Abstract
Polycomb group proteins are transcriptional repressors that are essential for normal gene regulation during development. Recent studies suggest that polycomb repressive complexes recognise and are recruited to through a range of different mechanisms, which involve transcription factors, CpG island elements and non-coding RNAs. Together with the realization that the interplay between polycomb repressive complexes 1 (PRC1) and 2 (PRC2) is more intricate than previously appreciated, this has increased our understanding of the vertebrate polycomb system at the molecular level.
Introduction
A remarkable feature of multicellular organisms is the capacity to create functionally unique cell types from an essentially invariant genome sequence shared by all cells in the organism. This diversity relies on the capacity of individual cell types to initiate and then maintain specific gene expression patterns during development. To achieve this, cellular signalling events are thought to regulate the activity of cell type-specific DNA binding transcription factors that function as master regulators of gene expression networks. Interestingly, however, genetic screens for factors involved in the regulation of gene expression and cell fate specification have also identified additional genes that seem to impact these processes through chromatin structure and histone modification. This is exemplified by the polycomb group genes, which were first identified in Drosophila melanogaster as regulators of Hox gene expression and normal developmental body plan specification1. Subsequently, orthologous genes were identified in vertebrate species where they also encode transcriptional repressors. Vertebrate polycomb group proteins are essential for normal gene regulation during embryonic development and are perturbed in a wide range of human cancers (reviewed in 2).
Since the initial identification of polycomb group genes, an immense amount of biochemical work has focussed on understanding how these chromatin-associated factors function. This has led to the discovery that polycomb group proteins usually belong to one of two multi-subunit protein complexes: Polycomb Repressive Complex 1 (PRC1) that adds a ubiquityl moiety to histone H2A at lysine 119 (H2AK119ub1) and Polycomb Repressive Complex 2 (PRC2) that catalyses the addition of one to three methyl groups to histone H3 at lysine 27, leading to H3K27me1, H3K27me2 and H3K27me3 (Box 1) (reviewed in 3).
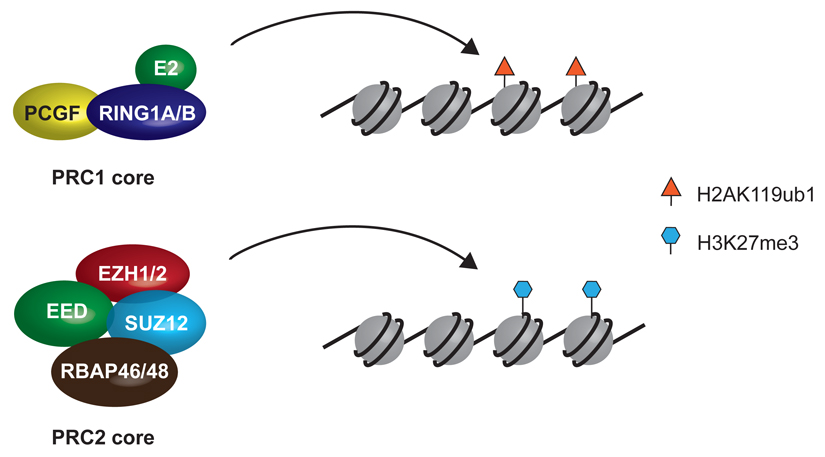
Box 1. Core PRC complexes and their chromatin modifying activities.
Polycomb repressive complexes 1 (PRC1) and 2 (PRC2) are comprise core protein components that are necessary for their respective enzymatic activities. PRC1 is a histone H2A lysine 119 ubiquitin ligase. In PRC1 two functionally equivalent yet mutually exclusive subunits, RING1A or RING1B, function as ubiquitin E3 ligases that guide the transfer of a ubiquityl moiety onto H2AK119 in chromatin. RING1A or RING1B dimerize with a PCGF subunit (of which there are six, PCGF1-6, in vertebrates). The PCGF components are required for the core enzymatic activity of the complex, and also define how the core complex interacts with auxiliary complex components to regulate its targeting to chromatin and enzymatic activity.
PRC2 is a histone H3 lysine 27 methyltransferase. The methyltransferase activity of PRC2 resides in the SET domain of two mutually exclusive proteins, EZH1 or EZH2. Alone these proteins exhibit little enzymatic activity in vitro, but when bound to EED, SUZ12, and RBAP46 or RBAP48 they form an active methyltransferase complex that is specific towards chromatin substrates. EED, SUZ12, and RBAP46 or RBAP48 contribute to the stability and structural integrity of PRC2, but they also regulate its enzymatic activity and support targeting to chromatin, either directly or via interaction with auxiliary complex components. Despite detailed characterization of the core PRC1 and PRC2 protein components, it remains unclear whether these complexes are stable entities inside cells or if they also display some capacity to assemble in a dynamic fashion on chromatin.
PRC1 and PRC2 usually co-occupy target sites in the genome, where their combined activities create ‘polycomb chromatin domains’ consisting of the polycomb group proteins themselves, H2AK119ub1, and H3K27me3. How vertebrate polycomb protein complexes are recruited to chromatin, how different PRC1 and PRC2 complexes function together in situ, and how polycomb chromatin domains actually repress transcription still remain poorly understood. Several recent excellent review articles have described the general features and functions of polycomb systems in different phyla3–7. In this ‘Progress Article’ we focus on exciting new advances that have begun to shed light on the molecular mechanisms that underpin recruitment of polycomb group protein complexes to target sites, the formation of polycomb chromatin domains, and the functional relevance this has for gene regulation in vertebrates.
