Abstract
Listeria monocytogenes must resist the deleterious actions of bile in order to infect and subsequently colonize the human gastrointestinal tract. The molecular mechanisms used by the bacterium to resist bile and the influence of bile on pathogenesis are as yet largely unexplored. This study describes the analysis of three genes—bsh, pva, and btlB—previously annotated as bile-associated loci in the sequenced L. monocytogenes EGDe genome (lmo2067, lmo0446, and lmo0754, respectively). Analysis of deletion mutants revealed a role for all three genes in resisting the acute toxicity of bile and bile salts, particularly glycoconjugated bile salts at low pH. Mutants were unaffected in the other stress responses examined (acid, salt, and detergents). Bile hydrolysis assays demonstrate that L. monocytogenes possesses only one bile salt hydrolase gene, namely, bsh. Transcriptional analyses and activity assays revealed that, although it is regulated by both PrfA and σB, the latter appears to play the greater role in modulating bsh expression. In addition to being incapable of bile hydrolysis, a sigB mutant was shown to be exquisitely sensitive to bile salts. Furthermore, increased expression of sigB was detected under anaerobic conditions and during murine infection. A gene previously annotated as a possible penicillin V amidase (pva) or bile salt hydrolase was shown to be required for resistance to penicillin V but not penicillin G but did not demonstrate a role in bile hydrolysis. Finally, animal (murine) studies revealed an important role for both bsh and btlB in the intestinal persistence of L. monocytogenes.
Listeria monocytogenes is a gram-positive food-borne pathogen that is the etiological agent of listeriosis, an infection that is a significant cause of mortality due to food-borne disease (38). In common with all food-borne pathogens, the bacterium must be capable of sensing and responding to the many stressful conditions encountered in the hostile environment of the human gastrointestinal tract. In particular, bile represents a major challenge to bacteria that survive and transit the stomach and enter the small intestine.
Bile is a digestive secretion that plays an essential role in the emulsification and solubilization of lipids. It is primarily composed of bile acids (12% by weight), which are present as sodium salts at physiological pH. The primary bile acids cholic acid and chenodeoxycholic acid are synthesized de novo in the liver from cholesterol and are typically secreted in the form of amino acid conjugates with an amide bond between the bile acid carboxyl group and the amino group of either glycine or taurine (28). Their amphipathic nature promotes a detergent action on particles of dietary fat that emulsifies fat globules, making them available for digestion by lipases. The ability to act as a detergent also confers antimicrobial properties on bile salts, and they form part of the body's physicochemical defense system by degrading lipid-containing bacterial and viral membranes (27).
It is generally accepted that the autochthonous microbiota are inherently bile resistant, as are common gram-negative pathogens (43, 44, 54, 55). However, compared to other physiologically relevant stresses, such as the low-pH stress of the stomach or the elevated osmolality of the gastrointestinal tract (reviewed in references 11, 20, and 48), bile research is still in its infancy, particularly at the molecular level. Loci implicated in the bile tolerance of gram-negative pathogens include genes for lipopolysaccharide biosynthesis, porins, and efflux pumps (reviewed in reference 25). Bile has been shown to play a key role in the pathogenesis of these organisms, and the disruption of bile tolerance loci often leads to impaired intestinal survival and reduced colonization (8, 33, 42). The molecular mechanisms used by gram-positive pathogens to resist bile and the influence bile has on their pathogenesis is poorly understood. The ability of several gram-positive indigenous bacteria to transform and modify bile salts has been noted. Bile salt hydrolysis is an important modification; enzymes termed bile salt hydrolases (BSHs) catalyze the hydrolysis of the amide bond, liberating the glycine/taurine moiety from the side chain of the steroid core (10, 15, 24, 36). Specific members of the genera Eubacterium and Clostridium can 7α-dehydroxylate unconjugated bile acids into secondary bile acids (13, 17, 56). This multistep pathway involves several proteins, including a bile acid dehydratase enzyme that catalyzes a dehydration step. The ecological significance of such modifications is unknown; however, it has been suggested that they may contribute to bile tolerance (14, 15). We have recently reviewed the available literature on all aspects of the interaction between bacteria and bile (5).
It is estimated that 2 to 10% of healthy adults shed L. monocytogenes (47), and it has been isolated from the human gallbladder (1, 9), indicating an inherent ability to tolerate high concentrations of bile. In addition, in vivo bioluminescence imaging experiments have revealed that the gallbladder is a major site of extracellular replication for the bacterium during murine infection (26). We previously examined the bile tolerance of several L. monocytogenes strains and observed a heterogeneity of resistances (6). Since strain LO28 was extremely tolerant of high levels of bile (capable of growth in 30% bovine bile), this strain was chosen for all subsequent investigations. We recently implemented a transposon-based approach to uncover some of the genetic loci underlying tolerance; interestingly, all of the identified loci are predicted to play putative roles in the preservation of membrane integrity or in the maintenance of homeostasis under stress conditions (7; M. Begley, C. Hill, and C. G. M. Gahan, unpublished data). Given that Glaser et al. (23) have published the genome sequence of L. monocytogenes EGDe, we were now in a position to specifically target annotated genes with putative roles in bile salt modifications, i.e., lmo2067, lmo0446, and lmo0754. Computational analyses predict that the protein encoded by lmo2067 is a member of the choloylglycine hydrolase family, and it exhibits greatest homology to the BSHs of Enterococcus and Lactobacillus spp. Indeed, a recent study by Dussurget et al. (18) clearly demonstrated that this locus encodes a functional BSH that contributes to survival of strain EGDe in the intestinal tract. The product of the second gene, lmo0446, has been annotated as a BSH or a penicillin V amidase (PVA). It is also predicted to belong to the choloylglycine hydrolase family, and it exhibits significant homologies to BSHs. The predicted protein encoded by the third gene, lmo0754, is weakly homologous to bile acid dehydratases.
In the present study, we investigated the distribution of these three genes across different Listeria species and used a combination of single-, double-, and triple-gene knockouts to investigate their contribution to the high bile tolerance of L. monocytogenes LO28. We also assessed the role of these systems in contributing to listerial tolerance of other environmental stresses (acid, salt, detergents and antibiotics), as well as during infection of an animal (murine) model. Finally, we examine the regulation of these genes and extend the work of both Dussurget et al. (18) and Sue et al. (51) by demonstrating an important role for σB in the regulation of bsh expression under anaerobic conditions and in modulating overall listerial resistance to bile.
MATERIALS AND METHODS
Media, chemicals, and growth conditions.
Bacterial strains and plasmids used in the present study are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani (LB) medium. L. monocytogenes strains were grown in brain heart infusion (BHI) broth (Oxoid) shaking at 37°C (aerobic growth conditions). When necessary, anaerobic conditions were created by using BBL anaerobic jars and the Merck Anaerocult A Gas Pak system, and the absence of oxygen was verified by using Anaerotest strips (Merck). Chloramphenicol, erythromycin, and rifampin were made up as concentrated stocks and added to media at the required levels. Bile acids (sodium salts of glycodeoxycholic acid [GDCA] and taurodeoxycholic acid [TDCA]; Sigma G3258 and T0875, respectively), penicillin G (benzylpenicillin; Sigma PEN-NA), and penicillin V (phenoxymethylpenicillinic acid; Sigma P-1376) were solubilized in water, and filter-sterilized stocks were added to cooled autoclaved media. Bovine bile (oxgall; Sigma B3883) was added to broth prior to autoclaving.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 Δlac U169 (Φ80 lacZΔM15)R17 recA1 endA1 gyrA96 thi-1 relA1 | Gibco-BRL |
| EC1000 | Kmr; MC1000 derivative carrying a single copy of pWV01 repA in glgB | 46 |
| EC101 | E. coli JM101 with repA from pWV01 integrated in the chromosome | 35 |
| L. monocytogenes | ||
| LO28 | Serotype 1/2c | P. Cossart |
| Δbsh | LO28 derivative with bsh deleted | This study |
| Δpva | LO28 derivative with pva deleted | This study |
| ΔbtlB | LO28 derivative with btlB deleted | This study |
| Δbsh Δpva | LO28 derivative with bsh and pva deleted | This study |
| Δbsh Δpva ΔbtlB | LO28 derivative with bsh, pva, and btlB deleted | This study |
| Δbsh pPL2::bsh | Δbsh with bsh integrated at tRNAArg-attB′ | This study |
| Δpva pPL2::pva | Δpva with pva integrated at tRNAArg-attB′ | This study |
| ΔbtlB pPL2::btlB | ΔbtlB with btlB integrated at tRNAArg-attB′ | This study |
| pORI19::prfA | LO28 derivative with an insertion in prfA | This study |
| pORI19::sigB | LO28 derivative with an insertion in sigB | This study |
| EGDe | Serotype 1/2a | W. Goebel |
| EGDeΔpva | EGDe derivative with pva deleted | This study |
| EGDeΔbtlB | EGDe derivative with btlB deleted | This study |
| EGDe pORI19::bsh | EGDe derivative with an insertion in bsh | This study |
| EGDe pORI19::prfA | EGDe derivative with an insertion in prfA | This study |
| EGDe pORI19::sigB | EGDe derivative with an insertion in sigB | This study |
| Plasmids | ||
| pKSV7 | Cmr; temperature sensitive | 49 |
| pORI13 | Emr; promoterless lacZ, Ori+, RepA− derivative of pWV01 | 46 |
| pORI19 | Emr; Ori+, RepA− derivative of pORI28 | 35 |
| pVE6007 | Cmr; temperature-sensitive derivative of pWV01 | 37 |
| pPL2 | Cmr; integrates at the PSA phage attachment site within a tRNAArg gene on the chromosome | 34 |
Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Kmr, kanamycin resistant.
