Abstract
The study of cellular glycosylation presents many challenges due, in large part, to the non-template driven nature of glycan biosynthesis and their structural complexity. Chemoenzymatic glycan labeling (CEGL) has emerged as a new technique to address the limitations of existing methods for glycan detection. CEGL combines glycosyltransferases and unnatural nucleotide sugar donors equipped with a bioorthogonal chemical tag to directly label specific glycan acceptor substrates in situ within biological samples. This article reviews the current CEGL strategies that are available to characterize cell-surface and intracellular glycans. Applications include imaging glycan expression status in live cells and tissue samples, proteomic analysis of glycoproteins, and target validation. Combined with genetic and biochemical tools, CEGL provides new opportunities to elucidate the functional roles of glycans in human health and disease.
Graphical abstract
Complex Glycosylation: Diversity and Challenges
Post-translational modifications are a major factor that distinguishes mammals from lower organisms. As stated by Venter and coworkers in their landmark report on the sequence of the human genome, “the finding that the human genome contains fewer genes than previously predicted might be compensated for by combinatorial diversity generated at the level of…post-translational modifications”1. Among the different types of post-translational modifications, glycosylation is the most chemically and biosynthetically complex.
The complexity of glycosylation arises from two factors: the diversity of the building blocks and the multiple ways in which oligosaccharides can be assembled from these building blocks within the cell. For instance, at least five stereocenters are found in a monosaccharide building block and in higher eukaryotes there are nine essential monosaccharides that can be linked to each other at multiple positions, in either an α- or β-glycosidic linkage, to form oligosaccharides2. The resulting linear or branched structures may be further modified by secondary modifications such as: phosphorylation, sulfation, or lipidation, just to name a few. Complex oligosaccharides are not assembled in a template-driven process like the biosynthesis of proteins or nucleic acids. Instead, oligosaccharides are assembled through stepwise, enzyme-catalyzed additions of monosaccharide building blocks in a spatiotemporally-controlled process within the secretory pathway. Accordingly, the ensemble of glycans contained within a particular cell, referred to as its ‘glycome’, is primarily governed by the spatial arrangement and temporal availability of nucleotide sugar donors, glycoprotein or glycolipid acceptors, as well as the glycosyltransferases and glycosidases within the secretory pathway3. This non-template driven biosynthesis generates enormous diversity and heterogeneity within the cellular glycome that enables diverse biological roles for glycans.
As a consequence of the non-template driven nature of glycan biosynthesis, the study of glycans is inherently challenging. In particular, genetically-encoded tags for tracking and visualizing proteins cannot be used for studying glycans. The detection of glycans using antibodies and lectins has well-documented drawbacks: glycan-binding antibodies can be difficult to raise and are often low affinity (mainly of the IgM isotype)4, while the majority of lectins lack stringent specificity5,6, making it difficult to study individual sectors of glycans. Chemical methods have long been used as alternative tools for the detection of glycans. However, many of these approaches are destructive. For example, periodic acid and alcian blue are widely used reagents for the detection of sialic acid and polysialic acid, respectively, but both cause permanent damage to the treated carbohydrate chains preventing downstream applications7.
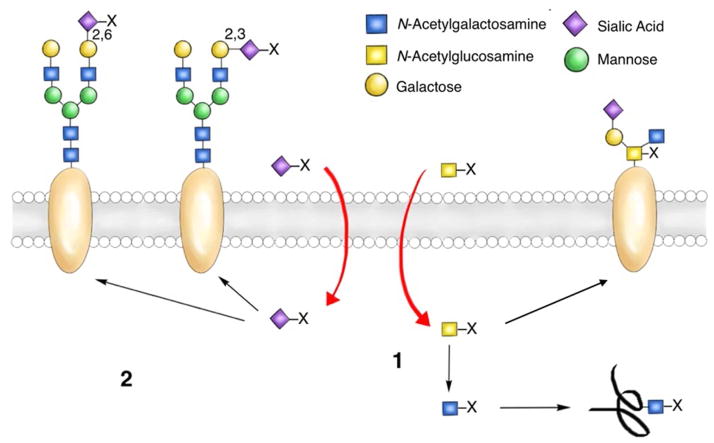
An alternative method for glycan detection relies on metabolic oligosaccharide engineering (MOE)8–10. Pioneered by Reutter and Bertozzi, MOE offers a method for glycan detection. Labeling and detecting sialylated8,11, fucosylated8,12–14, and mucin-type O-linked glycans8,15 in live cells and model organisms has become routine practice using this approach. Harnessing the glycan metabolic machinery to incorporate unnatural monosaccharide substrates to enable downstream imaging is a powerful application of this approach, however, two complications can be encountered (Figure 1). First, one metabolic precursor may enter multiple glycan biosynthetic pathways, be converted into distinct donor substrates, and get incorporated into more than one sector of glycans16. Second, linkage specificity cannot be controlled within the same sector of glycans due to the presence of multiple glycosyltransferases that transfer the same donor substrate to different acceptors or to the same acceptor substrate but form different linkages8–10. Consequently, MOE cannot be used to detect a unique class of higher order glycans, e.g. disaccharides or trisaccharides, which contains specifically linked monosaccharide building blocks. Neither can MOE be applied to analyse glycosylation patterns in human tissue samples; the feasibility of treating human patients with unnatural carbohydrate metabolic precursors, aside from 2-deoxy-2-(18F)fluoro-D-glucose17, has yet to be verified. Because peripheral higher order glycans, rather than monosaccharides, encode information for cell-surface receptor recognition18, there is an urgent and unmet need to develop methods for their detection.
Figure 1. Inherent limitations of metabolic oligosaccharide engineering.
(1) A metabolic precursor bearing an unnatural chemical tag (X) may be converted into different donor substrates and may enter multiple glycan biosynthetic pathways. (2) Linkage specificity cannot be controlled within the same sector of glycans.
Development of Chemoenzymatic Glycan Labeling: from Radioisotope Probes to Bioorthogonal Probes
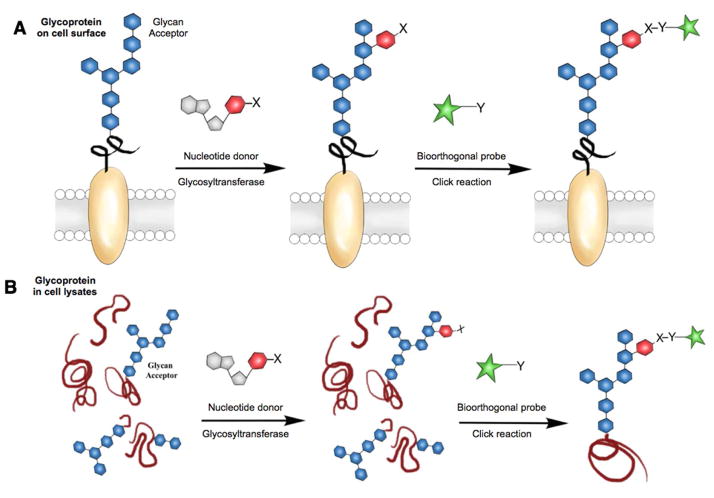
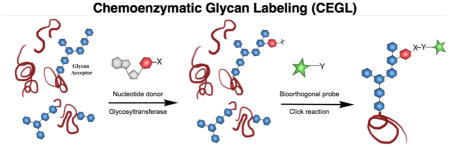
To address the unmet challenges encountered by using conventional glycan detection methods, a new approach has emerged, which we refer to here as chemoenzymatic glycan labeling (CEGL). It should be noted that this approach is also referred to as selective exoenzymatic labeling (SEEL) in some publications19–21; however we believe the use of CEGL is more appropriate for this review since it has a broader scope encompassing both intracellular and extracellular modifications, and it explicitly refers to its application for glycan detection. In this approach, a recombinant glycosyltransferase is used to transfer a monosaccharide analogue from a nucleotide sugar donor to a specific glycan acceptor directly on the cell surface or in cell lysates (Figure 2). The transferred monosaccharide is equipped with a reactive handle that can be further derivatized to incorporate a detection probe.
