Abstract
Background and Aim
The connection between gene polymorphisms of cytotoxic T-lymphocyte-associated protein 4 (CTLA4) and primary biliary cholangitis (PBC) is still vague and blurred. The purpose of this study is to precisely estimate the association of the polymorphisms of CTLA4 with the risk of PBC by using a meta-analysis.
Methods
PubMed and the Chinese National Knowledge Infrastructure (CNKI) database were used to search correlative literatures, and the documents which were about the relationships between the polymorphisms of CTLA4 (rs231775, rs231725, rs3087243, and rs5742909) and PBC were collected as of June 2016. The strength of correlation based on odds ratios (ORs) and its 95% confidence intervals (95%CIs) was computed by STATA.
Results
Generally, in rs231775, a significant risk was found in G allele, the value of OR was 1.32, and its 95%CI was 1.19 to 1.47. The same situation was found in A allele of rs231725, the value of OR was 1.33, and its 95%CI was 1.22 to 1.45. As genotypic level, different genotypic models were also found to have obvious relevance with PBC in rs231775 and rs231725. No obvious connections were found in other SNPs.
Conclusion
This study indicated that the polymorphisms of rs231775 and rs231725 would be the risk factors of PBC.
1. Introduction
Autoimmune diseases are resulted from the dysfunction of the immune system, which generate immune response to autoantigens. Primary biliary cholangitis (PBC) is one kind of specific autoimmune diseases, which can cause progression of fibrosis and cirrhosis in the liver and lead to liver failure finally [1–4]. There are still no special treatments for primary biliary cholangitis in the world. At present, the most efficient therapy is to use ursodeoxycholic acid for those who are in early period of primary biliary cholangitis. However, ursodeoxycholic acid could not still stop the progression of the disease. When the final stage of PBC occurred, the only therapy is liver transplantation. Up to now, the definite etiology of PBC is still not clear.
Cytotoxic T-lymphocyte-associated protein 4 (CTLA4) is expressed on the surface of activated T and transmits inhibitory signal, and it is also found on the surface of regulatory T cells. The functions of CTLA4 are to lower responses of T cell and maintain peripheral tolerance of T cell [5]. The abnormal costimulation between specific autoreactive T lymphocytes and CTLA4 in PBC patients causes the reaction of peripheral T lymphocyte not to be terminated, which might be one of the pathogens of PBC.
As to PBC, CD8+ T cells are important factors in the pathogenesis [6]. CD8+ T cells are sensitive to E2 components of pyruvate dehydrogenase complexes (PDC-E2) which are abnormally expressed on the surface of biliary epithelial cells (BECs) and would result in apoptosis for these epithelial cells and destruction of the small bile duct [7, 8]. As a coinhibitor signal, CTLA4 binds to CD80/CD86 on antigen-presenting cells (APCs) with higher affinity comparing with CD28 [9]. Binding with CD80/CD86 to deliver negative signal into T cells, CTLA4 can result T cell responses in inhibition or termination [10]. Then, CTLA4 can regulate immune suppression and peripheral tolerance in CD8+ T cells. Thus, CTLA4 could be involved in the regulation of pathological processes of PBC, which might be a therapeutic for PBC. Two studies demonstrated that treatment with CTLA4-Ig, which can reduce self-reactive T cell activation and liver inflammation significantly, could obviously reduce the level of portal inflammation and biliary cell damage in the mouse model [11, 12].
So, CTLA4 regulation plays an important role in the pathological process of PBC. Meanwhile, the results, which were shown in some researches about the treatment of CTLA4, have shown that CTLA4 plays a unique role in the pathogenesis and treatment of PBC.
Recently, genetic factors are deemed to be an important role in PBC, which is mainly in favor of familial clustering of PBC [13]. Recent study showed that PBC was significantly associated with some concrete gene polymorphisms [14]. Since PBC displays characteristics of autoimmunity, more and more studies concentrated in associations between genetic polymorphism and variations of autoimmunity.
There are several evidences to prove the connections between polymorphisms of CTLA4 and other autoimmune diseases in recent literatures [15–17]. In recent years, there are extensive researches about the links between CTLA4 and PBC. rs231775, rs231775, rs3087243, and rs5742909 are the most common four single-nucleotide polymorphisms (SNPs) to be widely studied [18–22]. Thus, with changes of the function in these SNPs, the possibility of PBC might be increased. Because of inconclusive connections between the polymorphisms of CTLA4 and the risks of PBC, those relative researches are necessary to be combined to conduct a meta-analysis. Several early systematic reviews which had been published mainly regarded the relationships between the polymorphisms in several SNPs and primary biliary cholangitis [23–25]. However, these studies either did not draw the clear conclusion or did not include some latest literatures. So, in this research, 16 studies are combined to analyze the correlation between the polymorphisms of CTLA4 and risks of PBC [1, 18–22, 26–35].
2. Materials and Methods
2.1. Study Selection and Data Extraction
We used PubMed and the China Knowledge Resource Integrated database up to June 2016, and related literatures about the relationships between the polymorphisms of CTLA4 and risks of PBC were researched on computer with retrieval words (“primary biliary cholangitis, PBC, cytotoxic T-lymphocyte antigen 4, Polymorphism, SNP, genetic variants”). At last, we found 26 studies contained relative contents about CTLA4 and PBC.
The following conditions should be met in studies: firstly, literature should be a case-control study; secondly, outcome was about primary biliary cholangitis; and thirdly, odds ratio and its 95% confidence interval should be estimated with adequate data in literatures. Exclusive criteria: insufficient information for data extraction. At last, this meta-analysis included 16 literatures after 10 literatures were excluded.
According to the inclusion conditions mentioned above, two researchers extracted data independently. The researchers gathered these data from each study: SNPs, name of the first author, date of publication, ethnicity, number of allele, and genotype. Diversities among researchers were solved with discussion.
3. Statistical Analysis
The intensity of associations between PBC risk and CTLA4 polymorphisms was assessed in a random-effect model or fixed-effect model by the estimated OR and its 95%CI. Assessing the difference between the CTLA4 polymorphisms and the PBC risk in Caucasian and Asian was also conducted by using subgroup analyzing. Z-test was used to compute the significant difference of pooled OR. The p value of Z-test was calculated to access significance. Because multiple comparisons were conducted in this study, the threshold of p values was corrected with formula 1 − (1 − p)1/n for the Bonferroni correction [36]. Q-test was used to assess heterogeneity and calculate I2 statistic. When p was less than 0.05 or I2 was more than 50%, the results among the studies indicated significant heterogeneity. In addition, possible publication biases were estimated by Begg's funnel plot and Egger's regression; p value was calculated to access bias. When p was less than 0.05, a publication bias was considered to be existence. The validity and reliability of a meta-analysis were evaluated by conducting sensitivity analysis [37]. All the statistics were performed by STATA 14 software.
