Abstract
Although the antiviral kinase PKR was originally characterized as a double-stranded RNA activated enzyme it can be stimulated by RNAs containing limited secondary structure. Single-stranded regions in such RNAs contribute to binding and activation but the mechanism is not understood. Here, we demonstrate that single-stranded RNAs bind to PKR with micromolar dissociation constants and can induce activation. Addition of a 5′-triphosphate slightly enhances binding affinity. Single-stranded RNAs also activate PKR constructs lacking the double-stranded RNA binding domain and bind to a basic region adjacent to the N-terminus of the kinase. However, the isolated kinase is not activated by and does not bind single-stranded RNA. Photocrosslinking measurements demonstrate that that the basic region interacts with RNA in the context of full length PKR. We propose that bivalent interactions with the double stranded RNA binding domain and the basic region underlie the ability of RNAs containing limited structure to activate PKR by enhancing binding affinity and thereby increasing the population of productive complexes containing two PKRs bound to a single RNA.
Introduction
Protein Kinase R (PKR) is a serine/threonine kinase which plays a critical role in the innate immunity pathway. PKR is synthesized in a latent form and is activated by duplex regions present within structured RNAs to undergo autophosphorylation. Stimulatory RNAs typically originate from viral infection but several endogenous RNAs have been identified as PKR activators1–5. PKR phosphorylates its primary cellular substrate, the α-subunit of eukaryotic initiation factor-2 (eIF2α), to inhibit protein synthesis and viral replication in infected cells6.
PKR is comprised of two tandem N-terminal dsRNA binding domains (dsRBD1 and dsRBD2) and a C-terminal kinase domain connected by an ~80 residue flexible linker (Fig. 1). The dsRBD recognizes an A-form RNA duplex with a site size of ~15 bp and primarily makes interactions with 2′-hydroxyls and the phosphodiester backbone7, 8. Both dsRBDs in PKR adopt a canonical αβββα fold9. The kinase domain has a bilobal architecture typical of protein kinases consisting of a smaller N-terminal lobe and larger C-terminal lobe connected by a flexible hinge region. The kinase domain co-crystallizes with eIF2α as a back to back dimer with an allosteric pathway linking the dimer interface to the active site10. Biochemical and biophysical data indicate that dimerization plays a critical role in the PKR activation mechanism11. Mutations to key residues within the dimer interface abolish activation12. Activation of PKR by simple dsRNAs requires a minimum of 30 bp13, 14 and is mediated by dimerization of the kinase domain15.
Figure 1.
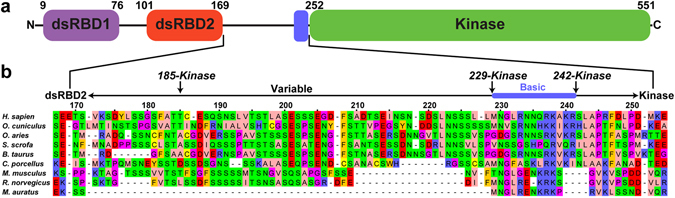
PKR domain schematic. (a) PKR domain organization. Canonical domain boundary positions are indicated above each domain. (b) Linker sequence alignment. The alignment was generated with ClustalW55 and prepared in JalView56. Residues are colored using the Zappo coloring scheme.
In addition to the canonical activators containing ≥30 bp dsRNA regions, PKR is stimulated by RNAs that contain limited secondary structure and include single-stranded regions or tertiary structure. Examples include a 17 bp stem loop within the 3′-UTR of TNF-α mRNA16, a pseudoknot within the 5′-UTR of IFN-γ mRNA4, 5, siRNAs containing short ssRNA overhangs17, 18, several snoRNAs induced under metabolic stress3, and certain bacterial RNAs19. Notably, short stem-loop RNAs with flanking ssRNA tails (ss-dsRNAs) have been identified as a potent PKR activation motif20, 21. Truncations to the ssRNA regions result in decreased binding affinity and activation potency, and complete deletion of either the 5′- or 3′- flanking tail abolishes activation22. Surprisingly, a duplex region as short as 10 nt with 15 nt 5′- and 3′- flanking tails binds two PKR monomers and functions as an activator. These observations imply that single-stranded regions can play a role in RNA-mediated activation of PKR. Although PKR does not contain a canonical ssRNA binding domain, interactions between ssRNAs and dsRBDs from other proteins has been reported23, 24. However, mutations in PKR within the dsRBDs that block dsRNA binding do not prevent photocrosslinking to an RNA with minimal secondary structure, suggesting that ssRNA binding may be mediated by other regions of the enzyme25. Potential interaction sites include a basic region adjacent to the N-terminus of the kinase (Fig. 1) that is important for kinase function26 as well as a basic cleft lying on the kinase domain27. A 5′-triphosphate (5′-ppp) is reported to be critical for PKR activation by RNAs with limited secondary structure, including the ss-dsRNA motif3, 19, 21, 28 (but see ref. 22). Duplex RNAs do not exhibit a triphosphate dependence so it is likely that ssRNA and the 5′-ppp bind to the same site21.
Here, we have directly probed ssRNA binding and activation of PKR. We demonstrate that ssRNAs can bind to PKR at both the dsRBD and the basic region. ssRNA mediated activation of the kinase domain requires the basic region. Our data support a model where PKR activation by RNAs is regulated in vivo by interaction with both duplex and single-stranded regions.
