Abstract
TNFα is a homotrimeric pro-inflammatory cytokine, whose direct targeting by protein biotherapies has been an undeniable success for the treatment of chronic inflammatory diseases. Despite many efforts, no orally active drug targeting TNFα has been identified so far. In the present work, we identified through combined in silico/in vitro/in vivo approaches a TNFα direct inhibitor, compound 1, displaying nanomolar and micromolar range bindings to TNFα. Compound 1 inhibits the binding of TNFα with both its receptors TNFRI and TNFRII. Compound 1 inhibits the TNFα induced apoptosis on L929 cells and the TNFα induced NF-κB activation in HEK cells. In vivo, oral administration of compound 1 displays a significant protection in a murine TNFα-dependent hepatic shock model. This work illustrates the ability of low-cost combined in silico/in vitro/in vivo screening approaches to identify orally available small-molecules targeting challenging protein-protein interactions such as homotrimeric TNFα.
Introduction
Protein-protein interactions (PPI) represent a large class of therapeutic targets that play a crucial role in biological processes. Despite their importance, they were considered intractable due to their large and flat topology compared to classical small molecule binding sites1. Considerable progress was achieved in the last decade since 27 PPIs have now been tackled by small molecules2, 3 including organometallic compounds4–6 and dendrimers7. PPIs are still considered to be a very challenging class of targets for therapeutic applications8, 9. Historically, PPI inhibitors are larger and more hydrophobic than “drug-like” orally available compounds10. Despite their “excessive” logP or molecular weight, some PPI inhibitors such as navitoclax11 (molecular weight 975 g.mol−1) or venetoclax12 (molecular weight 868 g.mol−1) are orally available. Among the different strategies devised to inhibit PPIs, directed allosteric modulation could provide a potential way forward for the most difficult targets13.
Tumor Necrosis Factor alpha (TNFα) is a homotrimeric cytokine of the immune system whose overproduction has been associated with several chronic inflammatory diseases such as rheumatoid arthritis, Crohn’s disease or psoriasis14. Clinically approved inhibitors of TNFα include monoclonal antibodies (infliximab, adalimumab) and soluble receptors of TNFα (etanercept). These biotherapies display several drawbacks including opportunistic infections15 and treatment resistance due to autoimmune reactions16 that could be addressed by small molecule modulators.
Identified in 2005, SPD304 constitutes a reference allosteric modulator of TNFα that inhibits its activity by disrupting TNFα homotrimeric form17. SPD304 cannot be used in vivo due to his high toxicity18. Despite many efforts5, 18–27 no orally available TNFα inhibitor has been identified so far.
In the present work, in order to identify allosteric modulators of TNFα, we targeted the binding site of SPD304 with a large compound collection through the use of in silico, in vitro and in vivo screening. We report the structure and properties of our best confirmed hit, compound 1, a high affinity small molecule inhibitor of TNFα that inhibits the activity of TNFα in vitro and is orally active in vivo in a reference TNFα-dependent murine model27–29. This work illustrates the ability of current virtual screening methods to identify high affinity orally available compounds targeting challenging PPIs such as TNFα.
Results and Discussion
In order to identify allosteric modulators of TNFα, we carried out a hierarchical in silico and in vitro screening of the top 0.2% scoring compounds of a collection of 700,000 commercially available compounds by targeting the binding pocket of SPD304 in TNFα identified by He et al.17 (Supplementary Fig. S1). This hierarchical protocol, described in Fig. 1a, led to the identification of compound 1, whose structure is depicted in Fig. 2a, and whose analytics are presented in Supplementary Fig. S2a–d.
Figure 1.

Flowchart of the screening protocol used in the study. 1st step. A collection of 700,000 drug-like commercially available compounds was screened in silico. Molecular docking was performed using Surflex-dock version 2.5. After visual inspection, a 1000 compound hit list was selected for experimental testing. 2 nd step: The inhibitory activity of the compounds composing the hit list was evaluated in vitro on human TNFα induced apoptosis on the L929 cell line. Top hit compounds displayed an IC50 between 1 and 100 µM. 3 rd step: 2D/3D similarity search methods were used to identify analogues of the top hits identified after step 2. Up to 100 analogues were found per top hit with a Tanimoto similarity score >0.6. 4 th step: As in step 2, their inhibitory activity was evaluated in vitro on human TNFα induced apoptosis on the L929 cell line. The 10 best compounds after these 4 steps were selected as candidates for in vivo evaluation on a murine model. 5 th step: The in vivo evaluation of the candidates was performed in the TNFα-dependent hepatic shock model triggered with LPS/D-Galactosamine via force-feeding. After this step, 1 in vivo active compound was selected. 6 th Step: Using 2D/3D similarity search methods, we searched in our large compound collection for new analogues of this best compound identified after step 5. Up to 500 analogues were identified and purchased from the chemical supplier. As in step 2, their inhibitory activity was evaluated in vitro on human and murine TNFα. The 9 best compounds were evaluated in vivo in our murine hepatic shock assay by force-feeding as described in step 5. The best compound identified after the 6th step is compound 1.
