SUMMARY
Orientia tsutsugamushi causes scrub typhus, a potentially fatal infection that afflicts one million people annually. This obligate intracellular bacterium boasts one of the largest microbial arsenals of ankyrin repeat-containing protein (Ank) effectors, most of which target the endoplasmic reticulum (ER) by undefined mechanisms. Ank9 is the only one proven to function during infection. Here, we demonstrate that Ank9 bears a motif that mimics the GRIP domain of eukaryotic golgins and is necessary and sufficient for its Golgi localization. Ank9 reaches the ER exclusively by retrograde trafficking from the Golgi. Consistent with this observation, it binds COPB2, a host protein that mediates Golgi-to-ER transport. Ank9 destabilizes the Golgi and ER in a Golgi-localization domain (GLD)-dependent manner and induces the ATF4-dependent unfolded protein response. The Golgi is also destabilized in cells infected with O. tsutsugamushi or treated with COPB2 siRNA. COPB2 reduction and/or the cellular events that it invokes, such as Golgi destabilization, benefit Orientia replication. Thus, Ank9/bacterial negative modulation of COPB2 might contribute to the bacterium’s intracellular replication. This report identifies a novel microbial GLD, links Ank9 to the ability of O. tsutsugamushi to perturb Golgi structure, and describes the first mechanism by which any Orientia effector targets the secretory pathway.
INTRODUCTION
Obligate intracellular bacterial pathogens use secreted protein effectors to commandeer host cell processes. During the co-evolution of intracellular microbes and their hosts, effectors acquired eukaryotic-like features, such as protein-protein interaction domains and trafficking signals, allowing them to co-opt vesicular trafficking networks, post-translational modification systems, and other pathways to remodel host cells into permissive niches. As knock out-complementation studies of essential genes in most obligate intracellular bacteria is infeasible, an effective way to discern an effector’s functions is to elucidate its intracellular trafficking patterns and binding partners when it is ectopically expressed and to correlate such findings with infection. The obligate intracellular bacterium, Orientia tsutsugamushi, is the causative agent of scrub typhus, a febrile illness that threatens one billion persons and afflicts at least one million annually in the Asia-Pacific, the most heavily populated region of the world (Watt et al., 2003). With a median 6% case fatality rate, the reemergence of scrub typhus in the Asia-Pacific and its spread to Africa and South America has raised public health concerns (Taylor et al., 2015, Weitzel et al., 2016). Despite its significance as a global health threat, the cellular microbiology of O. tsutsugamushi, particularly the host pathways that it hijacks, remains poorly understood.
The 33-residue ankyrin repeat is one of the most common protein protein-interaction motifs in nature [reviewed in (Voth, 2011)]. Long thought to be exclusive to eukaryotes, many viruses and intracellular bacteria were discovered to encode ankyrin repeat-containing proteins (Anks). Several microbial Anks have been identified as key virulence factors that target diverse host cell pathways to promote microbial survival [reviewed in (Voth, 2011)]. The O. tsutsugamushi Ikeda strain genome carries 47 Ank open reading frames (ORFs) (Nakayama et al., 2008). A recent analysis of 1,912 bacterial genomes determined that the Ikeda genome ranks in the top 0.3% in terms of its number of encoded Anks (Jernigan et al., 2014). The retention of such a large number of Ank genes over its reductive evolution implies their importance to O. tsutsugamushi. Of the 47 Ikeda Ank ORFs, 9 are pseudogenes (Nakayama et al., 2008). Of the remaining 38, 12 are single-copy and 26 are identical or near-identical paralogs of 8 representative anks, resulting in 20 unique ORFs that can be distinguished using PCR primers (VieBrock et al., 2014). Our laboratory confirmed that the Ikeda Anks are Type I secretion system substrates and all are transcriptionally expressed during infection of mammalian host cells (VieBrock et al., 2014). Sixteen Anks also carry another eukaryotic-like domain called the F-box (Beyer et al., 2015, Min et al., 2014), which recruits host ubiquitin ligase machinery [reviewed in (Lee et al., 2014)]. Thus, O. tsutsugamushi Anks utilize at least two different eukaryotic-like protein-protein interaction motifs. When ectopically expressed, 14 of the 20 Ikeda Anks localize to the endoplasmic reticulum (ER) (VieBrock et al., 2014). A motif that mediates specific targeting of an O. tsutsugamushi Ank to the secretory pathway or any subcellular locale has yet to be identified.
O. tsutsugamushi Ikeda Ank9 is an ER-tropic effector that has seven ankyrin repeats and a C-terminal F-box (Beyer et al., 2015, VieBrock et al., 2014). By virtue of its F-box, endogenous Ank9 interacts with SKP1 during infection of mammalian cells (Beyer et al., 2015), signifying it as the only Orientia effector verified thus far to function during infection. In this study, we demonstrate that Ank9 initially targets the Golgi apparatus using a previously uncharacterized Golgi localization domain (GLD) and subsequently retrograde traffics to the ER. Consistent with this finding, endogenous COPB2, a subunit of the COPI complex involved in Golgi-to-ER trafficking, was identified as an Ank9 binding partner. Ank9 negatively impacts the secretory pathway by destabilizing the Golgi and ER in succession, inducing ER stress, and inhibiting protein secretion. Orientia inhibits host cell protein secretion, indicating that Ank9 phenocopies cellular phenomena associated with infection. Neither ectopically expressed Ank9 nor Orientia infection reduces COPB2 cellular levels. However, as in Ank9 expressing cells, the Golgi is destabilized in both Orientia infected and COPB2 knockdown cells, suggesting that Ank9 interaction with COPB2 might be linked to the Golgi instability that occurs during infection. Moreover, Orientia replication is increased in COPB2 knockdown cells. These data reveal a novel microbial GLD, provide first insight into how an Orientia effector targets the secretory pathway, identify Ank9 as one of only a few known effectors that retrograde traffics from the Golgi to the ER, and underscore how a single effector uses multiple eukaryotic-like domains to maximize the number of host cell processes it co-opts. This study also confirms that O. tsutsugamushi can perturb Golgi structure and its replication is favored when the retrograde pathway is disrupted.
RESULTS
Ank9 localizes to and disrupts the morphology of the ER
Because Ank9 is the only O. tsutsugamushi effector proven to function during infection of host cells and is one of many Orientia Anks that localize to the ER by undefined mechanisms (Beyer et al., 2015, VieBrock et al., 2014), we sought to understand how it targets the secretory pathway. It was first examined whether Ank9 localizes to the ER membrane and/or lumen and if doing so affects the organelle’s morphology. Non-synchronized HeLa cell cultures were transfected to express GFP-Ank9 or GFP alone. At 16 h, the cells were screened with antibodies specific for the ER lumenal markers, calreticulin and protein disulfide isomerase (PDI), and the ER transmembrane proteins, calnexin and derlin-1, and examined using confocal microscopy. GFP-Ank9 colocalized with all four ER markers and pronouncedly disrupted ER morphology, fragmenting and condensing the organelle into Ank9-positive ER-derived structures that could be categorized into two distinct phenotypes, vesicles and rings (Fig. 1A). In control cells, GFP was distributed diffusely throughout the cytosol and nuclei, failed to colocalize with any ER marker, and, based on the immunolabeling patterns for all four ER markers, did not alter ER morphology (Fig. 1B). Thus, Ank9 localizes throughout the ER and destabilizes its structure.
Fig. 1.
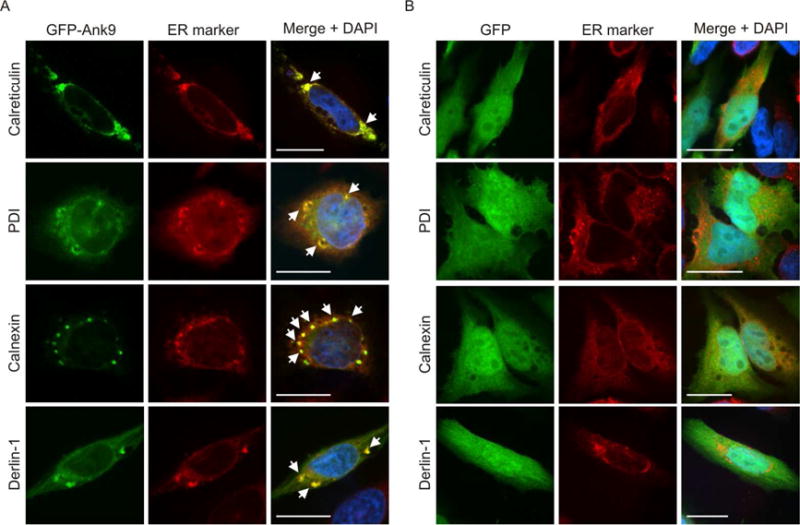
Ectopically expressed Ank9 localizes to and disrupts the morphology of the ER. HeLa cells expressing GFP-Ank9 (A) or GFP (B) were fixed and screened at 16 h post transfection with GFP antibody and antibody against one of the ER lumenal markers, calreticulin or PDI, or ER transmembrane proteins, calnexin or derlin-1, prior to examination by confocal microscopy. Representative fluorescence images of cells viewed for GFP, ER marker, and merged images plus DAPI, which stains the nucleus, are presented. White arrows denote representative points of GFP and ER marker signal colocalization. Because the cell cycles of the HeLa cells were not synchronized, the timing by which GFP-Ank9 localizes to and disrupts the ER could not be discerned. Results are representative of three independent experiments with similar results. Scale bars, 20 μm.
Ank9 also targets the Golgi apparatus
In the above analyses, we also observed cells wherein GFP-Ank9 displayed a perinuclear accumulation reminiscent of the Golgi. Because it was unknown if Ank9 or any other O. tsutsugamushi effector targets this organelle, cells expressing GFP-Ank9 or GFP were examined for colocalization with Golgi markers. Perinuclear GFP-Ank9 signal colocalized with Rer1 (ER-to-Golgi intermediate compartment [ERGIC] and cis-Golgi), GM130 (cis-Golgi), GolgB1 (cis- and medial-Golgi) and CellLight Golgi-red fluorescent protein (RFP; medial- and trans-Golgi) (Fig. 2A). GFP-Ank9 did not colocalize with LAMP-1-positive lysosomes (Fig. 2C). GFP failed to localize with any Golgi marker or LAMP-1 (Fig. 2, B and C). These data indicate that Ank9 specifically targets both the ER and Golgi.
Fig. 2.
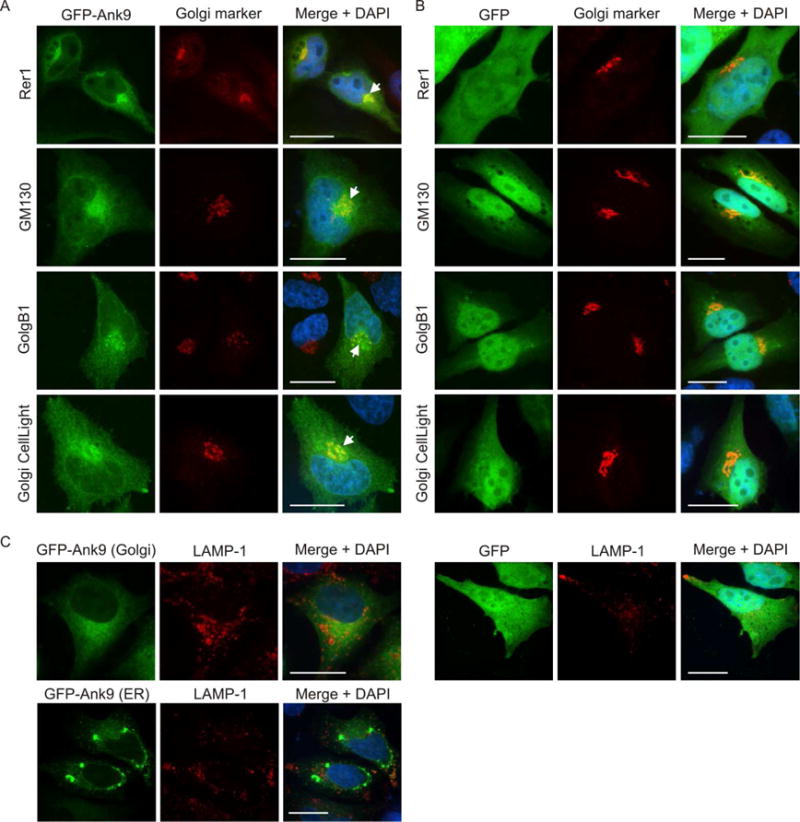
Ectopically expressed Ank9 localizes to the Golgi, but not to lysosomes. HeLa cells expressing GFP-Ank9 (A) or GFP (B) were fixed and screened at 16 h post transfection with GFP antibody and antibody against Rer1 (labels the ER-to-Golgi intermediate compartment and cis-Golgi), GM130 (labels the cis-Golgi), or GolgB1 (labels the cis- and medial-Golgi) prior to examination by confocal microscopy. Transfected cells were also incubated with CellLight Golgi-RFP, which labels the medial- and trans-Golgi, prior to fixation and confocal microscopic analyses. Representative fluorescence images of cells viewed for GFP, Golgi marker, and merged images plus DAPI are presented. White arrows denote representative points of GFP and Golgi marker signal colocalization. Because the cell cycles of the HeLa cells were not synchronized, the timing by which GFP-Ank9 localizes to the Golgi could not be discerned. (C) HeLa cells expressing GFP-Ank9 or GFP were also screened with antibody against LAMP-1, which labels lysosomes. Representative images depicting GFP-Ank9 exhibiting Golgi-like (Golgi) and ER-like (ER) localization patterns, as based on colocalization studies presented in Figure 2A and Figure 1A, respectively, are presented. Results are representative of at least three separate experiments having similar results. Scale bars, 20 μm.