Results and Discussion
PKR interaction with ssRNA
Based on the critical contribution of single-stranded regions to the binding and activation of PKR by ss-dsRNAs, we have investigated the interactions of PKR with isolated ssRNAs. We employed sedimentation velocity analytical ultracentrifugation in order to detect transient, lower affinity interactions that may not be reliably measured in gel shift29 or filter binding30 assays. Figure 2a shows a titration of a model ssRNA, U30, with PKR depicted as an overlay of g^(s*) sedimentation coefficient distribution functions. The homopolymeric U30 RNA was chosen to avoid base stacking and potential formation of secondary structure. Addition of PKR results in a decrease in the amplitude of the peak at 1.5S associated with free U30 and formation of a peak at higher sedimentation coefficient due to complex formation.
Figure 2.
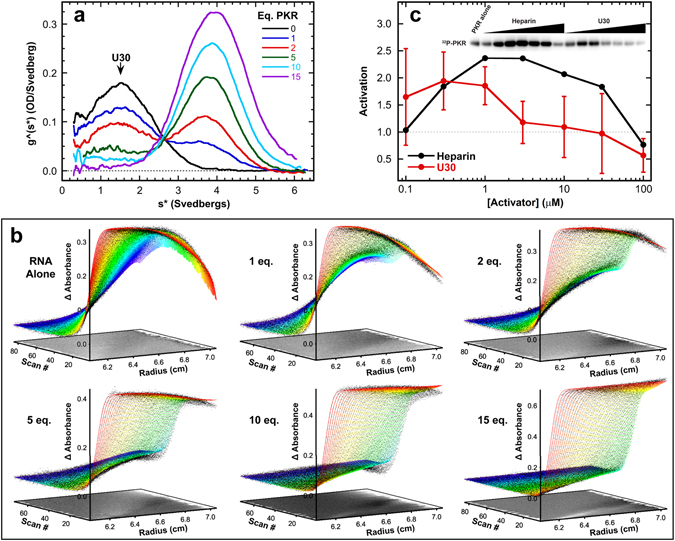
Interaction of PKR with ssRNA. PKR binding to U30 ssRNA was assayed by sedimentation velocity analytical ultracentrifugation. Measurements were performed in AU75 buffer at 20 °C and 50,000 rpm using absorbance detection at 260 nm. (a) Titration of U30 with PKR represented as an overlay of g^(s*) sedimentation coefficient distribution functions. The samples contained 1 µM U30 ( ) and 1 µM U30 plus 1 eq. (
) and 1 µM U30 plus 1 eq. ( ), 2 eq. (
), 2 eq. ( ), 5 eq. (
), 5 eq. ( ), 10 eq. (
), 10 eq. ( ), and 15 eq. PKR (
), and 15 eq. PKR ( ). The decrease in the U30 peak and appearance of the peak at higher S are due to complex formation. (b) Global analysis of the time difference curves. Scans within each data set were subtracted in pairs to remove time-invariant background noise and fit to a sequential 2:1 binding model using SEDANAL53. The data are indicated by points and the fit by solid lines. The residuals are plotted as a grayscale image in the x-y plane at z = 0. The best-fit parameters are in Table 1. (c) Activation of PKR by U30. 500 nM PKR was incubated with variable concentrations of U30 in AU75 buffer with 5 mM MgCl2 for 20 min at 32 °C. Samples were resolved by SDS-PAGE and 32P-PKR was quantified with a phosphorimager. The data are normalized to activation of PKR in the absence of activator. The error bars correspond to the standard deviation from three replicates. The inset shows a cropped image of the gel.
). The decrease in the U30 peak and appearance of the peak at higher S are due to complex formation. (b) Global analysis of the time difference curves. Scans within each data set were subtracted in pairs to remove time-invariant background noise and fit to a sequential 2:1 binding model using SEDANAL53. The data are indicated by points and the fit by solid lines. The residuals are plotted as a grayscale image in the x-y plane at z = 0. The best-fit parameters are in Table 1. (c) Activation of PKR by U30. 500 nM PKR was incubated with variable concentrations of U30 in AU75 buffer with 5 mM MgCl2 for 20 min at 32 °C. Samples were resolved by SDS-PAGE and 32P-PKR was quantified with a phosphorimager. The data are normalized to activation of PKR in the absence of activator. The error bars correspond to the standard deviation from three replicates. The inset shows a cropped image of the gel.
In order to define the interaction stoichiometry and obtain dissociation constants the sedimentation velocity profiles were subtracted in pairs to remove systematic noise and the resulting difference curves were fit to alternative association models. The data are well described by a 2:1 sequential binding model where two PKR monomers assembly on a single RNA (Fig. 2b). Similar to PKR interactions with duplex RNAs, the first PKR binds with the highest affinity, with a K d1 = 3.5 µM, and the second binds weaker, K d2 = 40 µM (Table 1). Nonspecific, sequential binding of multiple proteins to a nucleic acid lattice of identical sites, such as U30, is expected to become progressively weaker due to statistical effects31. Binding to U30 activates PKR autophosphorylation weakly (Fig. 2c) to a level about 1.5-fold above the background in the absence of RNA. As observed with dsRNA, higher ssRNA concentrations inhibit, consistent with dilution of PKR dimers by the excess nucleic acid. PKR binds about three-fold more strongly to a heteropolymeric ssRNA of the same length (Het30, Table 1). This enhancement may reflect weak secondary structure formation by the heteropolymeric sequence (Fig. S1) or some slight nucleotide preference.
Table 1.
