Abstract
The use of DNA sequencing to guide the discovery of natural products has emerged as a new paradigm for revealing chemistries encoded in bacterial genomes. A major obstacle to implementing this approach to natural product discovery is the transcriptional silence of biosynthetic gene clusters under laboratory growth conditions. Here we describe an improved yeast-based promoter engineering platform (mCRISTAR) that combines CRISPR/Cas9 and TAR to enable single-marker multiplexed promoter engineering of large gene clusters. mCRISTAR highlights the first application of the CRISPR/Cas9 system to multiplexed promoter engineering of natural product biosynthetic gene clusters. In this method, CRISPR/Cas9 is used to induce DNA double-strand breaks in promoter regions of biosynthetic gene clusters, and the resulting operon fragments are reassembled by TAR using synthetic gene-cluster-specific promoter cassettes. mCRISTAR uses a CRISPR array to simplify the construction of a CRISPR plasmid for multiplex CRISPR and a single auxotrophic selection to improve the inefficiency of using a CRISPR array for multiplex gene cluster refactoring. mCRISTAR is a simple and generic method for multiplexed replacement of promoters in biosynthetic gene clusters that will facilitate the discovery of natural products from the rapidly growing collection of gene clusters found in microbial genome and metagenome sequencing projects.
Keywords: promoter engineering, natural products, CRISPR/Cas9, TAR
Graphical abstract

Screening of microbial culture broth extracts has been an effective way to identify novel compounds that have proven useful as lead structures for the development of diverse therapeutics.1 As successful as this fermentation-based approach has been, extensive sequencing of bacterial genomes and metagenomes has revealed that these studies have likely failed to access the vast majority of bacterially encoded biosynthetic diversity.2–4 A key limiting factor in accessing the chemical diversity encoded in bacterial genomes is the fact that most natural product biosynthetic gene clusters remain silent in the laboratory setting.5 We and others have postulated that gene cluster promoter engineering, in which native promoters are replaced with well-characterized constitutive or inducible promoters, would provide a means to systematically activate transcriptionally silent gene clusters.6–8 We recently reported on the use of yeast homologous recombination in combination with small, synthetic promoter cassettes to transcriptionally activate silent natural product biosynthetic gene clusters.7 That system utilizes auxotrophic markers to select for promoter exchange events and provides an economically and technically simple means of replacing promoters in a gene cluster. However, multiplex promoter replacement using this method is not practical because of the inefficiency of native yeast recombination. In addition, the number of inserted promoter cassettes is limited by the number of auxotrophic markers available for selection.
The recombination efficiency in yeast has previously been shown to increase by as much as 4000-fold with the introduction of a DNA double-strand break (DSB) across a recombination site.9,10 The RNA-guided endonuclease Cas9 from the type-II clustered regularly interspaced short palindromic repeats (CRISPR) system provides a powerful tool for site-specific introduction of DSBs into DNA.11,12 As CRISPR/Cas9 has been successfully used for genome engineering in Saccharomyces cerevisiae through site-specific introduction of DSBs,13 we reasoned that yeast homologous recombination in combination with CRISPR/Cas9-directed cleavage at the promoter sites might provide an efficient system for multiplex promoter engineering of silent biosynthetic gene clusters. We presumed that the introduction of site-specific DSBs by CRISPR/Cas9 would not only increase the recombination efficiency but also provide effective counter-selection against native target promoter sequences, thereby eliminating the need for auxotrophic markers. Multiplex gene disruption with CRISPR in yeast has been reported previously using multiple gRNAs;14 however, this approach is difficult to adopt for the systematic activation of silent biosynthetic gene clusters because of the lengthy cloning protocol of multiple polymerase chain reaction (PCR)-amplified gRNAs into a CRISPR plasmid. Here we report the development of a single-marker multiplexed CRISPR/Cas9- and transformation-associated recombination (TAR)-mediated promoter engineering method that we have called mCRISTAR (for multiplexed-CRISPR-TAR). mCRISTAR provides a simple method for total refactoring of transcriptionally silent natural product biosynthetic gene clusters. This approach uses a CRISPR array15 instead of gRNAs to simplify the construction of a CRISPR plasmid through direct cloning of synthesized CRISPR arrays. The inefficiency of using CRISPR arrays for multiplex refactoring was overcome by including one auxotrophic marker selection in the mCRISTAR reaction. With this approach, a silent gene cluster targeted for refactoring is fragmented in yeast into distinct operons using the CRISPR/Cas9 system and reassembled by recombination using synthetic promoter cassettes containing gene-cluster-specific homology sequences (Figure 1). The mCRISTAR method allows simultaneous multiplexed incorporation of promoters upstream of each operon in a biosynthetic gene cluster, thus bypassing native transcriptional regulatory elements and inducing gene expression. By retaining native operon structures, this method enables complete transcriptional refactoring of gene clusters with minimal synthetic nucleic acid input, rendering the approach technically and economically scalable.
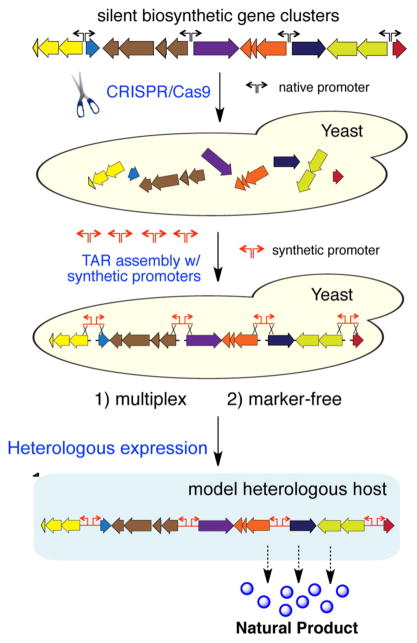
Figure 1.