Ank9 co-opts Golgi-to-ER retrograde trafficking
Localization of Ank9 to both the ER and Golgi suggested that it might traffic between the two organelles. Whether it does so in an anterograde or retrograde manner was unclear. Also, from the data generated thus far using Non-synchronized HeLa cells, it could not be discerned if the three subcellular localization phenotypes that GFP-Ank9 produces – perinuclear Golgi, destabilized ER-derived vesicles, destabilized ER-derived rings – occur concomitantly or in succession. Therefore, we subjected HeLa cells to serum starvation to synchronize the cell cycle prior to transfection and assayed for GFP-Ank9 subcellular localization at 4, 8, and 12 h. At 4 h, GFP-Ank9 signal was diffuse in the cytosol and also had accumulated in a perinuclear pattern reminiscent of the Golgi-associated phenotype (Fig. 3A). At 8 and 12 h, the cytosolic and perinuclear patterns were no longer apparent. Instead, GFP-Ank9 exhibited the vesicular pattern at 8 h and the ring-like pattern at 12 h. We hypothesized that upon being expressed in the cytosol, GFP-Ank9 first translocates to the Golgi and then retrograde traffics to the ER.
Fig. 3.
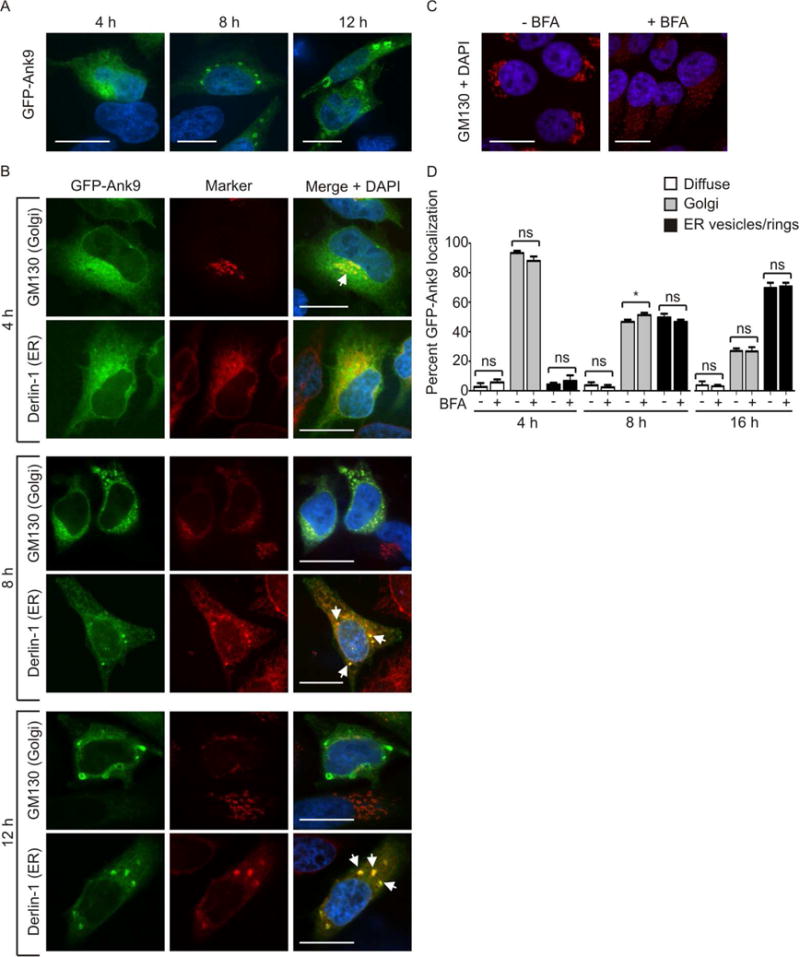
Ectopically expressed Ank9 exhibits temporal localization phenotypes that are consistent with Golgi-to-ER retrograde trafficking. Serum starved HeLa cells were transfected to express GFP-Ank9 (A and B). At 4, 8, or 12 h post transfection, the cells were fixed and examined by confocal microscopy (A–C). In some cases, fixed cells were screened with antibody against the cis-Golgi marker, GM130, or the ER protein, derlin-1 prior to confocal microscopic analyses (B). Representative fluorescence images of cells viewed for GFP; GM130 or derlin-1; and merged images plus DAPI are presented. White arrows in B denote representative points of GFP and organelle marker signal colocalization. (A) GFP-Ank9 exhibits subcellular localization patterns reminiscent of the Golgi at 4 h and destabilized ER at 8 and 12 h. (B) Immunofluorescent labeling confirms that GFP-Ank9 localizes to the Golgi at 4 h and subsequently traffics to the ER at 8 h and 12 h. Destabilization of GFP-Ank9-positive Golgi and ER is apparent at the 8 and 12 h time points. Representative cells exhibiting GFP-Ank9 Golgi like (Golgi) and ER-like (ER) subcellular localization patterns are denoted. (C and D) GFP-Ank9 does not require an intact Golgi to localize to the Golgi and ER. HeLa cells were treated with brefeldin A (BFA) or vehicle control. At 4 h, the cells were immunolabeled with GM130 antibody, stained with DAPI, and visualized using confocal microscopy to confirm that BFA destabilizes the Golgi (C). (D) Serum starved HeLa cells were treated with BFA- or vehicle control just prior to transfection with GFP-Ank9 plasmid. The cells were examined for GFP-Ank9 subcellular localization at 4, 8, and 16 h. Data are presented as the percentage of cells in which GFP-Ank9 signal was diffuse in the cytosol (Diffuse), localized to and disrupted the Golgi (Golgi), or localized to and altered the morphology of the ER to yield vesicular or ring-like structures (ER vesicles/rings). Results are representative of at least two separate experiments performed in triplicate, each yielding similar results. Scale bars, 20 μm. ns, not significant.
To test our hypothesis, the time course experiment was repeated except that GFP-Ank9 expressing cells were immunolabeled with Golgi-specific GM130 and ER-specific derlin-1 antibodies. At 4 h, perinuclear GFP-Ank9 signal predominantly colocalized with that of GM130 but not derlin-1 to yield the Golgi tropic phenotype (Fig. 3B). The Golgi and ER were intact at this time point. At 8 h, the Golgi was destabilized, as GFP-Ank9-positive, GM130-labeled vesicles were no longer ordered in a perinuclear stack-like arrangement typical of an intact Golgi but were instead fragmented and distributed throughout the cytosol. Also, at 8 h, transfected cells displayed distended, collapsed, and fragmented ER. As observed in Fig. 1A, the derlin-1 labeled ER-derived vesicles were GFP-Ank9-positive (Fig. 3B). At 12 h, the Golgi and ER were further fragmented. GFP-Ank9 localized to large vesicles or rings that were derlin-positive and GM130-negative. These results support that Ank9 traffics in a Golgi-to-ER retrograde manner and destabilizes both organelles. To determine if the Golgi must be intact for GFP-Ank9 to initially produce the perinuclear Golgi phenotype and then retrograde traffic to yield the ER-derived vesicles/rings phenotype and to quantify the percentages of cells exhibiting these phenotypes over the course of expression, immediately prior to transfection for expressing GFP-Ank9 HeLa cells were treated with brefeldin A to promote collapse of the Golgi (Sciaky et al., 1997). As expected, the Golgi was completely destabilized into GM130-immunolabeled vesicles (Fig. 3C). Yet, brefeldin A impeded neither the abilities of GFP-Ank9 to traffic to and disrupt the Golgi and ER nor the timing by which it does so (Fig. 3D).
Ank9 localization to and disruption of the Golgi and ER is F-box-independent
Aside from the ankyrin repeat domain, the F-box is the only known O. tsutsugamushi Ank functional domain (Beyer et al., 2015). To determine if the F-box is necessary for Ank9 to traffic to and perturb the structure of the Golgi and ER, GFP-tagged Ank9 lacking the F-box (Ank9ΔF-box) was expressed in HeLa cells and examined for colocalization with GM130 and derlin-1. Indistinguishable from the phenotypes exhibited by GFP-Ank9 (Figs. 1–3), GFP-Ank9ΔF-box localized to the Golgi and ER, altered the morphology of both, and failed to traffic to lysosomes (Fig. 4). Thus, the ability of Ank9 to traffic to and disrupt the structure of the Golgi and ER is not predicated on its F-box.
Fig. 4.
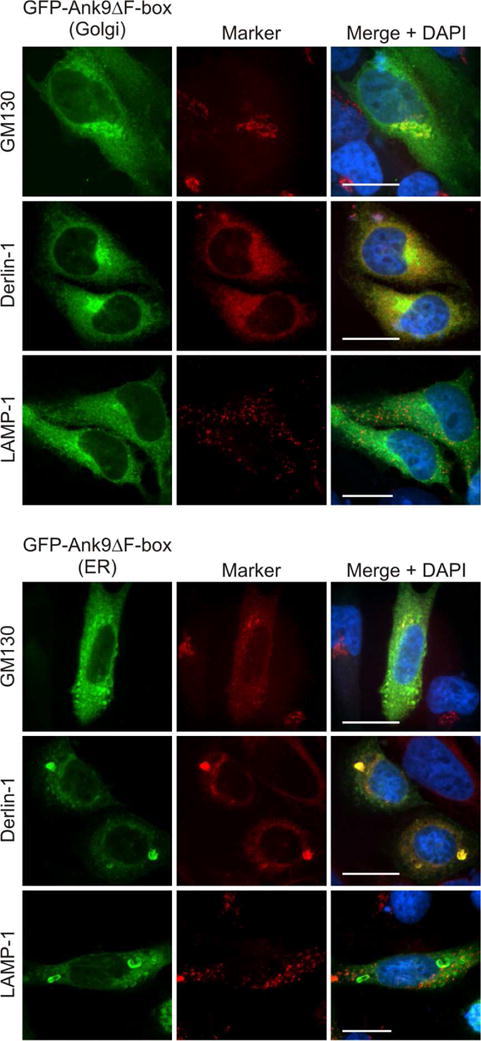
GFP-Ank9 localization to and perturbation of Golgi and ER morphology is F-box-independent. HeLa cells expressing GFP-Ank9ΔF-box were fixed, screened with GM130, derlin-1, and LAMP-1 antibodies, stained with DAPI, and examined by confocal microscopy. Representative fluorescence images of cells displaying GFP-Ank9ΔF-box Golgi-like (Golgi) and ER-like (ER) subcellular localization patterns viewed for GFP, organelle marker, and merged images plus DAPI are presented. Scale bars, 20 μm. Results shown are representative of three experiments with similar results.
