Summary
Despite the importance of molecular subtype classification of glioblastoma (GBM), the extent of extracellular vesicle (EV)-driven molecular and phenotypic reprogramming remains poorly understood. To reveal complex subpopulation dynamics within the heterogeneous intratumoral ecosystem, we characterized microRNA expression and secretion in phenotypically diverse subpopulations of patient-derived GBM stem-like cells (GSCs). As EVs and microRNAs convey information that rearranges the molecular landscape in a cell type-specific manner, we argue that intratumoral exchange of microRNA augments the heterogeneity of GSC that is reflected in highly heterogeneous profile of microRNA expression in GBM subtypes.
Keywords: cancer stem cells, subtypes, glioblastoma, GBM, cancer heterogeneity, microRNA, exosomes, extracellular vesicles
Highlights
-
•
MicroRNA signatures reveal tissue heterogeneity in defined glioblastoma subtypes
-
•
GSC EV/microRNA acts via cell-dependent targeting, propagating intratumoral heterogeneity
-
•
EV/microRNAs modify molecular landscape, acting in tumor anatomic sites
Godlewski and colleagues show that microRNAs, unlike long non-coding RNAs and protein-coding mRNAs, do not correlate with glioblastoma tissue subtype classification, and that heterogeneous expression of microRNAs observed in subtypes is further propagated by intratumoral exchange of extracellular vesicles (EVs). Cellular and EV microRNAs display cell-specific functions by targeting cell subpopulation-specific effectors, allowing cell adaptation to diverse tumor anatomic niches.
Introduction
Intercellular dialogue between tumor cells mediated by extracellular vesicles (EVs) is a powerful means of communication that facilitates exchange of active molecules (Bronisz et al., 2014, Skog et al., 2008). It is the linchpin of the molecular network, the invisible causeway on which the global cellular transcriptome hums. However, the culpability of EV communication for phenotypic and molecular diversity within heterogeneous tumors is not fully recognized.
Intratumoral heterogeneity and invasiveness are the key characteristics of glioblastoma (GBM), the most common and most aggressive primary brain malignancy in adults, with a median survival of 14.2 months (Johnson and O'Neill, 2012). The subpopulation of highly tumorigenic and therapy-resistant GBM stem-like cells (GSCs) (Schonberg et al., 2014) retains stem cell characteristics, including self-renewal and undifferentiated status, but also exhibits varying degrees of phenotypic and molecular polymorphism. Discovering whether the underlying cause of invasiveness is inherent or is a response to microenvironmental stimuli has important implications for better understanding of GBM pathobiology.
The signatures of protein-coding gene expression and somatic copy-number alterations have revealed the existence of several distinct subtypes among GBM patients, known as mesenchymal, proneural, neural, and classical according to The Cancer Genome Atlas (TCGA) database (Phillips et al., 2006). The classification is further complicated by the fact that individual tumors contain a spectrum of subtypes and hybrid cellular states (Patel et al., 2014) and that GSC subpopulations retain transcriptome heterogeneity (Mao et al., 2013). These findings indicate that tissue-based classification likely shows merely characteristics of the predominant cellular component. Importantly, tissue and GSC subtype classification was also demonstrated by signatures of non-protein-coding genes, such as long non-coding RNAs (Du et al., 2013, Mineo et al., 2016). Significantly, microRNAs have not been shown to predict GBM classification and prognosis by global signature to date, while being strongly implicated as functionally deregulated in GBM as individual molecules (Godlewski et al., 2015).
We aimed to highlight the magnitude of EV/microRNA-driven propagation of molecular and phenotypic diversity of GSCs. Using intracranial xenografts of patient-derived GSCs, we selected subpopulations of cells with distinct transcriptomes, displaying proliferative/nodular or migratory/invasive modes, which are associated with mesenchymal-like or proneural-like subtype, respectively. The highly heterogeneous expression profile of microRNAs in GBMs was separable into two unsupervised classes that partially overlapped with previously determined molecular subtypes, with both subclasses of GSCs displaying differential cellular and EV microRNA profiles. The analysis of microRNA/target networks provided evidence that EV/microRNAs are modifiers of both the molecular landscape and phenotype, acting via cell-dependent targeting that propagates GBM subtype heterogeneity.
