Abstract
PepV, a dipeptidase found in culture fluids of Streptococcus gordonii FSS2, was purified and characterized, and its gene was cloned. PepV is a monomeric metalloenzyme of approximately 55 kDa that preferentially degrades hydrophobic dipeptides. The gene encodes a polypeptide of 467 amino acids, with a theoretical molecular mass of 51,114 Da and a calculated pI of 4.8. The S. gordonii PepV gene is homologous to the PepV gene family from Lactobacillus and Lactococcus spp.
Streptococcus gordonii is an oral streptococcus that is important in both the formation of dental plaque and the production of infective endocarditis. Previous work with S. gordonii FSS2, a strain isolated from the blood of an endocarditis patient (10), has revealed a variety of cell-associated and extracellular glycosidolytic and proteolytic activities, thought to be important in the growth of this organism in the dental plaque and endocarditis vegetation environments (4, 5, 6, 11). In recent studies of extracellular proteinases made by this strain, an x-prolyl dipeptidyl peptidase (xPDPP) and an arginine aminopeptidase have been purified and characterized and the genes for both enzymes have been cloned and examined (4, 5). Here we report similar studies on a third such enzyme, S. gordonii PepV. S. gordonii PepV is a member of the PepV dipeptidase family, previously found in Lactobacillus and Lactococcus spp. (3, 7, 14, 15).
Materials and methods.
S. gordonii FSS2 (previously Streptococcus sanguinis FSS2 [6]) was described earlier (6, 10, 11). Other oral (viridans group) streptococci (see Fig. 3) were provided by Vincent Fischetti, Rockefeller University. S. gordonii FSS2 was grown with pH control as previously reported (4, 5).
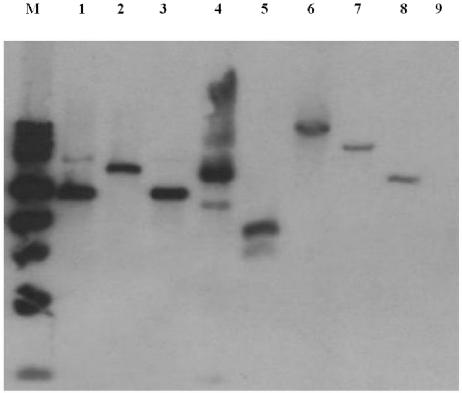
FIG. 3.
Presence of the PepV gene in viridans group streptococci, as shown by Southern blot analysis. Four micrograms of bacterial DNA was digested with HindIII and hybridized with the PepV probe. Lanes: M, molecular mass markers; 1, S. gordonii PK48; 2, S. gordonii PK48; 3, S. gordonii DL1; 4, S. gordonii PK2585; 5, S. parasanguinis PK2584; 6, S. mitis J27; 7, S. oralis J21; 8, S. oralis PK34; 9, S. salivarius ATCC 27945.
Enzyme assays measured both tripeptidase and dipeptidase activities of S. gordonii PepV (hereafter, PepV refers to S. gordonii PepV unless indicated otherwise; “S. gordonii” is added only when necessary to avoid ambiguity as to which version of PepV is meant). During purification (see Fig. 1) and most inhibitor studies (see Table 2), the tripeptidase activities of crude samples and purified PepV were measured in a two-step enzymatic reaction, with H-Ala-Phe-Pro-pNA (Sigma) as the substrate. PepV and the substrate (1 mM), both in assay buffer A (50 mM Tris, 1 mM CaCl2 [pH 7.8]), were incubated; products of this reaction were NH3+-Ala-COO- and NH3+-Phe-Pro-pNA. Excess S. gordonii xPDPP (100 ng) then was added to release the pNA reporter group, and A405 was measured in a plate reader. Since S. gordonii xPDPP is a serine proteinase (4), potential inhibition of PepV by serine class inhibitors (diisopropylfluorophosphate, phenymethylsulfonyl fluoride, 3,4-dichloroisocoumarin, and TLCK [Nα-p-tosyl-llysine chloromethyl ketone]; see Table 2) was examined in an independent dipeptidase assay that measured PepV-dependent release of ninhydrin-positive material using Leu-Gly as the substrate (2). Inhibitors were obtained from Sigma, Roche Molecular Biochemicals, or Calbiochem, except for Anstatin, which was a gift from Mirjana Grujic, Jozef Stefan Institute, Lubljana, Slovenia. For studies of cleavage specificity (see Table 1), hydrolysis of dipeptides, tripeptides, and higher peptides was measured in a high-performance liquid chromatography (HPLC) assay. The enzyme was incubated with the experimental peptide (enzyme-to-substrate ratio, 1:1,000) in 100 μl of 100 mM Tris, pH 7.8, for 2 h at 37°C, and the products were analyzed by HPLC.