Figure 2. Chemoenzymatic glycan labeling (CEGL).
In the first step, a monosaccharide equipped with a reactive group (X) is transferred from a nucleotide sugar donor to the target glycan acceptor on a glycoprotein either on the cell surface (A) or in cell lysates (B) by a glycosyltransferase. Subsequently, X is derivatized with a detection probe bearing a complementary reactive group (Y) via bioorthogonal click reaction.
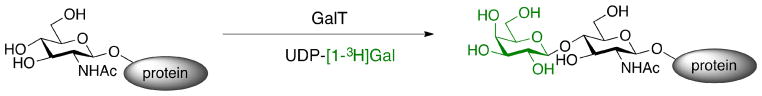
The basis for CEGL finds its roots in early studies by Paulson and coworkers who discovered that recombinant sialyltransferases catalyze the transfer of [14C]-sialic acid to desialylated erythrocytes22. This pioneering work demonstrated that recombinant glycosyltransferases can be used to directly transfer monosaccharides onto the cell surface from nucleotide sugar donors. Another early example of using enzyme-mediated labeling for the direct detection of glycans relied on a bovine milk galactosyltransferase to incorporate radiolabeled galactose onto accessible GlcNAc residues on lymphocytes using a UDP-[1-3H]galactose donor (Figure 3)23. Tracking the lability of the modified glycans, as well as their location within the cells, led to the serendipitous discovery of a Peptide:N-Glycosidase F (PNGaseF)-resistant, alkali labile, single O-linked GlcNAc modification located primarily in the cytoplasm and nucleus of the cells. This paradigm-breaking form of glycosylation has been shown to play essential roles in the modulation of signaling and transcription in response to cellular nutrients or stress23.
Figure 3. Discovery of intracellular O-GlcNAc via the transfer of radiolabeled UDP-Galactose23.
Galactosyltransferase I incorporates radiolabeled galactose from a UDP-[1-3H]Galactose donor onto an O-GlcNAc-modified protein.
Despite these early examples, the use of radioisotopes as tracing tools gradually diminished because of the inherent issues of working with these materials. Advantages associated with fluorescence-based detection, including high sensitivity and multiple transduction approaches, have made fluorescent probes obvious candidates to replace radioisotope probes; nevertheless, their use in CEGL awaited two key discoveries. The first breakthrough came from studies in glycan biosynthesis revealing that many glycan biosynthetic enzymes have a broad donor substrate scope24–27. The second key discovery was the development of bioorthogonal click chemistry reactions28–30. CEGL takes advantage of the promiscuity of glycosyltransferases for their nucleotide donors, which allows for the introduction of monosaccharide analogues bearing bioorthogonal chemical tags. However, the specificity of these enzymes towards their glycan acceptors is the key that allows their use in labeling specific sectors of higher-order glycans, enabling the tagged monosaccharides to be exploited during the second step of bioorthogonal click chemistry-enabled detection24,26. In 2004, Hsieh-Wilson and coworkers reported the first chemoenzymatic method for the detection of O-GlcNAc-modified proteins via the introduction of a galactose analogue bearing a ketone that can be derivatized into an biotin-bearing oxime using N-(aminooxyacetyl)-N′-(D-biotinoyl) hydrazine31.
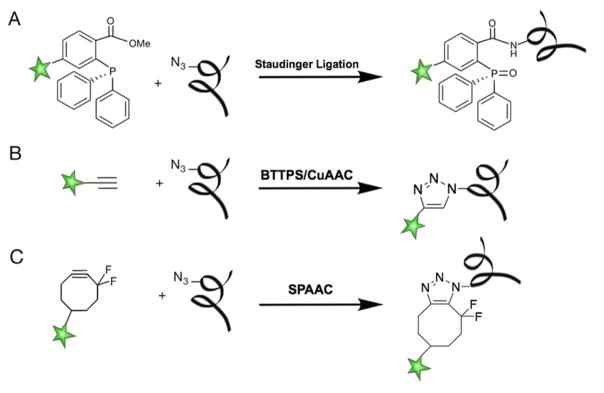
The timely development of bioorthogonal click chemistry provides a facile and biocompatible means to conjugate fluorescent dyes or biotin probes to the tagged glycans in their native environment or in crude cell lysates. In general, bioorthogonal probes need to be small to minimize any perturbations to the native glycan structure29,32,33 and inert under biological conditions. To monitor a biological process that takes place in a minute time scale, a bioorthogonal transformation must be selective and fast. Furthermore, to be performed in a living system, it necessarily needs to be nontoxic. To date, only a small number of reactions fulfill the stringent requirements of bioorthogonality, an in depth details discussion of which can be found in the excellent review by Finn and McKay34. A preeminent example of a biorthogonal reaction is the Staudinger ligation (Figure 4A), which was the first bioorthogonal reaction used to target biomolecules in living animals35,36. A few other reactions have been rendered bioorthogonal by the use of specifically designed reagents that circumvent their original limitations. The ligand-accelerated copper-catalyzed azide-alkyne cycloaddition (CuAAC - Figure 4B)37 is one of the reactions that has been modified to fulfill the conditions of bioorthogonality by the addition of a chelating ligand, such as BTTPS (3-(4-((bis((1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl)propyl hydrogen sulfate) or BTTAA (2-(4-((bis((1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl)acetic acid). The addition of the ligand has not only increased the reaction rates but also reduced the toxicity of copper, allowing the use of CuAAC in biological environments38–40.
Figure 4. Examples of bioorthogonal reactions.
(A) Staudinger Ligation25,36. (B) Ligand-accelerated Copper-catalyzed Azide-Alkyne cycloaddition (CuAAC)37. (C) Cu-free cycloaddition between azides and cyclooctynes known as Cu-free Click Chemistry or Strain-Promoted Alkyne-Azide Cycloadditions (SPAAC)41,84.
Today, the ligand-accelerated CuAAC and copper-free click chemistry, also known as Strain-promoted alkyne-azide cycloadditions (SPAAC - Figure 4C)41,42, remain the most popular click reactions, which serve as the standard conjugation methods of CEGL (Figure 4).
It is also worth noting that constant innovations in carbohydrate chemistry have increased the access to monosaccharide analogues equipped with a variety of chemical handles (e.g. alkyne, azide, aldehyde, diazirine). In parallel, the discovery of previously unknown glycosylation machineries in bacteria provides rich resources for kinases and phosphorylases that tolerate increasing levels of substrate modifications43–46, thus broadening the available toolkit for the preparation of tagged nucleotide mono- or di-phosphate donors available for the CEGL-based applications29,32,37,41,42.
