Abstract
IMPORTANCE
A substantial number of patients diagnosed with high-risk prostate cancer are at risk for metastatic progression after primary treatment. Better biomarkers are needed to identify patients at the highest risk to guide therapy intensification.
OBJECTIVE
To create a DNA damage and repair (DDR) pathway profiling method for use as a prognostic signature biomarker in high-risk prostate cancer.
DESIGN, SETTING, AND PARTICIPANTS
A cohort of 1090 patients with high-risk prostate cancer who underwent prostatectomy and were treated at 3 different academic institutions were divided into a training cohort (n = 545) and 3 pooled validation cohorts (n = 232, 130, and 183) assembled for case-control or case-cohort studies. Profiling of 9 DDR pathways using 17 gene sets for GSEA (Gene Set Enrichment Analysis) of high-density microarray gene expression data from formalin-fixed paraffin-embedded prostatectomy samples with median 10.3 years follow-up was performed. Prognostic signature development from DDR pathway profiles was studied, and DDR pathway gene mutation in published cohorts was analyzed.
MAIN OUTCOMES AND MEASURES
Biochemical recurrence-free, metastasis-free, and overall survival.
RESULTS
Across the training cohort and pooled validation cohorts, 1090 men were studied; mean (SD) age at diagnosis was 65.3 (6.4) years. We found that there are distinct clusters of DDR pathways within the cohort, and DDR pathway enrichment is only weakly correlated with clinical variables such as age (Spearman ρ [ρ], range, −0.07 to 0.24), Gleason score (ρ, range, 0.03 to 0.20), prostate-specific antigen level (ρ, range, −0.07 to 0.10), while 13 of 17 DDR gene sets are strongly correlated with androgen receptor pathway enrichment (ρ, range, 0.33 to 0.82). In published cohorts, DDR pathway genes are rarely mutated. A DDR pathway profile prognostic signature built in the training cohort was significantly associated with biochemical recurrence-free, metastasis-free, and overall survival in the pooled validation cohorts independent of standard clinicopathological variables. The prognostic performance of the signature for metastasis-free survival appears to be stronger in the younger patients (HR, 1.67; 95%CI, 1.12–2.50) than in the older patients (HR, 0.77; 95%CI, 0.29–2.07) on multivariate Cox analysis.
CONCLUSIONS AND RELEVANCE
DNA damage and repair pathway profiling revealed patient-level variations and the DDR pathways are rarely affected by mutation. A DDR pathway signature showed strong prognostic performance with the long-term outcomes of metastasis-free and overall survival that may be useful for risk stratification of high-risk prostate cancer patients.
Over 29000mendie from prostate cancer annually, despite the majority being diagnosed with clinically localized disease when cure is still likely.1,2 Therefore, there is a critical need to identify this subset with potentially lethal localized disease within the 220 000 men diagnosed each year. Current clinical paradigms using prostate-specific antigen (PSA) level, Gleason score, and tumor stage, provide guidance but are imperfect.3,4 Molecular biomarkers are an emerging strategy to identify patients with prostate cancer at high risk of metastatic progression from those at reduced or standard risk.5–7
Biomarker studies have evolved from single-gene, hypothesis-driven studies to unbiased gene biomarker nomination and signature development.7–10 Several analyses have taken a hybrid pathway-based approach focused on DNA repair pathways based on its central role in radiotherapy and chemotherapy response. These DNA repair pathway scores have prognostic and/or predictive power in lung and ovarian cancer.11,12 In prostate cancer, no such analyses have been performed, and a lack of gene expression data with long-term follow-up hampers biomarker studies in general.
We hypothesized that DNA damage and repair (DDR) pathways hold prognostic information in prostate cancer and developed a novel patient-level DDR pathway profiling approach to investigate this. We applied this approach to a large prostatectomy cohort with long clinical follow-up that allows for analysis of robust outcomes of biochemical recurrence free, metastasis-free, and overall survival. We assessed correlation between DDR pathways and molecular or clinical variables and used multivariate analysis to build and validate a prognostic signature biomarker combining the DDR pathways. Overall, this study investigates the potential for individualized DDR pathway profiling in the clinical management of prostate cancer.
Methods
Study Design and Tissue Samples
Tumor samples were from 4 published retrospective prostatectomy patient cohorts at the Mayo Clinic (MCI and MCII), Cleveland Clinic (CC), and Thomas Jefferson University (TJU).5–7,13 Informed consent protocols were approved by local institutional review boards. The MCI6 cohort was a nested case-control study with 545 men in matched triplets of metastatic progression, biochemical recurrence without metastatic progression, and no evidence of disease after prostatectomy. The MCII7 cohort was a metastatic progression case cohort study of 1010 men sampled to generate a final cohort of 232. The TJU5 cohort is comprised of 130 patients who underwent postoperative radiotherapy for pT3 or margin positive disease at prostatectomy. The CC cohort included 183 patients from a 3:1 no evidence of disease to metastatic progression sampling ratio from a case-control study of 2317 patients who underwent prostatectomy with preoperative PSA greater than 20 ng/mL, pT3, positive margins, or pathologic Gleason score of 8 or higher who did not receive adjuvant or neo adjuvant therapy.13 Treatment after prostatectomy, including androgen deprivation therapy and/or radiotherapy, was given for disease recurrence at the discretion of the treating physician in all cohorts. Patients in all cohorts were followed for multiple outcomes including biochemical recurrence, metastatic progression, and any death.
