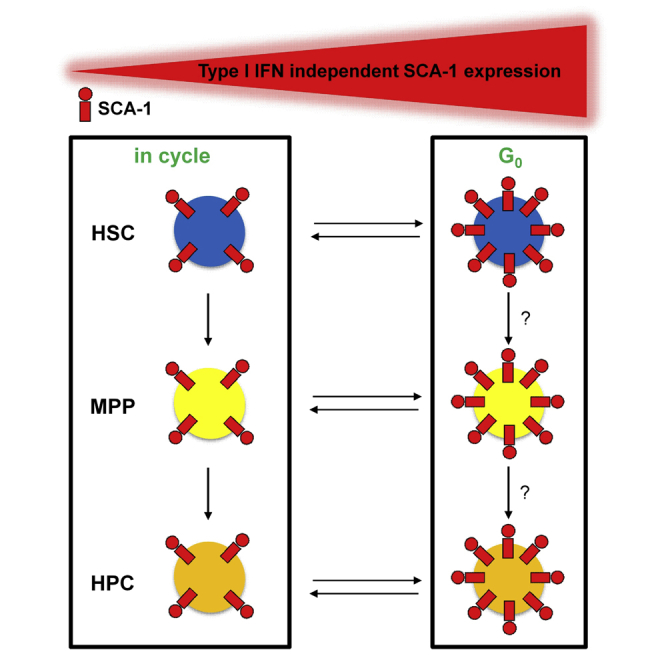
Summary
Blood cell generation depends on continuous cellular output by the sequential hierarchy of hematopoietic stem cell (HSC) and progenitor populations that all contain quiescent and actively cycling cells. Hematopoietic stem and progenitor cells (HSPCs) express the surface molecule Stem cell antigen 1 (SCA-1/LY6A). Using histone 2B-red fluorescent fusion protein label retention and cell-cycle reporter mice, we demonstrate that high SCA-1 expression (SCA-1hi) identifies not only quiescent HSCs but quiescent cells on all hierarchical levels within the lineage−SCA-1+KIT+ (LSK) population. Each transplanted SCA-1hi HSPC population also displayed self-renewal potential superior to that of the respective SCA-1lo population. SCA-1 expression is inducible by type I interferon (IFN). We show, however, that quiescence and high self-renewal capacity of cells with brighter SCA-1 expression at steady state were independent of type I IFN signaling. We conclude that SCA-1 expression levels can be used to prospectively isolate functionally heterogeneous HSPC subpopulations.
Keywords: hematopoeitic stem cells, hematopoeitic progenitor cells, type I interferon, hematopoietic stem and progenitor cell purification and analysis, hematopoietic stem and progenitor cell potential, hematopoietic stem and progenitor cell heterogeneity, cell cycle
Graphical Abstract
Highlights
-
•
High SCA-1 expression marks quiescent and potent hematopoietic progenitor cells
-
•
High SCA-1 expression by quiescent HSPCs is independent of type I IFN signaling
In this article, Gerbaulet and colleagues report on SCA-1 expression level as a marker for prospective isolation of quiescent cells with elevated repopulation potential not only in hematopoietic stem cells but also in progenitor populations. They show that high SCA-1 expression by quiescent hematopoietic stem and progenitor cells is independent of type I interferon signaling.
Introduction
The multi-potent and self-renewing hematopoietic stem cells (HSCs) at the top of the hierarchy give rise to hematopoietic progenitor cell (HPC) populations with gradually narrowing differentiation and self-renewal potential (Eaves, 2015). Murine HSCs and uncommitted progenitors express the glycosyl phosphatidylinositol-anchored cell surface protein Stem cell antigen 1 (SCA-1/LY6A) (Holmes and Stanford, 2007) and reside within the lineage−SCA-1+KIT+ (LSK) population (Okada et al., 1992, Purton and Scadden, 2007, Spangrude et al., 1988) of the bone marrow (BM).
Numerous studies have demonstrated heterogeneity within each of the hematopoietic stem and progenitor cell (HSPC) populations with regard to cell-cycle activity (Wilson et al., 2008), lineage specification (Dykstra et al., 2007, Müller-Sieburg et al., 2002), and repopulation activity (Foudi et al., 2008, Qiu et al., 2014). Multiple cell-surface markers allowing for prospective isolation of the quiescent subset of HSCs (Balazs et al., 2006, Grinenko et al., 2014, Kiel et al., 2005, Osawa et al., 1996, Shin et al., 2014, Sudo et al., 2012) have been proposed. However, a marker uniformly associated with quiescence among the LSK population has been lacking so far. Two recent studies (Säwén et al., 2016, Wilson et al., 2015) showed increased quiescence and repopulation activity of immunophenotypic HSCs with high expression of SCA-1 (SCA-1hi).