In this study, linkage disequilibrium (LD) was used to measure the association among these four CTLA4 SNPs after multiple comparison. The values of D' and r2 were calculated by SHEsis software (http://analysis.bio-x.cn/myAnalysis.php) [38]. When D' > 0.8 or r2 > 0.4, linkage disequilibrium could be considered.
4. Results
4.1. Literature Search
In total, the number of cases and controls were 4422 and 5210 in 16 studies, respectively (Table 1). As to CTLA4, the SNPs which were mostly consulted were rs231775, rs231725, rs3087243, and rs5742909. These SNPs were reported in 14, 6, 9, and 5 studies, respectively. The genotypes of controls were in line with Hardy-Weinberg equilibrium in most articles.
Table 1.
Basic characteristics of involved studies.
| SNP | First author | Year | Race | Case | Control | P(HWE) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | AA | AB | BB | A | B | AA | AB | BB | |||||
| rs231775 | Kosh | 2000 | C | 131 | 115 | 35 | 61 | 27 | 278 | 122 | 99 | 80 | 21 | 0.613 |
| Paulo | 2003 | C | 69 | 31 | 23 | 23 | 4 | 88 | 46 | 29 | 30 | 8 | 0.955 | |
| Fan | 2004 | A | 46 | 108 | 6 | 37 | 34 | 139 | 181 | 23 | 93 | 44 | 0.021 | |
| Yukiko | 2006 | A | 30 | 60 | 5 | 20 | 20 | 61 | 85 | 14 | 33 | 26 | 0.545 | |
| Eckart | 2007 | C | 206 | 154 | 58 | 90 | 32 | 243 | 161 | 78 | 87 | 37 | 0.149 | |
| Peter | 2007 | C | 389 | 243 | 40 | 29 | 4 | 497 | 285 | 54 | 35 | 10 | 0.716 | |
| Brian-3 | 2008 | C | 414 | 288 | 122 | 170 | 59 | 352 | 206 | 111 | 130 | 38 | 0.995 | |
| Raoul | 2008 | C | 313 | 203 | 95 | 123 | 40 | 407 | 165 | 145 | 118 | 23 | 0.883 | |
| Brian-2 | 2008 | C | 421 | 281 | 131 | 161 | 59 | 258 | 152 | 79 | 99 | 27 | 0.644 | |
| Erin | 2009 | C | 545 | 417 | 162 | 223 | 96 | 1562 | 934 | 493 | 577 | 178 | 0.661 | |
| Satoru | 2010 | A | 226 | 390 | 42 | 143 | 123 | 224 | 312 | 47 | 131 | 90 | 0.955 | |
| Yoshihiro | 2011 | A | 314 | 586 | 55 | 204 | 191 | 313 | 429 | 66 | 181 | 124 | 0.997 | |
| Mantaka | 2012 | C | 144 | 56 | 52 | 40 | 8 | 226 | 90 | 81 | 64 | 13 | <0.001 | |
| Li | 2013 | A | 180 | 444 | 20 | 140 | 152 | 312 | 438 | 49 | 214 | 112 | 0.001 | |
|
| ||||||||||||||
| rs231725 | Brian-3 | 2008 | C | 442 | 260 | 139 | 164 | 48 | 391 | 167 | 137 | 117 | 25 | 0.998 |
| Satoru | 2010 | A | 250 | 368 | 51 | 149 | 108 | 250 | 286 | 58 | 133 | 77 | 0.968 | |
| Brian-1 | 2010 | C | 1091 | 641 | 368 | 357 | 141 | 1032 | 490 | 350 | 332 | 79 | 0.984 | |
| Yoshihiro | 2011 | A | 351 | 549 | 68 | 214 | 168 | 347 | 395 | 81 | 185 | 105 | 0.977 | |
| Li | 2013 | A | 204 | 420 | 29 | 146 | 137 | 316 | 434 | 59 | 198 | 118 | 0.109 | |
|
| ||||||||||||||
| rs3087243 | Sabine | 2005 | C | 167 | 141 | 40 | 87 | 27 | 170 | 162 | 49 | 72 | 45 | 0.089 |
| Peter | 2007 | C | 222 | 168 | 59 | 104 | 32 | 301 | 251 | 82 | 137 | 57 | 0.987 | |
| Brian-3 | 2008 | C | 407 | 295 | 118 | 171 | 62 | 318 | 240 | 91 | 136 | 52 | 0.925 | |
| Brian-2 | 2008 | C | 400 | 302 | 117 | 168 | 66 | 234 | 176 | 70 | 94 | 41 | 0.358 | |
| Erin | 2009 | C | 602 | 360 | 198 | 205 | 78 | 1335 | 1161 | 362 | 613 | 273 | 0.656 | |
| Satoru | 2010 | A | 454 | 162 | 167 | 120 | 21 | 372 | 164 | 129 | 114 | 25 | 0.979 | |
| Yoshihiro | 2011 | A | 689 | 211 | 264 | 161 | 25 | 515 | 227 | 179 | 157 | 35 | 0.946 | |
| Mantaka | 2012 | C | 107 | 93 | 32 | 43 | 25 | 158 | 158 | 37 | 84 | 37 | 0.426 | |
| Li | 2013 | A | 430 | 194 | 159 | 112 | 41 | 492 | 258 | 170 | 152 | 53 | 0.048 | |
|
| ||||||||||||||
| rs5742909 | Fan | 2004 | A | 138 | 16 | 63 | 12 | 2 | 274 | 46 | 122 | 30 | 8 | 0.003 |
| Brian-3 | 2008 | C | 646 | 56 | 297 | 52 | 2 | 502 | 56 | 226 | 50 | 3 | 0.9 | |
| Raoul | 2008 | C | 477 | 39 | 220 | 36 | 2 | 509 | 63 | 226 | 56 | 4 | 0.803 | |
| Erin | 2009 | C | 852 | 110 | 377 | 99 | 5 | 2291 | 205 | 1055 | 183 | 10 | 0.509 | |
| Satoru | 2010 | A | 550 | 66 | 245 | 59 | 4 | 470 | 66 | 206 | 58 | 4 | 0.971 | |
| Li | 2013 | A | 541 | 83 | 246 | 49 | 17 | 644 | 106 | 288 | 68 | 19 | <0.001 | |
C: Caucasian; A: Asian; P(HWE): p value of Hardy-Weinberg equilibrium for control.
