Abstract
To facilitate further development of approximate quantum mechanical methods for condensed phase applications, we present a new benchmark dataset of intermolecular interaction energies in the solution phase for a set of 15 dimers, each containing one charged monomer. The reference interaction energy in solution is computed via a thermodynamic cycle that integrates dimer binding energy in the gas phase at the coupled cluster level and solute-solvent interaction with density functional theory; the estimated uncertainty of such calculated interaction energy is kcal/mol. The dataset is used to benchmark the performance of a set of semi-empirical quantum mechanical (SQM) methods that include DFTB3-D3, DFTB3/CPE-D3, OM2-D3, PM6-D3, PM6-D3H+, and PM7 as well as the HF-3c method. We find that while all tested SQM methods tend to underestimate binding energies in the gas phase with a root-mean-squared error (RMSE) of 2-5 kcal/mol, they overestimate binding energies in the solution phase with an RMSE of 3-4 kcal/mol, with the exception of DFTB3/CPE-D3 and OM2-D3, for which the systematic deviation is less pronounced. In addition, we find that HF-3c systematically overestimates binding energies in both gas and solution phases. As most approximate QM methods are parametrized and evaluated using data measured or calculated in the gas phase, the dataset represents an important first step toward calibrating QM based methods for application in the condensed phase where polarization and exchange repulsion need to be treated in a balanced fashion.
INTRODUCTION
There is currently a resurgent interest in semi-empirical quantum mechanical (SQM) methods due to the importance of balancing computational efficiency and accuracy in many chemical and biological applications.1–12 Due to approximations in SQM methods, such as NDDO13,14 and DFTB15–17 methods, they are often not quantitatively accurate as compared to high level ab initio calculations. For example, errors in thermochemical properties such as proton affinity and reaction energy are often in the range of 5-10 kcal/mol;7,18–21 a recent barrier height benchmark for enzyme models conducted by us found that errors can often reach the range of 10-15 kcal/mol.22 Thus systematic improvements of SQM methods are required to make them robust in mechanistic studies of reactive processes.
For non-covalent interactions, on the other hand, SQM methods augmented with empirical corrections2,7,23–31 yield results that can rival modern density functional theory (DFT) and ab initio methods (e.g., SCS-MP232) in accuracy.2,4 For instance, for the S66 dataset,33 which consists of 66 dimers and their gas-phase interaction energies calculated at the Coupled Cluster with Singles and Doubles with Perturbative Triples and Complete Basis Set extrapolation (CCSD(T)/CBS) level of theory, the Root Mean Squared Deviation (RMSD) interaction energy is kcal/mol for both DFT and a set of empirically corrected SQM methods.4 However, we note that empirically adjustable parameters of SQM methods are usually fitted to experimental data or high-level QM data in the gas phase.18,34,35 While datasets generated with high-level QM methods, such as CCSD(T)/CBS36 or SAPT,37 can be highly accurate, it is unclear whether parameters fitted to such data are transferable to condensed-phase systems, where both many-body polarization effects and exchange repulsion need to be treated in a balanced manner.23,38 For example, Yalmazer and Korth found the mean absolute deviation between interaction energies for 695 protein-ligand complexes computed at the PM6-DH+ and BP86-D2/TZVP levels of theory to be as large as 14 kcal/mol.39
In this work, we initiate the development of a dataset for intermolecular interaction energies in the solution phase, such that it is possible to calibrate and improve current SQM methods for condensed phase applications. The dataset is also useful for benchmarking various ab initio QM methods for condensed phase problems, as previous efforts focused on pair-wise interactions in the gas phase,33,40 although the clusters might have been obtained from biomolecules;36 there are also datasets that include solid crystals.2 As described in the section titled Dataset and computational methods, we extend the dataset of Řezáč and Hobza29 and construct a dataset that consists of small molecule dimers solvated by a water droplet; the intermolecular binding energy for the dimer solute is computed via a thermodynamic cycle that integrates CCSD(T) calculations for the dimer interaction in the gas phase and DFT calculations for the solute-solvent (water) interactions. For the SQM methods, we include DFTB341 and DFTB3-CPE,23 which improves the description of polarization via a chemical potential equalization model,42–44 several variants of the PM6 model24,28 and the OM2-D3 model;35 we also test the HF-3c model,45 which employs Hartree-Fock with a minimal basis along with several empirical corrections. Among those, we note that most empirical corrections are mechanical in nature (i.e., decoupled from the determination of the wavefunction or electron density), with the exception of DFTB3-CPE,23 in which the auxiliary response function is self-consistent with the DFTB3 density. Therefore, we expect that the DFTB3-CPE model is more likely to be transferable to the solution phase, although parameters were determined entirely based on gas phase systems. This was already evident from our recent analysis23 of “large” water clusters (which contain 6-17 water molecules46), for which empirically corrected PM6 models were found to give rather large errors in the binding energies, despite their excellent performance for dimeric systems.
In the following, we first describe the model systems and the computational approach we take to calculate the intermolecular binding energies in the solution phase. We then present the benchmark results for a set of SQM methods. We conclude with a few remarks and comments on future work.
