Normal and transformed motile cells follow a common trend in which size and contractile forces are negatively correlated with cell speed. However, tumorigenic factors amplify the preexisting population heterogeneity and lead some cells to exhibit biomechanical properties that are more extreme than those observed with normal cells.
Abstract
Tumor development progresses through a complex path of biomechanical changes leading first to cell growth and contraction and then cell deadhesion, scattering, and invasion. Tumorigenic factors may act specifically on one of these steps or have a wider spectrum of actions, leading to a variety of effects and thus sometimes to apparent contradictory outcomes. Here we used micropatterned lines of collagen type I/fibronectin on deformable surfaces to standardize cell behavior and measure simultaneously cell size, speed of motion and magnitude of the associated traction forces at the level of a single cell. We analyzed and compared the normal human breast cell line MCF10A in control conditions and in response to various tumorigenic factors. In all conditions, a wide range of biomechanical properties was identified. Despite this heterogeneity, normal and transformed motile cells followed a common trend whereby size and contractile forces were negatively correlated with cell speed. Some tumorigenic factors, such as activation of ErbB2 or loss of the βsubunit of casein kinase 2, shifted the whole population toward a faster speed and lower contractility state. Treatment with transforming growth factor β induced some cells to adopt opposing behaviors such as extremely high versus extremely low contractility. Thus tumor transformation amplified preexisting population heterogeneity and led some cells to exhibit biomechanical properties that were more extreme than those observed with normal cells.
INTRODUCTION
The malignant transformation of cells encompasses a complex sequence of events implicating many distinct pathways, making the process difficult to describe and categorize. Throughout the development of a tumor, abnormal biochemical signaling, abnormal cell growth, and changes in mechanical properties within the tumor are closely connected and interdependent. For example, cell stiffness promotes cell growth (Klein et al., 2009), which increases tissue density and mechanical pressure, which, in turn, trigger oncogenic pathways that further stimulate cell growth (Fernández-Sánchez et al., 2015). Moreover, the cross-talk between biochemical signaling and the mechanical environment evolve during tumor progression, so that defined biochemical or mechanical signals may have distinct consequences, depending on the stage of tumor development.
The mechanical properties of individual cells and the tumor tissue as a whole vary widely during tumor development, from the early cell adhesion and cell proliferation stages of tumor formation to the late cell dissociation and cell migration stages during tumor dissemination, when cells have become more transformed (Plodinec et al., 2012; Weder et al., 2014). Biochemical signals promoting cell contractility may have distinct effects on tumor cells at the early stages of tumor development (e.g., proliferating cells) in comparison with tumor cells at later stages (e.g., metastatic/invasive cells; Fritsch et al., 2010; Aguilar-Cuenca et al., 2014). Therefore the various cross-talks between signaling and mechanics at the distinct stages of tumor progression may underlie the heterogeneity of cell responses and explain many conflicting results on the effects of oncogenic factors, cell mechanics, and their interplay during tumor progression.
Although under specific conditions their roles have been clearly defined, the pleiotropic effects of the main oncogenic pathways are not universally observed in all tumors, preventing their use as prognostic markers. Conflicting results in carcinogenesis have been reported for thyroid hormones (Piekiełko-Witkowska, 2013), macrophage-colony-stimulating factor (Laoui et al., 2014), vascular endothelial growth factor (Zhan et al., 2009), epidermal growth factor (Nicholson et al., 2001), and fibroblast growth factor-2 (Korc and Friesel, 2009). Of note, transforming growth factor β (TGF-β), which has been described as a tumor suppressor and oncogenic factor (Kubiczkova et al., 2012), appears to affect cell contractility in a manner dependent on substrate stiffness (Marinković et al., 2012). Moreover, TGF-β appears to increase the contractility of melanoma cells (Cantelli et al., 2015) but decrease the contractility of muscle cells (Mendias et al., 2012). In addition, depending on substrate stiffness, TGF-β can increase or reduce the speed of cell migration (Wu et al., 2013), induce epithelial–mesenchymal transition (EMT), or induce apoptosis (Leight et al., 2012).
The effects of cell contractility on tumor progression remain to be fully elucidated. Tissue stiffness is a hallmark of cancer (Paszek et al., 2005) and an enhancer of tumor growth (Fernández-Sánchez et al., 2015). However, the observation that cell contractility promotes tumor growth (Samuel et al., 2011) contrasts with the repeated observation of cancer cells being softer than normal cells (Cross et al., 2007, 2008; Li et al., 2008; Fritsch et al., 2010; Jonas et al., 2011; Xu et al., 2012; Efremov et al., 2014). Nevertheless the mechanical landscape within a tumor is complex, as reflected by the variations in stiffness within tumor tissue (Plodinec et al., 2012).
The relationships between the magnitude of contractile forces and the metastatic potential of a given tumor cell may also vary with different types of tumor cells, the tumor-cell environment, and the types of measurements being made. Cell lines ranked with respect to increasing metastatic potential have been reported to produce traction forces that positively correlate (Jonas et al., 2011; Kraning-Rush et al., 2012) or negatively correlate with that potential (Munevar, 2001; Indra et al., 2011). However, this may reflect that metastatic potential is hard to quantify, and cell migration speed and depth of invasion into collagen matrices are often considered as good proxies for this potential. Nevertheless, migration speeds and invasive capacities have been positively correlated with stiffness and contractile forces in some studies (Rösel et al., 2008; Indra et al., 2011; Mierke et al., 2011a, b; Mierke, 2013) but negatively correlated in others (Swaminathan et al., 2011; Agus et al., 2013). The interpretation of these relationships is further complicated by the fact that two- and three-dimensional speeds may not necessarily be correlated (Indra et al., 2011; Meyer et al., 2012). Moreover, differences in the relationship between cell contraction and migration speed may well arise from the nonlinear relationship between cell speed and cell adhesion. There is an optimal adhesion strength promoting cell migration, and increases or decreases in adhesion strength from this optimum reduces cell speed (DiMilla et al., 1991; Gupton and Waterman-Storer, 2006). Again, the relationship between cell speed and cell adhesion can depend on the stage of the tumor (Weder et al., 2014) and the type of cell migration—mesenchymal or amoeboid (Friedl, 2004)—making it difficult to compare results generated in different experimental conditions.
The averaging of values in cell populations may also mask important cell heterogeneity that exists within a tumor (Lee et al., 2014; Mcgranahan and Swanton, 2015), such as differences in mechanical properties (Plodinec et al., 2012). Therefore, to better characterize the mechanical and biochemical relationships, it is necessary to measure all parameters on the same single cells and compare those values measured on individual cells or subgroups (Altschuler and Wu, 2010; Eberwine and Kim, 2015). Here we use a cell culture system based on micropatterned cell adhesion substrates as a platform to measure cell speed, cell size, and cell contractile forces under controlled conditions to compare the responses between individual normal cells and those submitted to tumorigenic factors.
