Abstract
Mitochondria are inherited maternally in most animals, but the mechanisms of selective paternal mitochondrial elimination (PME) are unknown. While examining fertilization in C. elegans, we observe that paternal mitochondria rapidly lose their inner membrane integrity. CPS-6, a mitochondrial endonuclease G, serves as a paternal mitochondrial factor that is critical for PME. The CPS-6 endonuclease relocates from the intermembrane space of paternal mitochondria to the matrix following fertilization to degrade mitochondrial DNA. It acts with maternal autophagy and proteasome machineries to promote PME. Loss of cps-6 delays breakdown of mitochondrial inner membranes, autophagosome enclosure of paternal mitochondria, and PME. Delayed removal of paternal mitochondria causes increased embryonic lethality, demonstrating that PME is important for normal animal development. Thus, CPS-6 functions as a paternal mitochondrial degradation factor during animal development.
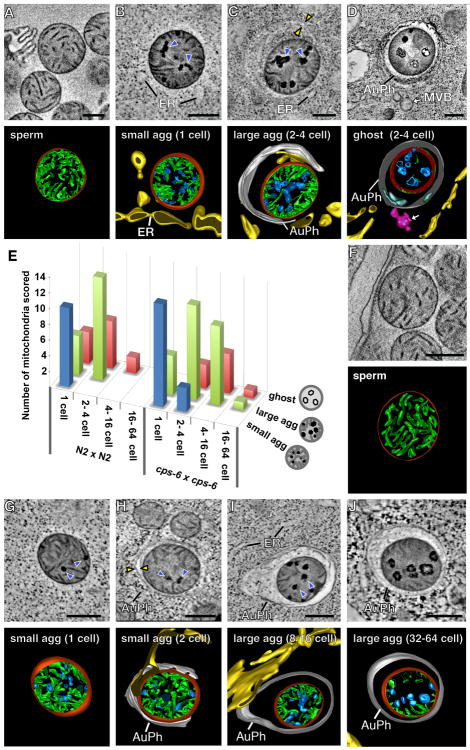
Mitochondria are critical for many cellular processes, including cellular respiration, apoptosis, and metabolism, and possess their own genome (mtDNA)(1, 2). However, only maternal mitochondria are passed on to progeny. Why and how paternal mitochondria are selectively eliminated during development is unclear (3, 4). To address these questions, we examined paternal mitochondria in C. elegans spermatozoa and embryos by electron microscopy (EM) and tomography. Mitochondria in wild-type (N2) spermatozoa are spherical (fig. S1A), with an average diameter of 464 ± 68 nm, and their cristae, formed by extensive infolding of the inner membrane, uniformly distribute in the matrix (Fig. 1A). Paternal mitochondria in N2 zygotes are readily distinguished from the tubular and thinner maternal mitochondria (with an average width of 238 ± 57 nm, fig. S1B). Notably, all paternal mitochondria in N2 zygotes have multiple dark aggregates (agg) in the matrix that form promptly after their entry into oocytes (Fig. 1, B and E, fig. S1, B to G, and movie S1). The double-layer membranes from autophagosomes have started to assemble around some paternal mitochondria (fig. S1B). We named paternal mitochondria containing small aggregates that lack nearby autophagosome membranes “small agg PM” (Fig. 1B). Those containing larger aggregates that are associated with autophagosome membranes are called “large agg PM” (Fig. 1C), and those with few cristae and enclosed in an autophagosome are called “ghost PM” (Fig. 1D). Many small agg PM arise independently of the autophagy machinery (Fig. 1, B and E, fig. S1, B to G, and movie S1). Large agg PM and ghost PM are observed in N2 zygotes, but mostly seen in 2- or 4-cell stage embryos (Fig. 1, C to E).
Fig. 1. Loss of cps-6 delays internal breakdown of paternal mitochondria following fertilization.