Available CEGL Methods and their Applications
The available CEGL methods, along with their unique features and documented applications, are summarized in Table 1 and Figure 5. With the exception of GalT1, which is mainly used to detect the intracellular O-GlcNAc-modified proteins, the enzymes listed in Table 1 have been tested and validated for the labeling of cell-surface glycans (Figure 6A). The development of biocompatible click reactions has enabled the utilization of these methods in live cells and organisms for characterizing their glycosylation patterns. The installation of biotin tags for glycoprotein pull-down from complex cell lysates is probably one of the most powerful applications enabled by CEGL. The enriched proteins can then be used for mass spectrometry-based analysis to generate a comprehensive profile of proteins of interest or for target validation to confirm the presence of a particular protein using Western blot.
Table 1.
Currently available enzymes for chemoenzymatic glycan detection.
| Glycosyltransferase | Acceptor | Donor | Applications | Ref |
|---|---|---|---|---|
| GalT1 (β1–4 Galatosyltransferase) | GlcNAc-O-R | UDP-GalNAc | Proteomics, In-gel fluorescence | 31 |
| 1–3 FucT (α1–3 Fucosyltransferase 3) | LacNAc | GDP-Fucose | Histology, In vivo imaging, FACS, Proteomics | 13,37,40, 47 |
| BgtA (Human blood group A antigen glycosyltransferase) | Fucα1–2Gal | UDP-GalNAc | In-gel fluorescence, WB, IHC, FACS | 48 |
|
WbwK (α1–2 Fucosyltransferase) BgtA |
Galβ1–3GalNAc | GDP-Fucose, UDP-GalNAc | FACS, WB | 49 |
| CgtA (β1–4 N-acetylgalactosaminyltransferase) | Neu5Acα2–3Gal | UDP-GalNAc | WB, In vivo imagining, FACS, Proteomics | 50 |
| ST6Gal1 (β Galactoside α2–6 sialyltransferase 1) | N-Glycans, mostly LacNAc | CMP-Sia | FACS, WB, Proteomics, In vivo imaging | 19–21 |
| ST3Gal1 (β Galactoside α2–3 sialyltransferase 1) | O-Glycans, mostly core 1 and 2 | CMP-Sia | FACS, WB, Proteomics | 21 |
WB - Western blot; FACS - Flow cytometry; IHC - Immunohistochemistry, CHoMP- CHemoenzymatic labeling of Membrane Polysaccharides.
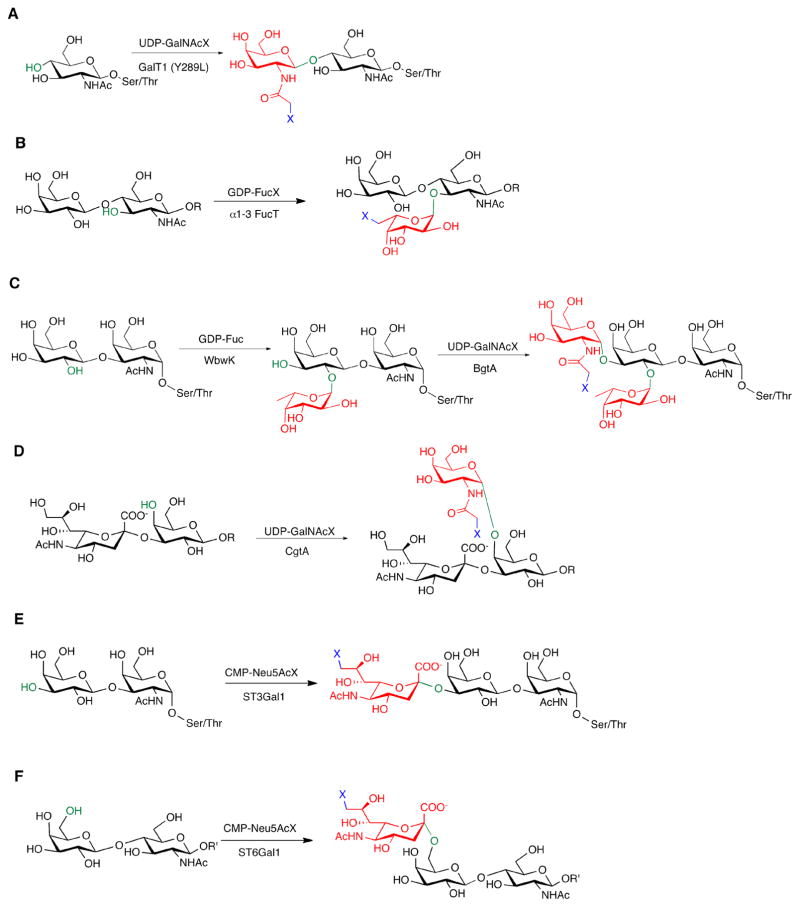
Figure 5. Available chemoenzymatic glycan detection methods.
Highlighted in green are the positions where the new linkages are created, shown in red are the new monosaccharides added, and in blue are the bioorthogonal reactive groups (X); R=linked oligosaccharides, R′=linked oligosaccharides specifically on N-glycans. (A) GalT1-enabled O-GlcNAc detection. (B) α1–3FucT-enabled LacNAc detection. (C) TF-Antigen detection by a tandem approach using WbwK and BgtA, and Fucα1–2Gal detection enabled by BgtA. (D) CgtA-enabled Neu5Acα2–3Gal detection. (E) A example of O-glycan detection enabled by ST3Gal1. (F) A example of N-glycan detection enabled by ST6Gal1.
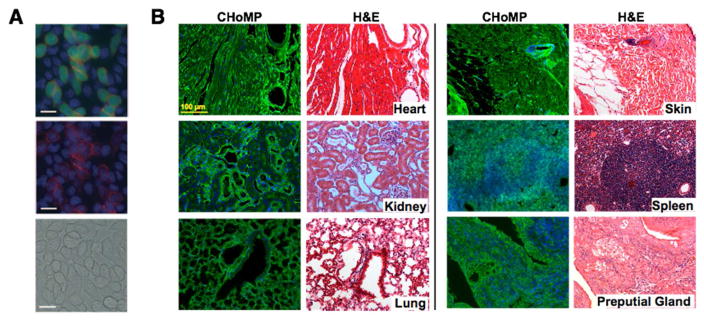
Figure 6. Chemoenzymatic glycan detection applications.
(A) Imaging of cell-surface glycans with LacNAc. Lec2 cells stained with chloromethyl fluorescein diacetate (CMFDA, green) and unstained Lec8 cells were mixed at a 1:5 ratio and were cultured for 3 days. The cells were treated with 500 mM GDP-FucAz and 30 mU α1–3 FucT for 15 min, then labeled with 20 mM DIFO-647 for 25 min. Top: Alexa Fluor 647 image merged with fluorescein (green) and Hoechst 33342 stain (blue); middle: Alexa Fluor 647 image merged with the Hoechst 33342 image; bottom: the bright-field image. Scale bars: 20 mm. Reprinted from 40 with permission from Wiley-VHC Verlag & Co © 2011. (B) CHoMP LacNAc labeling method applied to 5 μm, FFPE mouse heart, kidney, lung, skin, spleen, and preputial gland tissues, next to serial sections of the same tissues stained with H&E (Hematoxylin and eosin). Green: LacNAc staining; Blue: DAPI nuclear staining. Reprinted from 47 with permission from Wiley-VHC Verlag & Co © 2014.