Figure 2.

(a) Structure of compound 1. (b) Dissociation constants of TNFα/Compound 1. Determination of dissociation constants of TNFα/compound 1 complex, from gravimetric biosensor response, by using a “two-site binding” model. K d1 = 4.79 ± 1.12 µM and K d2 = 2.31 ± 1.03 nM. (c) Intrinsic Tryptophane Fluorescence. Intrinsic Tryptophan Fluorescence of 0.5 μM TNFα diluted in Phosphate Buffered Saline in the presence of DMSO alone, SPD304 (5–100 µM) in DMSO, compound 1 (5–100 µM) in DMSO or SPD304 (25 µM) + compound 1 (10–100 µM) in DMSO.
Dissociation constants and Ligand efficiency of compound 1 with TNFα
Dissociation constants of the TNFα/SPD304 and TNFα/compound 1 complexes were determined from gravimetric measurements using a Surface Acoustic Wave (SAW) biosensor30–32. On the TNFα/SPD304 complex, the phase shift variations (ΔΦ) suggested a “one-site binding” with a corresponding K d constant of 9.1 ± 1.1 µM which is consistent with the Kd values obtained by Papaneophytou et al. in the literature using a fluorescence binding assay (K d = 5.4 ± 0.2 μM)33. On the TNFα/compound 1 complex, the phase shift variations (ΔΦ) suggested a “two-site binding” (Fig. 2b). The corresponding equilibrium dissociation constants K d were respectively in the micromolar range (4.79 ± 1.12 µM) and in the nanomolar range (2.31 ± 1.03 nM) at room temperature. The corresponding ligand efficiencies of compound 1 (LE1 = 0.22 and LE2 = 0.37) are in the range of most of the protein-protein interaction inhibitors9.
Modification of the intrinsic tryptophan fluorescence profile of TNFα by compound 1
The intrinsic tryptophan fluorescence (ITF) of TNFα is modified by adding compound 1 in a dose dependent manner at the 20–100 µM range (Fig. 2c). The modification of the ITF profile by compound 1 is different to the one obtained with SPD30417 which is consistent with a mid micromolar affinity binding of compound 1 in an additional binding pocket at the surface of the homotrimer close to tryptophan residues.
Predicted binding modes of compound 1 on TNFα
The top-scoring binding mode of compound 1 predicted using Surflex-dock34 is illustrated in Fig. 3a. As expected from the highly hydrophobic surface of the binding pocket of SPD30417, 23, compound 1 displays numerous hydrophobic interactions with TNFα, its benzyl-piperazine moiety being deeply inserted in the primary binding pocket of SPD304 surrounded by Y232, Y264, Y179, L239 and L177. The phenyl-sulfonamide moiety of compound 1 displays additional hydrophobic interactions with an extension of the SPD304 binding pocket surrounded by L129, L43, L270, I268 and I127. This binding mode of compound 1, compared to the binding mode of SPD304, would not only hinder the positioning of the side chain of Y119 of the disrupted TNFα monomer17 but also the side chains of L57, L157 and V123 of the disrupted TNFα monomer.
Figure 3.

(a) Illustration of the top scoring binding mode of compound 1 with Surflex-dock in the TNFα binding site defined for the study (PDBID:2AZ5). (b) Alternate binding mode of compound 1 (green) predicted using Surflex-dock on the surface of TNFα dimer. Tryptophan residues are displayed in orange. (c) Secondary pocket on the structure of human dimeric TNFα. (d) Dual binding of compound 1 (yellow and green) predicted with Surflex-dock on the surface of the TNFα dimer co-cristallized with SPD304 displayed in purple (PDB ID: 2AZ5). Tryptophan residues are displayed in orange.
To illustrate the alternate binding mode of compound 1 on TNFα, we explored the whole surface of the structure of human trimeric TNFα (PDB ID: 1TNF) to identify an additional binding pocket at the surface of the homotrimer that contains tryptophan residues (Fig. 3b). After a careful visual inspection, a pocket has been identified at the junction of the three TNFα subunits that is conserved in the homodimeric structure cocristallized with SPD304 (PDB ID: 2AZ5). This secondary pocket illustrated in Fig. 3c could be the one we suspected after gravimetric studies (dual binding) and intrinsic tryptophan fluorescence measurements (alternate modification of the ITF profile) since it is the only one on the surface of TNFα that contains tryptophan residues (it contains one tryptophan residues per TNFα monomer). The top scoring binding mode of compound 1 on this new binding pocket on the surface of the TNFα dimer predicted using Surflex-dock is illustrated in Fig. 3d.
Activity of compound 1 on TNFα-TNFRI and TNFα-TNFRII interactions
We evaluated the inhibitory activity of compound 1 on the binding of TNFα to its receptors TNFRI and TNFRII using an ELISA assay. Compound 1 displayed IC50 of 37 µM on TNFα/TNFRI and 31 µM on TNFα/TNFRII (Fig. 4a). These values are comparable to the IC50 of 22 µM obtained with SPD304 on TNFα/TNFRI17.
Figure 4.