(A to D, F to J) Tomographic slice images and corresponding 3D models of a mitochondrion in an N2 (A) or cps-6(tm3222) (F) spermatozoon or a paternal mitochondrion in an N2 embryo (B to D) or a cps-6(tm3222) embryo (G to J) at the indicated stages. 3D models of autophagosomes (AuPh) and endoplasmic reticulum (ER) are shown. Mitochondrial membranes, cristae, and aggregates are colored in red, green, and blue, respectively. Dark aggregates and autophagosome membranes are indicated with blue and yellow arrowheads, respectively. Scale bars indicate 300 nm. (E) A histogram showing three classes of paternal mitochondria in different stage embryos from the indicated N2 cross (n=45) or cps-6(tm3222) cross (n=56).
In large agg PM, cristae are cleared from the central region as the aggregates enlarge in the matrix (Fig. 1C), which occurs before autophagosomes enclose paternal mitochondria. Once enclosed by autophagosomes, they lose matrix contents except for some remaining aggregates, but their outer membrane does not rupture until most of the cristae have disappeared (Fig. 1D and fig. S1H). These results suggest that paternal mitochondria are destroyed partly in embryos by self-initiated internal breakdown prior to the autophagosome assembly and degradation.
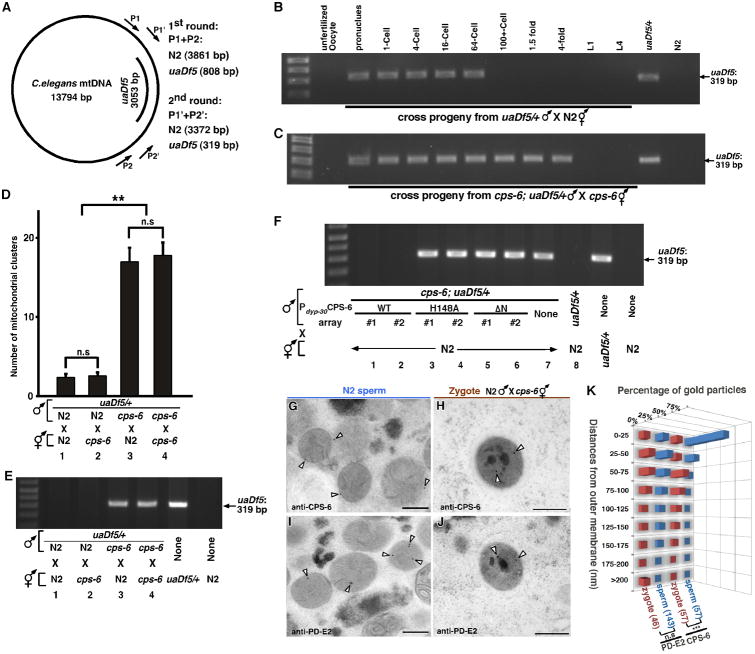
To identify intrinsic mitochondrial factors involved in PME, we performed an RNAi screen against 217 C. elegans nuclear genes predicted to encode mitochondrial proteins (table S1), using a sensitive polymerase chain reaction (PCR)-based method and a 3053 bp mtDNA deletion allele (uaDf5; Fig. 2A) to track the fate of mtDNA (5). uaDf5 mtDNA was detected in cross-progeny at all developmental stages from mating of N2 males with uaDf5/+ heteroplasmic hermaphrodites (fig. S2A)(5), but was only detected in early embryos and not in late embryos or larval cross-progeny from N2 hermaphrodites mated with uaDf5/+ males (Fig. 2B), indicating that PME is conserved in C. elegans (5-7). RNAi of the cps-6 gene, which encodes a homolog of human mitochondrial endonuclease G (8, 9), caused persistence of paternal uaDf5 mtDNA until the late stages of embryogenesis, a finding not observed in RNAi of other genes (fig. S2B and Materials and Methods). A 336 bp deletion (tm3222) in cps-6, which removes the catalytic site of CPS-6 (fig. S2C)(10), has the same effect as cps-6(RNAi), leading to persistence of paternal uaDf5 mtDNA throughout embryo development (Fig. 2C), whereas uaDf5 was detected only in 64-cell or earlier embryos in crosses between uaDf5/+ males and N2 hermaphrodites (Fig. 2B). These results indicate that cps-6 is associated with rapid removal of paternal mtDNA during early embryogenesis.