GalT1 for O-GlcNAc detection
Many human pathologies are marked by aberrant O-GlcNAcylation51–53. Therefore, the development of reliable methods for the profiling of proteins containing O-GlcNAc has attracted the attention of glycobiologists for over 30 years. Inspired by the work of Torres and Hart that employed GalT1 to incorporate radiolabeled UDP-[3H]-Gal onto GlcNAc residues, the Hsieh-Wilson method31 exploits a mutated form of β1–4 Galactosyltransferase (GalT1 Y289L) that tolerates unnatural substituents at the 2-acetamido position to enable the transfer of a GlcNAc bearing an unnatural ketone as a chemical handle (Figure 5A). This functional group can be subsequently reacted with an aminooxy-biotin probe for detection by streptavidin conjugates31,54. More recently, the substrate scope of this mutant GalT1 was expanded to include an azide-bearing UDP-GlcNAc as the nucleotide sugar donor, allowing the incorporation of a tag for detection through CuAAC, thus reducing non-specific labeling in crude cell lysates—a disadvantage associated with the ketone/aminooxy reactive pair55.
Subsequently, Hsieh-Wilson and colleagues developed a method to quantify the O-GlcNAc glycosylation stoichiometry and dynamics56. In this method, GalT1 is employed to install a ketogalactose residue to O-GlcNAcylated proteins, which is subsequently reacted with an aminooxy-functionalized polyethylene glycol (PEG) mass tag. In this fashion, tags with defined molecular masses are affixed to O-GlcNAcylated proteins and the stoichiometry of O-GlcNAc modifications can be resolved by SDS-PAGE by simple inspection of mass-shifted bands. O-GlcNAcylated proteins of low abundance can also be visualized by immunoblotting with antibodies for specific proteins of interest and quantified by the relative intensities of each band. This method enables the monitoring of O-GlcNAc levels of proteins ex vivo across different tissues, organs, or diseased states without the need for protein purification, or complex instrumentation. Furthermore, this method enables the quantification of changes in glycosylation on specific proteins. The use of GalT1-based CEGL led to the discovery that pharmacological inhibition of O-GlcNAcase induced significant changes to the glycosylation status of seven O-GlcNAc modified proteins, including those essential for synaptic function, in cultured cortical neurons from embryonic day-18 rats. Analogous to phosphorylation, the reversible O-GlcNAc glycosylation that occurs in neurons may have important roles in mediating neuronal communication57.
The Hsieh-Wilson method and antibody-based O-GlcNAc detection rely on mass spectrometry and immunoblotting to characterize O-GlcNAcylation in the proteome. Although useful for analyzing a small sample pool, these procedures are time-consuming, limited by the relatively low affinity and specificity of anti-O-GlcNAc antibodies58–60, and preferably detect high abundance proteins. To address these issues, Bertozzi and coworkers developed a fast and ultrasensitive detection method referred to as “Glyco-seek”61. Based on proximity ligation assay (PLA), Glyco-seek utilizes GalT1 and CuAAC to install a biotin tag to O-GlcNAc. By conjugating two complementary single-stranded DNA PCR amplicons to two antibodies—one binding to the protein of interest and the other to the biotinylated O-GlcNAc—the ssDNA can be joined by DNA ligase into a complete PCR amplicon to be detected by quantitative polymerase chain reaction (qPCR). This method can detect O-GlcNAcylated samples at levels as low as 0.5 amol (3 orders of magnitude more sensitive than Western blot)61.
α1–3 FucT for LacNAc detection
The chemoenzymatic method for the detection of type II LacNAc-containing glycans, developed by Wu and coworkers, was the first CEGL applied to detect glycans on the cell surface (Figure 5B)62. Many mammalian glycans associated with cell-surface receptors contain LacNAc at the penultimate or terminal position, with functions such as regulation of receptor dimerization and signaling. Such LacNAc-containing glycans have been reported to have an aberrant expression in colorectal carcinoma63. LacNAc is also a ligand for the galectin family of glycan binding proteins involved in mediating apoptosis, proliferation, and differentiation among other critical cellular processes64–66. The Wu method employs a Helicobacter pylori α1–3 fucosyltransferase (1–3 FucT) to transfer an azide or alkyne-bearing fucose to LacNAc on the cell surface, subsequent click ligation with fluorophores or biotin probes functionalized in a complementary fashion enables rapid and sensitive detection of the labeled LacNAc epitope.
LacNAc detection with 1–3 FucT was initially validated in various mutant CHO lines with different glycan expression patterns (Lec2, Lec8 and Lec12)40,67. When using this method to analyze glycosylation patterns of primary murine lymphocytes it was discovered that activated T and B-cells (CD44highCD62Llow, CD25+) express higher levels of LacNAc compared to their naive counterparts (CD44LowCD62Lhigh, CD25−)40. Chemoenzymatic LacNAc detection was also applied on zebrafish embryos to track glycan changes during zebrafish embryogenesis40.
The chemoenzymatic labeling of LacNAc has recently been translated into a histology technique capable of labeling glycans in tissue sections, which was termed CHoMP (CHemoenzymatic Histology of Membrane Polysaccharides; Figure 6B)28. It was discovered that conventional immunohistochemistry does not interfere with the chemoenzymatic labeling. Therefore, CHoMP can be applied to examine glycan expression patterns on select cell types in a tissue sample when combined with antibody-based staining of specific cell markers. Using this method to screen a lung adenocarcinoma microarray, a 13-fold decrease in LacNAc expression from normal lung to grade 1 adenocarcinoma was observed. This observation has recently been validated by Miramoto, Lebrilla and coworkers in a MS-based glycomics analysis68, suggesting that LacNAc may be used as an early detection marker for lung cancer. Although CHoMP has only been used for the detection of LacNAc, the same labeling principle is applicable to examine the distribution and disease-related changes of other glycan epitopes in tissue arrays or whole tissue sections by employing the glycosyltransferases listed in Table 1.
BgtA for Fucα1–2Gal detection
This method exploits a bacterial homologue of human blood group A antigen glycosyltransferase (BgtA) to transfer a GalNAc analogue to the C3 position of galactose within the Fucα1–2Gal acceptor (Figure 5C). Fucα1–2Gal is often found at the non-reducing terminus of cell-surface oligosaccharides, such as blood groups H1 and H2, the glycolipid Globo H, and the Lewis-type structures LeB and LeY 48. BgtA accepts unnatural donors (UDP-GalNAz or UDP-ketoGal) and is highly specific for the terminal Fucα1–2Gal epitope as found in a glycan array screening. This method was directly compared to the lectin UAEI and antibody A46-B/B10 based approaches, both of which are routinely used for detecting the Fucα1–2Gal epitope, with the chemoenzymatic method showing higher sensitivity in identifying proteins known to have this glycan epitope48.
BgtA-based CEGL was first applied to detect Fucα1–2Gal expression across different cancer and non-cancer cell lines. It was found that cancerous cells known to express GloboH (MCF-7, MDA-mb-231, LnCaP) express the highest level of Fucα1–2Gal while the non-cancerous cell PrEC had the lowest level. This observation suggests that the chemoenzymatic labeling Fucα1–2Gal glycans can be used to discriminate cancerous cells from normal cells48. Similar to the work described above, proteomic analysis of Fucα1–2Gal-containing glycoproteins was conducted from neuronal lysates48. Another application of CEGL is target validation. For example, Hsieh-Wilson et al. used BgtA-based CEGL to track fucosylation changes in wild-type, α1-2-fucosyltransferase FUT1 and FUT2 knockout mice69. This study led to the discovery that FUT2 is primarily responsible for Fucα(1-2)Gal biosynthesis in the mouse cortex.
