interfere with interactions of longer DNA fragments and with DSB repair proteins.
Materials and Methods
Plasmid and Genomic DNA Irradiation
Low-LET photon irradiation of pUC19 plasmid and genomic DNA was performed on an industrial Co-60 γ-irradiator at Neutron Products, Inc in Germantown, Maryland. Briefly, this is a high dose rate radiator capable of 20 kGy/hr. Genomic DNA samples were held in micro-centrifuge tubes and irradiated to 5 kGy.
High-LET neutron irradiation was performed at the TRIGA MARK-F neutron reactor at the Armed Forces Radiobiology Research Institute (AFRRI) in Bethesda, MD. This is a fission neutron reactor that can generate a neutron beam in either pulsed or steady-state mode. High neutron fields can be obtained by adjustment of the control rods of the reactor core. A bismuth enclosure selectively shields samples from the γ-rays produced by the core or from the walls of the exposure room, resulting in a neutron yield of 97% in the radiation fields. The fluence-weighted mean energy of the neutron beam is 0.71 MeV and the mean LET is 55 keV/μm. Details of the radiation characteristics and sample irradiations can be found in a previous publication (20).
The heavy-ion Argon irradiation was conducted at the NIRS-HIMAC charged-particle accelerator facility in Chiba, Japan. Samples of pUC19 plasmid and genomic DNA in buffer solution containing 10 mM HEPES and 1 mM MgCl2 were held in custom-designed, vacuum-tight cylindrical chambers with a diameter of 6 mm and height of 1 mm with a capacity of 28 μl solution were irradiated in a vacuum chamber at room temperature to doses of 5 and 10 kGy, respectively. The energy of the Argon ion was 390 MeV/nucleon and the LET was 99.5 keV/μm. The doses delivered to DNA samples were calculated as the product of the argon particle fluence and the LET of the particles multiplied by the time the beam was on. Details of the beam characteristics and dosimetry of the HIMAC facility can be found elsewhere (21,22).
Kinase Assay Oligonucleotide Preparation
The synthetic oligonucleotides used in the DNA-PK assays were synthesized by Operon Biotechnologies, Inc. (Huntsville, AL), and designed as random sequence blunt-ended duplexes. We further purified all of the oligonucleotides using 12% or 15% denaturing polyacrylamide gel electrophoresis (PAGE), and then determined the concentrations spectrophotometrically. Duplex DNA substrates were formed by mixing oligonucleotides with an equal amount of complementary oligonucleotide in a buffer containing 10 mM Tris-hydrochloride, pH 8.0, 1 mM EDTA, pH 8.0, and 100 mM NaCl. The mixture was heated at 100°C for 5 min, allowed to cool to room temperature for 3h, and then incubated at 4°C overnight.
DNA-PK kinase activity assays
DNA-PKcs and Ku proteins were purified by HPLC and stored in 50 mM Tris-HCl (PH 7.9), 1 mM EDTA, 0.02% Tween, 20% glycerol, 0.1 M KCl plus 1 μg/ml of a mixture (aprotinin, leupeptin, soybean trypsin inhibitor) and 10 μg/ml of PMSF. 50 ng/μl of DNA-PKcs and 20 ng/μl of Ku were used per 25 μl kinase assay.
Synthesized DNAs ranged from 14-36 bp, while PCR was used to produce 300-bp DNA fragments from pUC19. Radiation-generated DNA fragments were produced from genomic DNA that was irradiated to a dose of 5 kGy with neutrons or Co-60 γ-rays. The irradiated DNA fragments were separated by 20% nondenatured polyacrylamide gel electrophoresis and based on DNA molecular markers, short DNA fragments < 20 bp, 20-40 bp, and 40-100 bp were eluted from the gel and used in the DNA-PK kinase assay. A GST-p53 fusion protein (Santa Cruz, CA) was used as the kinase assay substrate in the presence of γ-32P-ATP. Typically, DNA-PKcs, Ku protein, and 20-50 pM double-stranded DNA fragments of various sizes were introduced in kinase buffer for reaction at 37°C for 30 mins. For competition assays, DNA-PKcs and Ku proteins were incubated with 14-mer or 20-mer at 37°C for 30 min first, 300 bp DNA fragments, p53, and γ-32P -ATP were then sequentially added in the reaction. Phosphorylated p53 proteins were separated by 7.5% SDS-PAGE and visualized by autoradiography. The intensities of the radiolabeled p53 bands were determined using an Alpha Imager 2000 analysis system and normalized to the intensity of the radiolabeled p53 band exhibited by the intrinsic Ku + DNA-PKcs kinase activity alone (i.e., when reactions were run with no short DNA fragments added).
Cellular transfection of short DNA fragments
MCF10A non-malignant human mammary epithelial cells were used as recipients of transfected DNA fragments. The cells were grown in P4-8F (Biological Search Faculty and Facility, Inc, Jamesville, MD) supplemental with 2% (v/v) heat-inactivated fetal bovine serum (Gibco-RBL), 5 mg/ml hydrocortisone, 100 U/ml penicillin G, and 100 U/ml streptomycin. Cells were maintained at 37% air and 5% CO2. Cell cultures were routinely screened for mycoplasma by a nucleic acid hybridization assay (Gen-Probe, San Diego, CA). Genomic short DNA (20-400 bp) fragments generated by γ-ray or neutrons were introduced to MCF-10A cells by using lipofectin according to the manufacture's instructions (Invitrogen). Linear plasmid DNA was used as a control. To monitor transfection efficacy, pGFP was co-transfected with the DNA fragments. After 4 days transfection, the GFP expression was determined using fluorescent microscopy. Greater than 50 % of the cells showed GFP expression. After transfection, the cells were passaged for 25-30 doublings, and a number of colonies were subcloned by performing a soft agar assay. Representative clonal cells were subjected to examination for genetic changes using comparative genomic hybridization (CGH) analysis.
Anchorage-independent Growth
Control and transfected MCF10A cells in cultures were trypsinized and resuspended at densities of 5 × 104 cells/ml in 2 ml of 0.35% Noble agar and overlaid in 60-mm dishes containing 0.6% agarose base. The media were replenished every 7 days, and viable anchorage-independent macroscopic colonies containing >50 cells (>0.2 mm) were observed at 21 days (23). Colonies selected for anchorage-independent growth assays were expanded for CGH assays.
