RABL2 interacts with the intraflagellar transport-B (IFT-B) complex and CEP19 in a mutually exclusive manner. A point mutation of RABL2 found in sperm motility–defective mice abolishes its binding to IFT-B but not to CEP19. A RABL2-defective Chlamydomonas strain exhibits a nonflagellated phenotype, suggesting a crucial role of RABL2 in ciliary assembly.
Abstract
Proteins localized to the basal body and the centrosome play crucial roles in ciliary assembly and function. Although RABL2 and CEP19 are conserved in ciliated organisms and have been implicated in ciliary/flagellar functions, their roles are poorly understood. Here we show that RABL2 interacts with CEP19 and is recruited to the mother centriole and basal body in a CEP19-dependent manner and that CEP19 is recruited to the centriole probably via its binding to the centrosomal protein FGFR1OP. Disruption of the RABL2 gene in Chlamydomonas reinhardtii results in the nonflagellated phenotype, suggesting a crucial role of RABL2 in ciliary/flagellar assembly. We also show that RABL2 interacts, in its GTP-bound state, with the intraflagellar transport (IFT)-B complex via the IFT74–IFT81 heterodimer and that the interaction is disrupted by a mutation found in male infertile mice (Mot mice) with a sperm flagella motility defect. Intriguingly, RABL2 binds to CEP19 and the IFT74–IFT81 heterodimer in a mutually exclusive manner. Furthermore, exogenous expression of the GDP-locked or Mot-type RABL2 mutant in human cells results in mild defects in ciliary assembly. These results indicate that RABL2 localized to the basal body plays crucial roles in ciliary/flagellar assembly via its interaction with the IFT-B complex.
INTRODUCTION
The centrosome is an organelle that is assembled from a pair of centrioles arranged in an L-shaped configuration and surrounded by a dense matrix of pericentriolar material. In interphase cells, the centrosome is located near the nucleus and serves as an organizing center for intracellular microtubules, and in the mitotic phase, bipolar mitotic spindles assemble on the duplicated centrosomes (Ishikawa and Marshall, 2011). When cells enter the G0 phase, one of the centrioles serves as the basal body beneath the plasma membrane to project the cilium. This hair-like structure is categorized into two types: motile cilia, which are present in limited types of cells, such as those of airway epithelial cells, ventricle ependymal cells, and sperm flagella; and immotile cilia (also called primary cilia), which are present in almost all cells and serve as cellular antennae to receive and transduce a wide variety of extracellular signals. The assembly and function of cilia require the selective trafficking of various proteins into and out of cilia, including tubulins and signaling proteins. Ciliary protein trafficking is mediated by the intraflagellar transport (IFT) machinery, often referred to as the IFT train, which is composed of the IFT-B complex, involved in anterograde trafficking, and the IFT-A complex, involved in retrograde trafficking (Ishikawa and Marshall, 2011; Sung and Leroux, 2013; Taschner and Lorentzen, 2016). Defects in the assembly and function of cilia, including those in the IFT machinery, cause various genetic diseases collectively called ciliopathies. These include Bardet–Biedl syndrome (BBS), Joubert syndrome (JBTS), Meckel syndrome, and nephronophthisis, with a wide spectrum of symptoms, such as polycystic kidney, retinal degeneration, obesity, infertility, and brain and skeletal malformation (Schwartz et al., 2011; Brown and Witman, 2014; Madhivanan and Aguilar, 2014). The molecular basis of these phenotypes is largely unknown.
Small GTPases belonging to the ARF/ARL and RAB families function as molecular switches by cycling between a GDP-bound, inactive state and a GTP-bound, active state and directly or indirectly regulate various cellular processes, including protein trafficking and cell division. Various ARF/ARL and RAB GTPases also play crucial roles in centrosomal functions and ciliary protein trafficking. These include ARF4, ARL2, ARL3, ARL6/BBS3, ARL13B/JBTS8, RAB8, RAB10, RAB11, RAB23, and RAB28 (Li and Hu, 2011; Lim et al., 2011). There are also RAB-like (RABL) GTPases, which are similar to RAB GTPases but lack a canonical C-terminal isoprenylation site found in the RAB GTPases (Rojas et al., 2012). Among them, RABL4 and RABL5, also known as IFT27/BBS19 and IFT22, respectively, are components of the IFT-B complex (Li and Hu, 2011; Lim et al., 2011; Katoh et al., 2016; see later discussion of Figure 8L); RABL4/IFT27/BBS19 has been proposed to serve as a functional linker between two multiprotein complexes involved in ciliary protein trafficking, namely, the IFT-B and BBSome (Eguether et al., 2014; Liew et al., 2014).
FIGURE 8:
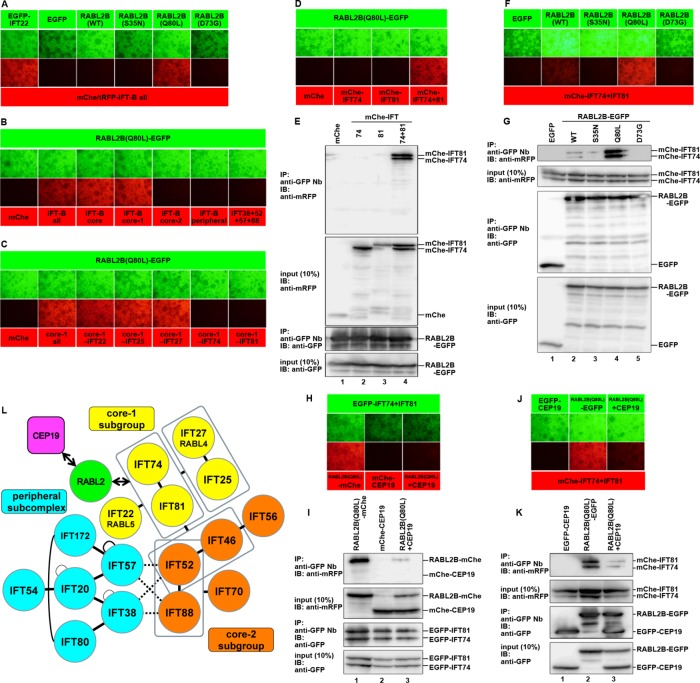
Mutually exclusive interactions of RABL2 with the IFT-B complex and CEP19. (A) Interaction of RABL2B(Q80L) with the IFT-B complex demonstrated by the VIP assay. An expression vector for EGFP-IFT22 (as a positive control), EGFP (as a negative control), or an EGFP-tagged RABL2B construct, as indicated, was cotransfected with expression vectors for all IFT-B subunits tagged with mChe or tRFP into HEK293T cells. Cell lysates were immunoprecipitated with GST–anti-GFP Nb prebound to glutathione–Sepharose beads and observed under a microscope as described in Materials and Methods. (B) Interaction of RABL2B(Q80L) with the IFT-B core 1 subunits. Lysates prepared from HEK293T cells coexpressing RABL2B(Q80L)-EGFP and all IFT-B, all core, all core 1, all core 2, all peripheral, or all connecting (IFT38/IFT52/IFT57/IFT88) subunits tagged with mChe or tRFP were processed for the VIP assay as described. (C) Interaction of RABL2B(Q80L) with IFT74 and IFT81 demonstrated by the subtractive VIP assay. Lysates of HEK293T cells coexpressing RABL2B(Q80L)-EGFP and all but one (as indicated) of the IFT-B core 1 subunits tagged with mChe were processed for the VIP assay. (D, E) Interaction of RABL2B(Q80L) with the IFT74–IFT81 dimer. Lysates from HEK293T cells coexpressing RABL2B(Q80L)-EGFP and mChe-IFT74 or mChe-IFT81 alone or both mChe-IFT74 and mChe-IFT81 were processed for VIP assay (D) or immunoblotting analysis with an anti-mRFP or anti-GFP antibody (E). (F, G) Interaction of RABL2 with the IFT74–IFT81 dimer in its GTP-bound state. HEK293T cells coexpressing mChe-IFT74+IFT81 and EGFP-tagged RABL2B(WT), RABL2B(S35N), RABL2B(Q80L), or RABL2B(D73G) were lysed and processed for the VIP assay (F) or immunoblotting analysis with an anti-mRFP or anti-GFP antibody (G). (H–K) Mutually exclusive interactions of RABL2 with the IFT-B complex and CEP19. Lysates prepared from HEK293T cells coexpressing EGFP-IFT74+81 and either RABL2B(Q80L)-mChe or mChe-CEP19 alone, or both RABL2B(Q80L)-mChe and mChe-CEP19 (H, I), or those coexpressing reciprocal combinations of the EGFP and mChe tags (J and K) were processed for the VIP assay (H, J) or immunoblotting analysis with an anti-mRFP or anti-GFP antibody (I, K). (L) Schematic representation of the overall architecture of the IFT-B complex and interactions of RABL2 with CEP19 and the IFT-B complex.
Here we focus on RABL2 for the following reasons: 1) the RABL2 gene is conserved in species that have motile cilia/flagella, and it was reported that the expression level of RABL2 is higher in ciliated than in nonciliated cells (Hoh et al., 2012); 2) male mice with a point mutation in RABL2 (Mot mice) were reported to be sterile as a consequence of severely compromised sperm motility (Lo et al., 2012), and RABL2 variants have been implicated in infertility in men (Jamsai et al., 2014); 3) RABL2 was reported to interact with the +TIP protein EB1 and some components of the IFT-B complex (Lo et al., 2012); 4) the results of a comprehensive yeast two-hybrid analysis (Rual et al., 2005) suggested a potential RABL2-interacting protein, C3ORF34, which was shown to localize to the centrosome and was therefore renamed as centrosomal protein of 19 kDa (CEP19; Jakobsen et al., 2011); and finally, 5) after we started this study, CEP19 deficiency in humans and mice was reported to cause sperm defects and morbid obesity (Shalata et al., 2013), both of which are typical symptoms of ciliopathies (Schwartz et al., 2011; Brown and Witman, 2014).
In this study, we show that RABL2 and CEP19 indeed interact with each other and colocalize to the mother centriole and the basal body. In addition, we show that CEP19 recruits RABL2 to the centriole and that CEP19 is recruited to the centrosome via its interaction with fibroblast growth factor receptor 1 oncogene partner (FGFR1OP, also known as FOP), which is another centrosomal protein. We then find that a mutant of Chlamydomonas reinhardtii with a disrupted RABL2 gene lacks flagella. Furthermore, we show that RABL2, in its GTP-bound state, can interact with the IFT-B complex via the IFT74–IFT81 dimer and that the interaction is abolished by the point mutation found in Mot mice. Furthermore, the exogenous expression of a GDP-bound or Mot-type RABL2 mutant significantly inhibits ciliogenesis. On the basis of these data, we propose that RABL2 localizes to the mother centriole/basal body, where it plays a crucial role in ciliary assembly by regulating IFT-B function in some way.