Sedimentation velocity analysis of PKR constructs binding to RNA.
| Protein | Nucleic Acid | K d1 (µM) | K d2 (µM) | RMSDa |
|---|---|---|---|---|
| Full length | U30 | 3.51 (3.49, 3.52) | 39.9 (39.4, 40.5) | 0.00662 |
| ppp-Het30 | 0.825 (0.761, 0.894) | 3.81 (3.49, 4.15) | 0.00876 | |
| Het30 | 1.07 (1.01, 1.12) | 10.8 (10.1, 11.6) | 0.00667 | |
| dsRBD | U30 | 8.42 (7.98, 8.88) | 52.3 (47.6, 57.7) | 0.00514 |
| ppp-Het30b | 5.36 (4.60, 6.31) | 9.66 (8.03, 11.6) | 0.00810 | |
| Het30b | 8.21 (7.38, 9.18) | 19.4 (16.3, 23.0) | 0.00701 | |
| 185-kinase | U30 | 31.9 (29.8, 34.2) | 68.9 (51.8, 97.5) | 0.00452 |
| 229-kinase | U30 | 1.91 (1.79, 2.05) | 7.87 (7.16, 8.64) | 0.00706 |
| ppp-Het30 | NDc | NDc | NDc | |
| Het30 | NDc | NDc | NDc | |
| ds30 | 3.80 (3.37, 4.33) | 3.27 (2.82, 3.77) | 0.00457 | |
| dT30 | 2.59 (2.44, 2.76) | 16.2 (14.5, 18.2) | 0.00630 | |
| 242-kinased | U30 | 90.0 (84.2, 96.5) | — | 0.00546 |
| ds30 | 19.9 (18.6, 21.4) | — | 0.00457 |
Parameters obtained by global nonlinear least square analysis of the sedimentation velocity data using a model of sequential binding of two proteins monomers. The values in parentheses represent the 95% joint confidence intervals obtained using the F-statistic.
aRoot mean square deviation in absorbance units.
bGood fits required that the sedimentation coefficients for the protein-RNA complexes be allowed to float to their best-fit values.
cNot determined. More than two protein monomers bind to the RNA and the data could not be reliably fit to an association model. Plots of the normalized g^(s*) distributions for these experiments are in Fig. S3.
d242-kinase data were fit to a 1:1 binding model.
These data provide the first direct evidence that PKR binds to and is activated by single-stranded RNAs and rationalizes previous reports19, 21, 22 that single-stranded regions present in structured RNAs contribute to PKR interactions. As in the case of duplex RNAs32, the bell-shape activation curve implies that ssRNAs induce PKR dimerization. The low extent of activation is likely a consequence of the relatively weak binding affinity and it is unlikely that PKR is significantly activated in vivo upon interaction with unstructured RNA.
Localization of ssRNA binding
We characterized U30 and Het30 binding to individual PKR domain constructs to define the region(s) responsible for interaction with ssRNA and the contribution of the 5′-ppp. The dsRBD (residues 1–184) binds U30 with affinities slightly reduced relative to the holoenzyme (Table 1), indicating that ssRNA can also bind to this canonical dsRNA binding site. However, there are other potential ssRNA binding sites in PKR. In the human enzyme, the dsRBD is separated from the kinase domain by an ~80 residue unstructured linker. The length of the linker is highly variable among PKR orthologs (Fig. 1b). This length variability arises within the N-terminal acidic portion of the linker, but a cluster of basic residues within the C-terminal portion is conserved. This portion of the linker is implicated in kinase function26 and ribosome association33 and represents a potential RNA interaction site. Alternatively, the heparin binding site is located on the kinase domain27, 34, 35 and represents another possible ssRNA binding motif. We prepared PKR domain constructs to probe the role of the linker, basic region, and isolated kinase domain in ssRNA binding. Each of the constructs is homogeneous and monomeric up to the highest concentration assayed (Fig. S2, Table S1).
Figure 3 shows titrations of U30 and ds30 RNAs with PKR kinase constructs that contain (229-kinase) or lack (242-kinase) the basic region. Addition of stoichiometric 229-kinase to U30 causes a decrease in the RNA peak and a shift of the distribution to higher sedimentation coefficient, indicating that this construct binds to ssRNA. The distribution at the highest protein concentration is bilobal due to the contribution of free 229-kinase at 2.8S and a protein:RNA complex which sediments at 4.7S. In contrast, addition of the 242-kinase construct lacking the basic region results in only a minimal decrease in the free RNA peak. The new feature which appears at 2.8 S is primarily due to absorbance of the free protein. The 229-kinase data fit well to the sequential 2:1 binding model with K d1 = 1.9 µM and K d2 = 7.9 µM. However, the 242-kinase binds very weakly and binding of a second protein was not detected. The data were fit to a 1:1 model (Table 1). Thus, the basic region mediates ssRNA binding to the C-terminal portion of PKR. Affinity is substantially reduced when the remainder of the linker is included in the 185-kinase construct. These measurements may be more representative of the affinity of the basic region for ssRNAs in the context of full length PKR.
Figure 3.