WbwK and BgtA for TF-Antigen detection
BgtA when used in a tandem fashion with the fucosyltransferase WbwK can be used to detect glycans containing terminal Galβ1–3GalNAc, also known as the Thomsen–Friedenreich (TF) antigen (Figure 5C). The TF antigen is highly expressed in various types of human carcinomas, and has been shown to contribute to tumor development, progression, and metastasis70. WbwK has very high substrate specificity towards the TF antigen; it transfers a fucose in a α1-2 linkage to the terminal galactose of the TF antigen. As mentioned above, the Fucα1–2Gal epitope can be targeted by BgtA to introduce a bioorthogonal chemical handle via the transfer of an unnatural UDP-GalNAc donor. Thus, a combination of BgtA and WbwK can be used to convert the disaccharide TF antigen to a tetrasaccharide containing a bioorthogonal functionality for detection49.
This approach was used to visualize TF antigen-modified glycoproteins on the surface of Jurkat cells by flow cytometry, as well as identify specific glycoproteins by proteomic analysis in MCF-7 cells. Furthermore, the combination of two distinct unnatural donors allowed the detection of two glycans on the same cell49. A ketoGal was introduced in an initial blocking step by BgtA, followed by WbwK and BgtA introducing GalNAz. Bioorthogonal ligation of the hydrazine/ketone and alkyne/azide pairs with different fluorophores allowed the simultaneous identification of Fucα1–2Gal and Galβ1–3GalNAc by flow cytometry.
CgtA for Neu5Acα2–3Gal detection
The Neu5Acα2-3Gal epitope can be detected using CgtA—a β1–4 N-acetylgalactosaminyltransferase from Campylobacter jejuni—based CEGL (Figure 5D). This epitope is known as the receptor for many infectious pathogens, including the influenza virus71. Abnormal expression of Neu5Acα2–3Gal has also been observed in many carcinomas72. CgtA accepts the unnatural UDP-GalNAz donor and is highly specific for the Neu5Acα2–3Gal epitope, with only background activity observed for Neu5Acα2–6Gal or desialylated substrates. The specificity of this method was validated using Neu5Acα2–3Gal containing fetal bovine fetuin as a model system50. Its application was then demonstrated by imaging cell-surface Neu5Acα2–3Gal epitopes and proteomic analysis of Neu5Acα2–3Gal modified proteins in human embryonic kidney 293T cells50.
ST6Gal1 for N-Glycan detection
Discovered by Boons and coworkers, sialyltransferase ST6Gal1 can be applied in situ on the cell surface to label a much broader acceptor subset of N-glycans instead of a specific oligosaccharide epitope (Figure 5E). ST6Gal1 adds α2–6-linked sialic acid and its analogs mainly to the terminal Galβ1–4GlcNAc disaccharide in N-linked glycans73. This method has been directly compared to MOE, proving that it has comparable sensitivity but much greater specificity for N-glycans than metabolic labeling19.
A recent report of a one-step ST6Gal1 CEGL strategy using CMP-Neu5Ac analogues conjugated at C5 or C9 with biotin showed significantly enhanced labeling efficiency compared to the two-step approach discussed above20. As demonstrated by proteomic analysis of HeLa cells labeled by the one-step vs. the two-step protocol, 294 proteins were identified at <1% false-discovery rate using the one-step protocol, whereas 174 proteins were identified with the two-step protocol. This one-step strategy was then applied to visualize and identify cell-surface N-linked glycoproteins after the treatment with chloroquine, a lysosomal inhibitor that suppresses protein degradation, to investigate how different glycoproteins respond to lysosomal disruption. This study identified 40 glycoproteins, including IGF2R (cation-independent mannose 6-phosphate receptor) and EPH2A (ephrin B receptor), that undergo internalization and degradation via the endolysosomal system, but accumulate inside cells when lysosomal function is compromised. By contrast, the turnover rate of another set of glycoproteins, including caveolin-1 and mucin-1, was not affected upon chloroquine treatment, suggesting the existence of a different cycling mechanism20.
ST3Gal1 for O-Glycan detection
Complementarily, the same team has developed ST3Gal1 as an efficient reagent to label O-glycans. ST3Gal1 transfers CMP-Neu5Ac and its analogues to Galβ1–3GalNAcαSer/Thr (TF antigen/core 1) and GlcNAcβ1–6(Galβ1–3)GalNAcαSer/Thr (core 2) in mucin type O-linked glycans (Figure 5F)74. As mentioned above, while the TF antigen is exposed in a variety of carcinomas, in healthy tissues core 1 and core 2 epitopes are generally elongated or capped by sialic acid2. A pre-treatment of healthy samples with sialidases (or other exoglycosidases) can therefore expose core 1 and core 2 epitopes to increase the sample pool of O-glycans that can be detected by ST3Gal1 CEGL21.
With both ST6Gal1 and ST3Gal1 in hand, Steet, Wells and coworkers conducted a direct comparison of CEGL with Ac4ManNAz-based MOE to identify N- and O-linked glycoproteins containing terminal sialic acid in human erythroleukemia cells (HEL) cells. To label sialylated glycoproteins via CEGL, HEL cells were first treated with neuraminidase to reveal cell-surface substrates to be modified by ST6Gal1 or ST3Gal1, in N- or O-glycans respectively. Following CEGL and SPAAC-based pull down a total of 103 sialylated glycoproteins were identified vs. 75 proteins identified by metabolic labeling21. A combination of ST6Gal1 and ST3Gal1 were also used to profile the expression of glycoproteins in undifferentiated and differentiated HEL21. It was found that in an undifferentiated state, cells express mostly O-glycoproteins, whereas phorbol myristate acetate (PMA)-activated cells undergo transition to express higher levels of N-glycoproteins21. This study also found that upon differentiation of HEL cells to adherent megakaryocytes, there was increased expression of sialylated cell adhesion molecules, such as the known megakaryocytic markers integrin β3 and CD4421.
Challenges and Future Direction
As a newcomer to the field of chemical glycobiology, CEGL-based methods are still under development and remain relatively few in number. One of the biggest challenges for the future expansion of CEGL is the identification of new glycosyltransferases with strict acceptor specificity but promiscuity for modified nucleotide sugar donors, which can be used for glycan labeling in a cellular system. In the past 30 years, numerous glycosyltransferases from both mammalian and bacterial organisms have been identified and used for the enzymatic synthesis of oligosaccharides75. Pilot studies from our own laboratory have discovered that many of the enzymes active to assemble oligosaccharides in test tubes do not exhibit activity on the cell surface or in complex cell lysates (Wu, P., unpublished results). In addition, the loose acceptor specificity of certain glycosyltransferases render them non-eligible for CEGL-based applications76,77. For example, H. pylori Hpβ3GlcNacT was identified as a GlcNAc transferase that prefers β-linked galactoside acceptor type II (Galβ1-4GlcNAc), but later found to use type I (Galβ1–3GlcNAc), type III (Galβ1–3GalNAcα), and type IV (Galβ1–3GalNAcβ) acceptors on both linear and branched glycans as well. Therefore, while this enzyme is very useful for the in vitro synthesis of oligosaccharides, it is of little use in CEGL for the detection of Galβ1–4GlcNAc-containing glycans78,79.
