Abstract
Zoonotic pathogens have the unique ability to cross the species barrier, causing disease in both humans and specific animal hosts. Streptococcus iniae is a zoonotic pathogen of both fish and humans, and the clinical presentations of S. iniae infections in fish and humans are very similar to those caused by various human-specific streptococcal pathogens. Virulence mechanisms required for infection by this pathogen of either host have yet to be determined. Using the previously reported zebrafish infectious disease model, we performed a large-scale screening to determine genes required for systemic infection. Screening 1,128 signature-tagged transposon mutants through the zebrafish model allowed identification of 41 potential mutants that were unable to survive within the host environment. Greater than 50% of the mutants that could be identified through homology searches were highly homologous to genes found in other human-specific streptococcal pathogens, while 32% were found to have no homology to any sequences found in the databases, suggesting as yet unknown gram-positive bacterial virulence factors. A large percentage of the insertions were found to be located in several putative capsule synthesis genes, an important virulence component for other systemic pathogens. Density gradient assays demonstrated that several of these putative capsule mutants have dissimilar buoyant densities, suggesting different levels of capsule synthesis. Putative capsule mutants were also less resistant to phagocytosis in whole-blood assays than wild-type S. iniae. Our initial large-scale characterization of S. iniae virulence highlights the importance of the capsule for successful infection.
Systemic infections caused by streptococcal pathogens can cause severe life-threatening disease in both immunocompromised and healthy individuals from birth to advanced age. In many cases, infections progress so rapidly that medical intervention is only minimally successful, if at all. Streptococcal pathogens are currently experiencing a reemergence in invasive human infections, aided in part by the evolution of antibiotic resistance and by the emergence of new invasive serotypes (51, 54). Although past and ongoing research has provided much information on the mechanisms used by systemic pathogens to cause disease, neither a complete picture nor a definitive strategy to combat these infections has emerged.
The best way to analyze a systemic infection is to examine it in the natural host. However, most streptococcal pathogens that cause systemic infections, such as Streptococcus pneumoniae, Streptococcus pyogenes, and Streptococcus agalactiae, are specific to humans and therefore cannot be experimentally analyzed in their natural host. In an effort to further define the virulence mechanisms used by systemic pathogens, we developed an infectious disease model using a streptococcal pathogen, Streptococcus iniae, and a natural host, the zebrafish (Danio rerio), so as to analyze the mechanisms of systemic infection in a natural host-pathogen system (38). S. iniae is an important pathogen in both marine animals and humans, causing systemic infection in both hosts. Symptoms of infection are very similar to infections caused by human-specific streptococcal pathogens, such as S. pyogenes, S. agalactiae, and S. pneumoniae (38). Both S. iniae and S. pyogenes can cause cellulitis in humans, especially after an abrasion to the skin, allowing access of the bacterium to the dermal layer. S. iniae, S. pneumoniae, and S. agalactiae are all capable of causing systemic infection leading to meningitis and bacteremia. Furthermore, a 16S rRNA phylogenetic tree of streptococcal species illustrates that S. iniae is a very close genetic relative to the human-specific pathogen S. agalactiae (26).
We have chosen the zebrafish infected with S. iniae as a model system to study host-pathogen interactions for multiple reasons. Zebrafish are natural hosts for S. iniae infection with disease characteristics that mimic those observed in infected fish grown in aquaculture (11). In addition, zebrafish have been used as a model for vertebrate development and are currently being used as a model for immune system development (2, 36, 60, 61). Recent advances in zebrafish research, including large-scale mutant screens and the completion of The Danio rerio Sequencing Project (The Sanger Center) (http://www.sanger.ac.uk/Projects/D_rerio/), have identified many large chromosomal segments that are conserved between zebrafish and mammals, making them ideal hosts for probing host response to bacterial infection (42). Reports also confirm that zebrafish possess well-developed immune systems with development mimicking that of humans (16, 22, 55, 57-59, 61), making the zebrafish highly advantageous for immunological study.
Several recent reports have provided insights into some of the pathogenic mechanisms of S. iniae. Fuller et al. (13) have shown that S. iniae subcutaneous infections in mice can lead to bacteremia. In addition, this group identified and characterized the genetic locus of the S. iniae cytolysin, which confers a beta-hemolytic phenotype. However, the cytolysin was not required for virulence in a murine model of bacteremia infection (14). Two serotypes of S. iniae originally recovered from diseased rainbow trout were analyzed by Zlotkin et al. (62) for their ability to survive in salmonid macrophages in vitro. This group found that S. iniae has the ability to invade and survive in fish phagocytes, causing the cell to undergo apoptosis. These researchers suggested that S. iniae may be adopting a “Trojan horse” strategy, allowing evasion of the immune system and intracellular transportation across the blood-brain barrier (62). However, additional research is needed to fully understand the mechanisms of pathogenesis and interactions this zoonotic pathogen has with its host, be it human or aquatic.
In this study, we utilized signature-tagged mutagenesis (STM) to identify genes necessary for full virulence of S. iniae using the zebrafish model host system. STM has proven to be a powerful technique that allows high-throughput analysis of bacterial mutants through an animal host (5, 8, 21, 24, 29, 35). Using the previously developed zebrafish animal model (38) allows us to analyze host-pathogen interactions in a model system that is identical to the host environment that S. iniae encounters in nature, in addition to being an ideal model to mimic human systemic infection (37, 38). With the coupling of STM and the zebrafish model, we have identified numerous genes involved in pathogenesis of S. iniae, most of which are conserved in gram-positive pathogens. Of 1,128 mutants examined, we identified 41 potential mutants that were attenuated for survival in the host environment. An additional 185 potential mutants with a more limited degree of attenuation were identified. A large percentage of insertions were found to be in multiple capsular polysaccharide synthesis genes, which have been shown to be important for virulence in many gram-positive systemic pathogens, including the human-specific streptococcal pathogens, S. agalactiae and S. pneumoniae. Here we report genes that are required for survival of S. iniae in zebrafish and examine how the capsule plays a role in virulence.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Plasmids were maintained in Escherichia coli Top10 cells (Invitrogen) and cultured in Luria-Bertani (LB) medium at 37°C. S. iniae strain 9117 (13) and derivatives were used for zebrafish infections. S. iniae was cultured in Todd-Hewitt medium (BBL) supplemented with 0.2% yeast extract (BBL) and 2% proteose peptone (BBL) (TP) and incubated in airtight conical tubes without agitation at 37°C. To produce solid medium, liquid medium was supplemented with 1.4% agar (BBL). Solid medium used for culturing S. iniae was incubated in an anaerobic environment (<0.03% O2, 10% CO2) generated by a commercial gas generator (GasPak; BBL). When required, erythromycin antibiotic was added to the TP or LB medium at the following concentrations: 2 μg/ml (TP-Erm2) for S. iniae and 750 μg/ml (LB-Erm750) for E. coli.
Construction of the pJDM-STM vector.
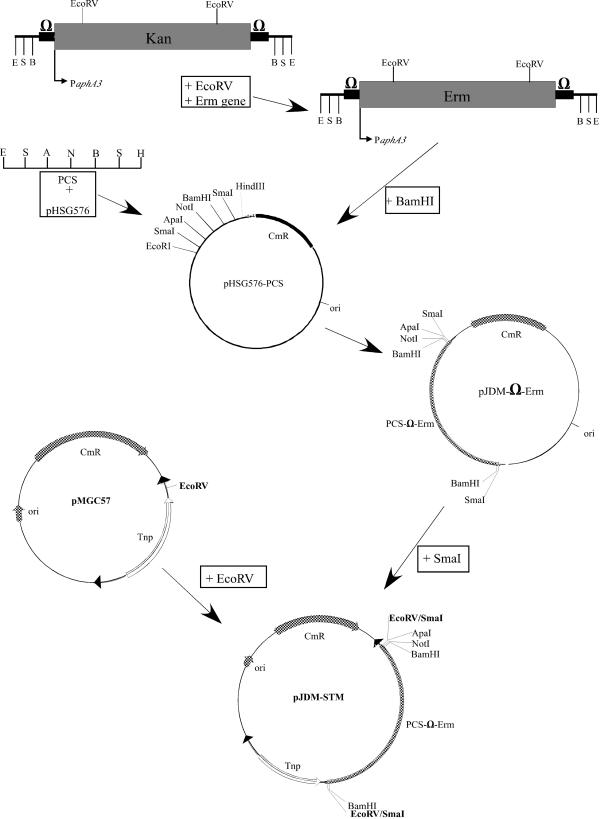
pJDM-STM carries an erythromycin gene within a modified Tn4001 transposon (33) with unique restriction sites for insertion of specific signature tags. Twelve specifically tagged pJDM-STM (Fig. 1) vectors were used to create a signature-tagged transposon library of S. iniae and were constructed as follows.
FIG. 1.
Construction of pJDM-STM vector. pJDM-STM was derived from pMGC57 which contains the modified Tn4001 transposon (34). The erythromycin resistance gene was placed within an omega cassette (39) and modified to contain unique ApaI and NotI restriction sites for insertion of specific signature tags. For details, see Materials and Methods. Sites for restriction enzymes are indicated as follows: B, BamHI; S, SmaI; E, EcoRI; N, NotI; A, ApaI; H, HindIII. PCS, polycloning site; Erm, erythromycin resistance gene; PaphA3; promoter for kanamycin resistance gene aphA3; Kan, kanamycin resistance gene; CmR, chloramphenicol resistance gene; ori, origin.