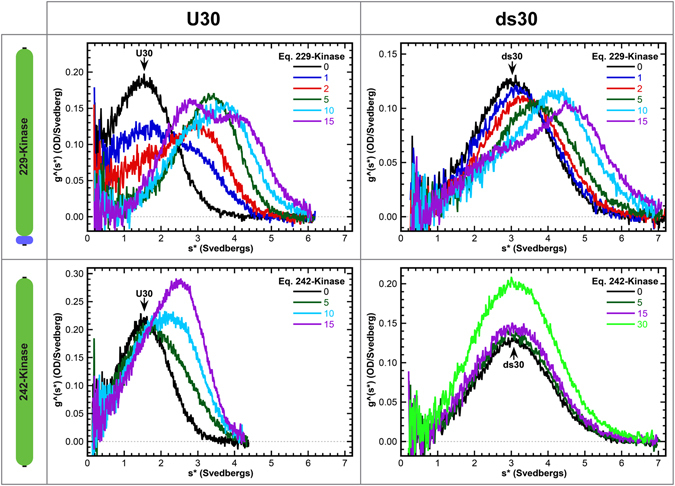
Interaction of the PKR kinase domain/basic region with RNA. Binding of 229-kinase and 242-kinase to U30 and ds30 were analyzed by sedimentation velocity. Measurements were performed in AU75 buffer at 20 °C and 50,000 rpm using absorbance detection at 260 nm. Each panel shows a titration of RNA with protein depicted as a g^(s*) sedimentation coefficient distribution. Addition of 229-kinase to U30 or ds30 causes a shift in the distributions to the right, indicating complex formation. In contrast, 242-kinase induces only a slight decrease in the peak corresponding to free U30, indicating a very weak interaction. The peak at 2.8 S at higher protein concentrations is due to free 242-kinase. The sedimentation coefficients of 242-kinase and ds30 are similar and the increase in the peak amplitude is primarily due to free protein. 229-kinase data were fit to a sequential 2:1 binding model and 242-kinase data were fit to a 1:1 binding model to extract the binding parameters displayed in Table 1.
Interestingly, 229-kinase also binds a 30 bp dsRNA and the dissociation constants are similar to those observed with U30. Thus, the basic region interaction is not specific for ssRNA. Indeed, 229-kinase also binds to a 30 nt ssDNA, dT30, albeit with reduced affinity indicating a small contribution from the 2′-OH (Table 1). The 242-kinase and ds30 have similar sedimentation coefficients (3.2 and 2.8S, respectively) so that the two species are not resolved by time derivative analysis. Although a discrete peak for the complex is not detected, the data are best described by a 1:1 binding model. The affinity is weaker than for 229-kinase.
Given previous reports indicating that a 5′-ppp contributes to PKR activation to certain RNAs with limited secondary structure3, 19, 21, 28, we probed the effect of this moiety on PKR binding to an ssRNA, Het30. The presence of a 5′-ppp enhances PKR binding, reducing K d2 by a factor of about 3 relative to the 5′-OH RNA (Table 1), but the binding affinities are still weaker than for a 30 bp dsRNA of the same length36. A similar magnitude effect of a 5′-ppp is observed for Het30 binding to the dsRBD. It was not possible to fit the data for binding of 229-kinase to Het30 and ppp-Het30 due to the formation of higher-order complexes (Fig. S3). However, comparison of the g^(s*) distributions at comparable protein concentrations with Het30 and ppp-Het30 reveals that the 5′-ppp slightly increases binding to both PKR domain constructs. Thus, the enhancement of ssRNA binding to PKR by the 5′-ppp is not associated with a distinct binding site, as previous suggested25 but is nonspecific and likely attributable to electrostatic interactions. A 5′-ppp also increases the affinity in the context of a ss-dsRNA containing a 15 bp stem and 15 nt 5′- and 3′-tails (15-15-15) but it is not required to detect kinase activation22. A 5′-ppp may play a role in PKR activation by RNAs with limited secondary structure3, 19, 21, 28 by increasing binding affinity and thereby increasing the maximum population of active PKR dimers. However, a 5′-ppp is not absolutely required for activation of PKR, or the 229-kinase (vide infra), by unstructured RNAs.
The weak dependence of binding affinity on a 5′-ppp is consistent with the absence of a distinct binding site for this moiety in PKR. The canonical cytosolic sensor for RNAs bearing a 5′-ppp is RIG-I. This protein contains a helicase domain which binds duplex RNA and a regulatory domain which binds 5′-ppp37. For RIG-I, the presence of a 5′-ppp increases the binding affinity by >100-fold relative to the 5′-OH form, but the enhancement is only 2.4-fold for the isolated helicase domain38. The latter is similar in magnitude to PKR and presumably represents the contribution of nonspecific electrostatic interactions. The antiviral proteins IFIT1 and IFIT5 recognize ssRNAs containing a 5′-ppp via a deep, positively-charged cavity39, 40. IFIT5 binds a 5′-ppp ssRNA with nanomolar affinity yet does not form a detectable complex with ssRNA bearing a 5′-OH41, 42.
Given the strong binding of ssRNA to 229-kinase we asked whether it could mediate activation. For comparison, we also assayed activation of the 242-kinase construct. The 229-kinase is activated by U30 with a bell-shaped activation curve (Fig. 4a and c). As observed for the full length enzyme, the maximum extent of activation by ssRNA is fairly weak, corresponding to about 3-fold over the basal level. Consistent with the lower ssRNA binding affinity, 242-kinase is not activated by U30.
Figure 4.