DATASET AND COMPUTATIONAL METHODS
Description of the dataset
The dataset presented herein consists of 15 different dimers motivated by biologically relevant molecules (see Table I) and they are embedded in a water droplet; three droplets that contain water molecules within 4, 6, and 8 Å of the solute are studied for each dimer. An example is shown in Fig. 1 for the formaldehyde ⋅ imidazolium dimer with a 4.0 Å thick water shell, which roughly corresponds to the first solvation shell. Table I lists the computed interaction energies for the 15 dimers in the gas phase and in the 8.0 Å water shell, which roughly corresponds to inclusion of up to the third solvation shell with a total of around 600 atoms. We limit discussion to the 8 Å systems in the main text, and results for smaller water shells are summarized in the supplementary material.
TABLE I.
Computed intermolecular binding energy in the gas-phase (g) and in an 8.0 Å water shell (aq) following the thermodynamic cycle in Fig. 2.a
| ID | Complex | b | |
|---|---|---|---|
| 1 | Acetate ⋅ methanol | −19.8 | −12.6 |
| 2 | Acetate ⋅ water | −21.1 | −15.3 |
| 3 | Acetate ⋅ methylamine | −11.5 | −0.5 |
| 4 | Methylammonium ⋅ formaldehyde | −19.1 | −11.9 |
| 5 | Methylammonium ⋅ methylamine | −28.6 | −22.6 |
| 6 | Methylammonium ⋅ methanol | −21.2 | −15.5 |
| 7 | Methylammonium ⋅ water | −18.5 | −13.8 |
| 8 | Guanidinium ⋅ formaldehyde | −18.1 | −13.0 |
| 9 | Guanidinium ⋅ methylamine | −20.2 | −16.0 |
| 10 | Guanidinium ⋅ methanol | −19.8 | −15.4 |
| 11 | Guanidinium ⋅ water | −17.5 | −14.0 |
| 12 | Imidazolium ⋅ formaldehyde | −16.4 | −11.7 |
| 13 | Imidazolium ⋅ methylamine | −26.0 | −23.1 |
| 14 | Imidazolium ⋅ methanol | −18.9 | −14.0 |
| 15 | Imidazolium ⋅ water | −16.5 | −12.1 |
Intermolecular binding energies are calculated using a combined CCSD(T)/DFT approach [see Eq. (3)]. The uncertainty is kcal/mol for and kcal/mol for . All energies are listed in units of kcal/mol.
Gas-phase interaction energies calculated at the CCSD(T)/CBS level are taken from the work of Řezáč and Hobza.29
FIG. 1.
An example for the dimer systems studied in the current work. The dimer between formaldehyde and imidazolium is studied in both the (a) gas phase and (b) solution phase, which is modeled by a water droplet that includes water molecules within a given distance (4, 6, or 8 Å) from the solute. The model in (b) contains water molecules within 4 Å from the solute. Discussion is limited to the 8 Å systems in the main text, and results for smaller water shells are summarized in the supplementary material.
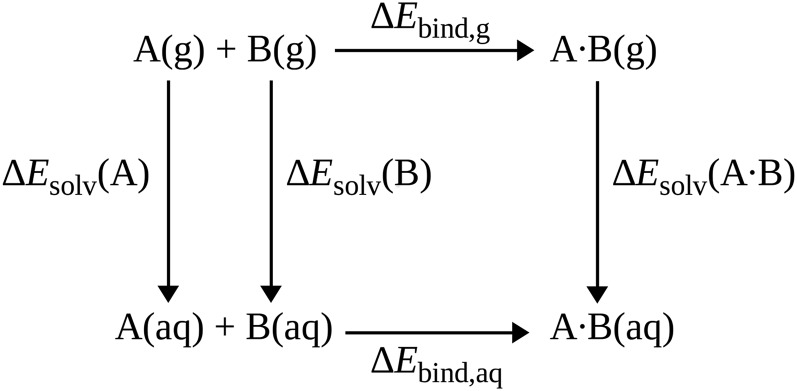
Calculation of reference binding energies in the solution phase
To compute the reference value for the binding energies in solution, we use an approach similar to the thermodynamic cycle (summarized in Fig. 2) used to compute binding free energies in solution.47,48 The dimer complex of monomers A and B is placed in a water droplet, and after equilibration with molecular dynamics (see below), the dimer complex and a surrounding shell of water molecules are extracted. The binding energy in the gas phase () is simply calculated as
| (1) |
For the values of , we use those calculated by Řezáč and Hobza29 at the level of CCSD(T)/CBS. For the “solvation energies” (note that no entropic contribution is considered in the current work), considering the large size of the water droplet, we compute these values at the DFT level (PBE049/def2-TZVP-D3),
| (2) |
where X is either one of the monomers A or B, or the complex A ⋅ B, and W is the water shell surrounding the complex. Note that the same geometry of W (extracted from the molecular dynamics snapshot for a solvated dimer AB) is used for the monomers (A and B) as for the dimer AB.
FIG. 2.
The thermodynamic cycle used to calculate the binding energy in the solution phase.
The final reference binding energy () is then calculated by combining the gas phase binding energy and the “solvation energies,”
| (3) |
The values of for each complex in the dataset are listed in Table I. For each dimer AB, only one solvent snapshot is used in the current work to estimate the “solvation energies” (although three water droplets of different sizes are examined, see the supplementary material) and this is done considering the large computational cost associated with DFT calculations for systems that consist of hundreds of atoms. The dataset can be systematically expanded by including additional snapshots for each dimer.