Fig. 2. CPS-6 relocates from the intermembrane space of paternal mitochondria to the matrix following fertilization to promote PME.
(A) A diagram of C. elegans mtDNA, the uaDf5 deletion, primers used in the nested PCR assays, and sizes of PCR products in N2 and uaDf5/+ animals. (B and C) Hermaphrodites and MTR-stained males were mated as indicated. Males also carried smIs42, an integrated Psur-5sur-5∷gfp transgene used to track cross progeny (see Fig. S2A). A single unfertilized oocyte and a single cross-fertilized embryo or larva (MTR or GFP positive) at the indicated stage was analyzed by PCR. uaDf5/+ and N2 hermaphrodites were controls. (D) Quantification of MTR-stained paternal mitochondrial clusters in 64-cell embryos from the indicated crosses with MTR-stained males. Means ± SEM; n=20 per cross. ** P < 0.0001 using unpaired Student's t-test. “n.s”, no significant difference. (E, F) Five cross-fertilized embryos (E) or transgenic embryos (F) at approximately 100-cell stage from the indicated crosses were analyzed by PCR. (G to J) Representative immuno-EM images of mitochondria in N2 spermatozoa and paternal mitochondria in zygotes from the indicated cross. CPS-6-specific and PD-E2-specific immunogold particles are marked with arrowheads. Scale bars: 300 nm. (K) Histogram of the distances of 15 nm immunogold particles from the mitochondrial membrane, illustrating CPS-6's movement after fertilization. Numbers in parentheses indicate the numbers of immunogold particles scored. *** P < 0.0001 using Mann-Whitney U test. cps-6(tm3222) was used in all figures.
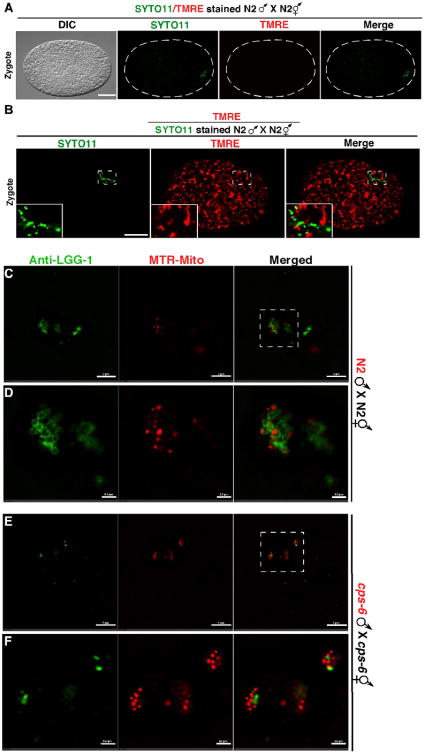
We performed microscopic analysis to monitor the disappearance of paternal mitochondria stained by Mitotracker Red (MTR), a mitochondrion-specific dye (5). When MTR-stained N2 males were mated with unstained N2 hermaphrodites, MTR-stained paternal mitochondria were seen in embryos before the 64-cell stage (fig. 3, A to G), indicating that PME occurs in concert with paternal mtDNA elimination (Fig. 2B). Conversely, loss of cps-6 resulted in persistence of MTR paternal mitochondria to around the 550-cell stage embryos (fig. S3, H to N), confirming that CPS-6 promotes rapid clearance of paternal mitochondria.
Fig. 3. Depolarization of paternal mitochondria following fertilization and autophagosome formation on paternal mitochondria.