RESULTS AND DISCUSSION
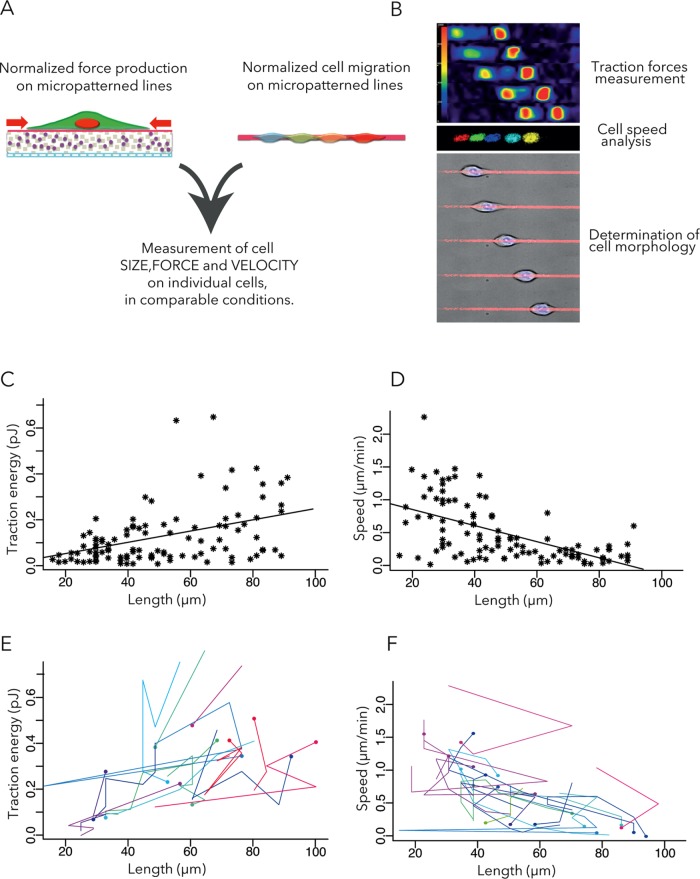
Cell shape changes in motile cells induce large variability in biophysical measurements. In cell culture, micropatterned lines of cell adhesion substrate have been shown to standardize cell behavior and better approximate to in vivo conditions than a planar coating of the cell adhesion substrate (Doyle et al., 2009; Provenzano et al., 2010). Moreover, the linear (fibril-like) organization of extracellular matrix proteins has been shown to be characteristic of the metastatic niche (Provenzano et al., 2006; Gritsenko et al., 2012; Lu et al., 2012; Artym et al., 2015). Therefore we evaluated cells in polyacrylamide hydrogels with a Young modulus of 10 kPa, which corresponds to the stiffness of breast tissue (Rzymski et al., 2011), which were micropatterned with 4-μm-wide lines made of collagen type I and fibronectin (Vignaud et al., 2014; Figure 1A). The experimental system was designed to measure the size, speed, and contractile forces of each individual cell (Bastounis et al., 2014; Figure 1B). Because cell shape is mostly linear and the production of traction forces limited to the micropatterned lines, we characterized cell geometry by measuring cell length. Tractional forces were measured using an automated approach based on an ImageJ plug-in we have developed previously by which individual cells are tracked and the energy required for the traction force field computed from the displacements of fluorescent beads embedded in the substrate (Martiel et al., 2015).
FIGURE 1:
Measurement of traction forces and migration speeds with respect to cell size. (A) Dynamic analyses of cell motion and traction forces exhibited by motile cells on micropatterned polyacrylamide hydrogels. (B) Examples of traction force field, nuclei displacements, and cell motion. (C) Contractile energy (measured by the energy required to deform the polyacrylamide hydrogel) vs. cell length. The line describes the linear regression. Pearson r = 0.39; 95% confidence interval (CI), 0.21–0.54. Seven independent experiments, 105 WT cells. (D) Cell speed vs. cell length. The line describes the linear regression. Pearson r = −0.57, 95% CI, −0.69 to −0.4. (E, F) Temporal variations of traction energy (E) and migration speed (F) with respect to cell length. Each color corresponds to a single cell. Dots correspond to initial time point and lines to temporal variations during the next 2 h. For clarity, only cells displaying traction energy variations > 0.2 pJ and cells displaying speed variations > 0.5 µm/min are shown.
The MCF10A cell line was derived from nontransformed human mammary epithelium (Debnath et al., 2003) and has been used in an established model of tumor transformation. In control conditions, the cells have the capacity to form acini-like structure in three-dimensional gels. However, the cells are sensitive to oncogenic factors such as TGF-β and ErbB2 activation, which can force cells to grow in nonproliferative conditions (Giunciuglio et al., 1995; Debnath et al., 2003; Seton-Rogers et al., 2004; Xue et al., 2013). At baseline, the traction energy (i.e., the mechanical energy used by the cell in substrate deformation) is positively correlated with the length of the cell (Figure 1C). These results are consistent with earlier studies showing a positive correlation between cell spreading area and the magnitude of traction forces (Wang et al., 2002; Tan et al., 2003; Reinhart-King et al., 2005; Califano and Reinhart-King, 2010; Mertz et al., 2012; Oakes et al., 2014). By contrast, the speed of a motile cell is inversely correlated with the length of the cell (Figure 1D). It was previously reported that cell spreading and cell speed are inversely correlated in the high-adhesion regime (Gupton and Waterman-Storer, 2006; Barnhart et al., 2011; Fuhrmann et al., 2015). The single-cell snapshots reported here were obtained by averaging the first two time points only, so that all plotted data points are truly independent. They reveal a high level of interindividual variance. However, it is also instructive to follow variations of size, force, and speed over time. Plotting the time evolution of single-cell biomechanical parameters reveals an additional level of intraindividual variance, which follows the same trend as the snapshots (Figure 1, E and F).
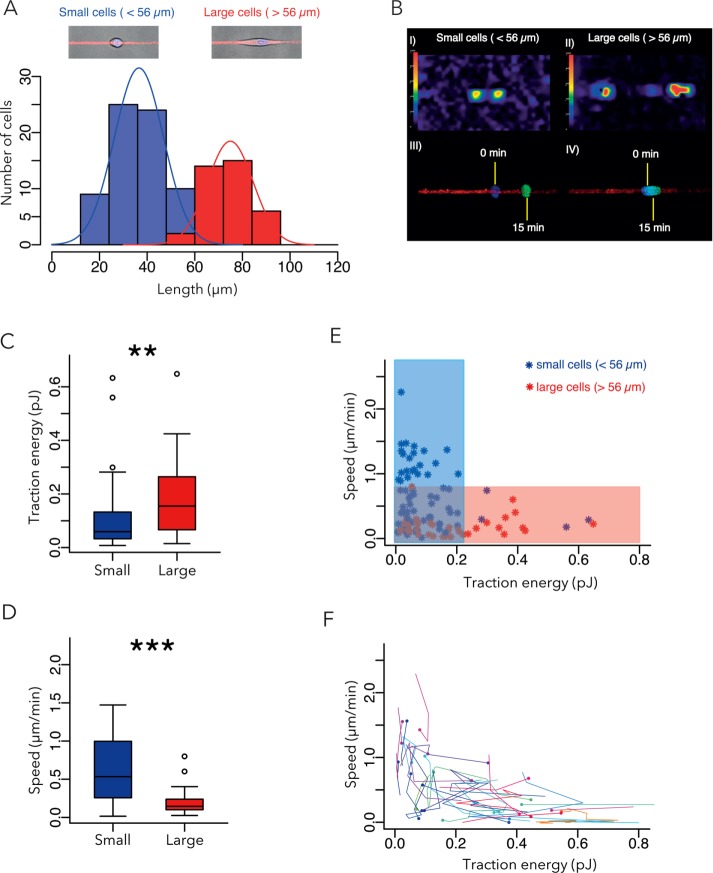
We then looked for a way to perform statistical comparisons of the traction force magnitudes and migration speeds of small and large cells. We chose the midpoint between the two main peaks in the distribution of cell lengths: 56 µm (Figure 2A). Strikingly, small cells exhibited lower contractile forces and were faster than large cells (Figure 2, B–E). A similar trend was visible when considering the temporal variations of these parameters for individual cells (Figure 2F). Thus the cells of different sizes appeared to have distinct biophysical properties, which later served as a reference in the study. Note that this conclusion and the others later would be identical if the distinction between small and large cells was based on median (46 µm) or average wild-type (WT) cell length (50 µm).