Tissue Preparation, RNA Extraction, and Microarray Hybridization
As described previously, formalin-fixed paraffin embedded (FFPE) prostatectomy samples were collected from the 4 cohorts.5–7 RNA extraction and microarray hybridization was performed in a CLIA (Clinical Laboratory Improvement Amendments)-certified, clinical operations laboratory (GenomeDx Biosciences, Inc). Purified total RNA was whole-transcriptome amplified using the WT-Ovation FFPE system (NuGen), fragmented and labeled using the Encore Biotin Module (NuGen), and hybridized to Affymetrix Human Exon (HuEx) 1.0STGeneChips (Affymetrix). Microarray normalization was done with single channel array normalization, and quality control was performed using Affymetrix Power Tools (Affymetrix, Inc). Gene expression was calculated using Affymetrix Core-level summaries for annotated genes as previously described. Microarray data are available with National Center for Biotechnological Information Gene Expression Omnibus accession numbers GSE46691, GSE62116, GSE72291, and GSE62667.5–7
Pathway Profiling and GSEA (Gene Set Enrichment Analysis)
We first ensured all expression values were positive by adding a small positive value (4) equal to the lowest negative expression value as previously described.13 Expression values were then median-scaled and ranked across all patients on a gene-by-gene basis. Individual patient gene rank profiles were used as input for the Gene Set Enrichment Analysis (GSEA) pre-ranked algorithm with 1000 permutations to generate individual patient DDR gene set profiles of normalized enrichment scores.14,15 The GSEA gene sets used represented the major DDR pathways in the standard curated pathway collections Bio-Carta, KEGG(Kyoto Encyclopedia of Genes and Genomes), and Reactome found at the GSEA mSigDB web site (http://www.broadinstitute.org/gsea/msigdb/index.jsp) (eTable 1 in the Supplement). Multiple gene sets for the same pathway were combined by using the mean to generate the following pathway scores: DNA damage checkpoint (Checkpoint), DNA repair (Repair), base excision repair (BER), double-stranded break repair (DSB), nonhomologous end-joining repair (NHEJ), mismatch repair(MMR), and nucleotide excision repair (NER); the homologous recombination gene sets were used individually (eTable 1 in the Supplement), and androgen receptor pathway activity was similarly assessed using the “ANDROGEN_RECEPTOR_SIGNALING_PATHWAY” GSEA gene set.
Mutation Analysis
Primary nonmetastatic prostate genomic profiles from published studies and The Cancer Genome Atlas unpublished data were analyzed using the R API from cBio Portal with a custom script (eMethods in the Supplement).
Statistical Analysis and Signature Generation
Correlation between DDR gene set enrichment and clinical and molecular variables was performed with the Spearman correlation. Univariate analysis (UVA) and multivariate analysis (MVA) for signature development and determining prognostic association were performed using Cox proportional hazards regression. For signature development, DDR pathway profile scores were used as continuous variables. A threshold for “high risk” was automatically chosen as the percentile that minimizes the UVA P value for metastatic progression in the training cohort MCI. “Cohort” was a stratification variable for analysis of the pooled validation cohorts (MCII, CC, and TJU) with different study designs, as described previously.13 Gleason score and PSA level were included as categorical variables based on risk group criteria: Gleason 6 and PSA less than 10 were considered low risk; Gleason 7 and PSA 10 to 20were considered intermediate risk; and Gleason 8 to 10 and PSA greater than 20 were considered high risk. Other clinical covariates included were lymph node invasion, extracapsular extension, seminal vesicle invasion, surgical margin status, and patient age. Outcomes were also analyzed by Kaplan-Meier analysis with log-rank test. Heatmaps were generated using R package heatmap.2 with default column scaling. All analyses were performed using R 2.15.
Results
Demographics
The demographics of the 1090 patients across 4 cohorts are listed in eTable 2 in the Supplement. The mean (SD) age at diagnosis was 65.3 (6.4) years. Commensurate with the high-risk nature of the cohorts, there are substantial rates of high-risk preoperative features, including 30% to 46% of patients with PSA greater than 10 ng/mL, 25%to 43% of tumors with a Gleason score of 8 to 10, and high rates of pathological high-risk features, including extracapsular extension in 41%to 81% and positive margins in 49%to 76%. Median follow-up across all cohortswas10.3 years, and metastatic progression rates varied among the cohorts from 8%to 39%. Thus, the large number of patients and long clinical follow-up make this is a powerful data set for exploring molecular biomarkers of disease progression risk in prostate cancer.
Pathway Profiling
We developed a novel patient-level pathway profiling approach to investigate the prognostic relevance of DDR pathways in prostate cancer. Our pathway profiling procedure is described in detail in thee Methods in the Supplement and depicted in Figure 1A and B. Briefly, the GSEA algorithm was used to convert gene expression data into DDR pathway expression data individually for each patient. Canonical DDR pathway gene sets from the curated collections BioCarta, KEGG, and Reactome were used as a fair representation of these pathways (eTable 1 in the Supplement). Separately, we found that the prevalence of mutations in the DDR gene set genes across 630 primary nonmetastatic prostate samples from published studies and The Cancer Genome Atlas unpublished data was less than2%for all genes except for ATM (3%) and TP53 (6.8%), indicating that the DDR pathways are negligibly disrupted by mutation (eTable 3 in the Supplement).