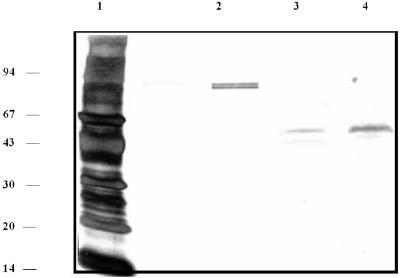
FIG. 1.
Purification of PepV seen by SDS-PAGE (silver stained). Lane 1, 5 μg of molecular mass markers (phosphorylase b, 94 kDa; bovine serum albumin, 67 kDa; ovalbumin, 43 kDa; carbonic anhydrase, 30 kDa; soybean trypsin inhibitor, 20 kDa; cytochrome c, 14 kDa); lanes 2 to 4, boiled, reduced samples of pure protein from their respective final chromatography steps: 500 ng of purified S. gordonii xPDPP from Gly-Pro Sepharose (lane 2); 100 ng of partially purified PepV from Gly-Pro Sepharose (lane 3), and 200 ng of purified PepV from Mono-P (lane 4).
TABLE 2.
Inhibition profile of S. gordonii PepV
| Inhibitorb | Concn or time | Residual activity (%)a |
|---|---|---|
| DFP | 5 mM | 90 |
| PMSF | 2 mM | 100 |
| 3,4-Dichloroisocoumarin | 2 mM | 97 |
| TLCK | 10 mM | 100 |
| Amastatin | 350 μM | 3 |
| Anstatin | 50 μM | 100 |
| Apstatin | 500 μM | 1 |
| Diprotin A | 500 μM | 98 |
| E-64 | 500 μM | 157 |
| Iodoacetamide | 5 mM | 100 |
| EDTA | 5 mM | 2 |
| EGTA | 5 mM | 6 |
| 1,10-o-Phenanthroline | 5 mM | 0 |
| Phosphoramidon | 1 mM | 110 |
| DTT | 5 mM | 215 |
| β-Mercaptoethanol | 5 mM | 130 |
| K+ | 5 mM | 76 |
| Ca2+ | 5 mM | 35 |
| Mg2+ | 5 mM | 136 |
| Mn2+ | 5 mM | 143 |
| Zn2+ | 5 mM | 9 |
| Urea | 1 mM | 16 |
| SDS | 1% | 1 |
| 100°C | 10 min | 0 |
Residual activity was expressed as the percentage of the Vmax of PepV with no inhibitor.
DFP, diisopropylfluorophosphate; PMSF, phenylmethylsulfonyl fluoride; DTT, dithiothreitol.
TABLE 1.
Cleavage specificity of S. gordonii PepV
| Peptidea | % Cleavageb |
|---|---|
| Ala-↓-Ala | 100 |
| Ala-↓-Phe | 100 |
| Pro-↓-Phe | 100 |
| Leu-↓-Gly | 100 |
| Met-↓-Tyr | 100 |
| Gly-↓-Gly-↓-Ala | 75 |
| Gly-↓-Pro-Ala | 20 |
| Ala-↓-Phe-Pro | 20 |
| Ala-↓-Pro-Gly | 6 |
Cleavage sites are indicated by arrows. Cleavage sites for tripeptides were deduced from the appearance on HPLC of a dipeptide peak corresponding to the decrease in the tripeptide. No such dipeptide peaks were seen for Gly-Gly-Ala, indicating that both peptide bonds could be cleaved. The following peptides were not cleaved: Ala-His (carnosine), Ala-Pro, Gly-Pro, Ile-Pro, Leu- Pro, Lys-Pro, Ser-Pro, Gly-Gly, Gly-Pro-Arg-Pro (fibrin inhibitory peptide), and Trp-Ala-Gly- Gly-Asp-Ala-Ser-Gly-Glu (sleep-inducing peptide).
The percent cleavage is defined as (1 − area of residual peptide peak after digestion/area of peptide peak in undigested control) × 100.