RESULTS
CEP19 binds to RABL2 via its C-terminal region
RABL2 is highly conserved in ciliated organisms, including humans, mice, zebrafish, and Chlamydomonas (Eliáš et al., 2016; Supplemental Figure S1A); humans have two virtually identical paralogues (Kramer et al., 2010; Eliáš et al., 2016), RABL2A and RABL2B (97% identity at the amino acid level), whereas mice have a single RABL2 gene. However, the RABL2 gene is not found in Drosophila melanogaster or Caenorhabditis elegans (Eliáš et al., 2016). Similarly, the CEP19 gene is conserved in humans, mice, zebrafish, and Chlamydomonas (Supplemental Figure S1B) but is not found in D. melanogaster or C. elegans; note that Chlamydomonas sequences deposited as two separate genes (XM_001690851 and XM_001690852) actually constitute a single CEP19 gene and that the Chlamydomonas protein predicted from the cDNA cloned by reverse transcription (RT)–PCR using a set of primers for the 5′- and 3′-terminal sequences of XM_001690851 and XM_001690852, respectively (Supplemental Table S3), has a large insertion in the middle region relative to the CEP19 protein of other species (Supplemental Figure S1B). Thus conservation of the RABL2 and CEP19 genes during evolution appears to have occurred in parallel.
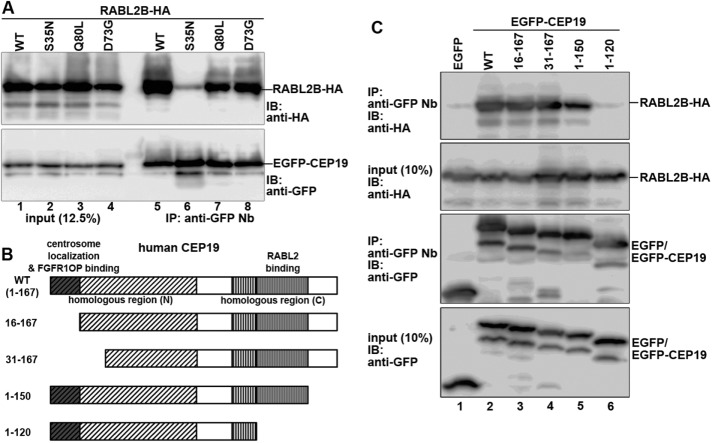
Although the database of a comprehensive yeast two-hybrid analysis suggested that RABL2 can interact with CEP19 (Rual et al., 2005), two-hybrid analyses often yield false-positive results. To verify the potential interaction between RABL2 and CEP19, we transiently coexpressed enhanced green fluorescent protein (EGFP)–CEP19 and RABL2B-HA in HEK293T cells and subjected lysates prepared from the cells to immunoprecipitation with a glutathione S-transferase (GST)-tagged anti-GFP nanobody (Nb); note that we used human RABL2B in this study because only RABL2B cDNA was obtained by PCR amplification of a human testis cDNA library using a set of primers with sequences common to both the RABL2A and RABL2B genes. As shown in Figure 1A, wild-type (WT) RABL2B-HA was coimmunoprecipitated with EGFP-CEP19 (lane 5). Because RABL2 has GTP-binding motifs that are conserved among the small GTPases, we then constructed RABL2B mutants according to previous studies for other GTPases: the Q80L mutant, which is predicted to be locked in a GTP-bound state, and the S35N mutant, which is predicted to be a GDP-bound form. We also constructed the RABL2B(D73G) mutant because a corresponding mutation in male mice (Mot mice) was reported to cause infertility (Lo et al., 2012). When we performed a coimmunoprecipitation assay using these RABL2 mutants, we found RABL2B(Q80L) to interact with CEP19 (Figure 1A, lane 7), similarly to RABL2B(WT) (lane 5), whereas RABL2B(S35N) did not interact with CEP19 (lane 6), indicating that RABL2 binds to CEP19 in its GTP-bound state. On the other hand, RABL2B(D73G) interacted with CEP19 (lane 8). These results indicate that the phenotype of male sterility in Mot mice is not caused by the inability of RABL2 to interact with CEP19.
FIGURE 1:
Interaction between RABL2 and CEP19. (A) Interaction of RABL2 with CEP19. HEK293T cells were transiently cotransfected with expression vectors for EGFP-CEP19 and RABL2B(WT)-HA or its mutant (S35N, Q80L, or D73G). At 24 h after the transfection, lysates were prepared from the transfected cells and immunoprecipitated with GST-fused anti–GFP Nb prebound to glutathione–Sepharose 4B beads. Proteins bound to the precipitated beads were subjected to SDS–PAGE and immunoblotting analysis using anti-HA or anti-GFP antibodies. (B) Schematic representation of the structures of CEP19 and its deletion constructs. (C) RABL2 interacts with the C-terminal region of CEP19. Lysates prepared from HEK293T cells transfected with expression vectors for RABL2B-HA and EGFP, or EGFP-tagged CEP19(WT) or its deletion construct, as indicated, were processed for immunoprecipitation with GST–anti-GFP Nb, followed by immunoblotting analysis, as described for A.
We then set out to determine the region of CEP19 that is responsible for its interaction with RABL2. To this end, we constructed various deletion mutants of CEP19 (Figure 1B) and coexpressed them with RABL2B, followed by the coimmunoprecipitation assay as described earlier. Truncation of up to the first N-terminal 30 amino acids of CEP19 did not affect its ability to bind to RABL2B (Figure 1C; compare lanes 3 and 4 with lane 2). When 17 amino acids were truncated from the C-terminus, the construct, CEP19(1-150), retained the ability to interact with RABL2B (lane 5). In considerable contrast, C-terminal truncation of 47 amino acid residues from the C-terminus, CEP19(1-120), abolished the RABL2B-binding ability of CEP19 (lane 6). These results indicate that the 30–amino acid region (residues 121–150) of CEP19 participates in its binding to RABL2; note that this region is highly conserved between human and Chlamydomonas (Supplemental Figure S1B).
RABL2 and CEP19 are colocalized to the centrosome and basal body
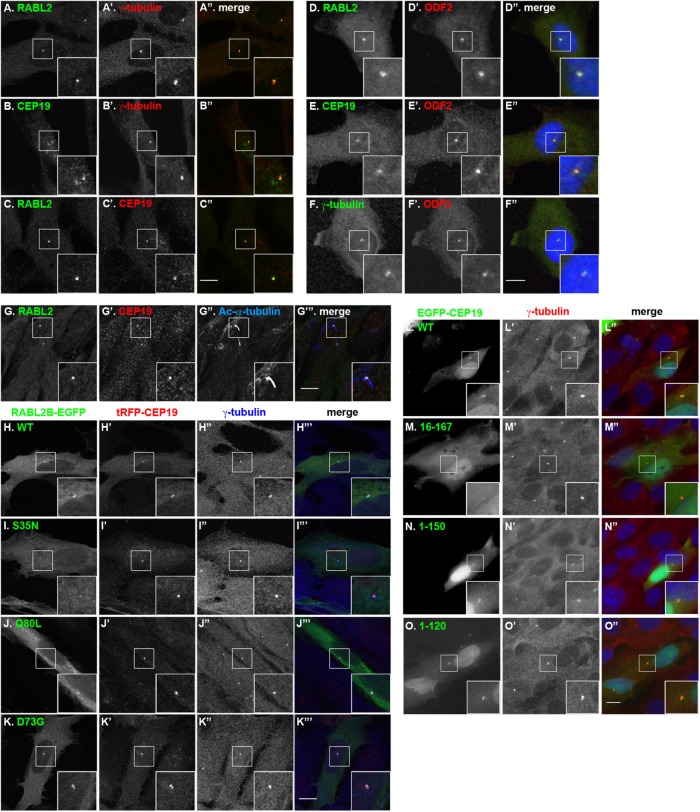
Although a previous study showed that CEP19 is localized to the centrosome (Jakobsen et al., 2011), the intracellular localization of RABL2 has not been examined. We therefore set out to compare the localization of RABL2 and CEP19 in human telomerase reverse transcriptase-immortalized retinal pigment epithelial (hTERT-RPE1) cells using antibodies against RABL2, CEP19, and γ-tubulin (a centrosomal marker). As shown in Figure 2, A–A′′ and B–B′′, RABL2 and CEP19 localized to one of two centriolar structures positive for γ-tubulin; note that although the anti-CEP19 antibody used in this study does not work well in immunoblotting experiments for an unknown reason (unpublished data), the antibody was able to detect the centriolar localization of the CEP19 protein in immunofluorescence analysis of hTERT-RPE1 cells, as the centriolar signals detected with the antibody are abolished in CEP19-knockout (KO) cells (see later discussion of Figure 5, A–C). When RABL2 and CEP19 were double stained for outer dense fiber protein 2 (ODF2), which is a marker for the mother centriole, the staining for both RABL2 (Figure 2, D–D′′) and CEP19 (Figure 2, E–E′′) was superimposed with the ODF2 staining, indicating that both RABL2 and CEP19 localize to the mother centriole. When hTERT-RPE1 cells were cultured under serum-starved conditions to induce ciliogenesis, RABL2 and CEP19 signals were found at the base of cilia depicted by staining for acetylated α-tubulin (Ac-α-tubulin; Figure 2, G–G′′′), indicating the colocalization of RABL2 and CEP19 to the basal body.
FIGURE 2:
Colocalization of RABL2 and CEP19 at the mother centriole and the ciliary base. (A–C) Localization of RABL2 and CEP19 at one of the centrioles. hTERT-RPE1 cells were double stained for RABL2 and γ-tubulin (A–A′′), CEP19 and γ-tubulin (B–B′′), or RABL2 and CEP19(C–C′′). (D–F) Colocalization of RABL2 and CEP19 with ODF2. hTERT-RPE1 cells were double stained for RABL2 and ODF2 (D–D′′), CEP19 and ODF2 (E–E′′), or ODF2 and γ-tubulin (F–F′′). (G–G′′′) Colocalization of RABL2 and CEP19 at the ciliary base. hTERT-RPE1 cells were cultured under serum-starved conditions for 24 h and processed for triple immunostaining for RABL2, CEP19, and Ac-α-tubulin. (H–K) Localization of exogenously expressed RABL2B and its mutants. hTERT-RPE1 cells were transiently cotransfected with expression vectors for tRFP-CEP19 and RABL2B(WT)-EGFP (H–H′′′) or its mutant, S35N (I–I′′′), Q80L (J–J′′′), or D73G (K–K′′′) and stained with an anti–γ-tubulin antibody. (L–O) Localization of CEP19 deletion constructs. hTERT-RPE1 cells were transfected with an expression vector for EGFP-tagged CEP19(WT) (L–L′′) or its deletion construct, as indicated (L–L′′, M–M′′, N–N′′, or O–O′′), and stained with an anti–γ-tubulin antibody. Insets. enlarged images of the boxed regions. Scale bars, 10 µm.
