Abstract
The multivalent pneumococcal conjugate vaccine is effective against both systemic disease and otitis media caused by serotypes contained in the vaccine. However, serotypes not covered by the present conjugate vaccine may still cause pneumococcal disease. To address these serotypes, and the remaining otitis media due to Streptococcus pneumoniae, efforts have been devoted to identifying protective protein antigens. Immunity to conserved surface proteins important for adhesion, nutrient acquisition, or other functions could result in a reduction of colonization and a lower disease potential. We have been searching for conserved surface-exposed proteins from S. pneumoniae that may be involved in pathogenesis to test as vaccine candidates. Here, an ∼20-kDa protein that has significant homology to a nonheme iron-containing ferritin protein from Listeria innocua and other bactoferritins was identified as pneumococcal protective protein A (PppA). We expressed and purified recombinant PppA (rPppA) and evaluated its potential as a vaccine candidate. The antibodies elicited by purified rPppA were cross-reactive with PppA from multiple strains of S. pneumoniae and were directed against surface-exposed epitopes. Intranasal immunization of BALB/c mice with PppA protein and either a synthetic monophosphoryl lipid A analog, RC529AF, or a cholera toxin mutant, CT-E29H, used as an adjuvant reduced nasopharyngeal colonization in mice following intranasal challenge with a heterologous pneumococcal strain. PppA-specific systemic and local immunoglobulin G (IgG) and IgA antibody responses were induced. The antisera reacted with whole cells of a heterologous S. pneumoniae type 3 strain. These observations indicate that PppA may be a promising candidate for inclusion in a vaccine against pneumococcal otitis media.
Infections with Streptococcus pneumoniae are a major cause of human diseases, such as otitis media, bacteremia, meningitis, and fatal pneumonia, worldwide (9). The rapid emergence of multidrug-resistant pneumococcal strains throughout the world has led to an increased emphasis on prevention of pneumococcal infections by vaccination (18). The presently available 23-valent pneumococcal capsular polysaccharide vaccine is not effective in children less than 2 years of age or immunocompromised patients, two of the major populations at risk for pneumococcal infection (14). A seven-valent pneumococcal polysaccharide-protein conjugate vaccine, recently licensed in the United States, was shown to be highly effective in infants and children against systemic pneumococcal disease caused by the vaccine serotypes and against cross-reactive capsular serotypes (36). However, parenteral immunization with the seven-valent vaccine was only 60% effective against serotype-specific otitis media (17), demonstrating the need for additional immunization strategies (e.g., intranasal [i.n.] immunization), additional noncapsular antigens, or both. Therefore, there is an immediate need for a cost-effective vaccine to cover most or all of the disease-causing serotypes of pneumococci. While this can be achieved by adding conjugates covering additional serotypes, some investigators have raised concerns over possible replacement of vaccine serotypes with nonvaccine serotypes or with other bacterial species (40). Thus, efforts to find noncapsular vaccine antigens that are conserved among all pneumococcal serotypes and effective against pneumococcal disease continue.
Protein antigens of S. pneumoniae have been evaluated for protective efficacy in animal models of pneumococcal infection. While numerous studies have used parenteral immunization to analyze pneumococcal protein vaccine candidates, since nasopharyngeal colonization is a prerequisite for otic disease, intranasal immunization of mice with pneumococcal proteins and appropriate mucosal adjuvants has also been used to study the mucosal antibody response and the effectiveness of protein vaccine candidates (8, 42). Some of the most commonly studied vaccine candidates include the PspA, PhpA, and CbpA proteins and the PsaA lipoprotein. Numerous studies have shown that PspA protein is a virulence factor (12, 27), but it is antigenically variable among pneumococcal strains. A recent study involving human adults has indicated that some antigenically conserved regions of a recombinant PspA variant may elicit cross-reactive PspA antibodies (29). PsaA (a 37-kDa divalent cation permease lipoprotein with similarity to other gram-positive adhesins), PhpA (a protein containing a unique histidine motif), and the surface-exposed choline binding protein CbpA are antigenically conserved and protective in mouse models of pneumococcal disease (35, 37, 43). While these protein antigens appear promising, it is possible that no one protein antigen will be effective against all pneumococcal serotypes. Thus, laboratories continue to search for additional candidates that are antigenically conserved and elicit antibodies that reduce colonization (important for otitis media), are protective against systemic disease, or both.
In the present study, we report the identification of a novel pneumococcal surface-exposed protein that is antigenically conserved among tested strains and elicits antibodies that are effective in reducing intranasal pneumococcal colonization in a mouse model. This low-molecular-weight protein is effective as a mucosal immunogen when mixed with either of two mucosal adjuvants and is a promising candidate for inclusion in an otitis vaccine formulation. (A portion of this work was presented at the 3rd International Symposium on Pneumococci and Pneumococcal Diseases, Anchorage, Alaska, May 2002.)
MATERIALS AND METHODS
S. pneumoniae strains.
S. pneumoniae strains utilized in this work were WU2, serotype 3 (obtained from Robert Austrian, University of Pennsylvania), serotypes 4, 5, 6B, and 7 (obtained from Gerald Schiffman, State University of New York, Brooklyn), and serotype 14 (ATCC 6314). S. pneumoniae CP1200, a nonencapsulated, highly transformable derivative of R36A, a rough variant of the virulent type 2 strain D39 (19), was also used. S. pneumoniae isolates were grown to log phase in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) with 0.5% yeast extract (Difco) at 37°C with aeration or on tryptic soy (Difco) blood agar plates. A spontaneous streptomycin-resistant mutant of S. pneumoniae serotype 3 (43) was used for challenge.
Extraction of surface-associated S. pneumoniae components.
Salt wash extracts of intact S. pneumoniae cells were made as described by Bernstein and Reddy (6). Bacteria were grown in 4 liters of Todd-Hewitt broth and harvested by centrifugation at 8,000 × g for 30 min. The pellets were suspended in ∼175 ml of phosphate-buffered saline (PBS) with the aid of a pipette and immediately centrifuged at 20,000 × g for 30 min. The wash was filtered through a 0.45-μm-pore-size filter (Nalgene, Rochester, N.Y.), dialyzed, and lyophilized.
Ion-exchange chromatography of surface-associated protein components.
The PBS extract of S. pneumoniae was dissolved in 10 mM Tris-HCl (100 ml), pH 7.6, and applied to a 50-ml DEAE-Sepharose CL-6B column. After washing the column with 2 column volumes (CV) of sample buffer, bound proteins were first eluted with 200 mM Tris-HCl, pH 7.6, followed by a linear NaCl gradient of 0 to 0.75 M (in 200 mM Tris-HCl, pH 7.6) over 300 ml. Column fractions were analyzed by use of a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Individual fractions containing substantial amounts of surface-associated proteins in the 18- to 22-kDa range were desalted using a Centricon SR3 concentrator (Amicon, Beverly, Mass.) and lyophilized.
N-terminal amino acid sequence analysis by PVDF blot excision.
