Abstract
Infections of dogs with virulent strains of Babesia canis are characterized by rapid onset and high mortality, comparable to complicated human malaria. As in other apicomplexan parasites, most Babesia virulence factors responsible for survival and pathogenicity are secreted to the host cell surface and beyond where they remodel and biochemically modify the infected cell interacting with host proteins in a very specific manner. Here, we investigated factors secreted by B. canis during acute infections in dogs and report on in silico predictions and experimental analysis of the parasite’s exportome. As a backdrop, we generated a fully annotated B. canis genome sequence of a virulent Hungarian field isolate (strain BcH-CHIPZ) underpinned by extensive genome-wide RNA-seq analysis. We find evidence for conserved factors in apicomplexan hemoparasites involved in immune-evasion (e.g. VESA-protein family), proteins secreted across the iRBC membrane into the host bloodstream (e.g. SA- and Bc28 protein families), potential moonlighting proteins (e.g. profilin and histones), and uncharacterized antigens present during acute crisis in dogs. The combined data provides a first predicted and partially validated set of potential virulence factors exported during fatal infections, which can be exploited for urgently needed innovative intervention strategies aimed at facilitating diagnosis and management of canine babesiosis.
Introduction
Apicomplexan parasites have evolved a plethora of targeted interactions with their host organisms and cellular machinery to elicit modification of gene expression and cellular functions. These modifications are critical for invasion and for establishment of intracellular niches for parasite replication, differentiation, and persistence, by securing nutrient import and mediating immune evasion1. As a result, host-parasite relationships in intracellular Apicomplexa can be viewed as co-adaptations driven by co-evolution of parasites with their hosts2. Most importantly, this complex interplay is directly linked to pathogenicity and virulence which characterize symptomatic infections with these protozoan parasites3–8.
Three genera of this phylum represent a triad of phylogenetically related hemoprotozoa: Plasmodium (human and animal malaria), Theileria (theileriosis in livestock), and Babesia (severe disease and death in cattle, horses, dogs, with some zoonotic potential). They are the causative agents of important human and/or animal diseases with high morbidity and mortality, responsible for significant economic burdens and healthcare challenges worldwide9. The clinically relevant part of the complex, heteroxenous Babesia life cycle takes place during asexual proliferation of parasites in red blood cells (RBC) of the mammalian intermediate host. This represents a distinct specialization of Babesia for growth in RBC, i.e. cells which lack organelles, as well as endocytic and exocytic membrane transport machinery10, 11.
Canine babesiosis is an emerging tick-borne disease of dogs with global distribution caused by morphologically distinctive Babesia species12–14. The geographical distribution of the disease caused by B. canis, the species responsible for a virulent form of canine babesiosis in Europe and Western Asia, appears to extend, as demonstrated by an increasing number of cases in areas outside of the established endemic regions15. The clinical spectrum of B. canis babesiosis is broad, ranging from apparently silent, mild disease to fulminant and often fatal presentations14. Indeed, in many respects canine babesiosis is highly comparable to clinical human malaria16, 17.
Analysis of correlated genomic, transcriptomic and proteomic data lays out the molecular underpinning for investigating the role of exported factors in disease manifestation and progression. However, experimental investigation of B. canis biology is limited by the current lack of cultivation techniques for propagation of this parasite in vitro. Here, we implemented short term in vitro cultivation of infected RBC in combination with whole genome sequencing, annotation, and comparative genomic characterization of a virulent Hungarian B. canis field isolate. By correlating RNA-seq and protein mass spectrometry data, we provide the first account of factors exported by B. canis blood stages with potential links to host-pathogen interaction and acute virulence.
Results
Genome sequence of a virulent Babesia canis strain
Until recently, only a few genes from canine Babesia species have been characterized and global genomic data were not available. To provide the molecular underpinning for targeted approaches to diagnosis, vaccination and clinical management of canine babesiosis we undertook complete sequencing, assembly, annotation and characterization of the genome of a virulent B. canis strain, designated BcH-CHIPZ. The BcH-CHIPZ isolate originated from the blood of a Swiss dog that was infected, presumably through the bite of an infected tick, during a trip to Hungary. The isolate was characterized by genotyping and identified as double B-type by restriction fragment length polymorphism (RFLP) analysis in the Bc28-multigene family and the 18S rRNA gene18. We implemented a next-generation high throughput approach (PacBio® SMRT) for sequencing BcH-CHIPZ genomic DNA prepared after enrichment of infected RBCs (iRBCs). Assembly of the parasite-specific sequence reads after elimination of all reads from host DNA and gap-filling yielded 43 high-quality scaffolds at 55-fold average coverage representing a total of 7′002′180 bases with a median weighted contig size (N50) of 185 kb. Hence, the size of the B. canis genome lies between the phylogenetically related B. microti (6.5 Mb) and the B. bovis (8.2 Mb) genomes19, 20. The main features of the annotated B. canis genome and a comparison with other apicomplexan genomes are presented in Table 1. The quality of this assembly, based on the presence of core eukaryotic genes21, 22, indicates the B. canis genome sequence to be 92.6% complete as compared with the genome of the T. gondii ME49 type II reference strain23 and consistent with values of other Babesia genome sequences, which lie between 91.7% and 95.6% (Supplementary Document S1). A robust annotation of species-specific gene models was based on the analysis of orthologues as well as on synteny in related species (described below). Furthermore, whole genome mapping approach allowed contiguous alignment of the B. canis scaffolds with the 4 B. bovis chromosomes. This indicated a high degree of synteny with B. bovis chromosomes (Fig. 1).
Table 1.
Genome characteristics of related apicomplexan hemoprotozoa (more information is provided in Supplementary Document S2).
| Features | Species | ||||
|---|---|---|---|---|---|
| B. canis [BcH-CHIPZ] | B. bovis [T2Bo] | B. microti [RI] | Th. annulata [Ankara] | P. falciparum [3D7] | |
| Genome size (Mb) | 7.0 | 8.2 | 6.5 | 8.4 | 23.3 |
| # of chromosomes | 4 | 4 | 4 | 4 | 14 |
| # of scaffolds | 43 | 12 | 3 | 7 | 14 |
| G + C content (%) | 45.8 | 41.5 | 36 | 32.5 | 19.4 |
| Protein coding genes | 3467 | 3706 | 3513 | 4082 | 5383 |
| Mean gene size (bp)1 | 1044 | 1503 | 1327 | 1602 | 2292 |
| % of coding regions | 51.7 | 68 | 73 | 73 | 53 |
| Gene density2 | 2020 | 2194 | 1816 | 2199 | 4374 |
1Excluding introns and UTRs; 2Genome size/number of protein coding genes.
Figure 1.
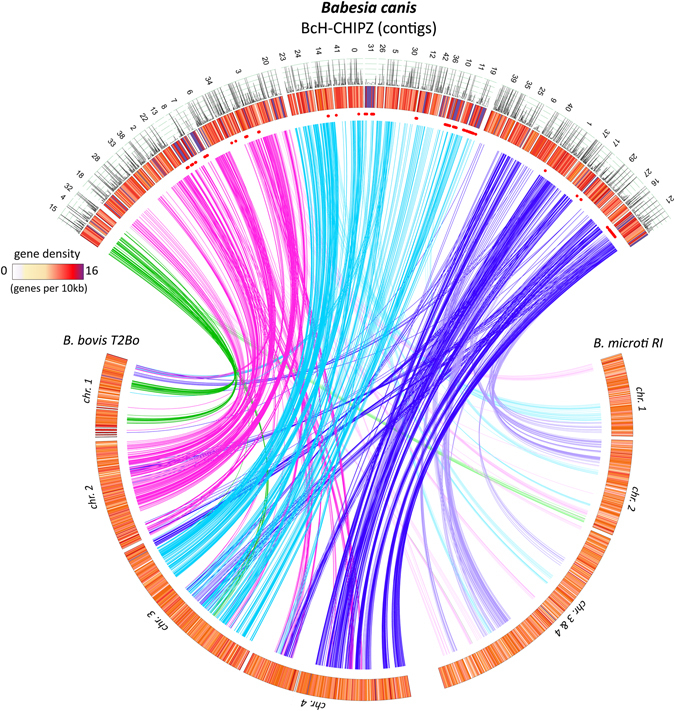
Genome characteristics of Babesia species. Assembled B. canis contigs (top arc) were arranged based on synteny to related Babesia species. The predicted arrangement of B. canis contigs on four chromosomes is indicated by colours of the lines linking conserved genes. Blood stage mRNA expression data for B. canis genes is included (line plot); gene density is colour coded and represented as number of genes per 10 kb (heatmap). Chromosomal distribution of B. canis VESA gene models is indicated with red dots.