Results
Intratumoral Architecture Is Imposed by Phenotype/Transcriptome-Diverse GSCs
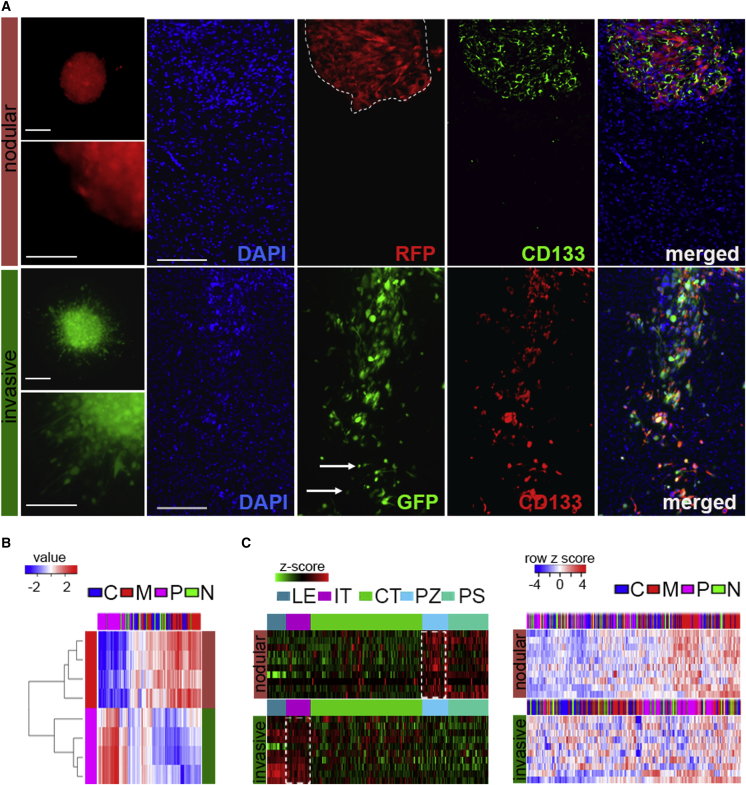
Recognizing molecular determinants which drive diverse GBM cell phenotypes would allow the identification of functional targets and provide much-needed insight into the phenotypic heterogeneity of GBM. Patient-derived GSCs revealed two major distinct subpopulations with nodular and invasive phenotypes by spheroid dispersal assay in vitro and intracranial xenografts in vivo (Figure 1A). The expression of GSC marker PROMININ 1 (CD133) showed significant upregulation in both subpopulations in vivo (Figure S1A), indicating that these cells retain a stem-like character in the brain microenvironment. The analysis of molecular and cellular function of genes deregulated in these two subpopulations showed proliferative and migratory modes of the transcriptome (Table S1 and Figure S1B). In fact, these genes' signatures, when queried with TCGA subtypes (Verhaak et al., 2010), clustered with mesenchymal or proneural subtypes (Figure 1B), indicating that these phenotypically diverse GSCs are characterized by distinct transcriptomic subtype classification. The phenotype-linked transcriptomics overlapped with tumor anatomic site, with mesenchymal-like/nodular signatures prevalent in perinecrotic zones and proneural-like/invasive signature in infiltrating areas of tumor (Figures 1C and S1C), suggesting that complex intratumoral architecture may arise from the co-existence of diverse GSCs within individual tumors.
Figure 1.
Intratumoral Architecture Is Imposed by Phenotype/Transcriptome-Diverse GSCs
(A) Patient-derived GSCs have distinct phenotypes in vitro and in vivo. Representative micrographs of GSC spheroids (left, n = 10 independent GSCs; scale bars, 100 μm and 50 μm) and GSC-derived intracranial tumors with CD133 immunostaining (right, n = 6 independent GSCs; scale bar, 150 μm). Nodular tumor and infiltrating tumor cells are indicated by the dashed line and arrows, respectively.
(B) Signature of genes with proproliferative or proinvasive function classifies GSC subpopulations. Gene expression (n = 8 independent GSCs, n = 4 per subclass) in selected categories of GSCs was queried with a gene signature retrieved from the TCGA GBM dataset, and identified by clustering as expression correlation analysis. C, classical; M, mesenchymal; P, proneural; N, neural.
(C) The phenotype-determining transcriptome overlap with tumor anatomic site-specific expression. The top ten genes in each category (n = 8 independent GSCs, n = 4 per subclass) (proproliferative or proinvasive) were queried with Ivy GAP database-based expression signature in different anatomic areas of GBM (left; LE, leading edge; IT, infiltrating tumor; CT, cellular tumor; PZ, perinecrotic zone; PS, pseudopalisading cells), or a gene signature retrieved from the TCGA GBM dataset and identified by clustering with subtype prediction (right; C, classical; M, mesenchymal; P, proneural; N, neural).
See also Figure S1.