Recruitment of polycomb complexes
In the Drosophila melanogaster genome, polycomb responsive elements (PREs) act as recruitment sites for polycomb repressive complexes. DNA binding transcription factors are thought to play an important role in bringing polycomb protein complexes to these sites (reviewed in4). However, attempts to define vertebrate PREs have proven largely unsuccessful and emerging evidence supports the idea that vertebrate polycomb complexes are directed to DNA by both locus-specific and more generalized targeting mechanisms.
Locus-specific targeting
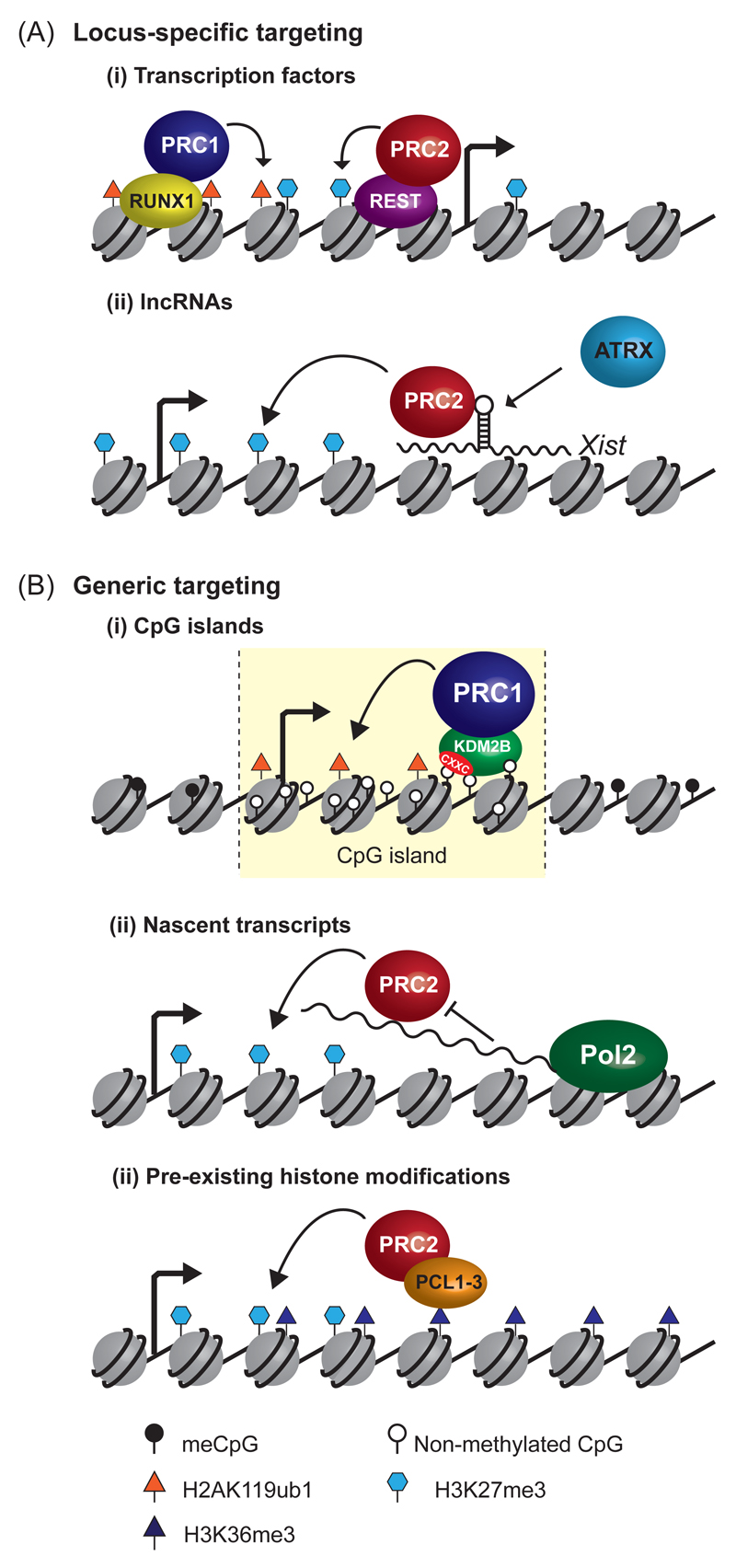
Inspired by observations in D. melanogaster, it has been proposed that vertebrate transcription factors might recruit polycomb repressive complexes to target sites in chromatin (Figure 1A). However, there are only a limited number of cases in which site-specific DNA binding transcription factors have been identified in unbiased biochemical isolations of polycomb complexes. Examples include E2F, MGA and MAX, which were found to interact with PRC18, 9. Thus, it has been brought into question whether interactions with transcription factors broadly underpin targeting. Candidate-based approaches have identified additional DNA binding factors, including REST, SNAIL and RUNX1 that interact with PRC1 or PRC2, but these factors seem to contribute to polycomb protein recruitment in only specific instances10–14. More recently, detailed proteomic profiling of polycomb protein complexes has identified novel interactors, including proteins that contain zinc finger domains (ZNF518A and ZNF518B15) that are often associated with DNA binding activity. Whether these newly identified proteins contribute to polycomb complex targeting remains to be determined.