4.2. Meta-Analysis
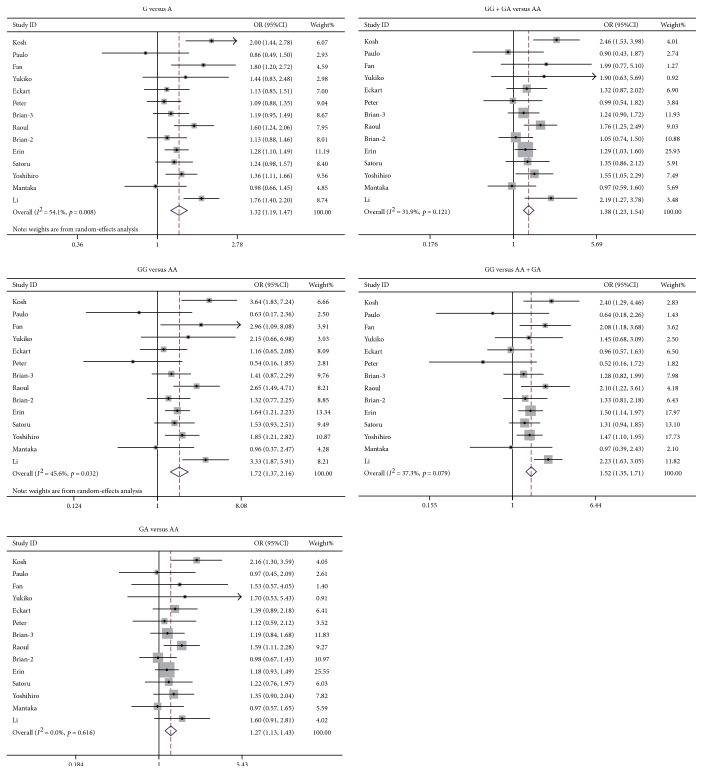
Obvious heterogeneity was identified in CTLA4 rs231775 polymorphism (G versus A: P(het) = 0.008, I2 = 54.1%; GG versus AA: P(het) = 0.032, I2 = 45.6%), rs3087243 polymorphism (GA versus GG: P(het) = 0.028, I2 = 53.5%; AA + GA versus GG: P(het) = 0.037, I2 = 51.2%), and rs5742909 polymorphism (T versus C: P(het) = 0.001, I2 = 69.4%; TC versus CC: P(het) = 0.01, I2 = 66.7%; and (TT + TC) versus CC: P(het) = 0.007, I2 = 69%). Therefore, we chose the random-effect model to generate extensive CIs in these genetic models, the rest of genetic models were used the fixed-effects model (Table 2).
Table 2.
The results of heterogeneity.
| SNP | Genetic model | I 2 (%) | P(het) | Effect model |
|---|---|---|---|---|
| rs231775 | G : A | 54.1 | 0.008 | Random |
| GG : AA | 45.6 | 0.032 | Random | |
| GA : AA | 0.0 | 0.616 | Fixed | |
| (GG + GA) : AA | 31.9 | 0.121 | Fixed | |
| GG : (AA + GA) | 37.3 | 0.079 | Fixed | |
|
| ||||
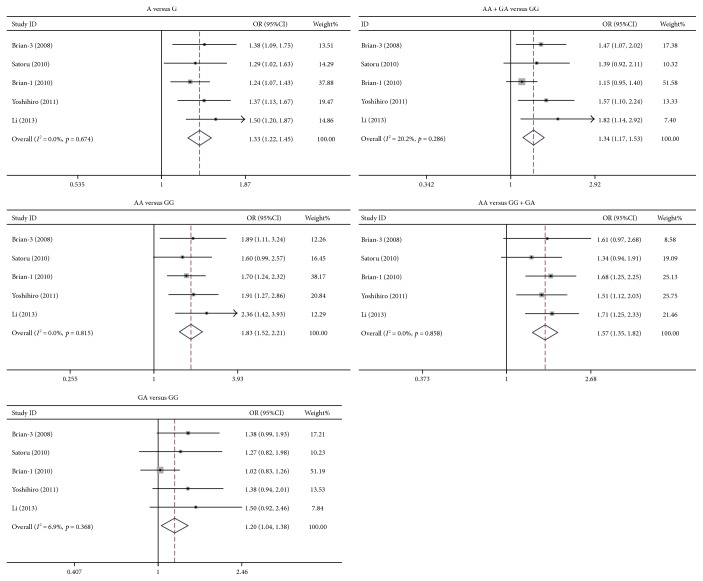
| rs231725 | A : G | 0.0 | 0.674 | Fixed |
| AA : GG | 0.0 | 0.815 | Fixed | |
| GA : GG | 6.9 | 0.368 | Fixed | |
| (AA + GA) : GG | 20.2 | 0.286 | Fixed | |
| AA : (GG + GA) | 0.0 | 0.858 | Fixed | |
|
| ||||
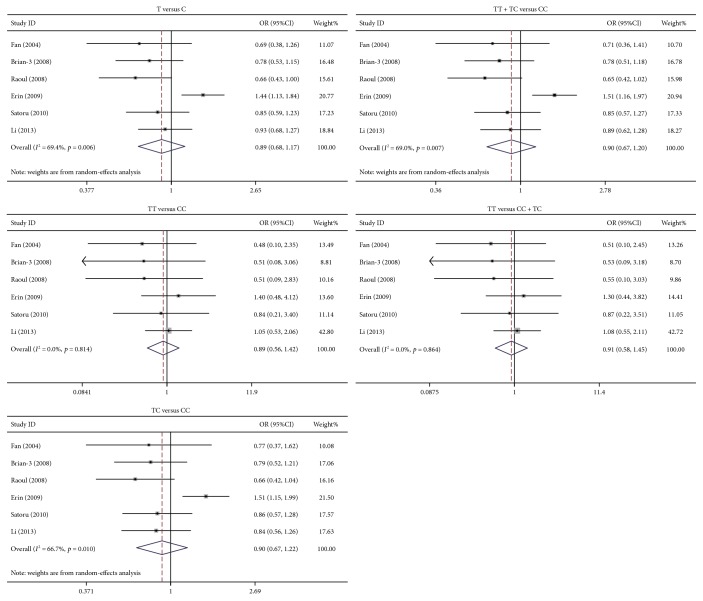
| rs3087243 | A : G | 37.9 | 0.116 | Fixed |
| AA : GG | 11.8 | 0.337 | Fixed | |
| GA : GG | 53.5 | 0.028 | Random | |
| (AA + GA) : GG | 51.2 | 0.037 | Random | |
| AA : (GG + GA) | 0.0 | 0.569 | Fixed | |
|
| ||||
| rs5742909 | T : C | 69.4 | 0.001 | Random |
| TT : CC | 0.0 | 0.814 | Fixed | |
| TC : CC | 66.7 | 0.01 | Random | |
| (TT + TC) : CC | 69.0 | 0.007 | Random | |
| TT : (CC + TC) | 0.0 | 0.864 | Fixed | |
P(het): p value of Q-test for heterogeneity test; I2: the proportion of total variation contributed among study variants; P(het) < 0.05 or I2 > 50% indicated significant heterogeneity, using a random model; otherwise, using a fixed model.