FIGURE 2:
Comparison of small- and large-cell biomechanical properties. (A) Size distribution of WT cells. Two distinct cell subpopulations of cells were identified on the basis of size: small (<56 µm in length; blue) and large (red; >56 µm in length). Same data set as in Figure 1 (seven independent experiments, 105 WT cells). (B) Representative examples of traction-force fields and corresponding nuclei displacements for small and large cells. (C) Contractile energy of small vs. large cells. (D) Speed of small vs. large cells. (E) Cell speed vs. contractile energy; colored regions distinguish the low-contraction-forces/high-speed phenotype (blue) from the high-contraction-force/low-speed phenotype (red). (E, F) Temporal variations of traction energy (E) vs. migration speed (F). Each color corresponds to a single cell. Dots correspond to initial time point, lines to temporal variations during the next 2 h. For clarity, only cells displaying traction energy variations >0.2 pJ and cells displaying speed variations >0.5 µm/min are shown.
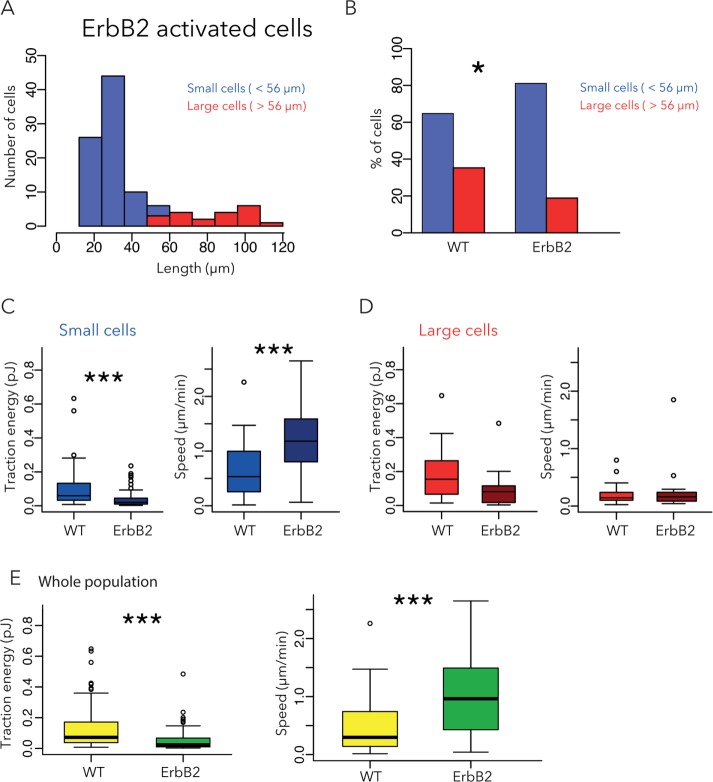
We then investigated the biophysical properties of a tumorigenic MCF10A cell line in which ErbB2 was constitutively active (Muthuswamy et al., 2001; Levental et al., 2009). These cells are characterized by an overexpression of the ErbB2 receptor tyrosine kinase, a receptor identified as an activator of proliferation and invasiveness in breast cancer cells (Feldner and Brandt, 2002; Brix et al., 2014). In contrast to WT cells, the majority of ErbB2 cells were round and small, with a peak length of 30 μm (Figure 3A), and only a minority of ErbB2 cells were large (>56 µm; Figure 3B). The average size of ErbB2 cells in the small-cell group was also smaller than that in the small-cell group of WT cells. These ErbB2 cells were also faster and displayed fewer contractile forces than WT cells (Figure 3C). These results are consistent with previous results showing that ErbB2 activates actin disassembly factors (Chen et al., 1996; Brandt et al., 1999) and adhesion remodeling (Spencer et al., 2000) and thereby reduces cell adhesion and stimulates cell migration (Feldner and Brandt, 2002). Of interest, the presence of small- and low-adhesive cells within surrounding tissues has been described as symptomatic of metastasis and associated with poor prognosis in urinary bladder carcinomas (Cheng et al., 2004). The average size of ErbB2 cells in the large-cell group was similar to that in the large-cell group of WT cells, and so were their traction energy and speed (Figure 3D). Therefore the trend relating small cell size, high speed, and low contractile forces observed in WT cells was apparent in ErbB2 cells, with an accentuation toward smaller and faster motile cells. Of interest, the consequences of ErbB2 activation for cell size, speed, and traction energy were also visible when comparing the entire population, although it could not be inferred that these differences stemmed from the small cells only (Figure 3E).
FIGURE 3:
Effect of ErbB2 activation on cell size, traction force, and migration speed. (A) Size distribution of ErbB2 cells. Small cells are shown in blue, large cells in red. Three independent experiments, 106 cells. (B) Comparison of the proportion of small (>56 µm; blue) and large cells (>56 µm; red) in WT and ErbB2 cells. (C, D) Contractile energy and migration speed of small (C) and large (D) cells. (E) Speed and traction energy of ErbB2-activated cells compared with WT cells. Data for WT cells are reproduced from Figure 2.