(a) Relative binding on TNFRI and TNFRII. Compound 1 inhibits the interaction between TNFα and its receptors TNFRI and TNFRII in a dose dependant manner. TNFRI IC50 = 37 μM, TNFRII IC50 = 31 μM. (b) Neutralization of TNFα and survival of L929 cells. Compound 1 inhibition of TNFα induced apoptosis in L929 cell line. Data represent neutralization of TNFα in presence of various concentrations of compound 1. IC50 = 12 μM (c) Neutralization of TNFα and survival of HEK cells. Compound 1 inhibition of the TNFα signaling pathway on HEK cells transfected with a reporter gene under the control of NF-κB. Data represent the neutralization of TNFα in the presence of different concentrations of compound 1. IC50 = 10 μM. (d) Inhibition of CXCL1 secretion in L929 supernatants in presence of various concentrations of compound 1 after stimulation with TNFα (5 ng/ml). (e) Inhibition of caspase 3 activity in presence of compound 1. (f) Inhibition of caspase 8 activity in presence of compound 1.
Activity of compound 1 on TNFα functional cellular models
Compound 1 inhibited the induction of apoptosis by TNFα in the L929 cell line with an IC50 of 12 µM (Fig. 4b and Supplementary Movie). In the same assay, SPD304 displayed a high cellular toxicity (no cells survived with more than 30 µM of SPD304). The cellular activity of compound 1 was confirmed on the TNFα signaling pathway, by using a HEK cell line expressing TNFRI and transfected with a reporter gene under the control of five NF-κb binding sites, where compound 1 displayed an IC50 of 10 µM (Fig. 4c). On the same assay, SPD304 displayed a high cellular toxicity (no cells survived with more than 10 µM of SPD304). Since TNFα and TNFβ share 30% sequence identity, the activity of compound 1 on TNFβ in the L929 cell line was also evaluated. Compound 1 inhibited the induction of apoptosis by TNFβ with an IC50 of 28 μM and a maximal survival of 40% at 100 μM (Supplementary Fig. S3).
Activity of compound 1 on CXCL1 expression, caspases and protein kinases related to the TNFα pathway
Compound 1 was tested for its ability to inhibit the production of CXCL1 on L929 cells since is triggered by TNFα. The supernatants from treated L929 cells with TNFα and compound 1 contained significantly less CXCL1 than the supernatants from untreated L929 cells as depicted in Fig. 4d. Compound 1 inhibited the secretion of CXCL1 at a similar level to the anti-TNFα antibody. We evaluated the ability of compound 1 to inhibit the activation of caspase 3 and caspase 8, widely used as readouts for TNFα signaling. Compound 1 blocked the activation of caspase 3 and caspase 8 (Fig. 4e,f), in a dose dependent manner. Compound 1 was tested for direct inhibition of caspases and kinases activities related to the TNFα pathway (IκB kinases (IKK), JNK1, p38 and Syk, caspase 3, caspase 8). Compound 1 displayed no significant effect on these proteins at 1 µM (Supplementary Fig. S4).
Activity of compound 1 on murine TNFα
Compound 1 inhibited both human and murine TNFα with similar IC50s on the TNFα-induced apoptosis assay on L929 cells (~10 µM).
In vivo activity of compound 1
The in vivo activity of compound 1 was evaluated in a TNFα dependent murine model, the LPS/D-Galactosamine induced shock assay29. As shown in Fig. 5a, compound 1 exhibited a fully protective effect with an oral administration of 5 mg of compound 1 per mouse eight hours before the induction of the shock (p < 4.10−3). The oral administration of 5 mg of compound 1 did not affect the mice serum level of TNFα induced after the shock (Fig. 5b) but diminished the mice serum level of AST/ALT which transcribes the damage induced to hepatocytes (Fig. 5c,d). We conducted hispathological and immunohistochemical analyses on the livers of the LPS/D-Galactosamine injected mice, treated with the vehicle (DMSO), compound 1 or etanercept (Fig. 5e,f). The mice that received only the vehicle displayed important liver damage and hemorrhage whereas the mice treated with compound 1 or etanercept (Fig. 5e) displayed reduced liver damage and hemorrhage. The immunohistochemical analysis of the mice livers treated with compound 1 or etanercept displayed a reduced expression of cleaved caspase 3 (Fig. 5f), which is consistent with a reduction of the liver apoptosis in these animals. Taken together, these results confirmed that compound 1 did not affect the production of TNFα but inhibited TNFα induced damage on the mice liver by preventing caspase 3 induced apoptosis of hepatocytes.
Figure 5.