Genetic manipulations and sequence analyses.
Sequencing upstream of the groESL operon revealed the presence of a gene (designated bsh) with significant homology to cbh, a Lactobacillus plantarum BSH gene. Inverse PCR was used to obtain the full sequence of the gene. Genomic DNA was digested with AccI. After self-ligation, a PCR was performed with primers CBH1 and CBH2, and the resulting product was purified with the Qiaex II gel extraction kit (Qiagen, Hilden, Germany) and cloned into the pCR2.1 vector by using the TOPO TA cloning kit (Invitrogen, San Diego, Calif.). Plasmids were isolated with the Qiagen QIAprep Spin Miniprep Kit. Nucleotide sequence determination was performed on an ABI 373 automated sequencer. Sequencing upstream of gadA revealed the presence of a BSH-PVA homolog. The remainder of the gene (designated pva) was cloned and sequenced by using a single-specific-primer-PCR approach. Genomic DNA was digested with EcoRI, cloned into similarily digested pORI13, and electroporated into E. coli EC1000 to create a random bank. Plasmids were purified and used as a template for PCRs with primers P1 or P3, situated on either side of the pORI13 multiple cloning site, and PBH3 (a primer situated at the 5′ end of the pva gene). A P1-PBH3 amplicon, ∼1.3 kb in size, was cloned into the pCR2.1 vector and sequenced. The bile acid dehydratase homolog (designated btlB for bile tolerance locus [see below]) was sequenced in L. monocytogenes LO28 by using primers based on the corresponding gene in L. monocytogenes EGDe (lmo0754). Various web-based programs were used to analyze sequences (http://psort.nibb.ac.jp/, http://www.sbc.su.se/∼miklos/DAS/maindas.html, http://.cbs.dtu.dk/services/TMHMM/, http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml, and www.ncbi.nlm.nih.gov/BLAST). Restriction enzymes, T4 DNA ligase, and PCR reagents were purchased from Boehringer Mannheim, Mannheim, Germany, and used according to the manufacturer's recommendations. PCR was carried out by using a Hybaid (Middlesex, United Kingdom) PCR express system. Colony PCRs were performed after the lysis of cells with IGEPAL CA-630 (Sigma).
Construction of L. monocytogenes deletion mutants.
Isogenic nonpolar deletion mutants of bsh, pva, and btlB with 783-, 804-, and 534-bp deletions, respectively, were created by using plasmid constructs generated by the splicing by overlap extension (SOEing) PCR procedure described by Horton et al. (29). In each case primers SOE AB and SOE CD (Table 2) were used to amplify regions flanking the sequence to be deleted. The resulting products were gel extracted, mixed in a 1:1 ratio, and reamplified with the SOE A and D primers. The product was digested with the appropriate restriction enzymes, cloned into the temperature-sensitive shuttle vector pKSV7, and transformed into E. coli DH5α. The resulting plasmids were electroporated into L. monocytogenes, and transformants were selected on BHI plates containing 10 μg of chloramphenicol/ml (BHI/Cm). Chromosomal integration of the plasmids at 42°C was selected for by serial passage of a transformant in prewarmed BHI/Cm broth and streaking onto prewarmed BHI/Cm plates. Plasmid excision and curing was brought about by continuous passage in BHI at 30°C and spreading at intervals on BHI agar at the same temperature. Replica plating onto BHI and BHI/Cm was used to select for the appropriate deletion events. The mutations were confirmed by using primers upstream of SOE A and downstream of SOE D. Double and triple mutants were created by sequential deletions.
TABLE 2.
PCR primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| CBH1 | TGGCAGGACTGAATTTCTCAGG |
| CBH2 | GGTAGCATCGTAATACAACGGG |
| P1 | TGTGCTGCAAGGCGATTAAG |
| P3 | GGTTGATAATGAACTGTGC |
| PBH3 | GCGCTAATTGTGTTGCTTGT |
| BSH F | GATTACTGCAGAGAATCAAT |
| BSH R | GAAACAGAATTCAATTTGGTA |
| PVA F | CTACAGACGGTGCCAATTATA |
| PVA R | TGGTTTAGCCATATACTCTCC |
| BtlB F | ACTGGGCATGGCTTTATTG |
| BtlB R | CGATCAATGCCGGGAAGA |
| CBH SOEA | GTTCCGCTCTAGAAAATGCATCA |
| CBH SOEB | CATTAAAAATCCCTCCTCAA |
| CBH SOEC | TTGAGGAGGGATTTTTAATGTTGGTGGGGGAAAATACG |
| CBH SOED | TGCAATAAGTCGACCAATAATGG |
| PVA SOEA | CACATTGGATCCGAACCGTCA |
| PVA SOEB | CATAATTTATTCTCTCCTTCAAA |
| PVA SOEC | TTTGAAGGAGAATAAATTATGGGTGACCCTGATTTCACACAAT |
| PVA SOED | GATAAGGAATTCTTGGATCTACAG |
| BtlB SOEA | GGTGAAAAGCTTCGAATGGTTT |
| BtlB SOEB | CATGTAATCTCCTCCGTATT |
| BtlB SOEC | AATACGGAGGAGATTACATGCCGGAGCAAGTGACTCA |
| BtlB SOED | GTATCTAGATTTTTCGTTACTGT |
| PVA pPL2R | CAATTCTTGGATCTAGAGATTCC |
| BtlB pPL2F | CACTTTCTAGAGAACTTGGCAG |
| BtlB pPL2R | CGTTACTGCAGTCTCTTTTTTG |
| PL102 | TATCAGACCTAACCCAAACCTTCC |
| PrfA F | AATCTGCAGTCCATCAGGAGTTTCTTTAC |
| PrfA R | ACGGATCCATCATGAATTTACAATACTAC |
| SigB F | GGTGAATTCAAACGGTTTTTACGTG |
| SigB R | CGCTCATCTAGAACAGGGAGAAC |
| L142 | GAGTGCTTAATGCGTTAG |
| U141 | TTGCTCTTCCAATGTTAG |
| MONO7F | GGCTAATACCGAATGATGAA |
| LisR | AAGCAGTTACTCTTATCCT |
Overhangs complementary to the corresponding SOEB primers are underlined. Restriction enzyme sites are in boldface.
Complementation of deletion mutants.
The site-specific phage integration vector pPL2 (34) was used for complementation of deletion mutants. The entire bsh, pva, and btlB genes and flanking regions were amplified from a parental L. monocytogenes LO28 genomic DNA preparation by using Pwo DNA polymerase (Roche) and primers (CBH SOEA and CBH SOED, PVA SOEA and PVA pPL2R, and BtlB pPL2F and BtlB pPL2R, respectively; Table 2) that were modified to contain restriction sites. Gel-extracted PCR products were cut and ligated to similarly digested pPL2 (since the plasmid does not have an XbaI-cut site, it was digested with SpeI which generates an identical 5′ overhang). The resulting ligation mixes were electroporated into freshly competent E. coli DH5α, and transformants were selected on Luria-Bertani plates containing 15 μg of chloramphenicol/ml. Plasmids were extracted with the Qiagen QIAprep Spin Miniprep kit and sequenced with primers T3 and T7 to ensure the validity of inserts. Then, 50 μl of each plasmid preparation was ethanol precipitated and resuspended in 5 μl of elution buffer (10 mM Tris-Cl [pH 8.5]) and transformed into freshly competent mutant cells. Wild-type cells were transformed with pPL2 containing no insert to use as a control. Transformants were selected on BHI plates containing 7.5 μg of chloramphenicol/ml. Integration of constructs at the appropriate chromosomal site was confirmed by PCR with a primer that anneals 5′ of the integration site tRNAArg-attB′ (PL102) and primers that anneal to the cloned fragments. The additional primer sets BSH F and BSH R, PVA F and PVA R, and BtlB F and BtlB R were used to specifically detect bsh, pva, and btlB, respectively. Each of these primer sets does not amplify a product in the relevant deletion mutant, e.g., BSH F and BSH R do not amplify a product in Δbsh.