GBM Subtypes Are Characterized by Highly Heterogeneous MicroRNA Profiles
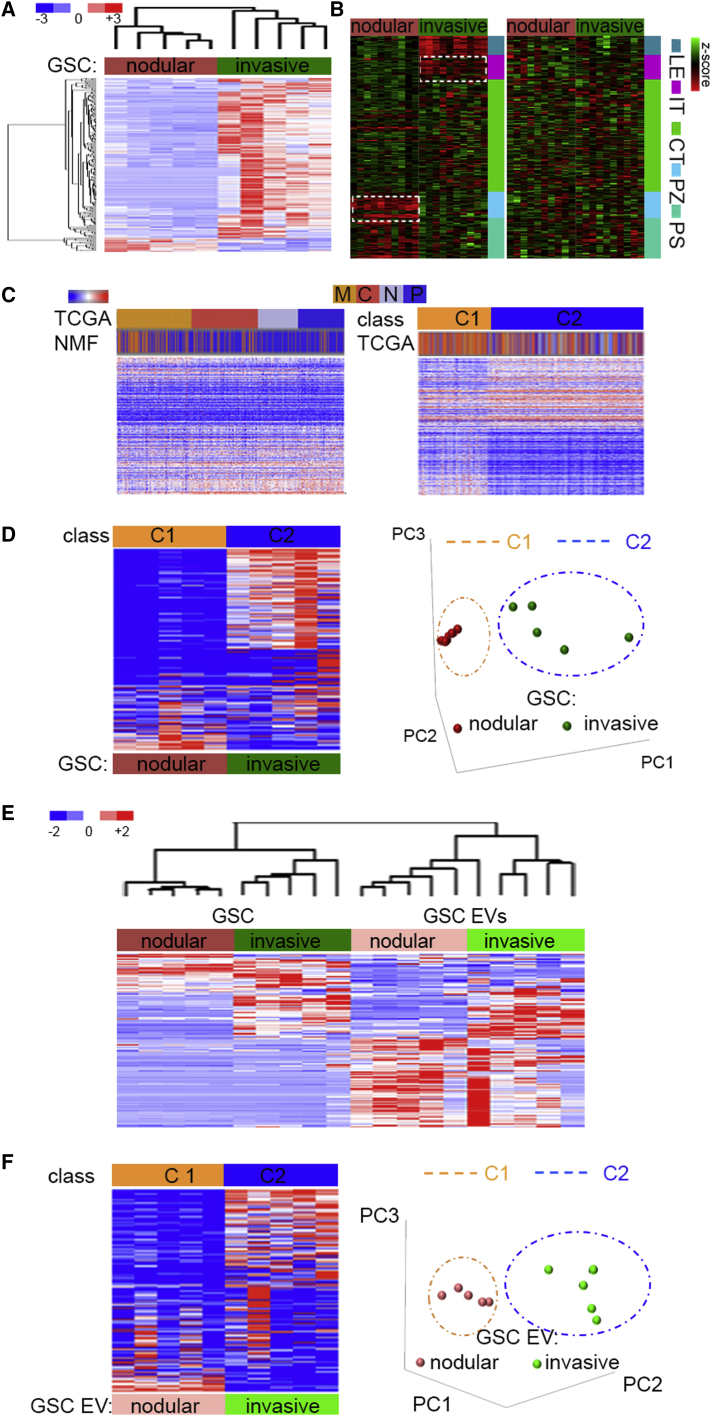
Global analysis of microRNA expression showed that the transcriptional subtype diversity observed in nodular and invasive GSCs is reflected by microRNA signatures (Figures 2A and S2A). The comprehensive analysis of subtype-specific microRNAs and their mRNA targets showed that the downregulated microRNAs in each GSC subpopulation negatively correlated with expression of their targets in a tumor anatomic site-dependent manner (Figure 2B [left panels] and Table S2). However, targets of highly expressed microRNA in these two GSC subpopulations did not show an anatomic site-dependent pattern (Figure 2B, right panels). The analysis of microRNA expression in GBM TCGA subtypes identified by protein-coding gene signatures revealed a highly heterogeneous pattern with no apparent clustering (Figures 2C [left] and S2B). However, unsupervised analysis of the GBM microRNA expression profiles by non-negative matrix factorization (NMF) (Brunet et al., 2004) identified two major clusters (C1 and C2) (Figure 2C, right; Figures S2C and S2D), which largely aligned with the molecular classes previously determined by the gene expression analyses (Figure 2D). C1 class (nodular) was significantly enriched in mesenchymal GBM tumors, whereas C2 class (invasive) exhibited enrichment in proneural and neural molecular groups (Figure S2E). The distinct profile of microRNA expression identified in GSC subpopulations in vitro, taken together with the observed negative correlation of microRNAs and their targets' expression in vivo, suggested tumor microenvironment-dependent regulation of microRNA expression and targeting.
Figure 2.
GBM Subtypes Are Characterized by Highly Heterogeneous MicroRNA Profiles
(A) MicroRNA profile distinguishes nodular and invasive GSCs. MicroRNA sets that vary coherently between GSCs (n = 10 independent GSCs, n = 5 per subclass) were identified by supervised clustering (fold >2, p < 0.05).
(B) MicroRNAs downregulated in GSC subpopulations (n = 10 independent GSCs, n = 5 per subclass) lack correlation with tumor anatomic site expression of their targets. IPA-based analysis of selected microRNA/mRNA target expression showed negative correlation (low microRNA/high target expression, left panels), and lack of correlation (high microRNA/low target expression, right panels). The top ten genes in each category were queried with Ivy GAP database-based expression signature in different areas of GBM (LE, leading edge; IT, infiltrating tumor; CT, cellular tumor; PZ, perinecrotic zone; PS, pseudopalisading cells). White dashed box indicates genes upregulated in PZ and LE zones.
(C) TCGA-classified GBM subtypes are characterized by highly heterogeneous profiles of microRNA expression. Hierarchical clustering of expression of 534 microRNAs in a core set of TCGA GBM samples (n = 173 patient samples) in supervised analysis (TCGA subtypes, top cluster) compared with unsupervised analysis (using NMF, bottom cluster) (left); and in supervised analysis (NMF-based classification into two classes C1 and C2, top cluster) versus unsupervised analysis (TCGA subtypes, bottom cluster) (right; C, classical; M, mesenchymal; P, proneural; N, neural; 200 microRNAs, false discovery rate [FDR] < 0.05).