Detection of genomic instability
Genomic instability in the transfected cells was assessed using CGH. CGH is a molecular cytogenetic screening technique that allows the identification of chromosomal imbalances (DNA gains and losses) in the test genome. Differentially labeled DNA from transfected cells and reference control DNA from the lymphocytes of a karyotypically normal donor were prepared using standard protocols. Comparative genomic hybridization (chromosomal CGH) analysis was performed using protocols that we have previously published (24). Quantitative evaluation of the hybridization was completed using a commercially available software package (Applied Imaging, Pittsburgh, PA, USA). Average ratio profiles were computed as the mean value of 8 ratio images, and were used to identify changes in chromosome copy number.
HeLa Cell Culture and Extraction
Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum, non-essential amino acids (10 mM), glutamine (100 mM), penicillin/streptomycin (10,000 U/ml), T4 DNA ligase, and Novex™ gels were obtained from Invitrogen (Carlsbad, CA).
HeLa cells were grown as monolayers at 37°C in DMEM containing 10% (v/v) fetal bovine serum, 1% (v/v) non-essential amino acids and 1% (v/v) penicillin/streptomycin. Cells were harvested by trypsinization and a representative flask was counted. Cells were washed two times by re-suspension in complete media followed by centrifugation at 800 × g for 5 min. The resulting cell pellet was washed two times with ice cold PBS in a similar manner. The cells were re-suspended in extraction buffer (10 mM HEPES, pH 7.9, 60 mM KCl, 1 mM DTT, 1 mM EDTA, 10 mM pefablock, 10 μg/ml aprotinin, 1.5 μg/ml leupeptin, 100 μg/ml bestatin) at a density of 8×107 cells/ml and lysed by the freeze/squeeze method using three cycles of freezing in a dry ice ethanol bath followed by immediate thawing at 37°C. After the third thaw cycle, cell debris was removed by centrifugation at 16000 × g for 30 minutes at 4°C. The supernatant constituted the HeLa whole cell extract (WCE) and was stored at −80°C in aliquots.
Oligonucleotide Based End-joining Competition Assay
The oligonucleotides used to prepare the duplex NHEJ substrate, and competitor duplexes that range in size from 10 bp to 56 bp, were purchased from Integrated DNA Technologies (IDT, San Diego, CA) and band purified from 12% denaturing PAGE gels in our laboratory. [γ–32P]-ATP (222 TBq/mmol) was purchased from Perkin-Elmer Biosciences (Boston, MA). The T4 polynucleotide Kinase (T4 PNK) used in [5′-32P] end labeling, and T4 DNA ligase, were obtained from Fermentas (Hanover, MD).
The 12 oligonucleotides presented in Table 1 constitute 6, paired, complementary oligos that upon annealing, form 6 duplex oligonucleotides ranging in size from 10 bp to 75 bp. All of the duplex oligos possess an unligatable blunt end (5′- and 3′-OH), and a ligatable (5′-PO4 and 3′-OH) self-complementary 4-base 5′ overhang. In all cases, duplex oligonucleotides were formed by annealing equimolar (5 pmol each) amounts of complementary upper and lower strand oligonucleotides in 500 mM Tris–HCl (pH 8.0) at a final vol. of 20 μl, and incubating at 90°C for 5 min, followed by gradual cooling to room temperature (RT). Duplex formation was confirmed in each case by 6% non-denaturing PAGE.
Table 1. Duplex Oligonucleotides Used in in vitro NHEJ Assays.
| a) 75 mer (Standard NHEJ-assay substrate) |
| 5′- TAG AGA CGG GAT GAG TGG AAT TAG GAC TGA GAC TAT GGT TGC TGA CTA ATC GAG ACC CAT CAT TAG CAT AGT TGC |
| 3′- ATC TCT GCC CTA CTC ACC TTA ATC CTG ACT CTG ATA CCA ACG ACT GAT TAG CTC TGG GTA GTA ATC GTA TCA ACG CAT G* |
| *Indicates 5′-32PO4 |
| Short Competitor Oligos |
| b) 56 mer duplex |
| 5′- AT TAG GAC TGA GAC TAT GGT TGC TGA CTA ATC GAG ACC CAT CAT TAG CAT AGT TGC |
| 3′- TA ATC CTG ACT CTG ATA CCA ACG ACT GAT TAG CTCTGG GTA GTA ATC GTA TCA ACG GTA C |
| c) 42 mer duplex |
| 5′- TAT GGT TGC TGA CTA ATC GAG ACC CAT CAT TAG CAT AGT TGC |
| 3′- ATA CCA ACG ACT GAT TAG CTC TGG GTA GTA ATC GTA TCA ACG GTA C |
| d) 32 mer duplex |
| 5′- GA CTA ATC GAG ACC CAT CAT TAG CAT AGT TGC |
| 3′- CT GAT TAG CTC TGG GTA GTA ATC GTA TCA ACG GTA C |
| e) 21 mer duplex |
| 5′- ACC CAT CAT TAG CAT AGT TGC |
| 3′- TGG GTA GTA ATC GTA TCA ACG GTA C |
| f) 10 mer duplex |
| 5′- G CAT AGT TGC |
| 3′- C GTA TCA ACG GTA C |
Our standard in vitro NHEJ assay (25) employs a 75 base pair duplex oligonucleotide (a 75 mer upper strand and a 5′-32P end labeled 79 mer lower strand) as the primary end-joining substrate (Table 1a), and HeLa whole cell extract (WCE) as the source of the NHEJ proteins. Typical reactions were run at 17°C for 90 min. and contained 50 mM Tris-HCl pH 7.6, 5 mM MgCl2, 1 mM ATP, 1 mM DTT, 5% polyethyleneglycol (PEG) 8000, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 10 μg/ml bestatin, 1 mM pefablock, 400 fmole 75-mer oligo-duplex substrate and 5 μg HeLa WCE protein. The reactions were stopped at 65°C for 15 min. Reactions were resolved in a 20% denaturing polyacrylamide gel run in 1× TBE (pH 8.4). Detection of end-joined products was achieved by phosphorimaging. Gels were visualized using a Fuji phosphorimager-2500 and analyzed by Image Gauge v3.45 software. Negative controls consisted of duplex 75-mer oligo alone without added protein, while positive controls for oligo dimer formation were produced by ligation using T4 DNA ligase (1U per reaction).
Size dependent competitive effects on end joining were assessed in the standard assay by the incorporation of unlabeled duplex oligonucleotides ranging in size from 10 bp to 56 bp (Table 1b-f). The competitor oligonucleotides were added in amounts of 0, 100, 200, 400, 800, and 1600 fmoles to the reaction mix containing the labeled 75-mer-substrate duplex on ice before initiation of the end joining reaction by addition of HeLa WCE. The relative amount of the end-joined 150-bp dimer product formed in the standard assay is a measure of the end-joining efficiency of the WCE mediated NHEJ reaction. In the presence of competitor duplex oligonucleotides, a reduction in the amount of 150-bp dimer product formed implicates competition between the primary 75-mer duplex substrate and the competitor oligonucleotides for proteins that constitute the putative NHEJ complex.