The conserved Ank9 N-terminus is necessary and sufficient for Ank9 Golgi localization, but is insufficient for Golgi-to-ER retrograde trafficking
We rationalized that the ability of Ank9 to modulate the secretory pathway is predicated on its initial localization to the Golgi. Therefore, the next step in mechanistically dissecting Ank9 was to determine how it targets the Golgi. As there is no one canonical Golgi localization domain (GLD), the BLOSUM (Blocks of amino acid substitution matrix) 62 algorithm, which scores alignments of evolutionary divergent protein sequences based on local alignments (Henikoff et al., 1992), was used to compare Ank9 against confirmed GLDs of numerous eukaryotic proteins and bacterial effectors that exhibit Golgi trafficking. The N-terminal 47 residues of Ank9 exhibited homology to a eukaryotic Golgi targeting sequence called the GRIP domain that is conserved among several golgins and is named after the first four in which it was identified (golgin-97, RanBP2α, Imh1p, and p230/golgin-245) (Munro et al., 1999, Kjer-Nielsen et al., 1999a, Kjer-Nielsen et al., 1999b, Lu et al., 2006). Of these four, Ank9 had the most sequence similarity to p230/golgin-245, a trans-Golgi network (TGN) membrane protein associated with non-clathrin coated vesicles that uses its C-terminal 42-amino acid GRIP domain to traffic from the cytosol to the Golgi (Kjer-Nielsen et al., 1999b, Kjer-Nielsen et al., 1999a). Figure 5A presents a schematic of Ank9 and an alignment of its N-terminus with the p230/golgin-245 GLD. As determined using the BLASTP algorithm, Ank9, especially its putative GLD-containing N-terminus, is highly conserved among several geographically diverse O. tsutsugamushi strains (Table 1, Supplementary Fig. S1, Fig. 5B). Five of these strains were isolated from human scrub typhus patients (Ikeda, Gilliam, Karp, Kato, Sido) and two from mammalian reservoirs (TA716, TA763) (Blacksell et al., 2008, Jiang et al., 2013).
Fig. 5.
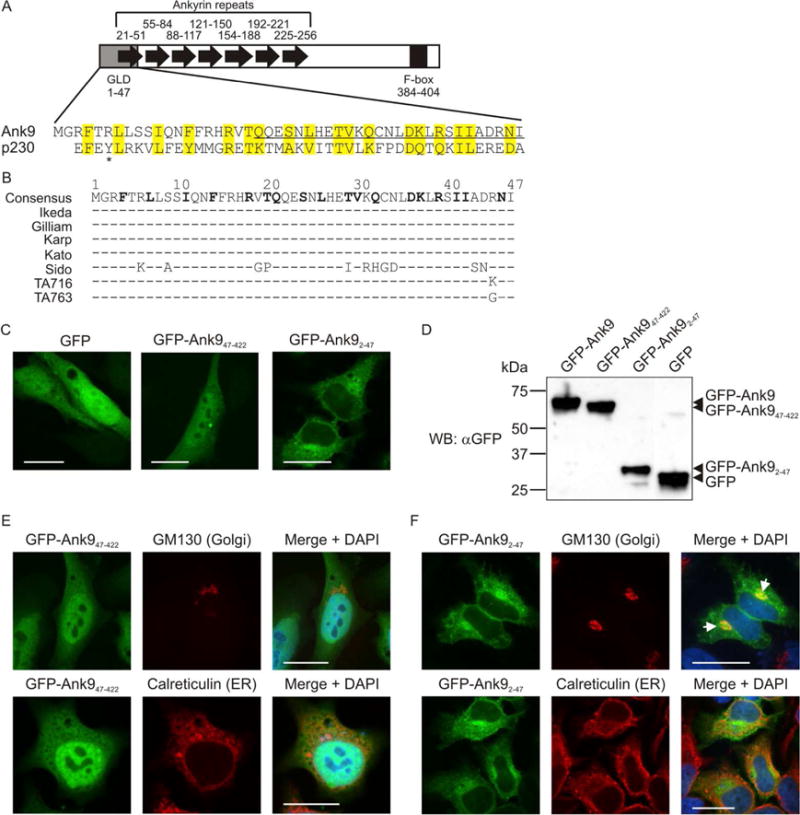
The conserved Ank9 N-terminus is both necessary and sufficient for Golgi localization, but is insufficient for Golgi-to-ER retrograde trafficking. (A) Schematic of Ank9. A gray rectangle denotes the Golgi localization domain (GLD). Black arrows denote individual ankyrin repeats. A black rectangle denotes the F-box. Numbers above or below each symbol represent the amino acids that make up the cognate motif. The Ank9 N-terminal GLD (amino acids 1–47) bears sequence similarity to the eukaryotic p230 GRIP domain, as shown in the amino acid alignment. Identical or similar amino acid residues are highlighted in yellow. An asterisk denotes the Y residue at position 4 of p230 that is conserved among eukaryotic GRIP domains. (B) Conservation of the Ank9 N-terminal GLD among O. tsutsugamushi strains. An alignment of amino acids 1 through 47 from seven O. tsutsugamushi Ank9 homologs is shown. Strain names are listed to the left of each sequence. Amino acid positions are denoted with numbers above the consensus sequence. Consensus sequence residues in bold text have identity/similarity to those of the p230 GRIP domain as shown in panel A. For each O. tsutsugamushi strain sequence, residues identical to the consensus sequence are represented by a dash (−) while non-identical amino acids are shown as their corresponding single letters. (C, E, and F) Ank9 residues 1–47 are sufficient and necessary for GFP-Ank9 to localize to the Golgi but are insufficient for GFP-Ank9 to retrograde traffic from the Golgi to the ER. HeLa cells transfected to express GFP, GFP-Ank947–422 (lacks the GLD), or GFP-Ank92–47 (GLD alone) were fixed and analyzed by confocal microscopy (C) or screened with antibody against the Golgi marker, GM130, or the ER marker, calreticulin, prior to confocal microscopic examination (E and F). Representative fluorescence images of cells viewed for GFP, GM130 or calreticulin, and merged images plus DAPI are presented. (E and F) Representative cells exhibiting GFP-Ank9 Golgi-associated (Golgi) and ER-associated (ER) subcellular localization patterns are denoted. White arrows in F designate representative points of GFP and GM130 signal colocalization. Scale bars, 20 μm. (D) Confirmation of expected protein sizes for GFP-Ank9, GFPAnk947–422, GFP-Ank92–47, and GFP. HeLa cell lysates of cells expressing GFP fusion proteins were analyzed by Western blot with GFP antibody. Arrowheads mark the expected apparent molecular weights for noted proteins. Results shown are representative of three independent experiments.
Table 1.
Amino acid similarity among O. tsutsugamushi Ank9 homologues
| Strain | Ank9 Homologue Annotation | GenBank Accession | Length | % Identitya | % Similaritya |
|---|---|---|---|---|---|
| Ikeda | OTT_0298 | BAG39756 | 422 | 100 | 100 |
| Gilliam | OTSGILL_1380 | KJV52595 | 451 | 86 | 91 |
| Karp | OTSKARP_0451 | KJV56569 | 422 | 85 | 90 |
| Kato | OTSKATO_0878 | KJV54885 | 422 | 96 | 98 |
| Sido | OTSSIDO_0886 | KJV97530 | 422 | 82 | 90 |
| TA716 | OTSTA716_0966 | KJV75930 | 422 | 86 | 92 |
| TA763 | OTSTA763_1177 | KJV74286 | 422 | 84 | 90 |
Identity and similarity based on NCBI BLASTP to str. Ikeda.
To determine if the Ank9 N-terminus confers Golgi targeting, subcellular localization of two Ank9 truncations N-terminally fused to GFP was assessed at 16 h post transfection. GFP-tagged Ank947–422, which lacks the putative GLD, was diffusely distributed throughout the cytosol and nucleus, similar to GFP (Fig. 5C). In contrast, GFP-tagged Ank92–47, which consists only of the putative GLD, displayed a perinuclear accumulation reminiscent of the Golgi tropic phenotype exhibited by GFP-Ank9. Screening Western blots of the corresponding transfected cell lysates with GFP antibody confirmed that the Ank9 truncations migrated at their expected sizes and, importantly, that the diffuse, GFP-like localization pattern of GFP-Ank947–422 was not due to cleavage of the protein to release free GFP (Fig. 5D). GFP-Ank92 47, but not GFP-Ank947–422 localized to the Golgi (Fig. 5, E and F). Neither GFP-Ank9 truncation localized to ER. The Golgi and ER remained intact in cells expressing either GFP-Ank947–422 or GFP-Ank92–47 (Fig. 5, E and F), even when the latter pronouncedly accumulated at the Golgi (Fig. 5F).
As a complementary approach, lysates of HeLa cells expressing GFP-tagged Ank9, Ank9ΔF-box, or Ank947–422 were subjected to density gradient fractionation. Western blots of successive fractions were screened with GFP, GM130, and calreticulin antibodies to confirm the Ank9 proteins’ subcellular trafficking patterns. HeLa cells that were untransfected or expressed only GFP were negative controls. Two post transfection time points, 4 and 16 h, were examined to determine if the duration that GFP or a GFP-fusion was overexpressed affected to which fractions it localized. GM130 and calreticulin were present in several fractions at both time points, but tended to be most abundant in fractions 5 to 8 and 6 to 8, respectively (Fig. 6). At 4 h, GFP exhibited no co-fractionation with GM130 or calreticulin but small proportions of GFP-tagged Ank9 and Ank9ΔF-box did, indicating that these two fusion proteins had begun to accumulate at the Golgi/ER-positive fractions. Based on data presented in Figure 3, we presume that GFP-Ank9 and GFP-Ank9ΔF-box had trafficked to the Golgi at 4 h. At 16 h, low levels of GFP were present in fractions additional to those observed at 4 h (Fig. 6). However, comparatively more GFP-Ank9 and GFP-Ank9ΔF-box were present in the fractions in which GM130 and calreticulin were most abundant, suggesting that both GFP fusions continued to accumulate at the Golgi and ER throughout the time course. Notably, GFP-Ank947–422 failed to accumulate in the GM130- and calreticulin-positive fractions at both time points, validating that the GLD is necessary for Ank9 to traffic to the Golgi and ER. Together, the data presented thus far demonstrate that the Ank9 N-terminal 47 residues contain a bona fide GLD, these residues alone are insufficient for targeting the ER, Ank9 translocation to the ER depends on its initial Golgi localization, and an Ank9 protein must contain the GLD and ankyrin repeat domain in order to disrupt Golgi and ER morphology.
Fig. 6.
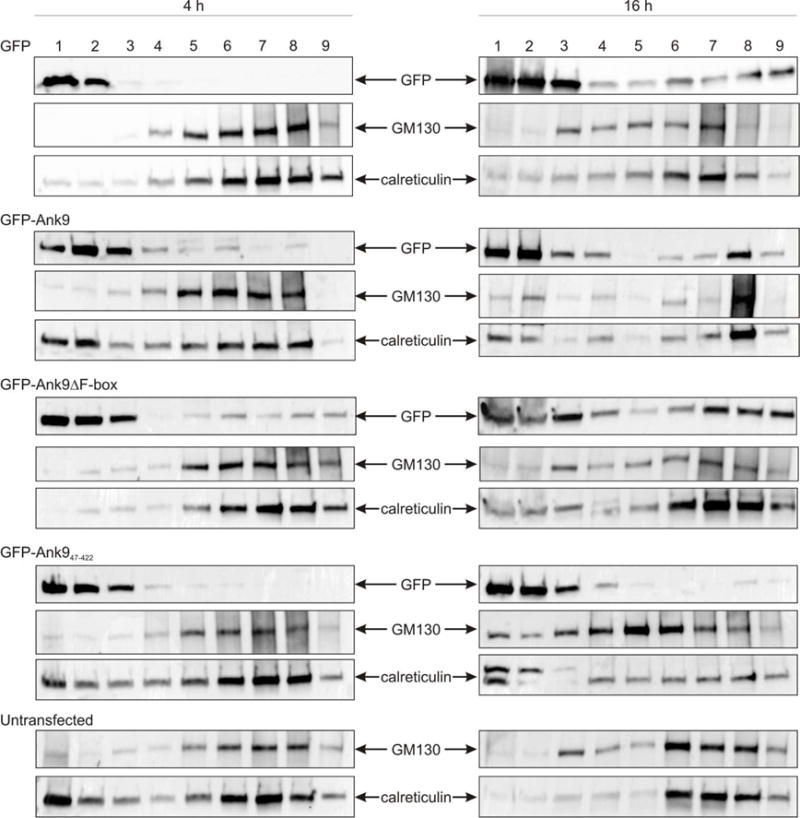
The Ank9 N-terminus is necessary for localization to Golgi- and ER-positive cellular fractions. Lysates of HeLa cells transfected to express GFP-tagged Ank9, Ank9ΔF-box, or Ank947–422 for 4 h or 16 h were subjected to density gradient fractionation. Western blots of nine successive fractions were screened with GFP, GM130, and calreticulin antibodies to confirm the Ank9 proteins’ subcellular trafficking patterns. Results are representative of two experiments with similar results.