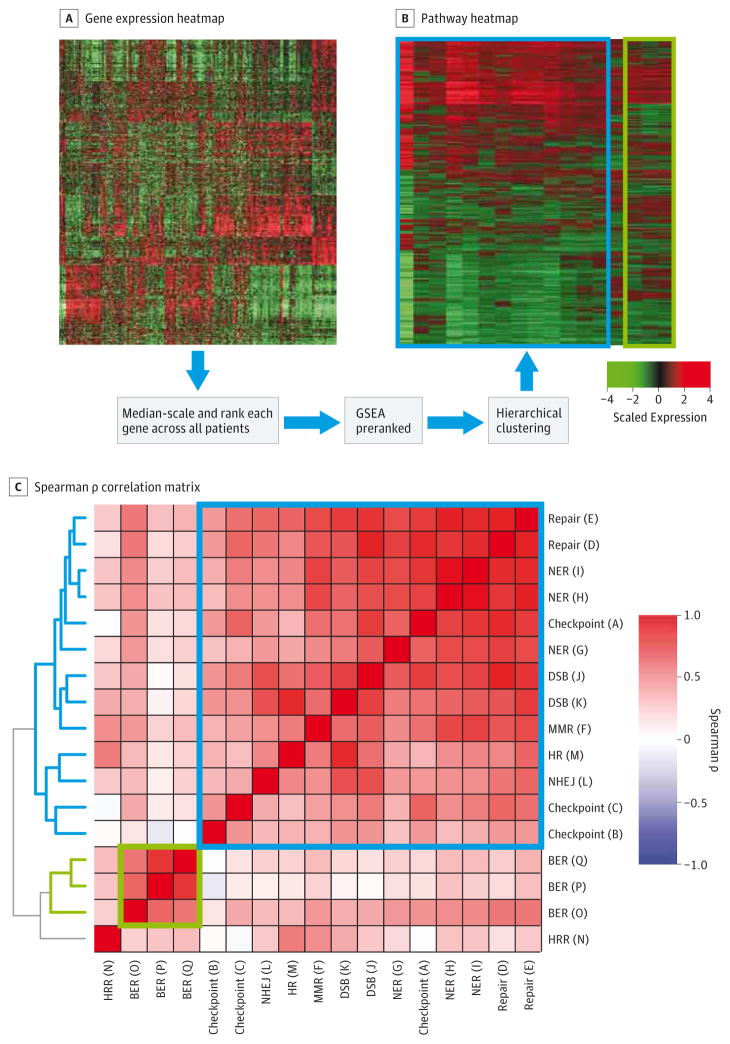
Figure 1. DDR Pathway Profiling Procedure.
A, Microarray gene expression data is converted into (B) pathway enrichment data. Expression is median-scaled and ranked across all samples gene-by-gene. B, Gene Set Enrichment Analysis preranked on gene ranks generates pathway enrichment NES, and hierarchical clustering generates a pathway heatmap. C, Spearman ρ correlation matrix of intercorrelation among the pathways across all samples. BCR indicates biochemical recurrence; BER, base excision repair; DDR, DNA damage and repair; DSB, double-strand break; GSEA, Gene Set Enrichment Analysis; HRR, homologous recombination repair; MMR, mismatch repair; NER, nucleotide excision repair; NES, Normalized Enrichment Score; NHEJ, nonhomologous end joining.
DNA damage and repair pathway profiling was applied across all 1090 patients. Unsupervised hierarchical clustering of patient-level DDR pathway enrichment profiles revealed distinct clusters of patterns of DDR pathway enrichment (Figure 1B). The gene sets representing the same pathway were highly correlated as expected (Figure 1C). There was also significant correlation among the pathways, though the 3 base excision repair gene sets followed an independent pattern. Of note, the HR pathway gene sets cluster distinctly, indicating that they capture disparate information. These findings confirmed our initial hypothesis that there is patient-level variation in DDR pathways which could carry prognostic information.
Gene Set Enrichment Correlation With Clinical and Molecular Variables
We then analyzed the correlation of DDR gene set enrichment with standard clinical and molecular variables (Figure 2) (eTable 1 and eFigure 1 in the Supplement). There was significant but weak correlation with Gleason score for 16 of 17 DDR gene sets (Spearman rho [ρ], range,0.09 to0.20), and with age for 7 of 17 gene sets (ρ, range, −0.07 to 0.24). Prostate specific antigen level was weakly correlated with only 3 of 17 gene sets (ρ, range, −0.07 to0.10). Thus, DDR gene sets are generally weakly correlated with clinical variables, indicating that DDR pathways can provide independent information.
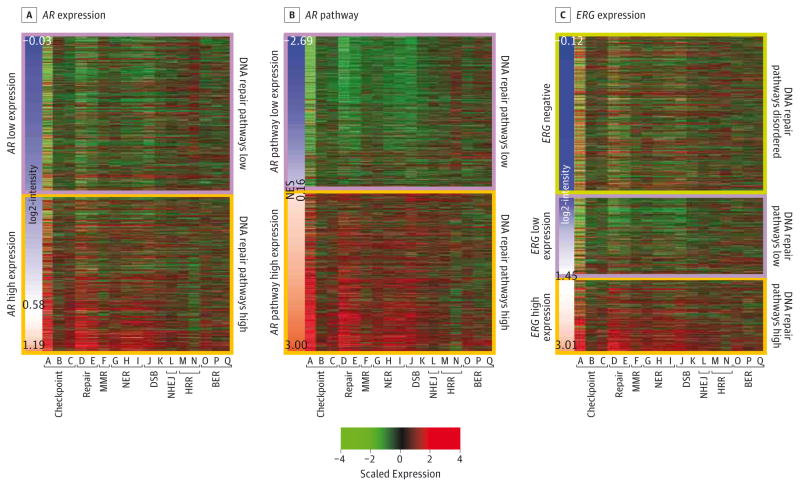
Figure 2. Correlation of DDR Pathway Profiles With Molecular Variables.