The results of this analysis in the association of CTLA4 polymorphisms (rs231775, rs231725, rs3087243, and rs5742909) with susceptibility to PBC are presented (Table 3).
Table 3.
Results of CTLA4 polymorphisms and PBC.
| SNP | Genetic model | Asian | Caucasian | Overall | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | p | OR | 95%CI | p | OR | 95%CI | p | ||
| rs231775 | G : A | 1.47 | 1.27, 1.72 | <0.0001 | 1.24 | 1.09, 1.42 | 0.001 | 1.32 | 1.19, 1.47 | <0.0001 |
| GG : AA | 2.10 | 1.55, 2.84 | <0.0001 | 1.51 | 1.12, 2.04 | 0.006 | 1.72 | 1.37, 2.16 | <0.0001 | |
| GA : AA | 1.39 | 1.08, 1.79 | 0.011 | 1.24 | 1.09, 1.42 | 0.001 | 1.27 | 1.13, 1.43 | <0.0001 | |
| (GG + GA) : AA | 1.65 | 1.30, 2.10 | <0.0001 | 1.31 | 1.16, 1.49 | <0.0001 | 1.38 | 1.23, 1.54 | <0.0001 | |
| GG : (AA + GA) | 1.66 | 1.40, 1.96 | <0.0001 | 1.39 | 1.18, 1.65 | <0.0001 | 1.52 | 1.35, 1.71 | <0.0001 | |
|
| ||||||||||
| rs231725 | A : G | 1.39 | 1.22, 1.57 | <0.0001 | 1.27 | 1.12, 1.44 | <0.0001 | 1.33 | 1.22, 1.45 | <0.0001 |
| AA : GG | 1.92 | 1.47, 2.49 | <0.0001 | 1.75 | 1.33, 2.29 | <0.0001 | 1.83 | 1.52, 2.21 | <0.0001 | |
| GA : GG | 1.38 | 1.07, 1.76 | 0.012 | 1.11 | 0.93, 1.33 | 0.236 | 1.20 | 1.04, 1.38 | 0.015 | |
| (AA + GA) : GG | 1.57 | 1.24, 1.99 | <0.0001 | 1.23 | 1.04, 1.46 | 0.014 | 1.34 | 1.17, 1.53 | <0.0001 | |
| AA: (GG + GA) | 1.52 | 1.27, 1.83 | <0.0001 | 1.66 | 1.29, 2.14 | <0.0001 | 1.57 | 1.35, 1.82 | <0.0001 | |
|
| ||||||||||
| rs3087243 | A : G | 0.78 | 0.68, 0.89 | <0.0001 | 0.83 | 0.76, 0.91 | <0.0001 | 0.81 | 0.75, 0.88 | <0.0001 |
| AA : GG | 0.66 | 0.49, 0.90 | 0.008 | 0.70 | 0.58, 0.85 | <0.0001 | 0.69 | 0.59, 0.81 | <0.0001 | |
| GA : GG | 0.76 | 0.63, 0.91 | 0.003 | 0.90 | 0.68, 1.21 | 0.491 | 0.84 | 0.70, 1.00 | 0.051 | |
| (AA + GA) : GG | 0.74 | 0.62, 0.88 | 0.001 | 0.86 | 0.66, 1.11 | 0.250 | 0.80 | 0.68, 0.94 | 0.008 | |
| AA : (GG + GA) | 0.74 | 0.55, 1.00 | 0.050 | 0.78 | 0.66, 0.92 | 0.004 | 0.77 | 0.67, 0.89 | 0.001 | |
|
| ||||||||||
| rs5742909 | T : C | 0.87 | 0.70, 1.08 | 0.206 | 0.92 | 0.55, 1.55 | 0.766 | 0.89 | 0.68, 1.17 | 0.417 |
| TT : CC | 0.90 | 0.51, 1.58 | 0.715 | 0.88 | 0.39, 1.99 | 0.762 | 0.89 | 0.56, 1.42 | 0.636 | |
| TC : CC | 0.84 | 0.64, 1.10 | 0.199 | 0.95 | 0.55, 1.64 | 0.846 | 0.90 | 0.67, 1.22 | 0.506 | |
| (TT + TC) : CC | 0.85 | 0.66, 1.09 | 0.200 | 0.93 | 0.53, 1.63 | 0.809 | 0.90 | 0.67, 1.20 | 0.463 | |
| TT : (CC + TC) | 0.93 | 0.53, 1.63 | 0.804 | 0.87 | 0.39, 1.97 | 0.743 | 0.91 | 0.58, 1.45 | 0.695 | |
OR: odd ratio; 95%CI: 95% confidence interval; p: p value of Z-test for significance test of OR; using the Bonferroni correction, p < 0.0127 means statistically significant.
The study identified that rs231775 polymorphism of CTLA4 was significantly associated with PBC susceptibility. The ORs (95%CIs) of G versus A, GG versus AA, GA versus AA, (GG + GA) versus AA, and GG versus (AA + GA) were 1.32 (1.19–1.47), 1.72 (1.37–2.16), 1.27 (1.13–1.43), 1.38 (1.23–1.54), and 1.52 (1.35–1.71), respectively. As to each model, the p value was below 0.0001 (Figure 1). The rs231725 polymorphism also showed significant association with PBC susceptibility. The ORs with 95%CIs of A versus G, AA versus GG, GA versus GG, (AA + GA) versus GG, and AA versus (GG + GA) were 1.33 (1.22–1.45), 1.83 (1.52–2.21), 1.20 (1.04–1.38), 1.34 (1.17–1.53), and 1.57 (1.35–1.82), respectively. As to each genetic model, the p value was below 0.05 (Figure 2). Nevertheless, no association was identified between rs3087243 (Figure 3) and rs5742909 (Figure 4) polymorphisms and PBC susceptibility. Subgroup analysis showed that both rs231775 and rs231725 showed significant association with PBC susceptibility for Asians and for Caucasians.
Figure 1.
Odds ratio with its 95% confidence interval of PBC linked with CTLA4 rs231775. The rhombus represented the pooled OR with 95%CI.
Figure 2.
Odds ratio with its 95% confidence interval of PBC linked with CTLA4 rs231725. The rhombus represented the pooled OR with 95%CI.
Figure 3.
Odds ratio with its 95% confidence interval of PBC linked with CTLA4 rs3087243. The rhombus represented the pooled OR with 95%CI.
Figure 4.
Odds ratio with its 95% confidence interval of PBC linked with CTLA4 rs5742909. The rhombus represented the pooled OR with 95%CI.