Genome-wide comparison of B. canis gene models with those of other Apicomplexa reveals species-specific genes
The B. canis gene models were grouped into orthologues in four representative apicomplexan parasites B. bovis, Th. annulata, P. falciparum, and T. gondii and the fully sequenced Babesia species B. bovis, B. bigemina, and B. microti using a Markov Cluster algorithm in OrthoMCL (Fig. 2A,B). The four Babesia species analysed here share a core set of 1216 pan-Babesia conserved orthologous groups (COG) (Supplementary Data S1). However, bovine and rodent Babesia genomes also share 1030 COGs which are not represented in the B. canis genome (Supplementary Data S2). The former and the pan-Babesia COGs together contain a high percentage of clusters with hypothetical proteins (46.9% and 38.1%, respectively), which is not surprising given the overall high proportions of proteins annotated as “hypothetical” (61.8% in B. bigemina, 49.9% in B. bovis, and 50.2% in B. canis). However, there was no specific enrichment with respect to protein domains and functions between the two groups. Functional classification of the 95 clusters unique to B. canis highlights potential species-specific innovations comprised of proteins annotated as “hypothetical” proteins (55.8%), surface proteins (proteins with VESA1-domains and merozoite surface antigen; 26.4%), a number of specified secreted proteins (9%), and various other proteins (8.6%) (Supplementary data S3). Analysis of the bovine Babesia shared orthologues revealed hypothetical proteins in 76.2% of the clusters analysed and predicted membrane proteins in 8.2% of the gene models. Prima facie, we interpret the diverged set of annotated genes in the different Babesia species as a reflection of the narrow and highly specific host range and a high degree of specialization for their respective vertebrate- and arthropod niches. This is based on the assumption that the diversity of encoded proteins is inversely correlated with the degree of specialization for different environments, niches, and lifestyles of a parasite, thus reflecting lineage-specific adaptations24, 25. Nevertheless, with 347 genes/Mb, the B. canis genome has a similar gene density as the B. bovis (372 genes/Mb) and the B. microti (354 genes/Mb) genomes, albeit some regions appear to have higher density than others (Fig. 1).
Figure 2.
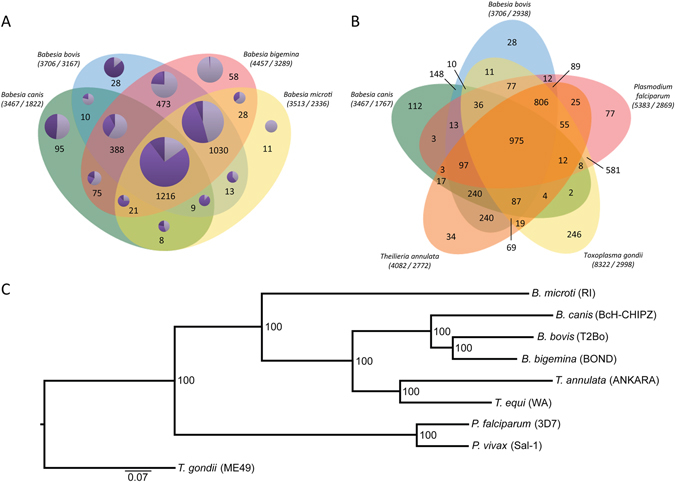
Comparative analyses of the Babesia canis BcH-CHIPZ genome. (A) Comparison of orthologous clusters in four Babesia species and (B) representative Apicomplexa species. The number of orthologous groups is indicated in the intersections. The total number of gene-models and clusters for each species is shown in brackets. Orthologous gene clusters in (A) are divided into three categories (pie chart colour code:  multigene families;
multigene families;  genes with annotations;
genes with annotations;  hypothetical genes). (C) Maximum likelihood phylogeny showing evolutionary relationship between selected Babesia, Theileria and Plasmodium species based on 975 shared orthologues in Apicomplexan species. The scale is in substitutions per site.
hypothetical genes). (C) Maximum likelihood phylogeny showing evolutionary relationship between selected Babesia, Theileria and Plasmodium species based on 975 shared orthologues in Apicomplexan species. The scale is in substitutions per site.
Despite significant species-specific features, analysis of orthologues revealed a considerable degree of synteny between the genomes of B. canis and B. bovis compared with the genome of the rodent B. microti (Fig. 1). This is also reflected in the phylogenetic analysis based on a core set of 975 shared genes in representative apicomplexan parasites (Fig. 2B,C).
Prediction of the B. canis exportome and analysis of mRNA expression levels
Exported proteins of apicomplexan parasites include virulence factors, i.e. parasite-specific proteins which interfere directly with host cell functions. As mature RBC lack de novo protein synthesis or trafficking pathways, membrane transport machinery for targeting these proteins via the cytoplasm to the iRBC surface and beyond are provided by the parasite. These parasite-induced cytoplasmic alterations of iRBC are critical for its development and directly linked to the severity of babesiosis26. As a first step to characterize parasite-secreted factors and surface proteins, we generated a predicted B. canis exportome based on the annotated BcH-CHIPZ genome. We used data mining tools to search for genes coding for predicted parasite surface- or iRBC-targeted factors. The criteria and prioritization for this selection were based on the presence of a canonical hydrophobic N-terminal signal peptide (cSP) sequence, an alternative non-classical secretion pathway (a/ncSP) signal, predicted transmembrane (TM) domain(s), a GPI anchor signal, or previously described domains of conserved apicomplexan secretory proteins and protein families (Supplementary Fig. S1; Supplementary Data S4). By this approach we generated a curated and parsed list of 509 genes coding for proteins that are predicted to be exported to the parasite surface and/or to the host, which corresponds to 14.6% of all B. canis gene models. This proportion is very similar to the 528 gene models (14%) representing the predicted B. bovis secretome27.
All Apicomplexa express members of gene families that are exported to the surface of invasive stages or the plasma membrane of iRBC where they interact with the host immune system28. Members of the Babesia variant erythrocyte surface antigen (VESA) family on the surface of iRBCs29 comprise the largest protein family in B. canis (103 gene models) in the predicted exportome (Fig. 3A). Most VESA genes cluster together on few contigs, unlike other gene families of the predicted exportome which are distributed more randomly (Fig. 1). Furthermore, VESA genes are in regions of the genome with high gene density, suggesting frequent gene duplication events consistent with a driving selection pressure on these proteins associated with immune-evasion. B. canis merozoite surface antigens (MSA; 2 gene models in B. canis) as well as Bc28 family members (20 gene models) are highly abundant on the merozoite surface but are also shed and interact with the iRBC membrane and the host’s immune system30, 31. Currently, two members of the Bc28 gene family are characterized as major merozoite surface antigens playing a critical function in the interaction of merozoites with RBC30, 32. The second-largest family of exported proteins comprises secreted DnaJ (Hsp40) chaperones (26 members). P. falciparum DnaJ proteins have demonstrated roles in remodelling of iRBCs as well as in pathogenesis33, 34. The so-called “secreted antigens” (SA-1 and SA-3, 24 members) comprise yet another family of genes coding for exported proteins. Some SA genes have been identified in other Babesia species35–38. pBLAST analysis showed that the B. canis SAs are highly homologous (21 of 24 proteins with e-values < 10E-5) to those of B. gibsoni, which also propagates in dogs, and to a lesser extent to SAs of B. bigemina (3 of 24 proteins with e-values < 10E-5), which infects bovines. This supports the idea that host-specific factors drive expansion and diversification of these orthologous gene families. Thus, this gene family appears to be expanded in Babesia species infecting canines. In contrast, genes coding for smORF and spherical body proteins (SBP) appear to be specific to B. bovis and are without orthologues in the B. canis genome27, 39. Accordingly, these protein families are also only represented in the bovine specific COGs. Proteins of the “secreted antigen family”, as the name indicates, are secreted beyond the iRBC and circulate in the blood of infected dogs but are otherwise not well characterized31. The predicted B. canis exportome also includes a group of 23 protein kinases. Secreted protein kinases and pseudokinases are known virulence factors in toxoplasmosis40–43. Kinases are also secreted into iRBCs in Plasmodium, where they are responsible inter alia for remodelling the host cell44, 45. However, functional data related to secreted kinases in Babesia are not available. Nevertheless, kinase inhibitors have been identified as potential novel drugs for the treatment of babesiosis in cattle46, 47.
Figure 3.