Lyophilized extracts were diluted to 1 mg of total protein/ml, combined 1:1 with 2× SDS sample loading buffer (0.25 M Tris-HCl [pH 6.8], 2% SDS, 10% β-mercaptoethanol, 30% glycerol, 0.01% bromophenol blue), and heated at 100°C for 5 min. Approximately 10 μg of total protein was loaded in each of 10 lanes on a 12-lane, 10- by 10- by 1-mm, 10- to 20%-gradient acrylamide-bisacrylamide gel (Zaxis, Hudson, Ohio). Molecular weight markers were loaded in the two outermost lanes of each side of the gel. Electrophoresis was carried out at a constant amperage of 50 mA for 1 h in Tris-glycine-SDS running buffer. The gel was then rinsed with distilled water and transferred to an Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, Mass.) using a semidry blotting system (Owl Separation Systems, Portsmouth, N.H.) at a constant amperage of 150 mA for 1 h. The resulting blot was stained with amido black (10% acetic acid, 0.1% amido black in distilled water) and destained in 10% acetic acid. The desired protein band was excised from all 10 lanes using a methanol-cleaned mini-Exacto knife and placed in the reaction cartridge of the Applied Biosystems (Foster City, Calif.) 477A protein sequencer. The N-terminal sequencer was then used under optimal blot conditions for 12 or more cycles (1 blank cycle, 1 standard cycle, and 10 or more cycles for desired residue identification). Phenylthiohydantoin (PTH) amino acid detection was done on the Applied Biosystems 120A PTH analyzer. The cycle data were collected both on an analog chart recorder and digitally via the instrument software. N-terminal amino acid assignment was performed by comparison of the analog and digital data to a standard set of PTH-amino acids and their respective retention times on the analyzer (cysteine residues are destroyed during conversion and are not detected).
Cloning and expression of the recombinant 20-kDa surface-associated protein.
The N-terminal sequence of the isolated protein was compared against both the National Center for Biotechnology Information's nonredundant database, available at www.ncbi.nlm.nih.gov, and as part of the public release of the S. pneumoniae genome (serotype 4), made available by The Institute for Genomic Research (www.tigr.org), using the BLAST algorithm developed by Altschul et al. (2). Subsequent DNA analyses of the corresponding open reading frame (ORF) in the S. pneumoniae genomic sequence and primers were designed using DNASTAR (Madison, Wisc.) Lasergene DNA and protein analysis software. Primers flanking the ORF were synthesized using an ABI 380A DNA synthesizer. To facilitate subcloning of the PCR product into the pET28a expression vector, restriction sites were included in the PCR primers. An NcoI site was included in the 5′ primer, which both allowed for the ligation into the NcoI site of the expression vector and also included an ATG start codon. To maintain the correct reading frame, two extra bases were included in the 5′ primer, resulting in the addition of a codon for leucine. A SalI site was included in the 3′ primer. The following primers were used: 5′, GGGGCCATGGCTGTAGAATTGAAAAAAGAA, and 3′, GGGGTCGACTAAACCAGGTGCTTGTCCAAGTTC. PCR was performed using a DNA Engine (MJ Research, Waltham, Mass.) thermal cycler with ReddyMix (ABgene; Epsom, Surrey, United Kingdom) according to standard conditions and examined on 0.7% agarose gels. A PCR fragment of the expected size was generated from strain CP1200 bacterial cells and ligated into pCR2.1 vector (Invitrogen, Carlsbad, Calif.). After transformation of Escherichia coli Top10F′ cells (Invitrogen), ampicillin-resistant transformants were screened by restriction digestion of plasmid DNA prepared by alkaline lysis (7). A plasmid containing the correct insert was identified, and the DNA sequence of the insert was determined.
The pppA gene was subcloned into the NcoI and SalI sites of pET28a and transformed into Top10F′ cells. A recombinant plasmid containing the pppA gene was selected, named pLP533, and transformed into BL21(DE3) cells (Novagen) for expression. BL21(DE3)(pLP533) was grown in SOB medium (28) supplemented with 30 μg of kanamycin (Sigma, St. Louis, Mo.)/ml to an optical density at 600 nm (OD600) of 0.6 and induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Boehringer Mannheim, Indianapolis, Ind.) for 2 to 4 h. Whole-cell lysates were prepared and electrophoresed on a 15% SDS-PAGE gel to confirm expression of the desired recombinant product.
Purification of the recombinant 20-kDa surface-associated protein.
Cultures of E. coli BL21(DE3)(pLP533) in 1 liter of SOB containing 30 μg of kanamycin/ml were incubated at 37°C with aeration until they reached an OD600 of 0.7, then IPTG was added to 0.8 mM, and the cultures were incubated at 37°C with shaking for an additional 3 h. Bacteria were harvested by centrifugation (15 min at 7,300 × g, 4°C), and the cell pellets were frozen at −20°C, thawed, and resuspended in 300 ml of 10 mM Na phosphate buffer, pH 6.0. The cell suspension was passed through a microfluidizer (Microfluidics Corporation, Newton, Mass.), and the lysate was centrifuged to remove cell debris (15 min, 16,000 × g, 4°C) and then to remove membranes (45 min, 200,000 × g, 4°C). The supernatant containing the recombinant PppA (rPppA) protein was diluted to 500 ml in 10 mM Na phosphate, pH 6.0, diafiltered with a 100,000-molecular-weight cutoff membrane (Millipore) against 1 liter of the same buffer (rPppA is retained due to oligomerization), and concentrated 2.5-fold. The retained protein was loaded onto a 70-ml ceramic hydroxyapatite column (Bio-Rad Laboratories, Hercules, Calif.) in 10 mM Na phosphate, pH 6.0, and the column was washed with 10 CV of loading buffer. Contaminating proteins were removed by washing the column with 10 CV of 108 mM Na phosphate, pH 6.0. Bound protein was eluted from the column using a 108 to 500 mM linear gradient of Na phosphate, pH 6.0, over 10 CV. Peak fractions were run on a 10-20% SDS-PAGE gel (Zaxis), and those containing the rPppA protein were pooled and stored at −20°C. Purified rPppA protein was analyzed for homogeneity by SDS-PAGE, and the protein concentration was determined. The approximate molecular size of soluble purified rPppA was determined on a Superose 12 10/300 GL size exclusion column (Amersham Biosciences, Piscataway, N.J) equilibrated with PBS. The column was calibrated using both high- and low-molecular-weight gel filtration calibration kits (Amersham) prior to 200 μg of rPppA being loaded onto the column. The molecular size of rPppA was calculated by comparison to elution profiles of the molecular weight standards.
Subcutaneous immunization of mice for Western blot analysis.
rPppA protein was used to generate polyclonal antisera in mice. Briefly, 10 μg of rPppA protein was mixed with 25 μg of monophosphoryl lipid A (MPL; Corixa Corporation, Hamilton, Mont.) and injected subcutaneously into 10 6- to 8-week-old Swiss-Webster mice (Jackson Laboratories, Bar Harbor, Maine). The mice were bled and vaccinated at week 0, given a booster at week 4, and then exsanguinated at week 6. The serum samples were pooled and used for further analysis.
SDS-PAGE and Western blotting.