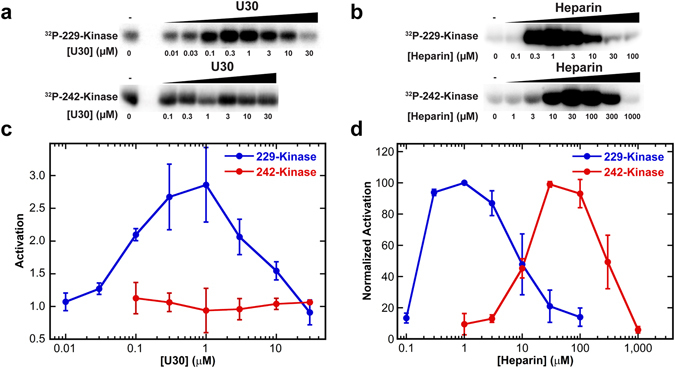
Activation of PKR kinase domain by ssRNA. Autophosphorylation reactions were carried out with a fixed concentration of protein and titrated with either U30 (a,c) or heparin (b,d). The basal (RNA-independent) activities of 229- and 242-kinase are much lower than the full length enzyme and are different from each other (Fig. S4). Thus, the protein concentrations were adjusted to give approximately equal extents of autophosphorylation in the absence of RNA: 1.5 µM 229-kinase and 5 µM 242-kinase. Phosphorimager scans of SDS-PAGE gels are shown in (a) and (b). The gels images are cropped to display the 32P kinase domain bands and the contrast settings within each gel are independently adjusted using using ImageQuant TL software to clearly show the intensity trends. The quantitation of 32P-incorporation is shown in (c) and (d). The error bars correspond to the standard deviation from three replicates. In (c), the activation by U30 is plotted relative to samples containing no activator. In (d), activation by heparin is normalized to the maximum signal because heparin is a potent activator and quantitation relative to the low signal in the absence of activator is not accurate. The ~100-fold increase in the heparin concentration required for maximal activation of the 242-kinase persists when the protein concentration is reduced to the same concentration used for 229-kinase (1.5 µM). Note that the maximum concentration of U30 assayed was 30 µM which is the peak of the 242-kinase heparin activation curve.
In a previous study we localized the heparin binding site to a pocket adjacent to helix αC within the kinase domain using an inactive construct corresponding to 242-kinase27. Figure 4b and d show that both 229- and 242-kinase are activated by heparin. The peak of maximal activation is shifted to about 100-fold lower heparin concentration for the 229-kinase implying that the basic region may also interact with heparin. Consistent with this interpretation, heparin binds more weakly to 242-kinase than to the full length enzyme27. The presence of the basic region contributes to heparin-mediated activation and possibly binding affinity. However, the heparin binding pocket on the kinase domain does not accommodate single-stranded RNA.
Our results indicate that PKR contains two RNA binding loci. We performed photocrosslinking assays to provide insight into how these regions interact with RNA in the context of the full length enzyme and 15-15-15, a potent ss-dsRNA activator. The ss-dsRNA was transcribed in the presence of 4-thiouridine triphosphate (s4U), resulting in incorporation of the modified nucleotide throughout the RNA (Fig. 5a), radiolabeled at the 5′-end and crosslinked to PKR by exposure to 365 nm light. Reactions were performed with PKR domain constructs and full length PKR constructs containing a TEV protease cleavage site located at position 185, 229, or 242. Following crosslinking, samples were cleaved by TEV protease, separated on SDS-PAGE and visualized by phosphorimaging. Crosslinking of the ss-dsRNA to wild type PKR is dependent on s4U and the adduct is not cleaved by TEV protease (Fig. 5b). The mutants containing TEV sites are efficiently cleaved by the protease giving two predominant products. When cleaved at positions 185 or 229 that are N-terminal to the basic region, most of the 32P-labeled RNA is associated with the C-terminal kinase domain. Cleavage at position 242 shifts the distribution so most of the label is attached to the N-terminal fragment containing the dsRBD and basic region. This switch in band intensity indicates that the basic region interacts with the ss-dsRNA in the context of full length PKR. The residual crosslinking of the C-terminal fragment may reflect the weak interactions observed between the 242-kinase construct and RNA.
Figure 5.
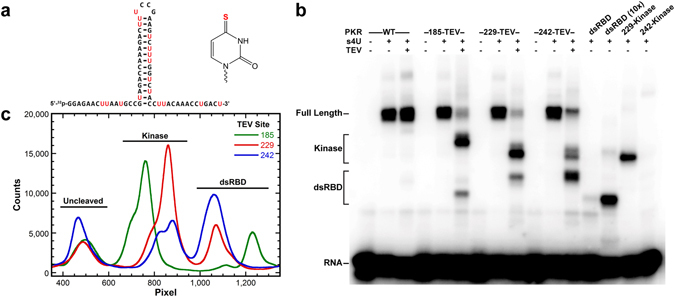
Crosslinking analysis of PKR binding to ss-dsRNA. (a) Secondary structure of the ss-dsRNA (15-15-15) and chemical structure of 4-thiouridine. Modified uridines are highlighted in red and the sulfur responsible for photochemical crosslinking is highlighted in red in the 4-thiouridine structure. (b) Phosphorimager analysis of 32P incorporation in RNA:protein complexes following crosslinking. Full length PKR constructs containing a TEV protease site and individual domains were crosslinked to radiolabeled ss-dsRNA. TEV cleavage was performed after the crosslinking reaction. The P1′ position of the cleavage site is indicated above the lanes. Samples contained 0.5 µM RNA and 1 µM protein. One sample of dsRBD contained 10 µM protein (labeled 10x). Lanes labeled as (−) s4U contained unmodified ss-dsRNA. (c) Quantitation of RNA:protein adducts following TEV cleavage. Distributions were created by vertical integration of data from panel b.
Crosslinking of the ss-dsRNA to the isolated domain constructs mirrors the trends observed with the full length protein. The kinase domain requires the basic region for crosslinking. Crosslinking of ss-dsRNA to the dsRBD is weak and requires addition of 10-fold molar excess of protein relative to the other samples to generate a similar amount of adduct. This low amount of product is not due to weak binding but likely reflect limited accessibility of s4U when the dsRBD is bound to dsRNA, its preferred binding site. The photoreactive thiol group lies in the major groove and dsRBDs do not interact with the major groove face of bases in the dsRNA helix7. The dsRBD binds duplex RNA strongly (intrinsic K d ~ 100 nM)31 whereas the affinity toward single-stranded RNA is in the low micromolar range (Table 1). Thus, the isolated dsRBD populates the single-stranded regions only after the duplex has been saturated, giving rise to more efficient crosslinking to s4U. In the context of the full length protein complex formation is driven by the high affinity interaction of the dsRBD with the duplex region. The crosslinking data indicate that the auxiliary interactions which stabilize the complex occur between the single-stranded tails and the basic region.