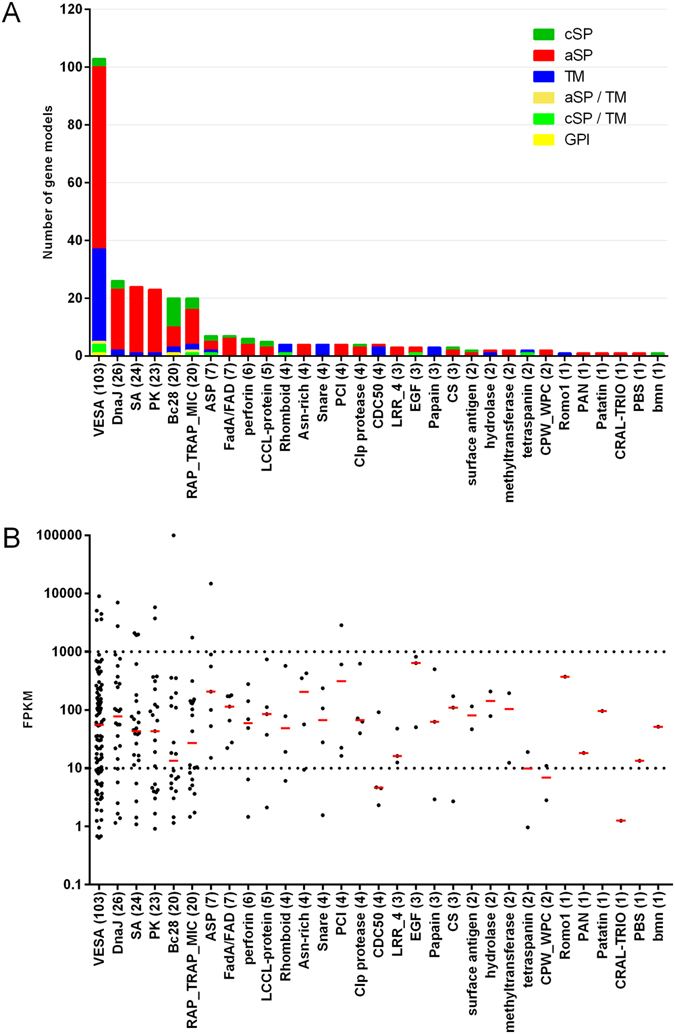
Exported protein families in Babesia canis. (A) In silico predicted exported protein families. Bar colours indicate the predicted route of secretion and/or type of membrane anchoring. cSP: canonical signal peptide; aSP: alternative (non-classical) secretory pathway; TM: transmembrane domain; GPI: predicted glycophosphatidylinositol anchor. (B) Transcript abundances of exported proteins. Each dot corresponds to the transcript abundance (FPKM) for each gene in the family during acute crisis in a splenectomised dog. Red bars indicate the median FPKM values in the presented protein family. FPKM: Fragments Per Kilobase of exon model per Million mapped reads.
To refine the predicted host-targeted B. canis exportome dataset we analysed the transcript levels of all 509 selected gene models in parasites harvested from a splenectomised dog. Evidence from a genome-wide RNAseq analysis showed transcription of 71.5% of the 509 exportome gene models (FPKM > 10; n = 364) (Fig. 3B). Remarkably, only a few members each of the five most abundant exported protein family genes (VESA-, DnaJ-, SA-, PK-, and Bc28- multigene families) with potential roles in antigenic variation and immune evasion were expressed at high levels (FPKM > 1000). The particular expression profile of the five gene families in this snapshot might be due to selection of a few specific phenotypes in the population at the peak of pathogenicity, but it should also be considered that the collected blood stage parasites do not represent a synchronously developing population. Nevertheless, this finding merits further investigation, in particular in the light of previous observations indicating that pathogenicity was associated with expression of a specific subset of VESA- and other exported genes in B. bovis infection26.
Further in silico characterization of the predicted B. canis exportome revealed a significant number of proteins (210/509) lacking a canonical hydrophobic signal sequence for co-translational insertion into the lumen of the endoplasmic reticulum, which is suggestive of non-classical secretion. This export pathway is generally known as ‘leaderless protein secretion’48, 49 and was described recently also in P. falciparum 50–52. We screened the B. canis exportome for additional motifs associated with protein trafficking into the host cell and beyond. First, we searched for matches to Plasmodium PEXEL export elements (RxLxE/Q/D) for trafficking beyond the parasitophorous vacuolar membrane (PVM)53, 54, which have been also identified in T. gondii and C. parvum 27, 55. Although Babesia builds a PVM during invasion of RBC it disappears within minutes and the parasite resides directly in the host cell cytoplasm56. A comprehensive comparative in silico analysis of protein export across 10 Apicomplexa species revealed PEXEL-like motifs (PLM) in B. bovis (but not in T. parva) with a role in retention of these proteins in so called spherical bodies and release in a cell-cycle dependent manner27. We found hits for the B. bovis PEXEL-like motifs within the first 100 N-terminal amino acids in 64.3% of cSP-containing proteins and only in 7.6% of predicted alternatively secreted B. canis proteins (Supplementary Data S4). Using a hidden Markov model to identify alternative motifs by de novo pattern discovery did not yield any additional hits.
Expression levels of stage-specific genes identifies asexual and sexual Babesia blood stage parasites at acute crisis
We collected biological material from three experimentally infected dogs, which all showed very similar rapid disease progression consistent with fatal babesiosis (described in ref. 57). The first mild clinical signs (e.g. lethargy) preceded acute crisis with early symptoms of septic shock by a maximum of 27 hours (Supplementary Fig. S3). In spite of the fulminant disease manifestation, a hallmark of canine babesiosis is a remarkably low parasitemia12, 58, 59 with typically <2% infected erythrocytes observed in the peripheral blood. The asexual erythrocytic stage of the parasite is entirely responsible for the morbidity and mortality associated with canine babesiosis. Morphological characterization of blood stage parasites revealed pleiomorphic forms in Giemsa-stained blood smears derived from a critically ill dog (Fig. 4A). Small and large ring stages were the most frequently detected forms, while only few pear-shaped forms could be identified. To characterize the parasite stages in iRBCs from experimentally infected animals on a molecular level in more detail, we used the high-throughput RNA-seq data to identify orthologous gene models with stage-specific expression in Plasmodium spp. We intersected the list of B. canis gene models with evidence for expression with a list of validated Plasmodium spp. stage-specifically expressed genes (e-value threshold of 10E-3) (Supplementary Data S5). We found evidence for RNA-seq reads (threshold FPKM > 10) mapping to 22 of 29 conserved merozoite, and 4 of 7 early gametocyte genes in a blood sample collected from a splenectomised dog (Fig. 4B). The higher mRNA abundance of merozoite- (proliferative stage) compared with early gametocyte-specific orthologues (sexual stage) during severe disease was consistent with the microscopy data and suggested only marginal numbers of circulating sexual stages in this peripheral blood sample. Nevertheless, some evidence for early gametocyte stages was found, which could indicate that acute clinical crisis in infected dogs might trigger development of sexual stages. For example in Plasmodium, gametocyte formation occurs in bone marrow and aggravates clinical symptoms and disease outcome60, 61. A potential bone marrow tropism could explain many typical clinical signs in canine babesiosis, e.g. leukopenia and thrombocytopenia, associated with poor outcome in acute infections57. B. canis orthologues of the 18 identified invertebrate stage-specific proteins (flagellar “late” gametocyte-, ookinete- and sporozoite genes) described for Plasmodium had FPKM values between 0 and 10.8, indicating that no or only insignificant amounts of these mRNAs were present in the sampled parasite population. Based on the degree of orthologue conservation and the microscopy data we interpret this as paucity of late gametocyte stages in the peripheral blood (Supplementary Data S5).
Figure 4.
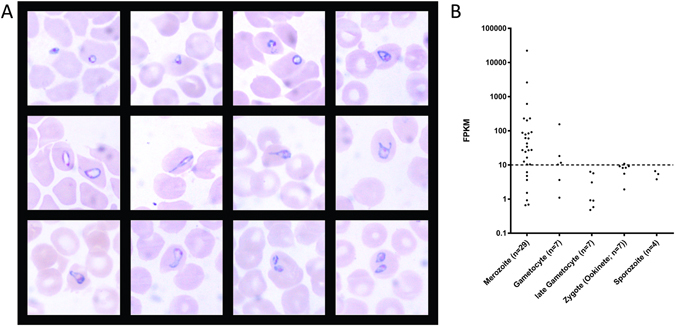
Specification of B. canis blood stages. (A) Representative Giemsa-stained blood smears from an experimentally infected dog with severe shock-like clinical signs showing typical pleiomorphic morphological appearance of B. canis parasites (parasitemia of 1.5% infected erythrocytes). (B) Transcriptome analysis of B. canis homologs of validated Plasmodium spp. stage-specific genes in a blood sample collected from an experimentally infected splenectomised dog with clinical signs consistent with acute canine babesiosis.
Shotgun proteomics and transcriptomics identify potential B. canis-specific secreted virulence factors during severe disease
Proteins involved in parasite-host interactions at acute crisis were identified by a comparative shotgun approach from B. canis infected and healthy host blood samples. Mass spectrometry datasets were generated from secreted soluble factors collected from pooled short-term cultures from three experimentally infected dogs at acute crisis and RBC cultures derived from healthy blood donors, as well as from RBC membrane fractions from experimentally infected animals and non-infected controls. We identified a set of abundant parasite-specific proteins in these fractions after filtering the data with the non-infected control sets. Based on their abundance in the iRBC and/or culture supernatant the identified secreted membrane-bound and soluble parasite proteins were considered candidate factors involved in host-pathogen interaction (Table 2; Supplementary Fig. S2).