The sidebar represents log2-intensity expression for (A) AR, (B) GSEA NES for AR pathways, and (C) ERG. Orange along the the sidebar color scale indicates higher expression and blue indicates lower expression; maximum, minimum, and average values are indicated. For the heatmap color scale, red indicates higher scaled expression and green indicates lower scaled expression. Gene set labels can be found in eTable 1 in the Supplement. BER, base excision repair; DDR, DNA damage and repair; DSB, double-strand break; GSEA, Gene Set Enrichment Analysis; HRR, homologous recombination repair; MMR, mismatch repair; NER, nucleotide excision repair; NES, Normalized Enrichment Score; NHEJ, nonhomologous end joining.
We also analyzed correlation with the prostate cancer driver genes AR and ERG (Figure 2) (eTable 1 in the Supplement). AR expression has varying levels of significant positive correlation with all gene sets (ρ, range, 0.12 to 0.54), consistent with literature showing AR signaling upregulates DDR pathways.16,17 As AR messenger RNA (mRNA) expression may not fully represent AR transcriptional activity, we also measured AR activity using GSEA, which analyzes many AR target genes in addition to PSA. Interestingly, correlation between AR activity and the gene sets was strongerc (ρ, up to 0.82). Correlation with ERG expression showed intermediate results, with 13 of 17 gene sets with significant correlations (ρ, range, 0.09 to 0.25).
Of note, the homologous recombination gene sets appeared to reflect a distinct pattern. The Reactome homologous recombination gene set M had a weak positive correlation with AR expression (ρ, 0.12), whereas the KEGG homologous recombination gene set N had a weak negative correlation (ρ, −0.22); results with the AR pathway were similarly disparate. We reviewed these 2 gene sets in detail and confirmed there are significant differences with 9 genes in common of 17 in Reactome homologous recombination gene set M and 28 genes in KEGG homologous recombination gene set N (eTable 4 in the Supplement). Interestingly, neither homologous recombination gene set was correlated with ERG expression (P > .10 for all). These results demonstrate that the complex interactions among the DDR pathways and prostate cancer molecular drivers that have been minutely dissected in vitro, can be assayed in vivo at the individual patient level for possible use in the clinical management of prostate cancer.
Prognostic Performance of Individual Pathways
We next analyzed the prognostic information that might be contained in the pathway profiles. Seven of the 9 DDR pathways had statistically significant associations with worse metastasis-free survival on UVA (P < .02 for all) (eTable 5 in the Supplement). To control for confounders, we examined the individual pathways in MVA. For example, the Checkpoint pathway, an average of Checkpoint gene sets A through C, had a statistically significant association with worse metastasis free survival (hazard ratio [HR], 1.24; 95%CI, 1.28–1.61). Gleason score and seminal vesicle invasion were also significantly associated with both outcomes (P < .05 for all), and 5 other clinical variables were not significant. Gleason score had the strongest association (HR, 2.42; 95%CI 1.04–5.62 for intermediate risk; and HR 6.59; 95%CI, 2.83–15.3 for high risk), whereas seminal vesicle invasion associations were of similar magnitude as the Checkpoint pathway. Repeating this MVA for each pathway individually revealed that 4 of 9 pathways were significantly associated with these outcomes (P < .05 for all).
DDR Pathway Signature
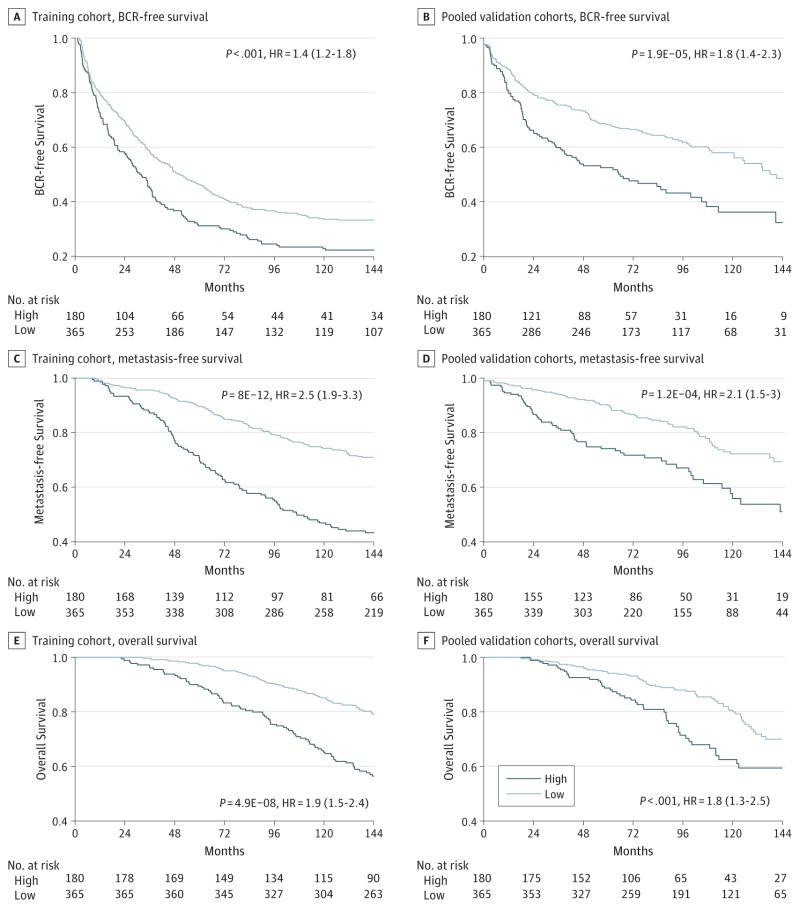
We sought to develop a prognostic signature biomarker based on combining the DDR pathway profiles consisting of a multivariate Cox model for metastasis-free survival in the training cohort MCI. As expected, this DDR pathway signature was significantly associated with metastasis-free survival in the training cohort (Figure 3A) but also had significant association with biochemical recurrence-free survival and overall survival (Figure 3C–E). This prognostic association was independent of standard clinical variables on MVA (eTable 6 in the Supplement). In the independent pooled validation cohorts, the DDR pathway signature was significantly associated with biochemical recurrence-free survival (HR, 1.8; 95% CI, 1.4–2.3) (Figure 3B), metastasis-free survival (HR, 2.1; 95% CI, 1.5–3.0) (Figure 3D), and overall survival (HR, 1.8; 95% CI, 1.3–2.5) (Figure 3F). Importantly, these findings were confirmed to be significant independent of standard clinicopathological variables on multivariate Cox analysis, with a concordance index of 0.71 for a model including the DDR pathway signature and the standard variables (Table 1) (eTable 7 in the Supplement). In support of the clinical use of these findings, we have constructed a nomogram for predicting metastasis-free survival at 10 years based on these models (eFigure 2 in the Supplement).