Results
High-LET argon irradiation generates a significantly larger fraction of short DNA fragments than low-LET γ photon irradiation
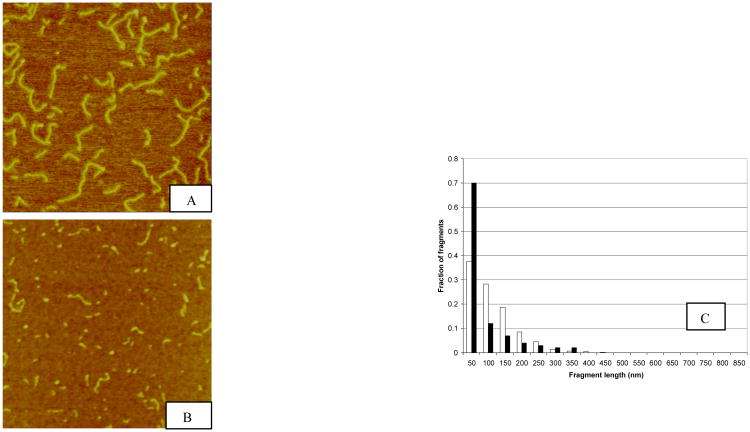
We have previously reported that neutron irradiation of pUC19 plasmid DNA (2864 bp) generates a significantly larger fraction of short DNA fragments in the 0-50 nm range (<147 bp) compared to photon irradiated plasmids (20,26). In subsequent experiments, we observed even greater quantities of short DNA fragments following plasmid irradiation by Argon ions at the HIMAC charged particle facility in Chiba, Japan. Figure 1 shows sample AFM images of pUC19 plasmids irradiated with 10 kGy Co-60 γ photons and 390 MeV/nucleon Argon ions, along with the corresponding DNA fragment size distributions. As shown, with Argon irradiation, nearly 70% of the DNA fragments generated are distributed in the 0-50 nm bin, whereas for Co-60 γ photon, the fraction of fragments generated is just over ∼35%.
Fig.1.
Sample AFM images of pUC19 DNA irradiated to 10 kGy Co-60 γ-photon (Panel A) and 390 MeV/nucleon Argon ion (Panel B) and the corresponding fragment size distributions (Panel C). The fragment size distributions were obtained by measuring the length of each fragment and group them into bins of 50-nm interval. The number of fragments in each bin was divided by the total number of fragments to yield the fractional number of fragments. 850 nm is the length of an intact pUC19 plasmid DNA molecule. In Panel C the black bars represent DNA fragment length distribution resulting from argon ion irradiation whereas white bars for Co-60 γ photon irradiation. Evident in this graph is the pronounced fraction of short DNA fragments generated by the high-LET Argon ions in the 50-nm bin.
The scavenging capacity of the buffer solution in which the DNA were irradiated was low in comparison to that in a cellular environment. Consequently, the majority of the DSBs formed, result from free radical induced damage. Compared to low-LET radiation, high-LET radiations induce densely and clustered free radical formation, which translates to clustered DSBs.
DNA-PK kinase activation is DNA fragment length dependent but not DNA end structure dependent
Based on our observation that high-LET radiation generates substantially larger amounts of short DNA fragments than low-LET radiation, we sought to investigate the biological relevance of these short DNA fragments by first looking at their binding interactions with Ku protein, the protein involved in the early response to DSBs in NHEJ. Using synthesized duplex oligonucleotides of various lengths as DNA substrates, we confirmed that Ku binding to DNA requires a minimum length of 14 bp to be effective (data not shown), a result consistent with previous publications (27,28).
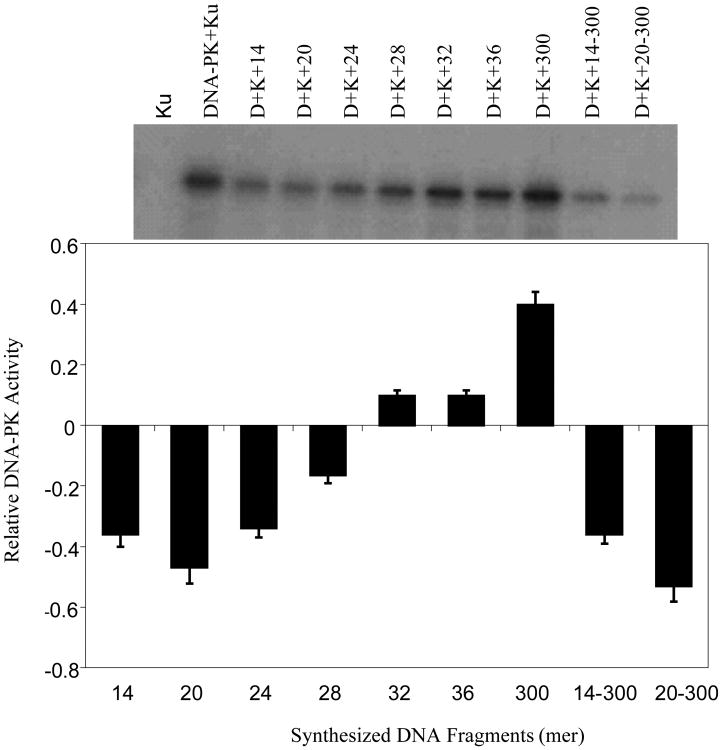
Since the cellular function of DNA-PK is largely through its kinase activity, we next tested purified DNA-PK complex for its ability to phosphorylate p53 in the presence of DNA fragments ranging from <20 bp to 300 bp. DNA-PK has been shown to phosphorylate a large number of targets, including Rpa32 (S4/8), Chk1 (S345), Chk2 (T68), Nbs1 (S343), ATM (T1981) and p53 (S15). Herein, we used recombinant p53 protein as a substrate for DNA-PK activity in the presence of γ-32P-ATP and various lengths of DNA: synthesized double-stranded oligos (14 mer, 20 mer, 24 mer, 28 mer, 32 mer, 36 mer, and 300 mer in length). Phosphorylated p53 was subsequently determined by imaging with an Alpha 2000 imager for intensity quantification. The relative DNA-PK kinase activity was determined by measuring intensities and normalizing to the reference intensity with DNA-PKcs + Ku alone with no oligos. As shown in Figure 2, the required DNA “foot-print” for DNA-PK kinase activation was ≥ 32 bp, and the kinase activity was significantly enhanced in the presence of the longer, 300-bp, fragments. Interestingly, preincubation of DNA-PK with 14-bp and 20-bp DNA fragments markedly inhibited the capacity of longer 300-bp fragments, added subsequently, to stimulate kinase activity. We interpret these data to show that short DNA fragments < 20 bp may bind to DNA-PK complexes in a non-productive manner, interfering with subsequent interactions between the enzyme and longer DNA fragments. These results are consistent with a report demonstrating that a minimum of 32 bp is required for DNA-PKcs binding and kinase activity activation (29).