Table 2.
List Babesia canis specific candidates from culture supernatant derived and membrane-bound fractions.
| Identification | GO-description1 | SP | TM | GPI | AS | Apicomplexan orthologue2 | Transcriptome evidence (FPKM) |
|---|---|---|---|---|---|---|---|
| Specific B. canis secreted soluble proteins: | |||||||
| BcH000240 | Hypothetical protein, conserved | 1 | 6 | 0 | 0 | no (0.7) | |
| BcH000493 | Hypothetical protein [VESA] | 0 | 0 | 0 | 1 | VESA (B. bovis BBOVII004140) | yes (673.5) |
| BcH001113 | Hypothetical protein, conserved | 0 | 0 | 0 | 0 | yes (544) | |
| BcH002928 | hypothetical protein, conserved | 1 | 0 | 0 | 0 | yes (64.2) | |
| BcH003445 | Secreted antigen 3 | 0 | 0 | 0 | 1 | Secreted antigen 3 (B. gibsoni BAH70325) | yes (26.4) |
| BcH000300 | Merozoite surface protein (Bc 28.2) | 1 | 0 | 0 | 0 | B. canis specific | no (4.5) |
| BcH002050 | SET domain-containing protein | 0 | 0 | 0 | 0 | SET- and MYND domain containing protein, putative (B. bigemina BBBOND0302680) | yes (147.6) |
| BcH002986 | hypothetical protein | 0 | 0 | 0 | 0 | yes (60.1) | |
| BcH003048 | Profilin | 0 | 0 | 0 | 0 | Polypeptide of profiling (B. bigemina BBBOND_0104160) | yes (100.6) |
| BcH002141 | Snare protein | 0 | 1 | 0 | 0 | Snare protein, putative (B. bigemina BBOND0107960) | yes (236.6) |
| BcH000974 | Hypothetical protein, conserved | 1 | 0 | 0 | 0 | VESA (B. bovis BBOV_III006920) | no (4.1) |
| BcH000821 | Spectrin repeat superfamily extracellular matrix binding | 0 | 1 | 0 | 0 | VESA (B. bovis BBOV_II004140) | yes (123.2) |
| BcH000175 | Hypothetical protein, conserved | 0 | 0 | 0 | 0 | yes (57.9) | |
| BcH002958 | Hypothetical protein [membrane protein] | 0 | 0 | 0 | 1 | Membrane protein, putative (B. bigemina BBBOND_0105580) | yes (56.4) |
| Specific B. canis host-RBC membrane presented proteins: | |||||||
| BcH000821 | Spectrin repeat superfamily extracellular matrix binding | 0 | 1 | 0 | 0 | VESA (B. bovis BBOV_II004140) | yes (123.2) |
| BcH000300 | Merozoite surface protein (Bc 28.2) | 1 | 0 | 0 | 0 | B. canis specific | no (4.5) |
| BcH000299 | Merozoite surface protein (Bc 28.1) | 1 | 1 | 1 | 0 | B. canis specific | yes (18.3) |
| BcH002699 | Histone h2a | 0 | 0 | 0 | 0 | 41-2-protein antigen precursor, putative (B. bigemina BBBOND_0103700) | yes (31.9) |
| BcH001932 | Hypothetical protein, conserved | 0 | 0 | 0 | 0 | (yes 12.5) | |
| BcH002268 | Hypothetical protein, conserved | 0 | 0 | 0 | 0 | yes (111.9) | |
| BcH001655 | VESA | 0 | 0 | 0 | 1 | VESA (B. bovis BBOV_IV002850) | yes (884.1) |
| BcH001448 | Hypothetical protein, conserved | 0 | 0 | 0 | 0 | no (0.9) | |
| BcH003376 | Secreted antigen 1 | 0 | 1 | 1 | 1 | Secreted antigen 1(B. gibsoni ALE14558) | yes (142.5) |
| BcH000391 | Hypothetical protein, conserved | 1 | 0 | 0 | 0 | yes (331.2) | |
| BcH000895 | Hypothetical protein, conserved | 0 | 0 | 0 | 0 | yes (26.4) | |
GO: gene ontology; SP: signal peptide; TM: transmembrane domain; GPI: AS: alternative secretion: GPI: glycophosphatidylinositol: VESA: variant erythrocyte surface antigen.
1Automated annotation by Blast2GO; [if no Blast2GO annotation could be assigned, best pBlast hit was provided in brackets].
2Other than hypothetical.
Not surprisingly, considering the limited understanding of apicomplexan pathogenicity determinants, the resulting B. canis dataset of exported proteins comprises 47.8% (11/23) proteins annotated as “hypothetical”. In addition, the following hits to functionally annotated proteins were identified: factors on cell surface membranes (from the parasite or the iRBC) which include 2 variant erythrocyte surface antigens (VESA) located on the surface of iRBC, a Babesia spp. membrane protein (homologous to B. bovis, B. microti, B. bigemina, Theileria annulata, T. equi, T. orientalis, T. parva, and Cytauxzoon felis), a spectrin repeat superfamily extracellular matrix binding protein with homology to B. bovis VESA, and 2 merozoite surface antigens of the Bc28 gene family. These proteins where shown to be involved in survival and virulence of several hemoprotozoa species10, 18, 19, 30, 62, 63. The dataset comprises SA1 and SA3, belonging to the above described third-largest exported family of proteins (secreted antigen family) suggesting a direct role in host-parasite interaction. The B. canis dataset also contains profilin, a secreted virulence factor described in P. falciparum and T. gondii with important roles in the process of crossing biological barriers during host cell invasion and egress64, 65, a SET-domain containing protein with putative function in lateral gene transfer in apicomplexans66, and a SNARE protein mediating membrane fusion of secretory vesicles with the plasma membrane67, 68. Furthermore, a secreted histone protein (classified as DNA-binding protein) was detected. Histone proteins were described to have moonlighting functions outside of the nucleus in P. falciparum, specifically at the interface between parasite and erythrocyte cytoplasm, i.e. the parasitophorous vacuole membrane69.
All hits in the proteomic dataset for which GO descriptions or blast homologies could be assigned were further validated using transcriptome evidence (threshold FPKM value > 10). Only Bc28.2, a known secreted B. canis-specific protein30 with a measured FPKM of 4.5, fell below this threshold. However, the correlation between transcript- and protein abundance is not linear and there is a complex relationship leading to notable discrepancies due to a range of factors such as protein- and mRNA turnover70. Taken together with additional evidence from the literature, these proteins are good candidates for factors that are presented to the host on the parasite or iRBC surface, or released in to the bloodstream and potentially involved in parasite persistence and virulence.
Discussion
Secreted soluble and membrane-targeted parasite proteins have important roles at the host-parasite interface and are frequently virulence and pathogenesis factors. Systematic investigation of these interactions in infections with potentially fatal blood-borne parasites, such as Babesia canis, which modify the host cell, its microenvironment, as well as host physiology in multiple ways, is a challenge. Moreover, these questions cannot be addressed with biological material from regular patients from an animal clinic and the parasite is difficult to cultivate. Hence, in this study, we used experimental infections of dogs in a controlled setting to gather data on disease progression, clinical parameters, and parasite development. We report the first genome and gene expression data of the apicomplexan B. canis with a focus on exported factors secreted during acute infection. Acute B. canis infection with fulminant disease progression is associated with high mortality and characterized by an excessive inflammatory response caused by protozoal sepsis. This is caused by dysregulation of pro- and anti-inflammatory mechanisms during the acute phase of the disease, resulting in shock and end-organ failure71–73 (Supplementary Fig. S3). Interestingly, fatal B. canis infections (natural and experimentally induced) are invariably associated with at most moderate non-haemolytic anaemia and very low parasitemia12, 57, 59. This strongly suggests that potent secreted parasite factors are responsible for the severe pathogenicity of canine babesiosis. Hence, these as yet uncharacterized, secreted virulence factors are prime targets for the development of vaccines but also diagnostic reagents as a prime goal in clinical research.
