Abstract
Bile acids play a critical role in regulation of glucose, lipid and energy metabolisms through activating the nuclear bile acid receptor farnesoid X receptor (FXR) and membrane G protein-coupled bile acid receptor-1 (Gpbar-1 aka TGR5) signaling. Paradoxical roles of FXR in regulation of glucose and lipid metabolism and metabolic disorder have been reported recently. Activation or inhibition of intestinal FXR signaling have been shown to improve insulin and glucose sensitivity and energy metabolism to prevent diabetes, obesity and non-alcoholic fatty liver disease (NAFLD). TGR5 has an anti-inflammatory function in the intestine and stimulates glucagon-like peptide-1 (GLP-1) secretion in the intestine to stimulate insulin secretion from the pancreas. The role of TGR5 in metabolism and metabolic regulation is not clear and warrants further study. FXR and TGR5 are co-expressed in the ileum and colon. These two bile acid-activated receptors may cooperate to stimulate GLP-1 secretion and improve hepatic metabolism. FXR and TGR5 dual agonists may have therapeutic potential for treating diabetes and NAFLD.
Keywords: Bile acid synthesis, CYP7A1, diabetes, drug therapy, Non-alcoholic fatty liver disease
Background
Bile acid synthesis
It is now well established that bile acids are signaling molecules that regulate lipid, glucose and energy homeostasis through activation of the bile acid nuclear receptor Farnesoid X receptor (FXR) and the membrane G-protein-coupled bile acid receptor 1 (Gpbar1, aka TGR5) [1]. Bile acid synthesis in the liver generates two primary bile acids, cholic acid (CA) and chenodeoxycholic (CDCA) (Figure 1) [2]. In mouse liver, CDCA is converted to α-and β-muricholic. Cholesterol 7α-hydroxylase (CYP7A1) is the first and rate-limiting enzyme in the classic bile acid synthesis pathway. Sterol 12α-hydroxylase (CYP8B1) catalyzes CA synthesis. Mitochondrial steroid 12α-hydroxylase (CYP27A1) catalyzes steroid side-chain oxidation, followed by side-chain cleavage reaction in the peroxisomes to cleave 2C-units from the side-chain to form C24 bile acids. In the alternative pathway, CYP27A1 initiates bile acid synthesis, followed by oxysterol 7α-hydroylase (CYP7B1) in most tissues. The oxysterol intermediates formed are transported to the liver for synthesis of mainly CDCA. Bile acids are re-conjugated to glycine and taurine and secrete into bile, store in the gallbladder and secreted into the intestinal tract after a meal to facilitate intestinal absorption of fats and lipid-soluble vitamins. In the intestine, gut microbial bile salt hydrolases (BSH) deconjugate taurine- and glycine-conjugated-bile acids, and bacterial 7α-dehydroxylases convert primary bile acids CA and CDCA to deoxycholic acid (DCA) and lithocholic acid (LCA), respectively, which are re-conjugated to glycine or taurine.
Figure 1. Bile acid synthesis and regulation.
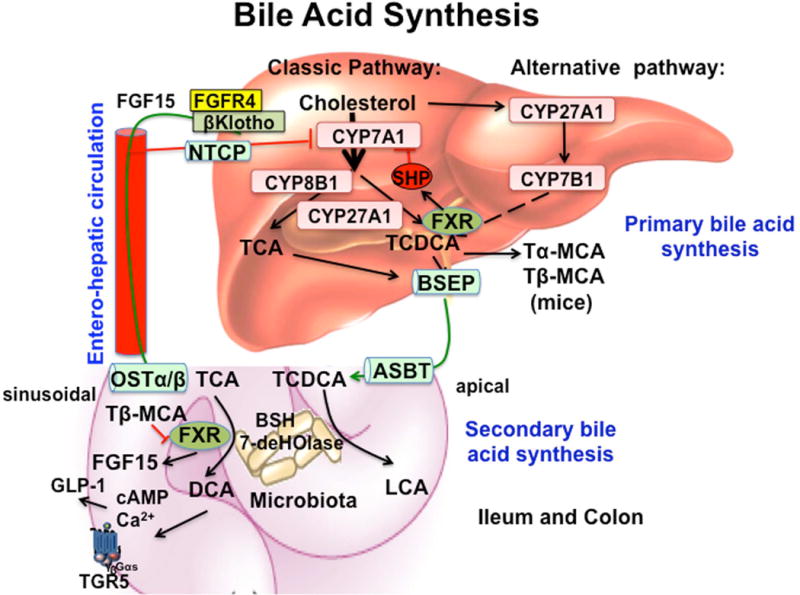