The aphA3 kanamycin gene from an Ω-kanamycin cassette (39) was removed by restriction digestion with EcoRV, leaving the strong aphA3 promoter intact. The erythromycin gene from pJRS233 (40) was treated to create blunt ends and ligated into the EcoRV sites of the Ω-kanamycin cassette. Insertion of the erythromycin gene in the correct orientation in regard to the aphA3 promoter of the Ω cassette was confirmed by PCR. This created an Ω-erythromycin cassette expressed from the aphA3 promoter.
The polycloning site of pHSG576 (53) was removed by digesting pHSG576 with EcoRI and HindIII and replaced with a DNA fragment containing restriction sites in the following order: EcoRI, SmaI, ApaI, NotI, BamHI, SmaI, and HindIII (Fig. 1). The Ω-erythromycin cassette was then placed into the BamHI site of this vector, creating pJDMΩERM. The Ω-erythromycin cassette along with the ApaI and NotI unique restriction sites was removed from pJDMΩERM by digestion with SmaI and placed into the EcoRV site of pMGC57 (33) creating pJDM-STM.
Signature-tagged transposons were made by ligating double-stranded 34-bp oligonucleotides (Table 1) into the unique ApaI and NotI sites within the transposon by the method of Lehoux et al. (29) and then sequenced to confirm insertion of the correct sequence. This procedure creates pJDM-STM vectors 1 through 12.
TABLE 1.
Signature tag sequences and primers
| Oligonucleotidea | Sequence (5′→3′) |
|---|---|
| STM-1L-fwd | GGCCGCCCGGGTTAAACGTTCAACTTGCAGGGCC |
| STM-1L-rev | CTGCAAGTTGAACGTTTAACCCGGGC |
| STM-1L-Tag | GGGCCCTGCAAGTTGAACG |
| STM-2L-fwd | GGCCGCCCGGGTTAAATAGCCTATAGCCTGGGCC |
| STM-2L-rev | CAGGCTATAGGCTATTTAACCCGGGC |
| STM-2L-Tag | GAGGGCCCAGGCTATAGGC |
| STM-3L-fwd | GGCCGCCCGGGTTAAAAGTCTCAAGTCTCGGGCC |
| STM-3L-rev | CGAGACTTGAGACTTTTAACCCGGGC |
| STM-3L-Tag | GGGCCCGAGACTTGAGAC |
| STM-4L-fwd | GGCCGCCCGGGTTAATAACGTGTAACGTGGGGCC |
| STM-4L-rev | CCACGTTACACGTTATTAACCCGGGC |
| STM-4L-Tag | GAGGGCCCCACGTTACACG |
| STM-5L-fwd | GGCCGCCCGGGTTAAACTGGTAACTGGTAGGGCC |
| STM-5L-rev | CTACCAGTTACCAGTTTAACCCGGGC |
| STM-5L-Tag | GGGCCCTACCAGTTACCAG |
| STM-6L-fwd | GGCCGCCCGGGTTAAGCATGTTGCATGTTGGGCC |
| STM-6L-rev | CAACATGCAACATGCTTAACCCGGGC |
| STM-6L-Tag | GGGCCCAACATGCAACATG |
| STM-7L-fwd | GGCCGCCCGGGTTAATGTAACCCCAATGTGGGCC |
| STM-7L-rev | CACATTGGGGTTACATTAACCCGGGC |
| STM-7L-Tag | GGGCCCACATTGGGGTTAC |
| STM-8L-fwd | GGCCGCCCGGGTTAAAATCTCGGCTCTAAGGGCC |
| STM-8L-rev | CTTAGAGCCGAGATTTTAACCCGGGC |
| STM-8L-Tag | GAGGGCCCTTAGAGCCGAG |
| STM-9L-fwd | GGCCGCCCGGGTTAATAGGCAAAACGGATGGGCC |
| STM-9L-rev | CATCCGTTTTGCCTATTAACCCGGGC |
| STM-9L-Tag | GGGCCCATCCGTTTTGCC |
| STM-10L-fwd | GGCCGCCCGGGTTAACAATCGTTGCTAACGGGCC |
| STM-10L-rev | CGTTAGCAACGATTGTTAACCCGGGC |
| STM-10L-Tag | GAGGGCCCGTTAGCAACG |
| STM-11L-fwd | GGCCGCCCGGGTTAATCAAGACCAGAACTGGGCC |
| STM-11L-rev | CAGTTCTGGTCTTGATTAACCCGGGC |
| STM-11L-Tag | GGGCCCAGTTCTGGTCTTG |
| STM-12L-fwd | GGCCGCCCGGGTTAACTAGTAGGATGATCGGGCC |
| STM-12L-rev | CGATCATCCTACTAGTTAACCCGGGC |
| STM-12L-Tag | GAGGGCCCGATCATCCTAC |
Forward (fwd) and reverse (rev) oligonucleotides were annealed to form the tag that was inserted between the NotI and ApaI sites on pJDM-STM vectors. STM-#L-Tag refers to the specific primer used in the PCR mixtures to determine the presence of a specific mutant clone in the pool.
Generation of S. iniae tagged library.
Mutagenesis utilized a previously reported Tn4001 transposon (33) placed within pJDM-STM that integrates randomly and in single copy into the streptococcal chromosome. The 12 pJDM-STM vectors were purified from E. coli using anion-exchange resin (Invitrogen) and used to transform S. iniae in 12 separate transformations. S. iniae cultures were grown overnight at 37°C in 10 ml of M9 medium supplemented with 20 mM glycine in airtight conical tubes without agitation. The 10-ml cell culture grown overnight was diluted into 100 ml of M9 medium (20 mM glycine) and incubated at 37°C until culture growth reached an optical density at 600 nm (OD600) of approximately 0.140 using a Beckman DU530 spectrophotometer. Cells were then centrifuged at 4°C, resuspended in 2 ml of spent supernatant, and placed at 43°C for 9 min. Eight milliliters of ice-cold sucrose (0.625 M, pH 4.0) was added to the heat-shocked culture. The bacteria were then centrifuged at 4°C and washed twice with 10 ml of cold sucrose. After the final wash, the bacterial pellet was resuspended in 0.6 ml of cold sucrose. Two hundred microliters of the competent cells were added to a microcentrifuge tube containing approximately 5 μg of DNA. Cells and DNA were immediately placed into a cold electro-cuvette and electroporated (Bio-Rad Gene Pulser) using the following settings: voltage at 2.5 kV, capacitance at 25 μF, and resistance at 600 Ω. Cells were immediately removed and placed into 10 ml of cold TP and incubated on ice for 45 min and then at 37°C for 1.5 h. Transformants were selected by plating on TP-Erm2 agar plates and incubated anaerobically at 37°C overnight. Individual transformants of S. iniae were then placed one per well in 96-well plates containing 200 μl of TP-Erm2. Plates (96-well plates) were incubated anaerobically at 37°C overnight and used to make freezer stocks that are kept at −80°C until screening. Following this format, a total of 12 libraries with 95 (1 well used as a medium negative control) mutants per library were prepared.
Preparation of streptococci.
Ten milliliters of TP-Erm2 was inoculated with 10 μl of freezer stock from one well of each of the 12 libraries and incubated at 37°C in airtight conical 15-ml tubes overnight to create pooled mutant bacteria for zebrafish infections. Cultures of streptococci grown overnight were diluted 1:100 in fresh TP-Erm2 and incubated at 37°C to allow cultures to reach mid-log phase. Streptococcal cells were washed once and resuspended in fresh TP to a concentration of 108 CFU/ml (OD600 of 0.225 on a Beckman DU530 spectrophotometer). Streptococcal cells were then diluted to 107 CFU/ml and kept on ice until they were injected. Unused infectious streptococcal culture was kept at 4°C for subsequent analysis of input clones by PCR. The final concentration of infectious bacteria was confirmed by serial dilution and plating on solid medium.
Zebrafish infection studies.
Prior to infection, zebrafish were kept under the established conditions reported previously (38). Groups of four anesthetized zebrafish were injected intramuscularly (i.m.) as outlined previously (38). Each fish was injected with 10 μl of a 107-CFU/ml culture, resulting in a dose of 105 CFU. This concentration is approximately 100-fold higher than the reported 50% lethal dose (LD50) of 103 CFU for infectious S. iniae given i.m. (38). Four fish per experiment were also mock infected with 10 μl of sterile TP. Injected zebrafish were placed in 400-ml glass beakers (with lids) containing 225 ml of sterilized double-distilled water supplemented with 60 mg of aquatic salts (Instant Ocean) per ml and placed in a glass front incubator at 29°C.
PCR and output analyses.
Twenty-four hours postinjection, zebrafish were euthanized in 100 ml of 320-μg/ml Tricaine (3-aminobenzoic acid ethyl ester; Sigma). Brains and hearts were aseptically removed by dissection from two fish of each mutant infection group and homogenized separately in 500 μl of phosphate-buffered saline (PBS). Homogenates were used to inoculate 10 ml of TP-Erm2 to amplify output mutants for subsequent PCR. Cultures were incubated at 37°C for 16 h.
To determine which mutant strains were present in both the pooled organ homogenate cultures and the pooled input cultures, genomic DNA was isolated from all cultures by the following method. After the streptococcal cells were harvested via centrifugation, cells were resuspended in 1 ml of Tris-EDTA buffer (TE) with 10 μl of mutanolysin (5,000 U/ml) and 25 μl of lysozyme (25 mg/ml) and incubated at 37°C for 1 h. Reaction mixtures were then centrifuged, and the supernatant was discarded. The pellet was subjected to a freeze-thaw cycle three times (one freeze-thaw cycle consists of 5 min at −80°C and 5 min at 37°C) and then resuspended in 500 μl of TE with 200 μl of 10% sodium dodecyl sulfate and 250 μl of Tween 20 lysing mix (2% Tween 20, 5% 1 M Tris [pH 8], 25% 0.2 M EDTA). Reaction mixtures were rocked end over end five times and placed at 65°C for 30 min. Ten microliters of RNase A (5 mg/ml) was then added to the reaction mixture, and the reaction mixture was incubated for 30 min at 37°C. Reaction mixtures were then added to Phase Lock Gel tube (catalog no. 0032 005.250; Eppendorf), and DNA was isolated according to the manufacturer's directions. Recovered DNA was resuspended in 50 μl of TE.