Whole-cell lysates were prepared by centrifuging equivalent numbers of pneumococcal cells, based on the OD600, in a microcentrifuge for 30 s. Pneumococcal cell pellets were resuspended in an appropriate volume of loading buffer and, where indicated, boiled for 5 min. Analysis was done on a 10% SDS-PAGE gel using the method of Laemmli (24). For Western analysis, proteins were transferred to nitrocellulose (Bio-Rad) using a Mini Transblot cell (Bio-Rad), and the blots were blocked at room temperature (RT) for 30 min in 5% nonfat milk-PBS (BLOTTO). Pooled mouse antisera were used at a 1:1,000 dilution in BLOTTO for 60 min, followed by 25-min washes in PBS-0.2% Tween 20. Goat anti-mouse immunoglobulin G (IgG) plus IgM conjugated to alkaline phosphatase (Biosource International, Camarillo, Calif.) was used to detect bound antibodies at a 1:1,000 dilution in BLOTTO. The blots were washed as previously described and detected with nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate) from Bio-Rad according to the manufacturer's directions.
Intranasal immunization of mice.
Mucosal immunization of mice via the intranasal route was performed as previously described (43). Briefly, 6-week-old pathogen-free BALB/c mice (Jackson Laboratories) were immunized with 5 μg of rPppA protein at weeks 0, 2, and 4 (10 animals per group). The adjuvant used was either 0.1 μg of CT-E29H, a genetically modified cholera toxin that is reduced in enzymatic activity and toxicity (38), or 10 μg of RC529AF, a synthetic MPL mimetic (Corixa) (3, 22, 31). The immunogen was slowly instilled into the nostril of each mouse in a 10-μl volume. Negative controls included either keyhole limpet hemocyanin (KLH) or Moraxella catarrhalis UspA2 protein (10), and serotype 3 pneumococcal polysaccharide conjugated to CRM197 protein (PnC-3) was used as a positive control (20). Serum samples were collected 1 week after the last immunization. Bronchoalveolar, nasal, and vaginal washes were collected as previously described (13). An enzyme-linked immunosorbent assay (ELISA) was used to analyze aliquots of wash samples from individual mice within each group for mucosal IgA and IgG antibodies.
Mouse intranasal challenge model.
S. pneumoniae intranasal challenge was performed as previously described (43). BALB/c mice were challenged 6 days after the last immunization with 1 × 105 to 2 × 105 CFU of serotype 3 Smr S. pneumoniae. Mice were anesthetized via intraperitoneal injection with 1.2 mg of ketamine HCl (Fort Dodge Laboratory, Fort Dodge, Iowa). The bacterial suspension was inoculated into the nostrils of anesthetized mice (10 μl per mouse). The actual dose of bacteria administrated was confirmed by plate counting. Four days after challenge, mice were sacrificed, and their noses were removed and homogenized in 3 ml of sterile saline with a tissue homogenizer (Ultra-Turax T25; Janke & Kunkel Ika-Labortechnik, Staufen, Germany). The homogenate was 10-fold serially diluted in saline and plated on streptomycin-containing tryptic soy agar plates. The plates were incubated overnight at 37°C, and the colonies were counted.
ELISA for rPppA antibodies.
Antibody titers against rPppA protein in pooled sera and mucosal washes were determined by ELISA. ELISAs were performed using rPppA (100 μl of a 5-μg/ml stock in PBS, pH 7.1, per well) to coat Nunc-Immuno PolySorp plates. Plates were coated overnight at 4°C. After blocking with 200 μl of BLOTTO for 1 h at RT, the plates were incubated with test sera serially diluted in blocking buffer for 1.5 h at RT. The plates were washed five times with PBS containing 0.1% Tween (PBS-T) and incubated with biotinylated goat anti-mouse IgG or IgA (Brookwood Biomedical, Birmingham, Ala.) (diluted 1:8,000 or 1:4,000 in PBS) for 1 h at RT. After five additional washes with PBS-T, the plates were incubated with streptavidin-conjugated horseradish peroxidase (Zymed Laboratory Inc., San Francisco, Calif.) (diluted 1:10,000 in PBS) for 1 h at RT. The plates were then washed five times with PBS-T and incubated 20 min with 100 μl of ABTS [2,2′-azinobis 3-ethylbenzthiazolinesulfonic acid] substrate (KPL, Gaithersburg, Md.), and then 100 μl of stopping solution (1% SDS) was added. Absorbance values were read at 405 nm using a VERSAmax microplate reader (Molecular Devices Corp., Sunnyvale, Calif.). The endpoint titers of test sera were the reciprocal of the highest dilution that resulted in an OD405 reading of 0.1. The mean background titers of test sera were quantified by absorbance values read at 405 nm on the wells that had all reagents except sera.
Immunoelectron microscopy of S. pneumoniae cells.
Immunoelectron microscopy of S. pneumoniae serotype 3 and 14 cells was performed as previously described (43). The cells were examined on a Zeiss 10C transmission electron microscope operating at 100 kV and photographed at a magnification of ×50,000.
DNA sequencing.
All sequencing reactions were performed with the Applied Biosystems Prism Dye Terminator cycle-sequencing core kit based on the Prism protocol supplied by the vendor. Approximately 1 μg of template DNA and 100 ng of primer were used for each cycling reaction. The reactions were cycled on the GeneAmp PCR Systems 2400 unit, purified using the Prism method, and analyzed on an ABI 373A DNA sequencer (Applied Biosystems).
Protein determination.
The concentration of protein during purification was determined by the method of Peterson (32). Protein concentration prior to immunization was determined using a bicinchoninic acid kit obtained from Pierce Chemicals (Northbrook, Ill.) and was used according to the manufacturer's directions. Bovine serum albumin was used as the protein standard.
Statistical methods.
Comparison of nasal colonization among groups was performed using the Tukey-Kramer test (11). Results were considered significant at P values of <0.05.
RESULTS
Identification of the 20-kDa surface-associated protein.
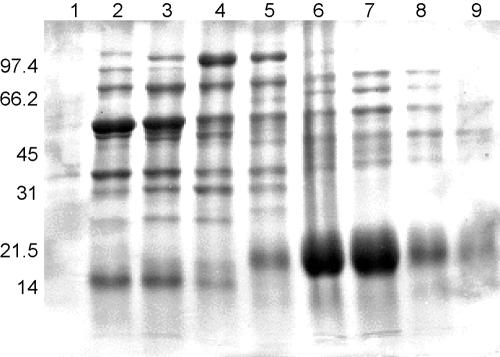
Bernstein and Reddy (6) previously reported that numerous putatively surface-exposed proteins are present in PBS washes of intact S. pneumoniae cells. To further examine these putative surface-exposed proteins, proteins in PBS washes of S. pneumoniae type 2 cells were separated by ion-exchange chromatography and analyzed by SDS-PAGE (Fig. 1). Fractions from this separation show large numbers of proteins present in the wash, with a prominent unique protein band migrating at approximately 20 kDa in fractions 14 to 15 (Fig. 1, lanes 6 and 7). This band was of interest since putative human mucin binding proteins have been reported in this general size range for S. pneumoniae. To identify the protein, the low-molecular-weight band was resolved on a preparative SDS-PAGE gel and transferred to a PVDF membrane, and the amino-terminal sequence of the protein was determined. A single predominant 20-residue amino-terminal sequence, VELKKEAVKDVTSLTKAAPV, was obtained from this band. Compared to the predicted amino acid sequences of ORFs in the S. pneumoniae genome of the Institute for Genomic Research, the N-terminal sequence had 100% correspondence with an amino-terminal portion of ORF SP1572 in the S. pneumoniae genome. This ORF sequence is mentioned in a publication by Pikis et al. (33) as being adjacent to the operon containing the dihydrofolate reductase gene responsible for trimethoprim resistance, but no function is ascribed to it.