While the function of the SCA-1 molecule remains enigmatic (Holmes and Stanford, 2007), it is well established that SCA-1 is strongly upregulated by type I (Essers et al., 2009, Pietras et al., 2014) and type II (de Bruin et al., 2014) interferon (IFN). SCA-1 seems to regulate cellular responses to type I IFN as loss of SCA-1 abrogates type I IFN-induced proliferation of HSCs (Essers et al., 2009, Walter et al., 2015).
In the present work, we show that type I IFN-independent high expression of SCA-1 identifies quiescent cells with elevated repopulation activity not only in the HSC compartment but also in LSK progenitor populations.
Results
High SCA-1 Expression Identifies Quiescent Cells Not Only in HSCs but Also in Progenitor Populations
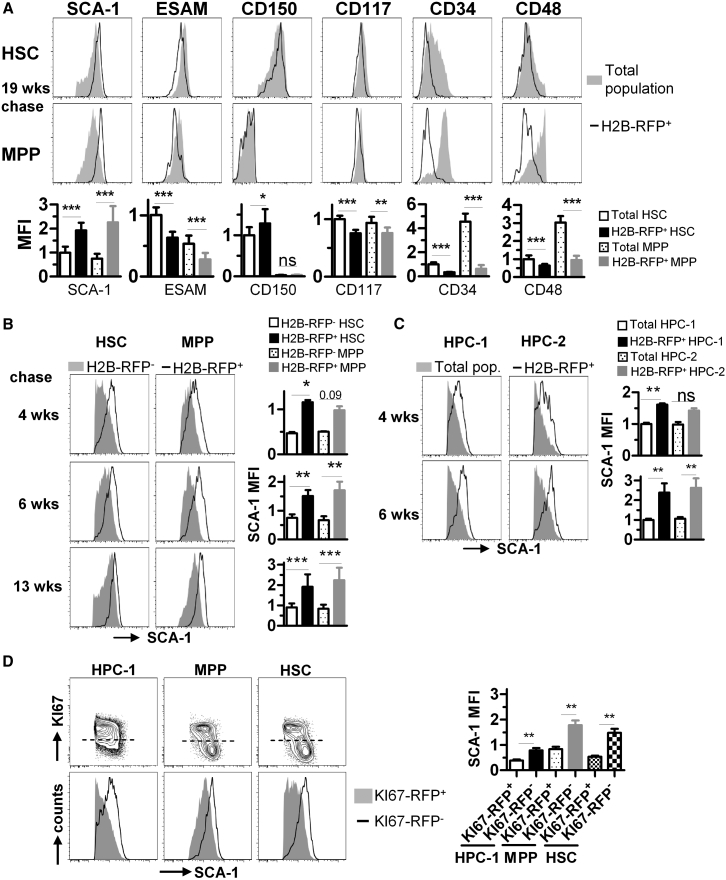
To identify the most quiescent HSPCs, we induced expression of histone 2B-red fluorescent fusion protein (H2B-RFP) by doxycycline (DOX) treatment of R26rtTA/Col1A1H2B-RFP mice (Egli et al., 2007) for 8 weeks and analyzed the BM after different chase periods for retention of H2B-RFP by flow cytometry (Figures S1A and S1B). In accordance with previous studies (Foudi et al., 2008, Qiu et al., 2014, Säwén et al., 2016, Wilson et al., 2008), we observed rapid dilution of H2B-RFP from HPCs (HPC-1, LSK CD48hiCD150−, HPC-2, LSK CD48hiCD150+) (Oguro et al., 2013) (see Figure S1C for gating), while a proportion of the HSC (LSK CD48−/loCD150+) and multipotent progenitor (MPP) populations (LSK CD48−/loCD150−) (Kiel et al., 2005) retained the label for up to 19 weeks (see Figure S1D). We compared the expression of surface antigens between quiescent H2B-RFP+ cells and their respective parental populations. Most prominently, we observed significantly higher expression of SCA-1 on label-retaining HSCs and MPPs after 4, 6, 13, and 19 weeks of chase (Figures 1A and 1B). Moreover, elevated SCA-1 expression was also detectable in both H2B-RFP+ HPC populations after 4 and 6 weeks of chase (Figure 1C). In addition, we found a significant downregulation of endothelial cell-specific adhesion molecule (ESAM) (Yokota et al., 2008), CD117 (Grinenko et al., 2014, Shin et al., 2014), CD34, and CD48 (Qiu et al., 2014) on 19-week label-retaining HSCs and MPPs, while CD150 (Beerman et al., 2010, Kiel et al., 2005, Morita et al., 2010) was slightly, but significantly, upregulated on H2B-RFP+ HSCs (Figure 1A).