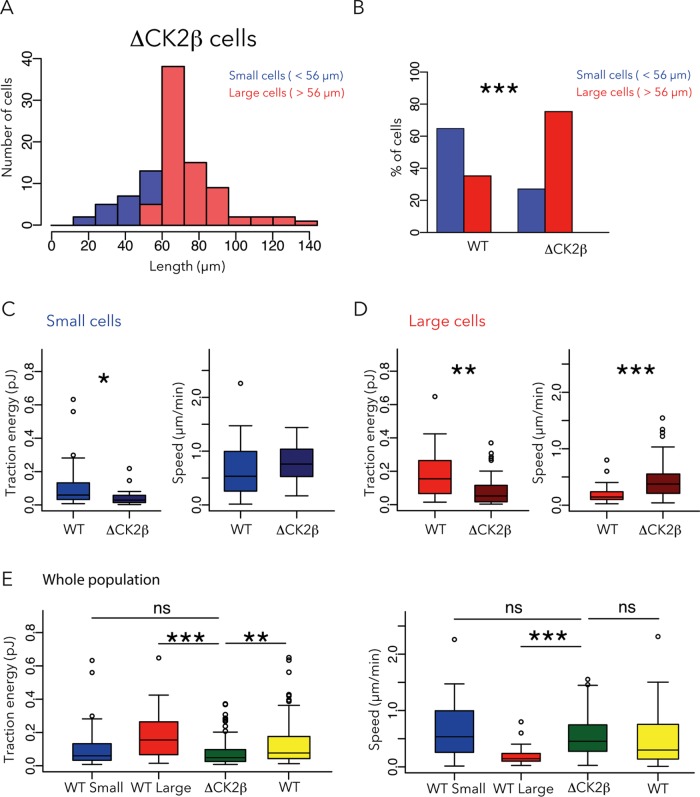
We then investigated the tumorigenic MCF10A cell line in which the β subunit of casein kinase 2 (CK2) is depleted (MCF10A ΔCK2β). These cells are antiapoptotic, prosurvival, and multi–drug-resistant and display EMT-like features (Deshiere et al., 2013; Vilmont et al., 2015). Indeed, CK2 contributes to maintain the epithelial phenotype and polarity (Canton and Litchfield, 2006). More precisely, two CK2 substrates (Snail1 and Foxc2), which are transcription factors, have to be phosphorylated to keep the epithelial phenotype (Golden and Cantley, 2015). The key point is that the regulatory subunit CK2β is mandatory for CK2-mediated phosphorylation of Snail1 and Foxc2 (Filhol et al., 2015). Consequently knocking down CK2β induces EMT and thus increases cell motility (Deshiere et al., 2013) and impairs polarity (Deshière et al., 2008, 2011). However, a direct role of CK2 depletion in cell metastasis and tissue invasion has yet to be identified. The distribution of ΔCK2β cell sizes fit into a single population with a peak size of 70 μm (Figure 4A) and resembled the elongated morphology of large WT cells. Indeed, ΔCK2β cells were larger than WT cells (Figure 4B). Small ΔCK2β cells were not so different from small WT cells (Figure 4C), and large ΔCK2β cells produced less traction forces and moved faster than WT (Figure 4D). Thus, in biomechanical terms, the entire population of ΔCK2β cells resembled the small WT cells rather than the large WT cells, that is, relatively high speed and low contractile forces (Figure 4E).
FIGURE 4:
Effect of CK2β removal on cell size, traction force, and migration speed. (A) Only a single population of ΔCK2β cells was identified based on size distribution. Six independent experiments, 101 cells. (B) Comparison of the proportion of small (>56 µm; blue) and large cells (>56 µm; red) in WT and ΔCK2β cells. (C, D) Contractile energy and migration speed of small (C) and large (D) ΔCK2β cells compared with WT cells. (E) Contractile energy and speed of the entire population of ΔCK2β cells compared with small, large, or entire population of WT cells. Data for WT cells are reproduced from Figure 2.
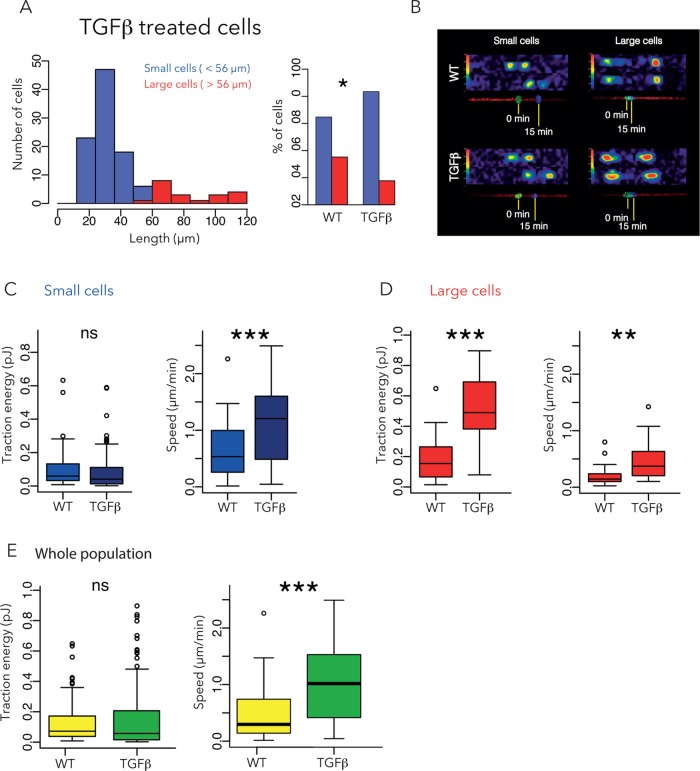
Finally, we investigated the effect of TGF-β on MCF10A cells by incubating the cells with 5 ng/ml TGF-β for 48 h before starting the experiment. In terms of morphology, TGF-β–treated cells resembled ErbB2 cells, with a majority of small cells (peak length ∼30 μm) and a minority of large cells (average length of 90 μm; Figure 5, A and B). In biomechanical terms, the small TGF-β–treated cells also resembled ErbB2 cells and exhibited significantly higher speed and (marginally) lower contractile forces than small WT cells (Figure 5C). However, the large TGF-β–treated cells differed from ErbB2 cells in that the former exhibited significantly higher speed and higher contractile forces than large WT cells (Figure 5D). Of note, and unlike ErbB2-expressing or ΔCK2β cells, compared with WT cells, the differences in contractile forces between TGF-β–treated and WT cells were masked when the entire populations of small and large cells were considered together (Figure 5E).
FIGURE 5:
Effect of TGF-β on cell size, traction force, and migration speed. (A) Size distribution of TGF-β-treated cells. Small cells are shown in blue, large cells in red. Five independent experiments, 114 cells. (B) Representative examples of traction force fields and corresponding nuclei displacements. (C, D) Contractile energy and migration speed of small (C) and large (D) TGFβ-treated cells compared with WT cells. (E) Contractile energy and speed of entire population, taking small and large cells together. Data for WT cells are reproduced from Figure 2.