Figure 3. Prognostic Performance of the DDR Pathway Signature.
Kaplan-Meier analysis of the DDR pathway signature for biochemical recurrence-free survival (A and B), metastasis-free survival (C and D), and overall survival (E and F) in the training cohort (A, C, and E) and the pooled validation cohorts (B, D, and F). Kaplan-Meier log-rank test results are shown. Patients at risk in each risk group are shown below each graph. BCR indicates biochemical recurrence.
Table 1.
Cox Regression Analysis of the DDR Pathway Signature in 545 Patients in the Pooled Validation Cohorts
| Characteristic | BCR-free Survival | Metastasis-free Survival | Overall Survival | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| HR (95% CI) | P Value | HR (95% CI) | P Value | HR (95% CI) | P Value | |
| UVA | 1.89 (1.44–2.48) | 5.01E-06 | 1.88 (1.33–2.67) | 3.91E-04 | 1.90 (1.32–2.72) | 5.15E-04 |
|
| ||||||
| MVA | ||||||
|
| ||||||
| DDR pathway signature (high risk vs low risk) | 1.71 (1.29–2.28) | <.001 | 1.56 (1.08–2.25) | .02 | 1.69 (1.16–2.47) | .006 |
|
| ||||||
| Age | 0.99 (0.97–1.01) | .21 | 0.99 (0.96–1.02) | .45 | 1.02 (0.99–1.05) | .20 |
|
| ||||||
| PSA | ||||||
|
| ||||||
| <10 (Low risk) | 1 [Reference] | 1 [Reference] | 1 [Reference] | |||
|
| ||||||
| 10–20 (Intermediate risk) | 1.47 (1.07–2.02) | .02 | 1.21 (0.80–1.84) | .37 | 0.87 (0.57–1.34) | .53 |
|
| ||||||
| >20 (High risk) | 1.83 (1.26–2.66) | .001 | 1.78 (1.09–2.91) | .02 | 1.31 (0.78–2.18) | .31 |
|
| ||||||
| Gleason score | ||||||
|
| ||||||
| 6 (Low risk) | 1 [Reference] | 1 [Reference] | 1 [Reference] | |||
|
| ||||||
| 7 (Intermediate risk) | 1.97 (0.98–3.94) | .06 | 8.84 (1.21–64.60) | .03 | 0.98 (0.47–2.07) | .96 |
|
| ||||||
| 8–10 (High risk) | 3.77 (1.85–7.70) | <.001 | 19.98 (2.71–147.35) | .003 | 2.06 (0.96–0.42) | .06 |
|
| ||||||
| SMS, + vs − | 1.13 (0.84–1.51) | .42 | 1.14 (0.79–1.64) | .49 | 1.30 (0.89–1.90) | .18 |
|
| ||||||
| SVI | 1.55 (1.16–2.09) | .003 | 1.46 (1.00–2.11) | .05 | 1.07 (0.73–1.57) | .72 |
|
| ||||||
| ECE | 1.44 (1.04–2.00) | .03 | 1.64 (1.04–2.57) | .03 | 1.54 (0.97–2.45) | .07 |
|
| ||||||
| LNI | 0.64 (0.38–1.07) | .09 | 1.07 (0.59–1.94) | .81 | 1.31 (0.68–2.54) | .42 |
Abbreviations: BCR, biochemical recurrence; DDR, DNA damage and repair; ECE, extracapsular extension; HR, hazard ratio; LNI, lymph node involvement; MVA, multivariate analysis; PSA, prostate-specific antigen; SMS, surgical margin status; SVI, seminal vesicle invasion; UVA, univariate analysis.
Older vs Younger Patients
A recent meta-analysis18 demonstrated that patients older than 70 years have decreased metastatic progression risk after radiotherapy for prostate cancer than younger patients, though a biological explanation remains unclear. We surmised that the lack of correlation between age and the DDR pathways (eTable 1 and eFigure 1C in the Supplement) indicated that the DDR pathways contain independent information and could shed light on this differential outcome by age. We analyzed the performance of the DDR pathway signature in patients younger than 70 years and 70 years and older in the pooled validation cohorts and found a difference (Table 2). The prognostic performance of the signature for metastasis-free survival appears to be stronger in the younger patients (HR, 1.67; 95% CI, 1.12–2.50) than in the older patients (HR, 0.77; 95% CI, 0.29–2.07) on multivariate Cox analysis. These results indicate that the DDR pathways play a different role in younger vs older patients, and that this may be responsible for the differential outcome after radiotherapy.
Table 2.