Construction of L. monocytogenes insertional mutants.
Plasmid pORI19 mutants were constructed as previously described (7, 35). Internal fragments of prfA and sigB were generated by PCR with primers PrfAF and PrfAR and SigBF and SigBR, respectively (Table 2), which were modified to contain restriction sites. The PCR products were cut and ligated to similarly digested pORI19. The resulting plasmids were transformed into E. coli EC101 and subsequently into L. monocytogenes harboring pVE6007. For each gene, one transformant was grown overnight in 10 ml of BHI broth prewarmed to 42°C (the nonpermissive temperature for pVE6007 replication in Listeria) and plated onto BHI containing 5 μg of erythromycin/ml. This selected for chromosomal integration at the point of homology with prfA and sigB. The mutations were confirmed by PCR analysis.
RNA procedures.
Cultures were inoculated from single colonies and grown overnight in BHI shaking at 37°C. The cultures were diluted 1:100 in BHI and grown either aerobically or anaerobically. Total RNA was extracted from exponential-phase (optical density at 600 nm of ca. 0.2) and stationary-phase (overnight) cultures by using the RNeasy kit (Qiagen). Samples were treated with the RNase-free DNase I set (Qiagen) according to the manufacturer's recommendations. The absence of DNA was confirmed by PCR. For reverse transcriptase PCR (RT-PCR) analysis, cDNA was synthesized from RNA by using Expand RT and a random primer p(dN)6 (Roche). Primers BSH F and R, PVA F and R, and BtlB F and R were used for PCR amplification of bsh, pva, and btlB cDNA, respectively. cDNA template was added to PCRs at levels which gave similar band intensities for control reactions performed with primers U141 and L142 (rrnA) (18). PCRs were carried out for 16, 22, 30, and 35 cycles to allow optimal quantitation of products. Densitometric analyses were performed by using Phoretix ID Advanced software. For ex vivo RNA experiments, 6-week-old BALB/c mice were orally infected with ca. 1010 L. monocytogenes LO28 cells that were grown overnight shaking in brain heart broth (Merck) at 37°C. An aliquot of the inoculum was flash frozen in an ethanol bath at −80°C. After 2 h mice were sacrificed and each entire intestine was removed and immediately placed in a sterile petri dish containing 1 ml of RNAprotect Bacteria Reagent (Qiagen). Intestinal contents were flushed out by using a sterile forceps and scissors and flash frozen. Approximately 5 min elapsed between the beginning of dissections and freezing. After thawing on ice, samples from three mice were pooled and centrifuged (10,000 rpm for 6 min), and pellets were resuspended in 500 μl of TE (10 mM Tris, 1 mM EDTA [pH 8.0]). In order to obtain higher RNA yields, total RNA was extracted by using the Macaloid method as outlined by Raya et al. (45). Briefly, the cell suspension was added to a 2-ml screw-cap microcentrifuge tube containing 0.5 g of 425- to 600-nm glass beads (Sigma), 50 μl of 10% sodium dodecyl sulfate (SDS), 500 μl of acid phenol-chloroform (5:1; Sigma), and 150 μl of a 2% Macaloid slurry (Bentone MA; Kronos), and cells were disrupted in a Mini-Beadbeater-8 (Biospec Products). After centrifugation at 13,200 rpm for 10 min, the aqueous supernatant was extracted with 500 μl of phenol chloroform, reextracted with 500 μl of chloroform-isoamyl alcohol (24:1; Sigma), precipitated with ethanol, and resuspended in 30 μl of TE. Mammalian RNA was removed with the MICROBEnrich kit (Ambion), and DNA was removed by using DNA-free (Ambion). RNA was extracted from the aliquot of the inoculum for comparison purposes. The RT reaction and ensuing PCR analyses were performed as described earlier except control PCRs were performed with L. monocytogenes-specific 16S RNA primers MONO7-F and LisR (50). For control purposes the entire procedure was also performed with intestines from noninfected mice.
Growth experiments.
For bile broth assays, overnight cultures were inoculated (3%) into BHI broth containing various concentrations of bile or bile acids. Cell growth was measured by performing viable cell counts by diluting cultures in one-quarter-strength Ringer solution and enumeration on BHI. To examine the pH dependency of bile salt toxicity, the effect of 5 mM GDCA and TDCA on cultures was investigated in BHI broth set at initial pH values ranging from 4.5 to 7.5 with 3 M lactic acid. Broth was inoculated (2%) with cells from an overnight culture and incubated anaerobically. Viable cell counts were performed at intervals. The pH was not controlled during the experiment. For all other growth experiments (acid, salt, and penicillin), overnight cultures were inoculated (2%) into BHI supplemented with the relevant stressor, and cell growth was measured spectrophotometrically over 24-h periods by using a Spectra Max 340 spectrophotometer (Molecular Devices, Sunnyvale, Calif.).
BSH plate assays.
The plate assay developed by Dashkevicz and Feighner (12) was used to examine bile hydrolase activity. Bacteria were grown on BHI agar and subsequently patch inoculated onto BHI or Man Rogosa and Sharpe (MRS) agar (Merck) supplemented with 0.5% (wt/vol) GDCA or TDCA. Plates were incubated anaerobically at 37°C. Strains with BSH activity were surrounded with white precipitate halos of unconjugated bile acids.
Virulence assays.
Spontaneous rifampin-resistant (Rifr) mutants of wild-type and bsh, pva, and btlB mutant strains were isolated by centrifuging 10 ml of overnight cultures, resuspending the cell pellets in 200 μl of phosphate-buffered saline (PBS) and plating them onto BHI supplemented with 50 μg of rifampin/ml (BHI/RIF). For each strain, four BALB/c mice were infected with ∼1010 cells by oral gavage. Excretion of viable Listeria was measured in fecal samples collected for 3 days postinfection. Samples were weighed, homogenized in PBS, diluted, and plated onto BHI/RIF. Bacterial numbers in the spleens were determined on the third day by spread plating homogenized organs onto BHI.
RESULTS
Identification and analysis of genes encoding putative bile-degrading enzymes.
As part of another study undertaken in our laboratory, a region upstream of groESL was sequenced in L. monocytogenes LO28 and revealed the presence of an open reading frame designated bsh (lmo2067 in L. monocytogenes EGDe) (21). Further analysis revealed a putative ribosomal binding site (AGGAG-14 nucleotides [nt]-ATG), which in turn is preceded by a consensus σB-dependent promoter binding site (GTTTTAN13GGGTAC-66 nt-ATG) and a putative PrfA box (TTAAAAATTTTTAA-147 nt-ATG). A 27-nt symmetric sequence followed by a stretch of T residues was identified 9 bp downstream of the stop codon and may function as a Rho-independent transcription termination signal. Further sequence analyses, using computer-aided approaches, suggest that bsh encodes an intracellular enzyme (designated BSH) that is a member of the choloylglycine hydrolase family (PSORT, Pfam02275, COG3049, ProDom009912). Searches for similarity and identity reveal that BSH shows most homology to BSHs of Enterococcus faecium, Enterococcus faecalis, and Lactobacillus plantarum (74, 70, and 66% identities, respectively). It also exhibits lower, yet significant, homology to the Bacillus sphaericus PVA (30% identity). This is in agreement with the previous annotation of EGDe lmo2067 as a BSH (23) and is consistent with the bile hydrolase activity ascribed previously to this locus (18).
A second gene, also annotated as a BSH, was designated pva (lmo0446 in L. monocytogenes EGDe). This gene encodes a putative choloylglycine hydrolase family enzyme, PVA (COG3049, Pfam02275, ProDom009912), which also shows homology to the B. sphaericus PVA (37% identity). The presence of an N-terminal sequence signal suggests an extracellular location (PSORT). Recently, members of the choloylglycine hydrolase enzyme family have been classified as N-terminal nucleophilic (Ntn) hydrolases, and the crystal structure of B. sphaericus PVA has been elucidated (52). The amino acids of the active site were identified as Cys-1 (thought to play an essential role in catalysis), Asp-20, Tyr-82, Asn-175, and Arg-228. Significantly, a CLUSTAL alignment of the L. monocytogenes BSH and PVA amino acid sequences with PVA of B. sphaericus showed that all five amino acids are conserved (not shown). No potential PrfA or σB consensus promoters were identified upstream of this locus.