We note that the solvated binding energy, , is not meant to estimate an average binding energy or binding free energy since the water molecules are not equilibrated when the molecules are removed. Rather is a measure of the binding interaction between the two molecules for a specific conformation of the molecular complex in the presence of surrounding water molecules.
For the SQM methods tested here, the same equation [Eq. (3)] is used, except that all components (gas phase binding and “solvation energies”) are computed at the same respective SQM method; the structures are not re-optimized, all reported values in Table II are based on single point energies at the SQM levels, and original binding energies are summarized in the supplementary material.
TABLE II.
The root mean squared error (RMSE) and mean error (Mean) to reference binding energy values for 7 methods on the C15 and solvated C15 datasets in units of kcal/mol. Note that the mean error of the null model is 0 kcal/mol by construction.
| Gas | Solution | |||
|---|---|---|---|---|
| Method | RMSE | Mean | RMSE | Mean |
| DFTB3-D3 | 5.0 | 3.6 | 3.9 | 0.6 |
| DFTB3/CPE-D3 | 1.9 | 0.9 | 2.2 | −0.6 |
| HF-3c | 4.4 | −3.6 | 7.0 | -6.6 |
| OM2-D3 | 2.1 | 1.8 | 1.9 | −1.4 |
| PM6-D3 | 2.1 | 0.9 | 3.5 | −2.6 |
| PM6-D3H+ | 2.2 | 1.3 | 3.7 | −3.0 |
| PM7 | 1.9 | 1.0 | 2.9 | −2.2 |
| Null | 3.8 | (0.0) | 4.9 | (0.0) |
Generation of solvated structures
The MP2-optimized structures from the dataset of 15 charged dimers from the work of Řezáč and Hobza29 (hereafter abbreviated as “C15”) are placed in a 20 Å radius water shell with the standard stochastic boundary condition,50 and MD simulations are carried out with the CHARMM program.51 The dimer is treated with DFTB3/3OB18 and water is treated with a modified TIP3P model;52,53 the internal structure of the dimer is held fixed during the molecular dynamics (MD) simulations, which are conducted for 1 ns with a time step of 2 fs. SHAKE54 is applied to constrain the structure of the water molecules, and non-bonded interactions are treated using extended electrostatics55 and a switching function56 for the van der Waals interaction between 8 and 12 Å. The structure from the very last step in the 1 ns MD simulation is used to construct the various water droplet models.
QM calculations
ORCA57 version 3.0.3 is used for all PBE0/def2-TZVP-D3 and HF-3c calculations. The RIJCOSX approximation is used for PBE0 to make the largest calculations computationally tractable. MOPAC201258 15.229L is used for PM6-D3 and PM7 calculations, while a developmental version of GAMESS59 is used for PM6-D3H+ calculations. MNDO200560 version 7.0 is used for the OM2 calculations, along with the DFT-D3 program61,62 for calculating the D3 dispersion energy term for OM2-D3. CHARMM51 version c41a1 is used for all DFTB3-D3 and DFTB3/CPE-D3 calculations and for the DFTB3/3OB/TIP3P MD simulations to generate the solvated dimer structures. We note that all corrections to the approximate QM methods4 discussed here, the D3 models for empirical dispersion, the H+ model for hydrogen-bonding interactions, and the “3c” correction for HF, are all mechanical in nature in that they are not coupled with the wavefunction/density determination. By contrast, the CPE correction is fully self-consistent with the determination of density in the DFTB3/CPE model.23
RESULTS
Uncertainty estimates for the reference binding energies
First, we estimate the uncertainty in the reference binding energies [, see Eq. (3)] that arises due to the choice of the method and basis set, using a standard propagation of error. For the gas phase binding energy, , the error in the CCSD(T)/CBS values is about 1.5% of the total interaction energies, which corresponds to 0.2 kcal/mol on average for the dataset.63 For the solvation energies, we estimate the uncertainty as follows: the calculations of are repeated for three cases (acetate ⋅ methanol, acetate ⋅ water, and guanidinium ⋅ formaldehyde complexes) with the smallest solvent shell (i.e., within 4 Å of the solute) with four additional functionals: B3LYP,64 M06-2X,65 TPSS,66 and PBE,67 and the basis sets def2-TZVP and def2-QZVP of these interaction energies are displayed in Table S7 of the supplementary material. In all cases, the differences in interaction energies between different DFT functionals are less than 1.1 kcal/mol. Likewise, the differences between results using the def2-TZVP and def2-QZVP basis sets are less than 1.0 kcal/mol.
Using the difference between the five different DFT functionals and the difference between def2-TZVP and def2-QZVP calculations as estimates for the uncertainty in choice of method and basis set, it is possible to use propagation of error to calculate an uncertainty estimate: under the assumption that these errors are random in nature, the total error is kcal/mol for the binding energies in solution (). Although such uncertainty is larger than the “chemical accuracy” (usually defined as kcal/mol), it is small compared to the precision of the SQM methods assessed in this work.