Figure 1. Getting polycomb repressive complexes to chromatin.
a | Locus-specific targeting of polycomb repressive complexes. PRC1 and PRC2 can associate with DNA binding transcription factors (top-left), such as RUNX1 and REST, which guide these complexes to chromatin. Similarly, interactions with long non-coding RNAs such as Xist function in chromosome- and locus-specific targeting of polycomb complexes (bottom-left). The chromatin remodelling protein ATRX may remodel the structure of Xist to achieve interaction with PRC2. Following recruitment to chromatin, PRC1 catalyses ubiquitylation of H2AK119 and PRC2 catalyses trimethylation of H3K27, as indicated by rounded arrows. The square arrows indicate transcription start sites.
b | Generic targeting of polycomb repressive complexes to gene regulatory elements. A variant PRC1 complex containsthe KDM2B protein. KDM2B has a zinc-finger CxxC DNA binding domain (red area) that specifically recognizes non-methylated CpG dinucleotides. This allows KDM2B to bind at CpG islands genome-wide, and contributes to PRC1 occupancy at these elements (top-right). PRC2 interacts with nascent RNA polymerase II (Pol II) transcripts at 5’-ends of genes, which may provide a mechanism to maintain repression at silent genes following stochastic transcription initiation events. Alternatively, at active genes, interaction of PRC2 with nascent RNA may constrain the catalytic activity of PRC2 and protect against polycomb-mediated repression (middle-right). A subset of PRC2 complexes contain PCL proteins which bind to H3K36me3, a modification associated with active transcription. This may enable PRC2 to bind at, or spread into, previously transcribed regions, catalysing H3K27me3 at these regions.
In addition to transcription factors, it has been suggested that long noncoding RNAs (lncRNAs) could recruit polycomb complexes to specific loci, however the generality of this as a targeting mechanism remains a topic of debate (reviewed in 16). This idea originated from the observation that the Xist lncRNA is required for localization of PRC217–20 to the inactivated X-chromosome during mammalian dosage compensation in a manner that relies on the sub-stoichiometric PRC2 component JARID221. Recent studies, aimed at understanding the relationship between PRC2 and Xist, have suggested that the interaction between Xist and the chromatin remodelling protein ATRX induces conformational changes within the Xist RNA which favours specific and direct interaction with PRC222 (Figure 1A). However, it has also been proposed that the relationship between Xist and PRC2 may be indirect. In support of an indirect interaction, super-resolution imaging studies have indicated that PRC2 is not intimately associated with Xist RNA on a Xist-inactivated chromosome23 and the Xist-binding protein SHARP was required to recruit PRC2 to Xist-coated chromosomes24. Nevertheless, there is additional evidence in favour of direct roles for lncRNAs in the recruitment of PRCs to chromatin. For example, PRCs are recruited to the Kcnq1ot1 locus on a paternally imprinted region of mouse chromosome 7 that transcribes lncRNAs25. Moreover,the lncRNA HOTAIR, which is expressed from the HOXC locus on human chromosome 12, seems to function in trans to target PRC2 to the HOXD locus on chromosome 226.
Although it is clear that in some specific cases transcription factors and lncRNAs contribute to locus-specific recruitment of polycomb repressive complexes, it seems unlikely that these targeting mechanisms are sufficient to create the widespread, and often tissue-specific, polycomb complex binding patterns observed in vivo.
Generic targeting
Important recent discoveries have shown that polycomb repressive complexes bind to regulatory sites across the genome through generic chromatin binding activities, with local chromatin features dictating residency and function at these sites.
In vertebrate genomes, the most universal and striking feature of polycomb-occupied sites is the presence of a CpG island (CGI). CGIs are approximately 1-2 kilobase regions of CpG-rich DNA that lack DNA methylation and typically encompass gene promoters27, 28. The striking correlation between polycomb protein occupancy and CGIs, coupled with the observation that artificial DNA sequences with a high CpG content are sufficient to nucleate polycomb group proteins in vivo29, has led to suggestions that CGIs may play a direct role in polycomb protein recruitment. Attempts to define a molecular link between polycomb complexes and CGIs have largely focussed on the KDM2B protein, which stably associates with PRC1. KDM2B encodes a ZF-CxxC DNA binding domain that specifically recognizes non-methylated CpG dinucleotides allowing KDM2B to bind to CGIs genome-wide30–32. Recent studies have demonstrated that KDM2B contributes to PRC1 occupancy at CGIs (Figure 1B). However, somewhat paradoxically, high-level PRC1 enrichment is only achieved at the most repressed sites, despite KDM2B residing at all CGIs. This suggests that the recruitment of PRC1 to CGIs by KDM2B, or the repressive activity of PRC1 at CGIs is regulated by additional mechanisms that permit the establishment of polycomb chromatin domains. Furthermore, KDM2B-independent mechanisms must also function at some CGIs to recruit or stabilize PRC1 binding, as removal of KDM2B does not lead to a complete loss of PRC1 at all CGI target sites30–33. Interestingly, it has been reported that the PRC2 component JARID2 preferentially recognises GC-rich DNA34. JARID2 could thus provide a complementary mechanism for targeting PRC2 to CGIs independently of KDM2B. Together these observations suggest that polycomb complexes can be recruited to all CGIs, though other mechanisms define whether this results in the formation of stable polycomb chromatin domains.