PCR was performed on genomic DNA from both input and output pools using each of the 12 primers listed in Table 1 as well as an internal primer for the erythromycin resistance gene, 3′ERM (5′CGTCAATTCCTGCATGTTTTAAGG3′). Reactions were performed in 96-well microtiter plates (Dot Scientific), using touchdown PCR with the following cycling parameters: (i) 5 min at 95°C; (ii) 20 cycles, with each cycle consisting of 30 s at 95°C, 30 s at 60°C (annealing temperature decreased 0.5°C/cycle), and 1 min at 72°C; (iii) an additional 10 cycles, with each cycle consisting of 30 s at 95°C, 30 s at 45°C, and 1 min at 72°C. PCR products were loaded on a 0.8% agarose gel and visualized by ethidium bromide staining.
Sequence analysis of tagged mutants.
Mutant strains that were not found in the output pools after zebrafish infection as determined by PCR analysis were prepared for subsequent sequencing by isolating chromosomal DNA using the Genomic Tip kit (QIAGEN). Sites of transposon insertion were determined by sequencing directly from the purified chromosomal DNA off the end of the inserted transposon into adjacent chromosomal DNA using primer 5′ Tn4001 Rev (5′CTTGGGTCATGTAAAAGTCCTCCTGGGTATG3′). This method not only allowed us to determine the exact site of transposon insertion but also confirmed that there was only a single insertion in the chromosome, as sequencing from double insertions on the chromosomal DNA results in a jumbled sequence. Chromosomal sequencing was performed by Fidelity Systems (Gaithersburg, Md.). Resultant sequences were compared to known sequences in public databases using the National Center for Biotechnology Information (NCBI) BLASTX software available at www.ncbi.nlm.nih.gov (3). All mutant strains were confirmed to be S. iniae by PCR using 16S rRNA primers specific for S. iniae (63).
Competitive assays.
Competitive assays were performed using a previously reported method with a few modifications (24). S. iniae 9117 (wild type) and mutant strains were cultured separately in TP at 37°C overnight. Cells from these cultures were diluted 1:100 and grown to mid-log phase (OD600 of 0.225). All cultures were washed once with PBS, and then bacterial concentrations were adjusted to 107 CFU/ml. Cells were mixed in a 1:1 ratio, pelleted, and then resuspended to a final concentration of 2 × 107 CFU/ml. Zebrafish were infected i.m. with 10 μl of this mixture, resulting in an infectious dose of 2 × 105 CFU. Infectious streptococci were serially diluted and plated on TP and TP-Erm2 to determine the input ratio of mutant to wild type.
After 24 h, hearts and brains were recovered from euthanized fish and homogenized, and the homogenates were plated onto TP and TP-Erm2 to determine the output ratio of mutant to the wild type. The competitive index (CI) was calculated by dividing the output ratio (mutant/wild type) by the input ratio (mutant/wild type). Eight infected fish were used in all competitive assays.
Gradients for measuring buoyant densities.
Linear hypotonic gradients were prepared in 10-ml plastic Falcon tubes using Percoll (Sigma) according to the manufacturer's instructions. First, 1.13-g/ml Percoll was diluted using double-distilled sterile water to make up gradients that covered a density range from 1.03 to 1.13 g/ml. After incubation overnight, 108 CFU of each bacterial strain was resuspended in 100 μl of PBS and gently added to the top of the gradient. Gradients were centrifuged at 4,000 × g for 4 h at 4°C in a swing bucket rotor. Bacteria banded at the level of the gradient at which the densities of the gradient and bacteria were equal.
Fractions were gently removed from gradients in 1-ml aliquots using a large-bore pipette so as not to disturb the gradient. Specific banding patterns of each strain based on the location in the gradient were recorded. Ten-microliter aliquots from each fraction were read on an Abbe refractometer to measure the refractive index. Using the refractometer, numerical values were assigned to each fraction, ranging from 1.03 to 1.13 g/ml. The 1.03- and 1.13-g/ml aliquots of Percoll were used as the endpoint standards.
Gentamicin protection assays.
RAW 264.7 murine macrophages (43) were seeded at ∼5 × 105 cells per well onto a tissue culture-treated glass coverslip (Thermonox; Nunc) in a 24-well plate using Dulbecco modified Eagle medium (DMEM) (Invitrogen) plus 10% fetal bovine serum (FBS) and incubated at 37°C in 5% CO2 overnight to allow adherence. Macrophages were washed three times with PBS, fresh DMEM plus FBS with ∼5 × 106 CFU of S. iniae (multiplicity of infection of 1) was added, and the plates were incubated at 37°C in 5% CO2 for 3 h. The plates were then washed five times with PBS. Fresh DMEM containing FBS supplemented with 100 μg of gentamicin per ml and 5 μg of penicillin per ml was added, and the plates were incubated for 1 h to kill any extracellular bacteria. Cultures incubated for 24 h in the presence of gentamicin, shown in the Intracellular + gentamicin columns in Table 3, were grown in fresh DMEM plus FBS with 10 μg of gentamicin per ml and 1 μg of penicillin per ml added. For those cultures incubated without gentamicin for 24 h, reported in the Intracellular + attached and Extracellular columns in Table 3, fresh DMEM plus FBS without antibiotics was added after the initial 1-h gentamicin treatment. Timing for infections began after the 1-h gentamicin treatment. The coverslips and media were removed at various time points to determine the number of surviving intracellular and extracellular bacteria. For enumeration of intracellular and attached bacteria, PBS-washed coverslips were placed in 5-ml portions of sterile water and vortexed vigorously. Aliquots were plated on TP agar plates in GasPak jars overnight at 37°C. To determine the number of extracellular surviving bacteria, 100-μl aliquots of medium were serially diluted and plated as described above. The number of bacteria counted for intracellular and attached and extracellular bacteria in Table 3 came from the same wells, i.e., these numbers added together equal the total number of bacteria surviving in that particular well. The wells of the intracellular and gentamicin columns in Table 3 had no viable extracellular bacteria.
TABLE 3.
S. iniae infection of murine macrophages
| Time (h) | No. of bacteria (CFU/ml)a
|
|||||
|---|---|---|---|---|---|---|
| 9117
|
G1-6
|
|||||
| Extracellularb | Intracellular + attachedc | Intracellular + gentamicind | Extracellularb | Intracellular + attachedc | Intracellular + gentamicind | |
| 2 | 4.30 × 102 ± 85 | 1.19 × 104 ± 2.11 × 103 | 1.13 × 104 ± 1.5 × 103 | 2.68 × 102 ± 51 | 2.08 × 104 ± 4.76 × 10 | 1.51 × 104 ± 3.85 × 103 |
| 4 | 6.70 × 103 ± 4.83 × 103 | 1.14 × 104 ± 8.41 × 103 | 9.21 × 103 ± 2.5 × 103 | 1.30 × 103 ± 6.14 × 102 | 1.01 × 104 ± 2.88 × 103 | 6.38 × 103 ± 1.31 × 103 |
| 24 | 3.72 × 107 ± 1.71 × 107 | 3.02 × 107 ± 8.48 × 106 | 0 | 1.37 × 107 ± 4.12 × 106 | 7.91 × 106 ± 1.01 × 106 | 0 |
All infections were performed with a 1-h incubation with gentamicin (100 μg/ml) and penicillin (5 μg/ml) to kill extracellular bacteria. Timing of infections began after this 1-h incubation. The medium was then replaced with either fresh medium without antibiotics (Intracellular + attached) or fresh medium containing 5 μg of gentamicin per ml and 1 μg of penicillin per ml for 24 h (Intracellular + gentamicin). Results are presented as means ± standard errors of the means from four macrophage infection experiments.
Extracellular numbers refer to the number of viable bacteria that have reemerged from the intracellular environment into the culture medium.
Intracellular + attached numbers refer to the viable intracellular bacteria and those that are strongly attached to the outside of the cell. Extracellular numbers and Intracellular + attached numbers come from the same well.
Intracellular + gentamicin numbers only refer to viable intracellular bacteria. No viable extracellular bacteria were recovered from these wells.
Whole-blood assays.
Survival in whole blood was performed using a modified version of the Lancefield bactericidal assay for S. pyogenes (27). Log-phase bacteria were washed and diluted in sterile PBS. One hundred CFU was added to 1-ml samples of fresh heparinized human whole blood in sterile glass tubes and incubated on a rocker for 1.5 h at 37°C. After incubation, 100 μl was removed from the tubes, and aliquots were plated for enumeration. The percent survival was calculated as the CFU recovered divided by the initial inoculum.
Statistical analysis.
Statistical significance between paired samples was determined using a two-tailed paired t test using the StatView statistical analysis software (SAS Institute Inc., Cary, N.C.).
RESULTS
Construction of signature-tagged S. iniae library.