Overview of the mCRISTAR method in yeast for activating silent natural product biosynthetic gene clusters in heterologous production hosts. We hypothesized that the combination of the CRISPR/Cas9 system and TAR-based DNA assembly would enable multiplexed, marker-free promoter engineering in yeast, thus significantly expanding the application of yeast homologous recombination-based promoter engineering into complex but transcriptionally silent natural product biosynthetic gene clusters.
RESULTS AND DISCUSSION
Selecting a CRISPR/Cas9 DNA Cleavage Approach for Multiplex Promoter Engineering
Starting with a biosynthetic gene cluster captured in an Escherichia coli:yeast:-Streptomyces shuttle vector, we envisioned that mCRISTAR would consist of site-specific cleavage of the cluster into individual operon fragments using a CRISPR/Cas9-mediated gene cluster disassembly process in yeast followed by reassembly of the cluster in yeast using TAR and pathway-specific promoter-bearing bridge DNAs. TAR efficiently assembles linear DNA fragments with as little as 40 bp homology sequences, permitting the facile synthesis (or production by PCR) of pathway-specific promoter-bearing bridge DNAs for use in the reassembly reaction. On the basis of published CRISPR/Cas9 methods, we considered three different ways of using the CRISPR/Cas9 system to assist with promoter engineering.12 First, DSBs could be created in vitro using purified Cas9 nuclease and synthetic single-guide RNAs (sgRNAs), which consist of crisprRNAs (crRNAs) fused with a transactivating crRNA (tracrRNA).16 Second, DSBs could be generated in vivo using sgRNAs, as is frequently done in genome-editing experiments.14,17 Third, DSBs could be generated in vivo using natural CRISPR arrays that are processed in vivo to produce mature crRNAs that, together with a tracrRNA and Cas9 nuclease, would induce DSBs at the target sites.15 Ultimately, we chose the third option because the use of natural CRISPR arrays would simplify the production of the CRISPR reagents needed for multiplex promoter engineering. With this approach, all of the required gene-cluster-specific mCRISTAR reagents (e.g., the CRISPR array and primers for generating gene-cluster-specific bridging promoter cassettes) can be easily synthesized, allowing the development of a simple, cost-effective, and scalable gene cluster refactoring platform.
Development of mCRISTAR Using Promoter Cassettes Containing Auxotrophic Markers
To evaluate the feasibility of mCRISTAR for multiplex promoter engineering in yeast, we used the environmental DNA (eDNA)-derived tetarimycin (Tam) gene cluster. The Tam cluster presents a useful model system for studying silent gene clusters as it is transcriptionally silent in Streptomyces albus but can be activated by overexpressing the pathway-specific SARP family transcriptional activator present in the gene cluster under the ermE* promoter.18 In our initial proof-of-concept experiments with the Tam cluster, we chose to use promoter cassettes containing auxotrophic markers7 to facilitate easy screening of colonies for successful promoter cassette insertions by simply repatching yeast colonies onto amino acid dropout plates. There are six commonly used genes for auxotrophic selection in yeast: URA3, LEU2, MET15, TRP1, HIS3, and LYS2.19 In our mCRISTAR method, one auxotrophic marker must be used to select for the plasmid carrying the gene cluster to be refactored and another auxotrophic marker must be used to select for the CRISPR plasmid. This leaves four markers available for use in promoter refactoring. In this study, potential promoter sites were identified using two criteria: (1) changes in gene directionality and (2) the presence of a large gap (>50 bp) between two neighboring genes. On the basis of gene directionality alone, the Tam gene cluster is predicted to contain seven putative operons (Figure 2a) controlled by four bidirectional (tam1–3) or unidirectional (tam4) promoters, meaning that the four remaining auxotrophic markers were sufficient to test the feasibility of multiplex promoter engineering using the Tam gene cluster.
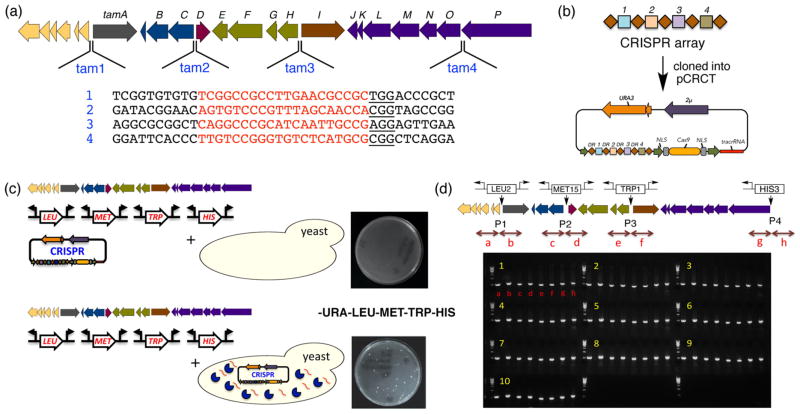
Figure 2.
Demonstration of multiplex promoter engineering in yeast using mCRISTAR. (a) Four CRISPR target sequences were selected from four promoter regions (tam1–4) in the Tam cluster. (b) A CRISPR array containing the four target sequences was synthesized and cloned into pCRCT. (c) Cotransformation of the Tam cluster, four gene-cluster-specific refactoring promoter cassettes, and the CRISPR plasmid failed to yield any colonies; however, transformation of the Tam cluster and promoter cassettes into yeast already containing pCRCT with the CRISPR array yielded a number of colonies. (d) PCR-based genotyping of 10 colonies using primer sets that produced amplicons bridging between the Tam cluster and the newly inserted promoter cassettes verified the correct insertion of all four promoter cassettes in Tam gene clusters present in these yeast.