FIGURE 5:
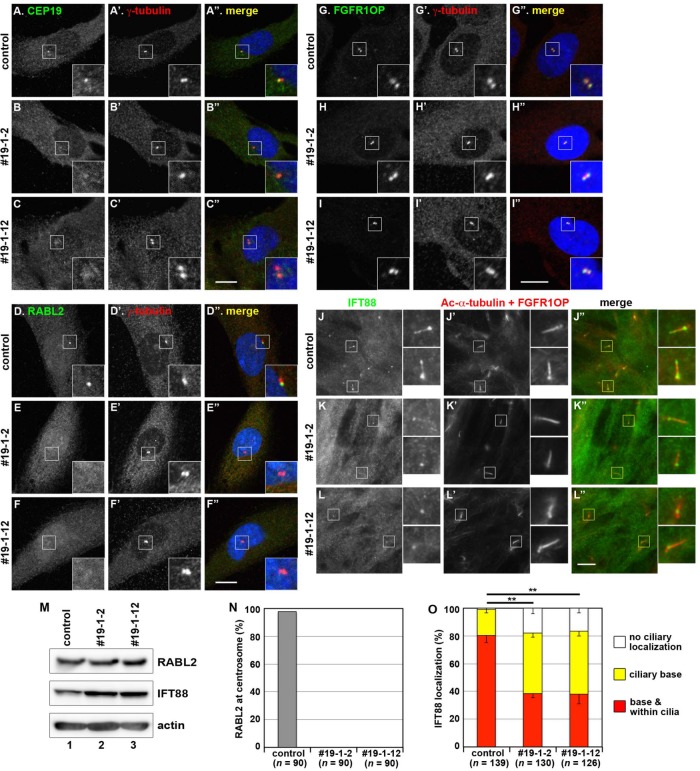
Loss of centriolar localization of RABL2 in CEP19-KO cells. Control RPE1 cells (A, D, G, J) and the CEP19-KO cell lines 19-1-2 (B, E, H, K) and 19-1-12 (C, F, I, L) were double immunostained for CEP19 (A–C), RABL2 (D–F), or FGFR1OP (G–I) and γ-tubulin (A′–I′) or triple immunostained for IFT88 (J–L) and Ac-α-tubulin plus FGFR1OP (J′–L′). Insets, enlarged images of the boxed regions. Scale bars, 10 µm. (M) Lysates prepared from control RPE1 cells (lane 1) or the CEP19-KO cell line 19-1-2 (lane 2) or 19-1-12 (lane 3) were processed for immunoblotting analysis using antibody against RABL2 (top), IFT88 (middle), or actin (bottom). (N) Cells with centrosomal RABL2 signals in the experiments in D–F were counted, and the percentages of RABL2-positive cells in each condition are shown as a bar graph. Values are means of three independent experiments. In each set of experiments, 30 cells were analyzed, and the total number of analyzed cells (n) is shown. (O) Localization of IFT88 in individual control and CEP19-KO cells was classified as “ciliary base + within cilia,” “mainly ciliary base,” and “no ciliary localization” and counted. The percentages of these populations are expressed as stacked bar graphs. Values are means ± SE of three independent experiments. In each set of experiments, 35–49 ciliated cells were observed, and the total number of ciliated cells observed (n) is shown. **p < 0.0001 (Pearson’s χ2 test).
When RABL2B(WT) fused to EGFP and CEP19 fused to TagRFP (tRFP; a monomeric red fluorescent protein [mRFP]) were transiently coexpressed in hTERT-RPE1 cells, these exogenously expressed proteins were colocalized on the centrosome (Figure 2, H–H′′′). Note that when RABL2B-EGFP was exogenously expressed in the absence of coexpressed CEP19, it showed a tendency to be distributed throughout the cytoplasm (e.g., see later discussion of Figure 9H); this was probably due to the limited level of endogenous CEP19. We also examined the localization of EGFP-tagged RABL2B(S35N), RABL2B(Q80L), and RABL2B(D73G). In parallel with their ability to interact with CEP19, RABL2B(Q80L) and RABL2B(D73G) were colocalized with tRFP-CEP19 on the centrosome (Figure 2, J–J′′′ and K–K′′′, respectively). By contrast, the distinct localization of RABL2B(S35N) to the centrosome was not observed (Figure 2, I–I′′′), although tRFP-CEP19 was localized to the centrosome. These observations suggest that RABL2 localizes to the centrosome via its interaction with CEP19, whereas the centrosomal localization of CEP19 is independent of RABL2.
FIGURE 9:
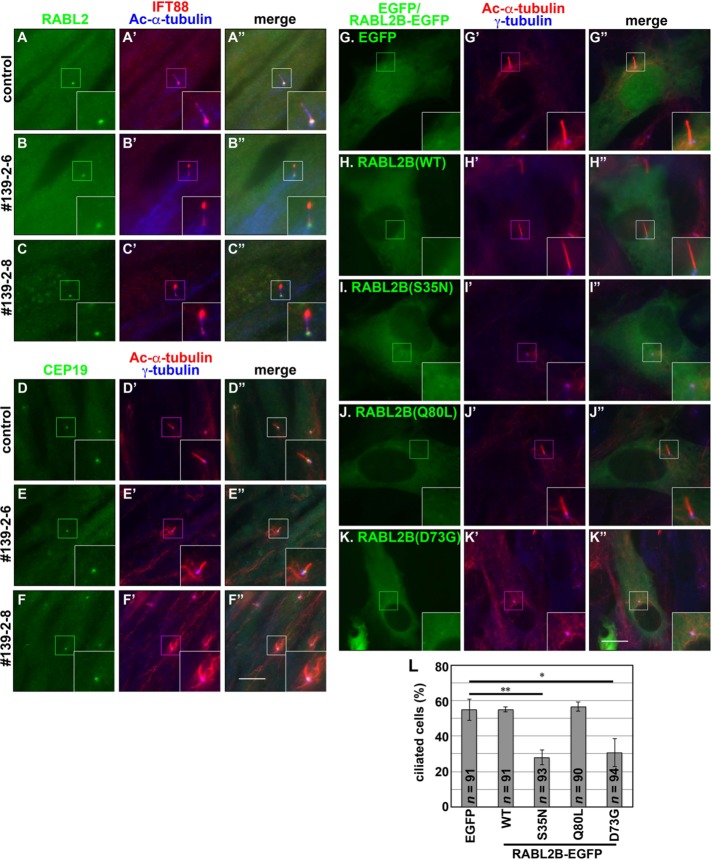
RABL2 participates in ciliary assembly. (A–F) RABL2 does not accumulate at the ciliary tip regardless of impaired retrograde trafficking. Control hTERT-RPE1 cells (A, D) or the IFT139-KO cell line 139-2-6 (B, E) or 139-2–8 (C, F) was cultured under serum-starved conditions to induce ciliogenesis and triple immunostained for RABL2 (A–C), IFT88, and Ac-α-tubulin (A′–C′), or for CEP19 (D–F), Ac-α-tubulin, and γ-tubulin (D′–F′). Insets, enlarged images of the boxed regions. Scale bars, 10 µm. (G–L) Exogenous expression of RABL2 mutants defective in IFT-B binding suppress ciliogenesis. hTERT-RPE1 cells were transfected with an expression vector for EGFP (G) or EGFP-tagged RABL2B(WT) (H), RABL2B(S35N) (I), RABL2B(Q80L) (J), or RABL2B(D73G) (K) and cultured for 12 h under normal conditions. The cells were then cultured under serum-starved conditions for 12 h and processed for immunostaining with antibodies against Ac-α-tubulin and γ-tubulin (G′–K′). (L) Ciliated cells in the experiments in G–K were counted, and percentages of ciliated cells are expressed as a bar graph. Values are means ± SE of three independent experiments. In each set of experiments, 30–33 cells were analyzed, and the total number of cells analyzed for each condition (n) is shown. **p < 0.005; *p < 0.02 (Student’s t test).
It is noteworthy that, unlike endogenous proteins, exogenously expressed RABL2 and CEP19 demonstrated a tendency to localize to both centrioles (see Discussion).
The N-terminal region of CEP19 is required for its localization to the centrosome and binding to FGFR1OP
We next investigated the region of the CEP19 protein that is required for its centrosomal localization, using the deletion mutants that were used for the RABL2-binding experiments (Figure 1, B and C). Like CEP19(WT) tagged with EGFP (Figure 2, L–L′′), the C-terminal deletion constructs EGFP-CEP19(1-150) and EGFP-CEP19(1-120) were localized to the centrosome when expressed in RPE1 cells (Figure 2, N–N′′ and O–O′′, respectively). By contrast, the N-terminal deletion construct EGFP-CEP19(16-167) did not show a distinct centrosomal localization but was distributed throughout the cytoplasm (Figure 2, M–M′′); we also attempted to examine the localization of EGFP-CEP19(31-167) but were unsuccessful because this construct was highly cytotoxic when expressed in RPE1 cells. These observations indicate that at least the N-terminal 15–amino acid region of CEP19 is essential for its localization to the centrosome and that RABL2 binding is not required for the centrosomal localization of CEP19.