(A, B) DIC and fluorescence images of zygotes from the indicated crosses are shown. TMRE had equal access to maternal and paternal mitochondria in the zygote (B). Scale bars represent 10 μm. (C to F) Zygotes from the indicated crosses with MTR-stained males were labeled with an antibody to LGG-1. Images were acquired using a Nikon SIM. Dash rectangles highlight the areas enlarged and shown below (D and F). Scale bars, 2 μm (C and E) and 0.5 μm (D and F).
CPS-6 was first identified as an apoptotic nuclease that translocates from mitochondria to the nucleus during apoptosis to mediate chromosome fragmentation (8, 9). A non-apoptotic role of CPS-6 in C. elegans has not been reported. We examined if CPS-6 is required paternally or maternally for PME using both the PCR assay and the microscopic assay (supplementary text) and found that a significant portion of paternal mitochondria and mtDNA persisted past the 64-cell embryonic stage when paternal cps-6 was defective (Fig. 2, D and E, mating 3, 4). By contrast, embryos without maternal cps-6 displayed normal PME (Fig. 2, D and E, mating 2). These results indicate that paternal CPS-6 is required to promote PME.
CPS-6 is imported into mitochondria through a mitochondrial targeting sequence (amino acids 1-21), because CPS-6ΔN, lacking this targeting sequence, localizes to the nucleus (8). Expression of CPS-6, but not CPS-6ΔN, in cps-6(tm3222) males through the ubiquitously expressed dpy-30 gene promoter rescued the defect in PME (Fig. 2F, mating 1, 2, 5-7; supplementary text), indicating that localization of CPS-6 in paternal mitochondria is required to mediate PME. Because expression of the nuclease-deficient CPS-6(H148A) mutant in cps-6(tm3222) males failed to rescue the PME defect (Fig. 2F, mating 3, 4), the nuclease activity of CPS-6 is essential for PME.
We examined how loss of cps-6 impacts PME by electron tomography. In cps-6(tm3222) zygotes, aggregates are still visible in paternal mitochondria, but are smaller and less than in N2 zygotes, and no ghost PM are detected (Fig. 1, B, E and G), indicating reduced and slower internal breakdown of paternal mitochondria. The autophagosome membranes start to assemble around paternal mitochondria in 2- or 4-cell cps-6(tm3222) embryos and complete enclosure by 16-cell embryos (Fig. 1, H and I), proceeding significantly slower than in N2 embryos, when autophagosome assembly starts earlier at the 1-cell stage and completes by 4-cell embryos (Fig. 1, C to E). Even after autophagosome enclosure, internal breakdown of paternal mitochondria is clearly delayed (Fig. 1, I and J), because a significant portion of cristae remained superficially intact and less than 40% of paternal mitochondria transited into ghost PM in 16-cell cps-6(tm3222) embryos (Fig. 1E). Some large agg PM even lingered on in 64-cell embryos (Fig. 1J), compared with 100% of paternal mitochondria either eliminated or becoming ghost PM by 4-cell N2 embryos (Fig. 1, D and E). Therefore, CPS-6 is important in mediating internal breakdown of paternal mitochondria and their enclosure by autophagosomes following fertilization.
Compromised mitochondria often show loss of membrane potential, which can be detected by tetramethylrhodamine ethyl ester (TMRE), a potential-sensitive mitochondrial dye. When N2 males prestained with TMRE and a nucleic acid dye SYTO11, which labeled sperm mitochondria and their mtDNA, respectively (fig. S4, A and B), were mated with N2 hermaphrodites, paternal mitochondria were still labeled by SYTO11 in N2 zygotes, but their TMRE staining was completely lost (Fig. 3A). In comparison, staining of paternal mitochondria by potential-insensitive MTR persisted (fig. S4B). When we mated SYTO11-stained N2 males with N2 hermaphrodites in the presence of TMRE, only maternal mitochondria were stained by TMRE and the SYTO11-positive paternal mitochondria were TMRE-negative (Fig. 3B). Therefore, paternal mitochondria are depolarized shortly after fertilization, preceding degradation of their mtDNA.