Plasmid pJDM-STM (Fig. 1), carrying a modified Tn4001 transposon (33), was constructed for signature-tagged mutagenesis of S. iniae strain 9117 (13). Twelve 34-bp oligonucleotide signature tags (Table 1) were incorporated into the unique ApaI and NotI sites of Tn4001 to create 12 individually tagged pJDM-STM vectors. The tagged vectors were used to create 12 libraries of S. iniae, each containing 95 transposon mutants with a specific tag as described in Materials and Methods. Southern analysis was used to confirm the randomness of transposon insertion in the mutant libraries using a DNA fragment of Tn4001 as a probe. One mutant of each tag type was selected for the analysis, which demonstrated that each mutant had a single transposon insertion in a unique site within the chromosome (data not shown). Signature-tagged analysis was performed using a modified version of the PCR technique first reported by Lehoux et al. (29) (see Materials and Methods) in which primers specific for each tag were used to determine the presence or absence of one of the 12 specific tags in a complex pool. Primers for each tag were tested for specificity by checking for cross-hybridization between tags during PCR. Touchdown PCR parameters were optimized to detect only the tags present in the testing pool and confirmed that no fragment was made when using primers for DNA not present in the testing pool (see Materials and Methods). No cross-hybridization was detected between tags (data not shown).
Screening of mutant clones.
The S. iniae signature-tagged mutant libraries were analyzed for attenuated virulence using the zebrafish infection model (38). We demonstrated previously that injection of S. iniae into the dorsal muscle of zebrafish resulted in a systemic infection. Streptococci could be isolated from the skin, heart, brain, and gall bladder within 24 to 44 h postinjection (38). However, in the present study, it was necessary to optimize the infection procedure, since we were screening multiple strains concurrently with the desired result that all the clones in the assay that could produce an infection did so simultaneously in all organs. We optimized the technique for pool size, inoculum level, time of bacterial recovery, and specific organs for bacterial recovery. In our infection model, this was particularly important, because we wished to identify mutants that were deficient for dissemination to the brain. Previous reports have suggested that passage through the blood-brain barrier may be a limiting step, thereby giving rise to clonal infections, which therefore are most likely not suitable for the application of STM (49).
Therefore, to determine the optimal infectious dose and recovery time of bacteria from infected organs, multiple concentrations of pooled bacteria were analyzed. Using a pool of 12 transposon mutants that were previously shown to produce a wild-type infection when injected separately, we tested doses of 106, 105, and 104 of total CFU by i.m. injection. A dose of 105 CFU proved to be the optimal dose, as all 12 mutants could be collected from both the heart and brain at 24 h, while 106 CFU killed the fish too rapidly (less than 24 h) and an infection dose of 104 did not allow all 12 mutants to disseminate to all organs. The infectious dose of 105 CFU chosen for the screening of mutant clones is approximately 100 times the LD50 for wild-type S. iniae 9117 strain in zebrafish (38).
Zebrafish were injected i.m., and infections were allowed to proceed for no longer than 24 h, as this was determined to be the optimal time for harvesting the organs of infected zebrafish. Previous experiments of timed infection trials showed that dissemination to all organs occurred between 22 and 26 h postinoculation (38). We confirmed this finding with the complex pool of 12 mutants showing that shorter infection times did not consistently reproduce a systemic infection, while longer infection times, at this inoculum, often proved to be lethal. Using the 105 concentration of inoculum, initial analyses with 12 tagged mutants confirmed that the 12 mutants injected into the dorsal muscle could subsequently be recovered from the brain within 24 h, thereby confirming that this pool size and infectious dose did not result in a clonal infection of the brain.
After experimental optimization, signature-tagged mutants were combined in pools of 12 with one mutant taken from each of the 12 libraries and injected into four zebrafish. At 24 h, the fish were examined for signs of infection. Two fish from each infection pool were euthanized, and streptococci were aseptically recovered from the hearts and brains of infected fish. The decision of which two fish were chosen for analysis was based on those displaying obvious symptoms of S. iniae disease, i.e., lethargy, swimming at the surface of the water, and any obvious signs of necrosis at the injection site. Recovered bacteria were grown in selective medium, and isolated chromosomal DNA was probed for specific tags using the PCR technique described in Materials and Methods.
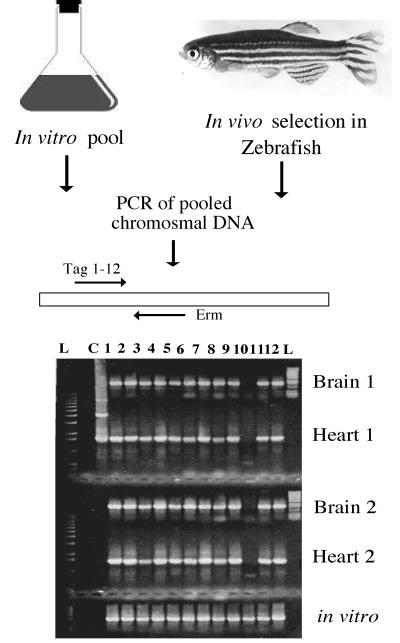
Using these optimal conditions, 1,128 transposon mutants were screened using the zebrafish animal model. Isolates with tags present in the in vitro input pool but absent from the in vivo output pools, as determined by the absence of a PCR fragment for that particular tag, were considered potential virulence mutants (Fig. 2). The isolates could be divided into five individual groups based on the profile of the missing tags. The first group included those isolates found to be missing from both the heart and brain of both fish examined (all four organs), which consisted of 41 potential virulence mutants. The other four groups included those missing from three of four organs of both fish (19 isolates), missing from only the brain of both fish (11 isolates), missing in both the heart and brain of only one fish (47 isolates), or missing in only one organ of one fish (108 isolates).
FIG. 2.
Signature-tagged mutagenesis strategy using the zebrafish model. Mutant strains are pooled in groups of 12 and grown to mid-log phase. An inoculum of 105 CFU of pooled bacteria is then injected into the dorsal muscle of zebrafish. After 24 h, fish are euthanized, and bacteria are recovered from homogenized hearts and brains of two fish (fish 1 and 2). Chromosomal DNA isolated from cultures is subjected to PCR to screen for the presence of all 12 signature tags (lanes 1 to 12) in both the in vitro input pool and in vivo output pool. The gel illustrates an experiment in which the PCR fragment for the STM-10 clone is missing in all four in vivo samples. Visualization on the gel of all five reaction mixtures is accomplished by loading the gel twice with the amplification products from the heart and brain samples, which are loaded into the same wells at different times. The DNA ladder (lane L) on the left of the gel and the positive control (lane C) amplified from an STM plasmid (extra bands in the control lane are the supercoiled forms of the plasmid) were loaded at the same time as the PCR products from the heart samples. Approximately 45 to 60 min after loading and electrophoresing the PCR products from the heart samples, the DNA ladder (L) on the right of the gel is loaded along with the PCR products from the brain samples. The in vitro amplification products are loaded at the same time as the PCR products from the brain samples.
Sequence analysis of virulence-attenuated clones.
The first group mentioned above, isolates missing from both organs of both fish, were given priority for further analysis because of their overall attenuation and because they represented 3.6% of all isolates screened in the zebrafish model. Of the 41 potential virulence mutants that were sequenced, 28 showed significant homology to genes encoding proteins found in other gram-positive pathogens (Table 2). A large portion of these mutants had insertions in open reading frames with high homology to genes coding for surface polysaccharide synthesis. Eight mutants had transposon insertions in open reading frames encoding products of unknown function from other gram-positive pathogens, and an additional 13 mutants showed no significant homology to any sequences in the databases (Table 2).
TABLE 2.
S. iniae sequences identified by signature-tagged mutagenesisa
| Classification | Mutant | Homologyb | Proposed functionc | % Identity (% homology)/spand |
|---|---|---|---|---|
| Cellular processes | F2-12e | Putative serine protease of the DegP/HtrA family (S. pyogenes, spyM18_2256) | Protease | 68(80)/224 |
| H6-4e | Putative serine protease of the DegP/HtrA family (S. pyogenes, spyM18_2256) | Protease | 76(90)/176 | |
| C7-5 | PurB (S. pyogenes, Spy0036, S. agalactiae, SAG0047) | Adenylosuccinate lyase | 94(98)/238 | |
| E7-1 | Putative dehydrogenase (S. pyogenes, spyM18_0608) | Nucleotide sugar dehydrogenase | 93(100)/48 | |
| G10-11 | Putative hydrolase (S. pyogenes, SpyM3_1547, S. agalactiae, SAG1678) | Haloacid, dehalogenase-like family | 73(85)/146 | |
| Cell envelope | F12-1 | EPS11J (S. thermophilus) | Capsule synthesis | 31(56)/180 |
| E4-5 | EPS11J (S. thermophilus) | Capsule synthesis | 42(62)/105 | |
| G6-8 | EPS11K (S. thermophilus) | Capsule synthesis | 50(69)/208 | |
| G1-6 | Hypothetical protein (B. thethaiotaomicron, BT1178) | Capsule synthesis | 30(57)/59 | |
| C6-1 | Hypothetical protein (B. thethaiotaomicron, BT1178) | Capsule synthesis | 24(48)/161 | |
| E6-9 | EPS5N (S. thermophilus) | Capsule synthesis | 32(52)/283 | |
| D5-5 | EPSU (S. thermophilus) | Capsule synthesis | 32(57)/236 | |
| B9-8 | FmhC (S. aureus) | Cell wall synthesis | 23(40)/256 | |
| Regulators | F5-11e | Putative transcription regulator of the TetR/AcrR family (S. mutans, SMU.1349) | Transcription regulator | 29(49)/171 |
| B9-11e | Putative transcription regulator of the TetR/AcrR family (S. mutans, SMU.1349) | Transcription regulator | 29(49)/171 | |
| Transport or binding | G8-2 | Putative sodium/decarboxylate symporter (S. pyogenes, SPy0324) | Ion transport | 55(68)/218 |
| H5-10 | ABC transporter/lantibiotic transport (Clostridium tetani, CTC00625) | ABC transporter | 64(82)/191 | |
| E12-6 | PTSf system, IIC component (S. agalactiae, SAG1899) | Phosphotransferase system | 79(82)/195 | |
| F10-12 | Mg transport, CorA family (Listeria monocytogenes, LMOh7858_0618) | Mg transport | 45(63)/172 | |
| Transposable elements | A6-2 | Transposase (Lactococcus lactis, umb1) | 98(100)/86 | |
| Conserved hypothetical | A6-6e | S. pyogenes, Spy2215 upstream of htrA | Unknown | 73(85)/159 |
| G8-10e | S. pyogenes, Spy2215 upstream of htrA | Unknown | 73(85)/159 | |
| A4-4e | S. agalactiae, GBS0237 similar to integrase | Unknown | 72(87)/33 | |
| B7-4e | S. agalactiae, GBS0237 similar to integrase | Unknown | 63(77)/88 | |
| D3-10 | S. pyogenes, Spy0316 | Unknown | 85(90)/216 | |
| F6-10 | S. pneumoniae, SP1706 | Unknown | 28(48)/226 | |
| G12-10 | S. agalactiae, gbs1976 | Unknown | 50(73)/52 | |
| H10-9 | Streptococcus mutans, SMU.1349 | Unknown | 36(60)/99 |
Thirteen sequences had no significant homology. DNA flanking the site of insertion showed no significant homology to any sequences found in the databases.