FIG. 1.
SDS-PAGE analysis of S. pneumoniae salt wash proteins after DEAE fractionation. Lanes: 1, fxn no. 8; 2, fxn no. 10, 3, fxn no. 11; 4, fxn no. 125, fxn no. 13; 6, fraction (fxn) no. 14; 7, fxn no. 15; 8, fxn no. 16; 9, fxn no. 17. Lanes 6 and 7 contain large amounts of ∼20-kDa protein that was sequenced. Migration of molecular mass standards (in kDa) is shown at the left.
BLASTP analysis of this ORF identified similarity to several proteins from streptococcal species, including the Streptococcus suis peroxide resistance protein Dpr (67% identity) and Streptococcus mutans DNA binding protein, and a nonheme iron-containing ferritin protein in Listeria innocua (44% identity), which may indicate similar functions in S. pneumoniae. Lower levels of similarity to many other bacterial proteins from these families were also identified, and the protein families appear to be widely distributed among bacteria.
The N-terminal amino acid sequence of the PppA protein begins at the eighth amino acid of ORF SP1572, implying either posttranslational processing of the gene product, use of a secondary translational start site adjacent to the N-terminal valine residue and removal of the methionine, or an incorrect assignment of the start site of ORF SP1572. Given the presence of an optimal S/D site, GAGGTXA, just 5′ to the second ATG of ORF SP1572, it is likely that the actual ORF start site is misidentified in the database.
Purification of recombinant PppA protein.
To obtain larger quantities of protein for further study, the gene encoding ORF 1572 was cloned and expressed behind the T7 polymerase promoter as described in Materials and Methods. All constructs were verified as being correct by DNA sequencing. Purification was aided by the solubility of the recombinant protein, as significant purification away from cellular membranes was achieved by differential centrifugation. In addition, oligomer formation (see Fig. 3B) was successfully utilized to remove the remaining low-molecular-weight contaminating proteins by diafiltration. The protein was purified to >95% homogeneity on a hydroxyapatite column (Fig. 2). Amino acid analysis of the purified protein closely matched the predicted values (data not shown), and Limulus amoebocyte lysate analysis showed very low levels of endotoxin (<0.01 endotoxin units/μg of protein). Purified, soluble rPppA had an approximate molecular size of 346 kDa in PBS as measured by size exclusion chromatography, which is equivalent to an oligomer of 17 rPppA subunits.
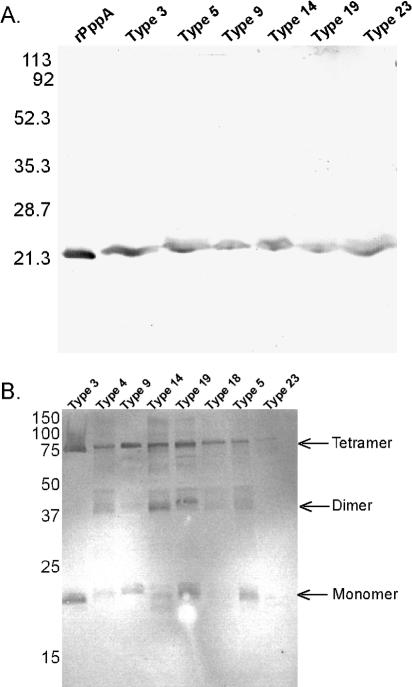
FIG. 3.
Western blot analysis of S. pneumoniae whole-cell lysates and rPppA probed with mouse anti-rPppA sera. (A) Heated and reduced whole-cell lysates from six serotype strains and 1 μg of purified rPppA as control. All strains show a reactive band that corresponds to a molecular mass of ∼21 kDa, the size of a PppA monomer. (B) Nonheated, reduced whole-cell lysates from eight various serotype strains. Reactive bands that equate to putative monomers, dimers, and tetramers are shown.
FIG. 2.
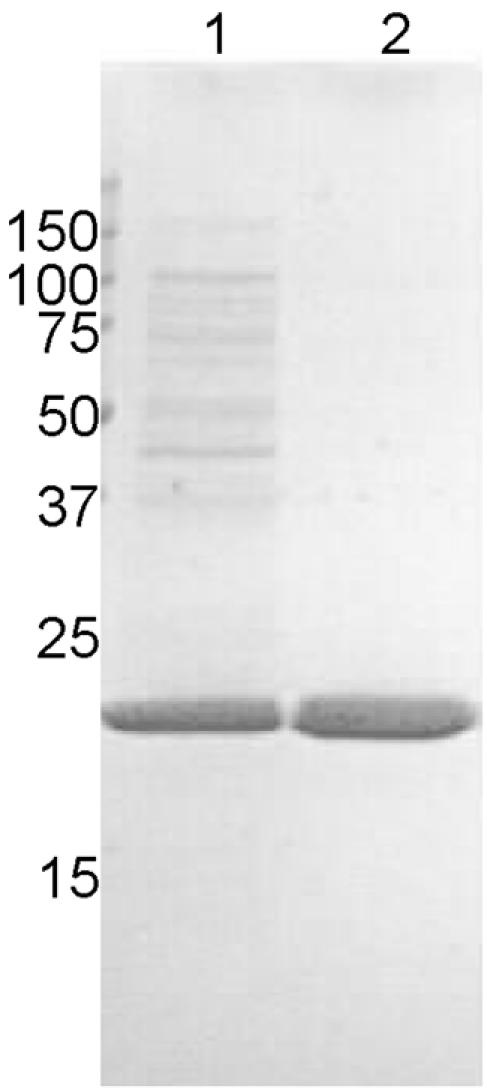
SDS-PAGE analysis of soluble E. coli whole-cell lysate and purified rPppA protein. Samples were loaded on a 15% SDS-PAGE gel and stained with Coomassie brilliant blue. Lanes: 1, molecular mass standards; 2, diafiltered cell lysate; 3, 5 μg of purified rPppA protein.
Reactivity of anti-rPppA sera.
Pooled polyclonal antisera to rPppA surface-associated protein were generated in Swiss-Webster mice to evaluate antigenic conservation of the protein among pneumococcal serotypes. Western blot analysis (Fig. 3A) shows that the protein is present in all of the strains tested and that the protein appears to be antigenically conserved. PppA protein appears to migrate anomalously slowly in SDS-PAGE gels, giving an apparent molecular size of >21 kDa (Fig. 3A).