We have tested this model by extending our previous study of ss-dsRNAs22 to analyze the binding contribution or single-stranded regions in the context of the isolated dsRBD. Binding affinities were obtained for 15-15-15 and a construct lacking the flanking tails, 0-15-0. The measurements are reported in Table 2 along with the previously-measured22 affinities for the full length protein. The relative binding affinities indicate that the tails predominantly interact with regions outside of the dsRBD. Removal of the tails decreases the affinity of full length PKR by about 35-fold but reduces dsRBD binding by only about two-fold. In the absence of the tails the two constructs bind about equally well, confirming that binding to duplex regions is mediated primarily by the dsRBD.
Table 2.
Sedimentation velocity analysis of PKR and dsRBD binding to ss-dsRNAs.
| Protein | RNAa | K d1 (nM) | K d2 (nM) | K d3 (nM) | RMSDb |
|---|---|---|---|---|---|
| PKR | 15-15-15 | 4 (2, 6) | 294 (252, 343) | — | 0.00821c |
| PKR | 0-15-0 | 138 (105, 178) | 1660 (1310, 2130) | — | 0.00906c |
| PKR | 15-5-15 | 118 (93, 150) | 853 (673, 1103) | — | 0.00667c |
| PKR | 0-5-0 | 5400 (4930, 5900) | — | — | 0.00460 |
| dsRBD | 15-15-15 | 111 (76, 160) | 190 (121, 279) | 2780 (2100, 3930) | 0.00358 |
| dsRBD | 0-15-0 | 218 (155, 300) | 1097 (961, 1249) | — | 0.00493 |
Parameters obtained by global nonlinear least square analysis of the sedimentation velocity data. The values in parentheses represent the 95% joint confidence intervals obtained using the F-statistic.
aThe three numbers indicate the length of the 5′-tail (nt), the stem (bp), and the 3′-tail (nt), respectively. Each of the RNAs contains a 5′-ppp and a tetraloop capping the stem sequence.
bRoot mean square deviation in absorbance.
cData from ref. 22.
The relatively weak binding of ssRNAs to PKR and the modest activation indicates that isolated single-stranded RNAs are unlikely to form a stable complex with PKR or to promote significant activation in vivo. However, these interactions provide a rationale for the contribution of single-stranded regions to PKR binding and activation in the context of structured RNAs. Invariably, potent RNA activators of PKR possess some secondary and tertiary structure43. Some of these RNAs lack the minimum 30 bp of duplex required to support PKR activation by simple dsRNAs but can form extended double stranded regions by coaxial stacking of shorter helices5 or by dimerization of self-complementary stem loops44, 45. Other RNA activators contain a single duplex as short as 10–15 bp21, 22 or two hairpins of 5 and 4 bp21, implying a substantial interaction of PKR with the unstructured regions. The crosslinking results demonstrate that both the dsRBD and the basic region-kinase domain interact with RNAs containing structured and unstructured regions. Despite the presence of these two RNA binding sites, sedimentation velocity data for full length PKR binding to ssRNAs and ss-dsRNAs only fit to a model where two monomers sequentially assemble onto a single RNA. The data do not fit well to alternative models which incorporate binding of multiple RNA ligands to a single protein (see supplement). The ~80 residue region separating the dsRBD and the basic region (Fig. 1) is intrinsically disordered and PKR adopts multiple conformations in solution46, 47. NMR data indicate that dsRBD2 can transiently interact with the kinase domain48, 49 suggesting a model in which PKR exists in an equilibrium between open and closed states50. In the closed conformation, the dsRBD and basic region/kinase domain would be in close proximity and capable of binding to a single RNA.
We propose that bivalent interactions underlie the ability of RNAs containing limited structure to activate PKR by enhancing binding affinity and thereby increasing the population of productive complexes containing two PKRs bound to a single RNA. In this model, a major contribution driving complex formation is the well-characterized interaction of duplex regions with the dsRBD31, 36. This interaction is stabilized by adjoining unstructured regions binding to the basic region. The bivalent interaction gives rise to strong (K d ~ 100 nM) binding even for ss-dsRNAs containing a stem as short as 5 bp. In addition to enhancing binding, the bivalent interactions may serve to orient the kinase domains to increase the propensity for the formation of active dimers15.
Methods
Protein expression and purification
PKR kinase domain constructs were created by inserting a TEV protease cleavage site (ENLYFQ↓G/S) upstream of the desired N-terminus. The 185- and 229-kinase domain constructs contain an extra glycine at the N-terminus. Wild-type PKR and constructs containing a TEV protease cleavage site were expressed and purified as previously described27. Following elution from the heparin sepharose column, the constructs containing a cleavage site were treated with AcTEV protease (Thermo Fisher Scientific) overnight at 4 °C. The kinase domain constructs were purified on hydroxyapatite (CHT ceramic hydroxyapatite, Bio-Rad Laboratories, Inc.). The dsRBD (residues 1–184) was expressed and purified as previously described31. All proteins were further purified by size exclusion chromatography on Superdex 75 or 200 HiLoad 16/60 columns (GE Healthcare) into AU75 buffer (75 mM HEPES (pH 7.5), 75 mM NaCl, 0.1 mM EDTA, 0.1 mM TCEP) immediately prior to use.