Several major secreted factors inducing acute pathogenicity or persistence of hemoprotozoan parasites have been described in P. falciparum and B. bovis 74. Following RBC invasion, B. bovis exports VESA proteins to the surface of the host cell modifying the iRBC membrane to induce cytoadherence of iRBC at capillary and postcapillary venous endothelia75–77. VESA expression is subject to rapid antigenic variation defined as the serial clonal replacement of the expressed VESA gene in daughter merozoites before invasion of a new RBC. This gives rise to antigenically distinct parasite populations, which complicates mounting of an effective host immune response. Antigenic variation, i.e. exclusive sequential expression of surface antigen family members is likely a sub-phylum-specific immune-evasion strategy in hematozoa documented in several Babesia and Plasmodium species including B. rodhaini, B. microti, B. bigemina, P. falciparum and P. vivax 8, 78. Not surprisingly, the largest exported protein family in B. canis consists of the variant erythrocyte surface antigen (VESA) proteins. Indeed, a recent study using comparative transcriptomics and proteomics of attenuated and virulent B. bovis strains showed a significantly increased diversity of upregulated VESA genes in virulent strains26. In the present study, we detected a high number (74/103) of transcribed VESA genes as well as their products in the B. canis secreted proteome. Hence, adherence of B. canis iRBC to endothelial cells is one explanation for the apparent discrepancy between the low parasitemia observed in peripheral blood samples and the severity of the disease, consistent with sequestration of parasites and removal from the peripheral circulation. Correspondingly, using parasites isolated from a peripheral blood sample for the transcriptomic and proteomic studies might introduce a bias with respect to the detection of important pathogenicity factors. Direct comparison of proteomic and transcriptomic data has to be interpreted with caution, since the RNA-seq data was generated from parasites isolated from a splenectomized animal. The peripheral blood of splenectomized individuals would present a dominance of a sub-population of mature asexual and immature sexual parasites in iRBCs that failed to sequester, because they are not expressing appropriate ligands for cytoadherence, as repeatedly demonstrated in human P. falciparum 79, 80. Modulation of the adhesive properties of iRBCs by the spleen has been discussed, although no underlying mechanism has been identified81, 82. For example, in splenectomized rhesus monkeys infected with Plasmodium knowlesi a vast reduction in the expression of SICAvar genes, corresponding to their immunovariant adhesins, was demonstrated83. In contrast, P. vivax which preferentially invades reticulocytes passage all asexual blood stages through the spleen and show spleen-specific cytoadherence important for the development of a chronic disease by encoding equivalent vir-genes8. Nevertheless, the responsible Plasmodium var/vir- and the Babesia ves genes differ in structure, sequence, and in their biochemical properties74. In contrast to in P. falciparum var genes, B. bovis appears to antigenically vary its surface by a combination of segmental gene conversion events from inactive ves genes into an actively transcribed locus with in situ gene switching (clonal activation of genes)84.
Shedding of extracellular (exosome-like) vesicles (EVs) as carriers for proteins, lipids and RNA for intra- and inter-species communication (reviewed in refs 85 and 86) was identified recently as a non-classical export pathway for a completely new class of important parasite-derived factors. In P. falciparum infections, the concentration of EVs in the blood is positively correlated with severe disease suggesting an important role in pathogenesis and immune modulation87. Strikingly, the B. canis BcH-CHIPZ genome encodes homologues of important structural proteins and enzymes frequently found in exosomes, including heat shock proteins (e.g. HSP70 and 90), tetraspanins and ALIX protein, TSG101/ESCRT protein, 14-3-3 protein, thioredoxin peroxidase, histones, and the RAB GTPases 5, 7, 11, as well as a RAB GDI85, 88, 89. Due to the mechanism for their biogenesis, exosomes often contain endosome-associated proteins, such as the SNARE proteins also detected in the B. canis culture supernatant. In P. falciparum, a SNARE protein was localized proximal to the plasma membrane where it is possibly involved in membrane trafficking events associated with the parasite’s food vacuole68. Hence, EVs in B. canis infections could be crucial vehicle carriers for the dissemination of pathogenicity factors that are linked to acute and severe disease, including coding and non-coding RNA which can interfere with host cell gene expression. In fact, secretion via EVs could account for many of the non-classically secreted proteins detected in the B. canis proteomic datasets.
We identified several proteins with moonlighting functions which have been identified as virulence factors in related Apicomplexa. For example profilin (pfn) was shown to have important roles in the process of crossing biological barriers during host cell invasion and egress, in addition to its classical role as a regulator of actin dynamics65. The structure of the P. falciparum pfn accommodates additional domains with as yet unknown functions, which are potentially involved in the parasite-host interplay64. Another interesting finding was a histone protein in the iRBC membrane fraction. In several Plasmodium species, a histone protein methylated at position L9 was detected in the PV membrane69. The conserved PVM localization in Plasmodium indicated that histones have evolved additional functions in invasion and interaction with the host. As Babesia are not contained within a PV, the potential moonlighting function of this protein will have to be further investigated. Furthermore, histones are commonly found in EVs90. We also detected a B. canis SET-domain containing protein (BcSET) in the secreted soluble fraction. SET proteins are involved in diverse mechanisms such as transcriptional regulation, enhancer function, mRNA splicing, DNA replication, and DNA damage response91. An exported SET exerting an immune-suppressive function by transcriptional silencing of host gene expression was described in the pathogen Bacillus anthraxis 92. Interestingly, deregulation of gene expression due to SET domain-containing proteins is correlated with unfavourable clinical outcomes in various forms of cancer93.
Members of the Bc28 and SA gene families are among the most prominent B. canis secreted factors, and have recently gained attention as candidates for innovative vaccine strategies and in diagnostics30, 31. Bc28 gene products are highly represented on the merozoite surface, but are also shed and can interact with the iRBC membrane30. Bc28 belong to a multigene family composed of polymorphic genes. Different B. canis strains in Europe are associated with different mortality rates and show genetic heterogeneity in Bc28.1, detectable by a PCR-RFLP test18. It has been suggested that this variable repertoire has evolved to allow the parasite to evade host immune responses, although merozoite surface proteins of Babesia spp. are thought to be the main targets of the adaptive host immune response32, 94. However, whether all members are directly involved in immune evasion and virulence, or even show stage-specific behaviour, will have to be investigated in more detail.
In the absence of reverse genetic techniques for B. canis, sequencing and annotation of a virulent B. canis strain together with the described parasite blood stage proteome and transcriptome provides a powerful underpinning for designing targeted intervention strategies such as the development of novel diagnostic tools, effective vaccines, and innovative drugs.
Methods
Parasites
Biological materials for the different experiments were collected from experimentally infected dogs. Animals were inoculated intravenously with 1 × 106 parasitized erythrocytes from a cryo-conserved field isolate, derived from a naturally infected Bernese mountain dog from Switzerland that had been exposed during a trip to Hungary. All experimentation with animals and protocols was in complete compliance with the strict Swiss animal welfare standards and regulations and approved by the responsible authorities (Veterinary Office of the Canton of Zurich; permission number 122/2012) previous to the study, including round the clock surveillance of the animals by a veterinarian. Whole blood was treated with citrate-phosphate-dextrose-adenine (12.3% CPDA-1, v/v) for anticoagulation under sterile conditions and prepared immediately after euthanasia. Blood samples for genomic and transcriptomic studies were collected from an experimentally inoculated splenectomised dog (4 years old). Splenectomy increased the final amount of parasites in the peripheral blood to approximately 4% infected erythrocytes. The biological material for the proteome analysis was collected from three experimentally infected adult beagles with a parasitemia of 1–1.75% (Supplementary Fig. S3). All samples were collected at the first signs of acute crisis detected by any clinical signs of acute shock or central nervous depression.
Genome sequencing, assembly, and annotation pipeline
Genomic DNA (gDNA) was prepared from purified and host leukocyte-depleted95 infected blood from an experimentally infected, splenectomised dog. High-throughput genome sequencing was performed on a PacBio Single Molecule Real Time (SMRT) sequencing platform96 run on 4 SMRT cells. 5 μg of input gDNA was used for library construction. Sequenced fragments were filtered to eliminate Canis familiaris reads and processed for assembly with PacBio read-alignment software BLASR97. The final draft of the genome was obtained using a two-step procedure. De novo assembly was initially performed using the HGAP2 algorithm98 and the outcome was further polished using the PacBio proprietary analysis package Quiver, a reference-guided consensus tool used as part of the SMRT analysis. The resulting draft was subjected to gap-closing and further scaffolding using PBJelly v14.1.1599. Genome annotation was performed using different sources of evidence-based prediction on the Maker2 pipeline100, on ab initio gene predictor algorithms101 with P. falciparum 3D7102, B. bovis T2Bo19, and T. annulata Ankara103 gene models as templates retrieved from EuPathDB release 26104, and on Trinity-assembled de novo transcriptome evidence105. Repetitive genomic elements were identified and masked from annotation with RepeatMasker using the full Repbase database106. Redundant transcripts potentially representing sequencing errors or genetic polymorphisms were clustered with the cd-hit software107. Non-overlapping sequences of the different evidence datasets were generated from the gff-files with BEDTools genome arithmetic software108. Functional annotation of the gene-predictions was achieved by gene ontology mapping through Blast2Go v3.1109 using an upper cut-off e-value of <0.0005.
Full methodological details are provided in the Supplementary Material.