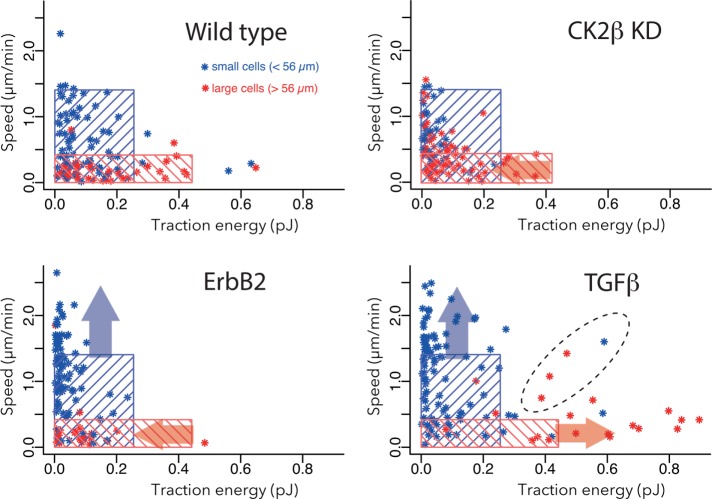
In a further analysis, we superimposed the limits that defined the groups of small and large WT cells on the graphs of cell speed versus contractile energy (Figure 6). In MCF10A-WT cells, the area defined by the blue rectangle set the limit for small cells (fast speed and low contractile forces), and the area defined by the red rectangle set the limit for large cells (low speed and high contractile forces). Although significantly biased toward low contractile forces, MCF10A ΔCK2β cells appeared to be within the limits defined by the WT cells (Figure 6). By contrast, 40% of small ErbB2 cells and 50% of small TGF-β cells were outside the limits defined by WT cells (blue arrows). Furthermore, almost 80% of large TGF-β–treated cells were outside the limits defined by WT cells (red arrows). Most of TGF-β–treated cells that exceeded the limit followed the expected trend, in that the contractile forces opposed the migration speed. However, 15% (3 of 20) of large TGF-β–treated cells exhibited relatively high speed in addition to high contractile forces (area defined by dashed line), suggesting that the mechanism relating shape regulation, force production, and cell motion was different in these cells than for all of other cells in this study.
FIGURE 6:
Heterogeneity of single-cell response to tumorigenic factors. Cell speed vs. contractile energy. Top, left graph: the blue rectangle includes 95% of the small WT cells, and the red zone includes 95% of the large WT cells. These data are reproduced from Figure 2E. In the other three graphs, representing the data from ΔCK2β cells and TGF-β–treated cells, the same blue and red rectangles defined by the WT-cell groups are superimposed. Blue arrows represent the tendency to shift small-cell behavior in the speed–force profile even outside the limit defined by small WT cells. Red arrows represent the tendency to shift large-cell behavior in the speed–force profile even outside the limit defined by large WT cells. The dashed line surrounds cells with relatively high speeds and contractile forces, and which, in addition to being outside the limits defined by WT cells, do not respect the global trend negatively relating cell speed and contractile energy.
Taken together, our results identified a large range of MCF10A cells size and a robust tendency relating cell mechanical and motile properties. The small cells exhibited higher speeds and lower contractile forces, and we suggest that these cells are likely to have high metastatic and invasive potential. By contrast, the large cells exhibited lower speeds and higher contractile forces, and we suggest that they are likely to have high proliferation capacities and support tumor growth and stiffness. Of interest, both normal and transformed cells followed a common trend in which size and contractile forces were negatively correlated with cell speed. These results were consistent with previous reports about size, force, and speed variations with substrate stiffness (Lo et al., 2000; Oakes et al., 2009). Tumorigenic factors did not perturb the force–speed relationship but instead shifted the location of the data population along a conserved trend and displaced cells toward extreme phenotypes. Given the relationship of ErbB2, CK2, and TGF-β to cancer, the differences in contractile forces and speed may reflect functional attributes that are relevant for transformed cells in vivo. The tumorigenic factor ErbB2 or depletion of CK2 shifted the whole population of cells toward a state of faster speed and lower contractility, with some cells outside the high-speed boundary set by normal cells. Therefore the balance in favor of cell dissemination versus cell growth in a metastatic tumor may reflect this overall shift in biomechanical properties (high to low contractile forces and low to high speeds). The response of MCF10A cells to TGF-β revealed that there might be further complexity and induction of opposite individual phenotypes within the population. TGF-β induced some cells outside both the high-speed/low-traction and high-contractility/low speed boundaries set by normal cells. In addition, a few cells stimulated by TGF-β seem to work outside of the general size-speed-force relationship and display both high speed and high contractility. Such complex outcomes, due to population heterogeneity and distinct effects on specific subgroups, might account for some of the conflicting conclusions that have been reported. Of greater importance, our results highlight the absolute necessity of considering the heterogeneity of individual tumor-cell biomechanical profiles when characterizing oncogenic factors.
MATERIALS AND METHODS
Experimental system
We made 22 × 22 mm patterned polyacrylamide hydrogels according to previously published methods (Vignaud et al., 2014). Briefly, hydrogels were placed on 20 × 20 mm silanized coverslips and patterned with 4.0-μm-wide, 1000-μm-long lines made of 20 µg/ml collagen type I (A1048301; Life Technologies) and 20 µg/ml fibronectin (F1141; Sigma-Aldrich).
Cells
Michigan Cancer Foundation (MCF10A) human mammary gland cells (CRL-10317; American Type Culture Collection) were maintained at 37°C and 5% CO2 in Lonza Mammary Epithelial Growth Medium (Lonza MEGM Bullet Kit CC3150 without gentamycin) in the presence of 100 ng/ml cholera toxin (C-8052; Sigma-Aldrich) and 0.5% antibiotic-antimycotic (15240062; Life Technologies). Stable gene silencing was accomplished by transduction with pLKO1 lentiviruses (Sigma-Aldrich) as previously described (Deshiere et al., 2013). The H-RasG12V retrovirus was a generous gift from M. Thomas (INSERM, U1239, Rouen; Herbet et al., 2012). For infection, MCF10A cells were plated into 24-well plates (5 × 104 in 500 µl of serum-supplemented growth medium). The next day, adherent cells were incubated with lentiviral particles (multiplicity of infection, 1–5) diluted in 250 µl of serum-supplemented medium containing 8 µg/µl Polybrene. After 4 h, 500 µl of medium was added to cultures, and transduction was maintained for 16 h before cells were washed and the medium changed. Puromycin selection started 36 h postinfection (at a concentration of 1 µg/ml) and was maintained during all cell culture.
Measurements of contractile energy were carried out using MCF10A WT cells, MCF10A-ErbB2 cells, MCF10A-ΔCK2β cells, and MCF10A WT cells after 48 h of incubation with TGF-β (5 ng/ml; Sigma-Aldrich). Before experimental setup, cells were maintained overnight on the micropatterned lines in an incubator (5% CO2, 95% humidity) and then assessed in terms of speed, morphology, and contractile energy.
Analysis of hydrogel stiffness
The elasticity of the polyacrylamide hydrogels was determined by atomic force microscopy using established procedures (Frey et al., 2007). Briefly, cantilevers (PT-GS with a borosilicate sphere tip of radius 2.5 μm and nominal spring constant of 0.06 N/m; Novascan Technologies) were mounted on a Bioscope Catalyst atomic force microscope (Bruker) and calibrated using the thermal noise method. Force–indentation curves were acquired for forces of 1–10 nN and fitted with a Hertz model to yield elastic moduli.
Measurement of cell length, contractile energy, and speed
Cell lengths were extracted from the traction force profiles by detecting the positions of force application sites. Contractile energy was expressed as the energy required to deform the 10-kPa polyacrylamide hydrogel (i.e., the traction force multiplied by the gel deformation), and measurements were carried out according to a previously published method using ImageJ (Martiel et al., 2015). Briefly, bead displacements were determined using particle image velocimetry with a window size of 3.96 µm. The corresponding contractile energy was estimated with the Fourier transform traction cytometry (FTTC) method. Projection of the exerted forces along the patterned line were then calculated and analyzed automatically in Python. Cell speed was determined by identifying the position of the cell nuclei (stained with Hoechst 33342 at 5 ng/ml) in consecutive pictures (15 min apart) with ImageJ. The first two time points were used to calculate cell speed.