Fig.2.
DNA-PK kinase activity inhibition by synthesized short DNA fragments. Recombinant p53 protein was incubated with DNA-PK in the presence of γ-32P-ATP and various lengths of DNA: synthesized double-stranded oligos (14 mer, 20 mer, 24 mer, 28 mer, 32 mer, 36 mer, and 300 mer in length). For competition assays, the shorter 14 and 20 bp fragments were incubated first and the longer fragments were added subsequently (14bp → 300bp or 20bp → 300bp). Phosphorylated p53 was determined by Western blotting and imaged with an Alpha 2000 imager for intensity quantification. The bar graph represents the relative DNA-PK kinase activity after normalization with the reference intensity (DNA-PK + Ku activity). Ku alone serves as a negative control. Error bars represent the standard error of the mean for three independent experiments.
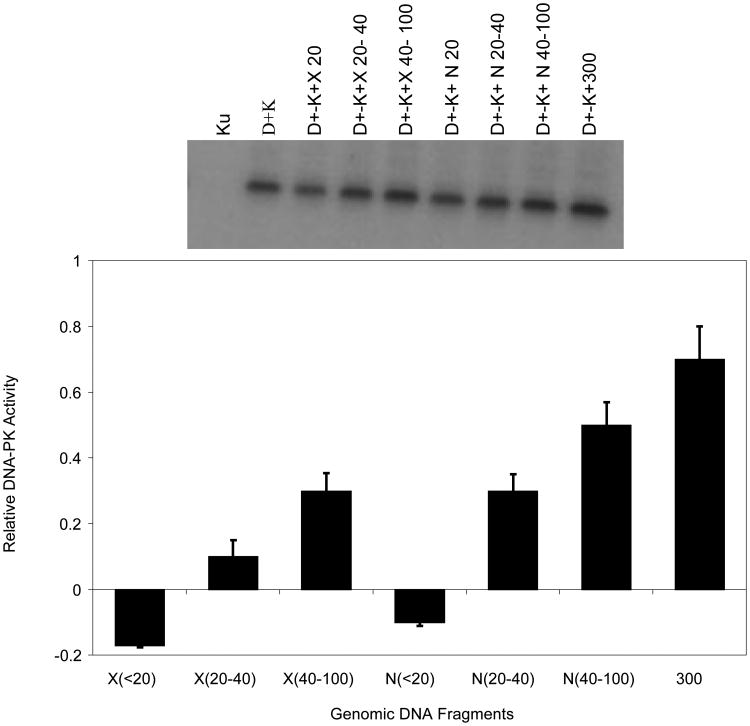
Radiation damaged DNA contains a variety of complex lesions that are also characterized by free radical damage to DNA bases and sugars, resulting in additional levels of complexity near broken ends (30,31,32,33). Therefore, we asked if broken ends caused by ionizing radiation affected DNA-PK activation. Genomic DNA samples were exposed to 5 kGy of Co-60 γ and 0.75 MeV fission-neutron irradiation and size fractionated on polyacrylamide gel electrophoresis to yield 10-100 bp DNA fragments. Extracted short DNA fragments permitted analysis of DNA-PK kinase assays with DNA fragments containing damaged end groups. As shown in Figure 3, results with radiation-generated DNA fragments are qualitatively similar to the observations obtained with synthetic DNA fragments, supporting the importance of DNA size dependency, but apparently not end-group chemistry, on inhibition of DNA-PK.
Fig. 3.
Comparison of kinase activations stimulated by fragments generated by Co-60 γ-rays and 0.75 MeV fission-neutron irradiation generated short DNA fragments. Recombinant p53 protein was incubated with DNA-PK in the presence of γ-32P-ATP and various lengths of DNA generated by either X-ray or Neutron. Phosphorylated p53 was determined by Western blotting and imaged with an Alpha 2000 imager for intensity quantification. The bar graph represents the relative DNA-PK kinase activity after normalization with the reference intensity (DNA-PKcs + Ku activity). Ku alone serves as a negative control. Error bars represent the standard error of the mean for three independent experiments. In the table D+K: DNA-PKcs + Ku; D+K+X (N) 20-40: DNA-PKcs+ Ku + photon (neutron) generated DNA fragments 20-40 bp in lengths.
The pattern of DNA-PK activation is consistent with observations using synthetic double-stranded DNA and Co-60 irradiated DNA. We interpret these in vitro observations to show that DNA-PK activity is inhibited by short DNA fragments (10-30 bp) and stimulated by larger DNAs. Furthermore, there are no appreciable differences among responses to synthetic dsDNAs, or irradiated DNAs using low-LET or high-LET radiations. These data suggest that the potential differences presented by the characteristics of DNA ends may be less significant for DNA-PK activation in vitro than the length of the DNA used for enzyme activation.