RNA preparation and RNA-seq data analysis
Total RNA was prepared from host leukocyte-depleted B. canis blood stages using the Zymo Direct-zol RNA MiniPrep kit (Zymo Research) including an on-column DNase I (QIAGEN) digest. Paired-end, stranded sequencing of the cDNA library was performed on 1 lane of an Illumina HiSeq 2000 sequencer (Illumina Inc.). Measurement of expression level was based on fragments per kilobase of exon model per million mapped reads (FPKM), normalized by the length of the gene. A gene was considered expressed if its FPKM was at least 10. Full methodological details are provided in the Supplementary Material.
Summary of computational genome analysis
The identification of potential homologues of B. canis genes to selected apicomplexan parasites (B. bovis T2Bo19, B. bigemina BOND39, B. microti RI20, T. annulata Ankara103, P. falciparum 3D7102, and T. gondii ME4923) was carried out by protein BLAST (Blast + executables v2.2.31) of each individual protein sequence. A e-value < 0.001 cut-off was considered appropriate. For comparative genomics analysis of species-specific genes and clustering of orthologues, OrthoMCL (v2.0.9) was used110, 111. Synteny analysis was performed in MCScanX112 by the pairwise determination of syntenic regions between species based on the order of orthologues from the OrthoMCL output with default values. B. canis contigs were arranged according to the syntenic relationship to B. bovis with ‘orderchr’ execution in Circos utilities. The graphical representation was made with Circos113. Whole genome phylogeny was based on the 975 identified OrthoMCL-orthologues shared among Apicomplexan species. Each of these gene models in 4 Babesia sp. (B. canis, B. bovis, B. bigemina, and B. microti), 2 Theileria sp. (T. annulata and T. equi), 2 Plasmodium sp. (P. falciparum and P. vivax) and Toxoplasma gondii was aligned using MAFFT v7114. Highly variable sites were trimmed using trimal115. A supermatrix of the alignments was concatenated using FASconCAT116. Based on this alignment a maximum likelihood phylogenetic tree was constructed using RAxML117, 118 based on amino acid input, gamma model, LG substitution matrix and 100 initial bootstrap (bootstrap percentage support is shown along the branching nodes), whereas the tree was rooted using T. gondii as an outgroup. Prediction of the B. canis exportome was based on several in silico analysis tools, databases, and manual curation (Supplementary Fig. S1). Full methodological details are provided in the Supplementary Material.
Proteomic studies
Proteomic analyses were based on secreted soluble Babesia antigens and membrane-bound B. canis proteins. Culture supernatant was collected after 18 h of short term in vitro parasite cultures of B. canis RBC in serum-free medium prepared from anticoagulated whole blood from experimentally inoculated dogs. Similarly, blood from a healthy canine blood donor was cultured and supernatant was processed as a control. Isolation of membrane bound proteins was based on anticoagulated, leucocyte-depleted blood. iRBC were concentrated by Percoll (GE Healthcare) gradient centrifugation119 and parasites were released from the iRBC by streptolysin-O mediated lysis120 and removed by an additional Percoll gradient centrifugation step121. Membrane proteins were collected by homogenization and repeated ultracentrifugation steps, mild sonication and acetone precipitation. The same protocol was used to collect proteins from uninfected erythrocyte membranes. All samples were collected and stored at −80 °C until further use.
Protein preparations were separated on one dimensional SDS-PAGE under sterile and reducing conditions. Each stained gel was cut into 8 equal sections and in-gel trypsin digestion was carried out for subsequent mass spectrometry analysis. Liquid chromatography-MS/MS was performed on a Q-exactive mass spectrometer (Thermo Scientific) equipped with a nanoAcquity UPLC (Waters Corporation).
Following peptide data acquisition, searches were performed using the MASCOT search program against a database built on the B. canis annotated genes with a concatenated decoy database supplemented with commonly observed contaminants and the Swissprot database to increase database size, or against NCBI database for Canis lupus familiaris. Identified hits were filtered in Scaffold Viewer V4 (Proteome Software, Portland, US) based on stringent parameters to reach false discovery rates of <5%. Furthermore, all datasets were corrected for obvious host- and environmental contaminants by an additional pBLAST search of the NCBI non-redundant protein sequences database with the single peptide hits.
Data Availability
The annotated B. canis genome will be made accessible via PiroplasmaDB hosted by EuPathDB (http://piroplasmadb.org/piro/).
Electronic supplementary material
Acknowledgements
We thank Dr. Sirisha Aluri, Dr. Peter Hunziker, Andrea Patrignani, Barbara Brändle and Armin Rüdemann for their excellent technical assistance. RME was a recipient of grants from the “Forschungskredit” of the University of Zurich (grant nos 55080506 and FK-13-053).
Author Contributions
R.M.E. participated in the overall design of the study, carried out a majority of the experiments, data computing and downstream analyses and drafted the manuscript. C.R. performed RNA purification, quality assessment, and coordinated the RNA Seq. G.R. coordinated gDNA Seq, performed genome assembly and annotation, and assisted in bioinformatics approaches. P.D. conceived the study and assisted in designing and planning of the study and supervised the infection experiments. ABH conceived the study, supervised data analyses and drafted the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-03445-x
Electronic supplementary material
Accession Codes: Sequences and data have been deposited for public access on the European Nucleotide Archive (http://www.ebi.ac.uk/ena; PRJEB15861) and on the ProteomeXchange platform via PRIDE (http://www.proteomexchange.org; PXD005117).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Goldberg DE, Cowman AF. Moving in and renovating: exporting proteins from Plasmodium into host erythrocytes. Nat Rev Microbiol. 2010;8:617–621. doi: 10.1038/nrmicro2420. [DOI] [PubMed] [Google Scholar]
- 2.Morrison DA. Evolution of the Apicomplexa: where are we now? Trends Parasitol. 2009;25:375–382. doi: 10.1016/j.pt.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Bradley PJ, Sibley LD. Rhoptries: an arsenal of secreted virulence factors. Curr Opin Microbiol. 2007;10:582–587. doi: 10.1016/j.mib.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haldar K, Mohandas N. Erythrocyte remodeling by malaria parasites. Curr Opin Hematol. 2007;14:203–209. doi: 10.1097/MOH.0b013e3280f31b2d. [DOI] [PubMed] [Google Scholar]
- 5.Hutchings CL, et al. New insights into the altered adhesive and mechanical properties of red blood cells parasitized by Babesia bovis. Mol Microbiol. 2007;65:1092–1105. doi: 10.1111/j.1365-2958.2007.05850.x. [DOI] [PubMed] [Google Scholar]
- 6.Maier AG, et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lobo CA, Rodriguez M, Cursino-Santos JR. Babesia and red cell invasion. Curr Opin Hematol. 2012;19:170–175. doi: 10.1097/MOH.0b013e328352245a. [DOI] [PubMed] [Google Scholar]
- 8.Singh V, Gupta P, Pande V. Revisiting the multigene families: Plasmodium var and vir genes. J Vector Borne Dis. 2014;51:75–81. [PubMed] [Google Scholar]
- 9.Lau AO. An overview of the Babesia, Plasmodium and Theileria genomes: a comparative perspective. Mol Biochem Parasitol. 2009;164:1–8. doi: 10.1016/j.molbiopara.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Gohil S, Kats LM, Sturm A, Cooke BM. Recent insights into alteration of red blood cells by Babesia bovis: moovin’ forward. Trends Parasitol. 2010;26:591–599. doi: 10.1016/j.pt.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Spielmann T, Gilberger TW. Critical Steps in Protein Export of Plasmodium falciparum Blood Stages. Trends Parasitol. 2015;31:514–525. doi: 10.1016/j.pt.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Schetters TP, et al. Different Babesia canis isolates, different diseases. Parasitology. 1997;115:485–493. doi: 10.1017/S0031182097001686. [DOI] [PubMed] [Google Scholar]
- 13.Boozer, A. L. & Macintire, D. K. Canine babesiosis. Vet Clin North Am Small Anim Pract33, 885–904, viii (2003). [DOI] [PubMed]
- 14.Irwin PJ. Canine babesiosis: from molecular taxonomy to control. Parasit Vectors. 2009;2:S4. doi: 10.1186/1756-3305-2-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halos L, et al. Questionnaire-based survey on the distribution and incidence of canine babesiosis in countries of Western Europe. Parasite. 2014;21:13. doi: 10.1051/parasite/2014015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyers F, et al. Canine babesiosis in South Africa: more than one disease. Does this serve as a model for falciparum malaria? Ann Trop Med Parasitol. 1998;92:503–511. doi: 10.1080/00034983.1998.11813308. [DOI] [PubMed] [Google Scholar]
- 17.Schetters TP, Eling WM. Can Babesia infections be used as a model for cerebral malaria? Parasitol Today. 1999;15:492–497. doi: 10.1016/S0169-4758(99)01566-5. [DOI] [PubMed] [Google Scholar]