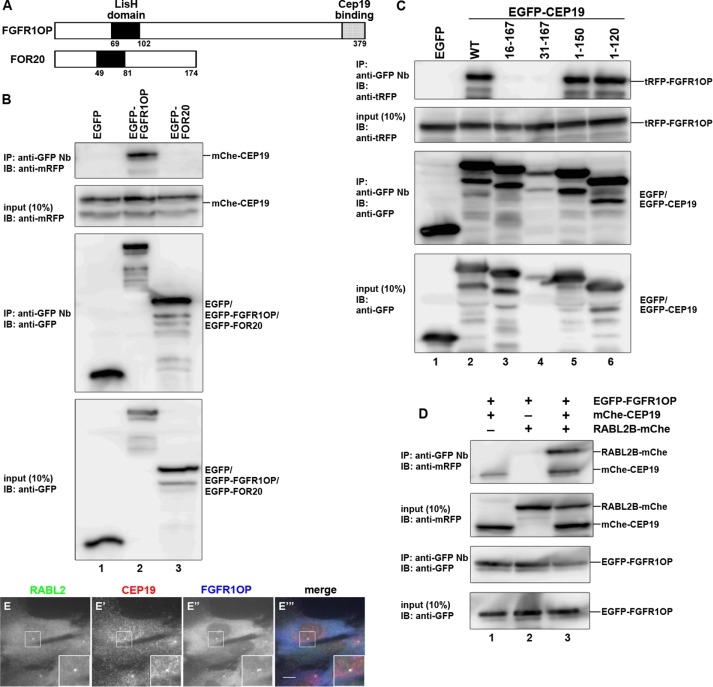
Because FGFR1OP was suggested to be localized to the centrosome and involved in ciliogenesis (Yan et al., 2006; Lee and Stearns, 2013) and three independent interactome analyses suggested an interaction between CEP19 and FGFR1OP (Rolland et al., 2014; Gupta et al., 2015; Huttlin et al., 2015), we then addressed the possibility that CEP19 and FGFR1OP associate with each other. We also analyzed whether FOR20 (also known as FOPNL) interacts with CEP19, because the N-terminal region of FGFR1OP is homologous to FOR20, particularly in the Lis1-homology (LisH) domain (Figure 3A), and FOR20 was also reported to be localized to the centrosome and crucial for ciliogenesis (Sedjaï et al., 2010). As shown in Figure 3B, mCherry (mChe)-tagged CEP19 was coimmunoprecipitated with EGFP-FGFR1OP but not with EGFP-FOR20 when expressed in HEK293T cells.
FIGURE 3:
Interaction between RABL2, CEP19, and FGFR1OP. (A) Schematic representation of the structures of FGFR1OP and FOR20. (B) Interaction of CEP19 with FGFR1OP. HEK293T cells were transiently cotransfected with expression vectors for mChe-CEP19 and either EGFP, EGFP-FGFR1OP, or EGFP-FOR20. At 24 h after the transfection, lysates were prepared from the transfected cells and immunoprecipitated with GST–anti-GFP Nb, followed by immunoblotting analysis with antibodies against mRFP or EGFP. (C) FGFR1OP interacts with the CEP19 N-terminal region. Lysates prepared from HEK293T cells transfected with expression vectors for tRFP-FGFR1OP and EGFP, or EGFP-tagged CEP19(WT) or its deletion construct, as indicated, were subjected to immunoprecipitation with GST–anti-GFP Nb, followed by immunoblotting analysis with anti-tRFP or anti-GFP antibodies. (D) Tripartite interaction of RABL2, CEP19, and FGFR1OP. hTERT-RPE1 cells were transfected with expression vectors for EGFP-FGFR1OP and either mChe-CEP19 or RABL2-mChe, or both mChe-CEP19 and RABL2-mChe. Lysates from the transfected cells were subjected to immunoprecipitation with GST–anti-GFP Nb, followed by immunoblotting analysis with anti-mRFP or anti-GFP antibodies. (E–E′′′) Colocalization of RABL2, CEP19, and FGFR1OP at one of the centrioles. hTERT-RPE1 cells were triple stained for RABL2, CEP19, and FGFR1OP. RABL2 and CEP19 were colocalized to one of the two FGFR1OP-positive centrioles. Insets, enlarged images of the boxed regions. Scale bar, 10 µm.
We then analyzed which region of CEP19 participates in FGFR1OP binding. As shown in Figure 3C, the C-terminal deletion constructs CEP19(1-150) and CEP19(1-120) interacted with FGFR1OP (lanes 5 and 6), like the WT construct (lane 2). In clear contrast, the N-terminal deletion constructs CEP19(16-167) and CEP19(31-167) lost the ability to interact with FGFR1OP (lanes 3 and 4). Thus the ability of CEP19 to interact with FGFR1OP is in parallel with its ability to localize to the centrosome, making it likely that CEP19 is recruited to the centrosome via its interaction with FGFR1OP.
To examine whether RABL2, CEP19, and FGFR1OP form a ternary complex, we coexpressed EGFP-tagged FGFR1OP with either CEP19 or RABL2B tagged with mChe or both mChe-CEP19 and RABL2B-mChe in HEK293T cells, immunoprecipitated cell lysates with GST-tagged anti-GFP Nb, and processed them for immunoblotting analysis with anti-mRFP antibodies that recognize mChe. As shown in Figure 3D, RABL2B-mChe was not coprecipitated with EGFP-FGFR1OP in the absence of mChe-CEP19 (lane 2). In striking contrast, RABL2B-mChe was robustly coprecipitated with EGFP-FGFR1OP in the presence of mChe-CEP19 (lane 3). These results demonstrate that the FGFR1OP–CEP19–RABL2 ternary complex can be formed in cells.
As shown in Figure 3, E–E′′′, FGFR1OP staining was found on both centrioles, as previously reported (Lee and Stearns, 2013). By contrast, both RABL2 and CEP19 were colocalized on one of the two FGFR1OP-positive centrioles. These observations suggest the existence of a determinant for the mother centriole–specific localization of CEP19 in addition to its binding to FGFR1OP (see Discussion).
FGFR1OP interacts with CEP19 through its C-terminal region
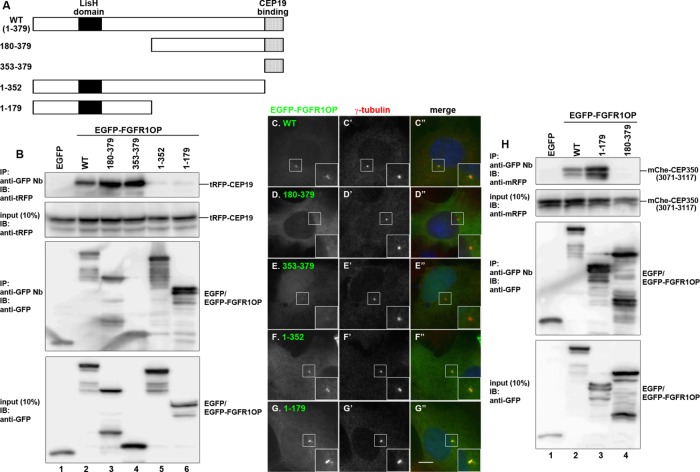
We then investigated the region of FGFR1OP involved in CEP19 binding. Previous studies indicated that the N-terminal region of FGFR1OP containing the LisH domain determines its centrosomal localization (Figure 4A; Mikolajka et al., 2006) and interacts with another centrosomal protein, CEP350 (also known as CAP350; Figure 4H) via its N-terminal region (Yan et al., 2006). However, the N-terminal region (residues 1–179) of FGFR1OP did not coimmunoprecipitate CEP19 (Figure 4B, compare lanes 6 and 2). Furthermore, the FGFR1OP construct FGFR1OP(1-352), lacking only the C-terminal 27 residues, did not retain the ability to interact with CEP19 (lane 5). Reciprocally, a construct containing the C-terminal 27 residues, FGFR1OP(353-379), demonstrated a robust interaction with CEP19 (lane 4), similarly to FGFR1OP(WT) (lane 2). When these FGFR1OP constructs tagged with EGFP were expressed in cells, the N-terminal constructs FGFR1OP(1-179) and FGFR1OP(1-352) (Figure 4, G–G′′ and F–F′′) were localized to the centrosome and interacted with the most C-terminal region of CEP350 (Figure 4H, lane 3), as previously reported (Mikolajka et al., 2006; Yan et al., 2006). It is thus unlikely that the centrosomal localization of FGFR1OP depends on its interaction with CEP19.
FIGURE 4:
FGFR1OP interacts with CEP19 and CEP350 via its N- and C-terminal regions, respectively. (A) Schematic representation of the structures of FGFR1OP and its deletion constructs. (B) FGFR1OP interacts with CEP19 via its C-terminal region. Lysates of HEK293T cells transfected with expression vectors for tRFP-CEP19 and EGFP, or EGFP-tagged FGFR1OP(WT) or its deletion construct, as indicated, were subjected to immunoprecipitation with GST–anti-GFP Nb, followed by immunoblotting analysis with anti-tRFP or anti-GFP antibodies. (C–G) Localization of FGFR1OP deletion constructs. hTERT-RPE1 cells were transfected with an expression vector for EGFP-tagged FGFR1OP(WT) (C–C”) or its deletion construct (D–D”, E–E”, F–F”, or G–G”) and stained with an anti–γ-tubulin antibody. Insets, enlarged images of the boxed regions. Scale bar, 10 µm. (H) FGFR1OP interacts with CEP350 via its N-terminal region. Lysates from HEK293T cells transfected with an expression vector for mChe-CEP350(3071-3117) together with that for EGFP, EGFP-tagged FGFR1OP(WT), or its deletion construct, as indicated, were subjected to immunoprecipitation with GST–anti-GFP Nb, followed by immunoblotting analysis with anti-mRFP or anti-GFP antibodies.
CEP19 is required for RABL2 localization to the centrosome
The data presented so far suggest that CEP19 determines the centrosomal localization of RABL2. To corroborate this, we established hTERT-RPE1 cell lines defective in CEP19, using the clustered regularly interspaced short palindromic repeats (CRISPR /Cas9 system with our original modifications, as described in Materials and Methods; experimental details were recently reported (Katoh et al., 2017; also see Funabashi et al., 2017; Hirano et al., 2017; Nozaki et al., 2017). PCR analysis of genomic DNA isolated from two established CEP19-KO cell lines, 19-1-2 and 19-1-12, demonstrated monoallelic and biallelic forward integration, respectively, of the donor knock-in vector into the CEP19 locus (Supplemental Figure S2A). Direct sequencing of the PCR products demonstrated that clone 19-1-2 has a 55–base pair insertion in one allele and donor vector integration in the other allele (Supplemental Figure S2B), and clone 19-1-12 has donor vector integration in both alleles (Supplemental Figure S2C).
In contrast to control RPE1 cells (Figure 5A), centrosomal signals were not detected with the anti-CEP19 antibody in the CEP19-KO cell lines 19-1-2 and 19-1-12 (Figure 5, B and C). In parallel with the loss of CEP19 signals, the centrosomal signals of RABL2 were abolished in the two CEP19-KO cell lines (Figure 5, E and F; also see Figure 5N); note that the level of the RABL2 protein was not substantially changed in the absence of CEP19 (Figure 5M, top). By contrast, the centrosomal localization of FGFR1OP was not affected by CEP19 deficiency (Figure 5, G–I), compatible with the finding that FGFR1OP constructs lacking the CEP19-binding region can localize to the centrosome (Figure 4, F and G).