The challenges in identifying glycosyltransferases with strict acceptor specificity for expanding the CEGL toolkit coincidentally reflect certain limitations of current methods. For example, it is known that besides O-GlcNAc GalT1 Y289L also acts, with less efficiency, on terminal galactoses on N-glycans 80. In addition, ST3Gal1 recognizes various O-glycan epitopes as acceptors. Nevertheless, each method presented here has included measures to ensure that the detection of the target glycans is specific. Lysates used for the detection of O-GlcNAc are subjected to PNGaseF digestion or cell fractionation prior to the GalT1 Y289L treatment to ensure removal of non-specific signals31. In other cases, acceptor promiscuity observed for ST3Gal1 has been harnessed to detect O-linked glycans in general19,21. Notably, all current CEGL methods rely on ex-vivo treatments, and their application in vivo has yet to be explored.
Besides the expansion of the CEGL toolkit for the detection of new glycan targets, coupling of current methods with new probes and imaging technologies is the natural direction to push forward this field. MOE has been coupled to state-of-the-art imaging technologies, producing several very interesting applications. For example, a combination of MOE with a genetically encoded protein (i.e. GFP) or peptide tag (LAP or LplA acceptor peptide) and FRET (Förster resonance energy transfer) has permitted the imaging of a single sialylated glycoprotein, such as GLUT4 and integrin αVβ381,82 on the cell surface in the presence of hundreds of other glycoproteins. Likewise, employing single-molecule tracking, Ovryn and Wu were able to analyze the trajectories of metabolically labeled N- and O-glycoproteins on the membrane of live cancer cells83. These imaging techniques can be readily coupled with CEGL to achieve linkage and protein specific glycosylation imaging.
Studies like those discussed in this review highlight the potential of CEGL for the development of new diagnostic tools for human disease. With the effort of many experienced chemical glycobiologists working tirelessly in the field, we expect in the near future the emergence of innovative tools to monitor the changes of specific glycoforms throughout the cell cycle or accompanying disease progression in real time.
Acknowledgments
The authors’ work highlighted in this review was performed at Albert Einstein College of Medicine with the financial support from the National Institutes of Health to P.W. (GM093282 and GM113046).
References
- 1.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Miklos GLG, Nelson C, Broder S, Clark AG, Nadeau J, Mckusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji R, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang ZY, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu SC, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, Mccawley S, Mcintosh T, Mcmullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers Y, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-stine J, Caulk P, Chiang Y, Coyne M, Dahlke C, Mays AD, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, Mcdaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The Sequence of the Human Genome. Science (80-) 2001;291:1304–1351. [Google Scholar]
- 2.Ajit V, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]
- 3.Freeze HH. Genetic defects in the human glycome. Nat Rev Genet. 2006;7:537–551. doi: 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- 4.Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray analysis of 24 lectins. Angew Chemie - Int Ed. 2006;45:3607–3610. doi: 10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein IJ, Poretz RD. Isolation, Physicochemical Characterization, and Carbohydrate-Binding Specificty of Lectins. In: Liener I, Sharon N, Goldstein IJ, editors. The Lectins: Properties, Functions, and Applications in Biology and Medicine. Academic Press; 1986. [Google Scholar]
- 6.Debray H, Decout D, Strecker G, Spik G, Montreuil J. Specificity of twelve lectins towards oligosaccharides and glycopeptides related to N-glycosylproteins. Eur J Biochem. 1981;117:41–55. doi: 10.1111/j.1432-1033.1981.tb06300.x. [DOI] [PubMed] [Google Scholar]
- 7.Kiernan JA. Histological and Histochemical Methods. 4. Scion Publishing; Bloxham: 2008. [Google Scholar]
- 8.Laughlin ST, Bertozzi CR. Imaging the glycome. Proc Natl Acad Sci. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dube DH, Bertozzi CR. Metabolic oligosaccharide engineering as a tool for glycobiology. Curr Opin Chem Biol. 2003;7:616–625. doi: 10.1016/j.cbpa.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Rouhanifard SH, Nordstrøm LU, Zheng T, Wu P. Chemical Probing of Glycans in Cells and Organisms. Chem Soc Rev. 2013;42:4284–4296. doi: 10.1039/c2cs35416k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mbua NE, Flanagan-steet H, Johnson S, Wolfert MA, Boons G, Steet R. Abnormal accumulation and recycling of glycoproteins visualized in Niemann – Pick type C cells using the chemical reporter strategy. Proc Nat Acad Sci. 2013;110:10207–10212. doi: 10.1073/pnas.1221105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc Natl Acad Sci. 2007;104:2614–2619. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B, Mock F, Wu P. Methods Enzymol. 1. Elsevier Inc; 2012. Imaging the glycome in living systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besanceney-Webler C, Jiang H, Wang W, Baughn AD, Wu P. Metabolic labeling of fucosylated glycoproteins in Bacteroidales species. Bioorganic Med Chem Lett. 2011;21:4989–4992. doi: 10.1016/j.bmcl.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR. Visualizing enveloping layer glycans during zebrafish early embryogenesis. Proc Nat Acad Sci. 2010;107:10350–10365. doi: 10.1073/pnas.0912081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyce M, Carrico IS, Ganguli AS, Yu S, Hangauer MJ, Hubbard SC. Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Nat Acad Sci. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phelps ME, Huang SC, Hoffman EJ, Selin C, Sokoloff L, Kuhl DE. Tomographic measurement of local cerebral glucose metabolic rate in humans with (F-18)2-fluoro-2-deoxy-D-glucose: validation of method. Ann Neurol. 1979;6:371–88. doi: 10.1002/ana.410060502. [DOI] [PubMed] [Google Scholar]
- 18.Gringhuis SI, den Dunnen J, Litjens M, van der Vlist M, Geijtenbeek TB. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat Immunol. 2009;10:1081–1088. doi: 10.1038/ni.1778. [DOI] [PubMed] [Google Scholar]