Figure 1.
Quiescent Hematopoietic Stem and Progenitor Cells Express High Levels of SCA-1
(A–C) R26rtTA/Col1A1H2B-RFP mice were DOX-pulsed, chased for either 4, 6, 13, or 19 weeks, and HSPC populations were analyzed by flow cytometry for retention of H2B-RFP (see also Figures S1A–S1D). (A) Representative histograms of BM HSCs (LSK CD48−/loCD150+, upper row) or MPPs (LSK CD48−/loCD150−, middle row) from mice (n = 9) chased for 19 weeks are shown. H2B-RFP+ label retaining cells (black lines) were overlaid on to the total parental population (solid gray histograms). Antigen median fluorescence intensities (MFI, lower row, normalized to the mean MFI of HSCs) of the total or H2B-RFP+ populations were determined and compared. (B) Representative histograms for SCA-1 expression of BM HSCs (left column) and MPPs (middle column) isolated from R26rtTA/Col1A1H2B-RFP mice chased for either 4 (upper row, n = 2), 6 (middle row, n = 4), or 13 weeks (lower row, n = 8) are shown and H2B-RFP+ (black lines) and H2B-RFP− cells (solid gray histograms) were overlaid. SCA-1 MFIs (right column, normalized to total HSC population) of H2B-RFP− and H2B-RFP+ HSPCs were compared. (C) Representative SCA-1 expression histograms of BM HPC-1 (LSK CD48hiCD150−, left column) or HPC-2 cells (LSK CD48hiCD150+, middle column) from R26rtTA/Col1A1H2B-RFP mice (as in B) are shown. H2B-RFP+ populations (black lines) were overlaid onto the respective parental population (solid gray histograms). SCA-1 MFI (right column, mean MFI of total HPC-1 set to 1) of total and H2B-RFP+ populations was calculated.
(D) BM of Ki67-RFPki/wt mice (n = 4, see also Figures S1E–S1G) was analyzed by flow cytometry. Representative contour plots of HPCs-1, MPPs, and HSCs are shown (upper row; dotted line shows threshold for KI67-RFP gating, set according to Ki67-RFPwt/wt control). Lower row compares SCA-1 expression between KI67-RFP+ cells (solid gray histograms) and KI67-RFP− (black lines) cells among the respective parental population. Right data plot quantifies SCA-1 MFI of HSPC populations separated by KI67-RFP expression. SCA-1 MFI was normalized to the mean MFI of the total HSC population, which was set to 1.
Means ± SD are shown throughout the figure. Significance was calculated by paired Student's t test and Bonferroni-Holm error correction for multiple testing. ∗p = 0.01–0.05, ∗∗p = 0.001–0.01, ∗∗∗p < 0.001; ns, not significant. See also Figure S1.
Next, we analyzed the BM of Ki67-RFPki/wt cell-cycle reporter mice (Basak et al., 2014), in which a KI67-RFP fusion protein faithfully reports quiescent (KI67-RFP−) and cycling (KI67-RFP+) HSPCs (Figure S1E). RFP expression did not alter hematopoiesis in this model as judged by HSPC compartment size and competitive transplantation (Figures S1F and S1G). Cells with bright SCA-1 expression were significantly enriched among quiescent KI67-RFP− HPC-1, MPP, and HSC populations, while cycling KI67-RFP+ HSPCs expressed lower SCA-1 levels (Figure 1D).
To identify alternative markers of HSPC quiescence, we correlated expression of CD201 (EPCR) and CD27 (Balazs et al., 2006, Vazquez et al., 2015, Wiesmann et al., 2000) to SCA-1 and KI67-RFP expression (Figures S1H and S1I). We found CD201 expression level to be extremely useful for prospective enrichment of KI67-RFP− HSPCs, and expression of CD201 and SCA-1 showed a strong positive correlation. In contrast, CD27 expression appeared to be independent of cell-cycle activity and SCA-1. We did not observe any link between ESAM and KI67-RFP expression among HSCs, as the latter uniformly expressed a high level of ESAM (Figure S1J), while ESAM expression was heterogeneous among HPC-1 and MPP and without correlation to KI67-RFP expression.