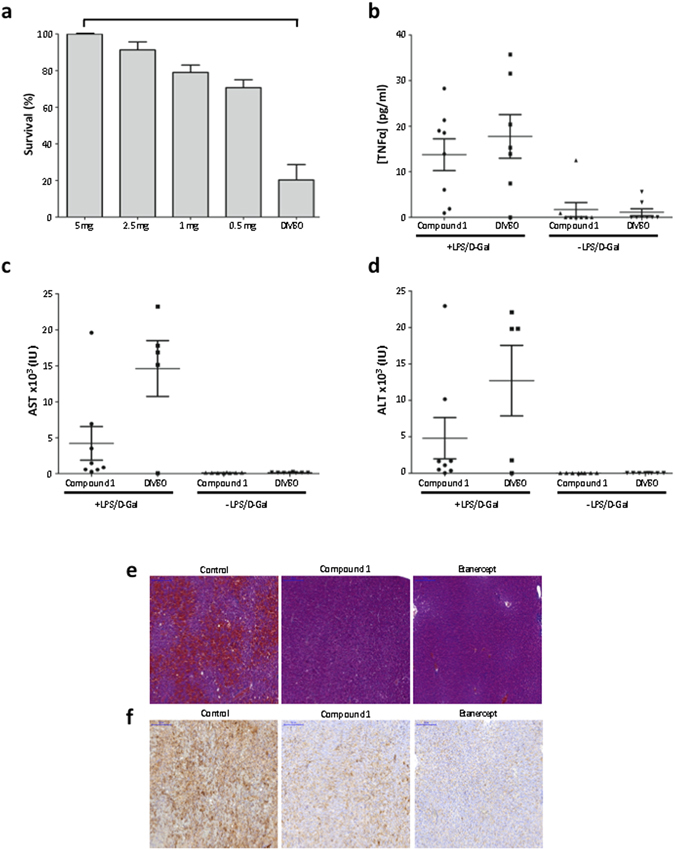
Effect of compound 1 in an in vivo murine model of LPS/D-Galactosamine induced shock. (a) Mice survival after force-feeding with different doses of compound 1 and an intraperitoneal injection of LPS/D-Galactosamine. Groups of eight mice were used. Values are mean ± s.e.m. (b) TNFα levels in mice sera three hours after the induction of the shock by LPS/D-Galactosamine. Mice were forced-fed with 5 mg compound 1 in DMSO. Groups of eight mice were used. Values are mean ± s.e.m. (c) Aspartate aminotransferase (AST) levels in mice sera eight hours after the induction of the shock. Mice were forced-fed with 5 mg compound 1 in DMSO. Values are mean ± s.e.m. (d) Alanine aminotransferase (ALT) levels in mice sera eight hours after the induction of the shock. Mice were forced-fed with 5 mg compound 1 in DMSO. Values are mean ± s.e.m. (e) Livers sections (H&E staining) from mice forced-fed with DMSO (control) or compound 1 or injected with etanercept and an intraperitoneal injection of LPS/D-Galactosamine. (f) Liver sections from mice forced-fed with DMSO (control) or compound 1 or injected with etanercept and an intraperitoneal injection of LPS/D-Galactosamine. Sections were incubated with an anti-cleaved caspase-3 antibody and revealed with an anti-rabbit antibody and DAB.
Conclusion
Using an in silico/in vitro/in vivo screening approach, we have identified compound 1 that binds TNFα with high affinity and inhibits its activity in vitro and in vivo via oral administration. Compound 1 physicochemical profile displays some drawbacks that could be addressed after a medicinal chemistry program, in particular a moderate solubility in aqueous solvents and a low bioavailability rendering necessary administration of several milligrams of compound per mouse. However, we could demonstrate that despite these drawbacks, compound 1 displayed a fully protective effect on an acute TNFα dependent mouse model in which large quantities of TNFα are released and inhibited by the compound. Compound 1 displayed a reduced inhibiting activity on TNFβ compared to TNFα. Targeting both TNFα and TNFβ could be beneficial for the treatment of rheumatoid arthritis35 since etanercept, a treatment of reference, displayed effective downregulation of both TNFα and TNFβ36, 37.
The next steps for the development of a drug based on the properties of compound 1 will be to improve its physicochemical profile, assess its full pharmacodynamics properties and evaluate its protective effect in a TNFα dependent chronic animal model closer to the physiopathology of chronic inflammatory diseases. This work illustrates a first step in the identification of orally available small molecule cytokine inhibitors that could be the basis for the development of alternate strategies to the biologics used for the treatment of chronic inflammatory diseases. In addition, this work highlights the ability of low-cost integrative in silico/in vitro/in vivo screening approaches to identify small-molecules targeting challenging protein-protein interactions such as homotrimeric TNFα.
Experimental Procedures
Materials, cell line and mice
Compounds were obtained from Chembridge (San Diego, CA, USA). Dimethyl Sulfoxide (DMSO), Lipopolysaccharide (LPS), TMB, Thiazolyl Blue Tetrazolium Bromide (MTT) and DAB were obtained from Sigma-Aldrich (Saint Quentin Fallavier, France). Human TNFα, human TNFβ and anti-TNFα antibody were obtained from R&D Systems (Lille, France). Anti-cleaved caspase 3 was obtained from Cell Signaling Technology (St Quentin en Yvelines, France), Anti-rabbit antibody coupled with HRP was obtained from Abcam (Paris, France) Dulbecco’s Modified Eagle and Phosphate Buffered Saline (PBS) were obtained from Pan Biotech (Brumath, France). Actinomycin D and D-Galactosamine were obtained from Fisher (Illkirch, France). L929 cell line has been grown in the Laboratory for years. HEK-Blue™ TNFα reporter cell line and QUANTI-Blue™ were obtained from InvivoGen (Toulouse, France). 7 weeks-old female Balb/C mice were obtained from Charles River Laboratories (L’Arbresle, France). Mice used in all experiments were handled according to the guidelines and protocols were approved by the ethical commitee of Paris Descartes University, France.