(D) GSC microRNA expression data reveals two profiles in TCGA-classified GBM subtypes. Hierarchical clustering (left) and principal component analysis (right) of microRNAs using 396 predictive microRNAs expressed in GSC subpopulations (n = 10 independent GSCs, n = 5 per subclass) and ordered based on gene subtype predictions using the core set of TCGA GBM samples (n = 173 patient samples) (NMF1 versus NMF2; 200 microRNAs, FDR < 0.05).
(E) GSCs and GSC EV microRNA profiles separate cells and EVs. Hierarchical clustering of expression of 692 microRNAs in GSCs (n = 10 independent GSCs, n = 5 per subclass) and GSC EVs (n = 10 EVs from independent GSCs, n = 5 per subclass) in unsupervised analysis.
(F) GSC EV microRNA expression data reveal two profiles in TCGA-classified GBM subtypes. Hierarchical clustering (left) and principal component analysis (right) of EV microRNAs using the curated list of 298 predictive microRNAs secreted in EVs released by distinct GSC subpopulations (n = 10 independent GSCs, n = 5 per subclass) and ordered based on gene subtype predictions using the core set of 173 TCGA GBM samples (n = 173 patient samples). FDR < 0.05.
See also Figure S2.
To validate whether the observed diversity of GSC cellular microRNAs was recapitulated by the microRNA composition of EVs released by these GSCs, we isolated, quantified, and characterized EVs. Interestingly, significant heterogeneity of EVs released by these two GSC subpopulations was observed regarding their size, shape, and expression of EV marker CD 63 molecule (CD63), but not the total number of particles or their RNA content (Figures S2F–S2J). Comprehensive analysis of cellular and EV microRNA in nodular and invasive GSCs showed that the microRNA signature separated GSCs and GSC EVs (Figures 2E, S2K, and S2L). The fact that cellular microRNA from two subpopulations of GSCs clustered together rather than with their own EV microRNA indicated that GSC EV microRNA profiles only partially mimic cellular microRNA expression, with sets of microRNAs enriched/depleted in EVs, and that subpopulation-specific EV microRNA signatures also exist (Figures S2M and S2N). Similar to GSC microRNA, GSC EV microRNA significantly separated into two classes which recapitulated the same classification as cellular microRNAs, despite the fact that different sets of microRNAs were identified in cells and EVs (Figure 2F). We thus hypothesized that in a setting as diverse as GBM, microenvironment-driven signaling may mediate dynamic transitions within tumor anatomic niches.
GSC EVs Support a Subpopulation-Specific Invasive Phenotype
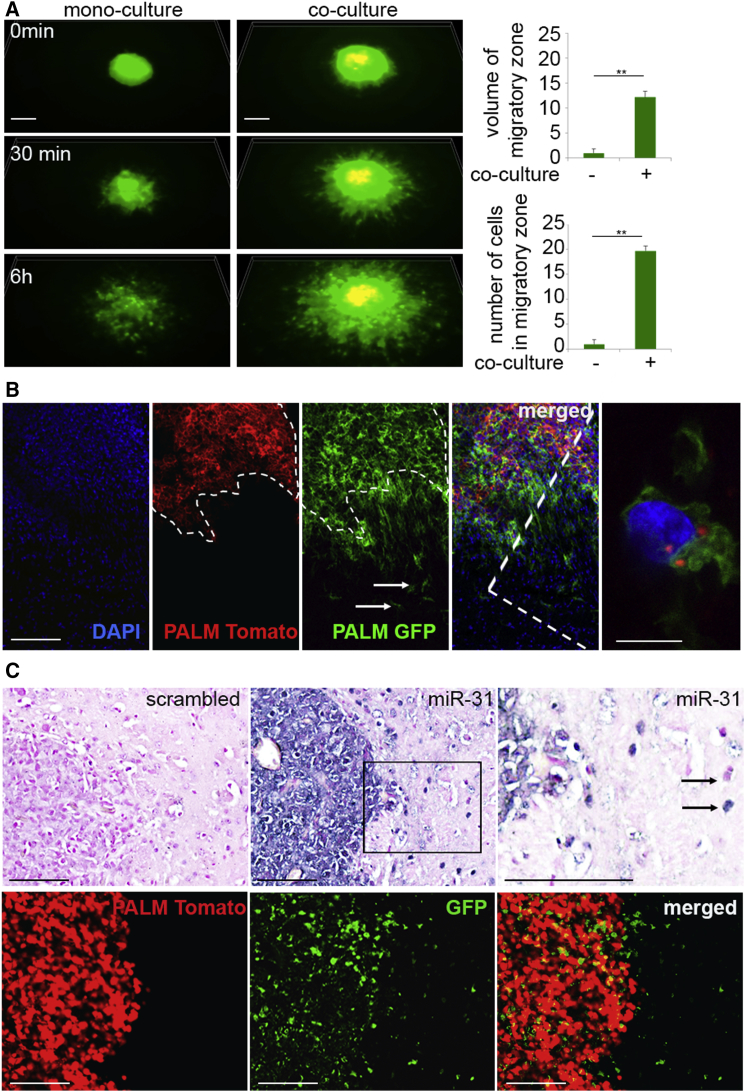
The treatment of invasive GSCs with EVs derived from nodular GSCs enhanced their migration in vitro, suggesting that EV exchange may contribute to cellular phenotype (Figure S3A and Movie S1). Thus, a heterogeneous sphere migration assay was performed to mimic co-existence of these subtypes within the tumor. Significantly, invasive cells migrated farther away from the spheroid core in the presence of other cells, either other invasive cells (Figure S3B) or, even more profoundly, in the presence of nodular cells (Figures 3A and S3C; Movie S2). Importantly, nodular cells remained within the spheroid regardless of co-culture variants. This phenotype was also recapitulated in vivo in a heterogeneous GSC intracranial xenograft model, showing subpopulation-specific invasive and nodular behavior (Figure 3B, left). As a proof of concept, we selected GBM-high microRNA-31 (miR-31), which was enriched in donor