A third locus designated btlB (for bile tolerance locus [see below]; lmo0754 in L. monocytogenes EGDe) was amplified and subsequently sequenced in L. monocytogenes LO28. A putative PrfA box (TTAAAAAGTGTTAC) was identified 365 bp upstream of the predicted start codon. This locus shows no significant homology to genes in the public databases and was annotated as being weakly homologous to bile acid dehydratase genes (baiE) (23). To date, these genes have only been identified and characterized in specific members of the genera Clostridium and Eubacterium. In both, the baiE genes are part of large operons with other genes involved in bile dehydroxylation (17), several regions upstream of the promoters are highly conserved (56), and the bile acid dehydratase enzymes possess lipocalin motifs that are thought to function as bile acid-binding sites (13). However, homologs of the remaining genes in the bile dehydroxylation pathway could not be found in the L. monocytogenes genome. In addition, neither the highly conserved promoter region nor the lipocalin motif could be located. Thus, although detailed computational analyses provided no further insight into a possible function for the encoded protein, it was one of only three loci identified by the European Listeria Genome Consortium as being possibly involved in bile responses and, as such, was deemed appropriate to be included in our investigation.
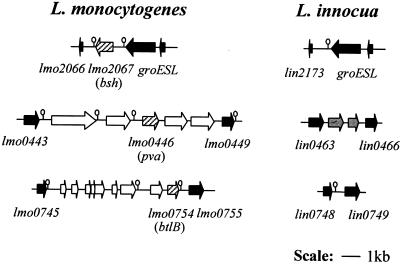
Comparison of the genomes of L. monocytogenes EGDe and the nonpathogenic strain L. innocua Clip 11262 revealed that all three genes (bsh, pva, and btlB) are specific to L. monocytogenes (23). Further analyses showed that the pva and btlB genes are part of islets present in L. monocytogenes but absent from L. innocua (See Fig. 1). The areas surrounding the bsh gene and pva and bltB gene islets are identical in both species; all upstream and downstream genes are orthologues of identical size. The “pva islet” consists of five genes (lmo0444 to lmo0448) and the “btlB islet” consists of nine as-yet-uncharacterized genes (lmo0746 to lmo07554). Notably, the bsh gene and pva and btlB islets have lower G+C contents than the flanking regions (36%, averages of 37 and 34%, respectively, as opposed to 39%), suggesting their recent acquisition through horizontal gene transfer from low G+C content bacteria.
FIG. 1.
Genomic organization of bsh, pva, and btlB regions. This figure was drawn approximately to scale by using L. monocytogenes EGDe and L. innocua Clip 11262 genome sequence data. The numbers are National Center for Biotechnology Information annotation numbers. Arrows indicate orientation of genes. Hairpins depict putative terminators. Black open reading frames are orthologs. Open reading frames present in L. monocytogenes but absent from L. innocua are in white. Those present in L. innocua but not L. monocytogenes are in gray. Hatched arrows represent the three genes examined in the present study.
Genetic distribution of bsh, pva, and btlB among Listeria species.
The observation that all three genes are absent from the sequenced L. innocua strain led us to examine, by PCR, the distribution of the genes among the six Listeria species, of which monocytogenes, ivanovii, and seeligeri are potentially pathogenic. In all cases, at least two sets of primers were used for each gene. In addition, negative results (i.e., the absence of bsh or of pva and btlB islets) were verified by amplifying products of expected sizes by using primers based upstream and downstream of bsh and of pva and btlB islets. bsh products were amplified in all L. monocytogenes, L. ivanovii, and L. seeligeri strains tested but not in strains of L. innocua, L. grayi, or L. welshimeri (Table 3). Primers based on pva generated products of the predicted size for 22 of the 45 L. monocytogenes strains tested (Table 3). Of the remaining strains, pva products were only obtained for four L. welshimeri strains. Finally, btlB primers generated products in all L. monocytogenes strains, two L. seeligeri strains, and one L. ivanovii strain (Table 3).
TABLE 3.
Genetic distribution of bsh, pva, and btlB genes among Listeria speciesa
| Listeria strains (n) | No. of strains with:
|
||
|---|---|---|---|
| bsh | pva | btlB | |
| L. monocytogenes (42) | 42 | 23 | 42 |
| L. innocua (5) | 0 | 0 | 0 |
| L. seeligeri (6) | 6 | 0 | 2 |
| L. ivanovii (5) | 5 | 0 | 1 |
| L. welshimeri (8) | 0 | 4 | 0 |
| L. grayi (3) | 0 | 0 | 0 |
The number of strains tested (n) is indicated. In each case, at least two sets of primers were used. Negative results were verified by amplifying PCR products of the expected size by using primers based upstream and downstream of bsh and of the pva and btlB islets.
Bile tolerance of bsh, pva, and btlB mutants.
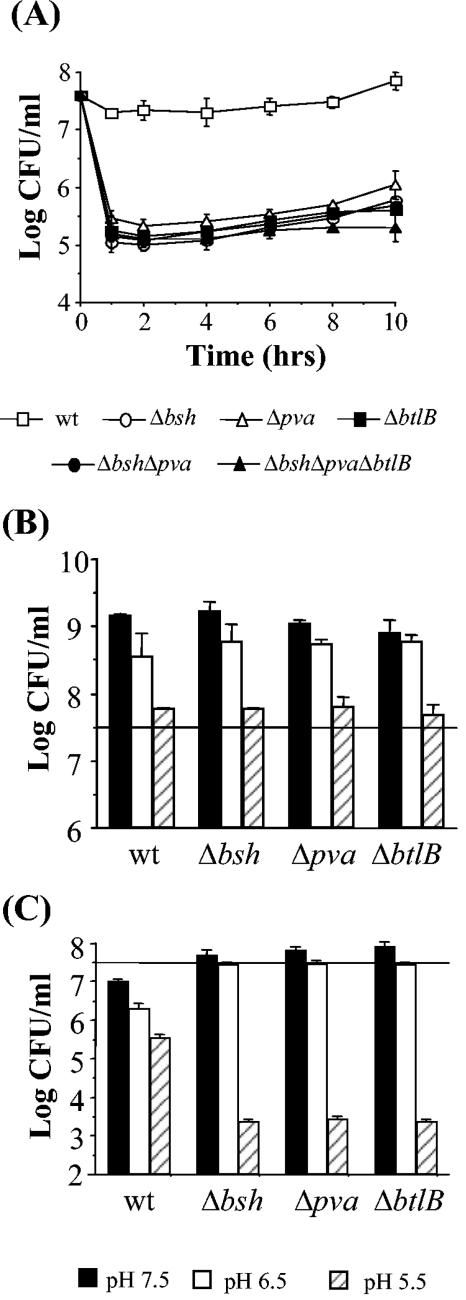
The splicing by overlap extension (SOEing) procedure was used to create appropriate plasmid constructs to generate isogenic in-frame deletion mutants of bsh, pva, and btlB in L. monocytogenes LO28. Double and triple mutants were created by sequential deletions. The bile tolerance of the resulting mutants was assessed by determining their survival in 30% oxgall (bovine bile). As expected from previous results (6), on initial exposure to the bile there was almost a half-log-unit reduction in numbers of the parent (Fig. 2A). However, Δbsh, Δpva, and ΔbtlB mutants are all significantly more sensitive (P < 0.05 as determined by the Student t test), demonstrating ∼2.5-log-unit reductions in numbers (Fig. 2A), indicating that all three genes play an important role in tolerating the acute toxicity of these high concentrations of bile. Interestingly, a subpopulation of each mutant survived the exposure to bile; however, when these survivors were recultured and reexposed to bile, they were not more resistant than the original deletion mutant cultures (data not shown). The integration vector pPL2 (34) was used to complement mutant strains by inserting the relevant wild-type gene as a single copy at the PSA bacteriophage attachment site. The bile tolerance of all complemented strains was comparable to the wild type, confirming that the bile-sensitive phenotype of Δbsh, Δpva, and ΔbtlB deletion mutants resulted from direct mutation of the loci in question (data not shown). Interestingly, multiple deletion of bsh, pva, or btlB did not result in an additive effect since double (Δbsh Δpva) and triple (Δbsh Δpva ΔbtlB) mutants were only as bile sensitive as single mutants (Fig. 2A). The type of bile used (oxgall) is a crude extract obtained from bovine gallbladders. Though it is composed of a mix of individual bile acids, oxgall's exact composition is unknown (Sigma technical support). In vivo bile acids are typically conjugated to either glycine (glycoconjugated) or taurine (tauroconjugated) before secretion into the duodenum (28). Survival of parent and mutants was therefore compared in BHI broth supplemented with a 5 mM concentration of a typical glycoconjugated bile acid (GDCA) and 5 mM concentration of a typical tauroconjugated bile acid (TDCA). The 5 mM concentration was chosen because this is considered the average concentration prevailing in the intestinal tract (14), and since a wide range of pHs may be encountered in vivo in the lumen the broth was adjusted to various pHs with lactic acid. Figure 2B and C show viable plate counts after 16 h of anaerobic incubation. As we have previously observed, TDCA did not prevent growth of the parent strain at any pH tested (6). Growth of the mutants was also not affected compared to the parent (Fig. 2B). Again, in agreement with our previous observations (6), GDCA inhibited the growth of wild-type cells (Fig. 2C). The toxicity of this bile acid increased at low pH, and at pH 5.5 a 2-log-unit reduction in numbers was observed. All mutants were significantly more sensitive at this pH (P < 0.05, as determined by the Student t test), each demonstrating ∼3.5-log-unit reductions in numbers. This observation reinforces the suggestion that all three genes play a role in tolerating the acute toxicity of bile. Again, double and triple mutants were only as sensitive as single mutants (data not shown). It was observed that mutant numbers were significantly higher than the parent in GDCA-supplemented broth at pH values above pH 5.5 (Fig. 2C). Similar results were observed when strains were patched onto a solid medium (BHI adjusted to the appropriate pH with lactic acid and supplemented with GDCA).