Although lncRNA-mediated targeting has been proposed as a locus-specific recruitment mechanism for polycomb complexes, it has been recently reported that PRC2 binds RNA promiscuously with little sequence specificity35, 36. This has led to several new suggestions as to how PRC2-RNA interactions may contribute to the generic recruitment or functionality of polycomb complexes at gene regulatory elements and genes. At repressed genes, binding of PRC2 to short abortive RNA transcripts may help to retain polycomb complexes and stabilize transcriptional repression during aberrant transcriptional initiation events37, 38. Alternatively, nascent transcripts produced from active genes may interact with PRC2 and provide a ‘decoy’ to block stable interactions between PRC2 and chromatin, thereby protecting the transcribed gene from repressive polycomb activity. This latter idea has received some support from the observation that interactions with nascent transcripts can constrain the histone methyltransferase activity of PRC239, 40. An alternative possibility is that PRC2 utilises interactions with nascent transcripts as a generalised targeting mechanism to increase residency around gene promoters (Figure 1B) and that other counteracting signals, including H3K4 and H3K36 methylation, prevent stable polycomb domain formation at active genes41. These emerging links between ncRNAs and PRC2 enzymatic activity seem to be evolutionarily conserved, as regulated bidirectional ncRNA expression from a D. melanogaster PRE acts as a switch to alter PRC2 activity and regulate gene expression42. Interestingly, vertebrate polycomb target sites often exhibit bidirectional transcription, so it will be important to understand if similar switch-like mechanisms are involved in regulating vertebrate PRC2 activity.
In addition to generic DNA and RNA binding activities, recent evidence suggests that certain polycomb repressive complexes recognise chromatin modifications placed by other histone modifying systems that are broadly associated with genes and regulatory elements. For example, the sub-stoichiometric PRC2 subunits Polycomblike 1-3 (PCL1, PCL2, PCL3) encode TUDOR domains that recognise the H3K36me3 modification43–46. H3K36me3 is typically associated with actively transcribed gene bodies, and during cellular differentiation, PCL proteins were proposed to facilitate spreading of PRC2 into these regions (Figure 1B). In addition, links have been identified between PRC2 and the H3K9 methylation systems. Most notably, biochemical interactions were identified between PRC2 and the H3K9 methyltransferases G9A and GLP, and loss of G9A resulted in impaired PRC2 recruitment at a subset of target sites15, 47.
Interplay between PRCs
Outlined above are simple examples of how PRC1 or PRC2 are individually recruited to target sites by locus-specific or generic targeting mechanisms. However, after their recruitment to chromatin, the functions of PRC1 and PRC2 are intimately related, as the enzymatic activity of each complex influences the other’s occupancy on chromatin and the full establishment of polycomb chromatin domains.
The prevailing hierarchical model
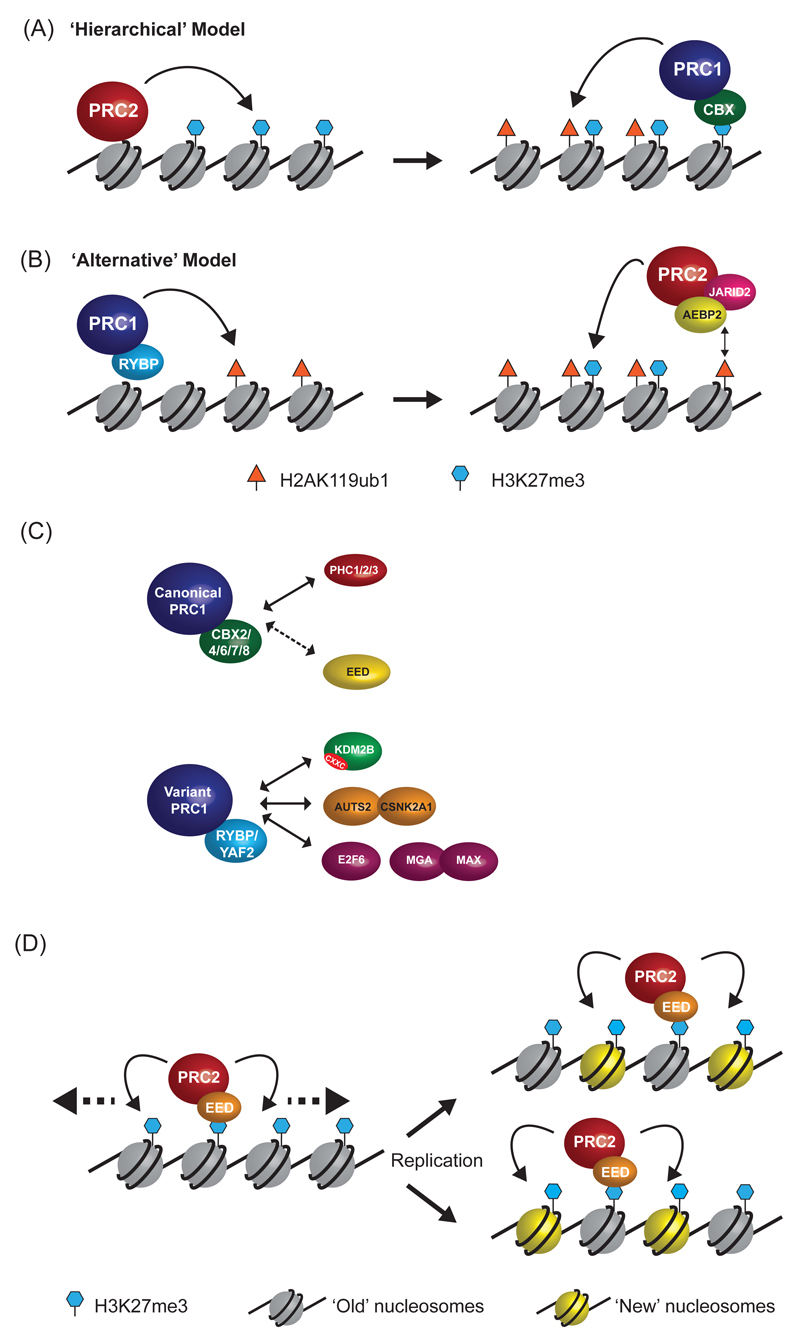
PRC1 and PRC2 typically co-localise at target sites throughout the genome. This has largely been attributed to a mechanism discovered over a decade ago in D. melanogaster which posits that de novo recruitment of PRC2 to target site catalyses H3K27me3, which is subsequently recognized by a chromobox (CBX)-containing protein in PRC1, leading to H2AK119ub1 placement and polycomb chromatin domain formation (Figure 2A)48, 49. This pathway is generally referred to as the ‘hierarchical’ recruitment mechanism and places PRC1 recruitment and activity downstream of PRC2 function. Based on the conservation of CBX proteins, and on the evidence that PRC1 binding to chromatin is sensitive to loss of PRC250, the hierarchical recruitment mechanism was largely adopted to explain polycomb chromatin domain formation in vertebrates. However, detailed studies of the relationship between PRC1 and PRC2 in vertebrates indicate that other mechanisms are involved in the formation of polycomb domains. For example, deletion of PRC2 in mouse embryonic stem cells led to a reduction, but not loss, of PRC1 proteins at target sites and had little effect on global levels of H2AK119ub151. These observations suggested that the relationship between PRC1 and PRC2 is more complex than originally envisaged.