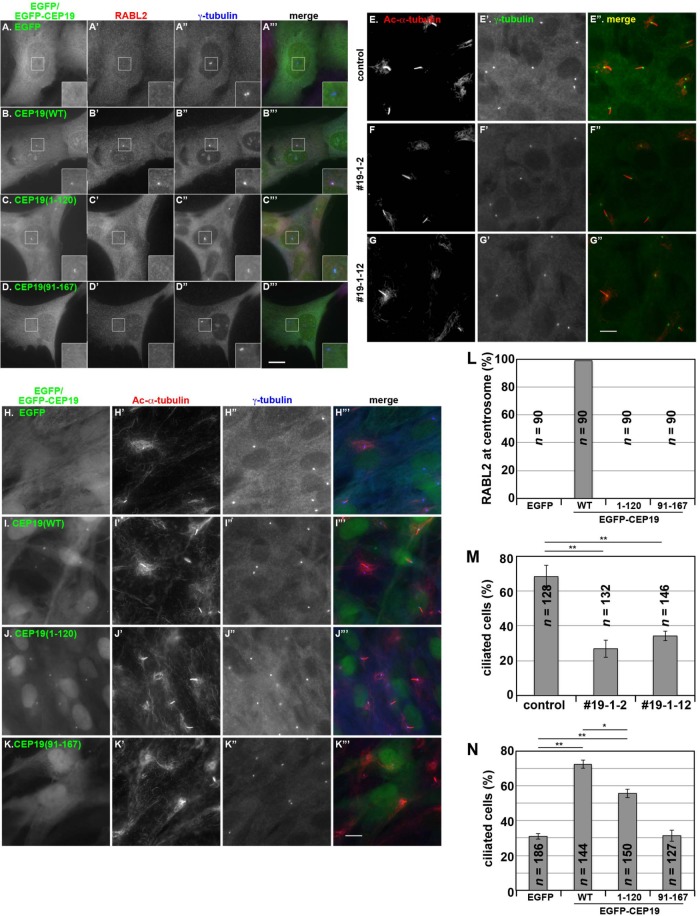
We then examined whether the centrosomal localization of RABL2 in CEP19-KO cells can be restored by the exogenous expression of CEP19 and its deletion mutants. When EGFP-CEP19(WT) was expressed in the 19-1-2 cells, the centriolar localization of RABL2 was almost completely recovered (Figure 6, B–B′′′; also see Figure 6L). In striking contrast, exogenous expression of EGFP-CEP19(1-120), which retains the ability to bind FGFR1OP and localizes to the centrosome but lacks RABL2 binding ability, or EGFP-CEP19(91-167), which cannot localize to the centrosome, did not recover the RABL2 localization (Figure 6, C–C′′′, D–D′′′, and L). We also attempted to establish CEP19-KO cells stably expressing CEP19(16-167) or CEP19(31-167) but were unsuccessful, probably due to the cytotoxic effects of these CEP19 constructs. These observations confirm that the RABL2 displacement observed in the CEP19-KO cells did not result from off-target effects and that CEP19 determines the centrosomal localization of RABL2.
FIGURE 6:
Decreased ciliogenesis efficiency in CEP19-KO cells. (A–D) Requirement of CEP19 for RABL2 centrosomal localization. CEP19-KO 19-1-2 cells stably expressing EGFP (A–A′′′), EGFP-CEP19(WT) (B–B′′′), EGFP-CEP19(1-120) (C–C′′′), or EGFP-CEP19(91-167) (D–D′′′) were double immunostained for RABL2 (A′–D′) and γ-tubulin (A′′–D′′). Insets, enlarged images of the boxed regions. (E–G) Ciliogenesis in CEP19-KO cells. Control RPE1 cells (E) or the CEP19-KO cell lines 19-1–2 (F) and 19-1-12 (G) were serum starved for 24 h and double immunostained for Ac-α-tubulin (E–G) and γ-tubulin (E′–G′). (H–K) Rescue experiments using CEP19 deletion constructs. CEP19-KO 19-1-12 cells stably expressing EGFP (H–H′′′), EGFP-CEP19(WT) (I–I′′′), EGFP-CEP19(1-120) (J–J′′′), or EGFP-CEP19(91-167) (K–K′′′) were serum starved for 24 h and double immunostained for Ac-α-tubulin (H′–K′) and γ-tubulin (H′′–K′′). Scale bars, 10 µm. (L) Cells with centrosomal RABL2 signals in the experiments shown in A–D were counted, and percentages of RABL2-positive cells are shown as a bar graph. Values are means of three independent experiments. In each set of experiments, 30 cells were analyzed, and the total number of analyzed cells (n) is shown. (M) Ciliated cells in the experiments in E–G were counted, and the percentages of ciliated cells are shown as a bar graph. Values are means ± SE of three independent experiments. In each set of experiments, 38–50 cells were analyzed, and the total number of analyzed cells (n) is shown. **p < 0.001 (Student’s t test). (N) Ciliated cells in the experiments in H–K were counted, and the percentages of ciliated cells are shown as a bar graph. Values are means ± SE of three independent experiments. In each set of experiments, 37–66 cells were analyzed, and the total number of analyzed cells (n) is shown. **p < 0.001; *p < 0.01 (Student’s t test).
Decreased ciliogenesis efficiency in CEP19-KO cells
When control RPE1 cells and CEP19-KO cells were cultured under serum-starved conditions to induce ciliogenesis followed by staining for Ac-α-tubulin, we noticed that the rate of ciliogenesis was decreased, although not abolished, in the CEP19-KO cells; ∼70% of control cells formed cilia, whereas ∼30% of the two CEP19-KO cell lines formed cilia (Figure 6, E–G; also see Figure 6M).
We then examined whether the decrease in ciliogenesis efficiency in CEP19-KO cells could be recovered by exogenous expression of CEP19. As shown in Figure 6, I–I′′′, and summarized in Figure 6N, ciliogenesis was significantly restored by the stable expression of EGFP-CEP19(WT) compared with the exogenous expression of EGFP used as a control (Figure 6, H–H′′′). By contrast, the expression of EGFP-CEP19(1-120) recovered ciliogenesis to a lesser extent (Figure 6, J–J′′′ and N). No significant recovery was observed when EGFP-CEP19(91-167) was expressed (Figure 6, K–K′′′ and N).
RABL2-deficient Chlamydomonas exhibits defects in flagellar assembly
To examine the role of RABL2 in ciliogenesis, we also attempted to establish RABL2-KO RPE1 cell lines. We obtained two independent cell lines in which both alleles of both the RABL2A and RABL2B genes were confirmed to be edited, and we confirmed the absence of RABL2 protein expression by immunoblotting; however, these cells underwent growth arrest at an early stage for unknown reasons, which made further analysis impossible (unpublished data). As an alternative to the functional analysis of RABL2, we turned our attention to C. reinhardtii, an organism that has often been used to study cilia/flagella. Of importance, the primary structure of RABL2 is highly conserved between vertebrates and Chlamydomonas (Supplemental Figure S1A).
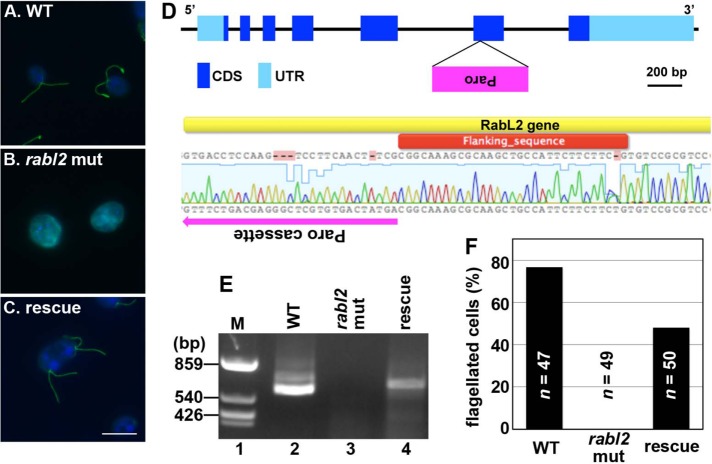
We searched the database of a genome-wide collection of Chlamydomonas mutants (Chlamydomonas Library Project [CLiP; www.chlamylibrary.org]; Li et al., 2016) and found a candidate of the rabl2 mutant (LMJ.RY 0402.205222). This mutant had no flagella and could not swim (compare Figure 7, A and B; also see Figure 7F and Supplemental Movie S1, left and middle). We then confirmed the insertion of the paromomycin resistance gene cassette in the RABL2 locus; we found an insertion causing gene disruption in exon 6 of the RABL2 gene (Figure 7D). We also confirmed by RT-PCR that the RABL2 transcript is not detectable in the rabl2 mutant (Figure 7E, lane 3)
FIGURE 7:
Chlamydomonas RABL2 mutant. (A–C) The C. reinhardtii wild-type (WT) strain (A; CC-4533 cw15 mt-) and rabl2 strain (B; LMJ.RY0402.205222) and the rabl2 strain transformed with a RABL2 expression vector (C) were stained with an anti–Ac-α-tubulin antibody and DAPI. (D) Schematic representation of the integration of the paromomycin (Paro) resistance gene cassette in the RABL2 locus (top) and an alignment of the mutant allele sequence determined by direct sequencing of the genomic PCR products with the reference sequence encompassing the coding sequence of exon 6 (bottom). The magenta arrow indicates the direction of Paro cassette integration. (E) RT-PCR analysis of RABL2 mRNA expression in the WT strain (lane 2), the rabl2 strain (lane 3), and the transformant (lane 4). Lane 1 is a DNA size marker (DdeI-digested pSP64 DNA). (F) Flagellated cells in the experiments in A–C were counted, and the percentages of flagellated cells are shown as a bar graph. The number of observed cells (n) is shown in the graph.
To exclude the possibility that the observed nonflagellated phenotype resulted from disruption of an unexpected locus, we performed a rescue experiment. We transformed the rabl2 strain with a RABL2 expression vector and confirmed the expression of RABL2 mRNA in the selected transformant by RT-PCR (Figure 7E, lane 4). The exogenous RABL2 expression in the rabl2 strain was confirmed to restore flagellar biogenesis (Figure 7, C and F) and the swimming behavior (Supplemental Movie S1, right). Thus the nonflagellated phenotype of the Chlamydomonas rabl2 strain indicates a crucial role of RABL2 in ciliary/flagellar assembly.
RABL2 interacts with CEP 19 and the IFT-B complex in a mutually exclusive manner
The foregoing data showing that the Chlamydomonas rabl2 mutant cannot form flagella suggest that the RABL2–CEP19 interaction is required for flagella formation in Chlamydomonas. However, if CEP19 functions upstream of RABL2, the observation that ciliogenesis of CEP19-KO RPE1 cells, although significantly decreased, was not completely abolished cannot be readily explained. We therefore addressed another possibility—that, via interacting with some downstream effector(s), RABL2 is in some way involved in the trafficking of flagellar/ciliary proteins, including the αβ-tubulin dimer, which is the building block of the flagellar/ciliary axoneme. One candidate downstream effector of RABL2 is the IFT-B complex, as it mediates intraciliary/intraflagellar trafficking of tubulins (Bhogaraju et al., 2013, 2014), and a previous study suggested that RABL2 interacts, directly or indirectly, with the IFT-B complex (Lo et al., 2012).