4.3. Calculation of Linkage Disequilibrium
Based on the values of r2, there was no obviously linkage disequilibrium among four SNPs (Table 4).
Table 4.
Results of linkage disequilibrium.
| rs231775 | rs231725 | rs3087243 | rs5742909 | |
|---|---|---|---|---|
| rs231775 | — | 0.908 | 0.964 | 0.942 |
| rs231725 | 0.030 | — | 0.924 | 0.921 |
| rs3087243 | 0.017 | 0.031 | — | 0.914 |
| rs5742909 | 0.020 | 0.074 | 0.030 | — |
Lower left areas are values of r2. Upper right areas are values of D'.
4.4. Sensitivity Analyses and Publication Bias
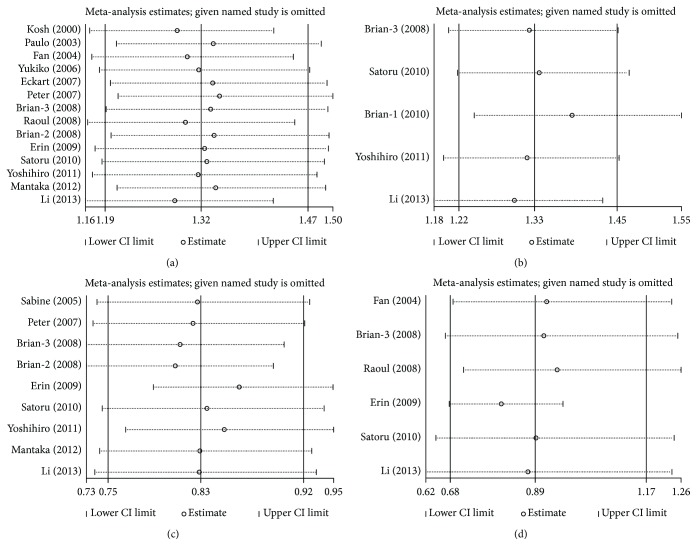
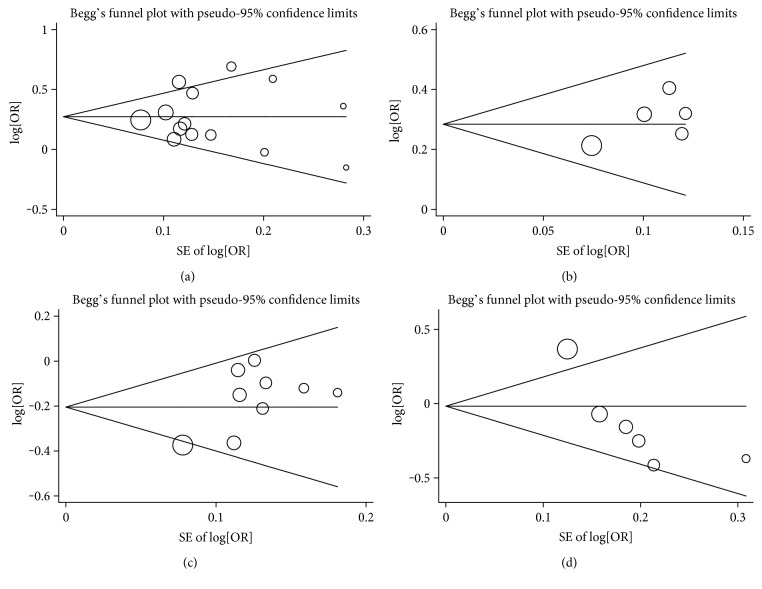
Sensitivity analysis was conducted by omitting each studies sequentially, suggesting that the results for the overall population were statistically stable and reliable (Figure 5). Publication bias was examined by using Egger's regression and Begg's funnel plot in our research. No obvious publication bias was identified. For Begg's test, p values of rs231775 (G versus A), rs231725 (A versus G), rs3087243 (A versus G), and rs5742909 (T versus C) were 0.661, 0.806, 0.466, and 0.060, respectively. For Egger's test, p values of genetic models mentioned above were 0.952, 0.186, 0.061, and 0.029, respectively (Figure 6).
Figure 5.
Sensitivity analysis for the effect of CTLA4 and PBC: (a) rs231755 (G versus A), (b) rs231725 (A versus G), (c) rs3087243 (A versus G), and (d) rs5742909 (T versus C).
Figure 6.
Publication bias for the influence of CTLA4 and PBC: (a) rs231755 (G versus A), (b) rs231725 (A versus G), (c) rs3087243 (A versus G), and (d) rs5742909 (T versus C).
5. Discussion
Multiple comparisons were conducted in this study. To minimize the type I error, the threshold of p values was corrected by the Bonferroni correction. The Bonferroni correction compensates for that increase by testing each individual hypothesis at a significance level [36]. Then, the threshold of p value for the Bonferroni correction can be calculated with the corresponding critical values 1 − (1 − p)1/n. There are four SNPs in this study for multiple comparisons, and the original threshold of p value was 0.05. So, after calculating with above formula (p = 0.05, n = 4), p < 0.0127 was considered statistically significant.
As to the polymorphisms of rs231775 and rs231725, significant connections were found to be associated with PBC in all 5 genetic models. For patients in cases, the frequencies of allele and genotype in rs231775 and rs231725 were increased more significantly than those in controls. As to allele, the results of rs231775 were similar to the results of five published meta-analyses by Eskandari-Nasab et al. [39], Huang et al. [23], Miyake et al. [24], Li et al. [25], and Chen et al. [14]. Li and Miyake indicated that the G allele might be connected with PBC as a risk factor. On the contrary, meta-analyses from Chen and Huang proposed that the relationship between G allele and susceptibility of PBC was observed only in Asian.
As to rs3087243, our analysis showed that both allele and genotype were negative associations with PBC in overall populations. For rs5742909, in codominant, dominant, and recessive models, there were no connections with susceptibility of PBC in Caucasian and Asian. These results were consistent with those in one meta-analysis, which was conducted by Li et al. [25], including 12 studies.
Through subgroup analysis, GG homozygosity of rs231775 and AA homozygosity of rs231725 were associated with the susceptibility to PBC both in Asians and in Caucasians. AA homozygosity of rs3087243 was protective against PBC in Asians and Caucasians. On the other hand, GA heterozygosity of rs231725 was associated with the susceptibility to PBC in Asians although it was not in Caucasians. GA heterozygosity of rs3087243 was protective against PBC in Asians although it was not in Caucasians. Thus, there may be a little different between Asians and Caucasians in the relationship between SNP polymorphism of CTLA4 and the susceptibility to PBC. In order to solve these problems, further studies in various ethnicities are required.