Figure 2. Beyond simple recruitment to more complex interactions.
a | In the ‘hierarchical’ model of polycomb complex function, PRC2 first binds to chromatin and places H3K27me3. It is then proposed that H3K27me3 is recognized by chromobox (CBX) proteins, which are a subunit of canonical PRC1 complexes, thereby allowing PRC1 to bind and mono-ubiquitylate H2AK119.
b | In the ‘Alternative’ model for polycomb recruitment, the initial event is the binding of variant PRC1 complexes (which contain RYBP instead of CBX) to chromatin. PRC1 then catalyses the mono-ubiquitylation of H2AK119, independently of PRC2 activity and H3K27me3. The H2AK119ub1 modification then promotes the recruitment of PRC2, possibly via direct recognition of H2AK119ub1 by the AEBP2-JARID2-PRC2 complex, and placement of the H3K27me3 mark.
c | PRC1 complexes functionally segregate into ‘canonical’ complexes that contain chromobox (CBX2/4/6/7/8) and polyhomeotic (PHC1/2/3) proteins, and a series of ‘variant’ complexes that contain RYBP (or the closely related YAF2 protein) and interact with proteins that are unique to individual variant PRC1 complexes (note that only selected proteins are shown here). Under some conditions the PRC2 subunit EED may interact with a CBX-containing canonical PRC1 complex (dashed arrow).
d | A model for the propagation of polycomb domains.Within the core PRC2 complex, the EED subunit is able to recognise H3K27me3 via its WD40 repeat domain. This interaction potentially recruits PRC2 to sites of pre-existing H3K27me3, as well as stimulating the enzymatic activity of PRC2. The EED-H3K27me3 interaction may facilitate spreading of H3K27me3 domains (left panel, dashed arrows) or copying of H3K27me3 onto newly incorporated histones (right panel, blue nucleosomes) during DNA replication, thereby stably propagating H3K27me3 domains in actively dividing cells.
A new twist in hierarchy: PRC1 recruits PRC2
Our understanding of polycomb systems has recently evolved with the surprising finding that PRC1 can recruit PRC2 to chromatin via a mechanism that involves recognition of H2AK119ub1 (Figure 2B). This alternative pathway was discovered using a cell-based system in which PRC1 was artificially recruited to a region of the genome devoid of genes and not normally occupied by polycomb proteins. Strikingly, de novo PRC1 binding to this site and monoubiquitylation of H2AK119 resulted in the subsequent recruitment of PRC2 and H3K27me3 deposition, creating a new polycomb chromatin domain52. A similar outcome was observed when PRC1 was artificially recruited to pericentric regions of the genome53.
This newly discovered recruitment mechanism seems to play a part in polycomb chromatin domain formation at natural target sites, as perturbation of PRC1 and loss of H2AK119ub1 caused a significant reduction in PRC2 binding and H3K27me3 across the genome. The precise mechanism by which PRC1-dependent H2AK119ub1 underpins de novo polycomb chromatin domain formation remains to be elucidated. However, in vitro studies have identified a PRC2 complex containing the auxiliary proteins AEBP2 and JARID2 that preferentially binds to and stimulates catalysis of H3K27me3 on chromatin containing H2AK119ub154 suggesting this may provide the molecular link between PRC1 activity on chromatin and recruitment of PRC2 and H3K27me3. Additional unexpected connections between the two polycomb complexes include the finding that PRC2 can interact directly with an alternative H2AK119ub1 E3 ligase, TRIM37, which is highly expressed in breast cancer cells carrying the 17q23 amplification55. Furthermore, EED, a core PRC2 subunit, has been reported to directly interact with PRC156 (Figure 2C). Together these observations indicate the potential for more functional overlap between PRC1 and PRC2 than previously appreciated.
Alternative roles for canonical PRC1
The observation that PRC1 can promote the recruitment of PRC2 to target sites on chromatin has placed a new focus on understanding how PRC1 complexes contribute to polycomb chromatin domain formation and function. Systematic biochemical purifications have revealed that PRC1 in vertebrates can be separated into ‘canonical’ PRC1 complexes, which have CBX proteins and presumably function as part of the hierarchical recruitment pathway, and the less well studied ‘variant’ PRC1 complexes, which lack CBX proteins and contain either RYBP or YAF2 (Figure 2C)8. Importantly, RYBP and YAF2 lack the capacity to bind H3K27me3, and can be recruited to chromatin in cells lacking functional PRC2, suggesting that variant PRC1 complexes must be recruited to chromatin via PRC2-independent mechanisms51. Furthermore, individual canonical and variant PRC1 complexes contain distinct protein subunits that likely contribute to target site recognition or catalysis8.