Ank9 interacts with endogenous COPB2 in a GLD- and F-box-independent manner
Yeast two-hybrid screening was performed to identify potential mammalian Ank9 interacting proteins. Several candidates were identified (Table 2), two of which, SKP1 and CUL1, we had previously verified to interact with Ank9 by virtue of its F-box (Beyer et al., 2015). Among the remaining candidates, COPB2 (coatomer protein complex subunit [COPI] beta 2) was the most conspicuous because it contributes to vesicle budding from the Golgi and Golgi-to-ER retrograde trafficking [reviewed in (Lorente-Rodríguez et al., 2011)], cellular roles that align with the Ank9 subcellular trafficking phenotype. To validate the interaction, FLAG-tagged Ank9, Ank947–422, Ank9ΔF-box, or the negative controls, FLAG-Ank4 or FLAG-bacterial alkaline phosphatase (BAP), were immunoprecipitated from transfected HeLa cells. Western blot analyses of the inputs and immunoprecipitates confirmed expression and pull down of each FLAG-tagged bait, respectively (Fig. 7A). Probing Western blots of immunoprecipitated proteins with COPB2 antibody revealed that all three FLAG-Ank9 proteins co-immunoprecipitated endogenous COPB2. FLAG-tagged BAP and Ank4, another ER-tropic Ank (VieBrock et al., 2014), precipitated little-to-no COPB2. From these data, it can be concluded that Ank9 binds endogenous COPB2 and that neither the GLD nor the F-box domain mediates this interaction.
Table 2.
Candidate Ank9 binding partners identified by yeast two-hybrid
| Protein | Name and Function | + clones |
|---|---|---|
| SKP1a | S-phase kinase associated protein 1; binds F-box in SCF1 ubiquitin ligase complex | 12/26 |
| ERC1 | ELKS/RAB6-interacting/CAST family member; regulatory unit of IKK complex, vesicle trafficking of endosomes to Golgi | 3/26 |
| COPB2 | Coatomer protein complex (COPI) subunit beta 2; vesicular trafficking, Golgi budding, Golgi-to-ER retrograde trafficking | 2/26 |
| PXR1b | Human peroximal targeting signal import receptor (PXR1) | 2/26 |
| EXOC1 | Exocyst complex component 1 (Sec3); subunit of exocyst complex targeting endosomes to plasma membrane | 1/26 |
| CUL1a | Cullin 1; core component of SCF1 ubiquitin ligase complex | 1/26 |
| FEM1B | Fem 1 homolog b (C. elegans); ankyrin repeat-containing protein, mediates apoptosis | 1/26 |
| PIAS4 | Protein inhibitor of activated STAT, 4 (PIASY); E3-type small ubiquitin-like modifier (SUMO) ligase, SUMO tethering factor, transcription co-regulator | 1/26 |
| SNRNP70 | Small nuclear ribonucleoprotein 70 kDa (U1); recognizes pre-mRNA 5′ splice site, essential for spliceosome assembly | 1/26 |
| SP110 | SP110 nuclear body protein; gene transcription activator | 1/26 |
| ZBTB16 | Zinc finger and BTB binding domain containing 16; transcription factor, involved in cell cycle progression | 1/26 |
Protein known to interact with Ank9 (Beyer et al., 2015).
(Italics) Prey sequences were from an untranslated region of the respective gene, therefore no protein function is listed.
Fig. 7.
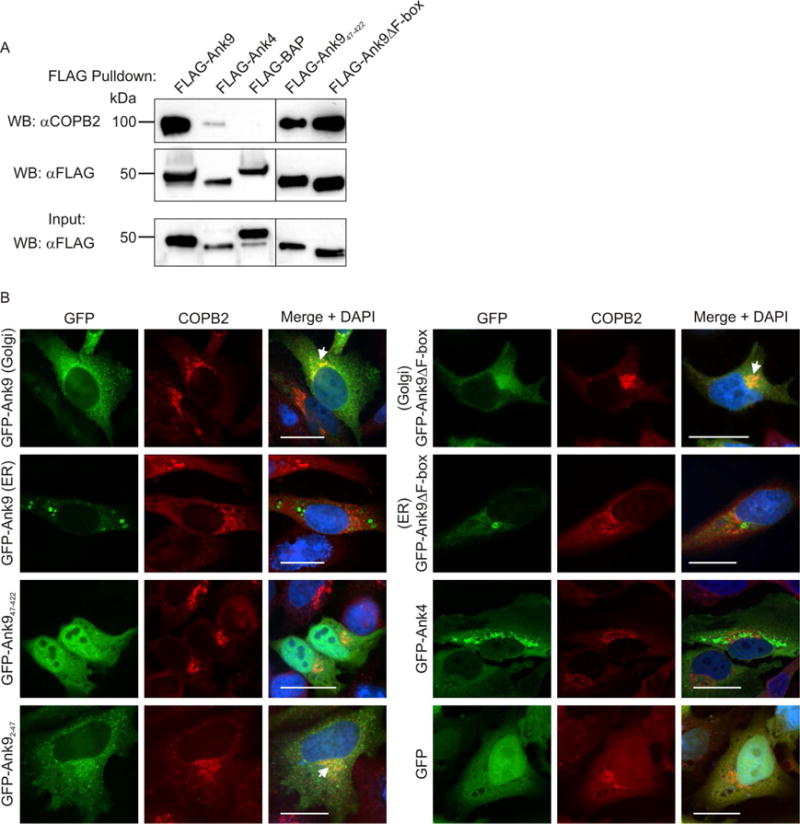
Endogenous COPB2 interacts with ectopically expressed Ank9 in a GLD- and F-box-independent manner and colocalizes with the Ank9 Golgi-associated but not ER-associated phenotype. (A) FLAG-Ank9 interacts with endogenous COPB2 in a GLD- and F-box-independent manner. Lysates of transfected HeLa cells expressing FLAG-tagged Ank9, Ank947–422 (lacks GLD), Ank9ΔF-box, Ank4, or BAP were incubated with FLAG antibody-conjugated agarose beads to immunoprecipitate FLAG-tagged proteins and co-precipitate interacting proteins. Resulting Western blots were probed with COPB2 antibody. Immunoprecipitation of FLAG-tagged proteins was verified by reprobing stripped blots with FLAG antibody. Expression of each FLAG-tagged protein of interest was confirmed by subjecting 3% of each input lysate to Western blotting using FLAG antibody. (B) GFP-Ank9 colocalizes with COPB2 in a GLD-dependent and F-box independent manner and only when Ank9 exhibits a Golgi-like localization pattern. HeLa cells expressing GFP tagged Ank9, Ank947–422, Ank92–47 (GLD only), Ank9ΔF-box, or the negative controls GFP-Ank4 or GFP alone were fixed, screened with COPB2 antibody, and examined by confocal microscopy. Representative fluorescence images of cells viewed for GFP, COPB2, and merged images plus DAPI are presented. Representative cells exhibiting GFP-Ank9 Golgi-associated (Golgi) and ER-associated (ER) subcellular localization patterns are denoted. White arrows designate representative points of GFP and COPB2 signal colocalization. Scale bars, 20 μm. Results shown are representative of three experiments with similar results.
Ank9 colocalization with COPB2 at the Golgi requires the GLD
We next determined if endogenous COPB2 colocalizes with GFP-tagged Ank9 or truncations thereof. COPB2 normally localizes to the Golgi cytosolic face as part of COPI vesicles (Stenbeck et al., 1993, Harrison-Lavoie et al., 1993) and, as such, concentrates at the Golgi when viewed by confocal microscopy (Hee-Kyung et al., 2015, Asada et al., 2004). GFP-Ank9, when exhibiting its Golgi tropic but not ER-tropic phenotype, and GFP-Ank92–47 each colocalized with COPB2 (Fig. 7B). Consistent with its inability to accumulate at the Golgi (Fig. 5E and Fig. 6), GFP-Ank947–422 failed to colocalize with the COPB2-immunolabeled organelle (Fig. 7B). When GFP-Ank9ΔF-box exhibited the Golgi-tropic phenotype, it colocalized with COPB2. Neither the GFP-Ank4 nor GFP controls colocalized with COPB2 (Fig. 7B).
Ank9 induces ER stress and impairs protein secretion
Given its tropism for the ER and Golgi and its propensity to alter the morphology of both organelles when ectopically expressed, it was next assessed if Ank9 invokes ER stress. As the F-box is the only Ank9 motif for which a bona fide function has been ascribed and F-box containing effectors have been shown to be important to the pathobiology of other intracellular bacteria (Price et al., 2009, Price et al., 2011, Ensminger et al., 2010, Lomma et al., 2010, Pilar et al., 2012, Voth et al., 2011), the role of the F-box in any Ank9 induced ER stress was also examined. A hallmark of ER stress is the increased expression of the unfolded protein response (UPR) transcription factors, ATF4 and/or XBP1, which activate expression of genes encoding ER chaperones and protein-folding enzymes (Lee et al., 2002) and genes involved in apoptosis, amino acid transport, and oxidative stress resistance, respectively (Urra et al., 2013, Harding et al., 2003). Western blots of whole cell lysates of HeLa cells expressing GFP-tagged Ank9, Ank9ΔF-box, or GFP were probed with antibody to ATF4 or XBP1. The densitometric signal intensity of each UPR transcription factor was compared to that of ß-actin for each sample. Compared to GFP alone, GFP-Ank9 stimulated a significant increase in the ratio of ATF4 to ß-actin (Fig. 8, A and B), but not XPB1 to ß-actin (Fig. 8, C and D). The ATF4 to ß-actin ratio in cells expressing GFP-Ank9ΔF-box was not statistically significant from that in cells expressing GFP-Ank9 or GFP only (Fig. 8B). Stripping and reprobing the blots with GFP antibody revealed that the expression of GFP alone was nearly twice that of GFP-Ank9 and GFP-Ank9ΔF-box and also that the levels of GFP-Ank9 and GFP-Ank9ΔF-box were comparable (Fig. 8, B and D). Thus, ATF4 induction is an F-box-independent function of Ank9 and does not merely result from ectopic overexpression of proteins. Based on the immunofluorescence data presented in Figures 4, 6, and 7, the residual ATF4 induction associated with GFP-Ank9ΔF-box expression was likely due to the fact that it retains the ability to traffic to and disrupt the Golgi and ER.
Fig. 8.
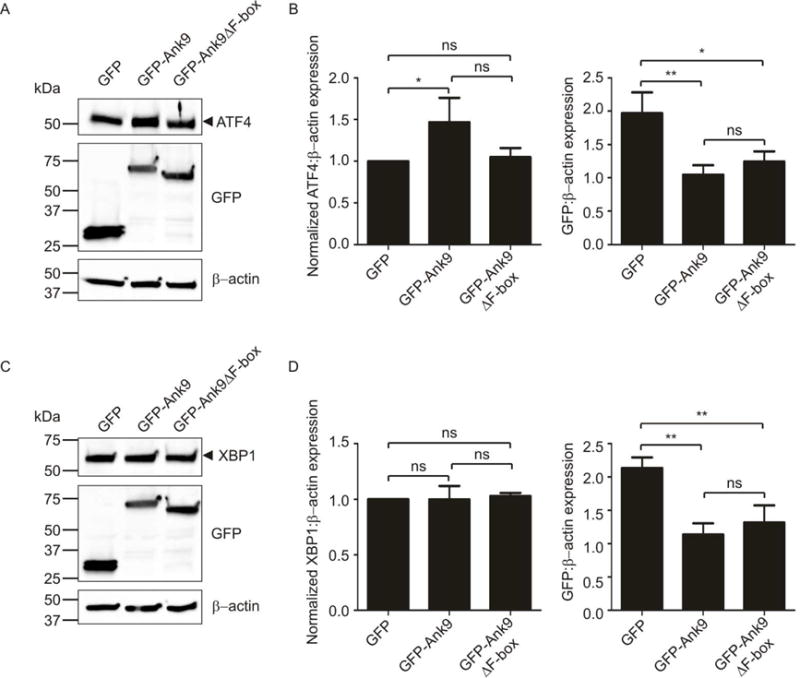
Ectopically expressed Ank9 induces ATF4, but not XBP1. Whole cell lysates of HeLa cells expressing GFP, GFP-Ank9, or GFP-Ank9ΔF-box were analyzed by Western blotting with antibodies to ATF4 (A) or XBP1 (C). The blots were stripped and reprobed with antibodies against GFP and β-actin to confirm expression of GFP-tagged proteins and as a loading control, respectively. Arrowheads mark the expected apparent molecular weights for noted proteins. Normalized ratios of ATF4 to β-actin (B), XBP1 to β-actin (D), and GFP to β-actin (B and D) were calculated using densitometry. Statistically significant (*P < 0.05; **P < 0.01) values are indicated. ns, not significant. Data are representative of three independent experiments.