SCA-1hi HSPCs Have High Repopulation Activity upon Transplantation
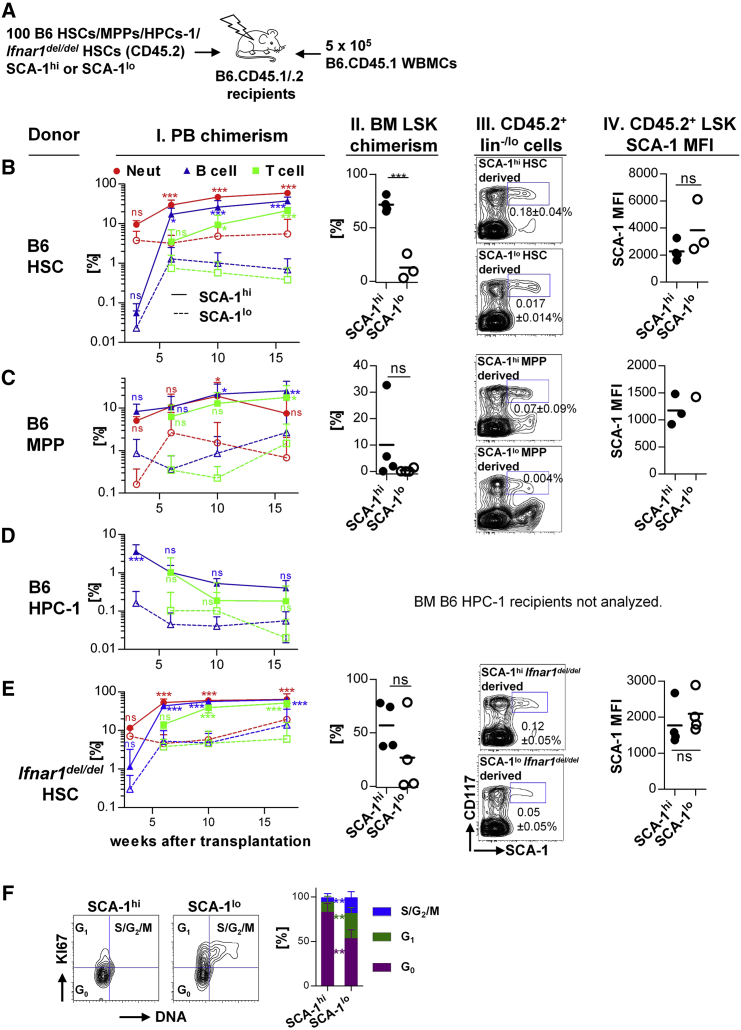
To investigate whether repopulation activity of donor HSPCs correlates with SCA-1 expression, we fractionated HSCs, MPPs, and HPCs-1 into either SCA-1hi or SCA-1lo populations (Figure S2A) and competitively transplanted these cells into lethally irradiated congenic recipients (Figure 2A). We observed a range of SCA-1 fluorescence intensity of approximately two decades among BM LSK cells and arbitrarily divided these SCA-1+ cells into two populations, in which the SCA-1lo fraction comprised events from the lower decade, while the SCA-1hi fraction consisted of events from the higher decade. The sorting gates were placed to discriminate SCA-1-negative outliers as well as avoiding overlap between SCA-1lo and SCA-1hi populations after sorting (Figures S2A and S2B).
Figure 2.
Type I IFN-Independent SCA-1 Expression Predicts Repopulation Activity
(A) HSPCs (either HSCs, MPPs, or HPCs-1 isolated from B6 mice or Ifnar1del/del HSCs, all CD45.2+) were fractionated according to SCA-1 expression (see Figure S2A) and 100 cells were transplanted together with 5 × 105 B6.CD45.1 competitor WBMCs into lethally irradiated B6.CD45.1/.2 recipients.
(B–E) Each row shows analysis of primary recipient mice transplanted with either B6 HSCs, B6 MPPs, B6 HPCs-1, or Ifnar1del/del HSCs (n = 3–4/group). (Column I) PB chimerism of recipient mice was monitored longitudinally (red circles, neutrophils; blue triangles, B cells; green boxes, T cells. Continuous line, filled symbols: SCA-1hi donor cells; dotted line, open symbols: SCA-1lo donor cells. Means ± SD are shown, significance calculated with repeated-measures two-way ANOVA with Bonferroni error correction). (Column II) BM analysis of recipient mice revealed LSK donor chimerism (individual recipients and means are shown, unpaired Student's t test, BM chimerism of B6 HPC-1 recipients was not analyzed due to complete lack of PB neutrophil reconstitution). (Column III) Representative contour plots of donor-derived (CD45.2+) lin−/lo cells are depicted. Frequency (mean ± SD) of the CD45.2+ LSK population among total WBMCs is shown. (Column IV) SCA-1 MFI of CD45.2+ LSK cells was calculated (individual recipients and means are shown, mice showing <0.001% of CD45.2+ LSK cells among total WBMCs were excluded from analysis, unpaired Student's t test). ∗p = 0.01–0.05, ∗∗p = 0.001–0.01, ∗∗∗p < 0.001; ns, not significant.