To examine whether the IFT-B complex can interact directly with RABL2, we took advantage of the visible immunoprecipitation (VIP) assay, which we recently developed to enable the convenient and flexible detection of protein–protein interactions (Katoh et al., 2015). Using this assay, we recently determined the overall architectures of the IFT-A and IFT-B complexes (Katoh et al., 2016; Hirano et al., 2017). The IFT-B complex can be divided into a core subcomplex, composed of 10 subunits, and a peripheral subcomplex, composed of 6 subunits (Katoh et al., 2016; Figure 8L). IFT-B subunits fused to mChe or tRFP were coexpressed with EGFP-tagged RABL2B(WT), RABL2B(S35N), RABL2B(Q80L), or RABL2B(D73G) in HEK293T cells, and lysates prepared from the cells were immunoprecipitated with GST–anti-GFP Nb prebound to glutathione–Sepharose beads. Subsequent observation under a microscope demonstrated the presence of robust red fluorescence signals on the precipitated beads when the IFT-B subunits were coexpressed with RABL2B(Q80L)-EGFP (Figure 8A).
We then set out to determine the IFT-B subunit(s) involved in the RABL2 interaction. When cells coexpressing mChe/tRFP-tagged core or peripheral subunits and RABL2B(Q80L)-EGFP were processed for the VIP assay, we observed red signals only when the mChe/tRFP-tagged core subunits were coexpressed (Figure 8B). When the core subunits were divided into two subgroups, IFT22/IFT25/IFT27/IFT74/IFT81 (core 1) and IFT46/IFT52/IFT56/IFT70/IFT88 (core 2; Figure 8L), only the core 1 subgroup resulted in intense red signals (Figure 8B). We then performed the subtractive VIP assay to determine the core 1 subunit(s) responsible for the RABL2 interaction. As shown in Figure 8C, red signals were greatly reduced when mChe-tagged IFT74 or IFT81 was omitted, suggesting that these two subunits are directly involved in the RABL2 interaction. Indeed, immunoprecipitation of RABL2B(Q80L)-EGFP resulted in red signals upon coexpression with mChe-tagged IFT74+IFT81 but not with IFT74 or IFT81 alone (Figure 8D), indicating that RABL2 interacts with a complex of IFT74 and IFT81, which are known to form a tight heterodimer through their coiled-coil regions, and directly interacts with tubulins (Bhogaraju et al., 2013). We next analyzed the interaction of wild-type RABL2 and its mutants with the IFT74–IFT81 dimer by the VIP assay. As was the case for all the IFT-B subunits fused to mChe, mChe-tagged IFT74+IFT81 interacted robustly with RABL2B(Q80L)-EGFP. In addition, mChe-IFT74+IFT81 demonstrated a weak interaction with RABL2B(WT)-EGFP (Figure 8F). By contrast, RABL2B(S35N)-EGFP did not result in red signals when it was coexpressed with mChe-IFT74+IFT81. Furthermore, virtually no red signals were detected when the Mot-type mutant RABL2B(D73G) was coexpressed with mChe-IFT74+IFT81. We then confirmed the VIP data by conventional immunoblotting analysis. RABL2B(Q80L)-EGFP coimmunoprecipitated mChe-tagged IFT74+IFT81 but not IFT74 or IFT81 alone (Figure 8E), and RABL2B(WT), but not RABL2B(S35N) or RABL2B(D73G), interacted weakly with mChe-IFT74+IFT81 (Figure 8G).
We then examined whether RABL2 can simultaneously interact with CEP19 and IFT74–IFT81. To this end, we coexpressed EGFP-tagged IFT74+IFT81 and RABL2B(Q80L)-mChe in the presence or absence of mChe-CEP19 and subjected the cell lysates to the VIP assay using GST–anti-GFP Nb. Red signals were detected when EGFP-IFT74+IFT81 and RABL2B(Q80L)-mChe were coexpressed (Figure 8H, left). Intriguingly, the red signals were abolished when mChe-CEP19 was additionally coexpressed (Figure 8H, right). The VIP data were confirmed by immunoblotting analysis. RABL2B(Q80L)-mChe was coimmunoprecipitated with EGFP-IFT74+IFT81 in the absence but not in the presence of coexpressed mChe-CEP19 (Figure 8I). Reciprocally, when RABL2B(Q80L)-EGFP was coexpressed with mChe-IFT74+IFT81 in the presence or absence of EGFP-CEP19, the amount of coimmunoprecipitated mChe-IFT74+IFT81 was greatly decreased by the coexpression of EGFP-CEP19 (Figure 8, J and K). Taken together, these results demonstrate that RABL2 is likely to interact, probably in its GTP-bound state, with CEP19 and the IFT-B complex in a mutually exclusive manner (Figure 8L).
Participation of RABL2 in ciliary assembly
The interaction data, together with the Chlamydomonas mutant phenotype, suggest two possible roles of RABL2 in ciliary function. One possibility is that, although RABL2 is localized mainly at the ciliary base in the steady state (Figure 2G), it is trafficked anterogradely as a cargo of the IFT-B complex during ciliogenesis. The other possibility is that RABL2 in some way regulates the IFT-B complex at the ciliary base. To address the first possibility, we used our recently established IFT139-KO RPE1 cells (Hirano et al., 2017). In the absence of IFT139 (an IFT-A subunit), anterograde ciliary trafficking mediated by the IFT-B complex still occurs, whereas retrograde trafficking mediated by the IFT-A complex is suppressed, resulting in accumulation of the IFT-B complex at the bulged ciliary tips (Figure 9, B′ and C′). Therefore this phenotype can be used to determine whether RABL2 is a cargo of the IFT-B complex. In the two independent IFT139-KO cell lines (139-2-6 and 139-2-8), RABL2 and CEP19 were found at the ciliary base (Figure 9, B and C, and E and F, respectively) as in control RPE1 cells (Figure 9, A and D), and neither of the proteins was colocalized with IFT88 (an IFT-B subunit) at the bulged tips. These observations suggest that RABL2 is not a cargo of the IFT-B complex, although it remains possible that RABL2 was once trafficked with the IFT train and then returned to the ciliary base by diffusion after dissociation from the IFT-B complex at the ciliary tips.
We then addressed the other possibility—namely, RABL2 in some way regulates the IFT-B complex. In view of the fact that CEP19 determines the localization of RABL2 at the ciliary base, we examined localization of IFT88 in CEP19-KO cell lines. In the majority of control RPE1 cells, IFT88 is found within cilia as well as at the ciliary base (Figure 5J; also see Figure 5O). In the CEP19-KO cell lines, however, the population of cells with IFT88 signals only at the ciliary base (namely, those without IFT88 signals within cilia) was significantly increased (Figure 5, K and L; also see Figure 5O), although the cellular level of the IFT88 protein was not substantially changed in the absence of CEP19 (Figure 5M, middle). These observations indirectly suggest that RABL2 somehow participates in recruitment of the IFT-B complex to the ciliary base and/or its entry into cilia (see Discussion).
We then anticipated that overexpression of RABL2B(S35N) or RABL2B(D73G) might have a dominant-negative effect on the assembly of cilia through affecting IFT-B function, because these mutants failed to interact with the IFT-B complex (Figure 8, F and G). Indeed, when expressed in hTERT-RPE1 cells, both RABL2B(S35N)-EGFP and RABL2B(D73G)-EGFP significantly suppressed ciliogenesis, as depicted by the staining for Ac-α-tubulin, compared with exogenously expressed EGFP or RABL2B(WT)-EGFP (compare Figure 9, I–I′′ and K–K′′ with G–G′′ and H–H′′; also see Figure 9L). Taken together with the phenotype of the Chlamydomonas rabl2 strain, it is likely that RABL2 is involved in ciliary assembly by regulating IFT-B function, which mediates the trafficking of tubulins and other ciliary proteins (Bhogaraju et al., 2013, 2014; see Discussion).
DISCUSSION
In this study, we demonstrated that RABL2 is recruited to the mother centriole/basal body via its interaction with CEP19, which is probably recruited to the centriole via its interaction with FGFR1OP. Of interest, endogenous RABL2 and CEP19 are localized on the mother centriole (Figure 2, D–F), whereas FGFR1OP is found on both centrioles (Figure 3, E–E′′′). These observations suggest that, besides FGFR1OP binding, other factor(s) on the mother centriole determine the localization of CEP19. In this context, it is noteworthy that exogenously expressed CEP19 localized to both centrioles (e.g., Figure 2L). Therefore it is also possible that some factor located at the daughter centriole restricts the access of CEP19 to FGFR1OP and that overexpressed CEP19 might titrate out the restricting factor.
CEP19-KO RPE1 cells lacked the centrosomal localization of RABL2 as well as exhibited mild ciliogenesis defects. On the other hand, the available Chlamydomonas rabl2 strain cannot form flagella. These results make it likely that the assembly of flagella in Chlamydomonas requires the interaction of RABL2 with proteins other than CEP19. We then unequivocally demonstrated that RABL2 interacts with the IFT-B complex via the IFT74–IFT81 heterodimer; the RABL2–IFT-B interaction was suggested by a previous study (Lo et al., 2012). Furthermore, we showed that RABL2 binding to IFT-B is mutually exclusive with that to CEP19. Although the IFT-B complex is involved in the anterograde trafficking of ciliary proteins (Ishikawa and Marshall, 2011; Sung and Leroux, 2013), our data using IFT139-KO and CEP19-KO cells suggest that RABL2 is unlikely to be trafficked as a cargo of IFT-B but might participate, directly or indirectly, in ciliary entry of the IFT-B complex at the ciliary base. In this context, it is noteworthy that the distribution patterns of IFT88 revealed by superresolution microscopy suggested accumulation of IFT particles at the ciliary base before entering cilia (Yang et al., 2015). It is therefore possible that RABL2 regulates recruitment or assembly of the IFT-B complex at the ciliary base.