Homology scores with a probability of ≤e−7 were considered significant. Homologies were found by using the NCBI BLASTX translating search program. The highest homology returned for each of our input sequences is listed in the table.
Based on functions assigned to homologous proteins returned in database searches.
Percent identity (percent homology) is at the amino acid level over the length of the span (number of amino acids) shown.
The following pairs of mutants were found to have insertions in the same locus: F2-12 and H6-4, F5-11 and B9-11, A6-6 and G8-10, and A4-4 and B7-4. However, all of these insertions occur within different regions of the protein.
PTS, phosphotransferase system.
Competitive assays with selected mutants.
Signature-tagged mutagenesis not only identifies mutant clones that are unable to survive in the host environment but also measures the ability to survive in vivo while competing for nutrients within a complex pool of clones. To rule out the possibility that the mutants identified in the screen had a general growth defect, in vitro growth assays were performed in both rich (TP) and minimal (M9) media. Only one mutant listed in Table 2 (F6-10) showed a growth defect in minimal medium, and all other mutants showed essentially wild-type growth in both minimal and rich media (data not shown).
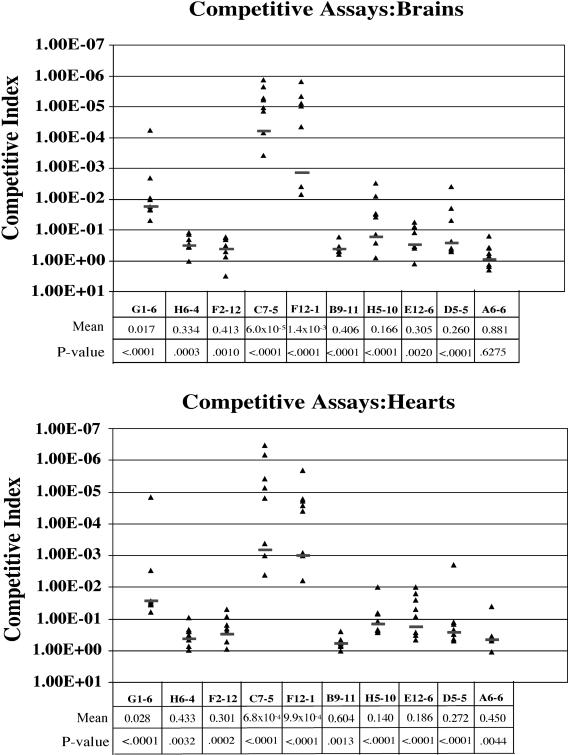
To quantify the attenuation of the mutants identified in the screen, 10 mutants were chosen for competitive assays with the wild-type strain as described in Materials and Methods. The 10 mutants were selected on the basis of the proposed protein functions suggested by their homology to other genes in the databases to examine the attenuation of a variety of different mutants recovered in our screen. The analyses were performed using eight fish per assay with the mutant culture mixed with the wild-type culture at a 1:1 ratio for a total i.m. injection of 2 × 105 CFU. Bacteria were aseptically recovered from both the heart and brain at 24 h postinjection by homogenization of the organs. A CI of less than 1 suggests that the virulence of the mutant clone was attenuated (Fig. 3).
FIG. 3.
Competitive assays of selected STM mutants. CIs are calculated against S. iniae wild-type strain 9117, as described in Materials and Methods. CI is defined as the output ratio (mutant/wild type) divided by the input ratio (mutant/wild type). A CI of less than 1 indicates a quantitative degree of attenuation in virulence of the mutant compared to the virulence of the wild type. CIs were determined individually for mutant strains recovered from both hearts and brains for each STM mutant assayed. Each triangle represents one individual fish experiment. The mean of all CI experiments is listed beneath each mutant label, along with the P value in comparison to the wild-type value. Black bars indicate mean values in eight individual fish experiments.
The data from the competitive assays illustrate that the STM screen was able to identify mutant strains with a wide range of attenuation, with only one mutant showing less than statistical significance in one organ (A6-6 brain). The other CIs ranged from extremely attenuated (C7-5, 6 × 10−5) to a minimal, but statistically significant, amount of attenuation (B9-11 heart, 0.604) demonstrating that the optimized STM screen was quite sensitive. Although there was as much as a twofold difference in the CIs of individual mutants isolated from the brain or heart, none of the differences were statistically significant.
Characterization of capsule mutants.
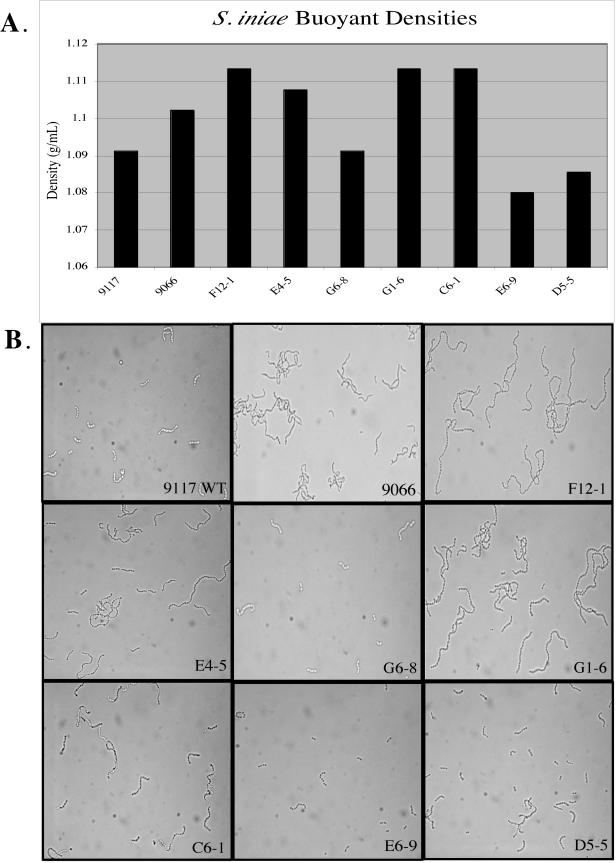
Five of the 41 attenuated clones identified in this group had insertions in open reading frames showing homology to surface polysaccharide synthesis genes from several gram-positive pathogens (Table 2). None of the insertions occurred at the same site, which suggests that the large number of mutations isolated at this locus was not the result of a hot spot for Tn4001 transposon insertion but that the capsular polysaccharide of S. iniae is of major importance to the pathogenicity of the bacterium in vivo. Three of the identified capsular polysaccharide mutants, E4-5, F12-1, and D5-5, showed a distinct phenotype when grown in broth, although there was no difference in their appearance when they were grown on solid medium. While wild-type S. iniae remains buoyant in broth, these mutants showed various degrees of aggregation. Capsule expression has been shown to greatly influence the buoyant density of bacteria (18).
Visualizing the aggregated cultures with light microscopy showed a striking difference in the morphology of the wild type and mutants. While wild-type S. iniae 9117 was found in chains of 4 to 10 cocci, aggregated mutants E4-5 and F12-1 were found in chains of 20 to 40 cocci (Fig. 4B). This phenotype is highly similar to the growth characteristics of a commensal strain of S. iniae, strain 9066 (Fig. 4B) (13). Mutant D5-5 shows various chain lengths and does not aggregate to the degree of the other two mutants but shows more aggregation than wild-type 9117. Two other mutants isolated in the STM screen, G1-6 and C6-1, also showed aggregation in broth culture as well as the long-chain phenotype, although sequence comparisons showed the strongest homology to a hypothetical gene from Bacteroides thetaiotaomicron (Table 2). However, the sequences on either side of this hypothetical gene in the B. thetaiotaomicron genome (accession no. AE016930, locus BT1178) were identified as genes involved in capsule synthesis.
FIG. 4.
Buoyant density profiles of S. iniae wild-type strains 9117 and 9066 and putative 9117 capsule mutants. A. Chart showing specific buoyant densities of wild-type and STM mutants in a Percoll gradient with endpoints of 1.03 to 1.13 mg/liter. Individual strains were added to the top of a gradient and centrifuged, facilitating separation to a specific density, which correlates to the amount of capsule expression. Strains with ample encapsulation have low buoyant density and will form bands in the lower concentration of the gradient, while strains with little or no encapsulation have high buoyant density and will separate out at a higher density. B. Photomicrographs of wild-type (WT) strain 9117 and putative capsule mutants illustrating the different chain lengths. Magnification, ×1,000.