The Cas9 endonuclease that originates from Streptococcus pyogenes specifically targets a DNA sequence for cleavage based on the sequence of a guide RNA or crRNA. A key requirement for a DNA target sequence is that the 3′ end of the target sequence must be adjacent to an NGG protospacer-adjacent motif (PAM) sequence (20 bp + “NGG”). Unique 20 bp sites fulfilling this condition were selected from each promoter region in the Tam cluster (Figure 2a) and used to create the CRISPR/Cas9 cut sites. Because of the wide discrepancy between the GC contents of the yeast genome (<40%) and the gene clusters (~70%) that we examined, we did not routinely check for homologous sites in the yeast genome. A CRISPR array of four CRISPR target sequences (tam1–4) with intervening direct repeat sequences was then synthesized and cloned into the iCas9/tracrRNA expression plasmid pCRCT to give pCRCT:Tam (Figure 2b).15 To test the feasibility of mCRISTAR for multiplex promoter engineering using pCRCT:Tam, four promoter cassettes carrying auxotrophic markers were generated by PCR using primers containing Tam1–4 promoter-region-specific homology sequences, and the Tam gene cluster was cloned on the E. coli:yeast:Streptomyces shuttle vector pTARa to give pTARa:Tam. Initially we simultaneously cotransformed the Tam-specific promoter cassettes pTARa:Tam and pCRCT:Tam into yeast and attempted to select for the insertion of all four promoter cassettes using synthetic composite (SC) LEU, MET, TRP, HIS, LYS, and URA dropout plates. Unfortunately, these experiments failed to produce any colonies, suggesting that cotransformation may not have provided sufficient time for expression of CRISPR elements from pCRCT:Tam. To address this issue, we first transformed pCRCT:Tam into yeast and selected transformants on SC URA dropout plates. Then, in a second transformation step, we introduced pTARa:Tam and the Tam-cluster-specific promoter cassettes into pCRCT:Tam-containing yeast that had been grown overnight in SC URA dropout medium. In this experiment, we observed colonies on dropout plates selective for the four auxotrophic markers carried on the promoter cassettes. No colonies appeared on the same selection plates in the control transformation with yeast that did not contain pCRCT:Tam, suggesting that in the two-step transformation protocol all four promoter cassettes were successfully inserted into the Tam cluster in a multiplexed CRISPR/Cas9-dependent manner (Figure 2c). The correct insertion of all four promoter cassettes was verified by genotyping 10 colonies using primer pairs designed to generate PCR amplicons only in the presence of the promoter-engineered Tam cluster (Figure 2d). The expression of the CRISPR system (i.e., Cas9, crRNA, and tracrRNA) prior to the cotransformation of a gene cluster with its corresponding promoter engineering cassettes in yeast appears to be essential for increasing the eficiency of the overall mCRISTAR method to the extent that enables multiplex promoter engineering.
Development of Single-Marker mCRISTAR
When promoter cassettes containing auxotrophic markers are used, a multiplexed approach to engineering of a gene cluster is limited by the number of markers available for selection. In practice this would limit the application of this method to natural product biosynthetic gene clusters that require five or fewer promoter cassettes for their activation. In an effort to overcome this limitation, we evaluated the feasibility of marker-free promoter engineering relying on the CRISPR/Cas9 system to counterselect against native promoter sequences and therefore to positively select for the insertion of promoter cassettes (Figure 3).
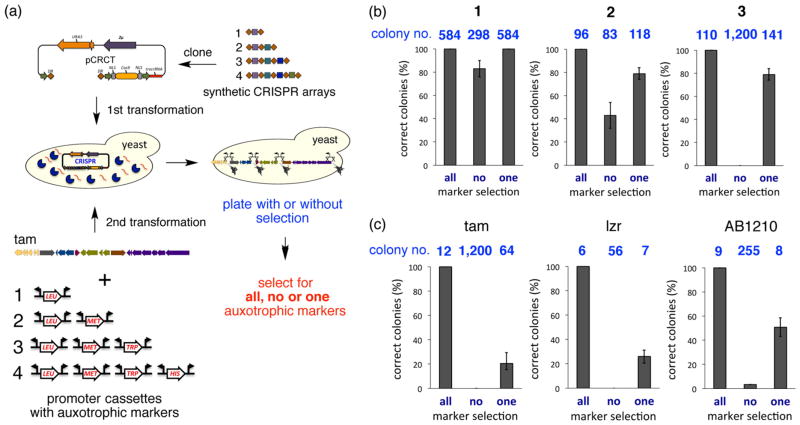
Figure 3.
Demonstration of single-marker multiplexed promoter engineering in yeast using mCRISTAR. (a) The efficiencies of simultaneous insertion of one, two, three, or four promoter cassettes with and without auxotrophic marker selections were compared using the Tam cluster. In these studies, transformants were plated on dropout media that selected for either all, one, or no auxotrophic markers. (b) The blue numbers indicate the average (n = 3) numbers of colonies that were obtained in mCRISTAR reactions with one, two, or three promoter cassettes using dropout media that select for all, none, or one of the auxotrophic markers used in each study. Bar graphs indicate the percentages of colonies from each of the three selection conditions that grew when they were patched on dropout media for all auxotrophic markers used in the experiment. (c) The same data as displayed in (b) are shown for mCRISTAR reactions with four promoter cassettes and either the Tam, Lzr, or AB1210 cluster.