(F) Representative contour plots of cell-cycle analysis of either SCA-1hi or SCA-1lo BM LSK cells isolated from Ifnar1del/del mice (n = 5). Right data plot shows frequencies of LSK cells in G0, G1, or S/G2/M phase (mean ± SD, paired Student's t test with Bonferroni-Holm error correction). ∗∗p = 0.001–0.01.
See also Figure S2.
SCA-1hi donor HSCs displayed durable multi-lineage repopulation of recipient peripheral blood (PB) and BM (Figure 2B, columns I and II; Figures S2B–S2D), while SCA-1lo HSCs either contributed to a much lower extent in primary (Figure 2B) and secondary recipients (Figure S2E) or were devoid of long-term and serial repopulation activity (Figure S2D). In contrast to HSCs, SCA-1hi MPPs did not harbor stable multi-lineage repopulation potential as evidenced either by a decline in PB neutrophil and BM chimerism at 16 weeks after transplantation (Figure 2C, columns I and II) or by secondary transplantation (Figure S2E). However, the intermediate-term repopulation of all leukocyte lineages by transplanted SCA-1hi MPPs was significantly enhanced compared with SCA-1lo donor MPPs. Likewise, SCA-1hi donor HPC-1 cells exhibited significantly elevated B-cell chimerism 3 weeks after transplantation in contrast to the corresponding SCA-1lo population (Figure 2D, column I).
The comparison of SCA-1 expression among donor-derived (CD45.2+) BM cells isolated from primary recipients of SCA-1hi or SCA-1lo donor cells (B6 HSCs or MPPs) revealed that SCA-1lo donor cells or their progeny can reacquire high expression of SCA-1 (Figures 2B and 2C, columns III and IV; Figure S2F).
Higher SCA-1 Expression and Increased Repopulation Activity of Quiescent HSCs Does Not Depend on Type I IFN Signaling
SCA-1 expression is strongly upregulated by type I IFN (Essers et al., 2009), and Ifnar1del/del mice, which lack type I IFN signaling (Kamphuis et al., 2006), showed on average decreased steady-state SCA-1 expression, albeit similar maximum levels (Figure S2G). The frequency of LSK cells was reduced in Ifnar1del/del mice, but the frequency as well as cell-cycle activity of HSCs was unchanged in comparison with control mice. However, there was still heterogeneity of SCA-1 expression levels within the various LSK HSPC populations of Ifnar1del/del mice. To investigate whether the superior repopulation activity of SCA-1hi HSPCs depends on type I IFN, we competitively transplanted either SCA-1hi or SCA-1lo Ifnar1del/del HSCs (Figure S2A). The PB repopulation activity of SCA-1hi Ifnar1del/del donor HSCs was significantly elevated compared with their SCA-1lo counterparts (Figure 2E, column I). However, the chimerism level of SCA-1lo Ifnar1del/del recipients started to rise at the end of primary transplantation (Figure 2E, columns I and II) and upon secondary transplantation (Figure S2E, columns I and II). SCA-1lo Ifnar1del/del derived LSK cells of recipients showed reacquisition of bright SCA-1 expression (Figures 2E and S2E, columns III and IV). At the end of secondary transplantation, PB and BM chimerism as well as SCA-1 expression of both donor cell entities were indistinguishable (Figure S2E, columns I-IV). However, as recipients of SCA-1hi Ifnar1del/del donor HSCs displayed significantly higher PB chimerism throughout primary transplantation and in the initial phase of secondary transplantation, we conclude that constitutive type I IFN signaling does not account for the superior repopulation potential of HSPCs with high SCA-1 expression. Moreover, cell-cycle analysis of LSK cells from Ifnar1del/del mice corroborated our finding that the increased quiescence of SCA-1hi HSPCs was type I IFN independent (Figure 2F).