Accuracy of semi-empirical quantum methods
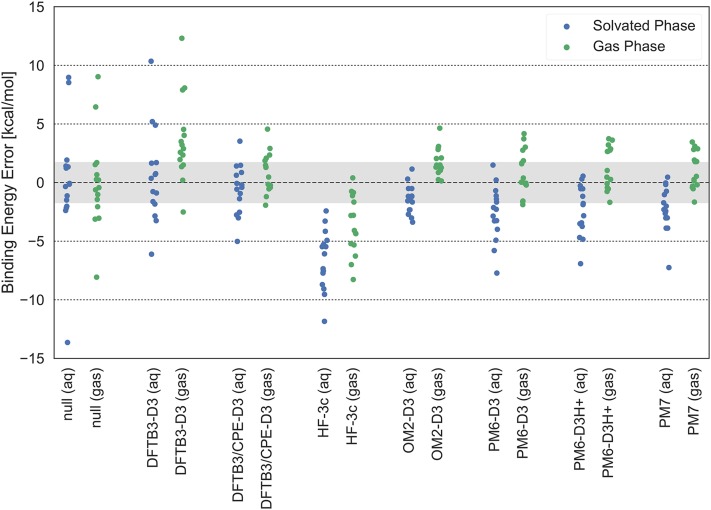
In this section, we use the dataset to benchmark the performance of the SQM methods DFTB3-D3, DFTB3/CPE-D3, OM2-D3, PM6-D3, PM6-D3H+, and PM7 as well as the HF-3c method in solution phase.14,23,28,34,35,41 An overview of the root-mean-squared error (RMSE) in binding energies and mean errors is summarized in Table II. Graphical overviews of the errors are presented in Figs. 3 and 4.
FIG. 3.
The deviation (error) between binding energies calculated using various semi-empirical methods and reference values (see Table I), for the set of 15 dimers in the gas phase (gas) or solution phase (aq). The gray line denotes an estimated kcal/mol uncertainty in the reference values in the solution phase. The gas-phase reference of CCSD(T)/CBS is expected to carry only a small uncertainty of kcal/mol for which the gray error bar is not representative. See text for the discussion of the null models.
FIG. 4.
Root-mean-squared error (RMSE) between binding energies calculated using various semi-empirical methods and reference values (see Table I), for the set of 15 dimers in the gas phase (gas) or solvated phase (aq). Error bars denote the 95% confidence interval, obtained via a boot-strapping procedure. See text for the discussion of the null models.
For DFTB3-D3, the error in the gas-phase is dominated by complexes with binding interactions with the nitrogen lone pair in methylamine. This problem has been noted previously and likely reflects the monopole approximation for lone pairs in charge-charge interactions in the current DFTB framework.23 On average, DFTB3-D3 exhibits systematic underbinding in the gas-phase for all complexes, except for the acetate ⋅ water complex, by 3.6 kcal/mol. In the solution phase, DFTB3-D3 does not systematically predict over- or underbinding, except for the complexes containing methylamine, which are still systematically predicted to underbind by 5-10 kcal/mol.
The polarization-corrected method DFTB3/CPE-D323 has been parametrized based on the gas-phase dataset (with the formaldehyde ⋅ imidazolium complex excluded) and is therefore by design more balanced and accurate for the gas-phase data. Encouragingly, this trend is maintained for the solution phase dataset DFTB3/CPE-D3, which exhibits the smallest mean error (−0.6 kcal/mol) among the SQM methods tested here, suggesting that the CPE correction is transferable to the solution phase. The RMSE values relative to the reference binding energies are comparable in the gas phase (1.9 kcal/mol) and solution phase (2.2 kcal/mol).
The three methods of the PMx family we benchmark here, PM6-D3, PM6-D3H+, and PM7, have been parametrized predominantly based on experimental heats of formation and they display a similar degree of accuracy in both the gas and solution phases. In all cases, the RMSE relative to the reference binding energies is around 2 kcal/mol in the gas-phase and around 3 kcal/mol in the solution phase; these are slightly larger, especially the solution phase values, than DFTB3/CPE-D3. All three PMx methods predict underbinding by around 1 kcal/mol in the gas phase and overbinding by between 2 and 3 kcal/mol in the solution phase. The use of the third-generation hydrogen-bond correction H+ (and slight reparametrization of the D3 parameters), in the case of PM6-D3H+, does not increase nor decrease the accuracy of the PM6 model when compared to PM6-D3. This presumably reflects the mechanical nature of the H+ model, which does not account for the missing many-body polarization in the PM6 method.
The orthogonalization-corrected OM2-D3 method shows very encouraging performance. In the gas phase, interaction energies are predicted to overbind by 1.8 kcal/mol on average, but with a small random error. In the solution phase, the RMSE of 1.9 kcal/mol is the lowest among all SQM methods tested here; although the mean error of −1.4 kcal/mol is slightly larger than that of DFTB3/CPE-D3, the accuracy of the methods does not differ with any statistical significance ().
The method tested with the least empiricism, HF-3c, which has only 9 adjustable parameters, is observed to overbind systematically in both the gas and solution phases. In fact, HF-3c systematically overbinds by −3.6 kcal/mol on average in the gas phase, and this error increases to −6.6 kcal/mol in the solution phase. These rather large errors suggest that parameters in the geometric counter-poise correction (gCP) and the short-range basis set-incompleteness correction (SRB) are not tuned adequately for the solution environment.