- 18.Carcy B, et al. Classification of Babesia canis strains in Europe based on polymorphism of the Bc28.1-gene from the Babesia canis Bc28 multigene family. Vet Parasitol. 2015;211:111–123. doi: 10.1016/j.vetpar.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 19.Brayton KA, et al. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 2007;3:1401–1413. doi: 10.1371/journal.ppat.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornillot E, et al. Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res. 2012;40:9102–9114. doi: 10.1093/nar/gks700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 22.Reid AJ, et al. Genomic analysis of the causative agents of coccidiosis in domestic chickens. Genome Res. 2014;24:1676–1685. doi: 10.1101/gr.168955.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gajria B, et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Templeton TJ, et al. The genome of Eimeria falciformis - reduction and specialization in a single host apicomplexan parasite. BMC Genomics. 2014;15:696. doi: 10.1186/1471-2164-15-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heitlinger, E., Spork, S., Lucius, R. & Dieterich, C. The genome of Eimeria falciformis-reduction and specialization in a single host apicomplexan parasite. BMC Genomics15, 696 (2014). [DOI] [PMC free article] [PubMed]
- 26.Pedroni MJ, Sondgeroth KS, Gallego-Lopez GM, Echaide I, Lau AO. Comparative transcriptome analysis of geographically distinct virulent and attenuated Babesia bovis strains reveals similar gene expression changes through attenuation. BMC Genomics. 2013;14:763. doi: 10.1186/1471-2164-14-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelle KG, et al. Shared elements of host-targeting pathways among apicomplexan parasites of differing lifestyles. Cell Microbiol. 2015;17:1618–1639. doi: 10.1111/cmi.12460. [DOI] [PubMed] [Google Scholar]
- 28.Spence PJ, et al. Vector transmission regulates immune control of Plasmodium virulence. Nature. 2013;498:228–231. doi: 10.1038/nature12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allred DR, Hines SA, Ahrens KP. Isolate-specific parasite antigens of the Babesia bovis-infected erythrocyte surface. Mol Biochem Parasitol. 1993;60:121–132. doi: 10.1016/0166-6851(93)90035-V. [DOI] [PubMed] [Google Scholar]
- 30.Yang YS, et al. Structural and functional characterization of Bc28.1, major erythrocyte-binding protein from Babesia canis merozoite surface. J Biol Chem. 2012;287:9495–9508. doi: 10.1074/jbc.M111.260745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou M, et al. Molecular identification and antigenic characterization of a merozoite surface antigen and a secreted antigen of Babesia canis (BcMSA1 and BcSA1) Parasit Vectors. 2016;9:257. doi: 10.1186/s13071-016-1518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carcy B, Precigout E, Schetters T, Gorenflot A. Genetic basis for GPI-anchor merozoite surface antigen polymorphism of Babesia and resulting antigenic diversity. Vet Parasitol. 2006;138:33–49. doi: 10.1016/j.vetpar.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 33.Rug M, Maier AG. The heat shock protein 40 family of the malaria parasite Plasmodium falciparum. IUBMB Life. 2011;63:1081–1086. doi: 10.1002/iub.525. [DOI] [PubMed] [Google Scholar]
- 34.Njunge JM, Ludewig MH, Boshoff A, Pesce ER, Blatch GL. Hsp70s and J proteins of Plasmodium parasites infecting rodents and primates: structure, function, clinical relevance, and drug targets. Curr Pharm Des. 2013;19:387–403. doi: 10.2174/138161213804143734. [DOI] [PubMed] [Google Scholar]
- 35.Jia H, et al. Identification of secreted antigen 3 from Babesia gibsoni. Clin Vaccine Immunol. 2009;16:944–948. doi: 10.1128/CVI.00087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia H, et al. Identification of a novel gene encoding a secreted antigen 1 of Babesia gibsoni and evaluation of its use in serodiagnosis. Am J Trop Med Hyg. 2006;75:843–850. [PubMed] [Google Scholar]
- 37.Luo Y, et al. Identification and characterization of a novel secreted antigen 1 of Babesia microti and evaluation of its potential use in enzyme-linked immunosorbent assay and immunochromatographic test. Parasitol Int. 2011;60:119–125. doi: 10.1016/j.parint.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Sevinc F, Cao S, Xuan X, Sevinc M, Ceylan O. Identification and expression of Babesia ovis secreted antigen 1 and evaluation of its diagnostic potential in an enzyme-linked immunosorbent assay. J Clin Microbiol. 2015;53:1531–1536. doi: 10.1128/JCM.03219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson AP, et al. The evolutionary dynamics of variant antigen genes in Babesia reveal a history of genomic innovation underlying host-parasite interaction. Nucleic Acids Res. 2014;42:7113–7131. doi: 10.1093/nar/gku322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saeij JP, et al. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature. 2007;445:324–327. doi: 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinfeldt T, et al. Phosphorylation of mouse immunity-related GTPase (IRG) resistance proteins is an evasion strategy for virulent Toxoplasma gondii. PLoS Biol. 2010;8:e1000576. doi: 10.1371/journal.pbio.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fleckenstein MC, et al. A Toxoplasma gondii pseudokinase inhibits host IRG resistance proteins. PLoS Biol. 2012;10:e1001358. doi: 10.1371/journal.pbio.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Etheridge RD, et al. The Toxoplasma pseudokinase ROP5 forms complexes with ROP18 and ROP17 kinases that synergize to control acute virulence in mice. Cell Host Microbe. 2014;15:537–550. doi: 10.1016/j.chom.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saeij JP, et al. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science. 2006;314:1780–1783. doi: 10.1126/science.1133690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim DC, Cooke BM, Doerig C, Saeij JP. Toxoplasma and Plasmodium protein kinases: roles in invasion and host cell remodelling. Int J Parasitol. 2012;42:21–32. doi: 10.1016/j.ijpara.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bork S, Das S, Okubo K, Yokoyama N, Igarashi I. Effects of protein kinase inhibitors on the in vitro growth of Babesia bovis. Parasitology. 2006;132:775–779. doi: 10.1017/S0031182006009917. [DOI] [PubMed] [Google Scholar]
- 47.Pedroni MJ, et al. Bumped kinase inhibitor prohibits egression in Babesia bovis. Vet Parasitol. 2016;215:22–28. doi: 10.1016/j.vetpar.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nickel W. The mystery of nonclassical protein secretion. A current view on cargo proteins and potential export routes. Eur J Biochem. 2003;270:2109–2119. doi: 10.1046/j.1432-1033.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 49.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 50.Lingelbach K, Przyborski JM. The long and winding road: protein trafficking mechanisms in the Plasmodium falciparum infected erythrocyte. Mol Biochem Parasitol. 2006;147:1–8. doi: 10.1016/j.molbiopara.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 51.Thavayogarajah T, et al. Alternative Protein Secretion in the Malaria Parasite Plasmodium falciparum. PLoS One. 2015;10:e0125191. doi: 10.1371/journal.pone.0125191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sudarsan R, Chopra RK, Khan MA, Sharma S. Ribosomal protein P2 localizes to the parasite zoite-surface and is a target for invasion inhibitory antibodies in Toxoplasma gondii and Plasmodium falciparum. Parasitol Int. 2015;64:43–49. doi: 10.1016/j.parint.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 53.Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 54.Hiller NL, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 55.Hsiao CH, Luisa Hiller N, Haldar K, Knoll LJ. A HT/PEXEL motif in Toxoplasma dense granule proteins is a signal for protein cleavage but not export into the host cell. Traffic. 2013;14:519–531. doi: 10.1111/tra.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Asada M, et al. Gliding motility of Babesia bovis merozoites visualized by time-lapse video microscopy. PLoS One. 2012;7:e35227. doi: 10.1371/journal.pone.0035227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eichenberger RM, Riond B, Willi B, Hofmann-Lehmann R, Deplazes P. Prognostic markers in acute Babesia canis infections. J Vet Intern Med. 2016;30:174–182. doi: 10.1111/jvim.13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schetters TP, et al. Systemic inflammatory responses in dogs experimentally infected with Babesia canis; a haematological study. Vet Parasitol. 2009;162:7–15. doi: 10.1016/j.vetpar.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 59.Furlanello T, Fiorio F, Caldin M, Lubas G, Solano-Gallego L. Clinicopathological findings in naturally occurring cases of babesiosis caused by large form Babesia from dogs of northeastern Italy. Vet Parasitol. 2005;134:77–85. doi: 10.1016/j.vetpar.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 60.Joice R, et al. Plasmodium falciparum transmission stages accumulate in the human bone marrow. Sci Transl Med. 2014;6:244re245. doi: 10.1126/scitranslmed.3008882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aguilar R, et al. Severity of anaemia is associated with bone marrow haemozoin in children exposed to Plasmodium falciparum. Br J Haematol. 2014;164:877–887. doi: 10.1111/bjh.12716. [DOI] [PubMed] [Google Scholar]