To study the efficiency of marker-free insertion using different numbers of promoter cassettes, CRISPR array sequences containing one (tam3), two (tam2 and tam3), three (tam1–3), or four (tam1–4) target sequences were synthesized, cloned into pCRCT, and transformed into yeast (Figure 3a). The Tam cluster was cotransformed with one, two, three, or four promoter exchange cassettes into yeast containing a pCRCT plasmid carrying a CRISPR array of corresponding complexity. In each case, transformants were plated on LYS and URA dropout media to select only for the presence of pTARa:Tam and pCRCT:Tam (Figure 3b,c). If the CRISPR/ Cas9 system worked as expected, counterselection by the Cas9 endonuclease against native promoter sequences would by default select for Tam clusters containing newly inserted synthetic promoter cassettes. Engineering attempts using only one or two promoter cassettes yielded 298 and 83 colonies, respectively, whereas well over 1000 colonies were observed in experiments using either three or four promoter cassettes. To rapidly screen colonies for the presence of synthetic promoter cassettes, 30–40 colonies were picked from each transformation plate and repatched on dropout media that would select for the specific collection of auxotrophic markers carried by the set of promoter cassettes used in that experiment. In the case of experiments using one or two promoter cassette insertions, 83% and 43% of the colonies that we screened contained the expected auxotrophic markers, respectively. In contrast, no colonies grew on dropout plates used to screen for insertions of all promoter cassettes when attempting three and four promoter cassette insertions, suggesting inefficient Cas9 counterselection in these experiments. To investigate the cause of the high background seen in the experiments using three or four promoter cassettes, we picked approximately 100 colonies from each transformation plate and patched these colonies on all four single amino acid dropout plates (LEU, MET, TRP, or HIS). Once again, no colonies grew on any of the dropout plates (Figure S5). It would appear that when CRISPR arrays with more than two target sequences are used, the efficiency of the CRISPR/Cas9 system to counterselect against target sequences drops dramatically, and therefore, only two promoter sites can be engineered simultaneously without any selection.
In an effort to ensure the insertion of at least one promoter cassette in experiments using multiple spacer sequences, we repeated the experiments involving insertion of three and four promoter cassettes, this time selecting transformants for pTARa:Tam and pCRCT:Tam (LYS and URA) plus one of the auxotrophic markers carried on a promoter cassette (Figure 3b,c). We observed a significant reduction in the number of colonies in experiments using either three (141 colonies) or four (64 colonies) promoter cassettes. The percentage of colonies with successful insertion of all of the promoter cassettes was assessed by patching 30–40 colonies on dropout plates that selected for auxotrophic makers on all of the promoter cassettes used in the experiments. In these experiments, 79% of the colonies from the three-promoter-cassette insertion experiment and 21% of colonies from the four-promoter-cassette insertion experiment grew on multiple amino acid dropout plates. This suggests that CRISPR/Cas9-mediated counterselection of multiple target sites using a CRISPR array is actually quite efficient in yeast as long as one can ensure that one cleavage event has occurred. Interestingly, we never observed any correctly refactored colonies when we tried the simultaneous insertions of more than four promoter cassettes.
The natural CRISPR/Cas9 system from S. pyogenes uses 30 bp spacer sequences, suggesting that longer spacer sequences might increase the mCRISTAR efficiency. To evaluate this possibility, we resynthesized the CRISPR array to contain four 30 bp target spacer sequences (tam1–4) while leaving the direct repeat sequences identical. The longer spacers were designed on the same principle as before (20 bp + “NGG”) but used a 30 bp target sequence (30 bp + “NGG”). When they are assembled as an array, the organization is the same except that the target sequences are 10 bp longer than before. We then repeated the four-promoter-cassette insertion experiment using the same protocol as used for the 20 bp target sequences. A significantly lower number of background colonies (30–60 colonies) was observed on the selection plate, allowing the marker-free insertion of four promoter cassettes. However, the percentage of correct colonies (~3%) was still too low to use this approach for refactoring studies (Figure S9). Our work indicates that CRISPR/Cas9-mediated promoter engineering enables the insertion of at least four promoter cassettes simultaneously using a single auxotrophic selectable marker with high efficiency. This should significantly increase the utility of yeast homologous recombination-based promoter engineering of natural product biosynthetic gene clusters. With the four commonly used auxotrophic genes available for selection, up to 16 bidirectional promoter cassettes (a total of 32 promoters) can be inserted into a biosynthetic gene cluster using four rounds of mCRISTAR.
Validation of Single-Marker mCRISTAR Using Additional Biosynthetic Gene Clusters
To test the general applicability of our multiplex gene cluster refactoring methods, we used the lazarimide (Lzr) biosynthetic gene cluster and a randomly selected aromatic polyketide gene cluster, AB1210 (Figure 3c). For each gene cluster, we performed an mCRISTAR experiment involving the insertion of four promoter cassettes. The CRISPR arrays containing four target sequences in four bidirectional promoter regions in the Lzr and AB1210 clusters (Table S1) were synthesized and cloned into pCRCT. The resulting pCRCT:lzr and pCRCT:AB1210 CRISPR plasmids were transformed into yeast. Plasmids carrying each gene cluster and four gene-cluster-specific promoter cassettes were then cotransformed into yeast carrying the appropriate CRISPR plasmid. Transformants were plated on dropout plates that would select for all four promoter cassettes, one promoter cassette, or no promoter cassettes. Although fewer colonies were observed in these experiments than in those using the Tam cluster, the same trends for successful promoter insertion were observed. When selecting for a single promoter cassette, 26% and 51% of the colonies were correctly engineered with all four promoter cassettes for the Lzr and AB1210 clusters, respectively. Interestingly, in the case of the AB1210 cluster, one out of 29 colonies (~3%) patched from the no-selection plates contained all four promoter cassettes.