Anti-rPppA sera reacted with bands corresponding to molecular masses of approximately 20, 40, and 80 kDa in unheated whole-cell lysates of S. pneumoniae run on an SDS-PAGE gel (Fig. 3B), while the major reactive species seen in heated samples is found at approximately 21 kDa (Fig. 3A). These data could result from incomplete unfolding of the 20-kDa protein in the pneumococcal whole-cell lysates, or the 20-kDa protein may exist as oligomers of ∼80 kDa on the pneumococcal surface. The absence of 17-mers may be due to the presence of SDS and β-mercaptoethanol in the sample buffer, or the protein may only exist as 17-mers in solution. Given that rPppA oligomerizes in solution, it is likely that the native protein also exists as an oligomer in S. pneumoniae. Anti-rPppA serum was also shown to react to the surface of both S. pneumoniae type 14 and type 3 using immunoelectron microscopy (Fig. 4). The preimmune serum shows no labeling of intact S. pneumoniae type 14 or type 3 cells, while the anti-rPppA serum shows moderate gold labeling on the surface of the bacteria for both type 3 and type 14. It is not known whether the relative amounts of the PppA protein on the surface of the two strains represent serotype-specific differences, experimental variation, or isolate-specific expression levels.
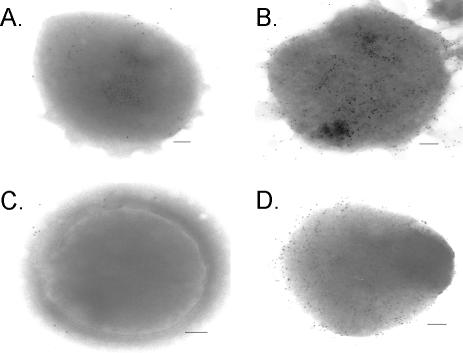
FIG. 4.
Transmission electron micrographs of S. pneumoniae whole cells surface labeled with mouse anti-rPppA sera. Bacteria were probed with either preimmune serum (A and C) or anti-rPppA serum (B and D). Bound antibodies were detected with 7-nm-gold-particle-labeled goat anti-mouse IgG sera. Panels A and B show S. pneumoniae type 3 cells, and panels C and D show S. pneumoniae type 14 cells. Bar, 100 nm.
Intranasal immunization.
Utilizing the potent mucosal adjuvant CT-E29H, BALB/c mice were immunized i.n. with rPppA. To assess specific antibody titers at the mucosal surface, bronchial, nasal and vaginal washes were obtained as described, and the rPppA ELISA titers of both IgG and IgA were determined. One week after the last booster immunization, strong, antigen-specific IgG and IgA antibody responses were elicited in both mucosal wash fluids (Table 1) and sera (Table 2) of mice immunized with rPppA plus CT-E29H. Negative control immunogen KLH plus CT-E29H elicited only background ELISA titers against the purified protein. While the i.n. immunization elicited measurable titers of rPppA-specific antibodies in all three wash fluids (Table 1), the nasal and vaginal washes contained much higher levels of specific IgA than IgG, while the reverse was true for the bronchial wash. These results demonstrate that i.n. immunization of rPppA with CT-E29H elicited both systemic and mucosal immune responses.
TABLE 1.
rPppA specific antibodies in BALB/c mice mucosal washes after i.n. immunization using CT-E29H as adjuvant.
| Antigenb | Bronchial Washa
|
Nasal Washa
|
Vaginal Washa
|
|||
|---|---|---|---|---|---|---|
| IgG | IgA | IgG | IgA | IgG | IgA | |
| rPppA | 143 | 8 | 43 | 268 | 97 | 1220 |
| KLH | <5 | <5 | <5 | <5 | <10 | <10 |
Pooled mucosal washes from 5 animals at week 5. Bronchial washes are 5-fold more dilute than Nasal and Vaginal washes.
Proteins administered at weeks 0, 2, and 4.
TABLE 2.
Analysis of pooled sera from BALB/c mice immunized i.n. with either CT-E29H or RC529AF as an adjuvant
| Antigen | Adjuvantc | ELISA titer of indicated antibody against:
|
||||
|---|---|---|---|---|---|---|
| rPppA protein in preimmune sera
|
rPppA protein in immune serab
|
Whole cells in immune seraa,b | ||||
| IgG | IgA | IgG | IgA | IgG | ||
| rPppA | CT-E29H | <100 | <100 | 362,673 | 22,027 | 1,274 |
| None | CT-E29H | <100 | <100 | <1,000 | <100 | <100 |
| rPppA | RC529AF | <100 | <100 | 308,622 | 20,775 | 2,456 |
| UspA2 | RC529AF | <100 | <100 | <1,000 | <1,000 | <100 |
| PnC-3 | RC529AF | <100 | <100 | <1,000 | <1,000 | 63,429 |
Whole-cell ELISA plates were coated with S. pneumoniae type 3 cells; titers were determined at an endpoint of 0.1 at OD405. Preimmune titers were all <100.
Mice (n = 10) were immunized at weeks 0, 2, and 4. Immune sera are pools of samples from bleeds performed at week 5.
CT-E29H was used at 0.1 μg per dose, and RC529AF was used at 10 μg per dose.
rPppA and UspA2 were used at 5 μg per dose, and PnC-3 was used at 1 μg per dose.
Additional i.n. immunization studies in BALB/c mice were performed using as adjuvants CT-E29H and RC529AF, a synthetic MPL analog that has been shown to have reduced pyrogenicity (16, 22, 23). In these studies, type 3 pneumococcal polysaccharide conjugate (20) was used as a positive control, and UspA2 was used as a nonrelated negative control. Serum ELISA titers were examined prior to i.n. challenge with type 3 pneumococci and represent serum immune status at the time of challenge. Intranasal immunization with rPppA plus either CT-E29H or RC529AF resulted in strong serum titers against rPppA (Table 2). As expected, animals immunized with UspA2, type 3 conjugate, or adjuvant alone had no detectable titer to rPppA. Whole-cell ELISA titers against the type 3 challenge strain were moderate in the groups immunized with the rPppA protein and higher in the animals immunized with the type 3 conjugate. However, the magnitude of the whole-cell response did not correlate with the level of reduction of bacteria in the nasopharynx seen in subsequent challenge experiments.
Pneumococcal colonization challenge model.
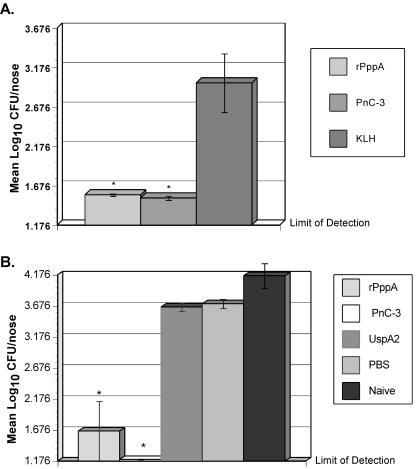
Our previously published pneumococcal intranasal challenge model (43) was used to evaluate the functional activity of immune responses elicited by immunization with rPppA protein. Mice immunized with rPppA without adjuvant had pneumococcal nasal colonization levels equivalent to animals that received PBS alone (data not shown). However, animals immunized with either rPppA plus CT-E29H or type 3 conjugate plus CT-E29H had significantly lower S. pneumoniae nasal colonization levels than sham (KLH plus CT-E29H)-immunized animals (Fig. 5A). These results were repeated and expanded in a subsequent experiment. Mice were immunized intranasally with either rPppA, type 3 conjugate, or UspA2 protein, all containing CT-E29H as an adjuvant, and challenged. To further determine any effects of adjuvant on colonization levels, PBS-plus-CT-E29H-immunized and naïve animals were also challenged. Nasopharyngeal colonization levels (Fig. 5B) show that both type 3 conjugate and rPppA significantly reduced colonization compared to that in control groups, with the type 3 conjugate-immunized animals having slightly lower levels than the rPppA animals, although the difference was not significant. No significant effects of CT-E29H were observed when comparing the PBS plus CT-E29H or UspA2 plus CT-E29H groups to naïve mice.