- 62.Foley M, Corcoran L, Tilley L, Anders R. Plasmodium falciparum: mapping the membrane-binding domain in the ring-infected erythrocyte surface antigen. Exp Parasitol. 1994;79:340–350. doi: 10.1006/expr.1994.1096. [DOI] [PubMed] [Google Scholar]
- 63.Lau AO, et al. Attenuation of virulence in an apicomplexan hemoparasite results in reduced genome diversity at the population level. BMC Genomics. 2011;12:410. doi: 10.1186/1471-2164-12-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kursula I, et al. Structural basis for parasite-specific functions of the divergent profilin of Plasmodium falciparum. Structure. 2008;16:1638–1648. doi: 10.1016/j.str.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 65.Plattner F, et al. Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe. 2008;3:77–87. doi: 10.1016/j.chom.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 66.Kishore SP, Stiller JW, Deitsch KW. Horizontal gene transfer of epigenetic machinery and evolution of parasitism in the malaria parasite Plasmodium falciparum and other apicomplexans. BMC Evol Biol. 2013;13:37. doi: 10.1186/1471-2148-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ayong L, Pagnotti G, Tobon AB, Chakrabarti D. Identification of Plasmodium falciparum family of SNAREs. Mol Biochem Parasitol. 2007;152:113–122. doi: 10.1016/j.molbiopara.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 68.Parish LA, Rayner JC. Plasmodium falciparum secretory pathway: characterization of PfStx1, a plasma membrane Qa-SNARE. Mol Biochem Parasitol. 2009;164:153–156. doi: 10.1016/j.molbiopara.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luah YH, Chaal BK, Ong EZ, Bozdech Z. A moonlighting function of Plasmodium falciparum histone 3, mono-methylated at lysine 9? PLoS One. 2010;5:e10252. doi: 10.1371/journal.pone.0010252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wastling JM, Armstrong SD, Krishna R, Xia D. Parasites, proteomes and systems: has Descartes’ clock run out of time? Parasitology. 2012;139:1103–1118. doi: 10.1017/S0031182012000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Udomsangpetch R, et al. Involvement of cytokines in the histopathology of cerebral malaria. Am J Trop Med Hyg. 1997;57:501–506. doi: 10.4269/ajtmh.1997.57.501. [DOI] [PubMed] [Google Scholar]
- 72.Clark IA, Budd AC, Alleva LM, Cowden WB. Human malarial disease: a consequence of inflammatory cytokine release. Malar J. 2006;5:85. doi: 10.1186/1475-2875-5-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matijatko V, Torti M, Schetters TP. Canine babesiosis in Europe: how many diseases? Trends Parasitol. 2012;28:99–105. doi: 10.1016/j.pt.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 74.Allred DR, Al-Khedery B. Antigenic variation and cytoadhesion in Babesia bovis and Plasmodium falciparum: different logics achieve the same goal. Mol Biochem Parasitol. 2004;134:27–35. doi: 10.1016/j.molbiopara.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 75.Allred DR, Cinque RM, Lane TJ, Ahrens KP. Antigenic variation of parasite-derived antigens on the surface of Babesia bovis-infected erythrocytes. Infect Immun. 1994;62:91–98. doi: 10.1128/iai.62.1.91-98.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O’Connor RM, Lane TJ, Stroup SE, Allred DR. Characterization of a variant erythrocyte surface antigen (VESA1) expressed by Babesia bovis during antigenic variation. Mol Biochem Parasitol. 1997;89:259–270. doi: 10.1016/S0166-6851(97)00125-4. [DOI] [PubMed] [Google Scholar]
- 77.Allred DR, et al. The ves multigene family of B. bovis encodes components of rapid antigenic variation at the infected erythrocyte surface. Mol Cell. 2000;5:153–162. doi: 10.1016/S1097-2765(00)80411-6. [DOI] [PubMed] [Google Scholar]
- 78.Allred DR, Al-Khedery B. Antigenic variation as an exploitable weakness of babesial parasites. Vet Parasitol. 2006;138:50–60. doi: 10.1016/j.vetpar.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 79.Saul A. The role of variant surface antigens on malaria-infected red blood cells. Parasitol Today. 1999;15:455–457. doi: 10.1016/S0169-4758(99)01534-3. [DOI] [PubMed] [Google Scholar]
- 80.Bachmann A, et al. Absence of erythrocyte sequestration and lack of multicopy gene family expression in Plasmodium falciparum from a splenectomized malaria patient. PLoS One. 2009;4:e7459. doi: 10.1371/journal.pone.0007459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ho M, Bannister LH, Looareesuwan S, Suntharasamai P. Cytoadherence and ultrastructure of Plasmodium falciparum-infected erythrocytes from a splenectomized patient. Infect Immun. 1992;60:2225–2228. doi: 10.1128/iai.60.6.2225-2228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pongponratn E, et al. Absence of knobs on parasitized red blood cells in a splenectomized patient in fatal falciparum malaria. Southeast Asian J Trop Med Public Health. 2000;31:829–835. [PubMed] [Google Scholar]
- 83.Lapp SA, et al. Spleen-dependent regulation of antigenic variation in malaria parasites: Plasmodium knowlesi SICAvar expression profiles in splenic and asplenic hosts. PLoS One. 2013;8:e78014. doi: 10.1371/journal.pone.0078014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Al-Khedery B, Allred DR. Antigenic variation in Babesia bovis occurs through segmental gene conversion of the ves multigene family, within a bidirectional locus of active transcription. Mol Microbiol. 2006;59:402–414. doi: 10.1111/j.1365-2958.2005.04993.x. [DOI] [PubMed] [Google Scholar]
- 85.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yanez-Mo M, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nantakomol D, et al. Circulating red cell-derived microparticles in human malaria. J Infect Dis. 2011;203:700–706. doi: 10.1093/infdis/jiq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 89.Silverman JM, Reiner NE. Exosomes and other microvesicles in infection biology: organelles with unanticipated phenotypes. Cell Microbiol. 2011;13:1–9. doi: 10.1111/j.1462-5822.2010.01537.x. [DOI] [PubMed] [Google Scholar]
- 90.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 91.Herz HM, Garruss A, Shilatifard A. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem Sci. 2013;38:621–639. doi: 10.1016/j.tibs.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mujtaba S, et al. Anthrax SET protein: a potential virulence determinant that epigenetically represses NF-kappaB activation in infected macrophages. J Biol Chem. 2013;288:23458–23472. doi: 10.1074/jbc.M113.467696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goo YK, et al. Development of a rapid immunochromatographic test using a recombinant thrombospondin-related adhesive protein of Babesia gibsoni. Vet Parasitol. 2012;190:595–598. doi: 10.1016/j.vetpar.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 95.Sriprawat K, et al. Effective and cheap removal of leukocytes and platelets from Plasmodium vivax infected blood. Malar J. 2009;8:115. doi: 10.1186/1475-2875-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 97.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 99.English AC, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215–225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 101.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pain A, et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science. 2005;309:131–133. doi: 10.1126/science.1110418. [DOI] [PubMed] [Google Scholar]
- 104.Aurrecoechea C, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38:D415–419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 107.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11 12 11–11 12 34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 110.Li L, Stoeckert CJ, Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fischer, S. et al. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics Chapter 6, Unit 6 12 11–19 (2011). [DOI] [PMC free article] [PubMed]
- 112.Wang Y, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kuck P, Meusemann K. FASconCAT: Convenient handling of data matrices. Mol Phylogenet Evol. 2010;56:1115–1118. doi: 10.1016/j.ympev.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 117.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 118.Le SQ, Dang CC, Gascuel O. Modeling protein evolution with several amino acid replacement matrices depending on site rates. Mol Biol Evol. 2012;29:2921–2936. doi: 10.1093/molbev/mss112. [DOI] [PubMed] [Google Scholar]
- 119.Figueroa JV, Buening GM, Kinden DA. Purification of the erythrocytic stages of Babesia bigemina from cultures. Parasitol Res. 1990;76:675–680. doi: 10.1007/BF00931086. [DOI] [PubMed] [Google Scholar]
- 120.Richier E, et al. Potent antihematozoan activity of novel bisthiazolium drug T16: evidence for inhibition of phosphatidylcholine metabolism in erythrocytes infected with Babesia and Plasmodium spp. Antimicrob Agents Chemother. 2006;50:3381–3388. doi: 10.1128/AAC.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rodriguez SD, Buening GM, Vega CA, Carson CA. Babesia bovis: purification and concentration of merozoites and infected bovine erythrocytes. Exp Parasitol. 1986;61:236–243. doi: 10.1016/0014-4894(86)90157-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The annotated B. canis genome will be made accessible via PiroplasmaDB hosted by EuPathDB (http://piroplasmadb.org/piro/).