While the total number of colonies and the percentage of colonies containing all of the desired promoter cassettes will likely vary from one experiment to the next, we believe the efficiency of the method outlined here is sufficient to provide a generally useful tool for multiplexed promoter engineering of gene clusters. At the efficiencies we observed, users would have to screen fewer than five yeast colonies to consistently identify a correctly refactored gene cluster. On the basis of our proof-of-principle studies, mCRISTAR should routinely enable the simultaneous insertion of two promoter cassettes without any selection and up to four promoter cassettes with selection for only one of the cassettes.
Construction of Marker-Free Promoter Cassettes for Use in the Heterologous Expression of Refactored Gene Clusters in Streptomyces sp
To facilitate the use of marker-free mCRISTAR for the heterologous expression of natural product biosynthetic gene clusters in Streptomyces sp., we constructed a set of promoter cassettes with orthogonal promoter and ribosome binding site (RBS) sequences (Figures 4a,b and S12). We believe that sequence orthogonality within the collection of promoters used for refactoring is important in preventing recombination between promoters during gene cluster refactoring.
Figure 4.
Construction of marker-free promoter cassettes. (a) Twenty-three sequence-orthogonal promoters reported to be active in Streptomyces were tested for activity using a Tam cluster/TamI (SARP) reporter system and a fixed RBS sequence. Each promoter was placed in front of the tamI gene cloned into the shuttle expression plasmid pIJ10257, and constructs were evaluated for the ability to induce the production of tetarimycin A in S. albus containing the Tam cluster. Fourteen promoters successfully produced tetarimycin A in this reporter system. (b) A library of sequence-orthogonal RBS sequences was created by randomizing the RBS in the ermE* promoter region of pIJ10257. This library was placed between the P16 promoter sequence and the start codon of the tamI gene cloned in pIJ10257. After the RBS library was transferred to S. albus containing the Tam cluster, colonies exhibiting the strongest yellow phenotypes were sequenced to identify a collection of functional sequence-orthogonal RBS sequences. (c) Seven marker-free bidirectional promoter cassettes were constructed by placing 100 bp of random sequence between two sequence-orthogonal promoters and RBS pairs. (d) The effect of promoter cassette size on the number of colonies observed in mCRISTAR reactions is shown.
To identify sequence-orthogonal promoters that function in our target expression host S. albus, 23 promoters (including 10 medium and 13 strong promoter sequences) were selected from a library of promoters that was reported to be recognized by the Streptomyces housekeeping sigma factor σ70.20 Each of these promoters was tested for its ability to activate the Tam gene cluster through expression of tamI, a Tam-cluster-specific SARP family transcriptional regulator (Figure 4a). To do this, the tamI gene was cloned into the E. coli:Streptomyces shuttle vector pIJ10257,21 and then the ermE* promoter (but no RBS) in pIJ12057 was replaced with each of these 23 promoters. The resulting constructs were tested for their ability to drive the transcription of tamI to a sufficient level to activate the Tam cluster in S. albus. Among the 23 promoters we examined, 15 promoters led to the successful production of tetarimycin A as determined by HPLC analysis of culture broth extracts (Figure 4a).
To generate a set of sequence-orthogonal RBSs, we selected one of the active promoter sequences (P16) and added to it a library of partially randomized RBSs based on the ermE* RBS found in the pIJ10257 plasmid (Figure 4b).21 This partially randomized 15 bp RBS library contains a fixed 6 bp Shine–Dalgarno consensus sequence (AGGAGG) flanked by 2 bp upstream and 7 bp downstream random sequences. The distance between the Shine–Dalgarno sequence and the start codon was kept at 7 bp. To find functional RBSs, the RBS library was created as a degenerate primer containing the P16 promoter, and this primer was used to amplify the tamI gene, which was then cloned into the pIJ10257 shuttle vector. The resulting constructs were transferred into S. albus harboring the Tam cluster, and colonies displaying a yellow phenotype, which is indicative of tetarimycin A production, were picked and sequenced.
Fourteen different promoters and RBSs were selected from the promoter activity and RBS screening studies and combined to create a collection of sequence-orthogonal promoters (Figure 4c). Seven unique, marker-free, bidirectional promoter cassettes were designed by appending two unique promoter sequences in opposite directions onto 100 bp random spacer sequences (Figure 4c). These promoter cassettes were synthesized and cloned into a TOPO vector for stable maintenance. We also created promoter cassettes in which the random spacer sequence was replaced with an auxotrophic marker gene sequence for use in multiplex promoter engineering with one auxotrophic marker. This new collection of promoter cassettes was tested in mCRISTAR using yeast containing pCRCT:Tam, one new LEU2-marker-containing promoter cassette, and three new marker-free promoter cassettes. Although the percentage of colonies containing all four cassettes was similar to that seen when the auxotrophic marker cassettes were used (~30%), the total number of colonies observed on the transformation plates was significantly lower with our marker-free cassettes than with cassettes containing auxotrophic markers. The most obvious difference between the two sets of promoter cassettes is their size: the marker-free promoter cassettes are <200 bp, while the auxotrophic marker cassettes are mostly >1000 bp. To evaluate the effect of cassette size on the efficiency of mCRISTAR, marker-free cassettes were resynthesized with 1000 bp of random sequence between the two promoters. These synthetic cassettes were used as templates in PCR reactions with primers designed to generate 200, 500, and 1000 bp Tam-cluster-specific promoter cassettes. The efficiency of mCRISTAR using the 1000 bp marker-free cassettes closely resembled that seen with the auxotrophic marker cassettes (Figure 4d), and therefore, larger promoter cassettes were selected for use in future single-marker mCRISTAR reactions.