In linkage disequilibrium, coefficients D' and r2 were frequently used. They have quite different characteristics and could be applied for different purposes. Typically, r2 is useful in the context of association studies, D' is the measure of choice to assess recombination patterns in a given population [40]. It is indicated that the two loci were not recombined and were in a complete linkage disequilibrium when the value of D' is 1. But the significance of values would be difficult to interpret when D' < 1. Meanwhile, when the sample size is small and the frequency of SNPs is low, the estimate of D' would be too large. In this case, even the sites of linkage equilibrium can get larger D' value, the actual meaning of the D' could easily be exaggerated. Then, the value of r2 could be more reliable under this condition. In this study, the frequency of genotypes in rs5742909 was much lower than the others (Table 1). Thus, the values of D' were much larger than the value of r2. So, r2 was chosen to assess the linkage disequilibrium. Based on the results, LD was not observed among four SNPs.
Heterogeneity may affect pooled results as one of possible factors. It can be categorized into heterogeneity of the genetic model and effect. In this study, a relatively moderate heterogeneity was heeded. Among 16 studies, HWE values of five studies were out. As to rs231775 and rs5742909, there were three and two studies to be out of HWE, respectively. Thus, we conducted the sensitivity analysis in all studies. In the analysis of rs231775, the results of I2values reduced when we removed the article by Li et al. [22]. The heterogeneity in Caucasians was larger than in Asians. This study showed that diversity of genetic ethnicities or methodological differences might be the sources of heterogeneity.
There are some characteristics in this meta-analysis. Comparing with other similar articles, we conducted four SNPs in one study, and each SNP included five different genetic models. We assessed subgroup by analyzing ethnicities and obtained more precise estimation of the relationships. We also performed sensitivity analysis to test the validity of the results.
To date, several genome-wide association studies (GWAS) and genome-wide meta-analysis on PBC have been performed. From these literatures [41–47], there were several genes to be identified as significant susceptibility loci for PBC. There were ethnic differences in genetic susceptibility loci such as TNFSF15, POU2AF1, IL12A, and IL12RB2 and common pathogenic pathways such as B cell differentiation, IL-12 signaling, and T cell activation.
As to GWAS, the relationship between mutations of SNPs and occurrence of disease might not be a necessity but a probability. So, a large number of samples should be analyzed for the association study between gene and disease. The number of cases which were enrolled in each publication of genome-wide association study of PBC was not enough comparing with the probability of gene mutation. Thus, some higher risk loci with lower mutation frequency could be concealed by lower risk loci with higher mutation frequency. Meanwhile, it was different from the GWAS that focused on the onset of disease, and the data of our meta-analysis might provide a point in the search for novel therapies that are urgently needed to improve outcomes for PBC patients.
On the one hand, GWAS efforts have focused on the identification of association of genetic variants with PBC, but not with specific properties of disease such as response of treatment [48]. As mentioned, the IL-12 pathway has been strongly implicated in the pathogenesis of PBC. The monoclonal antibody took the IL-12p40 subunit as the target and exerted its effect on both the IL-12/TH1 and IL-23/TH17 axes. While the monoclonal antibody has demonstrated therapeutic benefit in patients with Crohn's disease and psoriasis, none of the patients achieved the predefined primary endpoint of alkaline phosphatase reduction from baseline [10, 49]. Although, the data of CTLA4 polymorphism and the association between CTLA4 and PBC were not reported in the GWAS of PBC, CTLA4 was the main focus of PBC in many candidate gene studies, and certain benefit results were obtained as a therapeutic target from CTLA4. CTLA4-Ig has been developed as an exciting outcome in mouse model of PBC [11, 12]. Based on these studies, a new clinical study has been set to determine the effect of abatacept in PBC patients who have no response to UDCA (NCT02078882).
On the other hand, IL-12 signal pathway may be an important role for PBC through Th1/2 differentiation among these loci from GWAS, but CTLA4 could also impact Th1/2. Indeed, CTLA4-deficient mice and T cells were shown to be strongly trend a Th2 phenotype [50]. This is the control of Th1/Th2 differentiation, which was shown to depend both on the cytokine microenvironment and costimulatory signals [51]. It is evidenced that some gene loci could be potential risk for PBC in GWAS, but these findings still have not translated into clinic. Although, the polymorphism of CTLA4 could not be improved in GWAS, as to biliary cell damages in PBC, CTLA4 could influence the effect of these inflammatory cytokines, such as IL-12, and IL-23 [52].
In view of the publication of the GWAS of PBC, our meta-analysis might be quite basic. However, we have collected sufficient documentations that have ever been published and analyzed four SNPs of CTLA4 that have ever been reported in the publication of candidate gene studies. It should be said that this study was a more comprehensive meta-analysis of association between CTLA4 and PBC. Our findings might illustrate that relevant research could still be gained from the candidate gene investigation.
In this meta-analysis, there were still some limitations to be existed. First, except race, there were other factors to be concerned, which included age, gender, and alcohol habit. It would be useful to understand that different risk factors might interact with the development of PBC as genetic variations. Secondly, some studies suggested that patients in PBC would also suffer other autoimmune diseases. Since, these literatures which we included did not mention whether other diseases were excluded in those patients, these situations may introduce errors during analyzing.