Two bile acid synthesis pathways exist in the liver. The classic pathway is initiated by cholesterol 7α-hydroxylase (CYP7A1), while the alternative pathway is initiated by sterol 27-hydroxylase (CYP27A1). In humans, two primary bile acids are synthesized, cholic acid (CA) and chenodeoxycholic acid (CDCA). For synthesis of CA, sterol 12α-hydroxylase (CYP8B1) is needed. Mitochondrial CYP27A1 catalyzes the steroid side-chain oxidation reaction. In the alternative pathway, oxysterol 7α-hydroxylase (CYP7B1) is involved in the synthesis of oxysterol intermediates, which are converted to bile acids, mainly CDCA, in hepatocytes. In mice, CDCA is converted to tauro-α-muricholic acid (TαMCA) and then TβMCA as primary bile acids. Bile acids are secreted through the bile salt export pump (BSEP), stored in the gallbladder, and secreted into the gastrointestinal tract and reabsorbed through apical sodium-dependent bile salt transporter (ASBT) in the ileum. In the colon, TCA and TDCA are first de-conjugated by bacterial bile salt hydrolases (BSH), and dehydroxylated by bacterial 7α-dehydroxylases to form the secondary bile acids, deoxycholic acid (DCA) and lithocholic acid (LCA), respectively. In the ileum, the bile acid efflux transporter, sinusoidal organic solute transporter α/β (OSTα/β) effluxes bile acids and circulates bile acids to the liver via the portal vein to inhibit bile acid synthesis. Two mechanisms have been suggested to inhibit CYP7A1 and CYP8B1 gene transcription. In the liver, bile acids activate nuclear receptor farnesoid X receptor (FXR) to induce a negative nuclear receptor, small heterodimer partner (SHP) to inhibit CYP7A1 and CYP8B1 gene transcription. In the intestine, FXR induces fibroblast growth factor 15 (FGF15, or FGF19 in humans), which is circulated to the liver to activate plasma membrane FGF receptor 4 (FHFR4)/βKlotho signaling to inhibit CYP7A1 gene transcription. TGR5 (Gpbar-1) is expressed in the ileum and colon. TGR5 activates cAMP signaling to stimulate glucagon-like peptide-1 (GLP-1) secretion in enteroendocrine L cells. GLP-1 stimulates insulin secretion in pancreatic β cells to improve insulin sensitivity. Decreasing BSH activity in gut microbiota increases TβMCA, which antagonizes FXR activity to reduce FGF15/19 and stimulates bile acid synthesis in hepatocytes. Increasing BSH activity in gut microbiota reduces TMCA antagonism of FXR and stimulates FGF15/19 production to inhibit bile acid synthesis. Thus, the gut-to-liver axis plays a critical role in regulation of bile acid synthesis and metabolic homeostasis.
Regulation of bile acid synthesis
Bile acids are the end products of cholesterol catabolism in the liver. Bile acids facilitate biliary cholesterol and lipid secretion to form mixed micelles in the gallbladder. Bile acids are reabsorbed in the ileum and transported via enterohepatic circulation (EHC) to the liver to inhibit bile acid synthesis by transcriptional repression of CYP7A1 and CYP8B1 gene expression. EHC is highly efficient, occurs several time a day and recycles about 95% bile acids. EHC of bile acids facilitate absorption of nutrients to the liver to be metabolized and re-distributed to other tissues. Two mechanisms have been suggested for bile acid feedback regulation of bile acid synthesis. In the liver, bile acids activate FXR to induce a negative nuclear receptor small heterodimer partner (SHP) to inhibit CYP7A1 and CYP8B1 gene transcription. In the intestine, FXR induces fibroblast growth factor 15 (FGF15), which is transported to the liver to activate hepatic FGF receptor 4 signaling to inhibit CYP7A1 gene transcription.