Activation of the Silent Tam Gene Cluster Using Single-Marker mCRISTAR
To further the development of the mCRISTAR platform, we sought to more extensively engineer the Tam cluster (Figure 5). As mentioned above, when gene directionality alone is considered, the Tam cluster is predicted to contain seven putative operons (Figure 2a) controlled by four bi- or unidirectional promoters. The number of predicted transcriptional units is expanded to 10 if the presence of large (>50 bp) gaps between two neighboring genes is taken into consideration. These 10 putative transcriptional units are predicted to be controlled by eight bi- or unidirectional promoters (Figure 5b). Three of these are composed of single-transcription-factor genes (blue genes in Figure 5b) and are thus irrelevant for gene cluster refactoring. We set out to completely refactor the Tam cluster through the insertion of eight promoter cassettes [one bidirectional (tam1) and seven unidirectional (tam2–8)] using two rounds of mCRISTAR. To enable refactoring of all eight promoter sites, an additional CRISPR array containing target sequences in tam5–8 was designed, commercially synthesized, and cloned into pCRCT to give pCRCT:tam5–8. To avoid activation of tetarimycin A production by the TamI SARP family transcriptional activator, the reverse primer for the tam3 promoter region was designed to generate the 800 bp truncated tamI gene at the N-terminus.
Figure 5.
Refactoring of the silent Tam gene cluster using mCRISTAR. (a) The Tam cluster is normally transcriptionally silent in the heterologous host S. albus. Overexpression of pathway-specific SARP positive regulator by cloning of the SARP under the strong ermE* promoter activates the Tam cluster to produce tetarimycin A in S. albus. (b) To activate the Tam cluster using mCRISTAR without SARP overexpression, a total of eight promoter cassettes were inserted into the Tam cluster using two rounds of mCRISTAR. The promoter cassette used to replace the tam3 promoter region was designed to generate the 800 bp truncated tamI SARP transcriptional activator gene to eliminate its effect on the activation of the Tam cluster. The refactored Tam cluster containing eight promoter cassettes (tam1–8) successfully confers the production of tetarimycin A to S. albus.
In the first round of mCRISTAR, the Tam cluster was cotransformed with the LEU2-marker-containing promoter cassette (tam1) and three marker-free promoter cassettes (tam2–4) into yeast carrying the pCRCT:tam1–4 plasmid, and transformants were selected on SC LEU, LYS, and URA dropout plates. Ten colonies were picked from the transformation plate, and vector DNA was isolated from each culture for PCR-based genotyping using primer sets that generate amplicons bridging the Tam cluster and the newly inserted promoter cassettes. Eight of these colonies (80%) were found to contain the correct insertion of all four synthetic promoter cassettes (tam1–4). The Tam cluster from one yeast colony that tested positive for all four synthetic promoter cassettes was amplified in E. coli and used for a second round of mCRISTAR. In the second round of mCRISTAR, the Tam cluster already containing four promoter cassettes (tam1–4) was cotransformed with the MET15-marker-containing promoter cassette (tam5) and three marker-free promoter cassettes (tam6–8) into yeast harboring pCRCT:tam5–8 and plated on LEU, MET, LYS, and URA dropout plates. Genotyping of four colonies using primer sets that generate amplicons bridging between the gene cluster and the newly introduced promoter cassettes identified two colonies with the correct insertion of the four new promoter cassettes (tam5–8). This resulted in a Tam cluster that was successfully refactored with eight promoter cassettes using two rounds of mCRISTAR. Tam clusters refactored with eight promoter cassettes (tam1–8) were transformed into E. coli S17 and shuttled into S. albus via intergeneric conjugation. HPLC analysis of culture extracts of S. albus harboring the refactored Tam clusters identified the production of tetarimycin A. Cultures transformed with the native Tam cluster did not show any tetarimycin A production, while the eight-promoter-insertion construct showed tetarimycin A production nearly identical to that seen when the Tam cluster was activated through SARP gene expression (Figure 5b). In this case, the replacement of eight predicted promoter sites using two rounds of mCRISTAR was sufficient to activate the silent Tam cluster.
CONCLUSIONS
The mCRISTAR method combines the CRISPR/Cas9 system and TAR-based DNA assembly in yeast to enable efficient multiplexed engineering of natural product biosynthetic gene clusters. mCRISTAR highlights the first application of the CRISPR/Cas9 system to multiplex promoter engineering of natural product biosynthetic gene clusters. In a single round of mCRISTAR, four promoter cassettes can be simultaneously inserted into a gene cluster using a single auxotrophic marker selection, or two promoter cassettes can be inserted without any selection. In refactoring of a gene cluster with the reagents described here, up to 16 promoter cassettes, containing up to 32 promoters, can be inserted into a single natural product biosynthetic gene cluster using four auxotrophic markers commonly used in yeast. Additionally, an unlimited number of additional promoter cassettes can then be inserted two at a time using successive rounds of mCRISTAR without any auxotrophic selection. mCRISTAR minimizes the synthetic DNA input by maintaining natural operon structures, and the use of CRISPR arrays simplifies the construction of CRISPR plasmids. Two potential issues that could arise when using promoter refactoring to activate a silent gene cluster are difficulties identifying start codons and the need to balance promoter strengths to achieve optimal expression of all operons. With mCRISTAR, once a CRISPR array plasmid has been constructed, it can subsequently be used in conjunction with a variety of promoter cassettes designed to recombine at different potential start sites or carry different promoters, facilitating the exploration of multiple gene cluster refactoring options. Since the model gene clusters used in the development of mCRISTAR were type-II PKS and indolotryptoline gene clusters, it is not known how effective mCRISTAR will be at refactoring type-I PKS and NRPS gene clusters containing highly related domain sequences.
