Summary
Microglia, the immune cells of the brain, are crucial to proper development and maintenance of the CNS, and their involvement in numerous neurological disorders is increasingly being recognized. To improve our understanding of human microglial biology, we devised a chemically defined protocol to generate human microglia from pluripotent stem cells. Myeloid progenitors expressing CD14/CX3CR1 were generated within 30 days of differentiation from both embryonic and induced pluripotent stem cells (iPSCs). Further differentiation of the progenitors resulted in ramified microglia with highly motile processes, expressing typical microglial markers. Analyses of gene expression and cytokine release showed close similarities between iPSC-derived (iPSC-MG) and human primary microglia as well as clear distinctions from macrophages. iPSC-MG were able to phagocytose and responded to ADP by producing intracellular Ca2+ transients, whereas macrophages lacked such response. The differentiation protocol was highly reproducible across several pluripotent stem cell lines.
Keywords: human pluripotent stem cells, microglial differentiation, human microglia
Highlights
-
•
Efficient protocol to generate human microglia from PSCs
-
•
iPSC-derived microglia have ramified morphology and motile processes
-
•
Expression and cytokine profiles of iPSC-derived microglia resemble primary microglia
-
•
iPSC-derived microglia can phagocytose beads and respond to ADP
Douvaras and colleagues generated microglia from pluripotent stem cells using a chemically defined protocol through a myeloid progenitor. iPSC-derived microglia showed highly motile processes, were able to phagocytose and responded to ADP with calcium transients. Microglial identity was further confirmed by gene expression analysis comparing multiple iPSC-derived microglia samples with primary microglia and peripheral blood or other tissue-specific macrophages.
Introduction
Microglia are resident, tissue-specific macrophages that perform several critical roles in development and maintenance of the CNS (Hanisch and Kettenmann, 2007). They arise from primitive CD45+CX3CR1− myeloid progenitors in the yolk sac that differentiate to CD45+CX3CR1+ microglial progenitors and invade the developing brain before the emergence of definitive hematopoiesis (Ginhoux et al., 2010, Kierdorf et al., 2013, Schulz et al., 2012). In the healthy adult brain with an intact blood-brain barrier, microglia persist as a self-sustained population that is not replenished by circulating bone marrow-derived cells (Ajami et al., 2007, Ginhoux et al., 2010). “Resting” microglia are highly active as their processes continuously examine the entire brain for homeostatic disruptions (Nimmerjahn et al., 2005).
Microglia phagocytose pathogens and cell debris and remove toxic molecules and protein deposits, thus attenuating inflammation and promoting tissue regeneration and repair (Fenn et al., 2012, Napoli and Neumann, 2009). During development, microglia promote migration and differentiation of neural progenitors, neurogenesis, and oligodendrogenesis, and regulate synaptogenesis and synaptic plasticity through pruning (Aarum et al., 2003, Paolicelli et al., 2011, Ueno et al., 2013). Microglia can also contribute to pathological brain inflammation and disruption of the blood-brain barrier by releasing cytokines and neurotoxic molecules (Colton and Gilbert, 1987, Luheshi et al., 2011). Dysfunctional microglia have been linked to amyotrophic lateral sclerosis and Alzheimer's disease. Chronic activation of microglial cells is a possible trigger to the progression of multiple sclerosis and Parkinson's disease (Kierdorf et al., 2013), and defective phagocytosis and synaptic pruning have been implicated in schizophrenia and autism spectrum disorders (Ransohoff, 2016).
Although most of our knowledge regarding microglia derives from rodent studies, there are major interspecies differences, such as proliferation rate, adhesive properties, and expression of critical receptors (Smith and Dragunow, 2014). The analysis of primary human microglia is severely limited by tissue availability, especially from healthy individuals.
We developed a robust, reproducible protocol that generates microglia from human pluripotent stem cells (PSCs) in chemically defined conditions, by mimicking embryonic development. Human induced PSC (iPSC)-derived microglia (iPSC-MG) are similar by morphology, gene expression, and cytokine release profile to human primary microglia and are distinct from other tissue macrophages. Furthermore, they are able to phagocytose and generate intracellular Ca2+ transients in response to ADP.