Capsular phenotypes of group B streptococcus clinical isolates have been characterized on the basis of the inverse correlation between buoyant density and capsule thickness (48). Highly encapsulated strains were found to have a low density, while minimal encapsulation resulted in a high density using a linear, hypotonic density gradient. Using this technique (48), the seven putative capsule mutants mentioned above, along with wild-type S. iniae 9117 and the commensal strain, 9066, were analyzed for buoyant densities by centrifugation on a linear Percoll (Sigma) density gradient (Fig. 4A). As expected, the results of the density gradient illustrate that clones F12-1 and E4-5, as well as the commensal strain 9066, have much higher buoyant densities, correlating with less capsule production, than wild-type 9117 does. In addition, G1-6 and C6-1 also showed much higher buoyant densities, suggesting that these genes are also involved in capsule synthesis. Surprisingly, G6-8 exhibited the wild-type level of buoyant density, while E6-9 and D5-5 had buoyant densities lower than that of the wild type, which suggests that these strains may be highly encapsulated.
Group B streptococci, as well as other encapsulated strains, have a greater ability to avoid phagocytosis compared to nonencapsulated isolates (17). Previously, we showed that S. iniae has the ability to survive in zebrafish hepatocytes in tissue culture as well as a murine macrophage cell line (37). To determine whether the capsule mutants of S. iniae retain this ability to invade and survive in macrophages, we performed gentamicin protection assays, which assess the ability of bacteria to invade and survive in the intracellular environment. These experiments were designed to test overall survival in the intracellular environment up to 24 h (Table 3, Intracellular + gentamicin columns), as well as the ability of the intracellular bacteria to survive and reemerge from the intracellular environment to reestablish infection (Table 3, Intracellular + attached and Extracellular columns). The STM mutant G1-6 was used for these assays, as it showed one of the highest buoyant densities (Fig. 4A). Although there were some minor differences observed between the two strains tested, overall survival of the G1-6 mutant after 24 h with gentamicin-treated medium or reemergence and viability without gentamicin was not significantly different than that of wild-type strain (Table 3). Therefore, even though the capsule may limit phagocytosis, overall survival in this environment does not appear to be dependent on the capsule (Table 3).
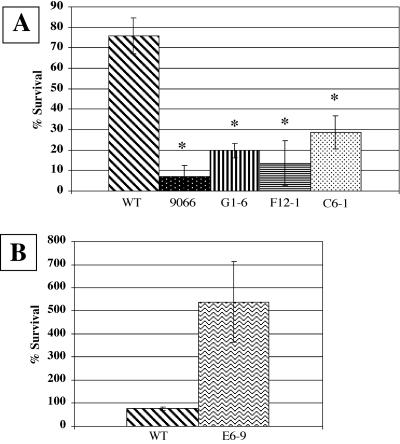
Fuller et al. (13) previously showed that the S. iniae 9066 commensal strain, which has a higher buoyant density than the virulent 9117 strain (Fig. 4), was more readily phagocytosed in fresh human whole blood, while strain 9117 was much more resistant to phagocytosis. This led us to question whether the 9117 capsule mutants would also be phagocytosed more readily. As shown in Fig. 5A, the capsule mutants G1-6, F12-1, and C6-1, which all have higher buoyant densities than the wild type, have lost the phagocytosis-resistant property displayed by the parent strain, 9117. While strain 9117 showed approximately 76% survival from phagocytosis, the three capsule mutants showed between 13 and 29% survival in whole blood. The phagocytosis results with the capsule mutants are very similar to what was observed for the 9066 commensal strain, which showed approximately 7% survival. The E6-9 mutant, which had a lower buoyant density than wild-type 9117, suggesting higher capsule production (Fig. 4), was also tested in the whole-blood assay. This mutant demonstrated even greater resistance to phagocytosis than the wild-type strain, as well as multiplication under these conditions (Fig. 5B). Taken together, these results suggest that the capsule may play an important protective role under these conditions, allowing greater survival in whole blood.
FIG. 5.
Whole-blood phagocytosis assay of S. iniae strains. The data are shown as percent survival of the indicated strain relative to the initial inoculum after 1.5 h at 37°C in fresh heparinized human whole blood. (A) Results with virulent strain 9117 (wild type [WT]), commensal strain 9066, and three 9117 capsular mutants, G1-6, F12-1, and C6-1. (B) Results comparing virulent strain 9117 (wild type [WT]) and mutant E6-9, which expresses excess capsule. Results are reported as means ± standard errors of the means (error bars) from six separate experiments. Values that are significantly different (P < 0.05) from the WT value are indicated by an asterisk.
DISCUSSION
Gram-positive pathogens are responsible for a large portion of systemic infections in humans, and many of these infections result in an alarmingly high incidence of mortality with very short incubation periods. We do not yet have a complete understanding of the factors in vivo that influence whether an infection is a superficial and self-limiting disease or progresses to a more serious, life-threatening disease. Furthermore, we do not understand what mechanisms allow a pathogen to survive, thrive, and disseminate in multiple tissue types. Identification and characterization of genes in both the bacterium and host that are specifically required for the establishment and subsequent dissemination of bacterial pathogens in systemic infections is integral to understanding the infection process. In this study, we have exploited the previously reported zebrafish infectious disease model (38) for studying a natural fish pathogen, which can also infect humans, to determine streptococcal genes required for systemic infection using signature-tagged mutagenesis.
This study represents the first genome-wide screen for virulence genes in S. iniae, as well as the first use of the zebrafish animal model for such a screen. We analyzed 1,128 S. iniae mutants in the zebrafish model and identified 41 potential mutants with attenuated survival in the host environment. In addition, some infections with several mutant pools demonstrated considerable variation of attenuation in virulence in the infected fish. The slight differences in the immune systems of individual zebrafish were most likely the cause of this variability, as the fish population is not inbred. However, the ability of the screen to identify genes with even a very small attenuation in virulence (which was not only statistically significant but also highly reproducible, even in an outbred animal model) validates the sensitivity of the screen.
While our main goal in this analysis was to determine the genes required for survival during a systemic infection by examination of hearts and brains for clearance of mutants, our method of inoculation by injection into the dorsal muscle also allows for identification of genes required for dissemination of the organism to an area which will allow systemic spread. Many previous STM screens bypassed this step by injecting mutants intraperitoneally (24, 28, 35) or intravenously (6), although much valuable information has come from these screens. By using the zebrafish model, we can capitalize on the natural relationship between the host and pathogen to observe a systemic infection that occurs through inoculation at a distant site, requiring the pathogen to encounter numerous host responses in a variety of tissues before disseminating systemically. Several of the mutants identified in this screen are currently being analyzed for tissue specificity in this systemic model of infection.
Although the S. iniae genome has not been sequenced, we have taken advantage of the multiple sequenced streptococcal genomes to postulate functions of our identified S. iniae mutants. Database searches revealed that 61% of the mutants identified in this screen show significant homology (a statistical significance threshold E value of <e−7) with genes from S. pyogenes, S. agalactiae, Streptococcus mutans, or S. pneumoniae, emphasizing the genetic relatedness between S. iniae and these other streptococcal species (data not shown). Recent findings reinforce this similarity, as S. iniae has multiple genetic homologs encoding virulence genes found in these other streptococcal species, including beta-hemolysin (14), CAMP factor, hyaluronidase, capsule, and C5a peptidase (M. N. Neely and J. D. Miller, unpublished results).
One protein that is highly conserved across all bacterial species is the DegP or HtrA serine protease. We isolated two independent insertions in an open reading frame with high homology to the gene encoding this protein. Two additional mutants (G8-10 and A4-4) have insertions in a region identified as being homologous to a conserved hypothetical protein in S. pyogenes (Spy2215) that is located directly upstream of the htrA gene in the S. pyogenes genome. The degP gene product has been shown to be involved in degradation of misfolded proteins in the bacterial cell (9, 25). degP knockout strains of E. coli are unable to grow at temperatures above 42°C as a result of this dependence on the protease to clear heat-damaged proteins (31). Neither one of our degP/htrA homolog mutants showed decreased growth due to elevated temperatures compared to the wild type (data not shown). In addition, after exposure to methyl viologen to test for sensitivity to oxidative stress, no growth inhibition of these mutants was observed, as has been reported previously for S. pyogenes (25) degP mutants (data not shown).
One virulence study with S. pyogenes demonstrated a temperature-dependent growth defect with an insertionally inactivated degP mutant and also showed that a functional DegP protease is necessary for full virulence in a murine model of streptococcal infection (25). In addition, an S. pneumoniae htrA mutant showed attenuated virulence in an extensive STM screen of a murine model of lung infection (19). Recently, however, Lyon and Caparon (32) suggested that this temperature sensitivity in S. pyogenes is a result of polar effects, as an in-frame deletion of htrA (degP) is not thermally sensitive. Therefore, although two separate mutations in the same locus were identified in our screen, suggesting its importance for virulence, and the degP homolog (H6-4) showed an approximately threefold decrease in virulence as shown by competitive assay (Fig. 3), the reason for the attenuation in virulence is not clear and is still under investigation.