As a statistical test, we also compare all models to a null model. The null model has one degree of freedom which we simply fit to the mean reference value. Thus the null model yields the binding energy −19.5 kcal/mol for complexes in the gas phase, and −14.1 kcal/mol for the solvated complexes. The RMSE for the null models is 3.8 kcal/mol for the gas-phase complexes and 4.9 kcal/mol for the solvated complexes. Using the statsig program,68 only the OM2 and DFTB3/CPE methods show up as being significantly different (with ) from the constructed null model for the solvated complexes—in both cases being more accurate than the null model. For the gas-phase reference data, only PM7 is significantly different from the null-model with , although the accuracy of PM7 is very comparable with an RMSE within few tenths of a kcal/mol to those of the methods DFTB3/CPE-D3, OM2-D3, PM6-D3, and PM6-D3H+. A detailed statistical comparison between the various methods is summarized in Tables S5 and S6 of the supplementary material.
We note that the acetate ⋅ water complex appears as a sort of an outlier with the largest negative binding energy error (i.e., predicted to bind too strong compared to the reference method) for all methods except OM2-D3 (see, e.g., Fig. 3). Since the error is systematic across all methods, it is entirely possible that the DFT reference is not accurate enough for this particular case—if the reference value had been 2-3 kcal/mol stronger, corresponding to 1-2 times the estimated uncertainly, the complex would not show up as a significant outlier. Despite this uncertainty, it is still apparent that most methods predict very strong overbinding for this complex—much larger than those that can be explained by errors in the reference binding energies.
CONCLUSION
We have developed a new dataset of intermolecular interaction energies for molecules in the solution phase, approximated by water shells that include up to the third solvation shell. The dataset is available under the terms of the CC0 license at https://github.com/Cuigroup/c15solv. The reference binding energies are computed using a thermodynamic cycle that integrates CCSD(T)/CBS binding energies in the gas phase and DFT solute-solvent interaction energies; the estimated uncertainty of the reference values is kcal/mol. The reference values are used to benchmark a set of SQM methods that include DFTB3-D3, DFTB3/CPE-D3, OM2-D3, PM6-D3, PM6-D3H+, and PM7 as well as the HF-3c method. Among these, only the two methods OM2 and DFTB3/CPE are better than the null model with statistical significance ().68 It is encouraging that the CPE-correction developed for DFTB3 is transferable to the solution phase since all parameters were developed based entirely on gas phase molecules. By contrast, the PMx family of methods appears to be somewhat more accurate in the gas phase but systematically predicts overbinding in the solution phase, and the addition of the H+ correction neither increases nor decreases the accuracy in the gas or the solution phase. These observations highlight the merits of developing systematic improvements of SQM methods that are explicitly coupled to the electronic density in a self-consistent manner.
We hope that the database will facilitate further calibration and developments of SQM methods and other approximate QM methods for the description of non-covalent interactions in the condensed phase. The thermodynamic cycle approach can also be used to estimate accurate binding interactions in heterogeneous environments such as proteins, nucleic acids, and lipid bilayers. Including additional snapshots will also help calibrate approximate QM methods for the description of entropic aspects of intermolecular interactions in the condensed phase. Further exploration for the relative orientations and separations of the solute molecules is also essential for ensuring continuous potential energy surfaces with proper geometries for key stationary points.31
SUPPLEMENTARY MATERIAL
See supplementary material for detailed benchmark data for gas-phase and different solution phase systems.
ACKNOWLEDGMENTS
This work is supported by NIH Grant No. R01-GM106443 to QC. Computational resources from the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by NSF Grant No. OCI-1053575, are greatly appreciated; computations are also supported in part by NSF through a major instrumentation Grant (No. CHE-0840494) to the Chemistry department.
REFERENCES
- 1.Hobza P. and Řezáč J., “Introduction: Noncovalent interactions,” Chem. Rev. 116, 4911–4912 (2016). 10.1021/acs.chemrev.6b00247 [DOI] [PubMed] [Google Scholar]
- 2.Brandenburg J., Hochheim M., Bredow T., and Grimme S., “Low-cost quantum chemical methods for noncovalent interactions,” J. Phys. Chem. Lett. 5, 4275–4284 (2014). 10.1021/jz5021313 [DOI] [PubMed] [Google Scholar]