Examining the function of individual PRC1 complexes in vivo has revealed that variant PRC1 complexes are proficient at catalysing H2AK119ub1 on chromatin, whereas canonical complexes catalyse little of this modification52. This is consistent with observations that RYBP-containing PRC1 variant complexes have enhanced H2AK119ub1 catalysis in vitro8. As a consequence of their restricted catalytic activity in vivo, canonical PRC1 complexes seem to have limited capacity to recruit PRC2 and form polycomb chromatin domains. One exception to this generality was recently reported: a CBX2-containing canonical PRC1 complex seems to have a very specific role in atypical polycomb chromatin domain formation by directly recognizing pericentromeric heterochromatin during early mouse development57.
Nevertheless, if canonical PRC1 complexes are usually limited in their capacity to deposit H2AK119ub1 in vivo, why are they recruited to polycomb chromatin domains at all? A hint as to possible functions came with the recent demonstration that a stable component of canonical PRC1 complexes, polyhomeotic 2 (PHC2), can auto-polymerise via its sterile-alpha motif (SAM) domain leading to chromatin compaction and gene silencing58–60. This is consistent with previous reports detailing a ubiquitin ligase-independent role for PRC1 in chromatin compaction61. Interestingly, inhibition of PHC2 polymerisation resulted in loss of canonical PRC1 binding to chromatin only at sites marked with H3K27me3, suggesting that both H3K27me3-CBX interactions and SAM domain polymerisation may have important structural roles in creating and translating repressive chromatin structures on chromatin59.
Propagation of polycomb domains
Biochemical studies have demonstrated that polycomb repressive complexes can bind the histone modifications that they themselves place. For example, the EED subunit of PRC2 core complex binds H3K27me3 via its WD40 repeat and this interaction seems to stimulate the catalytic activity of PRC2, thus forming an activity-based feedback loop62, 63. It has been proposed that this could promote the spreading of H3K27me3 along chromatin as well as ensure the propagation of H3K27me3 on newly replicated chromatin62 (Figure 2D). Based on the finding that PRC1, PRC2 and their chromatin modifying activities are more intimately linked than previously appreciated 52–54, it seems plausible that this robust series of PRC-dependent feedback mechanisms could underpin both the spreading and maintenance of polycomb chromatin domains once they are initially established. It is tempting to speculate that this would not only contribute to epigenetic maintenance of polycomb chromatin domains, but also to rigidly maintain gene expression states during cell division and development. A detailed understanding of the epigenetic nature of polycomb chromatin domains is an interesting and evolving area of polycomb biology.
Polycomb systems and gene regulation
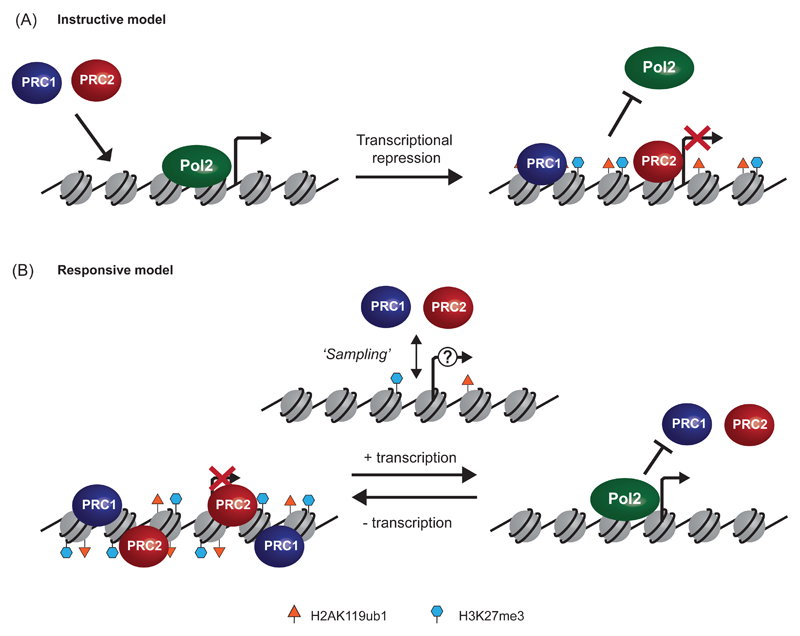
It is often suggested that vertebrate polycomb group proteins are recruited to target sites to actively drive transcriptional repression (Figure 3A). However, recent evidence suggests that this may not be the central modality connecting polycomb chromatin domains and gene repression. Firstly, kinetic analysis of gene expression in a cell culture model of Ras-induced transformation revealed that cessation of transcription preceded H3K27me3 acquisition64. Secondly, experiments in mouse embryonic stem cells demonstrated that small-molecule inhibitors that block transcription caused recruitment of PRC2 and H3K27me3 to previously active genes65. In light of these conceptually important findings, one interesting possibility is that polycomb systems may exploit generic targeting activities to constantly interface with or ‘sample’ gene regulatory elements and respond to the transcriptional state of individual genes. Within the context of this model, features of active transcription, for example the presence of RNA PolII or transcription-associated histone modifications, would decrease the residency time or catalytic activity of polycomb complexes at transcribed genes, meaning that full polycomb chromatin domain establishment would only occur at sites where transcriptional silencing has already been achieved (Figure 3B) (reviewed in 66).