We determined the localization of CPS-6 in paternal mitochondria using immuno-EM. CPS-6 immunogold particles were predominantly associated with the mitochondrial membrane in N2 spermatozoa (Fig. 2, G and K, and fig. S2G), in agreement with CPS-6's localization in mitochondrial intermembrane space (IMS). In zygotes from cps-6(tm3222) hermaphrodites mated with N2 males, CPS-6 immunogold particles were often located inside paternal mitochondria, away from the mitochondrial membrane (Fig. 2, H and K, and fig. S5). Because some paternal mitochondria had not been associated with autophagosomes (Fig. 2H), CPS-6 appeared to enter the matrix before the assembly of autophagosomes. The relocation of CPS-6 into the matrix following fertilization is clearly discerned when compared with the localization patterns of a mitochondrial matrix protein, the E2 subunit of pyruvate dehydrogenase (PD-E2, Fig. 2, I to K). Collectively, these different microscopy analyses provide strong evidence that paternal mitochondria are depolarized and damaged internally soon after fertilization, leading to the release of CPS-6 into the matrix to catalyze mtDNA degradation.
The autophagy and proteasome pathways promote PME in C. elegans (5-7). Both LGG-1, the worm LC3/Atg8 homolog necessary for autophagosome formation (11), and RAD-23, a ubiquitin receptor important for proteasomal degradation (5, 12), act maternally to promote PME (fig. S6, A and B, and supplementary text). Analyses of the double and the triple mutants among cps-6, lgg-1 and rad-23 indicate that cps-6, lgg-1, and rad-23 utilize distinct mechanisms (mitochondrial self-destruction, autophagy, and proteasomes, respectively) to coordinate swift and efficient PME (fig. S6, C to F).
Because loss of cps-6 slows down autophagosome formation and degradation of paternal mitochondria (Fig. 1), we further interrogated this issue by immunostaining and found that in N2 zygotes bright LGG-1 staining was seen clustering around MTR-stained paternal mitochondria near the site of sperm entry (fig. S4C), with 81% of paternal mitochondrial clusters co-localizing with LGG-1 autophagosomes (fig. S4E). By contrast, in cps-6(tm3222) zygotes, such co-localization dropped to 43% (fig. S4, D and E), indicating that loss of cps-6 reduces autophagosome formation on paternal mitochondria. Analysis using superresolution Structured Illumination Microscopy (SIM) revealed similar results. In N2 zygotes, the majority (77%) of paternal mitochondria were enclosed by LGG-1 autophagosomes, some (12%) were partially enclosed, and only 11% did not associate with (isolated) autophagosomes (Fig. 3, C and D, and fig. S4, F and H). By contrast, in cps-6(tm3222) zygotes, 51% of paternal mitochondria were isolated and only 29% and 20% of paternal mitochondria were enclosed and partially enclosed by autophagosomes, respectively (Fig. 3, E and F, and fig. S4, G and H). These findings indicate that the CPS-6 self-destruction process is important for efficient recruitment of autophagosomes to paternal mitochondria.