- 3.Gao J. L., Truhlar D. G., Wang Y. J., Mazack M. J. M., Loffler P., Provorse M. R., and Rehak P., “Explicit polarization: A quantum mechanical framework for developing next generation force fields,” Acc. Chem. Res. 47, 2837–2845 (2014). 10.1021/ar5002186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen A. S., Kubař T., Cui Q., and Elstner M., “Semiempirical quantum mechanical methods for noncovalent interactions for chemical and biochemical applications,” Chem. Rev. 116, 5301–5337 (2016). 10.1021/acs.chemrev.5b00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giese T. J., Chen H., Dissanayake T., Giambasu G. M., Heldenbrand H., Huang M., Kuechler E. R., Lee T. S., Panteva M. T., Radak B. K. et al. , “A variational linear-scaling framework to build practical, efficient next-generation quantum force fields,” J. Chem. Theory Comput. 9, 1417–1427 (2013). 10.1021/ct3010134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui Q. and Elstner M., “Density functional tight binding: Values of semi-empirical methods in an ab initio era,” Phys. Chem. Chem. Phys. 16, 14368–14377 (2014). 10.1039/c4cp00908h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dral P. O., Wu X., Sporkel L., Koslowski A., and Thiel W., “Semiempirical quantum-chemical orthogonalization-corrected methods: Benchmarks for ground-state properties,” J. Chem. Theory Comput. 12, 1097–1120(2016). 10.1021/acs.jctc.5b01047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui Q., “Perspective: Quantum mechanical methods in biochemistry and biophysics,” J. Chem. Phys. 145, 140901 (2016). 10.1063/1.4964410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghavachari K. and Saha A., “Accurate composite and fragment-based quantum chemical models for large molecules,” Chem. Rev. 115, 5643–5677 (2015). 10.1021/cr500606e [DOI] [PubMed] [Google Scholar]
- 10.Giese T. J., Chen H., Huang M., and York D. M., “Parametrization of an orbital-based linear-scaling quantum force field for noncovalent interactions,” J. Chem. Theory Comput. 10, 1086–1098 (2014). 10.1021/ct401035t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M., Giese T. J., Lee T.-S., and York D. M., “Improvement of DNA and RNA sugar pucker profiles from semiempirical quantum methods,” J. Chem. Theory Comput. 10, 1538–1545 (2014). 10.1021/ct401013s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giese T. J., Huang M., Chen H., and York D. M., “Recent advances toward a general purpose linear-scaling quantum force field,” Acc. Chem. Res. 47, 2812–2820 (2014). 10.1021/ar500103g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiel W., “Perspectives on semiempirical molecular orbital theory,” Adv. Chem. Phys. 93, 703–757 (1996). 10.1002/9780470141526.ch10 [DOI] [Google Scholar]
- 14.Stewart J., “Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters,” J. Mol. Model 19, 1–32 (2013). 10.1007/s00894-012-1667-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elstner M., Porezag D., Jungnickel G., Elsner J., Haugk M., Frauenheim T., Suhai S., and Seifert G., “Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties,” Phys. Rev. B 58, 7260–7268 (1998). 10.1103/physrevb.58.7260 [DOI] [Google Scholar]
- 16.Elstner M., Frauenheim T., and Suhai S., “An approximate DFT method for QM/MM simulations of biological structures and processes,” J. Mol. Struct.: THEOCHEM 632, 29–41 (2003). 10.1016/s0166-1280(03)00286-0 [DOI] [Google Scholar]
- 17.Gaus M., Cui Q., and Elstner M., “Density functional tight binding (DFTB): Application to organic and biological molecules,” Wiley Interdiscip. Rev.: Comput. Mol. Sci. 4, 49–61 (2014). 10.1002/wcms.1156 [DOI] [Google Scholar]
- 18.Gaus M., Goez A., and Elstner M., “Parametrization and benchmark of DFTB3 for organic molecules,” J. Chem. Theory Comput. 9, 338–354 (2013). 10.1021/ct300849w [DOI] [PubMed] [Google Scholar]
- 19.Gaus M., Lu X., Elstner M., and Cui Q., “Parameterization of DFTB3/3OB for sulfur and phosphorus for chemical and biological applications,” J. Chem. Theory Comput. 10, 1518–1537 (2014). 10.1021/ct401002w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nam K., Cui Q., Gao J., and York D. M., “Specific reaction parametrization of the AM1/d Hamiltonian for phosphoryl transfer reactions: H, O, and P atoms,” J. Chem. Theory Comput. 3, 486–504 (2007). 10.1021/ct6002466 [DOI] [PubMed] [Google Scholar]
- 21.Huang M., Giese T. J., and York D. M., “Nucleic acid reactivity: Challenges for next-generation semiempirical quantum models,” J. Comput. Chem. 36, 1370–1389 (2015). 10.1002/jcc.23933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kromann J. C., Christensen A. S., Cui Q., and Jensen J. H., “Towards a barrier height benchmark set for biologically relevant systems,” PeerJ 4, e1994 (2016). 10.7717/peerj.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen A. S., Elster M., and Cui Q., “Improving intermolecular interactions in DFTB3 using extended polarization from chemical-potential equalization,” J. Chem. Phys. 143, 084123 (2015). 10.1063/1.4929335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Řezáč J., Fanfrlík J., Salahub D., and Hobza P., “Semiempirical quantum chemical PM6 method augmented by dispersion and H-bonding correction terms reliably describes various types of noncovalent complexes,” J. Chem. Theory Comput. 5, 1749–1760 (2009). 10.1021/ct9000922 [DOI] [PubMed] [Google Scholar]