Image acquisition
Images were obtained using a spinning-disk confocal microscope (Nikon) with a 40× objective. Cells were imaged every 15 min for at least 2 h to control that cells were in a steady state, but image analyses for speed and contractile energy measurements were performed on the first two images.
Statistical analysis
Statistical analyses were performed using the statistical software R.
Cell length distributions and subgroups were defined from WT MCF10A cells. To distinguish the presence of subgroups, we tested the normality of length distributions for one, two, and three subpopulations. In each case, the separation of cell lengths in subpopulation clusters was done with a K-means algorithm. Each subpopulation was then tested for normality with Shapiro–Wilk normality test. For a single cluster or three clusters, the distributions failed the normality test (Shapiro–Wilk normality test, p < 0.0002 and p < 0.04 respectively), whereas two-means clustering generated two normal subpopulations (p > 0.1). Moreover, this clustering defined the threshold length (56 µm) separating small from large cells and was the median value between the longest small cell and the smallest large cell. Note that median length (46 µm) or average length (50 µm) of the whole population led to different groups of small and large cells but did not affect the conclusions about to their migration speeds and traction energies.
The comparison of two populations of cells based on the frequencies of cell-size phenotypes within these populations (Figure 3B) was carried out using Fisher’s exact test. Results of this test are represented on the graphs with the following thresholds: ns, p > 0.01; *p < 0.01, **p < 0.001, ***p < 0.0001.
The comparisons of populations of cells based on traction energies or speeds (i.e., between small and large WT cells, and between WT and other cell lines) were performed using the Mann–Whitney test. Distributions are represented in a box-plot graph, and results of this test are represented with the following thresholds: °p > 0.01, *p < 0.01, **p < 0.001, ***p < 0.0001.
The rectangular areas on the graphs of cell speed versus contractile energy were determined using 95 percentiles (threshold percentile values varied between 75 and 99 with little effect on the results) of speed and contractile energy data obtained from the WT cell subgroups (small and large, respectively). Fisher’s exact test was used to compare the number of outlying cells (out of WT rectangle domains; °p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.005).
Acknowledgments
We thank Laurent Blanchoin, Qingzong Tseng, and the entire CytoMorpho Lab for their great help and support all throughout the project. This work was supported by the Institut National du Cancer (INCA PLBIO2011 to O.F.C. and M.T.).
Abbreviations used:
- CK2
casein kinase 2
- TGF-β
transforming growth factor β
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-10-0694) on April 20, 2017.
REFERENCES
- Aguilar-Cuenca R, Juanes-García A, Vicente-Manzanares M. Myosin II in mechanotransduction: master and commander of cell migration, morphogenesis, and cancer. Cell Mol Life Sci. 2014;71:479–492. doi: 10.1007/s00018-013-1439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agus DB, Alexander JF, Arap W, Ashili S, Aslan JE, Austin RH, Backman V, Bethel KJ, Bonneau R, Chen W-C, et al. A physical sciences network characterization of non-tumorigenic and metastatic cells. Sci Rep. 2013;3:1449. doi: 10.1038/srep01449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference. Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym VV, Swatkoski S, Matsumoto K, Campbell CB, Petrie RJ, Dimitriadis EK, Li X, Mueller SC, Bugge TH, Gucek M, et al. Dense fibrillar collagen is a potent inducer of invadopodia via a specific signaling network. J Cell Biol. 2015;208:331–350. doi: 10.1083/jcb.201405099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart EL, Lee K-C, Keren K, Mogilner A, Theriot JA. An adhesion-dependent switch between mechanisms that determine motile cell shape. PLoS Biol. 2011;9:e1001059. doi: 10.1371/journal.pbio.1001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastounis E, Meili R, Alvarez-González B, Francois J, Del Álamo JC, RA Firtel, Lasheras JC. Both contractile axial and lateral traction force dynamics drive amoeboid cell motility. J Cell Biol. 2014;204:1045–1061. doi: 10.1083/jcb.201307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt BH, Roetger A, Dittmar T, Nikolai G, Seeling M, Merschjann A, Nofer JR, Dehmer-Möller G, Junker R, Assmann G, et al. c-erbB-2/EGFR as dominant heterodimerization partners determine a motogenic phenotype in human breast cancer cells. FASEB J. 1999;13:1939–1949. doi: 10.1096/fasebj.13.14.1939. [DOI] [PubMed] [Google Scholar]
- Brix DM, Clemmensen KKB, Kallunki T. When good turns bad: regulation of invasion and metastasis by erbb2 receptor tyrosine kinase. Cells. 2014;3:53–78. doi: 10.3390/cells3010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano JP, Reinhart-King CA. Substrate stiffness and cell area predict cellular traction stresses in single cells and cells in contact. Cell Mol Bioeng. 2010;3:68–75. doi: 10.1007/s12195-010-0102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantelli G, Orgaz JL, Rodriguez-Hernandez I, Karagiannis P, Maiques O, Matias-Guiu X, Nestle FO, Marti RM, Karagiannis SN, Sanz-Moreno V. TGF-β-Induced transcription sustains amoeboid melanoma migration and dissemination. Curr Biol. 2015;25:2899–2914. doi: 10.1016/j.cub.2015.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton DA, Litchfield DW. The shape of things to come: an emerging role for protein kinase CK2 in the regulation of cell morphology and the cytoskeleton. Cell Signal. 2006;18:267–275. doi: 10.1016/j.cellsig.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Chen P, Murphy-Ullrich JE, Wells A. A role for gelsolin in actuating epidermal growth factor receptor-mediated cell motility. J Cell Biol. 1996;134:689–698. doi: 10.1083/jcb.134.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Pan C-X, Yang XJ, Lopez-Beltran A, MacLennan GT, Lin H, Kuzel TM, Papavero V, Tretiakova M, Nigro K, et al. Small cell carcinoma of the urinary bladder. Cancer. 2004;101:957–962. doi: 10.1002/cncr.20456. [DOI] [PubMed] [Google Scholar]
- Cross SE, Jin Y-S, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- Cross SE, Jin Y-S, Tondre J, Wong R, Rao J, Gimzewski JK. AFM-based analysis of human metastatic cancer cells. Nanotechnology. 2008;19:384003. doi: 10.1088/0957-4484/19/38/384003. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Deshiere A, Duchemin-Pelletier E, Spreux E, Ciais D, Combes F, Vandenbrouck Y, Couté Y, Mikaelian I, Giusiano S, Charpin C, et al. Unbalanced expression of CK2 kinase subunits is sufficient to drive epithelial-to-mesenchymal transition by Snail1 induction. Oncogene. 2013;32:1373–1383. doi: 10.1038/onc.2012.165. [DOI] [PubMed] [Google Scholar]