FIG. 2.
Role of L. monocytogenes LO28 bsh, pva, and btlB genes in bile tolerance. Overnight cultures were inoculated into BHI supplemented with 30% oxgall (A) or BHI adjusted to various pHs and supplemented with either 5 mM TDCA (B) or 5 mM GDCA (C). Viable plate counts were performed at intervals (A) or after 16 h (B and C) by serial dilution in Ringer solution and enumeration on BHI agar. The horizontal lines in panels B and C represent the initial inoculum. The error bars represent the standard deviations of triplicate experiments.
General physiological analyses of mutants.
All mutants (Δbsh, Δpva, ΔbtlB, Δbsh Δpva, and Δbsh Δpva ΔbtlB) displayed growth rates (doubling times) similar to that of the wild-type when grown in BHI (pH 7) at 37°C, indicating that the disrupted genes were not necessary for growth under normal physiological conditions (data not shown). In addition, no obvious differences in cell or colony morphologies were observed between wild-type and mutant strains. The growth rate of the individual mutants was also comparable to that of the wild type under acidic conditions (BHI adjusted to pH 5.5 with 3 M lactic acid) and salt stress (BHI supplemented with 7% NaCl) (data not shown), confirming that the decreased survival observed in bile-supplemented broth was actually due to the presence of bile and did not result from associated pH or osmolarity effects.
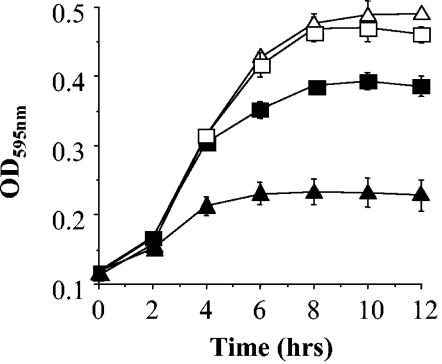
Disk diffusion assays were used to examine antibiotic resistances. Mutants exhibited resistance identical to that of the parent strain against all antibiotics tested (results not shown). Since pva was annotated as a putative PVA, the growth of Δpva was compared to the wild-type in BHI broth supplemented with various concentrations (ranging from 25 to 125 ng/ml) of penicillin G or V. It was observed that the mutant was significantly more sensitive to all concentrations of penicillin V examined but behaved identically to the parent with all levels of penicillin G tested. Figure 3 shows representative growth curves obtained. The ΔbtlB and Δbsh mutants were not affected in penicillin V or penicillin G resistance (data not shown).
FIG. 3.
Growth of L. monocytogenes LO28 wild-type (squares) and pva mutant (triangles) in BHI broth supplemented with 50 ng of penicillin G (open symbols) or 50 ng of penicillin V (closed symbols)/ml. Cell growth was measured spectrophotometrically by the determining optical density at 595 nm (OD595). The error bars represent the standard deviations of triplicate experiments.
Since bile acids function as biological detergents in vivo, the general detergent resistance of mutants was compared to the parent as outlined by De Smet et al. (15). Briefly, viable cell counts were determined after a 5-min exposure of strains to equivalent concentrations (critical micellar concentration plus 10% [wt/vol]) of four biological detergents: sodium dodecyl sulfate (anionic), Triton X-100 (nonionic), CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} (zwitterionic), and cetylpyridinium bromide (cationic). Mutants did not show any enhanced sensitivity (data not shown), implying that the three genes do not form part of a general response to detergent stress.
The growth of mutants were also compared to the parent under combined stress conditions (BHI broth adjusted to various pHs with lactic acid and supplemented with either various levels of salt or sodium dodecyl sulfate). Mutants behaved similarly to parent cells under all conditions examined (data not shown).
BSH activity of wild-type and bsh, pva, and btlB mutants.
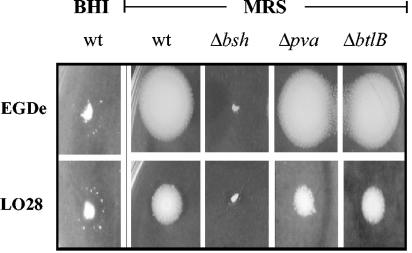
The agar plate assay developed by Dashkevicz and Feighner (12) was used to investigate BSH activity of the wild-type and mutants. In this assay, bacteria that possess BSH enzymes hydrolyze conjugated bile acids in the agar to the unconjugated form that precipitates at low pH, giving rise to a halo surrounding the colony. Patch inoculation of wild-type L. monocytogenes LO28 or EGDe (the strain used in the European Listeria genome sequencing project) onto BHI supplemented with a glycoconjugated bile acid GDCA resulted in the formation of identical translucent halos with small irregular precipitates, suggesting that both strains have similar BSH activity. Since it proved difficult to photographically document this phenomenon (Fig. 4), other media were investigated. Patch inoculation of the strains onto MRS supplemented with the same concentration of GDCA resulted in much more pronounced white precipitates; however, it was observed that the LO28 halo was smaller than that of EGDe. The reason for this difference on MRS is unclear, but it does not appear to be a consequence of the bile tolerance of each strain since LO28 is significantly more tolerant of bile than EGDe. In addition, halo sizes observed for eight L. monocytogenes strains randomly chosen from our laboratory culture collection did not correlate with their relative bile tolerance profiles (data not shown). Nevertheless, to confirm and accentuate results obtained for strain LO28, all genes in the present study were also mutated in strain EGDe, and these mutants were used solely for BSH plate assays. It was observed that only bsh mutants were incapable of GDCA hydrolysis (Fig. 4). However, since it was possible that pva could encode another BSH enzyme that specifically hydrolyzes tauroconjugated bile salts (similar to the BSH of Bacteroides vulgatus [31]), the agar plate assay was repeated with a tauroconjugated bile acid TDCA. It was again observed that only bsh mutants were incapable of hydrolysis (not shown), indicating that bsh encodes an enzyme that can deconjugate both glycoconjugated and tauroconjugated bile salts. Although PVA has been annotated as a BSH, loss of the pva gene does not affect hydrolysis of either the glyco- or tauroconjugated bile salts tested, demonstrating unambiguously that L. monocytogenes BSH activity results solely from the bsh gene. Deletion of btlB did not affect bile salt hydrolysis.
FIG. 4.
BSH activity of L. monocytogenes LO28 and EGDe bsh, pva, and btlB mutants. Strains were patch inoculated onto BHI or MRS agar supplemented with 0.5% (wt/vol) GDCA and incubated anaerobically for 48 h. Strains with BSH activity are surrounded with white precipitate halos of unconjugated bile acids.
Expression and regulation of bsh, pva, and btlB genes.
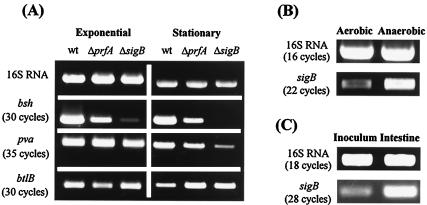
Expression of bsh, pva, and btlB was examined by using RT-PCR. cDNA was synthesized from RNA isolated from exponential- and stationary-phase cells grown anaerobically in BHI broth at 37°C. Control PCRs performed with rrn primers (16S RNA primers) confirmed that DNA concentrations were comparable in all samples (Fig. 5A). PCRs were then carried out with primers specific for bsh, pva, and btlB genes. All reactions were performed in triplicate and repeated at least once. It was observed that all three genes were expressed in wild-type cells during both exponential and stationary phases. Figure 5A shows the products obtained after 30 (bsh and btlB) or 35 cycles (pva). The intense band obtained for bsh suggests very high levels of transcription under the conditions examined. In contrast, a product was not amplified for pva until 35 cycles of PCR, indicating relatively low levels of transcription.