The interactions of the three RAB-like GTPases (RABL2, RABL4/IFT27/BBS19, and RABL5/IFT22) with the same IFT74–IFT81 heterodimer (Figure 8L) suggest pivotal roles of the heterodimer in the IFT-B complex. Indeed, in the architecture model we proposed of the IFT-B complex (Katoh et al., 2016) as well as in that proposed by others (Taschner and Lorentzen, 2016; Taschner et al., 2016), the IFT74–IFT81 heterodimer is responsible for the interactions with IFT22, the IFT25–IFT27 dimer, and the IFT46–IFT52 dimer. In addition and of greater importance, via their N-terminal regions, the IFT74–IFT81 dimer binds to and mediates trafficking of the αβ-tubulin dimer, which is the building block of the ciliary axoneme (Bhogaraju et al., 2013, 2014); this is compatible with the phenotype of the Chlamydomonas rabl2 strain and that of RPE1 cells with exogenous expression of the RABL2 mutants.
In this context, it is interesting to note that the IFT74–IFT81 dimer interacts more strongly with RABL2B(Q80L) than with RABL2B(WT) (Figure 8, F and G), whereas CEP19 binds to RABL2B(WT) and RABL2B(Q80L) with similar affinities (Figure 1A). It is also noteworthy that the interactions of RABL2 with IFT-B and CEP19 are mutually exclusive (Figure 8, H–K), whereas the RABL2–CEP19–FGFR1OP ternary complex can be formed (Figure 3D). A possible scenario is that 1) RABL2 localizes to the basal body/mother centriole via its constitutive interaction with CEP19, and 2) on its activation, RABL2 becomes able to interact with and somehow regulates the IFT-B complex, such as its recruitment to the ciliary base and its ciliary entry. To address this possibility, it is therefore an important issue to identify a putative guanine nucleotide exchange factor for RABL2.
Mot mice with a point mutation in the RABL2 gene (Lo et al., 2012), as well as CEP19-KO mice (Shalata et al., 2013), exhibit male sterility caused by a motility dysfunction of their sperm. However, in these mice, the formation of cilia and flagella does not appear to be severely affected. Our data showing that CEP19-KO RPE1 cell lines exhibit a mild defect in the formation of cilia are compatible with the phenotype of CEP19-KO mice. By contrast, the Chlamydomonas rabl2 strain lacks flagella. While this article was under review, male infertile patients with short sperm flagella were reported to have a splicing-site mutation in the RABL2B gene (Hosseini et al., 2017). Therefore the differences in the phenotype between sperm flagella of Mot mice, flagella of the Chlamydomonas rabl2 strain, and sperm flagella of infertile men may be attributed to differences between the effects of a point mutation (D73G) and the disruption of the RABL2 gene. Intriguingly, the D73G mutation abolishes or greatly decreases the interaction of RABL2 with the IFT-B complex (Figure 8, F and G). In marked contrast, the Mot-type mutation does not affect the RABL2–CEP19 interaction (Figure 1A), indicating that the phenotype of Mot mice is not due to a change in the interaction of RABL2 with CEP19. In view of the fact that Mot mice demonstrate a defect in sperm motility but not in the assembly of sperm flagella, at least at the light microscopic level (Lo et al., 2012), it is possible that the D73G mutation affects the trafficking of proteins other than tubulins. For example, motile cilia diseases are often caused by mutations in the genes of components of the outer dynein arm, inner dynein arm, and radial spoke (Praveen et al., 2015), some of which were reported to bind to the IFT-B complex (Ahmed et al., 2008; Ishikawa et al., 2014; Huttlin et al., 2015). Although our attempts to establish RABL2-KO mammalian cells have been unsuccessful, how RABL2 regulates the assembly and function of cilia/flagella is an important issue for future studies.
MATERIALS AND METHODS
Plasmids
Expression vectors for RABL2, CEP19, FGFR1OP, and CEP350 used in this study are listed in Supplemental Table S1. Expression vectors for the IFT-B subunits were described previously (Katoh et al., 2016; Nozaki et al., 2017).
Antibodies and reagents
Antibodies used in this study are listed in Supplemental Table S2. GST-tagged anti-GFP Nb was purified with glutathione–Sepharose 4B beads (GE Healthcare) from Escherichia coli (BL21 DE3) cells transformed with the corresponding vector, as described previously (Katoh et al., 2015, 2016). Polyethylenimine Max and Hoechst 33342 were purchased from Polysciences and Molecular Probes, respectively.
Coimmunoprecipitation assay and VIP assay
HEK293T cells (kindly provided by Hiroyuki Takatsu, Kyoto University) were plated on 6-cm dishes and cultured in DMEM with high glucose (Nacalai Tesque) supplemented with 10% fetal bovine serum (FBS). The cells were transfected with expression vectors for an EGFP fusion construct and a hemagglutinin (HA) fusion construct using Polyethylenimine Max and cultured for 24 h. The cells were then lysed in lysis buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]-KOH, pH 7.4, 100 mM KCl, 5 mM NaCl, 3 mM MgCl2, 0.5% Triton X-100, 10% glycerol, and 1 mM dithiothreitol [DTT]) containing a protease inhibitor cocktail (Nacalai Tesque). The cell lysates were immunoprecipitated with GST–anti-GFP Nb prebound to glutathione–Sepharose 4B beads, and bound proteins were subjected to SDS–PAGE and immunoblotting analysis using anti-HA, anti-mRFP, anti-tRFP, or anti-GFP antibodies as described previously (Katoh et al., 2015, 2016; Nozaki et al., 2017).
The VIP assay was performed as described previously (Katoh et al., 2015, 2016) with a slight modification; in this study, cells expressing EGFP-tagged and mChe-tagged proteins were lysed in the HMDEKN cell lysis buffer (10 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, 25 mM KCl, 0.05% NP-40; Cole et al., 1998).
Immunofluorescence analysis
The hTERT-RPE1 cells (CRL-4000; American Type Culture Collection) were cultured in DMEM/F-12 (Nacalai Tesque) supplemented with 10% FBS and 0.348% sodium bicarbonate. To induce ciliogenesis, cells were grown to 100% confluence on dishes or coverslips and starved for 24 h in Opti-MEM (Invitrogen) containing 0.2% bovine serum albumin. Expression vectors were transfected into the cells using X-tremeGENE9 DNA Transfection Reagent (Roche Applied Science).
Immunofluorescence analysis was performed as described previously (Takahashi et al., 2012; Nozaki et al., 2017). Cells were fixed with 3% paraformaldehyde at 37°C for 15 min, washed three times with phosphate-buffered saline (PBS), quenched with 50 mM NH4Cl for 10 min, washed three times with PBS, permeabilized with 0.1% Triton X-100 for 5 min, and washed three times with PBS. For the detection of endogenous RABL2, cells were fixed with 10% trichloroacetic acid on ice for 15 min. The fixed/permeabilized cells were blocked with 10% FBS and treated with antibodies diluted in 5% FBS. For the detection of γ-tubulin, antibodies were diluted with Can Get Signal immunostain (Toyobo). The stained cells were observed using an Axiovert 200 M microscope (Carl Zeiss) or an A1R-MP confocal laser-scanning microscope (Nikon).
Establishment of KO cell lines using the CRISPR/Cas9 system
Details of the knockout strategy by the CRISPR/Cas9 system using homology-independent DNA repair were recently reported (Katoh et al., 2017; also see Funabashi et al., 2017; Hirano et al., 2017; Nozaki et al., 2017). Single guide RNA (sgRNA) sequences targeting the human CEP19 gene (Supplemental Table S3) were designed using CRISPR Design (Hsu et al., 2013). A double-stranded oligonucleotide for the target sequence was inserted into the donor knock-in vector pDonor-tBFP-NLS-Neo (deposited in Addgene, ID 80766) and the all-in-one sgRNA expression vector pSpCas9(BB)-2A-Puro (Addgene plasmid 48139). hTERT-RPE1 cells cultured in a 12-well plate were transfected with 1 µg of the all-in-one vector and 0.25 µg of the donor vector using X-tremeGENE9 Transfection Reagent (Roche Applied Science). After selection in medium containing G418 (600 µg/ml), cells with nuclear tBFP signals were isolated. Genomic DNA isolated from the cloned cells was subjected to PCR using KOD FX Neo DNA polymerase (Toyobo). Three sets of primers (Supplemental Table S3) were used to distinguish the following three states of donor vector integration: forward integration (Supplemental Figure S1; lanes 3, 6, and 9), reverse integration (lanes 4, 7, and 10), and no integration with a small insertion or deletion (lanes 2, 5, and 8). Direct sequencing of the PCR products ensured the KO of both alleles of the CEP19 genes, with integration of the donor vector and/or a small deletion/insertion causing a frameshift (Supplemental Figure S1, B and C). Establishment of IFT139-KO cell lines was described previously (Hirano et al., 2017).
Preparation of cells stably expressing EGFP-tagged CEP19 and its deletion constructs
Lentiviral vectors were prepared as described previously (Takahashi et al., 2012; Nozaki et al., 2017). Briefly, either pRRLsinPPT-EGFP-CEP19(WT) or its mutant was transfected into HEK293T cells using Polyethylenimine Max along with packaging plasmids (pRSV-REV, pMD2.g, and pMDLg/pRRE; kind gifts from Peter McPherson, McGill University; Thomas et al., 2009). The culture medium was replaced 8 h after transfection and collected at 24, 36, and 48 h after transfection. The medium containing viral particles was passed through a 0.45-µm filter and centrifuged at 32,000 × g at 4°C for 4 h. Precipitated lentiviral particles were resuspended in Opti-MEM (Invitrogen) and stored at −80°C until use. Control cells and CEP19-KO cells expressing EGFP-CEP19(WT), EGFP-CEP19(1-120), or EGFP-CEP19(91-167) were prepared by adding the lentiviral suspension to the culture medium. These cells were used for immunofluorescence analysis.
Analysis of the Chlamydomonas rabl2 mutant
The C. reinhardtii wild-type (CC-4533 cw15 mt-) and rabl2 (LMJ.RY0402.205222) strains obtained from the CLiP (Li et al., 2016) were grown on a Tris-acetate-phosphate (TAP) agar plate and cultured in liquid TAP medium. To confirm the insertion of the paromomycin resistance gene cassette in the RABL2 locus, genomic PCR using the specific primers listed in Supplemental Table S3 was performed according to the manual provided by the CLiP.
An ∼2.1-kbp genomic DNA fragment encompassing the RABL2 gene (Cre17. g722350, from the initiation codon to the codon for the C-terminal amino acid) was amplified by PCR using the specific primers (CrRabL2-NS and CrRabL2-CAS) listed in Supplemental Table S3. An expression vector for Chlamydomonas RABL2 was constructed by inserting the resulting PCR product into the pIC2L-BCCPC-3 × HA vector (Oda et al., 2015). The rabl2 strain was transformed with the RABL2 expression vector, and the transformants were selected as described (Oda et al., 2015).