Inhibition of NHEJ is DNA fragment length and concentration dependent
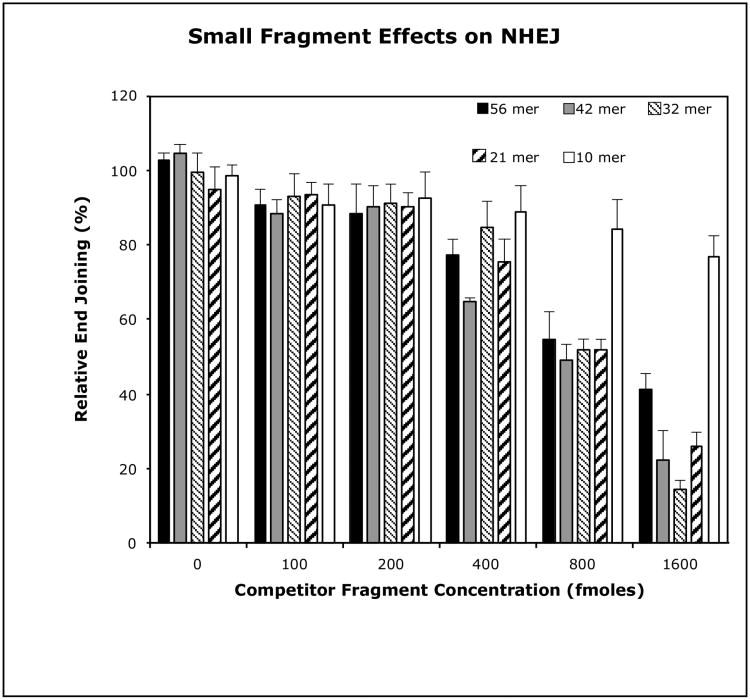
To confirm the hypotheses that short duplex DNA fragments may act as direct inhibitors of NHEJ, we examined the effect of 5 synthetic duplex oligonucleotides ranging in size from 10 bp to 56 bp on the in vitro NHEJ activity of human HeLa cell extracts (Fig. 4). This assay employs a duplex 75-mer oligo as the primary end joining substrate. This 75-mer possesses one non-ligatable unphosphorylated blunt end and a ligateable 5′-32P-labeled 4 nucleotide complementary overhang at the opposite end (Table 1a). Previously, we demonstrated that HeLa cell extract mediated production of the single detectable 150 bp ligated-dimer product from this substrate, is a function of the NHEJ pathway (25).
Fig. 4.
Inhibitory effects of small duplex DNA fragments on HeLa cell extract mediated NHEJ. The standard end-joining assay was run with 5 μg HeLa WCE and 400 fmoles 75 mer end-joining substrate per reaction. Inhibition was assessed in parallel reactions containing the short competitor duplex oligos at the concentrations indicated. The error bars represent the standard error of the mean for three independent experiments.
To investigate the effect of DNA fragments of varying size on the biologically significant end point of ligation by the NHEJ pathway, in vitro NHEJ was performed in the presence of increasing concentrations of each of the duplex competitor oligos (Table 1b-f). Oligos b-f range in size from 10 bp to 56 bp and have similar end configurations to those of the standard 75-mer end joining substrate, but possess self-complementary 4 nucleotide overhangs that are not complementary to that of the 75-mer. The molar ratios of the short competitor oligos to the 75-mer substrate oligo in each reaction were 0.25:1, 0.5:1, 1:1, 2:1 and 4:1.
A size dependency is seen for inhibition by the competitor fragments at high concentration, with the 21-mer, 32-mer, and 42-mer causing 75%, 86%, and 78% inhibition of end joining respectively. Each of these results is significantly greater than that produced by the 10mer or the 56mer. In fact, even at a molar ratio of 4:1, inhibition of the reaction by the 10-mer is only marginally significant with respect to its matched control, suggesting that fragments as short as 10 bp lack the capacity to interfere with NHEJ. In contrast, the NHEJ reaction is most sensitive to inhibition by the 42-mer fragment, which exhibits slight but significant inhibition (∼10%) with respect to its controls at molar ratios of 0.25:1 and 0.5:1, and nearly 40% inhibition with at a competitor:substrate molar ratio of 1:1. This result is of interest, since of the oligos used, 42 bp is the minimum size that is likely to permit binding of a complete NHEJ complex (34,35,36).
These results indicate that short duplex DNA fragments can act as direct competitive inhibitors of HeLa extract mediated NHEJ. We also observed a size and concentration dependency for this effect, with fragments greater than 10 bp and less than 56 bp being most effective in our assay, with as little as a 4:1 competitor to target substrate molar ratio being sufficient to cause severe inhibition of end joining.
Genomic instability initiated by transfected short DNA fragments
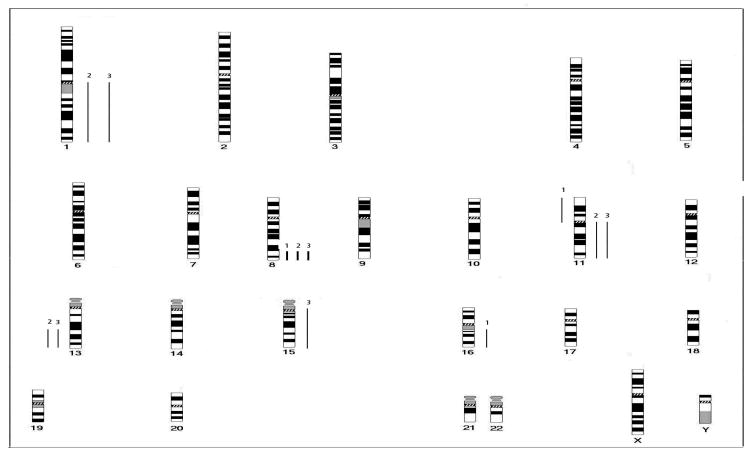
We next examined the potential consequences of persistent short DNA fragments on the integrity of the cellular genome. Reports of ionizing radiation causing genomic instability provide a possible link to radiation-induced carcinogenesis (37,38). We selected MCF10A cells as recipients for transfection with short DNA fragments (pooled 20-400bp fragments generated from 5 kGy of cobalt 60 γ-rays or 0.75 MeV fission neutrons). Cells were passaged for 25 doublings until morphologically transformed clones of MCF10A were observed. Soft agar assays confirmed loss of anchorage dependence in these transformed cells but not in the controls (Table 2). CGH analysis is an ideal method to detect overall genomic instability. CGH analysis of MCF10A cells showed no abnormalities. On the other hand, clones transfected with DNA fragments selected for analysis (MCF10A/X-Cl1, MCF10A/X-Cl2, MCF10A/X-Cl4) showed evidence of genomic instability including amplification at 8q23-qter (in all 3 clones), gain of 1q and 11q, and loss of 13q21-qter (in two of the clones). Additional changes were present in all the three clones. Figure 5 shows a summary of the changes detected in the three transfected clones.
Table 2.
Anchorage-independent colony formation of cells (MCF-10A) transfected with genomic DNA from AT5BIVA after irradiation (5 kGy of γ-ray and Neutron). In the Table, MCF-10A/gDNA-N (γ) designates MCF-10A cells transfected with neutron (γ-photon) irradiated genomic DNA fragments; CGH, Comparative Genomic Hybridization
| Cell line | Avg Foci# in 104 cells | Note |
|---|---|---|
| MCF-10A | 0 | |
| MCF-10A/lipofectin | 0 | |
| MCF-10A/gDNA-N | 21.5 | CL1 for CGH |
| MCF-10A/gDNA-γ | 31.4 | CL4 for CGH |
Fig. 5.