In silico screening
Structure preparation
The binding site has been defined at 4 Å around the co-crystallized SPD304 ligand in the structure of the TNFα dimer (PDB id: 2AZ5, Supplementary Fig. 1). Hydrogen atoms were added using Chimera38.
Compound collection
The 900,000 compounds Chembridge screening compound collection was retrieved from www.hit2lead.com. After an ADME-tox filtering using FAF-drugs239, 700,000 compounds were selected to constitute our commercially available drug-like compound collection.
Structure-based virtual screening
Molecular docking was performed using Surflex-dock version 2.534. Surflex-dock is based on a modified Hammerhead fragmentation/reconstruction algorithm to dock compounds flexibly into the binding site. The query molecule is decomposed into rigid fragments that are superimposed to the Surflex-protomol i.e., molecular fragments covering the entire binding site. The docking poses are evaluated by an empirical scoring function. The protomol generation step has been performed using the options proto_thresh = 0.5 and proto_bloat = 4. Screening step was performed using the option–pscreen.
Ligand-based virtual screening
Ligand-based virtual screening was performed using the 2D/3D similarity search methods implemented in the webservice provided by www.hit2lead.com. Analogues identifications were performed using 2D and 3D similarity search with a 60% Tanimoto-based 2D and 3D similarity cut-off. Compounds were selected for experimental tests after a careful visual inspection of the retrieved analogues.
Gravimetric measurements for the determination of dissociation constants
Gravimetric measurements were performed using surface acoustic wave (SAW) sensors. TNFα solutions were prepared by diluting 10 µg of lyophilized TNFα in 1 ml of PBS. They were kept in the freezer (at −18 °C) and put at room temperature just before their further grafting onto the gold surface area of the SAW sensor. We prepared stock solutions of 0.5 g/l of SPD304 in PBS and 0.1 g/l of compound 1 in HCl 0.1 M/H2O (1/1: v/v). pH was adjusted to ≈7 before the injection of the analyte of interest into the microfluidic channel of the detection system.
Phase shift variations (ΔΦ) versus compound concentration (C) were fitted using equation 1
where Φsat represents the maximum sensor’s response to the compound binding and Kd represents the equilibrium dissociation constant.
In the case of a two-site binding, equation 2 was used for fitting:
| 2 |
where Φsat1 and Φsat2 are related to the maximum sensor’s response for each binding site and Kd1 and Kd2 are their respective dissociation constants.
Measurement of TNFα intrinsic fluorescence
All samples were brought to 10 mM phosphate buffer, 0.05% Tween20, 1% DMSO. Fluorescence readings were made with a Quantamaster QM4CW spectrofluorimeter (Photon Technology International), by exciting TNFα at 290 nm and measuring the emission peaks at 322 nm. Compound 1 inner filter effects were corrected using absorbance measurements of compound alone solutions at 290 nm and 322 nm.
TNFα-TNFRI and TNFα-TNFRII binding ELISA
Microtiter plates were coated with 12.5 ng of TNFRI or TNFRII in 125 μl of PBS per well at 37 °C. The wells were washed, blocked with PBS/BSA 2% for three hours and washed as before. Serial dilutions of compounds were mixed with a fixed quantity of TNFα in PBS/BSA 1% and incubated two hours at 37 °C. 125 μl of the mix were added to the wells and plates were incubated overnight at 4 °C. Wells were washed incubated with 37.5 ng of TNFα biotinylated antibody in 125 μl of PBS/BSA 1% for two hours at 37 °C. Wells were washed and incubated with avidin-HRP (1:500) in 125 μl of PBS/BSA 1% for 30 minutes at 37 °C, 5% CO2. After washing, TMB solution was added to wells, quenched with 63 μl of 1 M H2SO4 solution. Absorbance was measured at 450 nm.
Neutralization of cellular TNFα induced apoptosis
80% confluent L929 cells were plated in flat bottom plates at 4 × 105 cells per well in 100 μl of DMEM medium containing 10% FBS, 2 mM L-Glutamine, 100 U/ml Penicillin − 100 μg/ml Streptomycin and incubated for one night at 37 °C, 5% CO2. TNFα or TNFβ (150 pg/ml), Actinomycin D (4 μg/ml) and the compounds at different concentrations (ranging from 100 μM to 0.8 μM) were mixed in 150 μl DMEM medium 1% FBS, 2 mM L-Glutamine, 100 U/ml Penicillin−100 μg/ml Streptomycin in U-bottom plates. After two hours incubation at 37 °C, 5% CO2, 100 μl of the mix was added to the plated cells and incubated at 37 °C, 5% CO2 for 24 hours. Supernatants were discarded and 100 μl of MTT at 0.5 mg/ml were added to wells. After two hours, supernatants were discarded and 200 μl of DMSO were then added. Plates were read at 570 nm with a spectrophotometer providing the optical density (OD) of each well. The percentage of neutralization of TNFα by a compound was calculated using equation 3:
| 3 |
An IC50 could be computed from the percentage of neutralization for each compound.