For RT-PCR analysis, total RNAs were extracted from the cells using an ISOGEN RNA extraction kit (Nippon Gene). RT-PCR using the specific primers (CrRabL2-NS and CrRabL2-G-Rv2) listed in Supplemental Table S3 was performed using a SuperScript III One-Step RT-PCR System (Invitrogen).
Immunofluorescence analysis was performed as described previously (Kubo et al., 2015). Briefly, Chlamydomonas cells attached to a coverslip were fixed and permeabilized with methanol at –20°C for 20 min, blocked with 10% FBS, and incubated with an anti–Ac-α-tubulin antibody, followed by staining with Alexa Fluor 488–conjugated anti-mouse immunoglobulin G and 4′,6-diamidino-2-phenylindole (DAPI). The coverslip was placed onto Mowiol mounting medium (Sigma-Aldrich), and the cells were observed under an Axiovert 200 M microscope. The swimming of Chlamydomonas cells was recorded using a Biozero BZ-8000 microscope (Keyence).
Supplementary Material
Acknowledgments
We are grateful to Atsuo Iida, Atsuko Sehara, Masafumi Hirono, and Hideya Fukuzawa for technical advice and assistance, Peter McPherson for providing vectors for lentiviral production, Helena Akiko Popiel for critical reading of the manuscript, Shohei Nozaki for valuable information, and Toshiyuki Oda for encouragement. This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas “Cilia and Centrosome” from the Ministry of Education, Culture, Sports, Science and Technology, Japan (25113514 and 15H01211 to K.N.); grants from the Japan Society for the Promotion of Science (15H04370 and 15K14456 to K.N. and 25860044 and 15K07929 to Y.K.); and grants from the Uehara Memorial Foundation and the Astellas Foundation for Research on Metabolic Disorders to K.N. and from the Takeda Science Foundation and the Uehara Memorial Foundation to Y.K.
Abbreviations used:
- BBS
Bardet–Biedl syndrome
- FBS
fetal bovine serum
- GST
glutathione S-transferase
- IFT
intraflagellar transport
- JBTS
Joubert syndrome
- KO
knockout
- Nb
nanobody
- PBS
phosphate-buffered saline
- RABL
RAB-like
- VIP
visible immunoprecipitation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-01-0017) on April 20, 2017.
REFERENCES
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 2008;183:313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Cajánek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, Lorentzen E. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 2013;341:1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Weber K, Engel BD, Lechtreck K-F, Lorentzen E. Getting tubulin to the tip of the cilium: one IFT train, many different tubulin cargo binding sites? Bioessays. 2014;36:463–467. doi: 10.1002/bies.201400007. [DOI] [PubMed] [Google Scholar]
- Brown JM, Witman GB. Cilia and diseases. BioScience. 2014;64:1126–1137. doi: 10.1093/biosci/biu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II–dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T, San Agustin JT, Keady BT, Jonassen JA, Liang Y, Francis R, Tobita K, Johnson CA, Abdelhamed ZA, Lo CW, Pazour GJ. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev Cell. 2014;21:279–290. doi: 10.1016/j.devcel.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliáš M, Klimeš V, Derelle R, Petrželková R, Tachezy J. A paneukaryotic genomic analysis of the small GTPase RABL2 underscores the significance of recurrent gene loss in eukaryote evolution. Biol Direct. 2016;11:5. doi: 10.1186/s13062-016-0107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi T, Katoh Y, Michisaka S, Terada M, Sugawa M, Nakayama K. Ciliary entry of KIF17 is dependent on its binding to the IFT-B complex via IFT46-IFT56 as well as on its nuclear localization signal. Mol Biol Cell. 2017;28:624–633. doi: 10.1091/mbc.E16-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GD, Coyaud É, Gonçalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SW, et al. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Katoh Y, Nakayama K. Intraflagellar transport-A complex mediates ciliary entry and retrograde trafficking of ciliary G protein-coupled receptors. Mol Biol Cell. 2017;28:429–439. doi: 10.1091/mbc.E16-11-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh RA, Stowe TR, Stearns T. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS One. 2012;7:e52166. doi: 10.1371/journal.pone.0052166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini SH, Sadighi Gilani MA, Meybodi AM, Sabbaghian M. The impact of RABL2B gene (rs144944885) on human male infertility in patients with oligoasthenoteratozoospermia and immotile short tail sperm defects. J Assist Reprod Genet. 2017;34:505–510. doi: 10.1007/s10815-016-0863-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ide T, Yagi T, Jiang X, Hirono M, Sasaki H, Yanagisawa H, Wemmer KA, Stainier DYR, Qin H, et al. TTC26/DYF13 is an intraflagellar transport protein required for transport of motility-related proteins into flagella. Elife. 2014;3:e01566. doi: 10.7554/eLife.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsai D, Lo JCY, McLachlan RI, O’Bryan MK. Genetic variants in the RABL2A gene in fertile and oligoasthenospermic infertile men. Fertil Steril. 2014;102:223–229. doi: 10.1016/j.fertnstert.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Michisaka S, Nozaki S, Funabashi T, Hirano T, Takei R, Nakayama K. Practical method for targeted disruption of cilia-related genes by using CRISPR/Cas9-mediated, homology-independent knock-in system. Mol Biol Cell. 2017;28:898–906. doi: 10.1091/mbc.E17-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Nozaki S, Hartanto D, Miyano R, Nakayama K. Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J Cell Sci. 2015;128:2351–2362. doi: 10.1242/jcs.168740. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Terada M, Nishijima Y, Takei R, Nozaki S, Hamada H, Nakayama K. Overall architecture of the intraflagellar transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J Biol Chem. 2016;291:10962–10975. doi: 10.1074/jbc.M116.713883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M, Backhaus O, Rosenstiel P, Horn D, Klopocki E, Birkenmeier G, Schreiber S, Platzer M, Hampe J, Huse K. Analysis of relative gene dosage and expression differences of the paralogs RABL2A and RABL2B by pyrosequencing. Gene. 2010;455:1–7. doi: 10.1016/j.gene.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Kubo T, Hirono M, Aikawa T, Kamiya R, Witman GB. Reduced tubulin polyglutamylation suppresses flagellar shortness in Chlamydomonas. Mol Biol Cell. 2015;26:2810–2822. doi: 10.1091/mbc.E15-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Stearns T. FOP is a centriolar satellite protein involved in ciliogenesis. PLoS One. 2013;8:e58589. doi: 10.1371/journal.pone.0058589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, et al. An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell. 2016;28:367–387. doi: 10.1105/tpc.15.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hu J. Small GTPases and cilia. Protein Cell. 2011;2:13–25. doi: 10.1007/s13238-011-1004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew GM, Ye F, Nager AR, Murphy JP, Lee JSH, Aguiar M, Breslow DK, Gygi SP, Nachury MV. The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev Cell. 2014;31:265–278. doi: 10.1016/j.devcel.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YS, Chua CEL, Tang BL. Rabs and other small GTPases in ciliary transport. Biol Cell. 2011;103:209–221. doi: 10.1042/BC20100150. [DOI] [PubMed] [Google Scholar]
- Lo JC, Jamsai D, O’Connor AE, Borg C, Clark BJ, Whisstock JC, Field MC, Adams V, Ishikawa T, Aitken RJ, et al. RAB-like 2 has an essential role in male fertility, sperm intra-flagellar transport, and tail assembly. PLoS Genet. 2012;8:e1002969. doi: 10.1371/journal.pgen.1002969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhivanan K, Aguilar RC. Ciliopathies: the trafficking connection. Traffic. 2014;15:1031–1056. doi: 10.1111/tra.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajka A, Yan X, Popowicz GM, Smialowski P, Nigg EA, Holak TA. Structure of the N-terminal domain of the FOP (FGFR1OP) protein and implications for its dimerization and centrosomal localization. J Mol Biol. 2006;359:863–875. doi: 10.1016/j.jmb.2006.03.070. [DOI] [PubMed] [Google Scholar]
- Nozaki S, Katoh Y, Terada M, Michisaka S, Funabashi T, Takahashi S, Kontani K, Nakayama K. Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J Cell Sci. 2017;130:563–576. doi: 10.1242/jcs.197004. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kikkawa M. Detailed structural and biochemical characterization of the nexin-dynein regulatory complex. Mol Biol Cell. 2015;26:294–304. doi: 10.1091/mbc.E14-09-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen K, Davis EE, Katsanis N. Unique among ciliopathies: primary dyskinesia, a motile cilia disorder. F1000Prime Rep. 2015;7:36. doi: 10.12703/P7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas AM, Fuentes G, Rausell A, Valencia A. The Ras protein superfamily: Evolutionary tree and role of conserved amino acids. J Cell Biol. 2012;196:189–201. doi: 10.1083/jcb.201103008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Schwartz R, Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedjaï F, Acquaviva C, Chevrier V, Chauvin J-P, Coppin E, Aouane A, Coulier F, Tolun A, Pierres M, Birnbaum D, Rosnet O. Control of ciliogenesis by FOR20, a novel centrosome and pericentriolar satellite protein. J Cell Sci. 2010;123:2391–2401. doi: 10.1242/jcs.065045. [DOI] [PubMed] [Google Scholar]
- Shalata A, Ramirez MC, Desnick RJ, Priedigkeit N, Buettner C, Lindtner C, Mahroum M, Abdul-Ghani M, Dong F, Arar N, et al. Morbid obesity resulting from inactivation of the ciliary protein CEP19 in humans and mice. Am J Hum Genet. 2013;93:1061–1071. doi: 10.1016/j.ajhg.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C-H, Leroux MR. The roles of evolutionary conserved functional modules in cilia-related trafficking. Nat Cell Biol. 2013;15:1387–1397. doi: 10.1038/ncb2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Kubo K, Waguri S, Yabashi A, Shin H-W, Katoh Y, Nakayama K. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci. 2012;125:4049–4057. doi: 10.1242/jcs.102913. [DOI] [PubMed] [Google Scholar]
- Taschner M, Lorentzen E. The intraflagellar transport machinery. Cold Spring Harb Perspect Biol. 2016;8:a028092. doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Weber K, Mourão A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, Lorentzen E. Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 2016;35:773–790. doi: 10.15252/embj.201593164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Ritter B, Verbich D, Sanson C, Bourbonnière L, McKinney RA, McPherson PS. Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem. 2009;284:12410–12419. doi: 10.1074/jbc.M809746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Habedanck R, Nigg EA. A complex of two centrosomal proteins, CAP350 and FOP, cooperates with EB1 in microtubule anchoring. Mol Biol Cell. 2006;17:634–644. doi: 10.1091/mbc.E05-08-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Su J, Wang W-J, Craige B, Witman GB, Tsou M-FB, Liao J-C. Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci Rep. 2015;5:14096. doi: 10.1038/srep14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.