We also isolated mutants with insertions in genes encoding proteins with basic metabolic functions, such as E7-1, encoding a putative nucleotide sugar dehydrogenase, and C7-5, encoding a putative adenylosuccinate lyase, which salvages purines from the host environment (1). In addition to the S. pyogenes adenylosuccinate lyase listed in Table 2, C7-5 also has very high homology to purB from S. pneumoniae strain R6 (Spr0056), which was recently determined to be required for virulence in a murine model of STM (28). The C7-5 mutant also had one of the most attenuated virulence phenotypes in our competitive assays (6.0 × 10−5 [Fig. 3]), although it did not display attenuated growth in vitro when it was grown on minimal medium. Although these genes are involved in basic metabolic functions, they have been found in multiple other STM screens for pathogenic bacteria (5, 28, 35) and may provide important information on the nutritional requirements of bacteria in vivo.
Exopolysaccharide (EPS) and capsular polysaccharide (CPS) play dominant roles in the pathogenesis of many gram-positive pathogens (4, 20, 45, 46, 50). The capsule has been suggested to be the single most important virulence factor in S. agalactiae pathogenesis (50). S. agalactiae polysaccharide capsule inhibits phagocytosis (15) as well as the deposition of complement component C3 and the activation of the alternative complement pathway (10, 41). Similarly, S. pneumoniae was shown to have a requirement for the capsule in colonization as well as systemic infection in a mouse model of pathogenesis (34). S. pyogenes hyaluronic acid capsule is also required for resistance to phagocytosis (56) and can act as an adhesin, binding to CD44 expressed on the surfaces of pharyngeal epithelial cells (7, 47). The importance of the S. pyogenes capsule for pharyngeal colonization and survival in the upper respiratory tract of a murine infection model was also demonstrated (23, 56). Inactivation of the two-component regulator CsrR/CsrS (CovR/CovS), which results in a highly encapsulated strain from derepression of capsule synthesis, has also been shown to have enhanced virulence in mouse models of soft tissue infection (12, 30, 44). Taken together, these studies highlight the importance of the capsule for streptococcal virulence.
The results reported here highlight that the capsular polysaccharide plays an important role in the pathogenesis of S. iniae as well, as 7 of 41 of the potential virulence mutants cleared by the host through the STM screen have insertions in multiple putative genes involved in capsular synthesis (Table 2). The transposon insertions all occurred either in different open reading frames involved in capsule synthesis or at different sites within the same open reading frame suggesting that this is not just a hot spot for Tn4001 transposon insertion. In addition, the screening of only 1,128 S. iniae mutants, which is less than what would be estimated to completely cover the genome, makes it unlikely that we would have saturated the system. Rather, it suggests that these genes are highly important for virulence of S. iniae in our systemic infection model and that inactivation or modification of any one of several genes within this region results in attenuation of virulence.
The capsule gene cluster that shares the strongest homology to multiple S. iniae putative capsule genes is the EPS operon of S. thermophilus (AF454496). Mutants D5-5, E6-9, G6-8, E4-5, and F12-1 all share homology with genes from the exopolysaccharide synthesis (EPS/CPS) operon of S. thermophilus (52), while the G1-6 and C6-1 mutants have insertions in different regions of a hypothetical gene in the capsule gene cluster of B. thetaiotaomicron. Both of these mutants also had the long-chain and aggregated phenotype observed in other putative capsule mutants from this screen that could be identified by homology as being involved in capsule synthesis. Our initial characterization of the G1-6 mutant showed a nearly 2-log-unit attenuation in virulence as measured by competitive assay (Fig. 3); however, this mutant was not significantly different from the wild type when tested for survival in macrophages (Table 3). However, the G1-6, F12-1, and C6-1 capsule mutants demonstrated a loss of resistance to phagocytosis in the whole-blood assay (Fig. 5), suggesting that the product of this gene (and possibly downstream genes) is required for survival during this phase of infection. The capsule mutants in this assay showed a somewhat higher resistance to phagocytosis than the 9066 commensal strain (13 to 29% compared to 7% for strain 9066) while also showing a higher buoyant density than the density of strain 9066 (Fig. 4). A higher buoyant density suggests less capsule production, which would imply that they should have less resistance to phagocytosis than 9066. However, 9066 is a different strain of S. iniae than 9117, and therefore, the capsule mutants generated from strain 9117 may be missing other genes required for survival under these conditions.
Surprisingly, the E6-9 capsule mutant showed the lowest buoyant density of any strain tested, including the parent strain (9117), suggesting that this strain has excess capsule on its surface. Consistent with this result is the greater resistance to phagocytosis demonstrated by this mutant in the whole-blood assays (Fig. 5). A homology search identified the disrupted gene in the E6-9 mutant as a putative polysaccharide transporter (eps5N of S. thermophilus; AAN63709). Given that mutants showing less capsule synthesis were identified as having attenuated virulence in our STM screen, a mutant having additional capsule might be expected to be more virulent than the parent strain and therefore should have been present in the output screen. However, similar to E6-9, capsule mutant D5-5 showed excess capsule production compared to the wild type (Fig. 4) but still showed significant attenuation in virulence in the competitive assays (Fig. 3). These results suggest that tight regulation of capsule expression must be maintained in vivo for a successful infection and that excess capsule may be as detrimental as too little capsule. Further studies are under way to characterize these mutants and the entire capsule operon of S. iniae to determine the roles of the genes in virulence.
Approximately 20% of the mutant isolates cleared by the host in this STM screen had insertions in sequences found in other gram-positive pathogens that were designated conserved hypothetical genes. An additional 32% of mutants found in this screen were found to have no significant homology to any other known bacterial gene products found in the sequence databases. These particular gene products may be involved in requirements specifically for infection in the fish host environment. Since S. iniae is a zoonotic pathogen, it may have alternative virulence mechanisms that are not found or required for infection by the human-specific gram-positive pathogens. Alternatively, since several other STM screens performed on human-specific gram-positive pathogens, such as S. agalactiae (24), Staphylococcus aureus (35), and S. pneumoniae (19, 28), have also identified a large percentage of genes involved in virulence with no known function or significant homology to genes in the databases, this likely serves to highlight that we still do not have a complete understanding of the multiple mechanisms involved in streptococcal pathogenesis. Further characterization of these unknown genes may provide new insights into alternative virulence strategies that may have relevance to multiple gram-positive pathogens.
In this study, we have identified numerous genes that are important for the virulence of the zoonotic pathogen S. iniae and determined that the majority of these genes are conserved in gram-positive pathogens. The application of an STM screen to the zebrafish animal model further validates the utility of this system as an effective and relevant model for the study of bacterial pathogenesis and host-pathogen interactions.
Acknowledgments
We thank Matthew Jackson, David Friedman, and Dave Hendrixson for critical reading of the manuscript. Special thanks also goes to Donna Runft for expert technical assistance.
This work was supported in part by the American Heart Association Scientist Development grant 0235039N and NIH-NIAID grant AI-52141.
Editor: V. J. DiRita
REFERENCES
- 1.Aimi, J., J. Badylak, J. Williams, Z. D. Chen, H. Zalkin, and J. E. Dixon. 1990. Cloning of a cDNA encoding adenylosuccinate lyase by functional complementation in Escherichia coli. J. Biol. Chem. 265:9011-9014. [PubMed] [Google Scholar]
- 2.Altmann, S. M., M. T. Mellon, D. L. Distel, and C. H. Kim. 2003. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J. Virol. 77:1992-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisno, A. L., M. O. Brito, and C. M. Collins. 2003. Molecular basis of group A streptococcal virulence. Lancet Infect. Dis. 3:191-200. [DOI] [PubMed] [Google Scholar]
- 5.Chiang, S. L., and J. J. Mekalanos. 1998. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonisation. Mol. Microbiol. 27:797-805. [DOI] [PubMed] [Google Scholar]
- 6.Coulter, S. N., W. R. Schwan, E. Ng, M. H. Langhome, H. D. Ritchie, S. Westbrock-Wadman, W. O. Hufnagle, K. R. Folger, A. S. Bayer, and C. K. Stover. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30:393-404. [DOI] [PubMed] [Google Scholar]
- 7.Cywes, C., I. Stamenkovic, and M. R. Wessels. 2000. CD44 as a receptor for colonization of the pharynx by group A Streptococcus. J. Clin. Investig. 106:995-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darwin, A. J., and V. L. Miller. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 32:51-62. [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Torres, M. L., and R. R. Russell. 2001. HtrA protease and processing of extracellular proteins of Streptococcus mutans. FEMS Microbiol. Lett. 204:23-28. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, M. S., M. R. Wessels, and C. J. Baker. 1993. Capsular polysaccharide regulates neutrophil complement receptor interactions with type III group B streptococci. Infect. Immun. 61:2866-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eldar, A., and C. Ghittino. 1999. Lactococcus garvieae and Streptococcus iniae infections in rainbow trout Oncorhynchus mykiss: similiar, but different diseases. Dis. Aquat. Org. 36:227-231. [DOI] [PubMed] [Google Scholar]