All of the required gene-cluster-specific mCRISTAR reagents (i.e., CRISPR arrays and primers for generating promoter cassettes) can be ordered commercially, allowing the development of an automated, scalable gene cluster refactoring platform. To assist with these efforts, we designed the mCRISTAR Webapp, which is a python package and a Web application that automates the generation of all sequences needed for the mCRISTAR gene cluster refactoring process. The mCRISTAR Webapp identifies the promoter regions from a GenBank-formatted file based on gene orientation and gaps between neighboring open reading frames. It then designs the two classes of nucleotide reagents that are required for mCRISTAR: the CRISPR array and the primers to amplify promoter cassettes with 40 bp homology arms. Built on top of the open source library Biopython,22 the mCRISTAR python package and Webapp can be found at www.mcristar.net. mCRISTAR and its accompanying Webapp provide a simple and cost-effective method for transcriptional activation of complex natural product biosynthetic gene clusters, which we believe should accelerate the discovery of novel bioactive metabolites identified from the rapidly growing collection of natural product biosynthetic gene clusters found in bacterial genomic and metagenomic sequence data.
METHODS
Yeast Strain, Plasmids, and Culture Conditions
A Dnl4 gene deletion yeast strain of S. cerevisiae BY4727 (ATCC no. 200889) with the genotype of MATα his3Δ 200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 dnl4::KanMX4 was used for all of the transformation experiments.7,19 The yeast strain was maintained on YPD agar plates containing the antibiotic G418 (200 μg/mL) and grown overnight at 30 °C (260 rpm) prior to transformation. DNA was transformed into the yeast strain using the LiAc/ss carrier DNA/PEG yeast transformation protocol.23 The positive selection of any genetic modification was made on the appropriate SC amino acid dropout medium purchased from Sunrise Science Products (San Diego, CA).
Construction of the Pathway-Specific CRISPR Plasmid
The pCRCT plasmid used for all of the CRISPR experiments was obtained from Addgene.15 CRISPR arrays containing target sequences were synthesized as gBlock fragments from Integrated DNA Technologies (IDT). Synthetic CRISPR arrays were cloned into pCRCT using golden gate cloning.24 Briefly, 1 μL of the gBlock fragment (10 ng/μL), 1 μL of pCRCT (100 ng/μL), 1.5 μL of T4 DNA ligase reaction buffer, 1.5 μL of 10×BSA, 1 μL of BsaI, 1 μL of T4 DNA ligase, and 8 μL of dH2O were mixed in a PCR tube. The cloning reaction was performed using a thermocycler as follows: 25 cycles of 3 m in at 37 °C and 4 min at 16 °C and then 1 cycle of 5 min at 50 °C and 5 min at 80 °C. A 5 μL aliquot of the reaction mixture was electroporated into E. coli EC100, and transformants were selected on LB agar plates containing 100 μg/ mL ampicillin. DNA was miniprepped from four transformants, and the correct incorporation of a CRISPR array into pCRCT was confirmed by Sanger sequencing.
mCRISTAR Protocol
A CRISPR plasmid containing a pathway-specific CRISPR array was transformed into the yeast strain BY4727 dnl4::KanMX using the LiAc/ss carrier DNA/ PEG yeast transformation protocol, and transformants were selected on SC URA dropout plates. After 2 days, a single colony was picked and restruck on the same dropout medium. For promoter engineering, silent gene clusters were TAR-cloned into the pTARa vector25 containing the LYS2 gene as an auxotrophic selectable marker.
Promoter cassettes with auxotrophic markers were amplified using primer sets containing 40 bp homology sequences targeting the gene cluster using the previously published PCR protocol.7 For transformations, 15 mL of SC URA dropout liquid medium was inoculated with a BY4727 dnl4::KanMX strain containing a CRISPR plasmid carrying a pathway-specific CRISPR array, and this culture was shaken overnight at 30 °C. A 10 mL aliquot of the overnight culture (OD 5.0 at 600 nm) was placed in fresh YPD medium (50 mL) containing 200 μg/ mL G-418 and incubated for 4 h (30 °C, 200 rpm). The culture (OD 2.0 at 600 nm) was harvested, washed with water twice, and resuspended in 1 mL of sterile water. A 100 μL aliquot of this cell suspension was pelleted by centrifugation and added to a transformation mixture containing 34 μL of DNA (10 μL of a gene cluster in pTARa and 5 μL of each promoter cassette), 50 μL of carrier DNA, 36 μL of 1 M LiAc, and 240 μL of PEG 3350 (50% w/v). The reaction mixture was heat-shocked at 40 °C for 40 min and plated on an appropriate SC dropout medium. The correct insertion of promoter cassettes was confirmed by patching colonies on dropout medium that selected for all of the auxotrophic markers. For PCR-based genotyping, colonies were picked, placed into 1.5 mL of the corresponding SC dropout liquid medium, and grown overnight. These overnight cultures were used for yeast DNA miniprep, which was performed by a lysis protocol using zymolyase (Zymo Research). The correct insertion of promoter cassettes was confirmed by PCR-based genotyping using primer sets that generate amplicons bridging between a gene cluster and newly inserted promoter cassettes using the same protocol as published previously.7
PCR Amplification of Cluster-Specific Promoter Cassettes
The marker-free promoter cassettes used in this study were synthesized as gBlock gene fragments from IDT. To generate cluster-specific promoter cassettes, forward and reverse primers were designed to contain 40 bp homology sequences to gene clusters, and PCR was performed to amplify marker-free promoter cassettes using the following PCR protocol: 50 μL PCR reactions contained 1 μL of TOPO vector template (10 ng/μL), 2.5 μL of each primer (5 μM), 25 μL of buffer G (Epicenter), 18.5 μL of water, and 0.5 μL of Taq polymerase. PCR was performed using the following protocol: initial denaturation (95 °C, 5 min), 36 cycles of denaturation (95 °C, 30 s), annealing (55 °C, 30 s), and extension (72 °C, 0.2 min), and a final extension (72 °C, 5 min). The resulting PCR products were column-purified and diluted in TE buffer to a concentration of 400 ng/μL prior to transformation.