PepV was purified from the culture fluid of a 15-liter stirred culture (pH was maintained at 7.5) in early stationary phase, at which time all glucose had been metabolized and approximately 25% of the total PepV activity was extracellular (data not shown). Cells were removed, and 80% ammonium sulfate was added to the culture fluid. The precipitate was collected, dialyzed, and chromatographed on DE-52. Active fractions were successively chromatographed on Superdex-75 HR 10/30, phenyl-Sepharose HP, Mono-Q HR 10/10, Cibacron Blue Sepharose CL-B6, Gly-Pro Sepharose, and Mono-P 5/5. Details of the purification scheme are available from the authors. Enzyme purity and molecular weight were assessed by Tris-HCl-Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12), silver staining (1), and gel filtration.
To clone the PepV gene, DNA from S. gordonii FSS2 was purified (Purgene; Gentra, Minneapolis, Minn.). The N-terminal peptide sequence of PepV (TIDFKAEVEKRREAL), obtained by Edman degradation, was used to search an S. gordonii database (ftp://ftp.tigr.org/pub/data/s_gordonii), resulting in identification of a 1,401-bp open reading frame encoding PepV. Subsequently, two PCR primers encoding the N and C termini were synthesized (5′-AGTGGATCCATGACAATTGATTCTAAAGC-3′ and 5′-TTTGGATCCTTATTTGATTAGTTCGTAG-3′) and used in PCR to obtain the full-length DNA fragment encoding PepV. A single 1,410-bp PCR product was obtained, gel purified, subcloned, and sequenced. Details of the cloning procedure and Southern blot analysis are available from the authors.
Results.
PepV copurified with the previously described S. gordonii xPDPP (4) through several steps and was isolated as a homogeneous protein by addition of a Mono-P chromatofocusing step, as shown in Fig. 1. The approximate molecular mass was 50 kDa by SDS-PAGE (Fig. 1). Gel filtration analysis (data not shown) indicated a molecular mass of 50 to 60 kDa, suggesting that native PepV is a monomer. Isoelectric focusing-PAGE and Mono-P standardization (not shown) indicated an approximate pI of 4.73. The cleavage specificity of PepV was examined with a variety of peptide substrates (Table 1). Several hydrophobic dipeptides were completely hydrolyzed, and a number of dipeptides with Pro in the P1′ position were not cleaved. Interestingly, tripeptide analogs (Gly-Pro-Ala and Ala-Pro-Gly) of two of such dipeptides (Gly-Pro and Ala-Pro) were partially degraded by PepV; the same was true for Gly-Gly and Gly-Gly-Ala. Larger peptides (fibrin inhibitory peptide and sleep-inducing peptide) were not cleaved even with prolonged incubation. Inhibitor studies (Table 2) indicate that PepV is inhibited by EDTA, EGTA, and 1,10-o-phenanthroline and thus is a metallopeptidase. The enzyme was sensitive to apstatin and amastatin (inhibitors of aminopeptidase P and glutamyl aminopeptidase/aminopeptidase A, respectively), but not to phosphoraminidon, an inhibitor of metalloendopeptidases.
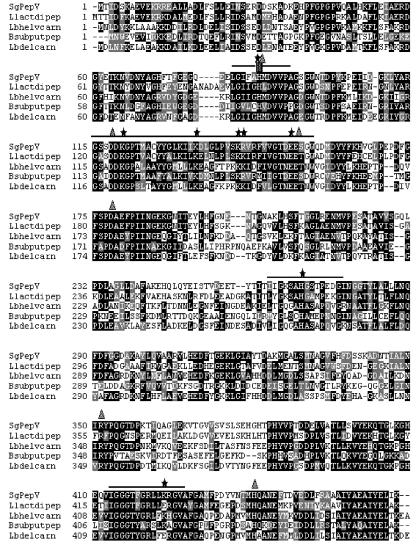
Gene cloning technology provided an open reading frame encoding a 467-amino-acid polypeptide with a theoretical molecular mass of 51,114 Da and a calculated pI of 4.8. A homology search for this polypeptide, done with National Center for Biotechnology Information TBLASTN, against the EMBL, DDBJ, GenBank, and PDB databases, indicated that PepV is highly conserved within the dinuclear peptidase M20 family in the MH clan of metallopeptidases. Previous members of this family have been found in lactic acid bacteria, where they are know as the PepV family (3, 7, 14, 15). The S. gordonii PepV gene has closest homology to PepV genes cloned from Lactococcus lactis (60% identity), Lactobacillus helveticus (49%), and Lactobacillus delbrueckii (45%), as well as a gene encoding a hypothetical dipeptidase from Bacillus subtilis (49%). Sequence alignments of these enzymes are shown in Fig. 2. The crystal structure of Lactobacillus delbrueckii PepV has been solved (8), and the active site includes a zinc-coordinating histidine (His 88) within the motif LGIIGHMDVVP (Fig. 2, box 1). By analogy, His 87 of PepV is likely to be the active-site residue. Furthermore an additional five residues of Lactobacillus delbrueckii PepV (Asp 119, Glu 154, Asp 177, Arg 351, and His 439) have been implicated in critical metal coordination and nucleophile stabilization within the catalytic pocket (8). PepV maintains all of these essential residues, which are marked in Fig. 2. Finally, the PepV sequence includes three additional regions (boxes 2, 3, and 4) that are conserved within the M20 family of metallopeptidases. PepV shows high identity with these signature sequences, namely, 69% for box 2, 57% for box 3, and 93% for box 4. Furthermore, nine charged residues that are invariant in the M20 family are conserved in PepV.