Discussion
We showed that high SCA-1 expression allows for prospective isolation of quiescent and potent hematopoietic progenitor cells. As previously reported, transplanted LSK CD48−/loCD150− MPPs did not exhibit long-term reconstitution, but showed robust intermediate-term contribution to all blood cell lineages (Oguro et al., 2013). We showed that the vast majority of this repopulation potential was confined to those cells which expressed the highest levels of SCA-1 within the MPP population. These cells were rarely dividing in situ, as evidenced by label retention characteristics similar to those of SCA-1hi HSCs as previously reported (Foudi et al., 2008, Säwén et al., 2016). Likewise, the B-lymphoid potential of the HPC-1 population was strongly enhanced among SCA-1hi cells. In addition, we confirmed the recent finding (Säwén et al., 2016, Wilson et al., 2015) that the majority of functional HSCs resided within the SCA-1hi LSK CD48−/loCD150+ population, while HSCs seemed to be rare or weak among those cells expressing lower amounts of SCA-1. However, SCA-1lo donor cells could give rise to progeny with a SCA-1hi phenotype, demonstrating plasticity of SCA-1 expression levels.
The BM lin−CD201+CD27+ population significantly overlaps with LSK cells in B6 mice and consists of early HSPCs. Lin−CD201+CD27+ cells have been proposed for alternative HSPC identification in mouse strains (e.g., BALB/c or NOD strains) or under stress conditions, which lack a distinct LSK immunophenotype (Vazquez et al., 2015). Our result that CD201 expression level correlates with quiescence of HSPCs similar to SCA-1 facilitates identification and isolation of quiescent subsets among lin−CD201+CD27+ HSPCs.
In contrast, ESAM, which was proposed to label HSCs under stress conditions in a cell-cycle-specific manner (Sudo et al., 2012), did not seem to be useful for prospective enrichment of quiescent HSPCs at steady state, as KI67-RFP and ESAM expression showed poor correlation.
We demonstrate that the properties of SCA-1hi cell populations were not a result of constitutive type I IFN signaling. The latter was surprising, as steady-state type I IFN partially accounted for SCA-1 expression (Figure S2G) and “tonic” (basal or constitutive) type I IFN signaling has been implied in HSC maintenance (Gough et al., 2012). However, our transplantation data revealed that SCA-1hi as well as SCA-1lo Ifnar1del/del HSCs outgrew wild-type (wt) competitor cells in secondary recipients, which may be explained either by a general competitive advantage of Ifnar1del/del HSCs over wt competitor HSCs or, due to lower overall SCA-1 expression in Ifnar1del/del mice, potent HSCs still reside in the SCA-1lo sorted population. The latter is very likely, as our SCA-1 sorting strategy for Ifnar1del/del donor cells separated only the brightest fraction of HSCs into the SCA-1hi subset, while SCA-1lo cells contained the majority of the cell population. Nevertheless, purified SCA-1hi Ifnar1del/del donor cells repopulated primary recipients faster and more robustly. Moreover, cell-cycle analysis of Ifnar1del/del HSPCs revealed that the increased quiescence of the SCA-1hi population was type I IFN independent.
Hematopoietic contribution, differentiation pattern, and the relationship between the HSPC subpopulations and diverse SCA-1 expression have not been addressed in situ so far. We speculate that discrepancies between recent studies reporting high (Sawai et al., 2016) or low (Busch et al., 2015, Schoedel et al., 2016, Sun et al., 2014) contribution of HSCs to steady-state hematopoiesis might reflect lineage tracing of exclusive HSC subpopulations that differ by SCA-1 expression and cell-cycle activity.
SCA-1 is already one of the most widely used markers for prospective isolation of murine HSPCs, and our finding that quiescent subpopulations are identified by differential SCA-1 expression easily allows for refined purification and analysis strategies.
Experimental Procedures
Mice
C57Bl/6 wt, B6.CD45.1, B6CD45.1/.2, Ifnar1del/del (Kamphuis et al., 2006), R26rtTA/Col1A1H2B-RFP (Egli et al., 2007) and Ki67-RFPki/wt (Basak et al., 2014) mice were housed at the Experimental Center, TU Dresden. R26rtTA/Col1A1H2B-RFP mice were induced by drinking water containing 1 mg/mL doxycycline (Applichem) and 1% sucrose ad libitum for 8 weeks.
All animal experiments were carried out in accordance with institutional guidelines and were approved by Landesdirektion Dresden Ref. No. 24-9168.11-1/2012-39.
Cell Preparation
Whole bone marrow cells (WBMCs) were isolated by crushing long bones with mortar and pestle using PBS/2% fetal calf serum (FCS)/2 mM EDTA and filtered through a 100-μm mesh. After erythrocyte lysis in NH4Cl buffer, cells were filtered through a 40-μm mesh. Hematopoietic lineage+ cells were removed with the lineage cell depletion kit (Miltenyi Biotec).