To sum up, this meta-analysis showed that the GG, GA genotype, and G allele of rs231775 and AA, GA genotype, and A allele of rs231725 in CTLA4 may be risk factors for PBC in Asians and Caucasians. AA, GA genotype, and A allele of rs3087243 may be negatively associated with PBC in overall populations, especially in Asian. There was no significant connection with PBC in rs5742909 of CTLA4. Not only the impact on cytokine regulation but also the benefit results as therapeutic target, to some extent, CTLA4 still plays a role which could not be completely ignored in PBC.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Poupon R., Ping C., Chrétien Y., et al. Genetic factors of susceptibility and of severity in primary biliary cirrhosis. Journal of Hepatology. 2008;49(6):1038–1045. doi: 10.1016/j.jhep.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 2.Jones D. E. Pathogenesis of primary biliary cirrhosis. Journal of Hepatology. 2003;39(4):639–648. doi: 10.1016/S0168-8278(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 3.Talwalkar J. A., Lindor K. D. Primary biliary cirrhosis. Lancet. 2003;362(9377):53–61. doi: 10.1016/S0140-6736(03)13808-1. [DOI] [PubMed] [Google Scholar]
- 4.Gershwin M. E., Mackay I. R. The causes of primary biliary cirrhosis: convenient and inconvenient truths. Hepatology. 2008;47(2):737–745. doi: 10.1002/hep.22042. [DOI] [PubMed] [Google Scholar]
- 5.Teft W. A., Kirchhof M. G., Madrenas J. A molecular perspective of CTLA-4 function. Annual Review of Immunology. 2006;24:65–97. doi: 10.1146/annurev.immunol.24.021605.090535. [DOI] [PubMed] [Google Scholar]
- 6.Yang G. X., Wu Y., Tsukamoto H., et al. CD8 T cells mediate direct biliary ductule damage in nonobese diabetic autoimmune biliary disease. Journal of Immunology. 2011;186(2):1259–1267. doi: 10.4049/jimmunol.1001597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kita H., Imawari M. Gershwin M. E. Cellular immune response in primary biliary cirrhosis. Hepatology Research. 2004;28(1):12–17. doi: 10.1016/j.hepres.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Kita H. Autoreactive CD8-specific T-cell response in primary biliary cirrhosis. Hepatology Research. 2007;37(Supplement 3):S402–S405. doi: 10.4103/ijo.IJO_437_16. [DOI] [PubMed] [Google Scholar]
- 9.Scalapino K. J., Daikh D. I. CTLA-4: a key regulatory point in the control of autoimmune disease. Immunological Reviews. 2008;223(1):143–155. doi: 10.1111/j.1600-065X.2008.00639.x. [DOI] [PubMed] [Google Scholar]
- 10.Carbone M., Lleo A., Sandford R. N., Invernizzi P. Implications of genome-wide association studies in novel therapeutics in primary biliary cirrhosis. European Journal of Immunology. 2014;44(4):945–954. doi: 10.1002/eji.201344270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhirapong A., Yang G. X., Nadler S., et al. Therapeutic effect of cytotoxic T lymphocyte antigen 4/immunoglobulin on a murine model of primary biliary cirrhosis. Hepatology. 2013;57(2):708–715. doi: 10.1002/hep.26067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka H., Yang G. X., Tomiyama T., et al. Immunological potential of cytotoxic T lymphocyte antigen 4 immunoglobulin in murine autoimmune cholangitis. Clinical and Experimental Immunology. 2015;180(3):371–382. doi: 10.1111/cei.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida O., Abe M., Furukawa S., et al. A familial case of autoimmune hepatitis. Internal Medicine. 2009;48(5):315–319. doi: 10.2169/internalmedicine.48.1533. [DOI] [PubMed] [Google Scholar]
- 14.Chen R. R., Han Z. Y., Li J. G., et al. Cytotoxic T-lymphocyte antigen 4 gene +49A/G polymorphism significantly associated with susceptibility to primary biliary cirrhosis: a meta-analysis. Journal of Digestive Diseases. 2011;12(6):428–435. doi: 10.1111/j.1751-2980.2011.00537.x. [DOI] [PubMed] [Google Scholar]
- 15.Kouki T., Sawai Y., Gardine C. A., Fisfalen M. E., Alegre M. L., DeGroot L. J. CTLA-4 gene polymorphism at position 49 in exon 1 reduces the inhibitory function of CTLA-4 and contributes to the pathogenesis of Graves’ disease. The Journal of Immunology. 2000;165(11):6606–6611. doi: 10.4049/jimmunol.165.11.6606. [DOI] [PubMed] [Google Scholar]
- 16.Howson J. M., Dunger D. B., Nutland S., Stevens H., Wicker L. S., Todd J. A. A type 1 diabetes subgroup with a female bias is characterised by failure in tolerance to thyroid peroxidase at an early age and a strong association with the cytotoxic T-lymphocyte-associated antigen-4 gene. Diabetologia. 2007;50(4):741–746. doi: 10.1007/s00125-007-0603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C. P., Jiang J. A., Wang T., et al. CTLA-4 and CD86 genetic variants and haplotypes in patients with rheumatoid arthritis in southeastern China. Genetics and Molecular Research. 2013;12(2):1373–1382. doi: 10.4238/2013.April.25.8. [DOI] [PubMed] [Google Scholar]
- 18.Walker E. J., Hirschfield G. M., Xu C., et al. CTLA4/ICOS gene variants and haplotypes are associated with rheumatoid arthritis and primary biliary cirrhosis in the Canadian population. Arthritis and Rheumatism. 2009;60(4):931–937. doi: 10.1002/art.24412. [DOI] [PubMed] [Google Scholar]
- 19.Joshita S., Umemura T., Yoshizawa K., et al. Association analysis of cytotoxic T-lymphocyte antigen 4 gene polymorphisms with primary biliary cirrhosis in Japanese patients. Journal of Hepatology. 2010;53(3):537–541. doi: 10.1016/j.jhep.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Juran B. D., Atkinson E. J., Larson J. J., et al. Carriage of a tumor necrosis factor polymorphism amplifies the cytotoxic T-lymphocyte antigen 4 attributed risk of primary biliary cirrhosis: evidence for a gene-gene interaction. Hepatology. 2010;52(1):223–229. doi: 10.1002/hep.23667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aiba Y., Nakamura M., Joshita S., et al. Genetic polymorphisms in CTLA4 and SLC4A2 are differentially associated with the pathogenesis of primary biliary cirrhosis in Japanese patients. Journal of Gastroenterology. 2011;46(10):1203–1212. doi: 10.1007/s00535-011-0417-7. [DOI] [PubMed] [Google Scholar]
- 22.Li Q., Wang B., Pan F., et al. Association between cytotoxic T-lymphocyte antigen 4 gene polymorphisms and primary biliary cirrhosis in Chinese population: data from a multicenter study. Journal of Gastroenterology and Hepatology. 2013;28(8):1397–1402. doi: 10.1111/jgh.12165. [DOI] [PubMed] [Google Scholar]
- 23.Huang Q., Shao F., Wang C., Qiu L. J., Hu Y. G., Yu J. H. Association between CTLA-4 exon-1 +49A>G polymorphism and primary biliary cirrhosis risk: a meta-analysis. Archives of Medical Research. 2011;42(3):235–238. doi: 10.1016/j.arcmed.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Miyake Y., Ikeda F., Takaki A., Nouso K., Yamamoto K. +49A/G polymorphism of cytotoxic T-lymphocyte antigen 4 gene in type 1 autoimmune hepatitis and primary biliary cirrhosis: a meta-analysis. Hepatology Research. 2011;41(2):151–159. doi: 10.1111/j.1872-034X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 25.Li M., Zheng H., Li T., Gao P., Zhang X. L., Liu D. W. Cytotoxic T-lymphocyte associated antigen-4 gene polymorphisms and primary biliary cirrhosis: a systematic review. Journal of Gastroenterology and Hepatology. 2012;27(7):1159–1166. doi: 10.1111/j.1440-1746.2012.07118.x. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal K., Jones D. E., Daly A. K., et al. CTLA-4 gene polymorphism confers susceptibility to primary biliary cirrhosis. Journal of Hepatology. 2000;32(4):538–541. doi: 10.1016/S0168-8278(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 27.Bittencourt P. L., Palacios S. A., Farias A. Q., et al. Analysis of major histocompatibility complex and CTLA-4 alleles in Brazilian patients with primary biliary cirrhosis. Journal of Gastroenterology and Hepatology. 2003;18(9):1061–1066. doi: 10.1046/j.1440-1746.2003.03091.x. [DOI] [PubMed] [Google Scholar]