Comparative summary karyogram showing chromosomal gains and losses in MCF10A/X-Cl1 (1), MCF10A/X-Cl2 (2), and MCF10A/X-Cl4 (3). Bars to the left side of the chromosome ideogram indicate losses, bars to the right side indicate gains. Bold bars on the right side indicate amplifications.
Discussion
Recent studies have focused on the need to investigate small DNA fragments resulting from DSBs formed in close proximity to one another, which are a hallmark of high-LET radiation as predicted by theoretical modeling based on Monte Carlo simulation and microdosimetry studies (39,40,41). Experimentally determined excesses of short DNA fragments of 0.1-2 kbp have been reported using pulsed field gel electrophoresis following irradiation of fibroblasts with iron ions (42). These data have been interpreted to be in good agreement with theoretical calculations based on treating the 30-nm chromatin fiber as the target for ionizing particles (43). We suggested that even smaller DNA fragments may have been overlooked in the reported experiments due to the inherent technical limitations of the experimental techniques. Our AFM investigations clearly show the presence of increased short DNA fragments following irradiation by high-LET particles (20,26). Consistent with our experimental observations, a recent study with Monte Carlo simulation has demonstrated production of short DNA fragments by high-LET radiation in a simulated cellular-like environment, and the fraction of short fragments increases with the LET of the radiation (44).
To investigate the potential molecular significance of short DNA fragments resulting from clustered DSBs in cellular responses to ionizing radiation, we studied interactions between several DNA repair proteins and short DNA fragments generated by ionizing radiation as well as with short synthetic dsDNA molecules in vitro and in cell extracts. We observed that the binding interactions of radiation-induced short DNA fragments (<14 bp) with these proteins are inefficient, a result consistent with previous reports (27, 28).
In both NHEJ and HR repair pathways, protein binding to damaged DNA-ends is the first step in the sequence of responses to DNA damaging agents. Following exposure to ionizing radiation, broken DNA ends are rapidly bound by proteins such as Ku and DNA-PKcs, which subsequently recruit other repair proteins such as DNA Ligase IV/XRCC4 to complete the end-joining reaction.
The broad relevance of Ku and DNA-PKcs has been shown in work from other laboratories investigating protein-DNA binding interactions, which support our hypothesis implicating small DNA fragments in faulty interactions with repair proteins (45,48,49,50). West et al. have examined the stimulation of DNA-PK kinase activity as a function of DNA concentration in the presence or absence of Ku, inhibition of DNA-PK kinase activity by Ku, and the minimum DNA length required for DNA-PKcs activation by Ku. They reported that DNA-PKcs alone can bind to DNA fragments as short as 18 bp and activate its kinase activity. However, in the presence of Ku, the DNA-PKcs kinase activity is not stimulated unless the fragment is at least 26 bp long (49). Similarly, Hammarsten and Chu have also reported DNA binding and activation of DNA-PKcs in the absence of Ku (29). They observed that in the absence of Ku, DNA-PKcs was able to bind to a 32-bp DNA fragment, but when Ku was added, it formed a complex with DNA at the expense of DNA-PKcs. Kinase activity occurred when bound to the 32-bp DNA, but the addition of Ku completely inhibited this activity. The use of longer DNA fragments resulted in a 5-fold stimulation of kinase activity. These results were interpreted to show that Ku was able to stimulate DNA-PK kinase activity only when the DNA fragments were large enough for Ku and DNA-PKcs to each contact DNA directly and form an active Ku-DNA-PKcs complex. Our observations, along with those of others using AFM, are consistent with these data (27,51)
The apparent discrepancy in the minimum DNA length required for DNA-PK kinase activation as reported by the above two laboratories remains to be resolved. However, consistent with Hammarsten and Chu's findings, other laboratories have implicated 32 bp as the minimum fragment length for DNA-PK activation (52). Using synthesized DNA fragments of 14 bp to 300 bp in size, we systematically studied p53 phosphorylation by DNA-PK kinase. As shown in Figure 2, kinase activity was not stimulated by fragments smaller than 32 bp. Activity was seen for fragments of 32 bp or longer, and increased by nearly four fold for 300-bp fragments. Furthermore, pre-incubation of DNA-PK with short DNA fragments of 14 bp and 20 bp inhibited kinase activity induced by the longer 300-bp fragments. We interpret these observations to suggest that short DNA fragments may result in non-productive binding with DNA-PK and consequently inhibit its interaction with longer DNA fragments. We performed similar experiments using size fractionated genomic DNA resulting from Co-60 γ-photon and neutron irradiations, as shown in Figure 3, no kinase activity was observed for fragments less than 20 bp for either photon or neutron irradiation generated DNA fragments. Kinase activity was observed for fragments in the 20-40 bp and 40-100 bp groups and longer fragments result in increased kinase activity. Since irradiation of DNA by low and high-LET radiations result in DSBs of different end structure complexity (30,31,32), our results demonstrate that DNA-PK kinase activity is mostly initiated by DNA fragments of certain lengths and is not influenced by the chemical structure of the DNA ends.
Our in vitro experiments investigating binding and biochemical activities induced by short DNA fragments are consistent with reports of other investigators (27,29). We confirmed the length dependence of Ku binding and DNA-PK kinase activation. Furthermore, we observed the inhibition of DNA-PK kinase activity by short DNA fragments in interactions with longer DNA molecules. Further support for the significance of short DNA fragments on cellular responses to radiation has been reported by Duterix and colleagues for which they show inhibition of DNA repair (52). They observed that short DNA fragments transfected into cells induce phosphorylation of histone H2AX, inhibit repair foci formation, and inhibit damage repair, consistent with our hypothesis that short DNA fragments may play an important role in modulating DNA damage repair. Another recent report has shown that high-LET radiation inhibits the Ku-dependent DNA repair pathway, presumably due to ineffective binding of two Ku proteins to both ends of a DNA fragment smaller than 40 bp (53). Using radiation induced and synthesized short DNA fragments, our experiments similarly demonstrated the length requirements of DNA fragments for effective interaction with Ku and DNA-PKcs. As shown in Figures 2 and 3, DNA-PK activity was only triggered by fragments longer than 32 bp and the activities were significantly greater for longer fragments (>300 bp). It is significant that the kinase activities associated with longer fragments were inhibited in the presence of short fragments (14 and 20 bp).
We advanced our investigation of DNA-DNA repair protein interactions to include short DNA fragments generated by ionizing radiations. We observed that the binding interactions of radiation-induced short DNA fragments (<14 bp) with these proteins are inefficient, a result consistent with that observed with synthesized short DNA fragments. Furthermore, as demonstrated in Figure 3, DNA-PK kinase activities were qualitatively similar to those stimulated by short synthetic DNA fragments, supporting the importance of DNA size dependency but not end group chemistry dependency for kinase activation. However, the question of biological significance remained to be addressed.