The gut microbiota plays a critical role in pathogen defense, immunity and nutrient harvest. Bile acids inhibit gut bacterial overgrowth and the gut microbiota controls bile acid metabolism by de-conjugation of bile acids by BSH and de-hydroxylation by 7α-dehydroxylases, thus controlling bile acid composition in the pool [3, 4]. The gut microbiota utilizes short chain fatty acids from dietary carbohydrates for energy metabolism and regulates glucose, lipid and energy metabolism in the liver, muscle and adipocytes. The gut microbiota also controls gut hormones/peptides release from the intestine, such as YY and glucagon-like peptide-1 (GLP-1). These hormones regulate insulin secretion from pancreatic β cells and in turn, glucose homeostasis. Dysbiosis has been associated with inflammatory bowel diseases, obesity and type 2 diabetes and non-alcoholic fatty liver disease, cirrhosis and liver cancer [5–8].
Bile acid synthesis is regulated by circadian rhythms and the fasting and refeeding cycle [1, 9]. In mice, Cyp7a1 and bile acid synthesis peaks right after entering the dark cycle and decreases in the mid-night. In humans bile acid synthesis peaks at 9 am and mid-afternoon [10–12]. Circadian rhythms are influenced by feeding and diets [13–15]. CYP7A1 expression rapidly increases after feeding and decreases after fasting, while CYP8B1 is stimulated by fasting and decreased by feeding [16]. Thus circadian expression pattern of these two regulatory enzymes determines the rate of bile acid synthesis and bile acid composition in the pool. Disruption of the circadian regulation of bile acid synthesis alters hepatic metabolism and homeostasis and contributes to diabetes and obesity [17].
Bile acid receptor regulation of hepatic metabolism
Bile acids regulate hepatic metabolism via activation of FXR and TGR5 signaling. The role of FXR in the regulation of hepatic lipid and glucose metabolism has been studied extensively. Activation of FXR by bile acids and agonists reduces serum triglycerides and improves glucose tolerance and insulin resistance in diabetic mice [18, 19]. However, more recent studies show the opposite effect in that deficiency of FXR improves glucose homeostasis [20] and activation of FXR lowers bile acid synthesis and induces obesity and diabetes [21]. Moreover, several recent studies show that deficiency of intestinal Fxr gene or antagonism of intestine FXR by the antioxidant tempol or intestine-selective FXR inhibitor Gly-MCA increases TMCA to antagonize intestinal FXR and decrease obesity and NAFLD [22–24]. In contrast, activation of intestinal FXR by a selective FXR agonist fexaramine (FEX) also improves diabetes and obesity in mice [25]. Thus the role and mechanism of FXR signaling in the regulation of lipid and glucose metabolism is controversial and not completely understood.
The role of TGR5 in the regulation of hepatic metabolism has not been studied in depth. TGR5 is widely expressed in many tissues including intestine, gallbladder, liver, and brain [26–28]. In the liver, TGR5 is expressed in Kupffer cells and sinusoidal endothelial cells, but not in hepatocytes [28, 29]. In the gastrointestinal tract, activation of TGR5 by bile acids and agonists protects intestinal barrier function, reduces inflammation, stimulates gallbladder refilling and GLP-1 secretion from enteroendocrine L cells [30], which increases postprandial insulin secretion from pancreatic β cells and improves insulin resistance [31]. GLP-1 is an intestinal incretin produced in L cells through processing of preproglucagon by prohormone convertase 1/3 (PC1/3) and is released in response to meal intake [32]. GLP-1 secretion is stimulated by nutrients in the intestinal lumen, such as carbohydrates, fats and proteins. The synthetic GLP-1 analog exendin-4 reduces hepatic steatosis by decreasing lipogenesis and inducing fatty acid oxidation [33]. Activation of TGR5 induces thyroid hormone deiodinase 2 to stimulate energy metabolism in brown adipose tissues [34]. Activation of TGR5 stimulates adenylyl cyclase to convert AYP to cAMP, which activates protein kinase A (PKA) to activate cAMP response element binding protein (CREB) and results in alleviation of obesity and hepatic steatosis in diet-induced obese mice [31]. Thus, further study of the role of FXR and TGR5 in hepatic metabolism and diabetes is needed.