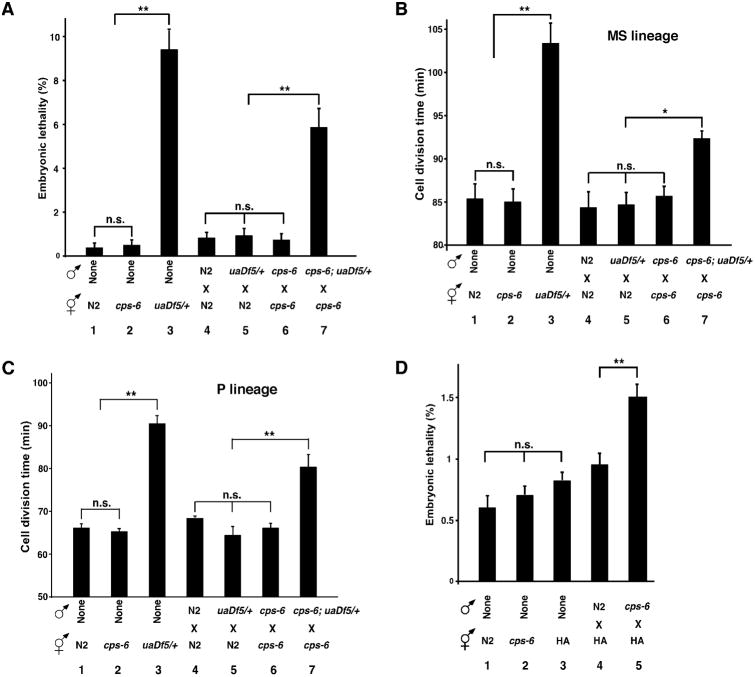
It has been suggested that the high rate of energy consumption during fertilization of an oocyte by many competing spermatozoa leads to increased oxidative damage and mutations in sperm mtDNA (13, 14). Failure to remove paternal mitochondria with mutated mtDNA can cause incompatibility with maternal mitochondria and the nuclear genome and adversely affect the fitness of animals (15-17). Comparison of N2 embryos with uaDf5/+ embryos, with four genes deleted in uaDf5 mtDNA (18), reveals a 23-fold increase in embryonic lethality from 0.4% to 9.4% (Fig. 4A, assays 1 and 3), indicating that the heteroplasmic presence of mtDNA mutations compromises embryo development. Delayed removal of uaDf5 paternal mitochondria in embryos by loss of cps-6 resulted in a lethality rate of 5.9%, 5-7 folds higher than that (0.7%) of cross-fertilized cps-6(tm3222) embryos or that (0.8-0.9%) of embryos with no persistent paternal mitochondria (Fig. 4A, assays 4-7, and supplementary text). Moreover, delayed clearance of uaDf5 paternal mitochondria slows down cell divisions, an energy-driven process, during C. elegans embryogenesis, because the average durations of cell divisions in two different cell lineages, the MS and the P lineages, were significantly prolonged in uaDf5/+ embryos and by delayed removal of uaDf5 paternal mitochondria (Fig. 4, B and C, fig. S7, and supplementary text). These results provide important evidence that delayed clearance of mutated paternal mitochondria leads to decreased fitness at the cellular and organismal levels and presents an evolutionary disadvantage.
Fig. 4. Delayed removal of paternal mitochondria increases embryonic lethality and cell division durations.
(A to D) The embryonic lethality rate (A and D) and the durations of cell divisions in the MS (B) and the P lineage (C) were scored in self-fertilized embryos (1-3) or cross-fertilized embryos from crosses (4-7) of the indicated genotypes. All males carried smIs42 and were stained with MTR to assist identification of zygotes (B and C). Means ± SEM; n >1000 embryos per cross at 25°C (A and D) and n=3 embryos per cross at 20°C (B and C). ** P < 0.001, * P < 0.05 using unpaired Student's t-test.
Next we examined the consequence of delayed removal of wild-type paternal mitochondria by mating two different C. elegans wild-type strains, the Bristol strain (N2) and the Hawaii strain (HA). Delayed removal of wild-type Bristol paternal mitochondria in HA embryos due to loss of paternal cps-6 also resulted in a significantly higher percentage of embryonic lethality than that seen in HA embryos without persistent paternal mitochondria (Fig. 4D, assays 4, 5, fig. S6G, mating 2, 4, and supplementary text). Therefore, delayed clearance of wild-type paternal mitochondria slightly different from maternal mitochondria also compromises animal development, suggesting that transmission of paternal mitochondria among different wild-type variants is evolutionarily disadvantageous.