nodular GSCs and their EVs (Figures S2M and S2N). Frequent exchange of EVs and EV microRNAs and their spread away from the cell of origin suggested both the existence of molecular hybrids and propagation of heterogeneity across the tumor (Figures 3B [right], 3C, and S3D). Together, these data suggested that EV exchange did not cause a phenotypic switch but rather facilitated formation of interdependent tumor organization. The analysis of relation between expression of microRNAs from selected classes (C1 and C2) and genes expressed differentially in tumor anatomic niches (perinecrotic/core zone versus infiltration/invasive zone) revealed a positive association in GBM tissue (Figures S3E and S3F). Thus, EV-mediated transfer of bioactive molecules leads to increased heterogeneity, allowing a more robust response to microenvironmental challenges and leading to increased cell survival (Ricklefs et al., 2016).
Figure 3.
GSC EVs Support Subpopulation-Specific Invasive Phenotype
(A) Heterogeneity of GSC spheroids promotes migration of invasive GSCs. Representative micrographs of GSC spheroids in monoculture (n = 3 independent experiments per three independent GSCs) (left) and heterogeneous co-culture (n = 3 independent experiments per three independent GSCs) (middle) in 3D time-lapse frames are shown. GFP-labeled invasive GSCs and PALM-Tomato-labeled nodular GSCs. Scale bar, 100 μm. Relative quantification of migratory zone volume (top right) and number of cells migrated out of spheroid core (bottom right) in mono- versus co-culture. ∗∗p < 0.01.
(B) Nodular and invasive phenotype of GSCs is recapitulated in vivo. Representative micrographs of co-implanted heterogeneous tumors (n = 6 independent experiments) are shown. GFP-labeled invasive GSCs and PALM-Tomato-labeled nodular GSCs. Nodular tumor burden and infiltrating tumor cells are indicated by dashed line and arrows, respectively. Intratumoral EV transfer between cells is shown on high-power magnification inset. Scale bars, 150 μm and 10 μm.
(C) GSC EV is transferred intratumorally. Representative micrographs of co-implanted heterogeneous tumors (n = 3 independent experiments) are shown. Scrambled or microRNA ISH (nodular specific miR-31) (top) and GFP-labeled invasive GSCs and PALM-Tomato-labeled nodular GSCs (bottom) from consecutive sections. Positive and negative microRNA detection is indicated by arrows. Scale bars, 100 μm.
See also Figure S3.
Transfer of EV-Encapsulated MicroRNAs Propagates GBM Heterogeneity
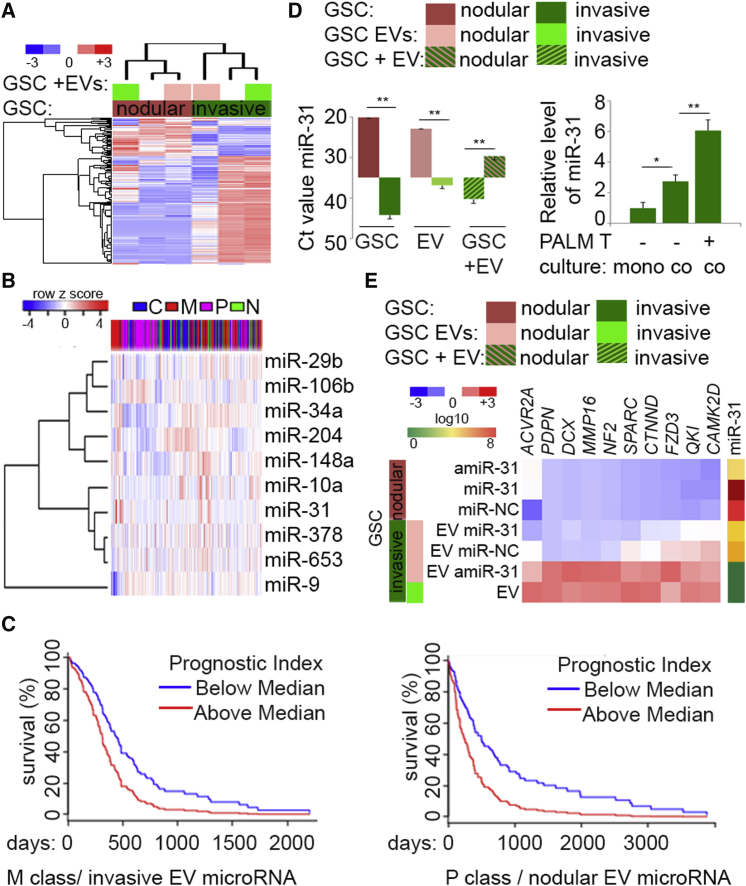
EVs are complex structures with cargo composed of multiple classes of molecules; thus, their uptake may result in EV-dependent alterations much broader than those caused by microRNA alone. It is likely impossible to “tease out” microRNA from other active molecules present in EVs. Transfer of EVs between GSC subpopulations resulted in shifts of clustering with significant global deregulation of microRNAs in each GSC subpopulation treated with EVs derived from other subpopulations (Figure 4A). The most enriched microRNAs in EV-treated cells (Figure S4A) showed a diverse pattern of expression in GBM TCGA subtypes (Figure 4B). However, EV microRNAs from invasive GSCs were associated with a significantly worse outcome in mesenchymal tumors. Conversely, EV microRNAs from nodular GSCs were associated with a significantly worse outcome in proneural but not mesenchymal tumor (Figures 4C and S4B). Nodular GSC- and EV-specific miR-31 (Figures S2M and S2N) was enriched in recipient invasive GSC upon exposure to nodular