- Deshiere A, Duchemin-Pelletier E, Spreux E, Ciais D, Forcet C, Cochet C, Filhol O. Regulation of epithelial to mesenchymal transition: CK2β on stage. Mol Cell Biochem. 2011;356:11–20. doi: 10.1007/s11010-011-0942-y. [DOI] [PubMed] [Google Scholar]
- Deshière A, Theis-Febvre N, Martel V, Cochet C, Filhol O. Protein kinase CK2 and cell polarity. Mol Cell Biochem. 2008;316:107–113. doi: 10.1007/s11010-008-9845-y. [DOI] [PubMed] [Google Scholar]
- DiMilla PA, Barbee K, Lauffenburger DA. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys J. 1991;60:15–37. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Kim J. Cellular deconstruction: finding meaning in individual cell variation. Trends Cell Biol. 2015;25:569–578. doi: 10.1016/j.tcb.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov YM, Lomakina ME, Bagrov DV, Makhnovskiy PI, Alexandrova AY, Kirpichnikov MP, Shaitan KV. Mechanical properties of fibroblasts depend on level of cancer transformation. Biochim Biophys Acta. 2014;1843:1013–1019. doi: 10.1016/j.bbamcr.2014.01.032. [DOI] [PubMed] [Google Scholar]
- Feldner JC, Brandt BH. Cancer cell motility—on the road from c-erbB-2 receptor steered signaling to actin reorganization. Exp Cell Res. 2002;272:93–108. doi: 10.1006/excr.2001.5385. [DOI] [PubMed] [Google Scholar]
- Fernández-Sánchez ME, Barbier S, Whitehead J, Béalle G, Michel A, Latorre-Ossa H, Rey C, Fouassier L, Claperon A, Brullé L, et al. Mechanical induction of the tumorigenic β-catenin pathway by tumour growth pressure. Nature. 2015;523:92–95. doi: 10.1038/nature14329. [DOI] [PubMed] [Google Scholar]
- Filhol O, Giacosa S, Wallez Y, Cochet C. Protein kinase CK2 in breast cancer: the CK2β regulatory subunit takes center stage in epithelial plasticity. Cell Mol Life Sci. 2015;72:3305–3322. doi: 10.1007/s00018-015-1929-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MT, Engler AJ, Discher DE, Lee J, Wang Y. Microscopic methods for measuring the elasticity of gel substrates for cell culture: microspheres, microindenters, and atomic force microscopy. Methods Cell Biol. 2007;83:47–65. doi: 10.1016/S0091-679X(07)83003-2. [DOI] [PubMed] [Google Scholar]
- Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr Opin Cell Biol. 2004;16:14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Fritsch A, Höckel M, Kiessling T, Nnetu KD, Wetzel F, Zink M, Käs JA. Are biomechanical changes necessary for tumour progression. Nat Phys. 2010;6:730–732. [Google Scholar]
- Fuhrmann A, Tlsty TD, Engler AJ. Metastatic state of cancer cells may be indicated by attachment strength. Biophys J. 2015;112:736–745. doi: 10.1016/j.bpj.2016.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunciuglio D, Culty M, Fassina G, Masiello L, Melchiori A, Paglialunga G, Arand G, Ciardiello F, Basolo F, Thompson EW. Invasive phenotype of MCF10A cells overexpressing c-Ha-ras and c-erbB-2 oncogenes. Int J Cancer. 1995;63:815–822. doi: 10.1002/ijc.2910630612. [DOI] [PubMed] [Google Scholar]
- Golden D, Cantley LG. Casein kinase 2 prevents mesenchymal transformation by maintaining Foxc2 in the cytoplasm. Oncogene. 2015;34:4702–4712. doi: 10.1038/onc.2014.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsenko PG, Ilina O, Friedl P. Interstitial guidance of cancer invasion. J Pathol. 2012;226:185–199. doi: 10.1002/path.3031. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Herbet M, Salomon A, Feige J-J, Thomas M. Acquisition order of Ras and p53 gene alterations defines distinct adrenocortical tumor phenotypes. PLoS Genet. 2012;8:e1002700. doi: 10.1371/journal.pgen.1002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra I, Undyala V, Kandow C, Thirumurthi U, Dembo M, Beningo KA. An in vitro correlation of mechanical forces and metastatic capacity. Phys Biol. 2011;8:15015. doi: 10.1088/1478-3975/8/1/015015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas O, Mierke CT, Käs JA. Invasive cancer cell lines exhibit biomechanical properties that are distinct from their noninvasive counterparts. Soft Matter. 2011;7:11488. [Google Scholar]
- Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, Levental I, Hawthorne E, Janmey PA, Assoian RK. Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr Biol. 2009;19:1511–1518. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korc M, Friesel RE. The role of fibroblast growth factors in tumor growth. Curr Cancer Drug Targets. 2009;9:639–651. doi: 10.2174/156800909789057006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraning-Rush CM, Califano JP, Reinhart-King CA. Cellular traction stresses increase with increasing metastatic potential. PLoS One. 2012;7:e32572. doi: 10.1371/journal.pone.0032572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiczkova L, Sedlarikova L, Hajek R, Sevcikova S. TGF-beta—an excellent servant but a bad master. J Transl Med. 2012;10:183. doi: 10.1186/1479-5876-10-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoui D, van Overmeire E, de Baetselier P, van Ginderachter JA, Raes G. Functional relationship between tumor-associated macrophages and macrophage colony-stimulating factor as contributors to cancer progression. Front Immunol. 2014;5:1–15. doi: 10.3389/fimmu.2014.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-CW, Lopez-Diaz FJ, Khan SY, Tariq MA, Dayn Y, Vaske CJ, Radenbaugh AJ, Kim HJ, Emerson BM, Pourmand N. Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc Natl Acad Sci USA. 2014;111:E4726–E4735. doi: 10.1073/pnas.1404656111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leight JL, Wozniak MA, Chen S, Lynch ML, Chen CS. Matrix rigidity regulates a switch between TGF-β1-induced apoptosis and epithelial-mesenchymal transition. Mol Biol Cell. 2012;23:781–791. doi: 10.1091/mbc.E11-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Lee GYH, Ong CN, Lim CT. AFM indentation study of breast cancer cells. Biochem Biophys Res Commun. 2008;374:609–613. doi: 10.1016/j.bbrc.2008.07.078. [DOI] [PubMed] [Google Scholar]