- 12.Engleberg, N. C., A. Heath, A. Miller, C. Rivera, and V. J. DiRita. 2001. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J. Infect. Dis. 183:1043-1054. [DOI] [PubMed] [Google Scholar]
- 13.Fuller, J. D., D. J. Bast, V. Nizet, D. E. Low, and J. C. de Azavedo. 2001. Streptococcus iniae virulence is associated with a distinct genetic profile. Infect. Immun. 69:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller, J. D., A. C. Camus, C. L. Duncan, V. Nizet, D. J. Bast, R. L. Thune, D. A. Low, and J. C. S. de Azavedo. 2002. Identification of a streptolysin S-associated gene cluster and its role in the pathogenesis of Streptococcus iniae disease. Infect. Immun. 70:5730-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson, R. L., M. K. Lee, C. Soderland, E. Y. Chi, and C. E. Rubens. 1993. Group B streptococci invade endothelial cells: type III capsular polysaccharide attenuates invasion. Infect. Immun. 61:478-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haire, R. N., J. P. Rast, R. T. Litman, and G. W. Litman. 2000. Characterization of three isotypes of immunoglobulin light chains and T-cell antigen receptor alpha in zebrafish. Immunogen 51:915-923. [DOI] [PubMed] [Google Scholar]
- 17.Hakansson, S., M. Granlund-Edstedt, M. Sellin, and S. E. Holm. 1990. Demonstration and characterization of buoyant-density subpopulations of group B Streptococcus type III. J. Infect. Dis. 161:741-746. [DOI] [PubMed] [Google Scholar]
- 18.Hakansson, S., S. E. Holm, and M. Wagner. 1987. Density profile of group B streptococci, type III, and its possible relation to enhanced virulence. J. Clin. Microbiol. 25:714-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1405. [PMC free article] [PubMed] [Google Scholar]
- 20.Hava, D. L., J. LeMieux, and A. Camilli. 2003. From nose to lung: the regulation behind Streptococcus pneumoniae virulence factors. Mol. Microbiol. 50:1103-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 22.Herbomel, P., B. Thisse, and C. Thisse. 1999. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126:3735-3745. [DOI] [PubMed] [Google Scholar]
- 23.Husmann, L. K., D. L. Yung, S. K. Hollingshead, and J. R. Scott. 1997. Role of putative virulence factors of Streptococcus pyogenes in mouse models of long-term throat colonization and pneumonia. Infect. Immun. 65:1422-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, A. L., K. M. Knoll, and C. E. Rubens. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 37:1444-1455. [DOI] [PubMed] [Google Scholar]
- 25.Jones, C. H., T. C. Bolken, K. F. Jones, G. O. Zeller, and D. E. Hruby. 2001. Conserved DegP protease in gram-positive bacteria is essential for thermal and oxidative tolerance and full virulence in Streptococcus pyogenes. Infect. Immun. 69:5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 27.Lancefield, R. C. 1962. Current knowledge of type-specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 28.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 29.Lehoux, D. E., F. Sanschagrin, and R. C. Levesque. 1999. Defined oligonucleotide tag pools and PCR screening in signature-tagged mutagenesis of essential genes from bacteria. BioTechniques 26:473-480. [DOI] [PubMed] [Google Scholar]
- 30.Levin, J. C., and M. R. Wessels. 1998. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A streptococcus. Mol. Microbiol. 30:209-219. [DOI] [PubMed] [Google Scholar]
- 31.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 171:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyon, W. R., and M. G. Caparon. 2004. Role for serine protease HtrA (DegP) of Streptococcus pyogenes in the biogenesis of virulence factors SpeB and the hemolysin streptolysin S. Infect. Immun. 72:1618-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an Rgg-like regulator in the transcription, secretion, and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magee, A. D., and J. Yother. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 69:3755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 36.Meijer, A. H., S. F. Gabby Krens, I. A. Medina Rodriguez, S. He, W. Bitter, B. Ewa Snaar-Jagalska, and H. P. Spaink. 2004. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol. Immunol. 40:773-783. [DOI] [PubMed] [Google Scholar]
- 37.Miller, J. D., and M. N. Neely. 2004. Zebrafish as a model host for streptococcal pathogenesis. Acta Trop. 91:53-68. [DOI] [PubMed] [Google Scholar]
- 38.Neely, M., J. Pfeifer, and M. G. Caparon. 2002. Streptococcus-zebrafish model of bacterial pathogenesis. Infect. Immun. 70:3904-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Casal, J., J. A. Price, E. Maugin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 41.Platt, M. W., N. Correa, Jr., and C. Mold. 1994. Growth of group B streptococci in human serum leads to increased cell surface sialic acid and decreased activation of the alternative complement pathway. Can. J. Microbiol. 40:99-105. [DOI] [PubMed] [Google Scholar]
- 42.Postlethwait, J. H., Y. L. Yan, M. A. Gates, S. Horne, A. Amores, A. Brownlie, A. Donovan, E. S. Egan, A. Force, Z. Gong, C. Goutel, A. Fritz, R. Kelsh, E. Knapik, E. Liao, B. Paw, D. Ransom, A. Singer, M. Thomson, T. S. Abduljabbar, P. Yelick, D. Beier, J. S. Joly, D. Larhammar, F. Rosa, M. Westerfield, L. I. Zon, S. L. Johnson, and W. S. Talbot. 1998. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 18:345-349. [DOI] [PubMed] [Google Scholar]
- 43.Ralph, P., and I. Nakoinz. 1977. Antibody-dependent killing of erythrocyte and tumor targets by macrophage-related cell lines: enhancement by PPD and LPS. J. Immunol. 119:950-954. [PubMed] [Google Scholar]
- 44.Ravins, M., J. Jaffe, E. Hanski, I. Shetzigovski, S. Natanson-Yaron, and A. E. Moses. 2000. Characterization of a mouse-passaged, highly encapsulated variant of group A streptococcus in in vitro and in vivo studies. J. Infect. Dis. 182:1702-1711. [DOI] [PubMed] [Google Scholar]
- 45.Rubens, C. E., R. F. Haft, and M. R. Wessels. 1995. Characterization of the capsular polysaccharide genes of group B streptococci. Dev. Biol. Stand. 85:237-244. [PubMed] [Google Scholar]
- 46.Rubens, C. E., M. R. Wessels, J. M. Kuypers, D. L. Kasper, and J. N. Weiser. 1990. Molecular analysis of two group B streptococcal virulence factors. Semin. Perinatol. 14:22-29. [PubMed] [Google Scholar]
- 47.Schrager, H. M., S. Alberti, C. Cywes, G. J. Dougherty, and M. R. Wessels. 1998. Hyaluronic acid capsule modulates M protein-mediated adherence and acts as a ligand for attachment of group A Streptococcus to CD44 on human keratinocytes. J. Clin. Investig. 101:1708-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sellin, M., S. Hakansson, and M. Norgren. 1995. Phase-shift of polysaccharide capsule expression in group B streptococci, type III. Microb. Pathog. 18:401-415. [DOI] [PubMed] [Google Scholar]
- 49.Shea, J. E., J. D. Santangelo, and R. G. Feldman. 2000. Signature-tagged mutagenesis in the identification of virulence genes in pathogens. Curr. Opin. Microbiol. 3:451-458. [DOI] [PubMed] [Google Scholar]
- 50.Spellerberg, B. 2000. Pathogenesis of neonatal Streptococcus agalactiae infections. Microbes Infect. 2:1733-1742. [DOI] [PubMed] [Google Scholar]
- 51.Stevens, D. L. 1999. The flesh-eating bacterium: what's next? J. Infect. Dis. 179:S366-S374. [DOI] [PubMed] [Google Scholar]
- 52.Stingele, F., J. R. Neeser, and B. Mollet. 1996. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J. Bacteriol. 178:1680-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeshita, S., M. Sato, M. Toba, W. Masahashi, and T. Hashimoto-Gotoh. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol or kanamycin-resistance selection. Gene 61:63-74. [DOI] [PubMed] [Google Scholar]
- 54.Thornsberry, C., P. Ogilvie, J. Kahn, and Y. Mauriz. 1997. Surveillance of antimicrobial resistance in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the United States in 1996-1997 respiratory season. The Laboratory Investigator Group. Diagn. Microbiol. Infect. Dis. 29:249-257. [DOI] [PubMed] [Google Scholar]
- 55.Trede, N. S., A. G. Zapata, and L. I. Zon. 2001. Fishing for lymphoid genes. Trends Immunol. 22:302-307. [DOI] [PubMed] [Google Scholar]
- 56.Wessels, M. R., and M. S. Bronze. 1994. Critical role of the group A streptococcal capsule in pharyngeal colonization and infection in mice. Proc. Natl. Acad. Sci. USA 91:12238-12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willett, C. E., A. Cortes, A. Zuasti, and A. G. Zapata. 1999. Early hematopoiesis and developing organs in the zebrafish. Dev. Dyn. 214:323-336. [DOI] [PubMed] [Google Scholar]
- 58.Willett, C. E., A. G. Zapata, N. Hopkins, and L. A. Steiner. 1997. Expression of zebrafish rag genes during early development identifies the thymus. Dev. Biol. 182:331-341. [DOI] [PubMed] [Google Scholar]
- 59.Yoder, J. A., and G. W. Litman. 2000. Immune-type diversity in the absence of somatic rearrangement. Curr. Top. Microbiol. Immunol. 248:271-282. [DOI] [PubMed] [Google Scholar]
- 60.Yoder, J. A., M. G. Mueller, S. Wei, B. C. Corliss, D. M. Prather, T. Willis, R. T. Litman, J. Y. Djeu, and G. W. Litman. 2001. Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian leukocyte receptor cluster. Proc. Natl. Acad. Sci. USA 98:6771-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoder, J. A., M. E. Nielsen, C. T. Amemiya, and G. W. Litman. 2002. Zebrafish as an immunological model system. Microbes Infect. 4:1469-1478. [DOI] [PubMed] [Google Scholar]
- 62.Zlotkin, A., S. Chilmonczyk, M. Eyngor, A. Hurvitz, C. Ghittino, and A. Eldar. 2003. Trojan horse effect: phagocyte-mediated Streptococcus iniae infection of fish. Infect. Immun. 71:2318-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zlotkin, A., H. Hershko, and A. Eldar. 1998. Possible transmission of Streptococcus iniae from wild fish to cultured marine fish. Appl. Environ. Microbiol. 64:4065-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]