FIG. 2.
Multiple sequence alignment of S. gordonii PepV (SgPepV) and bacterial homologues. Sequences of PepV from FSS2 and from Lactococcus lactis MG1363 (Llactdipep), Lactobacillus helveticus SBT2171 (Lbhelvcarn), Bacillus subtilis (Bsubputpep), and Lactobacillus delbrueckii subsp. lactis DSM7290 (Lbdelcarn) were aligned with the ClustalW multiple sequence alignment tool according to homology modeling (grey shading indicates similarity, and black shading indicates identity). Conserved box regions (boxes 1 to 4) are in sequential order and are denoted with single lines. Box, putative catalytic His residue within box 1; stars, conserved charged residues; triangles, residues implicated in the catalytic pocket from the crystal structure of Lactobacillus delbrueckii PepV.
The presence of the PepV gene in other oral streptococci is shown in Fig. 3. By Southern blot analysis using a PepV probe, a single copy of the gene was found in S. gordonii FSS2. Additionally, the gene was found in other S. gordonii strains (PK48, DL1, and PK2585) and in several species (Streptococcus mitis, Streptococcus oralis, and Streptococcus parasanguinis) that along with S. gordonii are members of the mitis group of viridans group streptococci (16). The PepV gene was not found in Streptococcus salivarius, which is not a member of the mitis group.
Discussion.
The biochemical results (Fig. 1; Tables 1 and 2) are consistent with the interpretation that PepV is a metalloclass aminopeptidase with specificity for hydrophobic dipeptides. The enzyme has weaker tripeptidase activity but has no activity on tested peptides with four or more amino acids, suggesting a size limitation for the specificity pocket. A novel feature is the resistance to cleavage of dipeptides with Pro in the P1′ position and the partial relief of that resistance when a third amino acid is added in the P2′ position (Table 1). Collectively, these results are comparable to those from PepV dipeptidases of Lactococcus lactis, Lactobacillus helveticus, and Lactobacillus delbrueckii (3, 7, 14, 15).
Genetic evidence (Fig. 3) indicates that in S. gordonii the PepV gene exists as a single copy and that this gene is conserved in the mitis group but not in at least one other group of the viridans group streptococci. The cloned gene for PepV (Fig. 2) reveals a protein sequence with significant homology (51% average identity) with PepV gene products of several lactic acid bacteria. The consensus sequence for the active site of PepV proteins from lactic acid bacteria (LGIIGHXDVVPAG) is present with slight modifications in S. gordonii PepV (Fig. 2, box 1). In S. gordonii PepV, the conserved IG in positions 4 and 5 of the consensus sequence is replaced by FA. Other notable features of PepV are the putative active-site His in box 1 and the strict conservation of charged residues in all four box motifs. The crystal structure of PepV from Lactobacillus delbrueckii has been elucidated (8), and there is substantial homology between PepVs of Lactobacillus delbrueckii and S. gordonii FSS2 (Fig. 3). Thus by analogy the active-site His and conserved charged residues in S. gordonii PepV also serve to stabilize zinc and the water nucleophile. These properties allow S. gordonii PepV to be included in the MH family of metallopeptidases.
This is the first report of the purification, characterization, and gene cloning of a PepV dipeptidase from the genus Streptococcus. The high degree of sequence conservation between S. gordonii PepV and PepV enzymes from Lactococcus and Lactobacillus spp. suggests the possibility of a common evolutionary gram-positive ancestor. The biological significance of the entire PepV subfamily is presently uncertain, particularly in view of the typically complex and redundant proteolytic systems of these bacterial groups (9, 13). Presumably S. gordonii PepV activity is useful for the growth of the mitis group of viridans group streptococci in dental plaque and heart valve vegetation environments. Future studies should focus on PepV expression and on the production of PepV knockouts in order to assess PepV's contributions to streptococcal growth and virulence in endocarditis.