- Lo C, Wang H, Dembo M, Wang Y. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinković A, Mih JD, Park J-A, Liu F, Tschumperlin DJ. Improved throughput traction microscopy reveals pivotal role for matrix stiffness in fibroblast contractility and TGF-β responsiveness. Am J Physiol Lung Cell Mol Physiol. 2012;303:L169–L180. doi: 10.1152/ajplung.00108.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiel J-L, Leal A, Kurzawa L, Balland M, Wang I, Vignaud T, Tseng Q, Théry M. Measurement of cell traction forces with ImageJ. Methods Cell Biol. 2015;125:269–287. doi: 10.1016/bs.mcb.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Mcgranahan N, Swanton C. Perspective biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Mendias CL, Gumucio JP, Davis ME, Bromley CW, Davis CS, Brooks SV. Transforming growth factor-beta induces skeletal muscle atrophy and fibrosis through the induction of atrogin-1 and scleraxis. Muscle Nerve. 2012;45:55–59. doi: 10.1002/mus.22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz AF, Banerjee S, Che Y, German GK, Xu Y, Hyland C, Marchetti MC, Horsley V, Dufresne ER. Scaling of traction forces with size of cohesive cell colonies. Phys Rev Lett. 2012;108:198101. doi: 10.1103/PhysRevLett.108.198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AS, Hughes-Alford SK, Kay JE, Castillo A, Wells A, Gertler FB, Lauffenburger DA. 2D protrusion but not motility predicts growth factor-induced cancer cell migration in 3D collagen. J Cell Biol. 2012;197:721–729. doi: 10.1083/jcb.201201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke CT. The integrin alphav beta3 increases cellular stiffness and cytoskeletal remodeling dynamics to facilitate cancer cell invasion. New J Phys. 2013;15:015003. [Google Scholar]
- Mierke CT, Bretz N, Altevogt P. Contractile forces contribute to increased GPI-anchored receptor CD24 facilitated cancer cell invasion. J Biol Chem. 2011b;286:34858–34871. doi: 10.1074/jbc.M111.245183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke CT, Frey B, Fellner M, Herrmann M, Fabry B. Integrin α5β1 facilitates cancer cell invasion through enhanced contractile forces. J Cell Sci. 2011a;124:369–383. doi: 10.1242/jcs.071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munevar S. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J. 2001;80:1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- Oakes PW, Banerjee S, Marchetti MC, Gardel ML. Geometry regulates traction stresses in adherent cells. Biophys J. 2014;107:825–833. doi: 10.1016/j.bpj.2014.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes PW, Patel DC, Morin NA, Zitterbart DP, Fabry B, Reichner JS, Tang JX. Neutrophil morphology and migration are affected by substrate elasticity. Blood. 2009;114:1387–1395. doi: 10.1182/blood-2008-11-191445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Piekiełko-Witkowska A. Molecular mechanism of thyroid hormone action in carcinogenesis. Thyroid Res. 2013;6:A48. [Google Scholar]
- Plodinec M, Loparic M, Monnier CA, Obermann EC, Zanetti-Dallenbach R, Oertle P, Hyotyla JT, Aebi U, Bentires-Alj M, Lim RYH, et al. The nanomechanical signature of breast cancer. Nat Nanotechnol. 2012;7:757–765. doi: 10.1038/nnano.2012.167. [DOI] [PubMed] [Google Scholar]
- Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, Keely PJ. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzano PP, Eliceiri KW, Inman DR, Keely PJ. Engineering three-dimensional collagen matrices to provide contact guidance during 3D cell migration. Curr Protoc Cell Biol 2010(Jun) 2010 doi: 10.1002/0471143030.cb1017s47. Chapter 10, Unit 10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart-King CA, Dembo M, Hammer DA. The dynamics and mechanics of endothelial cell spreading. Biophys J. 2005;89:676–689. doi: 10.1529/biophysj.104.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösel D, Brábek J, Tolde O, Mierke CT, Zitterbart DP, Raupach C, Bicanová K, Kollmannsberger P, Panková D, Vesely P, et al. Up-regulation of Rho/ROCK signaling in sarcoma cells drives invasion and increased generation of protrusive forces. Mol Cancer Res. 2008;6:1410–1420. doi: 10.1158/1541-7786.MCR-07-2174. [DOI] [PubMed] [Google Scholar]
- Rzymski P, Skórzewska A, Skibin´ska-Zielin´ska M, Opala T. Factors influencing breast elasticity measured by the ultrasound shear wave elastography - preliminary results. Arch Med Sci. 2011;7:127–133. doi: 10.5114/aoms.2011.20617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MS, Lopez JI, McGhee EJ, Croft DR, Strachan D, Timpson P, Munro J, Schröder E, Zhou J, Brunton VG, et al. Actomyosin-mediated cellular tension drives increased tissue stiffness and β-catenin activation to induce epidermal hyperplasia and tumor growth. Cancer Cell. 2011;19:776–791. doi: 10.1016/j.ccr.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seton-Rogers SE, Lu Y, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, Brugge JS. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc Natl Acad Sci USA. 2004;101:1257–1262. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer KSR, Graus-Porta D, Leng J, Hynes NE, Klemke RL. ErbB2 is necessary for induction of carcinoma cell invasion by ErbB family receptor tyrosine kinases. J Cell Biol. 2000;148:385–397. doi: 10.1083/jcb.148.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan V, Mythreye K, O’Brien ET, Berchuck A, Blobe GC, Superfine R. Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res. 2011;71:5075–5080. doi: 10.1158/0008-5472.CAN-11-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignaud T, Ennomani H, Théry M. Polyacrylamide hydrogel micropatterning. Methods Cell Biol. 2014;120:93–116. doi: 10.1016/B978-0-12-417136-7.00006-9. [DOI] [PubMed] [Google Scholar]
- Vilmont V, Filhol O, Hesse A-M, Couté Y, Hue C, Rémy-Tourneur L, Mistou S, Cochet C, Chiocchia G. Modulatory role of the anti-apoptotic protein kinase CK2 in the sub-cellular localization of Fas associated death domain protein (FADD) Biochim Biophys Acta. 2015;1853:2885–2896. doi: 10.1016/j.bbamcr.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Wang N, Ostuni E, Whitesides GM, Ingber DE. Micropatterning tractional forces in living cells. Cell Motil Cytoskeleton. 2002;52:97–106. doi: 10.1002/cm.10037. [DOI] [PubMed] [Google Scholar]
- Weder G, Hendriks-Balk MC, Smajda R, Rimoldi D, Liley M, Heinzelmann H, Meister A, Mariotti A. Increased plasticity of the stiffness of melanoma cells correlates with their acquisition of metastatic properties. Nanomed Nanotechnol Biol Med. 2014;10:141–148. doi: 10.1016/j.nano.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Wu T-H, Li C-H, Tang M-J, Liang J-I, Chen C-H, Yeh M-L. Migration speed and directionality switch of normal epithelial cells after TGF-β1-induced EMT (tEMT) on micro-structured polydimethylsiloxane (PDMS) substrates with variations in stiffness and topographic patterning. Cell Commun Adhes. 2013;20:115–126. doi: 10.3109/15419061.2013.833194. [DOI] [PubMed] [Google Scholar]
- Xu W, Mezencev R, Kim B, Wang L, McDonald J, Sulchek T. Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells. PLoS One. 2012;7:e46609. doi: 10.1371/journal.pone.0046609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Krishnamurthy K, Allred DC, Muthuswamy SK. Loss of Par3 promotes breast cancer metastasis by compromising cell-cell cohesion. Nat Cell Biol. 2013;15:189–200. doi: 10.1038/ncb2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan P, Wang J, Lv X, Wang Q, Qiu L, Lin X, Yu L, Song Y. Prognostic value of vascular endothelial growth factor expression in patients with lung cancer: a systematic review with meta-analysis. J Thorac Oncol. 2009;4:1094–1103. doi: 10.1097/JTO.0b013e3181a97e31. [DOI] [PubMed] [Google Scholar]