PB was drawn by retrobulbar puncture.
BM Transfer
B6.CD45.1/.2 recipient mice received a single dose of 9 Gy of total body irradiation (Yxlon Maxi Shot Source). Donor cells (purified CD45.2+ test cells mixed with B6.CD45.1 competitor WBMCs) were administered via intravenous injection into the retro-orbital sinus. For secondary transplantation, equal numbers of WBMCs from all primary transplanted individuals were pooled and 4 × 106 cells were transplanted into each lethally irradiated secondary recipient.
Flow Cytometry
Cells were stained for 30 min with fluorochrome-labeled antibodies (see Table S1) diluted in PBS/2% FCS/2 mM EDTA, washed twice, and analyzed or sorted on BD FACSAria or Miltenyi MACSquant flow cytometers. Data were analyzed using FlowJo 9.9.4 (Treestar). Negative and positive populations were identified by fluorescence-minus-one (FMO) controls. For gating of H2B-RFP+ and H2B-RFP− HSPC populations, the respective population from uninduced R26rtTA/Col1A1H2B-RFP mice served as background H2B-RFP controls. KI67-RFP+ and KI67-RFP− HSPCs were identified using antibody-stained HSPCs from Ki67-RFPwt/wt animals as negative controls.
For cell-cycle analysis, cells were incubated with antibodies against extracellular markers, fixated/permeabilized (BD Cytofix/Cytoperm kit), and intracellularly stained with anti-KI67 and FxCycle Violet DNA stain (Thermo Fisher Scientific). KI67− cells were gated according to FMO isotype controls.
Statistical Analysis
Statistical analysis was conducted with Prism 5 (GraphPad). Significance of results is indicated in the figures by ∗p = 0.01–0.05, ∗∗p = 0.001–0.01, ∗∗∗p < 0.001, and ns (not significant).
Author Contributions
M.N.F.M., A.G., K.B.S., A.H., and R.B. performed experiments and analyzed data. O.B. and H.C.C. provided Ki67-RFP mice. A.G. conceived the study and designed experiments. A.G. and A.R. wrote the manuscript.
Acknowledgments
This work was supported by the German Research Foundation (DFG) through grant SFB 655 B11 to A.R., by the Excellence Initiative of the German Federal and State Governments “Support the best” (ZUK64, to A.R.), and TU Dresden Medical Faculty “MeDDrive 2015” grant to A.G. The authors thank Livia Schulze, Christa Haase, Tobias Häring, and Christina Hiller for expert technical assistance.
Published: May 11, 2017
Footnotes
Supplemental Information includes two figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.04.012.
Supplemental Information
References
- Balazs A.B., Fabian A.J., Esmon C.T., Mulligan R.C. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood. 2006;107:2317–2321. doi: 10.1182/blood-2005-06-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak O., van de Born M., Korving J., Beumer J., van der Elst S., van Es J.H., Clevers H. Mapping early fate determination in Lgr5+ crypt stem cells using a novel KI67-RFP allele. EMBO J. 2014;33:2057–2068. doi: 10.15252/embj.201488017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I., Bhattacharya D., Zandi S., Sigvardsson M., Weissman I.L., Bryder D., Rossi D.J. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl. Acad. Sci. USA. 2010;107:5465–5470. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K., Klapproth K., Barile M., Flossdorf M., Holland-Letz T., Schlenner S.M., Reth M., Höfer T., Rodewald H.R. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- de Bruin A.M., Voermans C., Nolte M.A. Impact of interferon-γ on hematopoiesis. Blood. 2014;124:2479–2486. doi: 10.1182/blood-2014-04-568451. [DOI] [PubMed] [Google Scholar]
- Dykstra B., Kent D., Bowie M., McCaffrey L., Hamilton M., Lyons K., Lee S.J., Brinkman R., Eaves C. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- Eaves C.J. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood. 2015;125:2605–2613. doi: 10.1182/blood-2014-12-570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D., Rosains J., Birkhoff G., Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- Essers M.A.G., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., Trumpp A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- Foudi A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., Hock H. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 2008;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough D., Messina N., Clarke C., Johnstone R., Levy D. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36:166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinenko T., Arndt K., Portz M., Mende N., Günther M., Cosgun K.N., Alexopoulou D., Lakshmanaperumal N., Henry I., Dahl A. Clonal expansion capacity defines two consecutive developmental stages of long-term hematopoietic stem cells. J. Exp. Med. 2014;211:209–215. doi: 10.1084/jem.20131115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes C., Stanford W.L. Concise review: stem cell antigen-1: expression, function, and enigma. Stem Cells. 2007;25:1339–1347. doi: 10.1634/stemcells.2006-0644. [DOI] [PubMed] [Google Scholar]