Nucleic Acids
Synthetic oligoribonucleotides were obtained from GE Healthcare Dharmacon (U30, Het30) or TriLink Biotechnologies, Inc. (ppp-Het30), dT30 was obtained from IDT, Inc. ss-dsRNAs were prepared by in vitro transcription as previously described22. For RNAs containing 4-thiouridine, UTP was replaced with 4-Thio-UTP (TriLink Biotechnologies, Inc.) in the transcription reactions. All RNAs were purified by denaturing PAGE followed by electroelution in an Elutrap device (Schleicher and Schuell).
Analytical Ultracentrifugation
Protein-RNA interactions were assayed by sedimentation velocity analysis as previously described51 in a Beckman Coulter XL-I analytical ultracentrifuge with 260 nm absorbance optics at 20 °C and 50,000 rpm in AU75 buffer. g^(s*) sedimentation coefficient distributions were produced using the time derivative method implemented in DCDT+, version 2.3.252. Binding affinities and stoichiometries were extracted by global fits of multiple data sets to association models using SEDANAL, version 6.0153. It was not possible to fit for the sedimentation coefficients of some of the RNA-protein complexes due to cross-correlation with the binding constants. These parameters were fixed at physically reasonably values based on our experience that RNA complexes with PKR have frictional ratios (f/f 0) near 1.514, 22, 36. Frictional ratios, buffer densities, viscosities, and protein partial specific volumes were calculated using SEDNTERP, version 201112054. RNA partial specific volumes were fixed at 0.55 mL g−1.
UV Crosslinking
UV crosslinking reactions were carried out using RNAs containing 4-thiouridine (see SI Materials and Methods for detailed protocol). Briefly, 5′-32P-labeled RNAs were UV crosslinked to PKR constructs containing a TEV cleavage site, treated with TEV protease, and analyzed by SDS-page. The products were visualized on a Typhoon Phosphorimager.
Activity Assays
PKR autophosphorylation was monitored by incorporation of 32P from [γ-32P]ATP (Perkin-Elmer). Low molecular weight heparin (4–6 kDa) was obtained from Sigma-Aldrich. Samples contained a fixed concentration of PKR and a variable concentration of activator. The assay was performed in AU75 buffer plus 5 mM MgCl2 at 32 °C. Samples were prepared on ice then incubated at 32 °C for 10 minutes prior to initiation of phosphorylation by addition of ATP to a final concentration of 400 µM and 0.25 µCi/µL [γ-32P]ATP. Reactions were quenched after 20 min incubation with sample loading buffer and resolved by SDS-PAGE. The gels were dried and exposed to a phosphor screen followed by quantitation on a Typhoon phosphorimager using ImageQuant TL Software (GE Healthcare).
Electronic supplementary material
Acknowledgements
This work was supported by grant number AI-53615 from the NIH to J.L.C. We thank Cassie Zerbe for critical reading of the manuscript.
Author Contributions
C.B.M. and J.L.C. designed the experiments, C.B.M. performed biophysical and biochemical experiments, C.B.M. and J.L.C. wrote the manuscript and reviewed it.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-03047-7
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nussbaum JM, Gunnery S, Mathews MB. The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Research. 2002;30:1205–1212. doi: 10.1093/nar/30.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis S, Watson JC. In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3′ untranslated regions of human alpha-tropomyosin. Proc Natl Acad Sci USA. 1996;93:508–513. doi: 10.1073/pnas.93.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Youssef OA, et al. Potential role for snoRNAs in PKR activation during metabolic stress. Proc Natl Acad Sci USA. 2015;112:5023–5028. doi: 10.1073/pnas.1424044112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–232. doi: 10.1016/S0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 5.Cohen-Chalamish S, et al. Dynamic refolding of IFN-gamma mRNA enables it to function as PKR activator and translation template. Nat Chem Biol. 2009;5:896–903. doi: 10.1038/nchembio.234. [DOI] [PubMed] [Google Scholar]
- 6.Pindel A, Sadler A. The role of protein kinase R in the interferon response. J Interferon Cytokine Res. 2011;31:59–70. doi: 10.1089/jir.2010.0099. [DOI] [PubMed] [Google Scholar]
- 7.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masliah G, Barraud P, Allain FH-T. RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cell. Mol. Life Sci. 2013;70:1875–1895. doi: 10.1007/s00018-012-1119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Cole JL. Activation of PKR: an open and shut case? Trends Biochem Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey M, et al. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 13.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/MCB.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR Activation by dsRNA. J Mol Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain B, Hesler S, Cole JL. Regulation of PKR by RNA: Formation of Active and Inactive Dimers. Biochemistry. 2015;54:6663–6672. doi: 10.1021/acs.biochem.5b01046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osman F, Jarrous N, Ben-Asouli Y, Kaempfer R. A cis-acting element in the 3′-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 1999;13:3280–3293. doi: 10.1101/gad.13.24.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 18.Puthenveetil S, et al. Controlling activation of the RNA-dependent protein kinase by siRNAs using site-specific chemical modification. Nucleic Acids Research. 2006;34:4900–4911. doi: 10.1093/nar/gkl464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hull CM, Bevilacqua PC. Mechanistic Analysis of Activation of the Innate Immune Sensor PKR by Bacterial RNA. J Mol Biol. 2015;427:3501–3515. doi: 10.1016/j.jmb.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nallagatla SR, et al. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science (New York, NY) 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 22.Mayo CB, Wong CJ, Lopez PE, Lary JW, Cole JL. Activation of PKR by short stem-loop RNAs containing single-stranded arms. RNA. 2016;22:1065–1075. doi: 10.1261/rna.053348.