Secreted embryonic alcaline phosphatase reporter assay
80% confluent HEK-Blue™ TNFα were plated in flat bottom plates at 5 × 104 per well in 100 μl of DMEM containing 2 mM L-Glutamine, 100 U/ml Penicillin − 100 μg/ml Streptomycin and incubated at 37 °C, 5% CO2. Serial dilutions of compounds (ranging from 100 μM to 0.8 μM) were mixed with 400 pg/ml of human TNFα in DMEM containing 2% of FBS, 2 mM L-Glutamine, 100 U/ml Penicillin−100 μg/ml Streptomycin in U-bottom plates. After two hours of incubation at 37 °C, 5% CO2, 100 μl of the mix was added to the plated cells and incubated 24 hours at 37 °C, 5% CO2. 20 μl of supernatants were incubated for 3 hours with 180 μl of QUANTI-Blue™ to reveal secretion of phosphatase alcaline and plates were read at 620 nm with a spectrophotometer providing the optical density (OD).
Activation of Caspases 3 and 8 assays
Caspases 3 and 8 activation was determined using Caspase-Glo® 3 and Caspase-Glo-8® assays kits (Promega) according to the manufacter’s procedures. After 8 hours of treatment, plates were equilibrated at room temperature. 100 μl Caspase-Glo 3® and Caspase-Glo 8® reagent were added to each well. Luminescence was read on a FLUOstar OPTIMA reader.
Measurement of the concentration of CXCL1
Cells were plated at 2.5 × 105 cells/well in 24-well plates and treated as indicated. Eight hours later, supernatants were recovered and kept at −20 °C for later use. The concentration of murine CXCL1 was measured in supernatants with a dosage ELISA kit obtained from R&D Systems (Lille, France) according to the manufacter’s procedures.
Inhibition activity of kinases and caspases related to TNFα pathway
Kinases and caspases activities in presence of 1 µM of compound 1 were carried out by CEREP/Eurofins (Celle-Lévescault). The activity of compound 1 on the different kinases and caspases was compared to control values. Data are expressed as percentages of inhibition of control values.
LPS/D-Galactosamine induced lethal shock
7 weeks-old Balb/C mice were force-fed with 100 μl of a solution of DMSO containing various doses of compound 1 (5 mg down to 0.5 mg) eight hours before receiving an intraperitoneal injection of 200 μl of PBS containing 0.1 μg of LPS and 20 mg of D-Galactosamine. A control group (DMSO) was force-fed with 100 μl of a solution of DMSO alone eight hours before to be injected with 200 μl of LPS/D-Galactosamine solution. Mice survival was monitored for 48 hours after the injection of LPS/D-Galactosamine.
Histological and immunohistochemical studies of livers
7 weeks-old Balb/C mice were forced-fed with a solution of DMSO containing 5 mg of compound 1 eight hours before receiving an intraperitoneal injection of 200 μl of PBS containing 0.1 μg of LPS and 20 mg of D-Galactosamine. Eight hours after LPS/D-Galactosamine injection, mice were sacrificed by cervical dislocation and livers were harvested, fixed in 4% formalin and embedded in paraffin. Serial 4 μm sections were stained with hematoxylin and eosin or incubated with an anti-cleaved caspase 3 antibody (1:300) and revealed with an anti-rabbit antibody and DAB. Slides were scanned with a slide scanner (Perkin Elmer) and analyzed with Pannoramic viewer (3D HISTECH).
Cytokines quantification
7 weeks-old Balb/C mice were forced-fed with a solution of DMSO containing 5 mg of compound 1 eight hours before receiving an intraperitoneal injection of 200 μl of PBS containing 0.1 μg of LPS and 20 mg of D-Galactosamine. Three hours after LPS/D-Galactosamine injection, mice were anesthetized with ketamine/xylazine to recover blood by cardiac puncture. Sera were kept at −80 °C until assay.
TNFα in serum was quantified using U-CyTech biosciences kits (Utrecht, The Netherlands) according to manufacturer’s procedures.
AST/ALT quantification
7 weeks-old Balb/C mice were forced-fed with a solution of DMSO containing 5 mg of compound 1 eight hours before receiving an intraperitoneal injection of 200 μl of PBS containing 0.1 μg of LPS and 20 mg of D-Galactosamine. Eight hours after, mice were anesthetized by an injection of ketamine/xylazine to recover blood by cardiac puncture. Sera were kept at −80 °C until assay. Quantification of AST/ALT was performed using Architec ci 8200 analyser (Abbott) with reagent kits from Abbott.
Ethics
All study protocols and experimental procedures were approved by the Paris Descartes University ethical comittee and were carried out in accordance with the approved guidelines.
Statistics
One-tailed p-values were calculated using Fisher’s exact test.