- 19.Mbua NE, Li X, Flanagan-Steet HR, Meng L, Aoki K, Moremen KW, Wolfert MA, Steet R, Boons GJ. Selective exo-enzymatic labeling of N-glycans on the surface of living cells by recombinant ST6Gal I. Angew Chemie - Int Ed. 2013;52:13012–13015. doi: 10.1002/anie.201307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun T, Yu SH, Zhao P, Meng L, Moremen KW, Wells L, Steet R, Boons GJ. One-Step Selective Exoenzymatic Labeling (SEEL) Strategy for the Biotinylation and Identification of Glycoproteins of Living Cells. J Am Chem Soc. 2016;138:11575–11582. doi: 10.1021/jacs.6b04049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu SH, Zhao P, Sun T, Gao Z, Moremen KW, Boons GJ, Wells L, Steet R. Selective Exo-Enzymatic Labeling Detects Increased Cell Surface Sialoglycoprotein Expression upon Megakaryocytic Differentiation. J Biol Chem. 2016;291:3982–3989. doi: 10.1074/jbc.M115.700369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulson JC, Sadler E, Hill RL. Restoration Incorporation of Specific of Sialic Myxovirus Acid with Receptors to Asialoerythrocytes Pure Sialyltransferases. J Biol Chem. 1979;254:2120–2124. [PubMed] [Google Scholar]
- 23.Torres CR, Hart GW. Topography and Polypeptide Distribution of Terminal N-Acetylglucosamine Residues on the Surfaces of Intact Lymphocytes. J Biol Chem. 1984;259:2208–2217. [PubMed] [Google Scholar]
- 24.Paulson JC, Colley KJ. Glycosyltransferases. J Biol Chem. 1989;264:17615–17618. [PubMed] [Google Scholar]
- 25.Hang HC, Yu C, Pratt MR, Bertozzi CR. Probing glycosyltransferase activities with the Staudinger Ligation. J Am Chem Soc. 2004;126:6–7. doi: 10.1021/ja037692m. [DOI] [PubMed] [Google Scholar]
- 26.Beyer T, Sadler J, Rearick J, Paulson J, Hill R. Glycosyltransferases and their use in assessing oligosaccharide structure and structure-function relationships. Adv Enzym Relat Areas Mol Biol. 1981;52:23–175. doi: 10.1002/9780470122976.ch2. [DOI] [PubMed] [Google Scholar]
- 27.Palcic MM. Glycosyltransferases as biocatalysts. Curr Opin Chem Biol. 2011;15:226–33. doi: 10.1016/j.cbpa.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 28.Jewett JC, Bertozzi CR. Cu-free click cycloaddition reactions in chemical biology. Chem Soc Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baskin JM, Bertozzi CR. Bioorthogonal Click Chemistry: Covalent Labeling in Living Systems. QSAR Comb Sci. 2007;26:1211–1219. [Google Scholar]
- 30.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 31.Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. A Chemoenzymatic Approach toward the Rapid and Sensitive Detection of O-GlcNAc Posttranslational Modifications. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 32.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed Engl. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sletten EM, Bertozzi CR. From mechanism to mouse: A tale of two bioorthogonal reactions. Acc Chem Res. 2011;44:666–676. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKay CS, Finn MG. Click chemistry in complex mixtures: Bioorthogonal bioconjugation. Chem Biol. 2014;21:1075–1101. doi: 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 36.Saxon E, Bertozzi CR. Cell Surface Engineering by a Modified Staudinger Reaction. Science (80-) 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 37.Jiang H, Zheng T, Lopez-Aguilar A, Feng L, Kopp F, Marlow FL, Wu P. Monitoring Dynamic Glycosylation in Vivo Using Supersensitive Click Chemistry. Bioconjug Chem. 2014;25:698–706. doi: 10.1021/bc400502d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dehnert KW, Beahm BJ, Huynh TT, Baskin JM, Laughlin ST, Wang W, Wu P, Amacher SL, Bertozzi CR. Metabolic Labeling of Fucosylated Glycans in Developing Zebrafish. ACS Chem Biol. 2011;6:547–552. doi: 10.1021/cb100284d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, Hong S, Tran A, Jiang H, Triano R, Liu Y, Chen X, Wu P. Sulfated Ligands for the Copper(I)-Catalyzed Azide-Alkyne Cycloaddition. Chem - An Asian J. 2011;6:2796–2802. doi: 10.1002/asia.201100385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng T, Jiang H, Gros M, Soriano Del Amo D, Sundaram S, Lauvau G, Marlow F, Liu Y, Stanley P, Wu P. Tracking N-Acetyllactosamine on Cell-Surface Glycans In Vivo. Angew Chemie. 2011;123:4199–4204. doi: 10.1002/anie.201100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baskin JM, Prescher Ja, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ning X, Guo J, Wolfert MA, Boons GJ. Visualizing Metabolically-Labeled Glycoconjugates of Living Cells by Copper-Free and Fast Huisgen Cycloadditions. Angew Chem Int Ed Engl. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao G, Guan W, Cai L, Wang PG. Enzymatic route to preparative-scale synthesis of UDP-GlcNAc/GalNAc, their analogues and GDP-fucose. Nat Protoc. 2010;5:636–646. doi: 10.1038/nprot.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Yu H, Thon V, Chen Y, Muthana MM, Qu J, Hie L, Chen X. Donor substrate promiscuity of the N-acetylglucosaminyltransferase activities of Pasteurella multocida heparosan synthase 2 (PmHS2) and Escherichia coli K5 KfiA. Appl Microbiol Biotechnol. 2014;98:1127–1134. doi: 10.1007/s00253-013-4947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu H, Yu H, Karpel R, Chen X. Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorg Med Chem. 2004;12:6427–6435. doi: 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 46.Muthana MM, Qu J, Li Y, Zhang L, Yu H, Ding L, Malekana H, Chen X. Efficient one-pot multienzyme synthesis of UDP-sugars using a promiscuous UDP-sugar pyrophosphorylase from Bifidobacterium longum (BLUSP) Chem Commun. 2012;48:2728–2730. doi: 10.1039/c2cc17577k. [DOI] [PubMed] [Google Scholar]
- 47.Rouhanifard SH, Lopez-Aguilar A, Wu P. CHoMP: A chemoenzymatic histology method using clickable probes. Chem Bio Chem. 2014;15:2667–2673. doi: 10.1002/cbic.201402433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaubard JL, Krishnamurthy C, Yi W, Smith DF, Hsieh-Wilson LC. Chemoenzymatic probes for detecting and imaging fucose-α(1-2)-galactose glycan biomarkers. J Am Chem Soc. 2012;134:4489–92. doi: 10.1021/ja211312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q, Li Z, Duan X, Yi W. A Tandem Enzymatic Approach for Detecting and Imaging Tumor- Associated Thomsen – Friedenreich Antigen Disaccharide. J Am Chem Soc. 2014;136:12536–12539. doi: 10.1021/ja5054225. [DOI] [PubMed] [Google Scholar]
- 50.Wen L, Zheng Y, Jiang K, Zhang M, Kondengaden SM, Li S, Huang K, Li J, Song J, Wang PG. Two-Step Chemoenzymatic Detection of N - Acetylneuraminic Acid – α (2-3) -Galactose Glycans. J Am Chem Soc. 2016;138:11473–11476. doi: 10.1021/jacs.6b07132. [DOI] [PubMed] [Google Scholar]
- 51.Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA, Peters EC, Driggers EM, Hsieh-Wilson LC. Phosphofructokinase 1 Glycosylation Regulates Cell Growth and Metabolism. Science (80-) 2012;337:975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parker G, Taylor R, Jones D, McClain D. Hyperglycemia and Inhibition of Glycogen Synthase in Streptozotocin-treated Mice: Role of O-Linked N-Acetylglucosamine. J Biol Chem. 2004;279:20636–20642. doi: 10.1074/jbc.M312139200. [DOI] [PubMed] [Google Scholar]