A by-product of ER stress is the perturbation of cellular protein secretion [reviewed in (Trombetta et al., 2003)]. To further examine Ank9-induced ER stress and to confirm if the GLD contributes to the ability of the effector to induce ER stress, we assessed if ectopic expression of Ank9 alters secretory pathway function. Supernatants of HeLa cells co-expressing FLAG-tagged Ank9, Ank947–422, Ank9ΔF-box, Ank17, or BAP together with secreted luciferase were collected and assayed for relative luciferase activity. Ank17, which remains cytosolic when ectopically expressed (VieBrock et al., 2014), served as a non-ER targeting Ank control. Luciferase secretion by cells expressing FLAG-Ank9 was reduced by approximately 50% compared to cells expressing FLAG-BAP (Fig. 9A). Whereas deletion of the GLD (Ank947–422) eliminated the ability of FLAG-Ank9 to inhibit luciferase secretion, deletion of the F-box did not. Ank17 had no detrimental effect on luciferase secretion. Thus, Ank9 impairs mammalian cell protein secretion in a GLD-dependent, but F-box-independent manner.
Fig. 9.

Ank9 and O. tsutsugamushi each inhibit host cell protein secretion. (A) Ank9 inhibits host cell protein secretion in a GLD-dependent and F-box independent manner. Culture media from HeLa cells co-expressing FLAG-tagged BAP, Ank9, Ank947–422 (lacks GLD), or AnkΔF-box together with luciferase was assayed for the presence of secreted luciferase. Each condition was transfected in quadruplicate and each cell supernatant assayed in quadruplicate. (B) O. tsutsugamushi impairs host cell protein secretion. HeLa cells transfected to express a secreted luciferase reporter were mock infected or infected with O. tsutsugamushi at an MOI of 5, 17.5, or 35. Cell culture media was collected and analyzed for secreted luciferase activity. Relative luciferase activity in the media recovered from infected cells was normalized to that in media from mock infected cells. Data are representative of three experiments with similar results. Statistically significant (**P < 0.01; ***P < 0.001) values are indicated. ns, not significant. Data is representative of three independent experiments with similar results.
O. tsutsugamushi impairs host cell protein secretion
Because Orientia expresses Ank9, an effector that targets the secretory pathway, during infection of host cells, we next determined if the bacterium itself inhibits host cell protein secretion, HeLa cells were transfected to express the secreted luciferase reporter and infected with the Orientia at varying multiplicities of infection (MOI). At 24 h, the relative luciferase activity in the cell culture medium was measured and compared to mock infected cells (Fig. 9B). Luciferase secretion was reduced by approximately 50% in cells that had been infected at a MOI of 5 and nearly eliminated in cells infected at MOIs of 17.5 and 35. Thus, O. tsutsugamushi inhibits host cell protein secretion in a dose-dependent manner. Moreover, the bacterium and Ank9 phenocopy each other in their abilities to negatively impact the secretory pathway.
COPB2 levels are unchanged in cells ectopically expressing Ank9 or infected with O. tsutsugamushi
Ank9 carries a C-terminal F-box that binds SKP1 during infection and nucleates the SCF1 complex (Beyer et al., 2015), the role of which is to polyubiquitinate interacting proteins [reviewed in (Lee et al., 2014)]. Polyubiquitination can target proteins for degradation in the 26S proteasome, and other microbial F-box-containing Anks have been shown to promote F-box-dependent proteasomal destruction of their interacting partners (Lee et al., 2014, Price et al., 2009, Price et al., 2011). Knowing that Ank9 interacts with COPB2, we examined if Ank9 or O. tsutsugamushi facilitates a reduction in host cell COPB2 levels. Screening Western-blotted lysates of cells expressing FLAG-Ank9 versus FLAG-BAP or uninfected versus infected cells with FLAG and O. tsutsugamushi outer membrane protein A (OmpA) antibodies confirmed FLAG protein expression and infection, respectively (Fig. 10, A and C). Western blot analysis with COPB2 antibody revealed that COBP2 levels were unchanged in both respective pairs (Fig. 10, B and D).
Fig. 10.
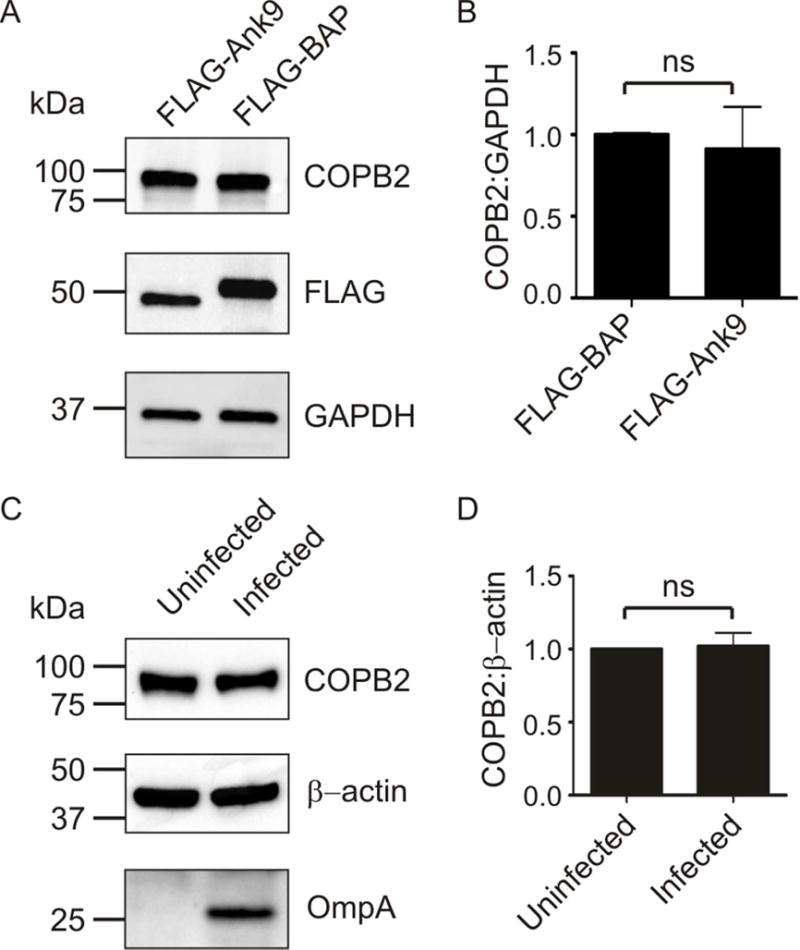
COPB2 levels are unchanged in cells expressing FLAG-Ank9 or infected with O. tsutsugamushi. Whole cell lysates of HeLa cells that were transfected to express FLAG-Ank9 versus FLAG-BAP (A) or that were uninfected or O. tsutsugamushi infected (C) were analyzed by Western blot with antibodies to detect COPB2, GAPDH or β-actin as a loading control, FLAG to confirm FLAG-tagged protein expression, and/or O. tsutsugamushi outer membrane protein A (OmpA). The mean ± SD COPB2 to GAPDH or β-actin ratios for pairs of lysates from at least three separate experiments were determined using densitometry (B and D). ns, not significant.
siRNA-mediated knockdown of COPB2 promotes Golgi destabilization and favors O. tsutsugamushi replication
As COPB2 is targeted but not reduced by Ank9 and COPB2 levels are unchanged in Orientia infected cells, we posited that the bacterium might negatively regulate COBP2 without degrading it. Furthermore, because the relevance of the Ank9-COPB2 interaction in Orientia infection could not be discerned using knock out-complementation of Ank9 due to the bacterium’s genetic intractability, we examined the importance of COPB2 to infection by siRNA knockdown. We rationalized that reducing COPB2 cellular levels would phenocopy any negative regulation of COPB2 that the bacterium might execute. HeLa cells were treated with COPB2 or non-targeting siRNA for 48 h, incubated with O. tsutsugamushi organisms at a MOI of 10, and the infection was allowed to proceed for 72 h. In COPB2 siRNA treated cells, COPB2 levels became progressively reduced throughout the time course (Fig. 11A) and the Golgi was destabilized (Fig. 11B). Western blot analysis with antibody against the O. tsutsugamushi outer membrane protein, TSA56 (56-kDa type-specific antigen), and qPCR analysis using primers targeting the bacterium’s 16S rRNA gene revealed that the Orientia DNA load was higher in COPB2 knockdown versus control cells at 48 and 72 h (Fig. 11, A and C). Thus, O. tsutsugamushi achieves a higher level of infection in cells in which COPB2 levels have been reduced.
Fig. 11.
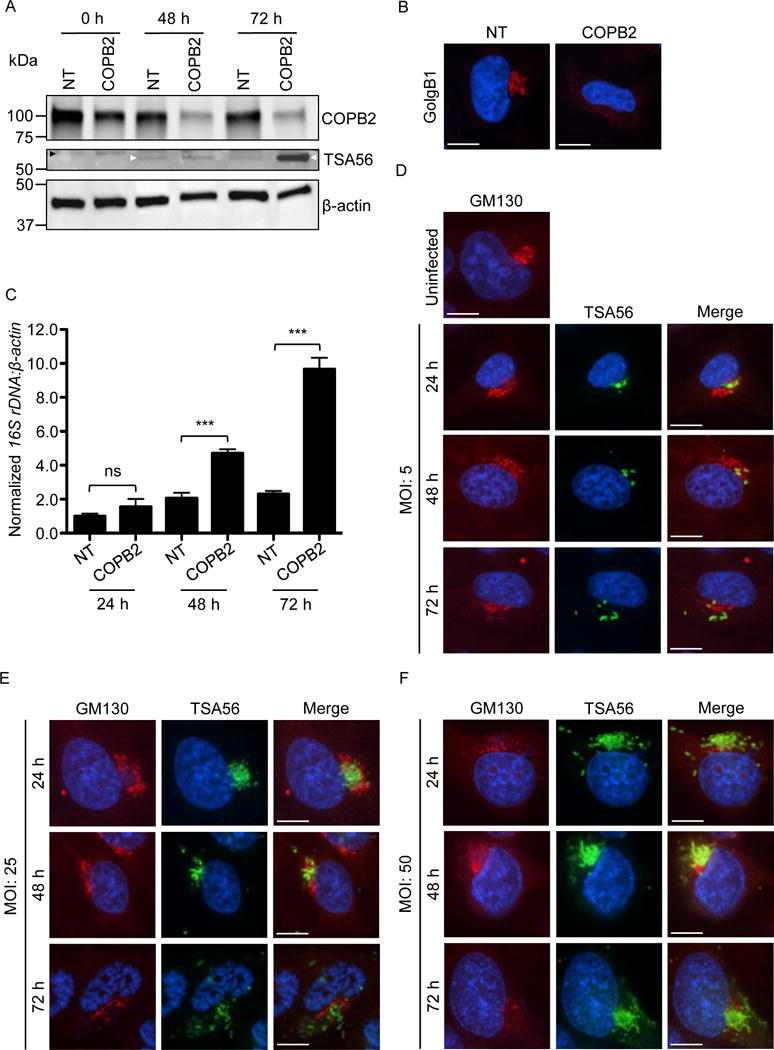
COPB2 knockdown increases Golgi instability and permissiveness to O. tsutsugamushi infection, and Orientia infection perturbs Golgi structure. (A–C) The Orientia load is higher and the Golgi is destabilized in cells in which COPB2 has been knocked down. (A) HeLa cells were treated with COPB2 or non-targeting (NT) siRNA for 48 h. The cells were infected with O. tsutsugamushi organisms (MOI of 10). Western blots of whole cell lysates generated at 0, 48, and 72 h post infection were screened with antibodies specific for COPB2, O. tsutsugamushi TSA56, and β-actin. White and black arrowheads denote the expected apparent molecular weights for TSA56 and a non-specifically recognized unknown HeLa cell protein, respectively. (B) At 48, NT and COPB2 siRNA treated uninfected HeLa cells were fixed, immunolabeled with GolgB1 antibody, stained with DAPI, and visualized by confocal microscopy. Results in A and B are representative of three experiments with similar results. (C) DNA isolated from HeLa cells that had been treated with NT or COPB2 siRNA for 48 h and infected with O. tsutsugaumushi for an additional 24, 48, or 72 h was analyzed by qPCR. Relative DNA levels of O. tsutsugamushi 16S rRNA gene were normalized to those of the human β-actin gene using the 2–ΔΔCT method. Results shown are the means ± standard deviation of triplicate samples and are representative of three independent experiments that yielded similar results. Statistically significant (***P < 0.001) values are indicated. ns, not significant. (D-F) O. tsutsugamushi infection promotes Golgi destabilization in bacterial dose- and time-dependent manners. HeLa cells infected with O. tsutsugamushi at MOIs of 5, 25, or 50 for 24, 48, or 72 h were fixed, immunolabeled with GM130 and TSA56 antibodies, stained with DAPI, and visualized by confocal microscopy. Results in D to F are representative of two experiments with similar results. Scale bars in B, D, E, and F are 10 μm.