The first important biological end-point lies in the demonstration that such fragments can act as direct inhibitors of DNA ligation mediated by the NHEJ pathway. As shown in our experiments, fragments 21-42 bp in length are effective inhibitors of NHEJ, with the 42-mer being the most effective (Fig. 4). Significant inhibition of the end-joining reaction was obtained at relative molar concentrations of this fragment to the end-joining substrate of as little as 1:1 and on up through 4:1. This interesting observation may be partially explained by a report that 40 bp is the smallest fragment size to which Ku proteins can bind to both ends of the fragment (53), resulting in more effective competition for this repair protein. In a cellular environment and at biologically relevant doses, although the absolute amount of small DNA fragments may be small compared to the total number of repair proteins, the relative local concentration of small fragments (especially in the context of chromatin) may be sufficiently high to result in competitive, non-productive binding of repair proteins and thus interfere with repair. High-LET radiation produces higher yields of small fragments in a small spatial region due to the clustered ionization effect, resulting in increased spatially localized concentrations of small fragments.
We also investigated the consequences of persistent short DNA fragments on the integrity of the cellular genome using CGH analysis and found that transfection of short fragments to MCF10A cells induced genomic aberrations in the cells. This provides additional evidence that short DNA fragments may cause genomic instability and could explain some aspects of their mechanism of action. Our results support the general discussion in a review by Hall and Hei on high-LET radiation-induced genomic instability (54).
Taken together, these data offer experimental evidence in support of short DNA fragments as an important ionizing radiation-induced lesion. The lesion may be produced by ionizations that result in DNA DSBs in close proximity along a DNA molecule. As demonstrated previously, more short DNA fragments are observed following high-LET radiation as compared to low-LET radiation. Potential biological relevance is suggested by the inability of NHEJ proteins to efficiently process these short DNA molecules and supported by the enhanced cellular genomic instability induced by such short DNA fragments. Further investigation would be required to fully assess the significance of short DNA fragments in cells exposed to typical therapeutic radiation doses.
In summary, our studies indicate that, in the process of DNA damage by ionizing radiation, short DNA fragments are generated and repair can be inhibited. Fragment sizes of interest may range from at least 14 bp to 32 bp. However, we believe that fragments of importance may also be longer than 32 bp. We hypothesize that these DNA fragments may interfere with, and hinder, biochemical processes that directly impact cellular survival and genetic stability after exposure to ionizing radiation, especially high-LET radiation.
Acknowledgments
We thank the staff at AFRRI in Bethesda, Maryland for assistance with neutron irradiation, and the staff of Accelerator Engineering Corporation contracting with NIRS-HIMAC, Chiba, Japan for Argon irradiation. Our experiment number at NIRS-HIMAC was 11P081. We are particularly indebted to Drs. Sheldon Datz and Barry Berman who helped with arranging irradiation at NIRS-HIMAC. We are saddened that both Drs. Datz and Berman have passed away due to illnesses. We also thank Drs. Yin Zhang for technical assistance and William Dynan for helpful discussion on Ku and DNA-PK. This research was supported in part by the Intramural Research Program of the NIH, through the Warren Grant Magnuson Clinical Center (TAW, SP), and by the Lombardi Comprehensive Cancer Center at Georgetown University Medical Center for AFM.
References
- 1.Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000;407:777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 2.Barendsen GW. The relationship between RBE and LET for different types of lethal damage in mammalian cells: biological and molecular mechanisms. Radiat Res. 1994;139:257–270. [PubMed] [Google Scholar]
- 3.Heilmann J, et al. DNA strand break induction and rejoining and cellular recovery in mammalian cells after heavy-ion irradiation. Radiat Res. 1993;135:46–55. [PubMed] [Google Scholar]
- 4.Roots R, et al. The formation of strand breaks in DNA after high-LET irradiation: a comparison of data from in vitro and cellular systems. Int J Radiat Biol. 1990;58:55–69. doi: 10.1080/09553009014551431. [DOI] [PubMed] [Google Scholar]
- 5.Kiefer J. Cellular and subcellular effects of very heavy ions. Int J Radiat Biol Relat Stud Phys Chem Med. 1985;48:873–892. doi: 10.1080/09553008514552041. [DOI] [PubMed] [Google Scholar]
- 6.Téoule R. Radiation-induced DNA damage and its repair. Int J Radiat Biol Relat Stud Phys Chem Med. 1987;51:573–589. doi: 10.1080/09553008414552111. [DOI] [PubMed] [Google Scholar]
- 7.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int J Radiat Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 8.Pastwa E, et al. Repair of radiation-induced DNA double-strand breaks is dependent upon radiation quality and the structural complexity of double-strand breaks. Radiat Res. 2003;159:251–261. doi: 10.1667/0033-7587(2003)159[0251:roridd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Ward JF. Some biochemical consequences of the spatial distribution of ionizing radiation produced free radicals. Radiat Res. 1981;86:185–195. [PubMed] [Google Scholar]
- 10.Ward JF. The yield of DNA double strand breaks produced intracellularly by ionizing radiation: a review. Int J Radiate Biol. 1990;57:1141–1150. doi: 10.1080/09553009014551251. [DOI] [PubMed] [Google Scholar]
- 11.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int J Radiat Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 12.Goodhead DT. Energy deposition stochastics and track structure: what about the target? Radiat Prot Dosimetry. 2006;122:3–15. doi: 10.1093/rpd/ncl498. [DOI] [PubMed] [Google Scholar]
- 13.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 14.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 18.Collis SJ, et al. The life and death of DNA-PK. Oncogene. 2005;24:949–961. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- 19.Budman J, Kim SA, Chu G. Processing of DNA for nonhomologous end-joining is controlled by kinase activity and XRCC4/ligase IV. J Biol Chem. 2007;282:11950–11959. doi: 10.1074/jbc.M610058200. [DOI] [PubMed] [Google Scholar]