Nucleotide sequence accession number.
The sequence for the PepV gene was deposited in GenBank under accession number AY496433.
Acknowledgments
This work was supported by grant DE009761 from the National Institute of Dental and Craniofacial Research.
We thank Vincent Fischetti and Mirjana Grujic for gifts of bacterial strains and Anstatin, respectively.
Editor: J. T. Barbieri
REFERENCES
- 1.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 2.Doi, E., D. Shibota, and T. Matoba. 1981. Modified colorimetric ninhydrin methods for peptidase assay. Anal. Biochem. 118:173-184. [DOI] [PubMed] [Google Scholar]
- 3.Dudley, E. G., A. C. Husgen, W. He, and J. L. Steele. 1996. Sequencing, distribution, and inactivation of the dipeptidase A gene (pepDA) from Lactobacillus helveticus CNRZ32. J. Bacteriol. 178:701-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein, J. M., A. Banbula, T. Kordula, J. A. Mayo, and J. Travis. 2001. Novel extracellular x-prolyl dipeptidyl-peptidase (DPP) from Streptococcus gordonii FSS2: an emerging subfamily of viridans streptococcal x-prolyl DPPs. Infect. Immun. 69:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein, J. M., D. Nelson, T. Kordula, J. A. Mayo, and J. Travis. 2002. Extracellular arginine aminopeptidase from Streptococcus gordonii. Infect. Immun. 70:835-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harty, D. W., J. A. Mayo, S. L. Cook, and N. A. Jacques. 2000. Environmental regulation of glycosidase and peptidase production by Streptococcus gordonii FSS2. Microbiology 146:1923-1931. [DOI] [PubMed] [Google Scholar]
- 7.Hellendoorn, M. A., B. M. D. Franke-Fayard, I. Mierau, G. Venema, and J. Kok. 1997. Cloning and analysis of the pepV dipeptidase gene of Lactococcus lactis MG1363. J. Bacteriol. 179:3410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jozic, D., G. Bourenkow, H. Bartunik, H. Scholze, V. Dive, B. Henrich, R. Huber, W. Bode, and K. Maskos. 2002. Crystal structure of the dinuclear zinc aminopeptidase PepV from Lactococcus delbrueckii unravels its preference for dipeptides. Structure 10:1097-1106. [DOI] [PubMed] [Google Scholar]
- 9.Kok, J. 1990. Genetics of the proteolytic system of lactic acid bacteria. FEMS Microbiol. Rev. 7:15-42. [DOI] [PubMed] [Google Scholar]
- 10.Manning, J. E., E. B. Hume, N. Hunter, and K. W. Knox. 1994. An appraisal of the virulence factors associated with streptococcal endocarditis. J. Med. Microbiol. 40:110-114. [DOI] [PubMed] [Google Scholar]
- 11.Mayo, J. A., H. Zhu, D. W. Harty, and K. W. Knox. 1995. Modulation of glycosidase and protease activities by chemostat growth conditions in an endocarditis strain of Streptococcus sanguis. Oral Microbiol. Immunol. 10:342-348. [DOI] [PubMed] [Google Scholar]
- 12.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 13.Tan, P. S., B. Poolman, and W. N. Konings. 1993. Proteolytic enzymes of Lactococcus lactis. J. Dairy Res. 60:269-286. [DOI] [PubMed] [Google Scholar]
- 14.Tan, P. S. T., M. Sasaki, B. W. Bosman, and T. Iwasaki. 1995. Purification and characterization of a dipeptidase from Lactobacillus helveticus SBT 2171. Appl. Environ. Microbiol. 61:3430-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vongerichten, K. F., J. R. Klein, H. Matern, and R. Plapp. 1994. Cloning and nucleotide sequence analysis of pepV, a carnosinase gene from Lactobacillus delbrueckii subsp. lactis DSM 7290, and partial characterization of the enzyme. Microbiology 140:2591-2600. [DOI] [PubMed] [Google Scholar]
- 16.Whiley, R. A., and D. Beighton. 1998. Current classification of the oral streptococci. Oral Microbiol. Immunol. 13:195-216. [DOI] [PubMed] [Google Scholar]