O. tsutsugamushi perturbs Golgi structure
Ank9, an effector that engages COPB2 and that O. tsutsugamushi expresses during infection, disrupts Golgi and ER structure. The ER is distended in O. tsutsugamushi infected cells (Yang et al., 2008). Whether the bacterium affects Golgi morphology was unknown. Therefore, HeLa cells were incubated with the bacterium at MOIs of 5, 25, or 50 and examined by immunofluorescence microscopy at 24, 48, and 72 h. O. tsutsugamushi organisms were consistently detected juxtaposed to the Golgi (Fig. 11, D–F). The Golgi in cells that had been infected with an MOI of 5 was intact at 24 h, but showed signs of structural perturbation at 48 h and 72 h as GM130-immunolabeled structures were more vesicular, spread out, and/or diffuse in signal. This trend was more pronounced in cells infected at the higher MOIs, with GM130-immunolabeled structures being more spread out and/or having diffuse signal at 24 h and becoming even more so at successive time points (Fig. 11, E and F). Thus, O. tsutsugamushi disrupts Golgi morphology in a dose- and time-dependent manner.
DISCUSSION
Its obligate intracellular lifestyle necessitates that O. tsutsugamushi co-opts host cellular pathways to facilitate its survival. An effective way to accomplish this task would be to utilize individual effectors that commandeer multiple eukaryotic processes. Data presented herein, together with our prior report (Beyer et al., 2015), delineate Ank9 as a multifunctional effector that is conserved among geographically diverse O. tsutsugamushi strains and employs at least three distinct domains that interface with/modulate several host cell proteins/processes – SKP1 and CUL1 (SCF1 ubiquitin ligase assembly), COPB2 (Golgi-to-ER retrograde vesicular trafficking), the UPR, and protein secretion. We propose the following model for Ank9 function. By virtue of its N-terminal GRIP-like GLD, Ank9 traffics to the Golgi, where it interacts with COPB2 to co-opt the COPI-mediated retrograde pathway and traffic to the ER. These events do not require the Golgi to be intact, as they occur in the presence of brefeldin A. Once at the ER, Ank9 invokes the ATF4-mediated UPR. Its actions at the ER and/or Golgi disrupt protein secretion. These events are Ank9-specific and do not merely result from the accumulation of overexpressed GFP-fusion protein, as ATF4 was not upregulated and Golgi and ER morphology were unaltered in cells ectopically expressing GFP, which accumulated to a level approximately twice that of GFP-Ank9.
O. tsutsugamushi replicates adjacent to the ER and Golgi (Kim et al., 2001), juxtaposing it at an ideal location for delivering secretory pathway targeting effectors. In infected host cells, the ER is distended (Yang 2008), Golgi structure is perturbed, and the secretory pathway is inhibited. The bacterium’s effect on Golgi stability is observable by 48 and 72 h in cells infected at an MOI as low as five. Ectopically expressed Ank9 phenocopies these aspects of Orientia infection. Within hours of accumulating at the Golgi and ER, GFP-Ank9 destabilizes both organelles in a highly reproducible temporal manner. We surmise that at the low level at which it is expressed during infection (Beyer et al., 2015), endogenous Ank9 modestly perturbs Golgi and ER morphology but that these phenotypes are exaggerated in cells overexpressing the effector. Support for this premise comes from the fact that the degree of Golgi destabilization and the rate at which it occurs both increase in a bacterial dose-dependent manner. COPB2 helps maintain Golgi stability, as the organelle is highly fragmented in COPB2 knockdown cells. Ank9 binds to COPB2, but neither overexpression of the effector nor O. tsutsugamushi infection reduces COPB2 levels. COPB2 reduction and/or the cellular events that it invokes, such as Golgi destabilization, benefit Orientia replication, as the bacterial load is significantly increased in COPB2 knockdown cells. Thus, Ank9/Orientia negative modulation of COPB2 and/or the resulting Golgi instability and inhibition of cellular protein secretion might contribute to the bacterium’s intracellular replication. An additional or alternative role for the Ank9-COPB2 interaction is that Ank9 could bind COPB2 solely to co-opt retrograde traffic and reach its ER destination where it targets a second interacting partner, perhaps one involved in maintaining ER homeostasis that, when degraded or modulated, invokes the ATF4-dependent UPR. Given that Ank9 bears seven ankyrin repeats, distinct tandemly-arranged subsets of these repeats could mediate interactions with different binding partners.
Without first localizing to the Golgi, Ank9 cannot properly traffic, disrupt Golgi and ER stability, or inhibit the secretory pathway. Neither the GLD nor the F-box is required for Ank9 to bind COPB2, which implicates the ankyrin repeat region or, albeit less likely, an unidentified protein-protein interaction domain. The ability of FLAG-Ank947–422 to co-precipitate COPB2 even though it could not traffic to the Golgi is presumably due to the fact that the interaction occurred during incubation in a whole cell lysate in which COPB2 and Ank9 would have opportunity to interact and compensate for the inability of Ank947–422 to traffic to the Golgi and access COPB2. To our knowledge, Ank9 is the first obligate intracellular bacterial effector identified to utilize a GLD and to also exhibit dual Golgi-ER localization. It is also the first microbial protein known to mimic the GRIP domain. By comparison, Salmonella Typhimurium effectors use a conserved N-terminal WEK(I/M)xxFF motif to target the Golgi (Brown et al., 2006). We deemed the Ank9 region as a GRIP-like GLD as opposed to a bona fide GRIP GLD for three reasons. First, GRIP domains are typically located at the C-termini of coiled-coil peripheral membrane proteins (Kjer-Nielsen et al., 1999a, Munro et al., 1999, Barr, 1999), while Ank9 lacks coiled-coils and has an N-terminal GLD. Second, eukaryotic GRIP domains have a conserved tyrosine at position 4 (Kjer-Nielsen et al., 1999a, Munro et al., 1999, Barr, 1999) (Fig. 5, asterisk), which the Ank9 sequence lacks. Third, whereas the GRIP domain specifically targets the TGN (McConville et al., 2002, Brown et al., 2001), Ank9 localizes throughout the cis-, medial-, and trans-Golgi. Despite these distinctions, the Ank9 GLD is similar enough to facilitate Golgi localization. It will be important to pinpoint which residues are essential for its function, as such knowledge will be useful for identifying other microbial Golgi-localizing effectors and stands to expand overall understanding of Golgi-targeting motifs.
Bacterial hijacking of host retrograde trafficking has been reported with exotoxins such as shiga toxin, cholera toxin, and Pseudomonas exotoxin A. While Pseudomonas exotoxin A and cholera toxin rely on a eukaryotic-like KDEL sequence for recognition by COPI vesicles and subsequent trafficking to the ER, Shiga toxin employs a COPI-independent, actin/microtubule-dependent mechanism for retrograde trafficking [reviewed in (Sandvig et al., 2010, Capitani et al., 2009)]. Ank9 retrograde trafficking to the ER is unique from these three exotoxins because it does not have a KDEL sequence and still co-opts COPI traffic, as inferred from its confirmed interaction and subcellular localization with COPB2. The Ank9-COPB2 association is likely short-lived, as GFP-Ank9-positive ER-derived vesicular structures are negative for COPB2. GFP-Ank947–422 is unable to localize to either the Golgi or the ER, while GFP-Ank92–47 localizes to the Golgi but not the ER. This indicates that the GLD is sufficient for Golgi targeting, but something more is needed for ER localization. GFP-Ank9ΔF-box includes the GLD, ankyrin repeats, and a unique region between the ankyrin repeat and F-box domains. As shown in this and our previous study (Beyer et al., 2015), GFP-Ank9ΔF-box localizes to and morphologically disrupts both the Golgi and ER akin to GFP-tagged full-length Ank9. Thus, the ankyrin repeat and/or unique region confers ER localization and Golgi/ER structural perturbation only when in the presence of the GLD, and the F-box plays no role in Ank9 subcellular trafficking or organelle disruption. Aside from binding SKP1 (Beyer et al., 2015), the role of the Ank9 F-box in infection remains unclear. As more Ank9 interacting partners are identified, it will be key to determine if the effector and Orientia promote their ubiquitination and/or proteasomal degradation.
This report signifies Ank9 as a single microbial protein that uses separate domains to co-opt multiple host cell processes and links O. tsutsugamushi infection related events to Ank9 function. Given that multifunctional Ank9 is but one of 20 distinct O. tsutsugamushi Anks (VieBrock et al., 2014), the number and diversity of cellular processes that this effector arsenal modulates is likely considerable. Moving forward, dissecting additional roles of not only Ank9 but also determining the functions of others in the bacterium’s Ank repertoire will be of paramount importance to better understand O tsutsugamushi molecular pathogenesis.
EXPERIMENTAL PROCEDURES
Cell culture, infection, and transfection
O. tsutsugamushi str. Ikeda was originally isolated from a human scrub typhus patient in Japan (Ohashi et al., 1996). HeLa human cervical epithelial cells were cultured and infected with O. tsutsugamushi str. Ikeda as previously described (Beyer et al., 2015). HeLa cells were seeded on coverslips of a 24-well plate such that 90–95% confluency was achieved the next day. Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used to transfect 0.4 μg of DNA per well per the manufacturer’s directions. Following transfection, the cells were incubated for 4, 8, or 12 h for time course studies, or 16 h for standard transfections prior to fixation. For transfection experiments of HeLa cells having synchronized cell cycles, HeLa cells were seeded to achieve 70–80% confluency the following day, and then serum starved for 24 h in Roswell Park Memorial Institute 1640 (RPMI) media (Thermo Fisher Scientific, Waltham, MA) without fetal bovine serum (FBS). Media was replaced with RPMI plus FBS to restore cell growth, and the cells were transfected 0–6 h later as above.
Plasmid constructs
N-terminal fusions of O. tsutsugamushi str. Ikeda Ank9 (OTT_0298) and Ank4 (OTT_0210) with GFP and FLAG (VieBrock et al., 2014), and pGFP-Ank9ΔF-box and pFLAG Ank9ΔF-box plasmids in which the C-terminal F-box motif is removed from Ank9 (Beyer et al., 2015) were made as previously described. pFLAG-BAP, which encodes the negative control, FLAG-tagged bacterial alkaline phosphatase (BAP), was purchased from Sigma-Aldrich (St. Louis, MO). pCMV-GLuc (New England Biolabs, Gardner, MA) used for expression of secreted Gaussia luciferase, was a kind gift from Dr. George A. Belov at the University of Maryland (College Park, MD). ank9 nucleotides 4 to 141 (corresponding to amino acids 2 to 47 comprising the GLD) were PCR amplified using primers listed in Table 3. The resulting amplicon was purified, digested with EcoRI and SalI, and the restriction product was ligated into pEGFP C1 (Donated by Dr. Marci Scidmore, Cornell University, Ithaca, NY) that had been digested with EcoRI and SalI to create pGFP-Ank92–47. ank9 lacking the first 138 nucleotides (amino acids 1–46) was PCR amplified using oligonucleotides listed in Table 3, digested, and ligated into the EcoRI and SalI sites of pEGFP–C1 and p3XFLAG-CMV-14 (Sigma Aldrich) to create pGFP-Ank947–422 and pFLAG Ank947–422, respectively. Nucleotide sequence corresponding to the O. tsutsugamushi Ikeda strain TSA56 gene (56-kDa type specific antigen; OTT_0945) consisting of nucleotides 313 to 609, 727 to 1035, and 1078 to 1260 encoding hydrophilic amino acid segments 105 to 203, 243 to 354, and 360 to 420, respectively, was synthesized by Biomatik (Wilmington, DE). Plasmid encoding a His-tagged version of the chimeric TSA56 sequence was generated by amplifying the chimeric TSA56 nucleotide sequence using primers listed in Table 3 and annealing the amplicons into the pET46 Ek/LIC vector (Novagen, EMD Millipore, Darmstadt, DE) per the manufacturer’s instructions.