Key Messages
FXR and TGR5 are co-expressed in intestinal endocrine L cells. It is possible that activation of FXR may stimulate GLP-1 secretion from L cells and contribute to increasing insulin secretion form pancreatic β cell to improve insulin sensitivity. To test this hypothesis, FXR and TGR5 agonists were used to study their effects on GLP-1 secretion in wild type, Fxr−/− and Tgr5−/− mice, and in enteroendocrine GLUTag and STC-1 cells. Oral gavage of FXR agonists OCA, GW4064 and FEX, and TGR5 selective agonist INT777 agonist stimulated GLP-1 secretion in wild type mice and GLUTag cells. Real time PCR assay showed that TGR5 mRNA expression level was induced in the intestine of wild type mice, but reduced in Fxr−/− mice, suggesting that FXR induced TGR5 gene transcription. Interestingly mRNA levels of PC1/3, which splices preproglucagon to glucagon, were also induced by OCA and GW4064 and FEX, and reduced in Fxr−/− mice. Immunoblot analysis showed OCA and FEX induced TGR5 and PC1/3 protein levels in mouse ileum. cAMP response element binding protein (CREB) reporter assay and site-directed mutagenesis identified a FXR response element located in the proximal promoter of human and mouse Tgr5 gene promoter. Electrophoretic mobility shift assay demonstrated FXR/RXR binding to this putative FXRE. Chromatinimmunoprecipitation assay (ChIP) demonstrated the occupancy of FXR/RXR and co-activator PGC-1α to the TGR5 promoter region containing the FXRE.
To further confirm the role of FXR in GLP-1 secretion, a GLP-1 secretion assay was performed in Fxr−/−, and Tgr5−/− mice. In Tgr5−/− mice, GLP-1 secretion was reduced ~50%, indicating some other factor (FXR) also involved in GLP-1 secretion. Oral gavage of FEX did not affect GLP-1 secretion in Tgr5−/− mice indicating TGR5 was required for GLP-1 secretion. In Fxr−/− mice, GLP-1 secretion was reduced ~50% because TGR5 expression was reduced in the absence of FXR. This study reports for the first time that activation of FXR induces Tgr5 and Pc1/3 gene transcription to stimulate GLP-1 processing and secretion from intestinal L cells during the postprandial state. FXR and TGR5 may coordinately regulate GLP-1 production and secretion.
Our data also showed that OCA and FEX have differential effects on hepatic glucose and lipid metabolism; FEX is more effective in inhibiting gluconeogenesis and improving glucose tolerance while OCA is more effective in stimulating fatty acid oxidation and improving lipid metabolism. This study also indicated that GLP-1 may directly stimulate insulin-like signaling in hepatocytes to inhibit gluconeogenesis and improve insulin and glucose sensitivity. FEX inhibits hepatic gluconeogenic gene expression and improves insulin and glucose tolerance and reduced serum cholesterol and hepatic triglycerides in diabetic db/db mice.
Conclusions
The gut-to-liver axis plays a critical role in regulating bile acid synthesis and FXR and TGR5 signaling in liver and intestine. Activation of FXR by bile acids in the intestine induces FGF15 to inhibit bile acid synthesis in the liver, whereas inhibition of BSH by gut bacteria or the antioxidant tempol increases T-β-MCA, which may antagonize FXR and FGF15 production to stimulate bile acid synthesis [22, 35]. In mice deficient of intestinal Fxr, FGF15 production is reduced and bile acid synthesis is up-regulated to stimulate hepatic FXR and improve glucose and insulin sensitivity, and thus prevent diet-induced NAFLD [23]. Intestine-selective inhibition of FXR by Gly-MCA also reduces intestine ceramide production and contributes to improving diabetes and obesity [24]. Disruption of the gut-to-liver axis by deleting or inhibiting intestinal FXR or treating with antibiotics, alters gut microbiota to increase ileum TMCA and bile acid synthesis to regulate glucose and lipid metabolism and prevent diet-induced diabetes and obesity. On the other hand, pharmacological activation of intestinal FXR may reduce TCA and increase LCA to stimulate FGF15 and GLP-1 production to improve hepatic metabolism and insulin and glucose sensitivity. Intestinal FXR and TGR5 signaling crosstalk may coordinately stimulate GLP-1 secretion to improve hepatic glucose and insulin sensitivity in diabetes. FXR and TGR5 dual agonists may be effective for treating liver-related metabolic diseases, diabetes and NAFLD.
Acknowledgments
Financial support: This study is supported by NIH grants DK44442 and DK58379.