GSC EVs (Figures 4D [right], S4C, and S4D), or after sorting from heterogeneous spheroids (Figure 4D, left), recapitulating exchange observed in vivo (Figure 3C). Interestingly, putative miR-31 targets were differentially expressed in both GSC subpopulations, indicating that different sets of targets exist in each subgroup of GSCs (Figure S4E). In fact, invasive GSC-specific miR-31 targets (Wong et al., 2015) were significantly downregulated upon treatment with EVs from nodular GSCs, but deregulation of miR-31 by transfection with an miR-31 mimic or inhibitor in donor nodular GSCs had no significant effect on their uniformly low expression (Figure 4E). The analysis of transcriptome of invasive GSCs treated with EVs derived from nodular GSC expressing either control or miR-31 inhibitor showed potent gene rearrangement with significant impact on miR-31 targets (Figure S4F). However, these targets did not show a tumor anatomic niche-specific pattern (Figure S4G), indicated that both cellular and EV microRNAs have a cell-specific function, targeting effectors existing exclusively in particular GSC subpopulations.
Figure 4.
Transfer of EV-Encapsulated MicroRNAs Propagates GBM Heterogeneity
(A) Exchange of EV between distinct GSCs shifts subpopulation-specific microRNA signatures. Unsupervised hierarchical clustering of expression of 307 microRNAs in non-treated GSCs (n = 4 independent GSCs, n = 2 per subclass) and EV-treated GSCs (n = 4 independent EVs, n = 2 per subclass) is shown.
(B) MicroRNAs upregulated upon EV uptake are diversely expressed in GBM. MicroRNA sets that are coherently upregulated in nodular and invasive GSCs (n = 4 independent GSCs, n = 2 per subclass) upon treatment with EVs (p < 0.05, fold >2) were queried with TCGA-classified GBM dataset and identified by clustering with subtype prediction. C, classical; M, mesenchymal; P, proneural; N, neural.
(C) Survival analysis in mesenchymal (left) and proneural (right) GBM subtypes based on the impact of the prognostic index of multiple microRNAs (miR-148a, miR-204, miR-34a, miR-106b, and miR-9 [left], and miR-31, miR-653, miR-378a, miR-29b, and miR-10a [right]) based on retrospective data extrapolated from the TCGA. For mesenchymal GBM (n = 125 patient samples), log-rank p = 0.004, Prognostic Index hazard ratio = 1.83, p = 0.004. For proneural GBM (n = 112 patient samples) log-rank p = 0.001, Prognostic Index hazard ratio = 2.09, p = 0.001.
(D) MiR-31 is EV-transferred between GSC subpopulations. Left: qPCR analysis of miR-31 in donor nodular GSCs (n = 3 independent GSCs), their EVs (n = 3 independent GSC EVs), and recipient invasive GSCs (n = 3 independent GSCs). Right: monoculture spheroid of GFP-tagged invasive GSC (mono-) and co-culture spheroids of GFP-tagged invasive GSCs and PALM-Tomato (PALM T) nodular GSCs were sorted for GFP-positive (co-culture negative) or double-positive (GFP and Tomato [co-culture positive]). Data (n = 3 independent experiments) are shown as the mean raw Ct value ± SD, ∗∗p < 0.01 (left); and as mean ± SD, ∗p < 0.05, ∗∗p < 0.01 (right).
(E) EV-miR-31 targets subclass GSC-specific genes. Nodular GSCs (n = 3 independent GSCs) were transfected with control (NC), microRNA mimic (miR-31), and microRNA inhibitor (amiR-31) (top three rows), and invasive GSC were treated with EVs derived from such nodular GSCs or their own EVs (n = 3 independent GSCs). qPCR analysis of selected targets and miR-31 is shown as hierarchical clustering and log10 assessed based on the value of expression, respectively.
See also Figure S4.