- 28.Fan L. Y., Tu X. Q., Cheng Q. B., et al. Cytotoxic T lymphocyte associated antigen-4 gene polymorphisms confer susceptibility to primary biliary cirrhosis and autoimmune hepatitis in Chinese population. World Journal of Gastroenterology. 2004;10(20):3056–3059. doi: 10.3748/wjg.v10.i20.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oertelt S., Kenny T. P., Selmi C., Invernizzi P., Podda M., Gershwin M. E. SNP analysis of genes implicated in T cell proliferation in primary biliary cirrhosis. Clinical & Developmental Immunology. 2005;12(4):259–263. doi: 10.1080/17402520500317859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanno Y., Rai T., Monoe K., et al. Possible association of cytotoxic T lymphocyte antigen-4 genetic polymorphism with liver damage of primary biliary cirrhosis in Japan. Fukushima Journal of Medical Science. 2006;52(2):79–85. doi: 10.5387/fms.52.79. [DOI] [PubMed] [Google Scholar]
- 31.Donaldson P., Veeramani S., Baragiotta A., et al. Cytotoxic T-lymphocyte-associated antigen-4 single nucleotide polymorphisms and haplotypes in primary biliary cirrhosis. Clinical Gastroenterology and Hepatology. 2007;5(6):755–760. doi: 10.1016/j.cgh.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Schott E., Witt H., Pascu M., et al. Association of CTLA4 single nucleotide polymorphisms with viral but not autoimmune liver disease. European Journal of Gastroenterology & Hepatology. 2007;19(11):947–951. doi: 10.1097/MEG.0b013e3282efa240. [DOI] [PubMed] [Google Scholar]
- 33.Juran B. D., Atkinson E. J., Schlicht E. M., Fridley B. L., Lazaridis K. N. Primary biliary cirrhosis is associated with a genetic variant in the 3′ flanking region of the CTLA4 gene. Gastroenterology. 2008;135(4):1200–1206. doi: 10.1053/j.gastro.2008.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juran B. D., Atkinson E. J., Schlicht E. M., Fridley B. L., Petersen G. M., Lazaridis K. N. Interacting alleles of the coinhibitory immunoreceptor genes cytotoxic T-lymphocyte antigen 4 and programmed cell-death 1 influence risk and features of primary biliary cirrhosis. Hepatology. 2008;47(2):563–570. doi: 10.1002/hep.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mantaka A., Goulielmos G. N., Koulentaki M., Tsagournis O., Voumvouraki A., Kouroumalis E. A. Polymorphisms of genes related to endothelial cells are associated with primary biliary cirrhosis patients of Cretan origin. Human Immunology. 2012;73(8):829–835. doi: 10.1016/j.humimm.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Goemana J. J., Solaric A. Multiple hypothesis testing in genomics. Statistics in Medicine. 2014;33(11):1946–1978. doi: 10.1002/sim.6082. [DOI] [PubMed] [Google Scholar]
- 37.Thakkinstian A., McElduff P., D’Este C., Duffy D., Attia J. A method for meta-analysis of molecular association studies. Statistics in Medicine. 2005;24(9):1291–1306. doi: 10.1002/sim.2010. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y. Y., He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Research. 2005;15(2):97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 39.Eskandari-Nasab E., Tahmasebi A., Hashemi M. Meta-analysis: the relationship between CTLA-4 +49 A/G polymorphism and primary biliary cirrhosis and type I autoimmune hepatitis. Immunological Investigations. 2015;44(4):331–348. doi: 10.3109/08820139.2014.1003651. [DOI] [PubMed] [Google Scholar]
- 40.Mueller J. C. Linkage disequilibrium for different scales and applications. Briefings in Bioinformatics. 2004;5(4):355–364. doi: 10.1093/bib/5.4.355. [DOI] [PubMed] [Google Scholar]
- 41.Hirschfield G. M., Liu X., Xu C., et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. The New England Journal of Medicine. 2009;360(24):2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X., Invernizzi P., Lu Y., et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nature Genetics. 2010;42(8):658–660. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mells G. F., Floyd J. A., Morley K. I., et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nature Genetics. 2011;43(4):329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juran B. D., Hirschfield G. M., Invernizzi P., et al. Immunochip analyses identify a novel risk locus for primary biliary cirrhosis at 13q14, multiple independent associations at four established risk loci and epistasis between 1p31 and 7q32 risk variants. Human Molecular Genetics. 2012;21(23):5209–5221. doi: 10.1093/hmg/dds359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J. Z., Almarri M. A., Gaffney D. J., et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nature Genetics. 2012;44(10):1137–1141. doi: 10.1038/ng.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura M., Nishida N., Kawashima M., et al. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. American Journal of Human Genetics. 2012;91(4):721–728. doi: 10.1016/j.ajhg.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cordell H. J., Han Y., Mells G. F., et al. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nature Communications. 2015;6:p. 8019. doi: 10.1038/ncomms9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gulamhusein A. F., Juran B. D., Lazaridis K. N. Genome-wide association studies in primary biliary cirrhosis. Seminars in Liver Disease. 2015;35(4):392–401. doi: 10.1055/s-0035-1567831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mousa H. S., Lleo A., Invernizzi P., Bowlus C. L., Gershwin M. E. Advances in pharmacotherapy for primary biliary cirrhosis. Expert Opinion on Pharmacotherapy. 2015;16(5):633–643. doi: 10.1517/14656566.2015.998650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bour-Jordan H., Grogan J. L., Tang Q., Auger J. A., Locksley R. M., Bluestone J. A. CTLA-4 regulates the requirement for cytokine-induced signals in T(H)2 lineage commitment. Nature Immunology. 2003;4(2):182–188. doi: 10.1038/ni884. [DOI] [PubMed] [Google Scholar]
- 51.Bour-Jordan H., Esensten J. H., Martinez-Llordella M., Penaranda C., Stumpf M., Bluestone J. A. Intrinsic and extrinsic control of peripheral T-cell tolerance by costimulatory molecules of the CD28/ B7 family. Immunological Reviews. 2011;241(1):180–205. doi: 10.1111/j.1600-065X.2011.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang C. Y., Leung P. S., Yang G. X., et al. Epitope-specific anti-nuclear antibodies are expressed in a mouse model of primary biliary cirrhosis and are cytokine-dependent. Clinical and Experimental Immunology. 2012;168(3):261–267. doi: 10.1111/j.1365-2249.2012.04577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]