- 20.Pang D, et al. Investigation of neutron-induced damage in DNA by atomic force microscopy: experimental evidence of clustered DNA lesions. Radiat Res. 1998;150:612–618. [PubMed] [Google Scholar]
- 21.Han ZB, et al. Relative biological effectiveness of accelerated heavy ions for induction of morphological transformation in Syrian hamster embryo cells. J Radiat Res. 1998;39:193–201. doi: 10.1269/jrr.39.193. [DOI] [PubMed] [Google Scholar]
- 22.Kanai T, et al. Irradiation of mixed beam and design of spread-out Bragg peak for heavy-ion radiotherapy. Radiat Res. 1997;147:78–85. [PubMed] [Google Scholar]
- 23.Kuettel MR, et al. Radiation-induced neoplastic transformation of human prostate epithelial cells. Cancer Res. 1996;56:5–10. [PubMed] [Google Scholar]
- 24.Santos SC, et al. Patterns of DNA copy number changes in sentinel lymph node breast cancer metastasis. Cytogenetic and Genome Research. 2008;122:16–21. doi: 10.1159/000151311. [DOI] [PubMed] [Google Scholar]
- 25.Datta K, Neumann RD, Winters TA. An in vitro nonhomologous end-joining assay using linear duplex oligonucleotides. Analytical Biochemistry. 2006;358:155–157. doi: 10.1016/j.ab.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 26.Pang D, et al. Spatial distribution of radiation-induced double-strand breaks in plasmid DNA as resolved by atomic force microscopy. Radiat Res. 2005;164:755–765. doi: 10.1667/rr3425.1. [DOI] [PubMed] [Google Scholar]
- 27.Yaneva M, Kowalewski T, Lieber LR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falzon M, Fewell JW, Kuff EL. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J Biol Chem. 1993;15:10546–10552. [PubMed] [Google Scholar]
- 29.Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci U S A. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta K, Neumann RD, Winters TA. Characterization of complex apurinic/apyrimidinic-site clustering associated with an authentic site-specific radiation-induced DNA double-strand break. Proc Natl Acad Sci U S A. 2005;102:10569–10574. doi: 10.1073/pnas.0503975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datta K, et al. Molecular analysis of base damage clustering associated with a site-specific radiation-induced DNA double-strand break. Radiat Res. 2006;166:767–781. doi: 10.1667/RR0628.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Datta K, et al. Determination and analysis of site-specific 125I decay-induced DNA double-strand break end-group structures. Radiat Res. 2007;167:152–166. doi: 10.1667/rr0629.1. [DOI] [PubMed] [Google Scholar]
- 33.Datta K, et al. Base damage immediately upstream from double-strand break ends is a more severe impediment to nonhomologous end joining than blocked 3′-termini. Radiat Res. 2010;175:97–112. doi: 10.1667/RR2332.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee KJ, et al. DNA ligase IV and XRCC4 form a stable mixed tetramer that functions synergistically with other repair factors in a cell-free end-joining system. J Biol Chem. 2000;275:34787–34796. doi: 10.1074/jbc.M004011200. [DOI] [PubMed] [Google Scholar]
- 35.Pascal JM. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 36.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: Structural and functional insights. Ann Rev Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang L, et al. Targeted and nontargeted effects of low-dose ionizing radiation on delayed genomic instability in human cells. Cancer Res. 2007;67:1099–1104. doi: 10.1158/0008-5472.CAN-06-3697. [DOI] [PubMed] [Google Scholar]
- 38.Sowa M, et al. Effects of ionizing radiation on cellular structures, induced instability and carcinogenesis. EXS. 2006;96:293–301. doi: 10.1007/3-7643-7378-4_12. [DOI] [PubMed] [Google Scholar]
- 39.Ottolenghi A, Merzagora M, Paretzke HG. DNA complex lesions induced by protons and alpha-particles: track structure characteristics determining linear energy transfer and particle type dependence. Radiat Environ Biophys. 1997;36:97–103. doi: 10.1007/s004110050060. [DOI] [PubMed] [Google Scholar]
- 40.Nikjoo H, et al. Goodhead, Quantitative modelling of DNA damage using Monte Carlo track structure method. Radiat Environ Biophys. 1999;38:31–38. doi: 10.1007/s004110050135. [DOI] [PubMed] [Google Scholar]
- 41.Campa A, et al. Simone and A. Tabocchini, DNA DSB induced by human cells by charges particles and gamma rays: Experimental results and theoretical approaches. Int J radiat Biol. 2005;81:841–854. doi: 10.1080/09553000500530888. [DOI] [PubMed] [Google Scholar]
- 42.Rydberg B. Clusters of DNA damage induced by ionizing radiation: formation of short fragments. II Experimental detection. Radiat Res. 1996;145:200–239. [PubMed] [Google Scholar]
- 43.Holly WR, Chatterjee A. Clusters of DNA damage induced by ionizing radiation: Formation of short DNA fragments. I. Theoretical Model. Radiat Res. 1996;145:188–199. [PubMed] [Google Scholar]
- 44.Alloni A, et al. A Monte Carlo study of the radiation quality dependence of DNA fragmentation spectra. Radiat Res. 2010;173:263–271. doi: 10.1667/RR1957.1. [DOI] [PubMed] [Google Scholar]
- 45.Hammel M, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burma S, Chen DJ. Role of DNA-PK in the cellular response to DNA double-strand breaks. DNA Repair (Amst) 2004;3:909–918. doi: 10.1016/j.dnarep.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 47.Kysela B, et al. Ku stimulation of DNA ligase IV-dependent ligation requires inward movement along the DNA molecule. J Biol Chem. 2003;278:22466–22474. doi: 10.1074/jbc.M303273200. [DOI] [PubMed] [Google Scholar]
- 48.Ramsden DA, Gellert M. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West RB, Yanevan M, Lieber MR. Productive and nonproductive complexes of Ku and DNA-dependent protein kinase at DNA termini. Mol Cell Biol. 1998;18:5908–20. doi: 10.1128/mcb.18.10.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinfeld M, et al. Interaction of DNA-dependent protein kinase and poly(ADP-ribose) polymerase with radiation-induced DNA strand breaks. Radiat Res. 1997;148:22–28. [PubMed] [Google Scholar]
- 51.Pang D, et al. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res. 1997;57:1412–1415. [PubMed] [Google Scholar]
- 52.Quanz M, et al. Small-molecule drugs mimicking DNA damage: a new strategy for sensitizing tumors to radiotherapy. Clin Cancer Res. 2009;15:1308–1316. doi: 10.1158/1078-0432.CCR-08-2108. [DOI] [PubMed] [Google Scholar]
- 53.Wang H, et al. The Ku-dependent non-homologous end-joining but not other repair pathway is inhibited by high linear energy transfer ionizing radiation. DNA Repair (Amst) 2008;7:725–733. doi: 10.1016/j.dnarep.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 54.Hall EJ, Hei TK. Genomic instability and bystander effects induced by high-LET radiation. Oncogene. 2003;22:7034–7042. doi: 10.1038/sj.onc.1206900. [DOI] [PubMed] [Google Scholar]