Electronic supplementary material
Acknowledgements
We would like to thank Prof. Jain for providing Surflex, and Chemaxon for providing the Marvin suite. H.G. and H.M. are recipients of CIFRE fellowships from Association Nationale de la Recherche et de la Technologie (ANRT). J.P. and N.B.N are recipients of fellowships from the French Ministère de l’Enseignement Supérieur et de la Recherche (MESR). We would like to thank O. De Oliveira for her technical support during the in vivo experiments; Prestwick Chemical and CMGPCE, CNAM for their support in analytical chemistry and the HistIM platform for technical support with the immunochemistry and histology experiments. This work was funded in part by grants from Conservatoire National des Arts et Métiers and from Peptinov SAS.
Author Contributions
H.G., N.B.N. and M.M. performed and analyzed the molecular modeling experiments. H.M., G.M. and L.D. performed and analyzed the biochemical and cellular assays. H.M., G.M., N.F., C.Z., B.B. and P.E. performed and analyzed the biophysical assays. H.M. and L.D. performed and analyzed the mice experiments. J.P., R.R. and J.L.S. participated in some experiments. M.M. and J.F.Z. conceived the whole study. H.M., M.M., B.B., P.E., C.Z., N.F., H.D. and J.F.Z. wrote the manuscript. All the authors reviewed the manuscript.
Competing Interests
J.F.Z., R.R., and H.D. are shareholders of Peptinov S.A.S. L.D., H.D., H.M., and G.M. are employed by Peptinov S.A.S.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-03427-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Matthieu Montes, Email: matthieu.montes@cnam.fr.
Jean-François Zagury, Email: zagury@cnam.fr.
References
- 1.Fuller JC, Burgoyne NJ, Jackson RM. Predicting druggable binding sites at the protein-protein interface. Drug Discov Today. 2009;14:155–161. doi: 10.1016/j.drudis.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Labbe CM, et al. iPPI-DB: an online database of modulators of protein-protein interactions. Nucleic Acids Res. 2016;44:D542–547. doi: 10.1093/nar/gkv982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basse, M. J., Betzi, S., Morelli, X. & Roche, P. 2P2Idb v2: update of a structural database dedicated to orthosteric modulation of protein-protein interactions. Database: the journal of biological databases and curation2016, doi:10.1093/database/baw007 (2016). [DOI] [PMC free article] [PubMed]
- 4.Zhong H, et al. An iridium(III)-based irreversible protein–protein interaction inhibitor of BRD4 as a potent anticancer agent. Chem. Sci. 2015;6:5400–5408. doi: 10.1039/C5SC02321A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang TS, et al. Identification of an Iridium(III)-Based Inhibitor of Tumor Necrosis Factor-alpha. J Med Chem. 2016;59:4026–4031. doi: 10.1021/acs.jmedchem.6b00112. [DOI] [PubMed] [Google Scholar]
- 6.Liu LJ, et al. Inhibition of the p53/hDM2 protein-protein interaction by cyclometallated iridium(III) compounds. Oncotarget. 2016;7:13965–13975. doi: 10.18632/oncotarget.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mignani S, El Kazzouli S, Bousmina MM, Majoral JP. Dendrimer space exploration: an assessment of dendrimers/dendritic scaffolding as inhibitors of protein-protein interactions, a potential new area of pharmaceutical development. Chem Rev. 2014;114:1327–1342. doi: 10.1021/cr400362r. [DOI] [PubMed] [Google Scholar]
- 8.Mullard A. Protein-protein interaction inhibitors get into the groove. Nat Rev Drug Discov. 2012;11:173–175. doi: 10.1038/nrd3680. [DOI] [PubMed] [Google Scholar]
- 9.Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 11.Park CM, et al. Discovery of an orally bioavailable small molecule inhibitor of prosurvival B-cell lymphoma 2 proteins. J Med Chem. 2008;51:6902–6915. doi: 10.1021/jm800669s. [DOI] [PubMed] [Google Scholar]
- 12.Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 13.Nussinov R, Tsai CJ. Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol Sci. 2014;35:256–264. doi: 10.1016/j.tips.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 15.Fleischmann RM, Iqbal I, Stern RL. Considerations with the use of biological therapy in the treatment of rheumatoid arthritis. Expert Opin Drug Saf. 2004;3:391–403. doi: 10.1517/14740338.3.5.391. [DOI] [PubMed] [Google Scholar]
- 16.Rubbert-Roth A, Finckh A. Treatment options in patients with rheumatoid arthritis failing initial TNF inhibitor therapy: a critical review. Arthritis Res Ther. 2009;11(Suppl 1):S1. doi: 10.1186/ar2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He MM, et al. Small-molecule inhibition of TNF-alpha. Science. 2005;310:1022–1025. doi: 10.1126/science.1116304. [DOI] [PubMed] [Google Scholar]