Abbreviations
- CYP7A1
cholesterol 7α-hydroxylase
- EHC
enterohepatic circulation
- FEX
fexaramine
- FXR
farnesoid X receptor
- GLP-1
glucagon-like peptide-1
- Gpbar-1
G protein-coupled bile acid receptor-1 (aka TGR5)
- NAFLD
non-alcoholic fatty liver disease
- TGR5
Takeda G protein couple receptor
Footnotes
Disclosure Statement
xxxxxxxx
References
- 1.Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66(4):948–83. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inagaki T, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103(10):3920–5. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones BV, et al. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105(36):13580–5. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aron-Wisnewsky J, et al. Gut microbiota and non-alcoholic fatty liver disease: new insights. Clin Microbiol Infect. 2013;19(4):338–48. doi: 10.1111/1469-0691.12140. [DOI] [PubMed] [Google Scholar]
- 6.Kakiyama G, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58(5):949–55. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joyce SA, Gahan CG. The gut microbiota and the metabolic health of the host. Curr Opin Gastroenterol. 2014;30(2):120–7. doi: 10.1097/MOG.0000000000000039. [DOI] [PubMed] [Google Scholar]
- 8.Ridlon JM, et al. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang JY, Miller WF, Lin GM. Regulation of cholesterol 7 alpha-hydroxylase in the liver. Purification of cholesterol 7 alpha-hydroxylase and the immunochemical evidence for the induction of cholesterol 7 alpha-hydroxylase by cholestyramine and circadian rhythm. J Biol Chem. 1990;265(7):3889–97. [PubMed] [Google Scholar]
- 10.Ferrell JM, Chiang JY. Circadian rhythms in liver metabolism and disease. Acta Pharm Sin B. 2015;5(2):113–22. doi: 10.1016/j.apsb.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galman C, Angelin B, Rudling M. Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology. 2005;129(5):1445–53. doi: 10.1053/j.gastro.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Lundasen T, et al. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J Intern Med. 2006;260(6):530–6. doi: 10.1111/j.1365-2796.2006.01731.x. [DOI] [PubMed] [Google Scholar]
- 13.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell metabolism. 2011;13(2):125–37. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106(50):21453–8. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatori M, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–60. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pathak P, Li T, Chiang JY. Retinoic Acid-related Orphan Receptor alpha Regulates Diurnal Rhythm and Fasting Induction of Sterol 12alpha-Hydroxylase in Bile Acid Synthesis. J Biol Chem. 2013;288(52):37154–65. doi: 10.1074/jbc.M113.485987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrell JM, Chiang JY. Short-term circadian disruption impairs bile acid and lipid homeostasis in mice. Cell Mol Gastroenterol Hepatol. 2015;1(6):664–677. doi: 10.1016/j.jcmgh.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe M, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113(10):1408–18. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, et al. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103(4):1006–11. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prawitt J, et al. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes. 2011;60(7):1861–71. doi: 10.2337/db11-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe M, et al. Lowering Bile Acid Pool Size with a Synthetic Farnesoid X Receptor (FXR) Agonist Induces Obesity and Diabetes through Reduced Energy Expenditure. The Journal of biological chemistry. 2011;286(30):26913–20. doi: 10.1074/jbc.M111.248203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4:2384. doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang C, et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest. 2015;125(1):386–402. doi: 10.1172/JCI76738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang C, et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun. 2015;6:10166. doi: 10.1038/ncomms10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang S, et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med. 2015;21(2):159–65. doi: 10.1038/nm.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maruyama T, et al. Identification of membrane-type receptor for bile acids (M-BAR) Biochem Biophys Res Commun. 2002;298(5):714–9. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 27.Kawamata Y, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 28.Keitel V, et al. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372(1):78–84. doi: 10.1016/j.bbrc.2008.04.171. [DOI] [PubMed] [Google Scholar]
- 29.Keitel V, et al. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 2007;45(3):695–704. doi: 10.1002/hep.21458. [DOI] [PubMed] [Google Scholar]
- 30.Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun. 2005;329(1):386–90. doi: 10.1016/j.bbrc.2005.01.139. [DOI] [PubMed] [Google Scholar]
- 31.Thomas C, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167–77. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mojsov S, et al. Preproglucagon gene expression in pancreas and intestine diversifies at the level of post-translational processing. J Biol Chem. 1986;261(25):11880–9. [PubMed] [Google Scholar]
- 33.Ding X, et al. Exendin-4, a glucagon-like protein-1 (GLP-1) receptor agonist, reverses hepatic steatosis in ob/ob mice. Hepatology. 2006;43(1):173–81. doi: 10.1002/hep.21006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe M, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 35.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–35. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]