Multivariate Cox Regression Analysis of the DDR Pathway Signature and Metastasis-Free Survival in 545 Patients in the Pooled Validation Cohorts Stratified by Age
| Characteristica | Age | |||
|---|---|---|---|---|
|
| ||||
| <70 Years | ≥70 Years | |||
|
|
|
|||
| HR (95% CI) | P Value | HR (95% CI) | P Value | |
| DDR pathway signature, high vs low risk | 1.67 (1.12–2.50) | .01 | 0.77 (0.29–2.07) | .61 |
|
| ||||
| PSA | ||||
|
| ||||
| <10 (Low risk) | 1 [Reference] | 1 [Reference] | ||
|
| ||||
| 10–20 (Intermediate risk) | 1.20 (0.76–1.89) | .43 | 2.10 (0.46–9.48) | .34 |
|
| ||||
| >20 (High risk) | 1.65 (0.97–2.80) | .06 | 3.73 (0.89–15.65) | .07 |
|
| ||||
| Gleason score | ||||
|
| ||||
| 6 (Low risk) | 0.13 (0.02–0.94) | .04 | 0.00 (0.00-∞) | >.99 |
|
| ||||
| 7 (Intermediate risk)b | 1 [Reference] | 1 [Reference] | ||
|
| ||||
| 8–10 (High risk) | 2.17 (1.46–3.23) | <.001 | 2.27 (0.81–6.36) | .12 |
|
| ||||
| SMS, + vs − | 1.07 (0.73–1.59) | .72 | 2.16 (0.70–6.62) | .18 |
|
| ||||
| SVI | 1.34 (0.90–2.02) | .15 | 2.01 (0.71–5.67) | .19 |
|
| ||||
| ECE | 1.77 (1.06–2.95) | .03 | 1.32 (0.40–4.35) | .65 |
|
| ||||
| LNI | 1.35 (0.72–2.54) | .35 | 0.25 (0.03–1.86) | .18 |
Abbreviations: DDR, DNA damage and repair; ECE, extra-capsular extension; HR, hazard ratio; LNI, lymph node involvement; PSA, prostate specific antigen; SMS, surgical margin status; SVI, seminal vesicle invasion.
The absolute values of each characteristic can be found in eTable2 in the Supplement.
Gleason 7 used as reference owing to a small number of patients with Gleason 6 in the 70 years or older subset (n = 4).
Analysis by Ethnicity
Ethnicity data was collected only for 2 of the 4 cohorts (313 patients), limiting these analyses. In this subset, the proportions of DDR signature low-risk vs high-risk patients were not different by ethnicity (Fisher exact test, P = .31) (eTable 8 in the Supplement). Comparison of mean DDR signature score (raw values) by ethnicity was similarly limited and did not show a clear difference (eFigure 3 in the Supplement).
Discussion
The genomics era has afforded an unprecedented view into tumor biology and the promise of personalized medicine. Unbiased expression studies have been used to identify prognostic and/or predictive biomarkers in the form of individual genes or signatures.7–10 However, these analyses generally under use the vast amount of biological pathway knowledge that has been accumulated over decades of research. There have been several attempts at hybrid approaches to develop biomarkers using large-scale gene expression data with guidance from pathway knowledge in lung and ovarian cancer.11,12 This approach has not been pursued in prostate cancer, in which long disease outcome intervals dictate that few data sets with the most meaningful outcomes—metastatic progression and overall survival—are available.
We present a pathway profiling approach in a large prostate cancer cohort with clinical follow-up long enough to capture meaningful outcomes. Our approach is based on GSEA, a well-established bioinformatics tool, and offers a novel method to build a pathway-based biomarker of metastatic progression risk in prostate cancer. We focused on DDR pathways because of their central role in response to chemotherapy and radiotherapy and found that there are clusters of distinct DDR pathway patterns in this high-risk prostate cancer cohort. These DDR pathway patterns are better correlated with the molecular variables AR and ERG than clinical variables (Gleason, PSA, age), indicating the DDR pathways can provide additional information independent of the clinical variables. Indeed, many of the individual pathways, and aDDR pathway signature, have statistically significant prognostic association with both metastasis-free and overall survival. Importantly, we also confirmed that the DDR pathways are rarely affected by mutation. These findings demonstrate that DDR pathway profiling is a promising tool in the management of prostate cancer, and we have built a nomogram that would ease its use.
DNA damage and repair pathway profiling also yielded interesting tumor biology insights that may have profound translational impacts. AR and ERG have links to DDR pathways,19,20 including direct functional interaction as shown by our group and others.16,17,21 These in vitro findings are reflected in the positive correlation between many DDR pathways and the AR pathway signature or ERG, thus supporting the validity of our pathway profiling approach. Clinically, sensitivity to PARP-1 inhibitors has been linked to homologous recombination deficiency from BRCA1 and BRCA2 mutations, but there are many roads to “BRCAness,” including overexpression of the prostate long noncoding RNA PCAT-1.22 Homologous recombination molecular deficiencies may be difficult to comprehensively assay. Therefore, detecting homologous recombination deficiency directly with a pathway profiling approach is particularly desirable as PARP-1 inhibitors gain traction in prostate cancer with the promising results in the Phase II TOPARP trial.23 The 2 homologous recombination gene sets used in this study showed disparate results, which is not surprising based on the fact that they were designed to reflect different aspects of homologous recombination as indicated by their names and confirmed by reviewing the member genes (eTable 1 in the Supplement). Homologous recombination gene set N is a more comprehensive representation, while homologous recombination gene set M focuses on replication-independent homologous recombination. The stronger positive correlation between homologous recombination gene set M may thus imply that AR preferentially uses replication-independent homologous recombination. The interplay between these gene sets needs to be studied further to accurately model the complexity of the homologous recombination pathway for use in patients.