Discussion
Our data indicated that phenotype-linked transcriptomics of GSCs overlapped with tumor anatomic site, with mesenchymal-like/nodular signatures prevalent in perinecrotic zones and proneural-like/invasive signature in infiltrating areas of tumor, suggesting that these GSCs both shape and adapt to microenvironmental conditions, and that complex intratumoral architecture likely arises from the co-existence of diverse GSCs within individual tumors (Patel et al., 2014).
The distinct microRNA profile identified in GSC subpopulations in vitro, and the negative correlation of microRNAs and their targets' expression in vivo, suggested tumor microenvironment-dependent regulation of microRNA expression and targeting. The concordance between gene and microRNA unsupervised expression analyses suggested that distinction between mesenchymal and proneural molecular subclasses might be at least partially driven by the microRNA expression signatures.
Our data strongly implicated that EV-mediated transfer of bioactive molecules leads to increased heterogeneity not due to passive transfer but via cell-specific targeting, as both cellular and EV microRNAs have a cell-specific function, targeting effectors existing exclusively in particular GSC subpopulations. EV-microRNA transfer between different subpopulations of tumor cells should be thus recognized as an important aspect of tumor intricacy that may propagate heterogeneity of GBM; thus, EV-microRNA secretion and uptake may be an additional trait of cellular adaptation into different anatomic niches. Recent evidence suggests that the microRNA repertoire in EVs only partially mirrors that of cellular microRNA and, in fact, its specific pattern may be surprisingly different from that of secreting cells (Koppers-Lalic et al., 2014, Skog et al., 2008). Our data strongly support the existence of an active mechanism of microRNA loading or, rather, the co-existence of diverse mechanisms, as global, non-random distribution of microRNA was detected in subclasses of GSC EVs. The complexity of solid tumors, including GBM, and their distinct pathophysiology relies on anatomic niches that transmit and receive signals through cellular and acellular mediators (Jones and Wagers, 2008). These components are highly reliant on each other and undergo constant architectural, phenotypic, and transcriptomic rearrangements depending on fluctuating microenvironmental contexts as the disease progresses. The brain tumor “ecosystem” is composed of distinct phenotypic and transcriptomic cell components, and our analysis of cellular and EV microRNA load discovered additional aspects of intratumoral diversity. EVs/microRNA as transcriptome and signaling communication tandem modulators arrange both molecular and phenotypic traits. We thus argue that observed highly heterogeneous profiles of microRNA expression in GBM and the co-existence of diverse subtypes and hybrid-stage cells within individual tumors is propagated by intratumoral exchange of microRNA.
Experimental Procedures
Human Specimens and Primary Cells
Tumor samples were obtained as approved by The Harvard Medical School (HMS) Institutional Review Board. Surgery was performed by E.A.C. and I.N. GSCs were obtained by dissociation of tumor samples and cultivated in stem cell-enriching conditions. The unique identity of cultured patient-derived cells (n = 12) was confirmed by short tandem repeat analysis (Kim et al., 2016).
Purification of EVs
The conditioned media were collected, and EVs were isolated by differential centrifugation and analyzed using a NanoSight.
MicroRNA/mRNA Expression Analysis
Nanostring microRNA technology was used to search for unique microRNA signatures in GSC and GSC EVs. Whole human genome oligo microarray was performed by Arraystar. The SBI's Exo-NGS service was used to build the Illumina NGS libraries followed sequencing using a 1 × 50-bp single-end Illumina HiSeq NGS and Maverix Analytic Platform.
In Vitro Assays
Nodular GSCs were labeled with either RFP or GFP (using the lentiviral pCDH vector), or PALM-Tomato (using the lentiviral CSCW2 vector [Lai et al., 2015]); and invasive GSCs were labeled with GFP or PALM-GFP.
For EV transfer, 3 × 105 GSCs/mL were maintained overnight in unsupplemented medium, followed by 24 hr of treatment with EVs.
For 3D spheroid dispersal assay, GSCs were dissociated to single cells using Accutase (Life Technologies), and plated at 200 (noninvasive GSCs) or 1,000 (invasive GSCs) cells/well in a 96-well plate for 48 hr. Spheroids were then transferred into collagen with unsupplemented medium or medium supplemented with EVs and analyzed after 0–6 hr or in time-lapse.
For co-culture assays, single-cell suspensions of co-culture (at ratio 1:4) were cultured for 48 hr before assay. Monocultured invasive GSC spheroids served as controls for co-cultured invasive/nodular spheroids followed by sorting for populations of pure PALM-Tomato (nodular), GFP (invasive), and double-positive (invasive with nodular PALM-Tomato EVs).
In Vivo Studies
Female athymic mice were purchased from Envigo. Mice were housed in the HMS animal facility in accordance with NIH regulations. Protocols were approved by the HMS Institutional Animal Care and Use Committee. Intracranial tumor injection was performed as described by Ricklefs et al. (2016). For GSC implantation and co-implantation experiments, either 1 × 103 nodular GSCs or 5 × 105 invasive GSCs, or both combined were used.