FIG. 5.
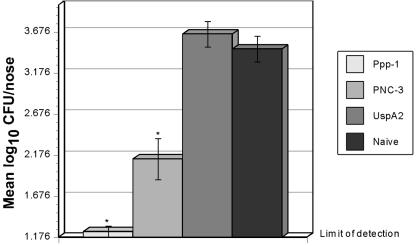
Nasopharyngeal colonization of mice intranasally vaccinated with antigens plus CT-E29H and challenged i.n. with 105 CFU of S. pneumoniae serotype 3. All vaccines contained CT-E29H as an adjuvant except for those given to the naïve (unimmunized) control group. Two experiments with differing controls and mice immunized with rPppA (5 μg) are shown. (A) Control groups immunized with KLH (5 μg) or PnC-3 (1 μg). (B) Control groups immunized with PnC-3 (1 μg), UspA2 (5 μg), or PBS. Pneumococci recovered from nasal tissues 4 days postchallenge are expressed as numbers of log10 CFU per nose ± standard errors of the means (SEM). An asterisk represents significant reduction compared to the appropriate control by Tukey-Kramer test (P < 0.05). (Note: SEM bars for the PNC-3 control group in panel B are not visible at this scale.).
An additional nasopharyngeal colonization experiment was performed using rPppA plus RC529AF as a mucosal adjuvant. Mice were immunized with rPppA, UspA2, or type 3 conjugate, all containing RC529AF. Previous experiments demonstrated that immunization with RC529AF had no significant effects on S. pneumoniae colonization (data not shown), so the UspA2 plus RC529AF group served as both adjuvant control and negative control. After challenge with S. pneumoniae type 3, the groups immunized with rPppA and type 3 conjugate again showed statistically significant reduced colonization compared to the sham-immunized or naïve controls (Fig. 6). Nasal colonization of the rPppA protein group was barely above detectable levels, almost 1 log lower than the type 3 conjugate group, although this difference was not statistically significant, being the inverse of results seen with CT-E29H as adjuvant.
FIG. 6.
Nasopharyngeal colonization of mice intranasally vaccinated with antigens plus RC529AF and challenged i.n. with 2 × 105 CFU of S. pneumoniae serotype 3. All vaccines contained RC529AF as an adjuvant except for those given to the naïve (unimmunized) control group. Mice were immunized with either rPppA (5 μg), UspA2 (5 μg), or PnC-3 (1 μg). Pneumococci recovered from nasal tissues 4 days postchallenge are expressed as numbers of log10 CFU per nose ± SEM. An asterisk represents significant reduction compared to the appropriate control by the Tukey-Kramer test (P < 0.05).
DISCUSSION
The success of the polysaccharide conjugate vaccines in preventing systemic pneumococcal disease, and their initial success in preventing pneumococcal otitis media (17, 36) has led to continued efforts to investigate common protein antigens as additives for a potentially more effective formulation for otitis media. Recent publications have utilized the published sequence of the pneumococcal genome (39) to search for potential vaccine candidates using algorithms that predict surface exposure and homology to known vaccine candidate molecules (34, 41). While these studies have identified numerous candidates, there are also proteins that are not identified by these methods, yet are surface exposed. Human nasopharyngeal mucins have been shown to be efficiently bound by surface-exposed proteins on S. pneumoniae (6), but the identities of the proteins responsible for these interactions have not been determined. The present studies began by isolating low-molecular-weight proteins present in PBS washes of S. pneumoniae to initiate efforts to identify mucin binding proteins. However, when examined using procedures outlined by Bernstein and Reddy (6), the major low-molecular-weight protein component present in the washes (Fig. 1) did not bind human nasopharyngeal mucin (data not shown). Since this protein was present in some abundance in the washes, and it appeared to be surface accessible, efforts to identify the protein and determine whether it was surface exposed were made.
N-terminal amino acid analysis allowed identification of the ORF. BLASTP analysis of the identified ORF, SP1572, showed it to have significant homology to known classes of proteins in bacteria: peroxide resistance proteins from S. suis, DNA binding protein from S. mutans, and a nonheme iron-containing ferritin protein, possibly indicating similar functions in S. pneumoniae. While the protein families identified are widely distributed among bacterial species, it is unknown whether they are surface exposed in these species and thus whether antibodies to the S. pneumoniae protein might react with the surface of other bacteria. The possibility of a S. pneumoniae vaccine reacting with the surface of other strains of bacteria must be taken into account when deciding on the use of the rPppA protein as a vaccine component. The highest level of homology detected by the BLASTP analysis is 67% identity, and the identity rapidly falls off when compared to proteins from other bacterial species and genera, thus reducing the potential for antibody cross-reactivity. However, since cross-reactivity of anti-rPppA antibodies cannot be ruled out, the effects of the antibodies on human commensal bacterial species must be evaluated prior to human studies.
The immunoelectron microscopy data (Fig. 4) show that there is some PppA protein located on the S. pneumoniae cell surface, but the modest labeling of the antibodies does not appear to correlate with the large quantity of material found in the PBS washes. This may be due to the solubility of PppA in PBS leading to its extraction during the treatment of the bacteria for immunomicroscopy, since antisera against rPppA protein have strong reactivity in the whole-cell ELISA to S. pneumoniae (Table 1). It is not known how the PppA protein makes it to the pneumococcal surface since it does not appear to have a conventional signal sequence. However, similar results have been found for other bacterial proteins, including the Helicobacter pylori urease (15) and the streptococcal enolase (5, 30), PavA (21), PhtA, and glyceraldehyde-3-phosphate dehydrogenase (4) proteins, some of which are vaccine candidates.
To be considered a good vaccine candidate, the PppA protein must be immunogenic, surface exposed, and conserved, and it must elicit functional antibodies. The purified rPppA protein is highly soluble without detergents and was purified to greater than 90% homogeneity from E. coli (Fig. 2). Analysis of anti-rPppA sera demonstrated that the protein is antigenically conserved (Fig. 3) among S. pneumoniae serotype strains and appeared to show oligomer formation in whole-cell lysates. Cloning and sequencing of pppA genes from 10 additional S. pneumoniae strains also demonstrated that the pppA gene is conserved at the DNA and amino acid sequence levels as well, with amino acid sequence identity ranging from 92 to 97% (data not shown).
Since otitis media is a mucosal surface disease, mucosal immunization is the most logical route for vaccination against otitis. A potentially important feature of mucosal immunization is the ability to elicit both mucosal antibodies and serum antibodies, particularly against a pathogen that causes both systemic and mucosal diseases. Intranasal immunization using rPppA with either of the two mucosal adjuvants resulted in serum ELISA titers against both purified protein and whole S. pneumoniae cells (Table 2) and high specific-antibody titers in mucosal washes after i.n. immunization with CT-E29H (Table 1), demonstrating that true mucosal immunization had been achieved.