Figure 3. Polycomb systems and gene regulation.
a | An instructive model for polycomb complex-mediated silencing would posit that newly recruited polycomb complexes lead to polycomb chromatin domain formation, which then directs repression of transcription by RNA Polymerase II (Pol II) at the associated gene.
b | A responsive model would posit that polycomb complexes constantly ‘sample’ chromatin at regulatory elements via generic targeting modalities in order to respond to the transcriptional state of the associated gene. Transcriptional cessation would lead to the subsequent establishment of polycomb chromatin domains that protect against low-level or stochastic reactivation signals. However, in response to active transcription, the presence of Pol II or other features of transcriptional initiation would block polycomb domain establishment.
What would be the purpose of establishing polycomb chromatin domains at already silenced genes? One possibility is that following transcriptional silencing, polycomb systems form repressive chromatin domains to limit the potential for stochastic reactivation events in inappropriate tissues. This general concept is consistent with observations in D. melanogaster, where polycomb group genes are important for the maintenance, but not the establishment, of gene expression programmes67. Interestingly, in pluripotent cells, polycomb-occupied gene promoters also typically exhibit low levels of the transcriptionally permissive histone modification H3K4me3, which is placed by trithorax group proteins68, 69. This modification is thought to result from the capacity of the trithorax system to generically ‘sample’ regulatory elements in a manner analogous to polycomb proteins. Given that polycomb and trithorax are two opposing chromatin-based gene regulatory systems, these constant sampling activities at gene regulatory elements might form the basis for a chromatin-encoded bistable switch that helps to regulate the transition between, and then stable maintenance of, chromatin states that are either permissive or repressive to transcription (discussed in detail in 4, 27, 66, 70).
Although polycomb chromatin domains are predominantly found at silenced genes, recent evidence has implicated polycomb proteins and their chromatin modifications in active transcription and other gene regulatory functions. This is the case for a variant PRC1 complex containing the AUTS2 protein, which recruits Casein Kinase 2 (CSNK2A) to counteract PRC1 ubiquitin ligase activity and repression71. Similarly, in erythroid lineages, a PRC2 complex containing an alternative catalytic core associated with regions of the genome characterized by chromatin modifications associated with active transcription (H3K4me3 and H3K27ac) and promoted gene expression72. Furthermore, it was recently demonstrated that PRC2-dependent H3K27 mono-methylation is enriched in the bodies of transcribed genes, while the H3K27 dimethyl state covers much of the genome, including non-genic locations, possibly as a mechanism to block aberrant function of enhancer elements73. These interesting recent observations highlight an incomplete understanding of the precise mechanisms by which polycomb systems regulate gene expression, and elucidating such mechanisms remains a key challenge.
Conclusion and future directions
A complex series of interactions between PRC1 and PRC2, chromatin, and transcription are essential for normal polycomb chromatin domain formation and function. Although transcription factor and lncRNA-based mechanisms contribute to polycomb complex targeting to specific chromatin regions, recent evidence suggests that polycomb complexes also constantly engage regulatory elements throughout the genome and establish repressive polycomb chromatin domains in a manner that is often responsive to the transcriptional state of associated genes. These emerging principles prompt alternative ways to think about the vertebrate polycomb system, and further experiments will be required to fully understand the molecular mechanisms underlying PRC function. Importantly, there is little evidence that the chromatin modifications placed by the polycomb systems directly repress transcription, suggesting that components of the polycomb protein complexes, such as PHC, may induce transcriptional inhibition by directly modifying chromatin structure.
Understanding the precise molecular mechanisms by which polycomb protein complexes repress gene expression remains a central and outstanding question in the field. A cryo-EM structure of the PRC2 holocomplex74 and the recently solved atomic structure of the PRC1 core complex bound to a nucleosome75 have provided exciting new insights into the interactions that occur within polycomb complexes and also with their chromatin substrates at high-resolution. These and other new discoveries will contribute to the understanding of how polycomb systems regulate gene expression programs during normal multicellular development. They will also pave the way for new approaches to therapeutic interventions in human diseases, including cancer, where polycomb systems are frequently perturbed.
Acknowledgments
Work in the Klose lab is supported by the Wellcome Trust, the Lister Institute of Preventive Medicine, and Exeter College, University of Oxford. NRR is supported by a Junior Research Fellowship at St John’s College, University of Oxford. We would like to thank Dr Emilia Dimitrova and Dr Sarah Cooper for constructive comments on the manuscript.
Author biographies
Rob Klose is Wellcome Trust Senior Research Fellow and Professor of Cell and Molecular Biology in the Department of Biochemistry at the University of Oxford. He carried out his doctoral work with Adrian Bird at the University of Edinburgh, UK, and his postdoctoral studies with Yi Zhang at the University of North Carolina, Chapel Hill, USA. His laboratory studies how chromatin modification and architecture contribute to gene regulation.
Neil Blackledge is a senior post-doctoral researcher in the group of Rob Klose. His doctoral studies were carried out with Ann Harris at Northwestern University, Chicago, USA. His post-doctoral work is aimed at understanding the role of CpG islands in gene regulation, with a particular focus on the polycomb repressive system.
Nathan Rose is a Junior Research Fellow in the Department of Biochemistry and St John’s College, University of Oxford. His doctoral studies were carried out with Christopher Schofield at the University of Oxford, and his postdoctoral work with Rob Klose focuses on the biochemistry of polycomb repressive complexes.