Data Analysis
Functional bioinformatics analyses were performed using Qiagen's Ingenuity Pathway Analysis (IPA; www.qiagen.com/ingenuity). Experimental and clinical data were analyzed using the GBM-BioDP (Celiku et al., 2014). Clinical data were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/). Gene expression in the various anatomical regions of glioblastoma was analyzed using the Ivy Glioblastoma Atlas Project (http://glioblastoma.alleninstitute.org/).
Level 3 microRNA expression data (unc.edu_GBM.H-miRNA_8 × 15K.Level_3.1.8.0) from 479 glioblastomas were obtained from TCGA (Brennan et al., 2013). An unpaired, two-tailed t test was used to compare two groups. One-way ANOVA, followed by Bonferroni's test, was conducted to test for significance among multiple groups. Fisher’s exact test was used to identify associations between unsupervised microRNA classes and previously determined GBM subtypes. p < 0.05 was considered significant.
Author Contributions
All authors agree to be accountable for all aspects of the work.R.F.-L. analyzed TCGA data. Y.S.T., A.K.R., and F.R. performed in vitro and in vivo assays. M.M. characterized EV cargo. M.O.N. performed microscope analysis. E.S., H.L., R.W., and R.B. assisted with writing the manuscript and interpretation of data. I.N. acquired patients’ specimens. E.A.C. acquired patients’ specimens and assisted with writing the manuscript. J.G. and A.B. conceived and designed the overall work, analyzed and interpreted data, and wrote the manuscript.
Acknowledgments
This research was supported by NCI P01 CA069246, NCI P01 CA069246-20 (both to E.A.C.), and NCI 1R01 CA176203-01A1 (to J.G.).
Published: May 18, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, two tables, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.04.024.
Contributor Information
Jakub Godlewski, Email: jgodlewski@bwh.harvard.edu.
Agnieszka Bronisz, Email: abronisz@bwh.harvard.edu.
Accession Numbers
The accession number for the gene microarray reported in this paper is GEO: GSE89501.
Supplemental Information
References
- Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronisz A., Wang Y., Nowicki M.O., Peruzzi P., Ansari K.I., Ogawa D., Balaj L., De Rienzo G., Mineo M., Nakano I. Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res. 2014;74:738–750. doi: 10.1158/0008-5472.CAN-13-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet J.P., Tamayo P., Golub T.R., Mesirov J.P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celiku O., Johnson S., Zhao S., Camphausen K., Shankavaram U. Visualizing molecular profiles of glioblastoma with GBM-BioDP. PLoS One. 2014;9:e101239. doi: 10.1371/journal.pone.0101239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Fei T., Verhaak R.G., Su Z., Zhang Y., Brown M., Chen Y., Liu X.S. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski J., Krichevsky A.M., Johnson M.D., Chiocca E.A., Bronisz A. Belonging to a network—microRNAs, extracellular vesicles, and the glioblastoma microenvironment. Neuro Oncol. 2015;17:652–662. doi: 10.1093/neuonc/nou292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.R., O'Neill B.P. Glioblastoma survival in the United States before and during the temozolomide era. J. Neuro Oncol. 2012;107:359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- Jones D.L., Wagers A.J. No place like home: anatomy and function of the stem cell niche. Nat. Rev. Mol. Cell Biol. 2008;9:11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Ezhilarasan R., Phillips E., Gallego-Perez D., Sparks A., Taylor D., Ladner K., Furuta T., Sabit H., Chhipa R. Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-kappaB-dependent manner. Cancer Cell. 2016;29:201–213. doi: 10.1016/j.ccell.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers-Lalic D., Hackenberg M., Bijnsdorp I.V., van Eijndhoven M.A., Sadek P., Sie D., Zini N., Middeldorp J.M., Ylstra B. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- Lai C.P., Kim E.Y., Badr C.E., Weissleder R., Mempel T.R., Tannous B.A., Breakefield X.O. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 2015;6:7029. doi: 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P., Joshi K., Li J., Kim S.H., Li P., Santana-Santos L., Luthra S., Chandran U.R., Benos P.V., Smith L. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl. Acad. Sci. USA. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo M., Ricklefs F., Rooj A.K., Lyons S.M., Ivanov P., Ansari K.I., Nakano I., Chiocca E.A., Godlewski J., Bronisz A. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep. 2016;15:2500–2509. doi: 10.1016/j.celrep.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Ricklefs F., Mineo M., Rooj A.K., Nakano I., Charest A., Weissleder R., Breakefield X.O., Chiocca E.A., Godlewski J., Bronisz A. Extracellular vesicles from high-grade glioma exchange diverse pro-oncogenic signals that maintain intratumoral heterogeneity. Cancer Res. 2016;76:2876–2881. doi: 10.1158/0008-5472.CAN-15-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberg D.L., Lubelski D., Miller T.E., Rich J.N. Brain tumor stem cells: molecular characteristics and their impact on therapy. Mol. Aspects Med. 2014;39:82–101. doi: 10.1016/j.mam.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J., Wurdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.K., Fatimy R.E., Onodera C., Wei Z., Yi M., Mohan A., Gowrisankaran S., Karmali P., Marcusson E., Wakimoto H. The cancer genome atlas analysis predicts MicroRNA for targeting cancer growth and vascularization in glioblastoma. Mol. Ther. 2015;23:1234–1247. doi: 10.1038/mt.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.