Table 3.
Oligonucleotides used in this study
| Designationa | Sequence (5′ to 3′) |
|---|---|
| β-actin F | AGAGGGAAATCGTGCGTGAC |
| β actin R | CAATAGTGATGACCTGGCCGT |
| ank9 4F | ATCGGAATTCgGGGAGATTCACCAG |
| ank9 141R | ATCGGTCGACTCAGATGTTCCTATCGGCGATAATG |
| ank9 139F | ACGTGAATTCgATCAATATCAATGCCCTGAATG |
| ank9 1266R | ATCGGTCGACTCAGTCGCAGATG |
| Ot 16S 911F | GTGGAGCATGCGGTTTAATTCGATGATC |
| Ot 16S 1096R | TAAGAATAAGGGTTGCGCTCGTTGC |
| Ot TSA56 313F | GACGACGACAAGATAGTTGGCAAAGGCAAAGTGGATAGTCG |
| Ot TSA56 1260R | GAGGAGAAGCCCGGTTAGCTCAGATCAAATTCGGTTTCTTTTTTAGACACTTC |
F and R refer to primers that bind to the sense and antisense strand, respectively. Underlined nucleotides correspond to EcoRI (GAATTC) and SalI (GTCGAC) restriction sites used in cloning; bold residues indicate a stop codon; small letters indicate nucleotides added to maintain frame; italicized nucleotides denote ligase-independent cloning tails.
Immunofluorescence labeling and confocal microscopy
Transfected or O. tsutsugamushi infected HeLa cells on coverslips were fixed and immunofluorescently labeled as described (VieBrock et al., 2014). His-tagged chimeric TSA56 protein was expressed and purified by immobilized metal-affinity chromatography as previously described (Miller et al., 2011) and supplied to New England Peptide (Gardner, MA) for generating rabbit antiserum. Coverslips were incubated with primary antibodies to detect TSA56 (1:1000), calnexin (C-terminus, 1:100; Enzo Life Sciences, Farmingdale, NY), calreticulin (1:250; Sigma-Aldrich), Rer1 (1:1000; Sigma-Aldrich), derlin-1 (1:400; Sigma-Aldrich), protein disulfide isomerase (PDI; 1:500; Sigma-Aldrich), GolgB1 (1:100; Sigma-Aldrich), GM130 (1:250; BD Biosciences, San Jose, CA), LAMP-1 (2 μg/mL; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), COPB2 (1:1000; Bethyl Laboratories, Montgomery, TX) and FLAG (1:1000; Sigma-Aldrich). Alexa Fluor 594-conjugated goat anti-rabbit IgG or goat anti-mouse IgG (1:1000; Invitrogen) were used as secondary antibodies prior to mounting with ProLong gold antifade plus 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). CellLight Golgi-RFP was used according to the manufacturer’s directions (Invitrogen). Slides were examined and images were acquired by spinning disk confocal microscopy using a BX51 microscope (Olympus, Center Valley, PA) affixed with an Olympus disk spinning unit and an ORCA-R2 CCD camera (Hammamatsu, Japan). Spinning disk images were processed using the Slidebook 5.0 software package (Intelligent Imaging Innovations, Denver, CO).
In silico analyses
The NCBI protein Basic Local Alignment Search Tool (BLASTP; http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to search for O. tsutsugamushi str. Ikeda Ank9 (OTT_0298) homologs within the annotated proteomes of nine O. tsutsugamushi strains: human isolates Boryong (South Korea), Gilliam (Burma), Karp (New Guinea), Kato (Japan), Sido (Australia), UT76 (Thailand), and UT144 (Thailand); ground squirrel isolate TA716 (Thailand); and brown spiny rat isolate TA763 (Thailand) (Jiang et al., 2013, Blacksell et al., 2008). GenBank accession numbers and gene annotations are listed in Table 1. Proteins were considered an Ank9 homolog if they had >80% coverage and >60% sequence similarity as determined by the BLAST algorithm. Amino acid sequence alignments were constructed using Geneious R6 v.6.1.8 software (http://www.geneious.com/), and similarity between Ank9 and p230 was determined using the default Geneious alignment algorithm.
Yeast two-hybrid
ULTImate yeast two-hybrid analysis was performed by Hybrigenics Services (Paris, France) (http://www.hybrigenics-services.com). The coding sequence for mammalian codon optimized O. tsutsugamushi str. Ikeda ank9 (amino acids 2–422) was PCR amplified and cloned into the pB27 construct as an N-terminal fusion with LexA (N-LexA-Ank9-C). After confirming sequence fidelity, the construct was introduced into yeast as bait and screened by mating with yeast bearing a randomly primed Human Placental cDNA library (prey). As O. tsutsugamushi infects multiple mammalian host cell types, the placental library offered the best representation of the human proteome for identifying putative Ank9 interactions. Positively selected clones were isolated and the corresponding prey fragments were PCR amplified, sequenced, and identified using the NCBI GenBank Database (http://blast.ncbi.nlm.nih.gov/genbank) and bioinformatics tools (BLAST). Predicted biological score (PBS) was calculated to assess the reliability of each interaction, ranging from the highest probability of specificity (A score) to the lowest probability of specificity (E score) between two proteins (Rain et al., 2001).
FLAG pulldown assays
Precipitation of FLAG-tagged proteins was performed as previously described (Beyer et al., 2015). Briefly, cells were lysed in lysis buffer (20 mM Tris, pH 7.4, 0.5 M NaCl, 0.7% Tween 20, with protease inhibitor [Roche Diagnostics, Germany]) and incubated overnight with FLAG affinity agarose resin (Sigma-Aldrich). The resin was washed 3–5 times in wash buffer (50 mM Tris, pH 7.4, 400 mM NaCl with protease inhibitor) prior to elution of proteins with 2X SDS sample buffer. Samples were resolved by SDS-PAGE and transferred to nitrocellulose, which was screened with antibodies to detect COPB2 (1:10000) and FLAG (1:1000). Pulldown lysate inputs (5%) were used to confirm FLAG protein expression. To verify that Ank9 truncated proteins were of the expected sizes, cells were lysed in RIPA buffer. Each lysate (30 μg) was subjected to SDS-PAGE and Western blotting with GFP antibody. For assessment of COPB2 levels in lysates of uninfected versus O. tsutsugamushi-infected or FLAG-BAP versus FLAG-Ank9 expressing HeLa cells, cells were lysed in RIPA buffer or 50 mM HEPES, pH 7.5 with 1 mM dithiothreitol (DTT; Thermo Fisher Scientific) and quantified using Protein Assay Reagent (Bio-Rad, Hercules, CA). Lysates (15–30 μg) were Western blotted and screened with antibodies against COPB2, β-actin, GAPDH, and O. tsutsugamushi OmpA (Beyer et al., 2015), each at a 1:1000 dilution. Densitometry was used to calculate COPB2:β-actin ratios using the ChemiDoc Touch imager (Bio-Rad) and ImageLab software (Bio-Rad).
Brefeldin A treatment and scoring of GFP-Ank9 phenotypes
Brefeldin A (eBioscience, San Diego, CA) at a final concentration of 3.0 μg per mL was added to freshly changed primary growth media of HeLa cell coverslips just prior to transfection. After Lipofectamine transfection of pGFP-Ank9 DNA (0. 4 μg per well of a 24-well plate), triplicate samples of cells were incubated for 4, 8, or 16 h prior to fixation and mounting with DAPI. At least 100 GFP-positive cells were counted for each triplicate sample. Cells were examined by spinning disc confocal microscopy and classified as: “diffuse” whereby GFP signal was distributed evenly throughout the cell cytoplasm; “Golgi” when the GFP signal was distributed throughout the cytoplasm but with a perinuclear accumulation that was verified herein using immunolabeling to correspond to Golgi localization; or “ER vesicles/rings” when the GFP signal manifested as large aggregates or rings in the cytoplasm verified herin via immunolabeling to be ER-derived.
Density gradient centrifugation
A T-75 flask of near-confluent HeLa cells was transfected with 12 μg of pEGFP, pGFP-Ank9, pGFP-Ank9ΔF-box, or pGFP-Ank947–422. A confluent flask of untransfected HeLa cells was used as a control. At 4 h or 16 h post transfection, the cells were washed twice with cold PBS prior to processing for density gradient centrifugation and trichloroacetic acid precipitation as previously described (Truchan et al., 2016). Equal volumes of fractions 1 to 9 were immunoblotted using GM130 (1:500, BD Biosciences), calreticulin (1:4000, Sigma Aldrich), and GFP (1:1000, Thermo Fisher) antibodies.
Determination of ATF4 and XBP1 induction
Confluent HeLa cell cultures grown in T-25 flasks were transfected with 4 μg of plasmid to facilitate expression GFP, GFP-Ank9, or GFP-Ank9ΔF-box. At 18 h, the cells were dislodged by scraping, pelleted at 500 × g, and washed twice with phosphate buffered saline (PBS). Cell pellets were lysed using radioimmunoprecipitation assay (RIPA) buffer [described in (Troese et al., 2011)] on ice for 45 min, centrifuged at 16,000 × g for 10 min, and the supernatants were quantified using Protein Assay Reagent (Bio-Rad). For each sample, 30 μg of lysate was resolved by SDS-PAGE as described (VieBrock et al., 2014) and Western blotted with antibodies against ATF4 (1:1000; (Cell Signaling Technology, Beverly, MA) or XBP1 (1:1000; Cell Signaling Technology) diluted in 5% (vol/vol) bovine serum albumin (BSA) in tris-buffered saline with 0.5% Tween-20 (TBS-T). Blots were subsequently screened with anti β-actin (1:2500, Santa Cruz Biotechnology, Dallas, TX) and anti-GFP, and densitometry was performed as described above.
Luciferase secretion assay
HeLa cells seeded into wells of 24-well plates were co-transfected with 0.4 μg of endotoxin free pCMV-GLuc, for expression of secreted Gaussia luciferase, and 0.4 μg endotoxin free pFLAG-BAP, pFLAG-Ank9, pFLAG-Ank9∆F-box, or pFLAG-Ank947–422 using Lipofectamine 2000. Each transfection was performed in triplicate. At 18 h, the culture media was removed from each well and replaced with fresh media. The cells were incubated for an additional 6 hours, then the media was collected. The luciferase activity of each collected media sample was measured in triplicate using the BioLux Gaussia luciferase assay (New England Biolabs) and a Victor 3 plate reader (Perkin Elmer, Waltham, MA).
Assessment of the effect of COPB2 knockdown on Golgi stability and the O. tsutsugamushi DNA load and the effect of Orientia infection on Golgi stability
HeLa cells were seeded on 12-mm coverslips and grown to approximately 85% confluency. ON-TARGETplus human COPB2 SMARTpool siRNA or non-targeting siRNA (GE Dharmacon, Lafayette, CO) was added to a final concentration of 0.025 μM in culture media. After 48 h, the cells were immunolabeled with GolgiB1 antibody, stained with DAPI, and examined by spinning disk confocal microscopy. To determine the effect of O. tsutsugamushi infection on Golgi integrity, siRNA was added to HeLa cells grown in 6-well plates. To generate O. tsutsugamushi inocula, infected (≥ 90%) HeLa cells were mechanically disrupted using glass beads in culture medium. At 48 h post transfection, COPB2 or control siRNA treated HeLa cells were incubated with 200 μL of inoculum for 1 h to achieve an MOI of 10, after which the cells were washed, fresh media was added, and incubation continued for an additional 47 h. The cells were dislodged by scraping and pelleted at 5000 × g for 5 min prior to DNA collection using the DNeasy Blood and Tissue kit (QIAGEN, Valencia, CA, USA). qPCR was performed on triplicate samples consisting of 100 ng genomic DNA, SsoFast Evagreen Supermix (Bio-Rad) and primers targeting genes encoding O. tsutsugamushi 16S rDNA (OTT_RNA006) and human β-actin (Table 3). qPCR assays were performed using the CFX96 real-time PCR detection system (Bio-Rad). Relative infection load levels were normalized to the DNA levels of the host cell β-actin gene using the 2–ΔΔCT method (Livak et al., 2001). To determine the effect of O. tsutsugamushi infection on Golgi integrity, HeLa cells seeded on 12-mm coverslips were infected with O. tsutsugamushi organisms at MOIs of 5, 25, and 50. At 24, 48, and 72 h, coverslips were fixed, immunolabeled with GM130 and TSA56 antibodies, stained with DAPI, and examined by spinning disk confocal microscopy.