- Kamphuis E., Junt T., Waibler Z., Forster R., Kalinke U. Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood. 2006;108:3253–3261. doi: 10.1182/blood-2006-06-027599. [DOI] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz Ö.H., Iwashita T., Yilmaz O.H., Terhorst C., Morrison S.J. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Morita Y., Ema H., Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J. Exp. Med. 2010;207:1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Sieburg C.E., Cho R.H., Thoman M., Adkins B., Sieburg H.B. Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood. 2002;100:1302–1309. [PubMed] [Google Scholar]
- Oguro H., Ding L., Morrison S.J. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S., Nakauchi H., Nagayoshi K., Nishikawa S., Miura Y., Suda T. In vivo and in vitro stem cell function of C-KIT- and SCA-1-positive murine hematopoietic cells. Blood. 1992;80:3044–3050. [PubMed] [Google Scholar]
- Osawa M., Hanada K., Hamada H., Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Pietras E.M., Lakshminarasimhan R., Techner J.-M., Fong S., Flach J., Binnewies M., Passegué E. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 2014;211:245–262. doi: 10.1084/jem.20131043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton L.E., Scadden D.T. Limiting factors in murine hematopoietic stem cell assays. Cell Stem Cell. 2007;1:263–270. doi: 10.1016/j.stem.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Qiu J., Papatsenko D., Niu X., Schaniel C., Moore K. Divisional history and hematopoietic stem cell function during homeostasis. Stem Cell Reports. 2014;2:473–490. doi: 10.1016/j.stemcr.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai C., Babovic S., Upadhaya S., Knapp D., Lavin Y., Lau C., Goloborodko A., Feng J., Fujisaki J., Ding L. Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity. 2016;45:597–609. doi: 10.1016/j.immuni.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säwén P., Lang S., Mandal P., Rossi D., Soneji S., Bryder D. Mitotic history reveals distinct stem cell populations and their contributions to hematopoiesis. Cell Rep. 2016;14:2809–2818. doi: 10.1016/j.celrep.2016.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoedel K., Morcos M., Zerjatke T., Roeder I., Grinenko T., Voehringer D., Göthert J., Waskow C., Roers A., Gerbaulet A. The bulk of the hematopoietic stem cell population is dispensable for murine steady-state and stress hematopoiesis. Blood. 2016;128:2285–2296. doi: 10.1182/blood-2016-03-706010. [DOI] [PubMed] [Google Scholar]
- Shin J.Y., Hu W., Naramura M., Park C.Y. High C-KIT expression identifies hematopoietic stem cells with impaired self-renewal and megakaryocytic bias. J. Exp. Med. 2014;211:217–231. doi: 10.1084/jem.20131128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude G.J., Heimfeld S., Weissman I.L. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Sudo T., Yokota T., Oritani K., Satoh Y., Sugiyama T., Ishida T., Shibayama H., Ezoe S., Fujita N., Tanaka H. The endothelial antigen ESAM monitors hematopoietic stem cell status between quiescence and self-renewal. J. Immunol. 2012;189:200–210. doi: 10.4049/jimmunol.1200056. [DOI] [PubMed] [Google Scholar]
- Sun J., Ramos A., Chapman B., Johnnidis J.B., Le L., Ho Y.J., Klein A., Hofmann O., Camargo F.D. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez S.E., Inlay M.A., Serwold T. CD201 and CD27 identify hematopoietic stem and progenitor cells across multiple murine strains independently of KIT and SCA-1. Exp. Hematol. 2015;43:578–585. doi: 10.1016/j.exphem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520:549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- Wiesmann A., Phillips R.L., Mojica M., Pierce L.J., Searles A.E., Spangrude G.J., Lemischka I. Expression of CD27 on murine hematopoietic stem and progenitor cells. Immunity. 2000;12:193–199. doi: 10.1016/s1074-7613(00)80172-7. [DOI] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Wilson N., Kent D., Buettner F., Shehata M., Macaulay I., Calero-Nieto F., Sanchez Castillo M., Oedekoven C., Diamanti E., Schulte R. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell. 2015;16:712–724. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T., Oritani K., Butz S., Kokame K., Kincade P.W., Miyata T., Vestweber D., Kanakura Y. The endothelial antigen ESAM marks hematopoietic stem cells throughout life. Blood. 2008;112:727. doi: 10.1182/blood-2008-07-167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.