FIG. 5.
(A) Analysis of expression and regulation of bsh, pva, and btlB genes by using RT-PCR. cDNA was synthesized from RNA isolated from L. monocytogenes LO28 wild-type and prfA and sigB mutant cells grown anaerobically to exponential and stationary phases in BHI broth at 37°C. (B) Expression of sigB in cells grown to exponential phase either aerobically or anaerobically in BHI broth at 37°C. (C) Expression of sigB during murine infection. cDNA was synthesized from RNA that was extracted from intestines 2 h after oral infection with 1010 L. monocytogenes LO28 cells. In all cases, control PCRs were performed with 16S RNA primers to confirm identical template DNA levels. The number of PCR cycles are shown in parentheses.
Sequence analysis revealed that the bsh gene is preceded by a consensus binding site for PrfA, the global regulator of virulence gene expression in Listeria spp. Indeed, Dussurget et al. (18) demonstrated a role for PrfA in transcription of the L. monocytogenes EGDe bsh gene during exponential growth. A consensus PrfA box was also identified upstream of the btlB gene, prompting us to determine the effect of inactivating prfA on expression of all three genes. We determined that bsh transcription was reduced in an LO28 prfA mutant during exponential phase (∼1.8-fold) (Fig. 5A); however, this reduction did not appear to be as dramatic as that reported by Dussurget et al. (18) for strain EGDe. The differences between studies may be a result of variances in experimental conditions (in our study cultures were grown anaerobically, whereas Dussurget et al. grew cultures microaerophilically) or differences between strains LO28 and EGDe. Levels of bsh transcript were also lower in the prfA mutant in stationary phase (∼2-fold), suggesting that PrfA regulates transcription of this gene during both stages of growth. Transcription of btlB was reduced in a prfA mutant in exponential phase (∼1.5-fold), whereas stationary-phase expression of btlB was moderately increased in the prfA mutant (Fig. 5A). Transcription of pva was not affected by mutating prfA.
Given the role of σB in regulation of bsh (51), we examined expression of the three genes in a sigB mutant. Although pva is not preceded by a σB consensus sequence, transcription of the gene was reduced significantly (∼4-fold) in stationary-phase cultures of the sigB mutant. It is possible that the σB binding site for pva differs from that of known listerial σB-regulated genes or, alternatively, that pva transcription is not controlled directly by σB but rather indirectly by a member of its regulon. In the present study, btlB transcription was not reduced by inactivating sigB but was marginally increased (∼1.2-fold) in stationary phase relative to the wild type (Fig. 5A). Transcription of bsh was markedly reduced during exponential phase (∼7.5-fold) and eliminated during stationary phase. Although roles for both PrfA (18) and σB (51) in the regulation of bsh have been demonstrated previously, our data suggest a greater role for σB in the regulation of this locus under anaerobic conditions that mimic the intestinal environment. This evidence is supported by bile hydrolysis (Fig. 6A) and bile resistance (Fig. 6B) assays outlined below.
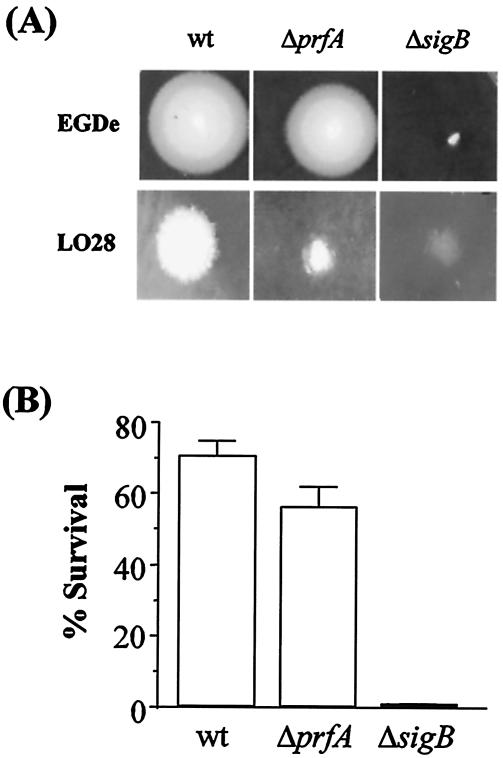
FIG. 6.
(A) BSH activity of prfA and sigB mutants. Strains were patched onto MRS agar supplemented with 0.5% (wt/vol) GDCA and then incubated anaerobically for 48 h. White precipitate halos indicate BSH activity. (B) Bile tolerance of the wild type and of prfA and sigB mutants. Overnight cultures were inoculated into BHI (pH 6) supplemented with 5 mM GDCA and incubated anaerobically at 37°C. Viable plate counts were performed after 4 h. The error bars represent the standard deviations of triplicate experiments.
Expression of sigB under anaerobic conditions and during murine infection.
The role of σB in the regulation of bsh in exponential phase was unexpected, since sigB is generally not expressed to high levels during exponential phase (3). Further RT-PCR analyses revealed that sigB transcript levels were increased in exponential phase by the anaerobic conditions used in our experiments (Fig. 5B). To our knowledge, this is the first report of sigB induction by anaerobiosis. Although it has been shown that σB regulates loci that contribute to the survival of L. monocytogenes in the intestine (30, 51), expression of sigB during infection has not been investigated to date. To address this, we examined sigB expression in ex vivo L. monocytogenes cells isolated directly from the intestine of orally infected mice. The data clearly indicate that sigB is upregulated within the intestine during the early stages of colonization (Fig. 5C). PCR products were not amplified with any of the primer pairs (16S or sigB) when the entire experiment was repeated on RNA extracted from the intestines of noninfected mice, confirming the validity of our observations.
BSH activity and bile tolerance of prfA and sigB mutants.
Since PrfA and σB were shown to regulate transcription of bsh, the affect of inactivating either of these genes on BSH activity was investigated by using the agar plate assay. Although halo size of both LO28 and EGDe prfA mutants were measurably smaller than those of the corresponding wild-type on agar supplemented with either GDCA (Fig. 6A) or TDCA (not shown), halos were not observed for the sigB mutants (Fig. 6A). This is consistent with transcriptional studies and confirms that σB plays a greater role in bsh regulation than does PrfA under the conditions tested. The bile tolerances of the prfA and sigB mutants were assessed by determining their survival in broth adjusted to pH 6 and supplemented with 5 mM GDCA. Figure 6B shows that the prfA mutant was significantly more sensitive to bile than the parent (56% versus 71% survival). However, mutating sigB was more detrimental to cells and survival of the sigB mutant was over 2-log-units lower than the parent (0.65% versus 71%). The growth of mutants was not affected in broth at pH 6 without bile (data not shown), confirming that the observed affects were due to exposure to bile and not pH effects.
Investigation of the role of bsh, pva, and btlB in gastrointestinal persistence.
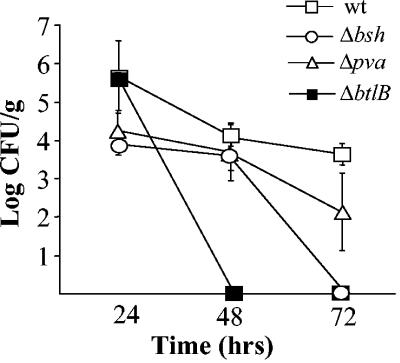
The persistence of wild-type and mutants in the murine intestinal tract was assessed by measuring excreted bacteria in fecal samples for 3 days after oral inoculation. In order to detect Listeria from the background flora, all strains were marked with rifampin. These rifampin-resistant (Rifr) strains behaved in a manner identical to that of the non-drug-marked strains under all conditions examined (growth in BHI broth alone, growth in BHI supplemented with various levels of salt or adjusted to various pHs with lactic acid, and survival in BHI supplemented with 30% oxgall; data not shown). The virulence potential of the Rifr parent strain was comparable to that of the non-drug-marked parent (data not shown). Figure 7 shows that numbers of the wild-type were reduced by 1-log-unit over the 72 h of the experiment. Although numbers of Δpva were slightly lower, differences were not statistically significant (Student t test). In contrast, ΔbtlB and Δbsh cells were not detected after 48 and 72 h, respectively. This represents 4-log-unit reductions in numbers of each strain demonstrating that BSH and BtlB play a significant role in the persistence of L. monocytogenes LO28 in the murine gastrointestinal tract. Significantly, bacterial numbers in the spleens of inoculated mice after 72 h mirrored these observations in that Δbsh and ΔbtlB were not recovered from spleens, whereas the wild-type and Δpva were isolated at levels of 2.7 × 103 and 3.5 × 103 CFU/organ, respectively.