Statistical analyses
One-way analysis of variance with Tukey’s post-test was performed using the Prism 5.0 software package (Graphpad, San Diego, CA) to assess statistical significance (set at P < 0.05). For densitometry data, ANOVA analyses using Prism 5.0 was applied.
Supplementary Material
Fig. S1. Alignment of Ank9 homologs from seven O. tsutsugamushi strains. Full-length amino acid sequences of Ank9 homologs are aligned beneath the consensus sequence. A dash (−) indicates sequence identity with the Ank9 consensus, while a letter denotes variation. Underlined regions correspond to the seven ankyrin repeats of Ikeda Ank9. Residues in bold text correspond to the Ikeda Ank9 F-box domain. Numbers above the alignment denote amino acid positions.
Acknowledgments
This work was supported by National Institutes of Health grants R03 AI101666, R21 AI103606 (J.A.C.), and R56 AI123306 (J.A.C.), K12 GM093857 (A.R.B.); American Heart Association (AHA) Grant-in-Aid 13GRNT16810009 (J.A.C.), AHA Predoctoral Fellowship 13PRE16840032 (L.V.), the VCU Presidential Research Quest Fund (J.A.C.), and George and Lavinia Blick Scholar funds (J.A.C.), and VCU Office of Postdoctoral Services (A.R.B.).
Footnotes
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
References
- Asada M, Irie K, Yamada A, Takai Y. Afadin- and α-actinin-binding protein ADIP directly binds β′-COP, a subunit of the coatomer complex. Biochemical and Biophysical Research Communications. 2004;321:350–354. doi: 10.1016/j.bbrc.2004.06.143. [DOI] [PubMed] [Google Scholar]
- Barr FA. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr Biol. 1999;9:381–384. doi: 10.1016/s0960-9822(99)80167-5. [DOI] [PubMed] [Google Scholar]
- Beyer AR, VieBrock L, Rodino KG, Miller DP, Tegels BK, Marconi RT, Carlyon JA. Orientia tsutsugamushi Strain Ikeda Ankyrin Repeat-Containing Proteins Recruit SCF1 Ubiquitin Ligase Machinery via Poxvirus-Like F-box Motifs. J Bacteriol. 2015;197:3097–3109. doi: 10.1128/JB.00276-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacksell SD, Luksameetanasan R, Kalambaheti T, Aukkanit N, Paris DH, McGready R, et al. Genetic typing of the 56-kDa type-specific antigen gene of contemporary Orientia tsutsugamushi isolates causing human scrub typhus at two sites in north-eastern and western Thailand. FEMS Immunol Med Microbiol. 2008;52:335–342. doi: 10.1111/j.1574-695X.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- Brown DL, Heimann K, Lock J, Kjer-Nielsen L, van Vliet C, Stow Jennifer L, Gleeson Paul A. The GRIP Domain is a Specific Targeting Sequence for a Population of trans-Golgi Network Derived Tubulo-Vesicular Carriers. Traffic. 2001;2:336–344. doi: 10.1034/j.1600-0854.2001.002005336.x. [DOI] [PubMed] [Google Scholar]
- Brown NF, Szeto J, Jiang X, Coombes BK, Finlay BB, Brumell JH. Mutational analysis of Salmonella translocated effector members SifA and SopD2 reveals domains implicated in translocation, subcellular localization and function. Microbiology. 2006;152:2323–2343. doi: 10.1099/mic.0.28995-0. [DOI] [PubMed] [Google Scholar]
- Capitani M, Sallese M. The KDEL receptor: New functions for an old protein. FEBS Letters. 2009;583:3863–3871. doi: 10.1016/j.febslet.2009.10.053. [DOI] [PubMed] [Google Scholar]
- Ensminger AW, Isberg RR. E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect Immun. 2010;78:3905–3919. doi: 10.1128/IAI.00344-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Harrison-Lavoie KJ, Lewis VA, Hynes GM, Collison KS, Nutland E, Willison KR. A 102 kDa subunit of a Golgi-associated particle has homology to beta subunits of trimeric G proteins. Embo J. 1993;12:2847–2853. doi: 10.1002/j.1460-2075.1993.tb05946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hee-Kyung A, Yong Won K, Hye Min L, Inhwan H, Hyun-Sook P. Physiological Functions of the COPI Complex in Higher Plants. Mol Cells. 2015;38:866–875. doi: 10.14348/molcells.2015.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan KK, Bordenstein SR. Ankyrin domains across the Tree of Life. PeerJ. 2014;2:e264. doi: 10.7717/peerj.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Paris DH, Blacksell SD, Aukkanit N, Newton PN, Phetsouvanh R, et al. Diversity of the 47-kD HtrA Nucleic Acid and Translated Amino Acid Sequences from 17 Recent Human Isolates of Orientia. Vector Borne and Zoonotic Diseases. 2013;13:367–375. doi: 10.1089/vbz.2012.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Ihn KS, Han SH, Seong SY, Kim IS, Choi MS. Microtubule- and dynein-mediated movement of Orientia tsutsugamushi to the microtubule organizing center. Infect Immun. 2001;69:494–500. doi: 10.1128/IAI.69.1.494-500.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Teasdale RD, van Vliet C, Gleeson PA. A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr Biol. 1999a;9:385–388. doi: 10.1016/s0960-9822(99)80168-7. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, van Vliet C, Erlich R, Toh BH, Gleeson PA. The Golgi-targeting sequence of the peripheral membrane protein p230. J Cell Sci. 1999b;112(Pt 11):1645–1654. doi: 10.1242/jcs.112.11.1645. [DOI] [PubMed] [Google Scholar]
- Lee EK, Diehl JA. SCFs in the new millennium. Oncogene. 2014;33:2011–2018. doi: 10.1038/onc.2013.144. [DOI] [PubMed] [Google Scholar]
- Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lomma M, Dervins-Ravault D, Rolando M, Nora T, Newton HJ, Sansom FM, et al. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 2010;12:1272–1291. doi: 10.1111/j.1462-5822.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- Lorente Rodríguez A, Barlowe C. Entry and Exit Mechanisms at the cis-Face of the Golgi Complex. Cold Spring Harbor Perspectives in Biology. 2011;3 doi: 10.1101/cshperspect.a005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Tai G, Wu M, Song H, Hong W. Multilayer interactions determine the Golgi localization of GRIP golgins. Traffic. 2006;7:1399–1407. doi: 10.1111/j.1600-0854.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- McConville MJ, Ilgoutz SC, Teasdale RD, Foth BJ, Matthews A, Mullin KA, Gleeson PA. Targeting of the GRIP domain to the trans-Golgi network is conserved from protists to animals. European Journal of Cell Biology. 2002;81:485–495. doi: 10.1078/0171-9335-00268. [DOI] [PubMed] [Google Scholar]
- Miller DP, McDowell JV, Bell JK, Marconi RT. Crystallization of the factor H-binding protein, FhbB, from the periopathogen Treponema denticola. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:678–681. doi: 10.1107/S1744309111011298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min CK, Kwon YJ, Ha NY, Cho BA, Kim JM, Kwon EK, et al. Multiple Orientia tsutsugamushi ankyrin repeat proteins interact with SCF1 ubiquitin ligase complex and eukaryotic elongation factor 1 alpha. PLoS One. 2014;9:e105652. doi: 10.1371/journal.pone.0105652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Nichols BJ. The GRIP domain - a novel Golgi-targeting domain found in several coiled-coil proteins. Curr Biol. 1999;9:377–380. doi: 10.1016/s0960-9822(99)80166-3. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Yamashita A, Kurokawa K, Morimoto T, Ogawa M, Fukuhara M, et al. The Whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008;15:185–199. doi: 10.1093/dnares/dsn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi N, Koyama Y, Urakami H, Fukuhara M, Tamura A, Kawamori F, et al. Demonstration of Antigenic and Genotypic Variation in Orientia tsutsugamushi Which Were Isolated in Japan, and Their Classification into Type and Subtype. Microbiol Immunol. 1996;40:627–638. doi: 10.1111/j.1348-0421.1996.tb01120.x. [DOI] [PubMed] [Google Scholar]
- Pilar AV, Reid-Yu SA, Cooper CA, Mulder DT, Coombes BK. GogB is an anti-inflammatory effector that limits tissue damage during Salmonella infection through interaction with human FBXO22 and Skp1. PLoS Pathog. 2012;8:e1002773. doi: 10.1371/journal.ppat.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CT, Al-Khodor S, Al-Quadan T, Santic M, Habyarimana F, Kalia A, Kwaik YA. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 2009;5:e1000704. doi: 10.1371/journal.ppat.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CT, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science. 2011;334:1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Sandvig K, Torgersen ML, Engedal N, Skotland T, Iversen TG. Protein toxins from plants and bacteria: Probes for intracellular transport and tools in medicine. FEBS Letters. 2010;584:2626–2634. doi: 10.1016/j.febslet.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Sciaky N, Presley J, Smith C, Zaal KJ, Cole N, Moreira JE, et al. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. The Journal of cell biology. 1997;139:1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenbeck G, Harter C, Brecht A, Herrmann D, Lottspeich F, Orci L, Wieland FT. Beta’-COP, a novel subunit of coatomer. Embo J. 1993;12:2841–2845. doi: 10.1002/j.1460-2075.1993.tb05945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AJ, Paris DH, Newton PN. A Systematic Review of Mortality from Untreated Scrub Typhus (Orientia tsutsugamushi) PLoS Negl Trop Dis. 2015;9:e0003971. doi: 10.1371/journal.pntd.0003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troese MJ, Kahlon A, Ragland SA, Ottens AK, Ojogun N, Nelson KT, et al. Proteomic analysis of Anaplasma phagocytophilum during infection of human myeloid cells identifies a protein that is pronouncedly upregulated on the infectious dense cored cell. Infect Immun. 2011;79:4696–4707. doi: 10.1128/IAI.05658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta ES, Parodi AJ. Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- Truchan HK, VieBrock L, Cockburn CL, Ojogun N, Griffin BP, Wijesinghe DS, et al. Anaplasma phagocytophilum Rab10-dependent parasitism of the trans-Golgi network is critical for completion of the infection cycle. Cell Microbiol. 2016;18:260–281. doi: 10.1111/cmi.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urra H, Dufey E, Lisbona F, Rojas-Rivera D, Hetz C. When ER stress reaches a dead end. Biochim Biophys Acta. 2013;1833:3507–3517. doi: 10.1016/j.bbamcr.2013.07.024. [DOI] [PubMed] [Google Scholar]
- VieBrock L, Evans SM, Beyer AR, Larson CL, Beare PA, Ge H, et al. Orientia tsutsugamushi ankyrin repeat-containing protein family members are Type 1 secretion system substrates that traffic to the host cell endoplasmic reticulum. Front Cell Infect Microbiol. 2014;4:186. doi: 10.3389/fcimb.2014.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth DE. ThANKs for the repeat: Intracellular pathogens exploit a common eukaryotic domain. Cell Logist. 2011;1:128–132. doi: 10.4161/cl.1.4.18738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, et al. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol. 2011;193:1493–1503. doi: 10.1128/JB.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt G, Parola P. Scrub typhus and tropical rickettsioses. Curr Opin Infect Dis. 2003;16:429–436. doi: 10.1097/00001432-200310000-00009. [DOI] [PubMed] [Google Scholar]
- Weitzel T, Dittrich S, Lopez J, Phuklia W, Martinez-Valdebenito C, Velasquez K, et al. Endemic Scrub Typhus in South America. N Engl J Med. 2016;375:954–961. doi: 10.1056/NEJMoa1603657. [DOI] [PubMed] [Google Scholar]
- Yang L, Zhao Z, Li B, Liu Y, Feng Y. Electron-microscopic observation of mouse spleen tissue infected with Orientia tsutsugamushi isolated from Shandong, China. Journal of electron microscopy. 2008;57:169–174. doi: 10.1093/jmicro/dfn017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Alignment of Ank9 homologs from seven O. tsutsugamushi strains. Full-length amino acid sequences of Ank9 homologs are aligned beneath the consensus sequence. A dash (−) indicates sequence identity with the Ank9 consensus, while a letter denotes variation. Underlined regions correspond to the seven ankyrin repeats of Ikeda Ank9. Residues in bold text correspond to the Ikeda Ank9 F-box domain. Numbers above the alignment denote amino acid positions.