References
- 1.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 2.Scelfo A, Piunti A, Pasini D. The controversial role of the Polycomb group proteins in transcription and cancer: how much do we not understand Polycomb proteins? FEBS J. 2014 doi: 10.1111/febs.13112. [DOI] [PubMed] [Google Scholar]
- 3.Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–24. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15:340–56. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14:853–64. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 6.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–55. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 7.Grossniklaus U, Paro R. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb Perspect Biol. 2014;6:a019331. doi: 10.1101/cshperspect.a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–56. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa H, Ishiguro K-i, Gaubatz S, Livingston DM, Nakatani Y. A Complex with Chromatin Modifiers That Occupies E2F- and Myc-Responsive Genes in G0 Cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 10.Herranz N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–81. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietrich N, et al. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 2012;8:e1002494. doi: 10.1371/journal.pgen.1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold P, et al. Modeling of epigenome dynamics identifies transcription factors that mediate Polycomb targeting. Genome Res. 2013;23:60–73. doi: 10.1101/gr.142661.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren X, Kerppola TK. REST interacts with Cbx proteins and regulates polycomb repressive complex 1 occupancy at RE1 elements. Mol Cell Biol. 2011;31:2100–10. doi: 10.1128/MCB.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu M, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45:330–43. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier VK, et al. Functional proteomic analysis of repressive histone methyltransferase complexes PRC2 and G9A reveals ZNF518B as a G9A regulator. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.M114.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brockdorff N. Noncoding RNA and Polycomb recruitment. Rna. 2013;19:429–42. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohlmaier A, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva J, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–95. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 19.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 20.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–9. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 21.da Rocha ST, et al. Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol Cell. 2014;53:301–16. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Sarma K, et al. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell. 2014;159:869–83. doi: 10.1016/j.cell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerase A, et al. Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc Natl Acad Sci U S A. 2014;111:2235–40. doi: 10.1073/pnas.1312951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 26.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackledge NP, Klose R. CpG island chromatin: a platform for gene regulation. Epigenetics. 2011;6:147–52. doi: 10.4161/epi.6.2.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendenhall EM, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genetics. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farcas AM, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He J, et al. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15:373–84. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b Recruits Polycomb Repressive Complex 1 to CpG Islands and Regulates H2A Ubiquitylation. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Boulard M, Edwards JR, Bestor TH. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat Genet. 2015;47:479–85. doi: 10.1038/ng.3272. [DOI] [PubMed] [Google Scholar]
- 34.Li G, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–80. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davidovich C, et al. Toward a Consensus on the Binding Specificity and Promiscuity of PRC2 for RNA. Mol Cell. 2015 doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–7. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–64. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell. 2014;55:171–85. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014;28:1983–8. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitges FW, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–41. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 42.Herzog VA, et al. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat Genet. 2014;46:973–81. doi: 10.1038/ng.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai L, et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell. 2013;49:571–82. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin S, et al. Tudor domains of the PRC2 components PHF1 and PHF19 selectively bind to histone H3K36me3. Biochem Biophys Res Commun. 2013;430:547–53. doi: 10.1016/j.bbrc.2012.11.116. [DOI] [PubMed] [Google Scholar]
- 45.Brien GL, et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol. 2012;19:1273–81. doi: 10.1038/nsmb.2449. [DOI] [PubMed] [Google Scholar]
- 46.Musselman CA, et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol. 2012;19:1266–72. doi: 10.1038/nsmb.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mozzetta C, et al. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol Cell. 2014;53:277–89. doi: 10.1016/j.molcel.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 48.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–8. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, et al. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–46. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 51.Tavares L, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–78. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–59. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper S, et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014;7:1456–70. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalb R, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol. 2014;21:569–71. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 55.Bhatnagar S, et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516:116–120. doi: 10.1038/nature13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao Q, et al. The central role of EED in the orchestration of polycomb group complexes. Nat Commun. 2014;5:3127. doi: 10.1038/ncomms4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tardat M, et al. Cbx2 Targets PRC1 to Constitutive Heterochromatin in Mouse Zygotes in a Parent-of-Origin-Dependent Manner. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 58.Gambetta Maria C, Müller J. O-GlcNAcylation Prevents Aggregation of the Polycomb Group Repressor Polyhomeotic. Dev Cell. 2014;31:629–639. doi: 10.1016/j.devcel.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 59.Isono K, et al. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell. 2013;26:565–77. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 60.Kim CA, Gingery M, Pilpa RM, Bowie JU. The SAM domain of polyhomeotic forms a helical polymer. Nat Struct Biol. 2002;9:453–7. doi: 10.1038/nsb802. [DOI] [PubMed] [Google Scholar]
- 61.Eskeland R, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–64. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 63.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–7. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hosogane M, Funayama R, Nishida Y, Nagashima T, Nakayama K. Ras-induced changes in H3K27me3 occur after those in transcriptional activity. PLoS Genet. 2013;9:e1003698. doi: 10.1371/journal.pgen.1003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riising EM, et al. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell. 2014;55:347–60. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 66.Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. Chromatin sampling--an emerging perspective on targeting polycomb repressor proteins. PLoS Genet. 2013;9:e1003717. doi: 10.1371/journal.pgen.1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 68.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 69.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–8. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 70.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–38. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gao Z, et al. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature. 2014;516:349–54. doi: 10.1038/nature13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu J, et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol Cell. 2015;57:304–16. doi: 10.1016/j.molcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferrari KJ, et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell. 2014;53:49–62. doi: 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 74.Ciferri C, et al. Molecular architecture of human polycomb repressive complex 2. Elife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–6. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]