Refactoring of the Tam Cluster Using Two Rounds of mCRISTAR
The Tam cluster in pWEB-TNC was recloned into pTARa containing LYS2 as a selectable marker to allow replication in yeast (see the Supporting Information for details). For the first transformation, one promoter cassette (tam1) containing a LEU2 marker and three marker-free promoter cassettes (tam2–4) were amplified to contain 40 bp homology sequences to the Tam cluster, transformed together with pTARa:Tam into yeast containing an appropriate CRISPR array, and selected on a SC LEU, LYS, and URA dropout agar plate. After 3 days, four colonies were picked and grown in the same liquid dropout medium. These 1.5 mL aliquots of overnight cultures were used for yeast DNA miniprep, which was performed by a lysis protocol using zymolyase (Zymo Research). The correct insertion of all four promoter cassettes (tam1–4) was confirmed by PCR-based genotyping using primer sets that would generate amplicons bridging between the gene cluster and promoter cassettes. The Tam cluster with all four promoters correctly inserted was used for the second round of mCRISTAR, which was done in the same way using the refactored Tam cluster (pTARa:Tam1–4), one promoter cassette (tam5) containing a MET15 marker, and three marker-free promoter cassettes (tam6–8). PCR-based genotyping confirmed the insertion of all eight promoter cassettes into the Tam cluster.
HPLC Analysis of Tetarimycin A Production from Refactored Tam Gene Clusters
Tam gene clusters refactored with either four (pTARa:tam1–4) or eight (pTARa:tam1–8) promoter cassettes were transformed into E. coli S17, transferred into S. albus via intergeneric conjugation, and incorporated into its genome using ΦC31 integrase. S. albus harboring refactored Tam clusters was grown in 100 mL (2 × 50 mL) of R5A medium for 7 days (30 °C, 250 rpm). The cultures were extracted with ethyl acetate, and the production of tetarimycin A was confirmed by HPLC–MS analysis (reversed-phase C18 analytical column, 4.6 mm × 150 mm, 5% to 100% acetonitrile/water gradient, 30 min).
Supplementary Material
Acknowledgments
This work was supported by NIH (U01 GM110714-1A1 and AI110029 to Z.C.-P.).
Footnotes
Author Contributions
H.-S.K., Z.C.-P., and S.F.B. designed the experiments; H.-S.K. conducted the experiments; Z.C.-P. built the mCRISTAR Webapp; H.-S.K. and S.F.B. wrote the paper.
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.6b00080.
Supplementary methods and discussion, Figures S1–S13, and Tables S1–S11 (PDF)
References
- 1.Demain AL, Sanchez S. Microbial drug discovery: 80 years of progress. J Antibiot. 2009;62:5–16. doi: 10.1038/ja.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–21. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discovery. 2015;14:111–29. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 4.Lok C. Mining the microbial dark matter. Nature. 2015;522:270–3. doi: 10.1038/522270a. [DOI] [PubMed] [Google Scholar]
- 5.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–23. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 6.Luo Y, Huang H, Liang J, Wang M, Lu L, Shao Z, Cobb RE, Zhao H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun. 2013;4:2894. doi: 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montiel D, Kang HS, Chang FY, Charlop-Powers Z, Brady SF. Yeast homologous recombination-based promoter engineering for the activation of silent natural product biosynthetic gene clusters. Proc Natl Acad Sci U S A. 2015;112:8953–8. doi: 10.1073/pnas.1507606112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao Z, Rao G, Li C, Abil Z, Luo Y, Zhao H. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold. ACS Synth Biol. 2013;2:662–9. doi: 10.1021/sb400058n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci U S A. 2003;100:14994–9. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–45. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 11.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–39. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–43. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakociunas T, Bonde I, Herrgard M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–22. doi: 10.1016/j.ymben.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Bao Z, Xiao H, Liang J, Zhang L, Xiong X, Sun N, Si T, Zhao H. Homology-Integrated CRISPR–Cas (HI-CRISPR) System for One-Step Multigene Disruption in Saccharomyces cerevisiae. ACS Synth Biol. 2015;4:585–94. doi: 10.1021/sb500255k. [DOI] [PubMed] [Google Scholar]
- 16.Karvelis T, Gasiunas G, Siksnys V. Programmable DNA cleavage in vitro by Cas9. Biochem Soc Trans. 2013;41:1401–6. doi: 10.1042/BST20130164. [DOI] [PubMed] [Google Scholar]
- 17.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–26. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallifidas D, Kang HS, Brady SF. Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J Am Chem Soc. 2012;134:19552–5. doi: 10.1021/ja3093828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–32. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Seghezzi N, Amar P, Koebmann B, Jensen P, Virolle MJ. The construction of a library of synthetic promoters revealed some specific features of strong Streptomyces promoters. Appl Microbiol Biotechnol. 2011;90:615–23. doi: 10.1007/s00253-010-3018-0. [DOI] [PubMed] [Google Scholar]
- 21.Hong HJ, Hutchings MI, Hill LM, Buttner MJ. The role of the novel Fem protein VanK in vancomycin resistance in Streptomyces coelicolor. J Biol Chem. 2005;280:13055–61. doi: 10.1074/jbc.M413801200. [DOI] [PubMed] [Google Scholar]
- 22.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–3. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 24.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden Gate Shuffling: A One-Pot DNA Shuffling Method Based on Type IIs Restriction Enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JH, Feng Z, Bauer JD, Kallifidas D, Calle PY, Brady SF. Cloning large natural product gene clusters from the environment: piecing environmental DNA gene clusters back together with TAR. Biopolymers. 2010;93:833–44. doi: 10.1002/bip.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.