Our pathway profiling approach and our findings have several important similarities and differences with existing literature. Consistent with our findings, overexpression of DNA repair genes has been shown to associate with metastatic progression across several other cancer types.24 Intriguingly, we found that the DDR pathways appear to play a slightly disparate role in patients depending on age, though a larger cohort of patients 70 years or older would be required to fully explore and identify the difference. This is consistent with, and provides some explanation for, the findings of 2 studies18,25 totaling more than 8000 patients, which show that older patients with prostate cancer have better outcomes after radiotherapy than younger patients. While there are no concrete explanations for this finding, it may be reasonable to hypothesize that older patients have an increased competing risk of death which precludes disease progression, although Hamstra et al18 show a difference in prostate cancer–specific mortality, indicating that other mechanisms must be present. However, there are intriguing basic research findings that aging results not only in joint stiffness but also perhaps in stiffening at the cellular level that reduces metastatic potential.26 These results indicate that further work must be done to fully understand these age-related differences. Our approach differs from2 recent efforts— the recombination proficiency score and the DNA repair pathway focused score —in that we profiled across entire pathways, rather than focusing on representative genes. Hazard ratios of our DDR pathway signature for high metastatic progression risk and overall survival are larger than for either the recombination proficiency score or the DNA repair pathway focused score, 11,12 consistent with our hypothesis that there is prognostic information in the pathways en bloc.
While pathway profiling represents a novel approach, there are limitations. The exact meaning of pathway gene overexpression is ambiguous and is not a direct marker of pathway activity. Most notably, there may be a significant discrepancy between DDR gene expression and enzymatic activity, and unfortunately high-throughput analysis of enzymatic activity is not yet possible. There are also limitations of the underlying retrospective cohorts, which do not have uniform or randomized treatment or follow-up, though we attempt to account for these differences with cohort stratification and inclusion of clinical and pathologic variables in MVA. It would be reasonable to hypothesize that DDR pathway profiles might differ by ethnicity or between patients with and without a family history. Unfortunately, family history data was not collected, and, as noted in the results section, ethnicity data was collected only for 2 of the 4 cohorts. In this subset of patients, the proportions of DDR signature low-risk vs high-risk patients were not different by ethnicity, and comparison of mean DDR signature score (raw values) by ethnicity was similarly limited and did not show a clear difference. Thus, prognostic association by ethnicity was not possible with the limited numbers. However, within these limitations, the distinct pathway pattern clusters and the prognostic performance of our composite pathway biomarker are provocative.
Conclusions
Our novel patient-level DDR pathway profiling approach revealed distinct DDR pathway clusters and demonstrated that the DDR pathway profiles have prognostic association with metastatic progression and overall survival. Thus, DDR pathway profiling may be useful for identifying prostate cancer patients who have increased risk of metastatic progression after radical prostatectomy. As a biomarker of disease progression, DDR pathway profiling may be used to select patients for earlier treatment intensification with approaches such as adjuvant radiation or systemic therapy and may represent an avenue toward improved personalization of therapy for prostate cancer patients.
Supplementary Material
Highlights.
Question
Can 9 DNA damage and repair (DDR) pathways using 17 gene sets from prostatectomy samples identify better biomarkers for patients at the highest risk to guide therapy intensification?
Findings
We developed a novel patient-level Gene Set Enrichment Analysis–based pathway profiling approach and applied it to high-risk prostatectomy tumor samples from 1090 men using DDR pathways. The DDR pathway profiles correlate with AR and ERG levels and the AR pathway molecular variables and do not correlate with clinical variables including age, Gleason score, or prostate-specific antigen level. A DDR pathway signature trained in a cohort of 545 patients significantly associated with biochemical recurrence-free, metastasis-free, and overall survival in 545 additional patients pooled from 3 validation cohorts.
Meaning
DNA damage and repair pathway profiling may be a useful tool in the management of high-risk prostate cancer.
Acknowledgments
Funding/Support: This research was supported by the Prostate Cancer Foundation (Challenge Grants awarded to Dr Feng, Dr Knudsen, Dr Rubin, Dr de Bono, Dr Evans, and Dr Tomlins ) and the Evans Foundation.
Footnotes
Conflict of Interest Disclosures: Dr Davicioni is President, CSO, and Director of GenomeDx Biosciences Inc and holds patents or intellectual property with GenomeDx Biosciences Inc. Dr Chang is an employee of PFS Genomics, a joint venture with GenomeDx Biosciences. Drs Karnes, Feng, and Zhao have received funds for travel, accommodations, or expenses from GenomeDx Biosciences Inc. Dr Nguyen has worked as a consultant for GenomeDx Biosciences Inc. Drs Den and Klein have received funds for research from GenomeDx Biosciences Inc. All other authors declare no conflicts of interest.
Role of the Funder/Sponsor: The Prostate Cancer Foundation and the Evans Foundation had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Correction: This article was corrected online January 28, 2016, for an incorrectly expanded acronym in the abstract.
Supplemental content at jamaoncology.com
Author Contributions: Drs Evans and Zhao are coprimary authors of this study. Dr Feng had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Dr Evans and Dr Zhao contributed equally to this work.
Study concept and design: Evans, Zhao, Dicker, Rubin, de Bono, Knudsen, Feng.
Acquisition, analysis, or interpretation of data: Evans, Zhao, Chang, Tomlins, Erho, Sboner, Schiewer, Spratt, Kothari, Klein, Den, Dicker, Karnes, Yu, Nguyen, Davicioni, Feng.
Drafting of the manuscript: Evans, Zhao, Chang, Dicker, Rubin, Davicioni, Feng.