- 53.Erickson JR, Pereira L, Wang L, Han G, Ferguson A, Dao K, Copeland RJ, Despa F, Hart GW, Ripplinger CM, Bers DM. Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature. 2013;502:372–376. doi: 10.1038/nature12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-wilson LC. Direct In-Gel Fluorescence Detection and Cellular Imaging of O - GlcNAc-Modified Proteins. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rexach JE, Rogers CJ, Yu SH, Tao J, Sun YE, Hsieh-Wilson LC. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat Chem Biol. 2010;6:645–651. doi: 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-wilson LC. Probing the dynamics of O -GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 58.Ma J, Hart GW. O-GlcNAc profiling: from proteins to proteomes. Clin Proteomics. 2014;11:8. doi: 10.1186/1559-0275-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isono T. O-GlcNAc-Specific Antibody CTD110.6 Cross-Reacts with N-GlcNAc2-Modified Proteins Induced under Glucose Deprivation. PLoS One. 2011;6:e18959. doi: 10.1371/journal.pone.0018959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mond JJ, Lees A, Snapper CM. T Cell-Independent Antigens Type 2. Annu Rev Immunol. 1995;13:655–692. doi: 10.1146/annurev.iy.13.040195.003255. [DOI] [PubMed] [Google Scholar]
- 61.Robinson PV, Tsai C, de Groot AE, McKechnie JL, Bertozzi CR. Glyco-seek: Ultrasensitive Detection of Protein-Specific Glycosylation by Proximity Ligation Polymerase Chain Reaction. J Am Chem Soc. 2016;138:10722–10725. doi: 10.1021/jacs.6b03861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W, Hu T, Frantom PA, Zheng T, Gerwe B, Soriano D, Garret S, Seidel RD, Wu P. Chemoenzymatic synthesis of GDP- L -fucose and the Lewis X glycan derivatives. Proc Nat Acad Sci. 2009;106:16096–16101. doi: 10.1073/pnas.0908248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ichikawa T, Nakayama J, Sakura N, Hashimoto T, Fukuda MN, Fukuda MN, Taki T. Expression of N-acetyllactosamine and 1,4-Galactosyltransferase (4GalT-I) During Adenoma-Carcinoma Sequence in the Human Colorectum. J Histochem Cytochem. 1999;47:1593–1601. doi: 10.1177/002215549904701211. [DOI] [PubMed] [Google Scholar]
- 64.Wu C, Thalhamer T, Franca RF, Xiao S, Wang C, Hotta C, Zhu C, Hirashima M, Anderson AC, Kuchroo VK. Article Galectin-9-CD44 Interaction Enhances Stability and Function of Adaptive Regulatory T Cells. Immunity. 2014;41:270–282. doi: 10.1016/j.immuni.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inohara H, Akahani S, Raz A. Galectin-3 stimulates cell proliferation. Exp Cell Res. 1998;245:294–302. doi: 10.1006/excr.1998.4253. [DOI] [PubMed] [Google Scholar]
- 66.Muglia C, Mercer N, Toscano Ma, Schattner M, Pozner R, Cerliani JP, Gobbi RP, Rabinovich Ga, Docena GH. The glycan-binding protein galectin-1 controls survival of epithelial cells along the crypt-villus axis of small intestine. Cell Death Dis. 2011;2:e163. doi: 10.1038/cddis.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanley P. Chinese Hamster Ovary Cell Mutants with Multiple Glycosylation Defects for Production of Glycoproteins with Minimal Carbohydrate Heterogeneity. Mol Cell Biol. 1989;9:377–383. doi: 10.1128/mcb.9.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruhaak LR, Taylor SL, Stroble C, Nguyen UT, Parker EA, Song T, Lebrilla CB, Rom WN, Pass H, Kim K, Kelly K, Miyamoto S. Differential N-glycosylation patterns in lung adenocarcinoma tissue. J Proteome Res. 2015;14:4538–4549. doi: 10.1021/acs.jproteome.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wibowo A, Peters EC, Hsieh-Wilson LC. Photoactivatable Glycopolymers for the Proteome-Wide Identification of Fucose- α (1-2)-Galactose Binding Proteins. J Am Chem Soc. 2014;136:9528–9531. doi: 10.1021/ja502482a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu LG. The oncofetal Thomsen-Friedenreich carbohydrate antigen in cancer progression. Glycoconj J. 2007;24:411–20. doi: 10.1007/s10719-007-9034-3. [DOI] [PubMed] [Google Scholar]
- 71.Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Investig. 2007;87:851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varki A. Sialic acids in human health and disease. Trends Mol Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olio FD, Bologna Á, Giacomo VS, Sperimentale P. The sialyl- a 2, 6-lactosaminyl-structure: Biosynthesis and functional role. Glycoconj J. 2001;17:669–676. doi: 10.1023/a:1011077000164. [DOI] [PubMed] [Google Scholar]
- 74.Blixt O, Han S, Liao L, Zeng Y, Hoffmann J, Futakawa S, Paulson JC. Sialoside Analogue Arrays for Rapid Identification of High Affinity Siglec. J Am Chem Soc. 2008;130:6680–6681. doi: 10.1021/ja801052g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu H, Chen X. One-pot multienzyme (OPME) systems for chemoenzymatic synthesis of carbohydrates. Org Biomol Chem. 2016;14:2809–2818. doi: 10.1039/c6ob00058d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khedri Z, Muthana MM, Li Y, Muthana SM, Yu H, Caoa H, Chen X. Probe sialidase substrate specificity using chemoenzymatically synthesized sialosides containing C9-modified sialic acid. Chem Commun. 2012;48:3357–3359. doi: 10.1039/c2cc17393j. [DOI] [PubMed] [Google Scholar]
- 77.Sugiartoa G, Laua K, Qua J, Lia Y, Lima S, Mua S, Amesa JB, Fishera AJ, Chena X. A Sialyltransferase Mutant with Decreased Donor Hydrolysis and Reduced Sialidase Activities for Directly Sialylating LewisX. ACS Chem Biol. 2012;7:1232–1240. doi: 10.1021/cb300125k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, Xue M, Sheng X, Yu H, Zeng J, Thon V, Chen Y, Muthana MM, Wang PG, Chen X. Donor substrate promiscuity of bacterial β1–3-N-acetylglucosaminyltransferases and acceptor substrate flexibility of β1–4-galactosyltransferases. Bioorg Med Chem. 2016;24:1696–1705. doi: 10.1016/j.bmc.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peng W, Pranskevich J, Nycholat C, Gilbert M, Wakarchuk W, Paulson JC, Razi N. Helicobacter pylori B1,3-N-acetylglycosaminyltransferase for versatile synthesis of type 1 and type 2 poly-LacNAcs on N-linked, O-linked and I-antigen glycans. Glycobiology. 2012;22:1453–1464. doi: 10.1093/glycob/cws101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boeggeman E, Ramakrishnan, Boopathy Kilgore C, Khidekel N, Hsieh-Wilson LC, Simpson JT, Qasba PK. Direct Identification of Nonreducing GlcNAc Residues on N- Glycans of Glycoproteins Using a Novel Chemoenzymatic Method. Bioconjug Chem. 2013;18:806–814. doi: 10.1021/bc060341n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haga Y, Ishii K, Hibino K, Sako Y, Ito Y, Taniguchi N, Suzuki T. Visualizing specific protein glycoforms by transmembrane fluorescence resonance energy transfer. Nat Commun. 2012;3:907. doi: 10.1038/ncomms1906. [DOI] [PubMed] [Google Scholar]
- 82.Haga Y, Suzuki T. Use of transmembrane FRET to investigate the internalization of glycosylated proteins. In: Ivanov AI, Walker JM, editors. Exocytosis and Endocytosis. 2. Vol. 1174. Humana Press; 2014. pp. 225–30. [DOI] [PubMed] [Google Scholar]
- 83.Jiang H, English BP, Hazan RB, Wu P, Ovryn B. Tracking Surface Glycans on Live Cancer Cells with Single-Molecule Sensitivity. Angew Chemie Int Ed. 2015;54:1765–1769. doi: 10.1002/anie.201407976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Copper-free click chemistry in living animals. Proc Natl Acad Sci. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]