- 25.Korth M., Pitoňák M., Řezáč J., and Hobza P., “A transferable H-bonding correction for semiempirical quantum-chemical methods,” J. Chem. Theory Comput. 6, 344–352 (2010). 10.1021/ct900541n [DOI] [PubMed] [Google Scholar]
- 26.Řezáč J. and Hobza P., “A halogen-bonding correction for the semiempirical PM6 method,” Chem. Phys. Lett. 506, 286–289 (2011). 10.1016/j.cplett.2011.03.009 [DOI] [Google Scholar]
- 27.Korth M., “Third-generation hydrogen-bonding corrections for semiempirical QM methods and force fields,” J. Chem. Theory Comput. 6, 3808–3816 (2010). 10.1021/ct100408b [DOI] [PubMed] [Google Scholar]
- 28.Kromann J. C., Christensen A. S., Steinmann C., Korth M., and Jensen J. H., “A third-generation dispersion and third-generation hydrogen bonding corrected PM6 method: PM6-D3H+,” PeerJ 2, e449 (2014). 10.7717/peerj.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Řezáč J. and Hobza P., “Advanced corrections of hydrogen bonding and dispersion for semiempirical quantum mechanical methods,” J. Chem. Theory Comput. 8, 141–151 (2012). 10.1021/ct200751e [DOI] [PubMed] [Google Scholar]
- 30.Řezáč J., Riley K. E., and Hobza P., “Benchmark calculations of noncovalent interactions of halogenated molecules,” J. Chem. Theory Comput. 8, 4285–4292 (2012). 10.1021/ct300647k [DOI] [PubMed] [Google Scholar]
- 31.Miriyala V. M. and Rezac J., “Description of non-covalent interactions in SCC-DFTB methods,” J. Comput. Chem. 38, 688–697 (2017). 10.1002/jcc.24725 [DOI] [PubMed] [Google Scholar]
- 32.Antony J. and Grimme S., “Is spin-component scaled second-order moller-plesset perturbation theory an appropriate method for the study of noncovalent interactions in molecules?,” J. Phys. Chem. A 111, 4862–4868 (2007). 10.1021/jp070589p [DOI] [PubMed] [Google Scholar]
- 33.Řezáč J., Riley K. E., and Hobza P., “S66: A well-balanced database of benchmark interaction energies relevant to biomolecular structures,” J. Chem. Theory Comput. 7, 2427–2438 (2011). 10.1021/ct2002946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart J. J. P., “Optimization of parameters for semiempirical methods V: Modification of NDDO approximations and application to 70 elements,” J. Mol. Model. 13, 1173–1213 (2007). 10.1007/s00894-007-0233-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scholten M., “Semiempirische verfahren mit orthogonalisierungskorrekturen: Die OM3 Methode,” Ph.D. thesis, Heinrich-Heine-Universität, Düsseldorf, 2003. [Google Scholar]
- 36.Marshall M. S., Burns L. A., and Sherrill C. D., “Basis set convergence of the coupled-cluster correction, delta(CCSD(T))(MP2): Best practices for benchmarking non-covalent interactions and the attendant revision of the S22, NBC10, HBC6, and HSG databases,” J. Chem. Phys. 135, 194102 (2011). 10.1063/1.3659142 [DOI] [PubMed] [Google Scholar]
- 37.Parker T. M., Burns L. A., Parrish R. M., Ryno A. G., and Sherrill C. D., “Levels of symmetry adapted perturbation theory (SAPT). I. Efficiency and performance for interaction energies,” J. Chem. Phys. 140, 094106 (2014). 10.1063/1.4867135 [DOI] [PubMed] [Google Scholar]
- 38.Giese T. J. and York D. M., “Many-body force field models based solely on pairwise coulomb screening do not simultaneously reproduce correct gas-phase and condensed-phase polarizability limits,” J. Chem. Phys. 120, 9903–9906 (2004). 10.1063/1.1756583 [DOI] [PubMed] [Google Scholar]
- 39.Yilmazer N. D. and Korth M., “Comparison of molecular mechanics, semi-empirical quantum mechanical, and density functional theory methods for scoring protein–ligand interactions,” J. Phys. Chem. B 117, 8075–8084 (2013). 10.1021/jp402719k [DOI] [PubMed] [Google Scholar]
- 40.Jurečka P., Šponer J., Černý J., and Hobza P., “Benchmark database of accurate (MP2 and CCSD(t) complete basis set limit) interaction energies of small model complexes, DNA base pairs and amino acid pairs,” Phys. Chem. Chem. Phys. 8, 1985–1993 (2006). 10.1039/b600027d [DOI] [PubMed] [Google Scholar]
- 41.Gaus M., Cui Q., and Elstner M., “DFTB3: Extension of the self-consistent-charge density-functional tight-binding method (SCC-DFTB),” J. Chem. Theory Comput. 7, 931–948 (2011). 10.1021/ct100684s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.York D. M. and Yang W., “A chemical potential equalization method for molecular simulations,” J. Chem. Phys. 104, 159–172 (1996). 10.1063/1.470886 [DOI] [Google Scholar]
- 43.Giese T. J. and York D. M., “Improvement of semiempirical response properties with charge-dependent response density,” J. Chem. Phys. 123, 164108 (2005). 10.1063/1.2080007 [DOI] [PubMed] [Google Scholar]
- 44.Kaminski S., Giese T. J., Gaus M., York D. M., and Elstner M., “Extended polarization in third-order SCC-DFTB from chemical-potential equalization,” J. Phys. Chem. A 116, 9131–9141 (2012). 10.1021/jp306239c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sure R. and Grimme S., “Corrected small basis set Hartree-Fock method for large systems,” J. Comput. Chem. 34, 1672–1685 (2013). 10.1002/jcc.23317 [DOI] [PubMed] [Google Scholar]