Critical revision of the manuscript for important intellectual content: Evans, Zhao, Tomlins, Erho, Sboner, Schiewer, Spratt, Kothari, Klein, Den, Dicker, Karnes, Yu, Nguyen, de Bono, Knudsen, Davicioni, Feng.
Statistical analysis: Evans, Zhao, Chang, Erho, Sboner, Yu, Davicioni.
Obtained funding: Tomlins, Knudsen, Davicioni, Feng.
Administrative, technical, or material support: Tomlins, Erho, Spratt, Kothari, Den, Dicker, Karnes, Yu, Davicioni.
Study supervision: Spratt, Dicker, Nguyen, de Bono, Feng.
References
- 1.Fossati N, Trinh QD, Sammon J, et al. Identifying optimal candidates for local treatment of the primary tumor among patients diagnosed with metastatic prostate cancer: a SEER-based study. Eur Urol. 2015;67(1):3–6. doi: 10.1016/j.eururo.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 2.Mahmood U, Levy LB, Nguyen PL, Lee AK, Kuban DA, Hoffman KE. Current clinical presentation and treatment of localized prostate cancer in the United States. J Urol. 2014;192(6):1650–1656. doi: 10.1016/j.juro.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albertsen PC, Hanley JA, Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. JAMA. 2005;293(17):2095–2101. doi: 10.1001/jama.293.17.2095. [DOI] [PubMed] [Google Scholar]
- 4.Lu-Yao GL, Albertsen PC, Moore DF, et al. Outcomes of localized prostate cancer following conservative management. JAMA. 2009;302(11):1202–1209. doi: 10.1001/jama.2009.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Den RB, Feng FY, Showalter TN, et al. Genomic prostate cancer classifier predicts biochemical failure and metastases in patients after postoperative radiation therapy. Int J Radiat Oncol Biol Phys. 2014;89(5):1038–1046. doi: 10.1016/j.ijrobp.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakagawa T, Kollmeyer TM, Morlan BW, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One. 2008;3(5):e2318. doi: 10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190(6):2047–2053. doi: 10.1016/j.juro.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubin MA, Zhou M, Dhanasekaran SM, et al. alpha-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287(13):1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 9.Koutalellis G, Stravodimos K, Avgeris M, et al. L-dopa decarboxylase (DDC) gene expression is related to outcome in patients with prostate cancer. BJU Int. 2012;110(6 Pt B):E267–E273. doi: 10.1111/j.1464-410X.2012.11152.x. [DOI] [PubMed] [Google Scholar]
- 10.Wehbi NK, Dugger AL, Bonner RB, Pitha JV, Hurst RE, Hemstreet GP., III Pan-cadherin as a high level phenotypic biomarker for prostate cancer. J Urol. 2002;167(5):2215–2221. [PubMed] [Google Scholar]
- 11.Pitroda SP, Pashtan IM, Logan HL, et al. DNA repair pathway gene expression score correlates with repair proficiency and tumor sensitivity to chemotherapy. Sci Transl Med. 2014;6(229):229ra42. doi: 10.1126/scitranslmed.3008291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang J, D’Andrea AD, Kozono D. A DNA repair pathway-focused score for prediction of outcomes in ovarian cancer treated with platinum-based chemotherapy. J Natl Cancer Inst. 2012;104(9):670–681. doi: 10.1093/jnci/djs177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prensner JR, Zhao S, Erho N, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15(13):1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin JF, Schiewer MJ, Dean JL, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov. 2013;3(11):1254–1271. doi: 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polkinghorn WR, Parker JS, Lee MX, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3(11):1245–1253. doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamstra DA, Bae K, Pilepich MV, et al. Older age predicts decreased metastasis and prostate cancer-specific death for men treated with radiation therapy: meta-analysis of radiation therapy oncology group trials. Int J Radiat Oncol Biol Phys. 2011;81(5):1293–1301. doi: 10.1016/j.ijrobp.2010.07.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karanika S, Karantanos T, Li L, Corn PG, Thompson TC. DNA damage response and prostate cancer: defects, regulation and therapeutic implications. Oncogene. 2015;34(22):2815–2822. doi: 10.1038/onc.2014.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ta HQ, Gioeli D. The convergence of DNA damage checkpoint pathways and androgen receptor signaling in prostate cancer. Endocr Relat Cancer. 2014;21(5):R395–R407. doi: 10.1530/ERC-14-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner JC, Ateeq B, Li Y, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19(5):664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prensner JR, Chen W, Iyer MK, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74(6):1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mateo J, Hall E, Sandhu S, et al. Antitumour activity of the PARP inhibitor olaparib in unselected sporadic castration-resistant prostate cancer (CRPC) in the TOPARP trial. [Accessed Nov 18, 2014];2014 https://www.webges.com/cslide/library/esmo/browse/search/ohR#9f9K03b9.
- 24.Sarasin A, Kauffmann A. Overexpression of DNA repair genes is associated with metastasis: a new hypothesis. Mutat Res. 2008;659(1–2):49–55. doi: 10.1016/j.mrrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Proust-Lima C, Taylor JM, Williams SG, et al. Determinants of change in prostate-specific antigen over time and its association with recurrence after external beam radiation therapy for prostate cancer in five large cohorts. Int J Radiat Oncol Biol Phys. 2008;72(3):782–791. doi: 10.1016/j.ijrobp.2008.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribeiro AJ, Khanna P, Sukumar A, Dong C, Dahl KN. Nuclear stiffening inhibits migration of invasive melanoma cells. Cell Mol Bioeng. 2014;7(4):544–551. doi: 10.1007/s12195-014-0358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.