A colonization model rather than a systemic disease model (e.g., CBAN/xid mice) was chosen to evaluate functional activity of immunization with rPppA since otitis media is not an invasive disease. Intranasal immunization was used with either of two mucosal adjuvants to examine possible adjuvant critical effects on the functional activity of the immune response. The mouse challenge data demonstrate that the rPppA protein is reproducibly able to elicit functional immune responses (Fig. 5 and 6) when administered in formulation with either of the mucosal adjuvants, thus broadening the options for future mucosal immunization studies. Since uncertainties exist surrounding the use of enterotoxin-derived mucosal adjuvants, such as CT-E29H, for intranasal immunization (25), especially in children, the mucosal adjuvant RC529AF was also chosen for evaluation. The ability of the RC529AF plus rPppA or PnC-3 to elicit functional antibody responses in these studies is very encouraging. Mixtures of RC529AF and nontypeable Haemophilus influenzae and M. catarrhalis proteins have also recently been shown to elicit functional antibodies in nasopharyngeal challenge models (26), making this adjuvant an attractive choice for clinical evaluation.
Given the lack of a noncapsular typing system for S. pneumoniae and the large number of protein vaccine candidates shown to be effective in S. pneumoniae animal models, it may be difficult to determine whether any one candidate can truly be effective against all strains. The pppA gene used as the source of protein for these studies was isolated from a derivative of a serotype 2 strain, and heterologous challenges were performed with a serotype 3 strain. While the rPppA protein is sequentially and antigenically conserved among tested strains and is effective in reducing nasopharyngeal colonization in murine models, the amount of surface-exposed PppA protein may argue against its use as a stand-alone vaccine against S. pneumoniae. Even though the extent to which rPppA will protect against multiple pneumococcal serotypes is presently unknown, studies with other conserved pneumococcal proteins have shown protection against several serotypes (1, 43). The value of rPppA as a component of a protein-based pneumococcal vaccine will be partially dependent upon its level of protection against multiple pneumococcal serotypes, and experiments to evaluate the efficacy of rPppA against additional heterologous serotypes are planned.
Although the efficacy of rPppA immunization in animal models of systemic disease has not yet been determined, the success of the pneumococcal conjugate vaccine will make testing a protein candidate for efficacy against human systemic disease very difficult. Therefore, the most likely use of any protein-based vaccine against S. pneumoniae will first be directed against otitis media. Combining protein candidates that elicit differing types of functional antibodies (i.e., blocking attachment, opsonizing, clumping bacteria together, etc.) may be the best approach to reduce or eliminate S. pneumoniae-mediated otitis media. While nutritional acquisition may be a function of the PppA protein in S. pneumoniae and the mechanism of action of anti-PppA antibodies is not known, denial of nutrient acquisition is probably not sufficient by itself to be effective in the murine colonization model, given the mucosal nature of the model. Data on the actual function of the pppA gene in the pneumococcus will enhance the ability to explore this question. Construction of pppA knockout mutants will be critical to this determination.
It is sensible that a mucosal immunization approach should be the first one tested for protein-based vaccines against otitis media. Experiments to test combination vaccines containing antigens from S. pneumoniae, nontypeable H. influenzae, and M. catarrhalis in the mouse nasopharyngeal colonization model to look for interference or enhancement of the functional immune responses are planned.
Acknowledgments
We thank Elena Novikova and Gary Zlotnick for help with size exclusion chromatography of rPppA.
Editor: J. N. Weiser
REFERENCES
- 1.Adamou, J. E., J. H. Heinrichs, A. L. Erwin, W. Walsh, T. Gayle, M. Dormitzer, R. Dagan, Y. A. Brewah, P. Barren, R. Lathigra, S. Langermann, S. Koenig, and S. Johnson. 2001. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect. Immun. 69:949-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Baldridge, J. R., C. W. Cluff, J. T. Evans, M. J. Lacy, J. R. Stephens, V. G. Brookshire, R. Wang, J. R. Ward, Y. M. Yorgensen, D. H. Persing, and D. A. Johnson. 2002. Immunostimulatory activity of aminoalkyl glucosaminide 4-phosphates (AGPs): induction of protective innate immune responses by RC-524 and RC-529. J. Endotoxin Res. 8:453-458. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, S., M. Rohde, and S. Hammerschmidt. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 72:2416-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann, S., D. Wild, O. Diekmann, R. Frank, D. Bracht, G. S. Chhatwal, and S. Hammerschmidt. 2003. Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49:411-423. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, J. M., and M. Reddy. 2000. Bacteria-mucin interaction in the upper aerodigestive tract shows striking heterogeneity: implications in otitis media, rhinosinusitis, and pneumonia. Otolaryngol. Head Neck Surg. 122:514-520. [DOI] [PubMed] [Google Scholar]
- 7.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briles, D. E., E. Ades, J. C. Paton, J. S. Sampson, G. M. Carlone, R. C. Huebner, A. Virolainen, E. Swiatlo, and S. K. Hollingshead. 2000. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect. Immun. 68:796-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler, J. C., E. D. Shapiro, and G. M. Carlone. 1999. Pneumococcal vaccines: history, current status, and future directions. Am. J. Med. 107:S69-S76. [DOI] [PubMed] [Google Scholar]
- 10.Chen, D., J. C. McMichael, K. R. VanDerMeid, D. Hahn, T. Mininni, J. Cowell, and J. Eldridge. 1996. Evaluation of purified UspA from Moraxella catarrhalis as a vaccine in a murine model after active immunization. Infect. Immun. 64:1900-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conover, W. J., and R. Iman. 1982. Analysis of covariance using the rank transformation. Biometrics 38:715-724. [PubMed] [Google Scholar]
- 12.Crain, M. J., W. D. Waltman II, J. S. Turner, J. Yother, D. F. Talkington, L. S. McDaniel, B. M. Gray, and D. E. Briles. 1990. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect. Immun. 58:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cutter, D., K. W. Mason, A. P. Howell, D. L. Fink, B. A. Green, and J. W. St. Geme III. 2002. Immunization with Haemophilus influenzae Hap adhesin protects against nasopharyngeal colonization in experimental mice. J. Infect. Dis. 186:1115-1121. [DOI] [PubMed] [Google Scholar]