FIG. 7.
Role of bsh, pva, and btlB in persistence in the murine gastrointestinal tract. Excretion of rifampin-marked strains was measured in fecal samples collected 24, 48, and 72 h after oral infection of BALB/c mice. The error bars represent the standard deviations of triplicate experiments.
DISCUSSION
The publication of the genome sequences of both the pathogenic strain L. monocytogenes EGDe and the nonpathogenic strain L. innocua Clip 11262 (23) represented a major step toward the identification and characterization of specific genetic loci that contribute to survival of L. monocytogenes in the human gastrointestinal tract. Recent work has demonstrated that extraluminal growth of L. monocytogenes occurs in the gallbladders of listeria-infected mice and suggests that mechanisms to survive bile salts significantly contribute to the ability of the bacterium to cause infection (26). Only three loci, namely, bsh, pva, and btlB, were described by the European Listeria Genome Consortium as being possibly involved in bile responses, and these loci are absent from L. innocua (23). Of these, bsh was previously analyzed by Dussurget et al. (18). In the present study we extend their observations and investigate all three genes for possible roles in bile tolerance and persistence of L. monocytogenes in the murine gastrointestinal tract. Furthermore, we analyze the role for the alternative sigma factor σB in the physiological resistance of bile and examine sigB expression under anaerobic conditions in vitro and during murine infection.
Previous examination of bsh in L. monocytogenes EGDe demonstrated that it encodes a functional BSH enzyme and is involved in bacterial persistence in the intestinal tract (18). In addition to corroborating these findings in strain LO28, we examined its exact role in bile tolerance and further explored the regulation of its expression. Although several BSH enzymes hydrolyze only either glyco- or tauroconjugated bile salts (2, 16, 22, 31), we demonstrate here that the L. monocytogenes enzyme can hydrolyze both. The ability of the enzyme to deconjugate different moieties of bile salts undoubtedly increases its effectiveness during infection of the gastrointestinal tract. We show that BSH is essential for resisting the toxicity of glycoconjugates at low pH. Interestingly, deletion of bsh did not affect general detergent resistance or resistance to other physiologically relevant stresses such as low pH or elevated osmolarity, implying that the enzyme is not part of a general detergent shock response or, indeed, a general stress response. In contrast, of 12 bile tolerance loci recently identified by using a random mutagenesis approach, 11 were found to play a role in resistance to other environmental stresses (6, 7; Begley et al., unpublished). Overall, the results indicate that unlike many other bile tolerance loci, BSH plays a highly specific role in resisting the acute bactericidal shock exerted by bile salts.
As demonstrated previously by Dussurget et al. (18) and corroborated here, the enzyme plays a key role in the ability of L. monocytogenes to colonize the intestinal tract. We have determined that the gene is found in Listeria species that are potentially pathogenic but absent from nonpathogenic species, suggesting that bsh may not be required outside the host and is only important during infection or gut colonization. It is likely that BSH enzymes of the indigenous intestinal microflora also contribute to bile tolerance and colonization. Indeed, it has been noted that within a given species BSH activity is found in strains isolated from the gastrointestinal tracts of mammals (Bifidobacterium spp., Lactobacillus acidophilus, Lactobacillus gasseri, Lactobacillus johnsonii, some strains of Lactobacillus plantarum, etc.), whereas organisms isolated from environments from which bile salts are absent such as fermented milk preparations and vegetables (Lactococcus lactis, Lactobacillus delbrueckii, Lactobacillus helveticus, Streptococcus thermophilus, etc.) exhibit no detectable BSH activity (19, 40, 53).
Previous work has determined that bsh is regulated by the principal regulator of virulence gene expression, PrfA (18), and by the general stress responsive sigma factor, σB (51). However, in paired experiments we determined that σB plays a more significant role in regulation of BSH than did PrfA. Bsh transcription and BSH activity were detectable in stationary-phase cells of a prfA mutant but were eliminated completely in a sigB mutant. Furthermore, the sigB mutant was exquisitely sensitive to bile salts in vitro. To our knowledge, this is the first description of a direct physiological role for sigB in microbial bile tolerance. The data presented here support the view that BSH is coregulated by both σB and PrfA. Nadon et al. (41) have recently demonstrated that expression of PrfA is codependent upon σB activity, whereas many PrfA-regulated genes also possess σB promoter regions (39), suggesting a close interdependency between the two systems. In addition, we have demonstrated that expression of sigB is upregulated during infection of the murine gastrointestinal tract, possibly through growth under anaerobic conditions. The present study therefore supports a model in which anaerobiosis (and possibly other environmental signals) can activate the σB regulon within the intestine, preparing invading bacteria for resistance to further environmental stress and possibly priming the pathogen for PrfA-dependent systemic disease. It is undoubtedly significant that many σB regulated genes are involved in survival of stresses encountered within the gastrointestinal tract (18, 30, 51) or are responsible for cell invasion (30) and that many of these genes are coregulated by PrfA (39).
The product of the second gene under investigation was initially annotated as a putative PVA or BSH. PVAs hydrolyze penicillin V to yield 6-aminopenicillinic acid, which is widely used in the industrial production of semisynthetic antibiotics (32). Their microbial role has not been elucidated, but since they show significant homologies to BSHs it has been suggested that they may hydrolyze bile salts (10, 52); however, to date this hypothesis has never been satisfactorily proven. We demonstrate here that the L. monocytogenes PVA homolog does not hydrolyze glyco- or tauroconjugated bile salts but does play a role in survival of bile salts. Furthermore, the pva mutant demonstrated significantly increased sensitivity to penicillin V, but not penicillin G, suggesting that PVA activity is associated with this locus. On the basis of these data, we propose that currently the most accurate annotation of the locus is as a PVA. The role of the gene in bile tolerance in the absence of bile hydrolysis is intriguing and requires further investigation.
The pva gene is part of a five-gene islet that is only present in half of the L. monocytogenes strains examined. Sequence analyses revealed that this islet has a lower G+C content than flanking regions, suggesting recent acquisition. It is likely that the penicillin resistance afforded by the pva gene imparts a selective advantage under some as-yet-undefined conditions to strains that possess the islet, and this is the subject of further ongoing research in our laboratory. In the present study, we demonstrate that the pva gene product is not required for colonization of the murine gastrointestinal tract. In addition, we have demonstrated that this locus is absent from some strains isolated from the human intestine and some strains associated with human disease, indicating that the locus may be dispensable for the pathogenesis of some strains (data not shown). However, pva is involved in bile tolerance, and its transcription is at least partially regulated by σB. Interestingly, another gene in the pva islet (lmo0448/gadE) was identified during our transposon-based screening approach for identifying bile-sensitive mutants (6), suggesting that loci within the islet may be important at some level for adaptation of specific strains.
The third gene analyzed in the present study was initially annotated as a putative bile acid dehydratase gene. However, classical bile acid dehydratase activity requires a number of additional enzymes that also contribute to the process (13, 17, 56). These enzymes are apparently absent from the genome of L. monocytogenes (23). However, a mutation in this locus renders cells sensitive to the effects of bile salts, an effect that is specific for bile salts and is not related to general stress resistance. Given that this locus is specifically involved in bile resistance in the absence of other enzymes in the bile acid dehydratase pathway, we propose the nomenclature, bile tolerance locus B (btlB). Interestingly, this gene only seems to be present in potentially pathogenic species of Listeria, including all L. monocytogenes strains examined and some L. seeligeri and L. ivanovii strains. Indeed, the btlB mutant is exquisitely sensitive to conditions in the murine gastrointestinal tract and is rapidly eliminated from this environment. The data indicate that this locus encodes a potential virulence factor required for persistence of pathogenic strains within the intestinal tract and subsequent systemic disease.
In conclusion, we have demonstrated roles for bsh, pva, and btlB in the bile tolerance of L. monocytogenes. In the course of the present study, we have unequivocally demonstrated that L. monocytogenes possesses only one BSH enzyme and that an enzyme initially annotated as having possible BSH activity is actually involved in penicillin V resistance. In addition, we have highlighted the requirement for both bsh and btlB in listerial persistence in the murine intestinal tract. Finally, we have uncovered a significant role for the alternative sigma factor σB as a central regulator of the listerial bile stress response, a response that may be triggered by anaerobic conditions within the host gastrointestinal tract.
Acknowledgments
We acknowledge funding received from the Irish Government under National Development Plan 2000-2006 and through funding of the Alimentary Pharmabiotic Centre by the Science Foundation of Ireland Centres for Science Engineering and Technology scheme. R.D.S. is funded by Embark and ESCMID/FEMS research fellowships.