- 18.Alexiou P, et al. Rationally designed less toxic SPD-304 analogs and preliminary evaluation of their TNF inhibitory effects. Archiv der Pharmazie. 2014;347:798–805. doi: 10.1002/ardp.201400198. [DOI] [PubMed] [Google Scholar]
- 19.Choi H, Lee Y, Park H, Oh DS. Discovery of the inhibitors of tumor necrosis factor alpha with structure-based virtual screening. Bioorg Med Chem Lett. 2010;20:6195–6198. doi: 10.1016/j.bmcl.2010.08.116. [DOI] [PubMed] [Google Scholar]
- 20.Leung CH, et al. Structure-based repurposing of FDA-approved drugs as TNF-alpha inhibitors. ChemMedChem. 2011;6:765–768. doi: 10.1002/cmdc.201100016. [DOI] [PubMed] [Google Scholar]
- 21.Mancini F, et al. Inhibition of tumor necrosis factor-alpha (TNF-alpha)/TNF-alpha receptor binding by structural analogues of suramin. Biochem Pharmacol. 1999;58:851–859. doi: 10.1016/S0006-2952(99)00150-1. [DOI] [PubMed] [Google Scholar]
- 22.Alzani R, et al. Suramin induces deoligomerization of human tumor necrosis factor alpha. J Biol Chem. 1993;268:12526–12529. [PubMed] [Google Scholar]
- 23.Chan DS, et al. Structure-Based Discovery of Natural-Product-like TNF-alpha Inhibitors. Angew Chem Int Ed Engl. 2010;49:2860–2864. doi: 10.1002/anie.200907360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung CH, et al. A metal-based inhibitor of tumor necrosis factor-alpha. Angew Chem Int Ed Engl. 2012;51:9010–9014. doi: 10.1002/anie.201202937. [DOI] [PubMed] [Google Scholar]
- 25.Buller F, et al. Discovery of TNF inhibitors from a DNA-encoded chemical library based on diels-alder cycloaddition. Chem Biol. 2009;16:1075–1086. doi: 10.1016/j.chembiol.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Hu Z, et al. Japonicone A antagonizes the activity of TNF-alpha by directly targeting this cytokine and selectively disrupting its interaction with TNF receptor-1. Biochem Pharmacol. 2012;84:1482–1491. doi: 10.1016/j.bcp.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 27.Ma L, et al. A novel small-molecule tumor necrosis factor alpha inhibitor attenuates inflammation in a hepatitis mouse model. J Biol Chem. 2014;289:12457–12466. doi: 10.1074/jbc.M113.521708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galanos C, Freudenberg MA, Reutter W. Galactosamine-induced sensitization to the lethal effects of endotoxin. Proc Natl Acad Sci USA. 1979;76:5939–5943. doi: 10.1073/pnas.76.11.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freudenberg MA, Galanos C. Tumor necrosis factor alpha mediates lethal activity of killed gram-negative and gram-positive bacteria in D-galactosamine-treated mice. Infect Immun. 1991;59:2110–2115. doi: 10.1128/iai.59.6.2110-2115.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitakis M, Dellaporta A, Gizeli E. Measurement of two-dimensional binding constants between cell-bound major histocompatibility complex and immobilized antibodies with an acoustic biosensor. Biophys J. 2008;95:4963–4971. doi: 10.1529/biophysj.108.132118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergaoui Y, Zerrouki C, Fourati N, Fougnion J, Abdelghani A. Antigen-antibody selective recognition using LiTaO3 SH-SAW sensors: investigations on macromolecules effects on binding kinetic constants. Eur Phys J Appl Phys. 2011;56:13705–13709. doi: 10.1051/epjap/2011100225. [DOI] [Google Scholar]
- 32.Fourati, N. & Zerrouki, C. Immunosensing with surface acoustic wave sensors: toward highly sensitive and selective improved piezoelectric biosensors. New sensors and processing chain, Wiley ISTE ISBN: 978-1-84821-626-6 (2014).
- 33.Papaneophytou CP, Mettou AK, Rinotas V, Douni E, Kontopidis GA. Solvent Selection for Insoluble Ligands, a Challenge for Biological Assay Development: A TNF-alpha/SPD304 Study. ACS medicinal chemistry letters. 2013;4:137–141. doi: 10.1021/ml300380h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 35.Calmon-Hamaty F, Combe B, Hahne M, Morel J. Lymphotoxin alpha revisited: general features and implications in rheumatoid arthritis. Arthritis Res Ther. 2011;13:232. doi: 10.1186/ar3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neregard P, et al. Etanercept decreases synovial expression of tumour necrosis factor-alpha and lymphotoxin-alpha in rheumatoid arthritis. Scandinavian journal of rheumatology. 2014;43:85–90. doi: 10.3109/03009742.2013.834964. [DOI] [PubMed] [Google Scholar]
- 37.Buch MH, et al. True infliximab resistance in rheumatoid arthritis: a role for lymphotoxin alpha? Ann Rheum Dis. 2004;63:1344–1346. doi: 10.1136/ard.2003.014878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Lagorce D, Sperandio O, Galons H, Miteva MA, Villoutreix BO. FAF-Drugs2: free ADME/tox filtering tool to assist drug discovery and chemical biology projects. BMC Bioinformatics. 2008;9:396. doi: 10.1186/1471-2105-9-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