- 14.Douglas, R. M., J. C. Paton, S. J. Duncan, and D. J. Hansman. 1983. Antibody response to pneumococcal vaccination in children younger than five years of age. J. Infect. Dis. 148:131-137. [DOI] [PubMed] [Google Scholar]
- 15.Dunn, B. E., and S. H. Phadnis. 1998. Structure, function and localization of Helicobacter pylori urease. Yale J. Biol. Med. 71:63-73. [PMC free article] [PubMed] [Google Scholar]
- 16.Evans, J. T., C. W. Cluff, D. A. Johnson, M. J. Lacy, D. H. Persing, and J. R. Baldridge. 2003. Enhancement of antigen-specific immunity via the TLR4 ligands MPL adjuvant and Ribi.529. Expert Rev. Vacc. 2:219-229. [DOI] [PubMed] [Google Scholar]
- 17.Fireman, B., S. B. Black, H. R. Shinefield, J. Lee, E. Lewis, and P. Ray. 2003. Impact of the pneumococcal conjugate vaccine on otitis media. Pediatr. Infect. Dis. J. 22:10-16. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein, F. W., and J. Garau. 1997. 30 years of penicillin-resistant S. pneumoniae: myth or reality? Lancet 350:233-234. [DOI] [PubMed] [Google Scholar]
- 19.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henriksen, J. L., L. C. Preheim, and M. J. Gentry. 1997. Vaccination with protein-conjugated and native type 3 capsular polysaccharide in an ethanol-fed rat model of pneumococcal pneumonia. Alcohol Clin. Exp. Res. 21:1630-1637. [PubMed] [Google Scholar]
- 21.Holmes, A. R., R. McNab, K. W. Millsap, M. Rohde, S. Hammerschmidt, J. L. Mawdsley, and H. F. Jenkinson. 2001. The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol. 41:1395-1408. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. A., D. S. Keegan, C. G. Sowell, M. T. Livesay, C. L. Johnson, L. M. Taubner, A. Harris, K. R. Myers, J. D. Thompson, G. L. Gustafson, M. J. Rhodes, J. T. Ulrich, J. R. Ward, Y. M. Yorgensen, J. L. Cantrell, and V. G. Brookshire. 1999. 3-O-desacyl monophosphoryl lipid A derivatives: synthesis and immunostimulant activities. J. Med. Chem. 42:4640-4649. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, D. A., C. G. Sowell, C. L. Johnson, M. T. Livesay, D. S. Keegan, M. J. Rhodes, J. T. Ulrich, J. R. Ward, J. L. Cantrell, and V. G. Brookshire. 1999. Synthesis and biological evaluation of a new class of vaccine adjuvants: aminoalkyl glucosaminide 4-phosphates (AGPs). Bioorg. Med. Chem. Lett. 9:2273-2278. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lang, D. 10January2002, posting date. Safety evaluation of toxin adjuvants delivered intranasally. NIAID Division of Microbiology and Infectious Diseases. [Online.] http://www.niaid.nih.gov/dmid/enteric/intranasal.htm.
- 26.Mason, K. W., D. Zhu, C. A. Scheuer, J. C. McMichael, G. W. Zlotnick, and B. A. Green. 2004. Reduction of nasal colonization of nontypeable Haemophilus influenzae following intranasal immunization with rLP4/rLP6/UspA2 proteins combined with aqueous formulation of RC529. Vaccine 22:3449-3456. [DOI] [PubMed] [Google Scholar]
- 27.McDaniel, L. S., J. S. Sheffield, P. Delucchi, and D. E. Briles. 1991. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect. Immun. 59:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics, p. 203. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Nabors, G. S., P. A. Braun, D. J. Hermann, M. L. Heise, D. J. Pyle, S. Gravenstein, M. Schilling, L. M. Ferguson, S. K. Hollingshead, D. E. Briles, and R. S. Becker. 2000. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine 18:1743-1754. [DOI] [PubMed] [Google Scholar]
- 30.Pancholi, V., and V. A. Fischetti. 1998. α-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 31.Persing, D. H., R. N. Coler, M. J. Lacy, D. A. Johnson, J. R. Baldridge, R. M. Hershberg, and S. G. Reed. 2002. Taking toll: lipid A mimetics as adjuvants and immunomodulators. Trends Microbiol. 10:S32-37. [DOI] [PubMed] [Google Scholar]
- 32.Peterson, G. L. 1977. A simplification of the protein method of Lowry et al. which is more generally applicable. Anal. Biochem. 83:346-356. [DOI] [PubMed] [Google Scholar]
- 33.Pikis, A., J. A. Donkersloot, W. J. Rodriquez, and J. Keith. 1998. A conservative amino acid mutation in the chromosome-encoded dihydrofolate reductase confers trimethoprim resistance in Streptococcus pneumoniae. J. Infect. Dis. 178:700-706. [DOI] [PubMed] [Google Scholar]
- 34.Rigden, D. J., M. Y. Galperin, and M. J. Jedrzejas. 2003. Analysis of structure and function of putative surface-exposed proteins encoded in the Streptococcus pneumoniae genome: a bioinformatics-based approach to vaccine and drug design. Crit. Rev. Biochem. Mol. Biol. 38:143-168. [DOI] [PubMed] [Google Scholar]
- 35.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819-829. [DOI] [PubMed] [Google Scholar]
- 36.Shinefield, H. R., and S. Black. 2000. Efficacy of pneumococcal conjugate vaccines in large scale field trials. Pediatr. Infect. Dis. J. 19:394-397. [DOI] [PubMed] [Google Scholar]
- 37.Talkington, D. F., B. G. Brown, J. A. Tharpe, A. Koenig, and H. Russell. 1996. Protection of mice against fatal pneumococcal challenge by immunization with pneumococcal surface adhesin A (PsaA). Microb. Pathog. 21:17-22. [DOI] [PubMed] [Google Scholar]
- 38.Tebbey, P. W., C. A. Unczur, J. A. Peek, D. Zhu, N. A. LaPierre, E. D. Phillips, A. R. Ibraghimov, B. A. Green, J. H. Eldridge, and G. E. Hancock. 2000. Effective mucosal immunization against respiratory syncytial virus using a genetically detoxified cholera holotoxin, CT-E29H. Vaccine 18:2723-2734. [DOI] [PubMed] [Google Scholar]
- 39.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 40.Veenhoven, R., D. Bogaert, C. Uiterwaal, C. Brouwer, H. Kiezebrink, J. Bruin, E. IJzerman, P. Hermans, R. de Groot, B. Zegers, W. Kuis, G. Rijkers, A. Schilder, and E. Sanders. 2003. Effect of conjugate pneumococcal vaccine followed by polysaccharide pneumococcal vaccine on recurrent acute otitis media: a randomised study. Lancet 361:2189-2195. [DOI] [PubMed] [Google Scholar]
- 41.Wizemann, T. M., J. H. Heinrichs, J. E. Adamou, A. L. Erwin, C. Kunsch, G. H. Choi, S. C. Barash, C. A. Rosen, H. R. Masure, E. Tuomanen, A. Gayle, Y. A. Brewah, W. Walsh, P. Barren, R. Lathigra, M. Hanson, S. Langermann, S. Johnson, and S. Koenig. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69:1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto, M., D. E. Briles, S. Yamamoto, M. Ohmura, H. Kiyono, and J. R. McGhee. 1998. A nontoxic adjuvant for mucosal immunity to pneumococcal surface protein A. J. Immunol. 161:4115-4121. [PubMed] [Google Scholar]
- 43.Zhang, Y., A. W. Masi, V. Barniak, K. Mountzouros, M. K. Hostetter, and B. A. Green. 2001. Recombinant PhpA protein, a unique histidine motif-containing protein from Streptococcus pneumoniae, protects mice against intranasal pneumococcal challenge. Infect. Immun. 69:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]