- 46.Leverentz H. R., Qi H. W., and Truhlar D. G., “Assessing the accuracy of density functional and semiempirical wave function methods for water nanoparticles: Comparing binding and relative energies of (H2O)16 and (H2O)17 to CCSD(T) results,” J. Chem. Theory Comput. 9, 995–1006 (2013). 10.1021/ct300848z [DOI] [PubMed] [Google Scholar]
- 47.Simonson T., Archontis G., and Karplus M., “Free energy simulations come of age: Protein-ligand recognition,” Acc. Chem. Res. 35, 430–437 (2002). 10.1021/ar010030m [DOI] [PubMed] [Google Scholar]
- 48.Deng Y. Q. and Roux B., “Computations of standard binding free energies with molecular dynamics simulations,” J. Phys. Chem. B 113, 2234–2246 (2009). 10.1021/jp807701h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adamo C. and Barone V., “Toward reliable density functional methods without adjustable parameters: The PBE0 model,” J. Chem. Phys. 110, 6158 (1999). 10.1063/1.478522 [DOI] [Google Scholar]
- 50.Brooks C. L. III and Karplus M., “Deformable stochastic boundaries in molecular dynamics,” J. Chem. Phys. 79, 6312–6325 (1983). 10.1063/1.445724 [DOI] [Google Scholar]
- 51.Brooks B. R., Brooks C. L. III, Mackerell A. D., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S. et al. , “CHARMM: The biomolecular simulation program,” J. Comput. Chem. 30, 1545–1614 (2009). 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L., “Comparison of simple potential functions for simulating liquid water,” J. Chem. Phys. 79, 926–935 (1983). 10.1063/1.445869 [DOI] [Google Scholar]
- 53.A. D. MacKerell, Jr., Bashford D., Bellott M., R. L. Dunbrack, Jr., Evenseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S. et al. , “All-atom empirical potential for molecular modeling and dynamics studies of proteins,” J. Phys. Chem. B 102, 3586–3616 (1998). 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- 54.Ryckaert J.-P., Ciccotti G., and Berendsen H. J. C., “Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes,” J. Comput. Phys. 23, 327–341(1977). 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- 55.Stote R. H., States D. J., and Karplus M., “On the treatment of electrostatic interactions in biololecular simulation,” J. Chim. Phys. 88, 2419–2433 (1991). [Google Scholar]
- 56.Steinbach P. J. and Brooks B. R., “New spherical-cutoff methods for long-range forces in macromolecular simulation,” J. Comput. Chem. 15, 667–683 (1994). 10.1002/jcc.540150702 [DOI] [Google Scholar]
- 57.Neese F., “The ORCA program system,” Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2, 73–78 (2012). 10.1002/wcms.81 [DOI] [Google Scholar]
- 58.Stewart J. J. P., MOPAC2012, Stewart Computational Chemistry, Colorado Springs, CO, USA, 2009.
- 59.Schmidt M. W., Baldridge K. K., Boatz J. A., Elbert S. T., Gordon M. S., Jensen J. H., Koseki S., Matsunaga N., Nguyen K. A., Su S. et al. , “General atomic and molecular electronic structure system,” J. Comput. Chem. 14, 1347–1363 (1993). 10.1002/jcc.540141112 [DOI] [Google Scholar]
- 60.Thiel W., MNDO2005, Max-Planck-Institut für Kohlenforschung: Mülheim an der Ruhr, Germany, 2005.
- 61.Grimme S., Antony J., Ehrlich S., and Krieg H., “A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu,” J. Chem. Phys. 132, 154104 (2010). 10.1063/1.3382344 [DOI] [PubMed] [Google Scholar]
- 62.Grimme S., Ehrlich S., and Goerigk L., “Effect of the damping function in dispersion corrected density functional theory,” J. Comput. Chem. 32, 1456–1465 (2011). 10.1002/jcc.21759 [DOI] [PubMed] [Google Scholar]
- 63.Řezáč J. and Hobza P., “Describing noncovalent interactions beyond the common approximations: How accurate is the “gold standard,” CCSD(t) at the complete basis set limit?,” J. Chem. Theory Comput. 9, 2151–2155 (2013). 10.1021/ct400057w [DOI] [PubMed] [Google Scholar]
- 64.Becke A. D., “Density-functional thermochemistry. III. The role of exact exchange,” J. Chem. Phys. 98, 5648–5652 (1993). 10.1063/1.464913 [DOI] [Google Scholar]
- 65.Zhao Y. and Truhlar D. G., “The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: Two new functionals and systematic testing of four M06-class functionals and 12 other functionals,” Theo. Chem. Acc. 120, 215–241 (2008). 10.1007/s00214-007-0310-x [DOI] [Google Scholar]
- 66.Tao J., Perdew J. P., Staroverov V. N., and Scuseria G. E., “Climbing the density functional ladder: Nonempirical meta-generalized gradient approximation designed for molecules and solids,” Phys. Rev. Lett. 91, 146401 (2003). 10.1103/physrevlett.91.146401 [DOI] [PubMed] [Google Scholar]
- 67.Perdew J. P., Burke K., and Ernzerhof M., “Generalized gradient approximation made simple,” Phys. Rev. Lett. 77, 3865–3868 (1996). 10.1103/physrevlett.77.3865 [DOI] [PubMed] [Google Scholar]
- 68.Jensen J. H., “Which method is more accurate? or errors have error bars,” PeerJ Preprints 5, e2693v1 (2017). 10.7287/peerj.preprints.2693v1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for detailed benchmark data for gas-phase and different solution phase systems.