Editor: V. J. DiRita
REFERENCES
- 1.Allerberger, F., B. Langer, O. Hirsch, M. P. Dierich, and H. P. Seeliger. 1989. Listeria monocytogenes cholecystitis. Z. Gastroenterol. 27:145-147. [PubMed] [Google Scholar]
- 2.Aries, V., and M. J. Hill. 1970. Degradation of steroids by intestinal bacteria. I. Deconjugation of bile salts. Biochim. Biophys. Acta 202:526-534. [DOI] [PubMed] [Google Scholar]
- 3.Becker, L. A., M. S. Çetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begley, M. 2003. Physiology and genetics of bile tolerance in Listeria monocytogenes. Ph.D. thesis. National University of Ireland, University College Cork, Cork, Ireland.
- 5.Begley, M., C. G. M. Gahan, and C. Hill. The interaction between bacteria and bile. FEMS Microbiol. Rev., in press. [DOI] [PubMed]
- 6.Begley, M., C. G. M. Gahan, and C. Hill. 2002. Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 68:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Begley, M., C. Hill, and C. G. M. Gahan. 2003. Identification and disruption of btlA, a locus involved in bile tolerance and general stress resistance in Listeria monocytogenes. FEMS Microbiol. Lett. 218:31-38. [DOI] [PubMed] [Google Scholar]
- 8.Bina, J. E., and J. J. Mekalanos. 2001. Vibrio cholerae tolC is required for bile resistance and colonization. Infect. Immun. 69:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briones, V., M. M. Blanco, A. Marco, N. Prats, J. F. Fernandez-Garayzabal, G. Suarez, M. Domingo, and L. Dominguez. 1992. Biliary excretion as possible origin of Listeria monocytogenes in fecal carriers. Am. J. Vet. Res. 53:191-193. [PubMed] [Google Scholar]
- 10.Christiaens, H., R. J. Leer, P. H. Pouwels, and W. Verstraete. 1992. Cloning and expression of a conjugated bile acid hydrolase gene from Lactobacillus plantarum by using a direct plate assay. Appl. Environ. Microbiol. 58:3792-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dashkevicz, M. P., and S. D. Feighner. 1989. Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl. Environ. Microbiol. 55:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson, J. A., D. H. Mallonee, I. Björkhem, and P. B. Hylemon. 1996. Expression and characterization of a C24 bile acid 7α-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J. Lipid Res. 37:1258-1267. [PubMed] [Google Scholar]
- 14.De Boever, P., and W. Verstraete. 1999. Bile salt deconjugation by Lactobacillus plantarum 80 and its implication for bacterial toxicity. J. Appl. Microbiol. 87:345-352. [DOI] [PubMed] [Google Scholar]
- 15.De Smet, I., L. Van Hoorde, M. Vande Woestyne, H. Christiaens, and W. Verstraete. 1995. Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Bacteriol. 79:292-301. [DOI] [PubMed] [Google Scholar]
- 16.Dickinson, A. B., B. E. Gustafsson, and A. Norman. 1971. Determination of bile acid conversion potencies of intestinal bacteria by screening in vitro and subsequent establishment in germ-free rats. Acta Pathol. Microbiol. Scand. 79:691-698. [DOI] [PubMed] [Google Scholar]
- 17.Doerner, K. C., F. Takamine, C. P. LaVoie, D. H. Mallonee, and P. B. Hylemon. 1997. Assessment of fecal bacteria with bile acid 7α-dehydroxylation activity for the presence of bai-like genes. Appl. Environ. Microbiol. 63:1185-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, C. Buchreiser, P. Glaser, P. Cossart, et al. 2002. Listeria monocytogenes bile salt hydrolase is a prfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 19.Elkins, C. A., S. A. Moser, and D. C. Savage. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 147:3403-3412. [DOI] [PubMed] [Google Scholar]
- 20.Gahan, C. G. M., and C. Hill. 2003. Relationship between stress adaptation and virulence in foodborne pathogenic bacteria, p. 213-245. In A. E. Yousef, and V. K. Juneja (ed.), Microbial stress adaptation and food safety. CRC Publishing Co., Inc., Boca Raton, Fla.
- 21.Gahan, C. G. M., J. O'Mahony, and C. Hill. 2001. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect. Immun. 69:3924-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilliland, S. E., and M. J. Speck. 1977. Deconjugation of bile acids by intestinal lactobacilli. Appl. Environ. Microbiol. 33:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-Del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. Mata Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 24.Grill, J. P., F. Schneider, J. Crociani, and J. Ballongue. 1995. Purification and characterization of conjugated bile salt hydrolase from Bifidobacterium longum BB536. Appl. Environ. Microbiol. 61:2577-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunn, J. S. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2:907-913. [DOI] [PubMed] [Google Scholar]
- 26.Hardy, J., K. P. Francis, M. DeBoer, P. Chu, K. Gibbs, and C. H. Contag. 2004. Extracellular replication of Listeria monocytogenes in the murine gallbladder. Science 303:851-853. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann, A. F. 1999. Bile acids: the good, the bad, and the ugly. News Physiol. Sci. 14:24-29. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann, A. F., and K. J. Mysels. 1992. Bile acid solubility and precipitation in vitro and in vivo: the role of conjugation, pH and Ca2+ ions. J. Lipid Res. 33:617-626. [PubMed] [Google Scholar]
- 29.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-534. [PubMed] [Google Scholar]
- 30.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawamoto, K., I. Horibe, and K. Uchida. 1989. Purification and characterization of a new hydrolase for conjugated bile acids, chenodeoxycholyltaurine hydrolase, from Bacteroides vulgatus. J. Biochem. 106:1049-1053. [DOI] [PubMed] [Google Scholar]
- 32.Kovacikova, G., W. Lin, and K. Skorupsk. 2003. The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome. J. Bacteriol. 185:4825-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lacroix, F. J., A. Cloeckaert, O. Grepinet, C. Pinault, M. Y. Popoff, H. Waxin, and P. Pardon. 1996. Salmonella typhimurium acrB-like gene: identification and role in resistance to biliary salts and detergents and role in murine infection. FEMS Microbiol. Lett. 135:161-167. [DOI] [PubMed] [Google Scholar]
- 34.Lauer, P., M. Y. N. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundeen, S. G., and D. C. Savage. 1992. Multiple forms of bile salt hydrolase from Lactobacillus sp. strain 100-100. J. Bacteriol. 174:7217-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mead, P. S., L. Slutsker, V. Dietz, L. F. Mc Caig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milohanic, E., P. Glaser, J.-Y. Coppée, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchreiser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 40.Moser, S. A., and D. C. Savage. 2001. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in Lactobacilli. Appl. Environ. Microbiol. 67:3476-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfjammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pope, L. M., K. E. Reed, and S. M. Payne. 1995. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect. Immun. 63:3642-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prouty, A. M., and J. S. Gunn. 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68:6763-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raya, R., J. Bardowski, P. S. Andersen, S. D. Ehrlich, and A. Chopin. 1998. Multiple transcriptional control of the Lactococcus lactis trp operon. J. Bacteriol. 180:3174-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders, J. W., G. Venema, J. Kok, and K. Leenhouts. 1998. Identification of a sodium chloride regulated promoter in Lactococcus lactis by single-copy chromosomal fusion with a reporter gene. Mol. Gen. Genet. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 47.Schlech, W. F. 2000. Foodborne listeriosis. Clin. Infect. Dis. 31:770-775. [DOI] [PubMed] [Google Scholar]
- 48.Sleator, R. D., and C. Hill. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26:49-71. [DOI] [PubMed] [Google Scholar]
- 49.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 50.Somer, L., and Y. Kashi. 2003. A PCR method based on 16S rRNA sequence for simultaneous detection of the genus Listeria and the species Listeria monocytogenes in food products. J. Food Prot. 66:1658-1665. [DOI] [PubMed] [Google Scholar]
- 51.Sue, D., K. J. Boor, and M. Wiedmann. 2003. σB - dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiology 149:3247-3256. [DOI] [PubMed] [Google Scholar]
- 52.Suresh, C. G., A. V. Pundle, H. Siva Raman, K. N. Rao, J. A. Brannigan, C. E. McVey, C. S. Verma, Z. Dauter, E. J. Dodson, and G. G. Dodson. 1999. Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members. Nat. Struct. Biol. 6:414-416. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka, H., H. Hashiba, J. Kok, and I. Mierau. 2000. Bile salt hydrolase of Bifidobacterium longum: biochemical and genetic characterization. Appl. Environ. Microbiol. 66:2502-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells, J. E., and P. B. Hylemon. 2000. Identification and characterization of a bile acid 7α-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7α-dehydroxylating strain isolated from human feces. Appl. Environ. Microbiol. 66:1107-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]