In this study, we show that soon after fertilization, paternal mitochondria are depolarized and lose their inner membrane integrity, which apparently marks them for degradation by autophagy (19, 20). The inner membrane breakdown probably triggers the entry of the intermembrane CPS-6 into the matrix of paternal mitochondria to degrade mtDNA, which encodes 12 mitochondrial proteins, two rRNAs, and 22 tRNAs, that are essential for normal functions and maintenance of mitochondria (1, 13, 18). Degradation of mtDNA is detrimental and accelerates breakdown of paternal mitochondria, and probably, externalization of signals recognized by the autophagy or proteasome machinery (19, 20), leading to PME (fig. S8). Consistent with this model, loss of paternal cps-6 delays internal breakdown of paternal mitochondria and their enclosure and degradation by the autophagy machinery. Interestingly, delayed removal of either mutant or slightly different wild-type paternal mitochondria results in increased embryonic lethality in heteroplasmic animals, likely due to incompatibility in cellular signaling between the mitochondrial and the nuclear genomes (15, 17). This provides evidence that persistence of paternal mitochondria compromises animal development and may be the impetus for maternal inheritance of mitochondria. Lastly, the non-apoptotic function of CPS-6 in PME expands the complex roles of Endonuclease G in apoptosis, mitochondrial maintenance, and spermatogenesis (9, 21, 22).
Supplementary Material
Acknowledgments
We thank T. Blumenthal, R. Poyton, and K. Krauter for comments and H. Zhang for strains and antibodies. This work was supported by a March of Dimes grant (#1-FY14-300) and NIH grants (GM59083, GM79097 and GM118188) to D.X. and Research Grants Council of Hong Kong (AoE/M-05/12) to B.H.K. The present address of Q.H.Z. is: The first affiliated hospital, Biomedical Translational Research Institute, Jinan University, Guangzhou, 510630.
References and Notes
- 1.Calvo SE, Mootha VK. Annu Rev Genomics Hum Genet. 2010;11:25. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X. Genes Dev. 2001;15:2922. [PubMed] [Google Scholar]
- 3.Birky CW., Jr Proc Natl Acad Sci USA. 1995;92:11331. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Elazar Z. Science. 2011;334:1069. doi: 10.1126/science.1215480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Q, Li H, Xue D. Cell Res. 2011;21:1662. doi: 10.1038/cr.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al Rawi S. Science. 2011;334:1144. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 7.Sato M, Sato K. Science. 2011;334:1141. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 8.Parrish J, et al. Nature. 2001;412:90. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- 9.Li LY, Luo X, Wang X. Nature. 2001;412:95. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 10.Lin JL, et al. J Biol Chem. 2012;287:7110. doi: 10.1074/jbc.M111.316075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia K, Levine B. Adv Exp Med Biol. 2010;694:47. doi: 10.1007/978-1-4419-7002-2_5. [DOI] [PubMed] [Google Scholar]
- 12.Madura K. Trends Biochem Sci. 2004;29:637. doi: 10.1016/j.tibs.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Wallace DC. Environ Mol Mutagen. 2010;51:440. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 14.Cummins JM. Human Reproduction. 2000;15:92. doi: 10.1093/humrep/15.suppl_2.92. [DOI] [PubMed] [Google Scholar]
- 15.Sharpley MS, et al. Cell. 2012;151:333. doi: 10.1016/j.cell.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz M, Vissing J. N Engl J Med. 2002;347:576. doi: 10.1056/NEJMoa020350. [DOI] [PubMed] [Google Scholar]
- 17.Lane N. Bioessays. 2011;33:860. doi: 10.1002/bies.201100051. [DOI] [PubMed] [Google Scholar]
- 18.Tsang WY, Lemire BD. Biochem Cell Biol. 2002;80:645. doi: 10.1139/o02-135. [DOI] [PubMed] [Google Scholar]
- 19.Ashrafi G, Schwarz TL. Cell Death Differ. 2013;20(1):31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Klionsky DJ. Autophagy. 2011;7:297. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDermott-Roe C, et al. Nature. 2011;478:114. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLuca SZ, O'Farrell PH. Dev Cell. 2012;22:660. doi: 10.1016/j.devcel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.