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barraud P, et al. An extended dsRBD with a novel zinc-binding motif mediates nuclear retention of fission yeast Dicer. EMBO J. 2011;30:4223–4235. doi: 10.1038/emboj.2011.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gredell JA, Dittmer MJ, Wu M, Chan C, Walton SP. Recognition of siRNA asymmetry by TAR RNA binding protein. Biochemistry. 2010;49:3148–3155. doi: 10.1021/bi902189s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toroney R, Hull CM, Sokoloski JE, Bevilacqua PC. Mechanistic characterization of the 5′-triphosphate-dependent activation of PKR: lack of 5′-end nucleobase specificity, evidence for a distinct triphosphate binding site, and a critical role for the dsRBD. RNA. 2012;18:1862–1874. doi: 10.1261/rna.034520.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SB, Green SR, Mathews MB, Esteban M. Activation of the double-stranded RNA (dsRNA)-activated human protein kinase in vivo in the absence of its dsRNA binding domain. Proc Natl Acad Sci USA. 1994;91:10551–10555. doi: 10.1073/pnas.91.22.10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson E, Pierre-Louis WS, Wong CJ, Lary JW, Cole JL. Heparin Activates PKR by Inducing Dimerization. J Mol Biol. 2011;413:973–984. doi: 10.1016/j.jmb.2011.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dauber B, et al. Influenza B virus ribonucleoprotein is a potent activator of the antiviral kinase PKR. PLoS Pathog. 2009;5:e1000473. doi: 10.1371/journal.ppat.1000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nature Protocols. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc Natl Acad Sci USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ucci JW, Cole JL. Global analysis of non-specific protein-nucleic interactions by sedimentation equilibrium. Biophys Chem. 2004;108:127–140. doi: 10.1016/j.bpc.2003.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kostura M, Mathews MB. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol Cell Biol. 1989;9:1576–1586. doi: 10.1128/MCB.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu S, Kumar KU, Kaufman RJ. Identification and requirement of three ribosome binding domains in dsRNA-dependent protein kinase (PKR) Biochemistry. 1998;37:13816–13826. doi: 10.1021/bi981472h. [DOI] [PubMed] [Google Scholar]
- 34.Patel RC, Stanton P, Sen GC. Role of the amino-terminal residues of the interferon-induced protein kinase in its activation by double-stranded RNA and heparin. J Biol Chem. 1994;269:18593–18598. [PubMed] [Google Scholar]
- 35.Fasciano S, Hutchins B, Handy I, Patel RC. Identification of the heparin-binding domains of the interferon-induced protein kinase. PKR. 2005;272:1425–1439. doi: 10.1111/j.1742-4658.2005.04575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Husain B, Mukerji I, Cole JL. Analysis of high-affinity binding of protein kinase R to double-stranded RNA. Biochemistry. 2012;51:8764–8770. doi: 10.1021/bi301226h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu C, et al. The structural basis of 5′ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vela A, Fedorova O, Ding SC, Pyle AM. The thermodynamic basis for viral RNA detection by the RIG-I innate immune sensor. Journal of Biological Chemistry. 2012;287:42564–42573. doi: 10.1074/jbc.M112.385146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 40.Abbas YM, Pichlmair A, Górna MW, Superti-Furga G, Nagar B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katibah GE, et al. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc Natl Acad Sci USA. 2014;111:12025–12030. doi: 10.1073/pnas.1412842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katibah GE, et al. tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Mol Cell. 2013;49:743–750. doi: 10.1016/j.molcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hull CM, Bevilacqua PC. Discriminating Self and Non-Self by RNA: Roles for RNA Structure, Misfolding, and Modification in Regulating the Innate Immune Sensor PKR. Acc. Chem. Res. 2016;49:1242–1249. doi: 10.1021/acs.accounts.6b00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinicke LA, et al. RNA dimerization promotes PKR dimerization and activation. J Mol Biol. 2009;390:319–338. doi: 10.1016/j.jmb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinicke LA, Bevilacqua PC. Activation of PKR by RNA misfolding: HDV ribozyme dimers activate PKR. RNA. 2012;18:2157–2165. doi: 10.1261/rna.034744.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanoudenhove J, Anderson E, Krueger S, Cole JL. Analysis of PKR structure by small-angle scattering. J Mol Biol. 2009;387:910–920. doi: 10.1016/j.jmb.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna SA, et al. Molecular framework for the activation of RNA-dependent protein kinase. J Biol Chem. 2007;282:11474–11486. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- 48.Gelev V, et al. Mapping of the auto-inhibitory interactions of protein kinase R by nuclear magnetic resonance. J. Mol. Biol. 2006;364:352–363. doi: 10.1016/j.jmb.2006.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nanduri S, Rahman F, Williams BR, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson E, Cole JL. Domain stabilities in protein kinase R (PKR): evidence for weak interdomain interactions. Biochemistry. 2008;47:4887–4897. doi: 10.1021/bi702211j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong CJ, Launer-Felty K, Cole JL. Analysis of PKR-RNA interactions by sedimentation velocity. Meth Enzymol. 2011;488:59–79. doi: 10.1016/B978-0-12-381268-1.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 53.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 54.Laue, T. M., Shah, B., Ridgeway, T. M. & Pelletier, S. L. In Analytical Ultracentrifugation in Biochemistry and Polymer Science (eds Rowe, A. J., Horton, J. C. & Harding, S. E.) 90–125 (Royal Society of Chemistry, Cambridge, England, 1992).
- 55.McWilliam H., et al. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Research. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