Results
Differentiation of Human PSCs to Myeloid Progenitors
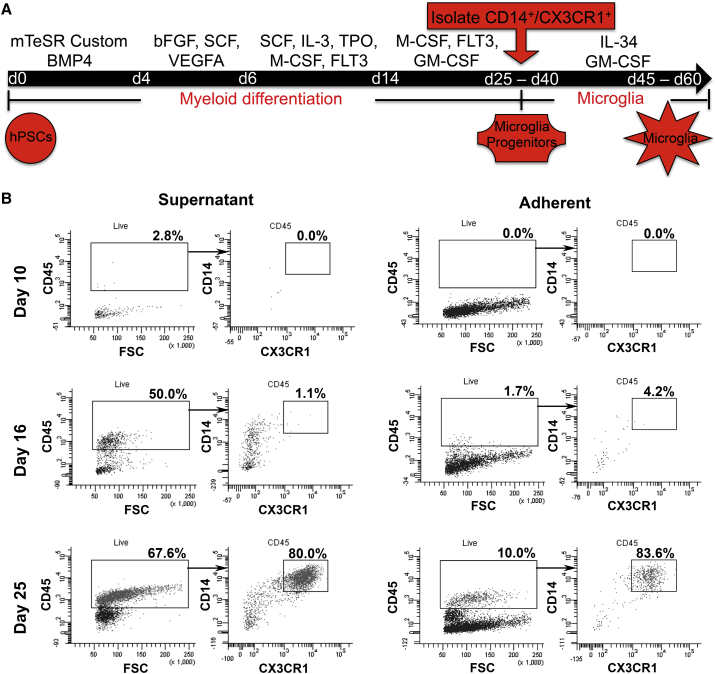
As microglial cells arise from myeloid progenitors in the yolk sac during development, we established a serum- and feeder-free protocol to differentiate human PSCs toward the myeloid lineage (Figure 1A). Building upon previous studies (Yanagimachi et al., 2013), we induced primitive streak-like cells through bone morphogenetic protein 4 (BMP4) signaling to obtain KDR+CD235a+ primitive hemangioblasts (Sturgeon et al., 2014) (Figure S1A). CD45+CX3CR1− microglial progenitors appeared in the supernatant by day 16, while CX3CR1 was upregulated between days 20 and 25. In contrast, the adherent population contained only a small fraction of CD45+CX3CR1+ progenitors. Interestingly, CD14 was upregulated in a subset of CD45+ cells around day 16, before the appearance of CX3CR1 (Figure 1B). Between days 25 and 50, 82% ± 5% of the CD14+ cells co-expressed CX3CR1. The protocol's efficiency to generate microglial progenitors, based on CD14 expression, was 68% ± 4% across 16 lines (Figure S1D). Microglial progenitors were continuously generated in the supernatant for up to 1 month and were isolated every week with an average yield of 224 ± 42 × 103 cells per isolation, per 100 × 103 PSCs plated. Microglial progenitors were isolated either via fluorescence-activated cell sorting (FACS) (Figures S1B and S1C) or magnetic bead separation for further differentiation or long-term storage. Thawed progenitors retained their differentiation capacity, with a post-thaw viability of 57% ± 5%.
Figure 1.
PSC Differentiate to Microglia through Myeloid Progenitors
(A) Diagram depicting the major steps of the microglial differentiation protocol.
(B) Kinetics of CD45, CX3CR1, and CD14 expression between day 10 and day 25 in the adherent and supernatant fractions.
See also Figure S1.
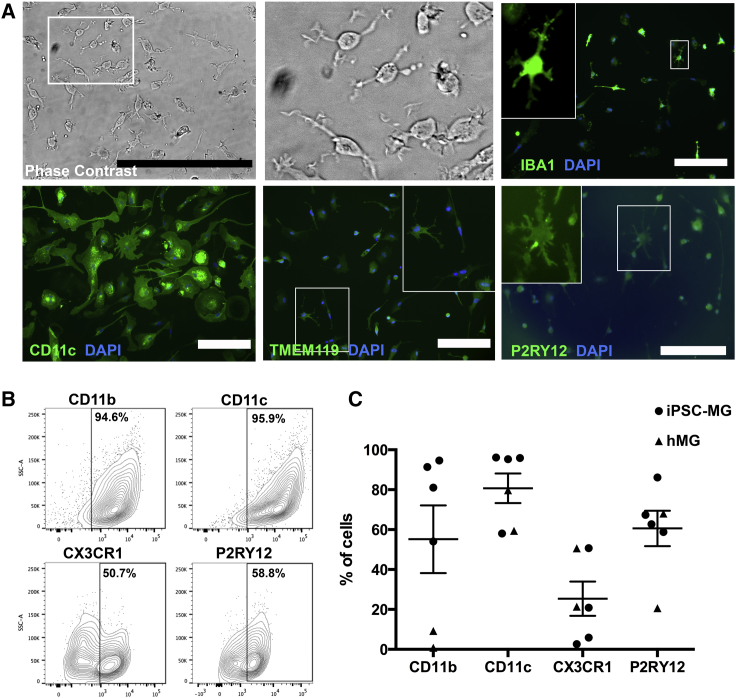
iPSC-MG Express Typical Microglial Markers and Show Highly Motile Processes
Interleukin-34 (IL-34) and granulocyte macrophage colony-stimulating factor (GM-CSF) stimulation (Ohgidani et al., 2014) of plated microglial progenitors resulted in iPSC-MG, extending highly motile processes, constantly scanning the microenvironment (Movie S1), similarly to microglia in vivo (Davalos et al., 2005, Nimmerjahn, 2012). iPSC-MG expressed known markers such as IBA1, CD11c, TMEM119, P2RY12, CD11b, and CX3CR1 (Figures 2A–2C). Fetal human primary microglia (hMG) were used for comparison (Figure S2).
Figure 2.
Characterization of iPSC-MG
(A) Panel of representative images of iPSC-MG in phase contrast and after immunofluorescent labeling for IBA1, CD11c, TMEM119, and P2RY12. White boxes indicate the areas of the magnified insets. Scale bars, 50 μm (phase contrast) and 200 μm (all other images).
(B) Representative flow-cytometry plots for typical microglial surface antigens in iPSC-MG.
(C) Dot plot showing the percentage of total cells expressing the microglial surface antigens shown in (B) across four independent iPSC-MG (depicted as circles) and two hMG samples (depicted as triangles). Error bars denote mean ± SEM.
See also Movie S1 and Figure S2 for characterization of hMG.
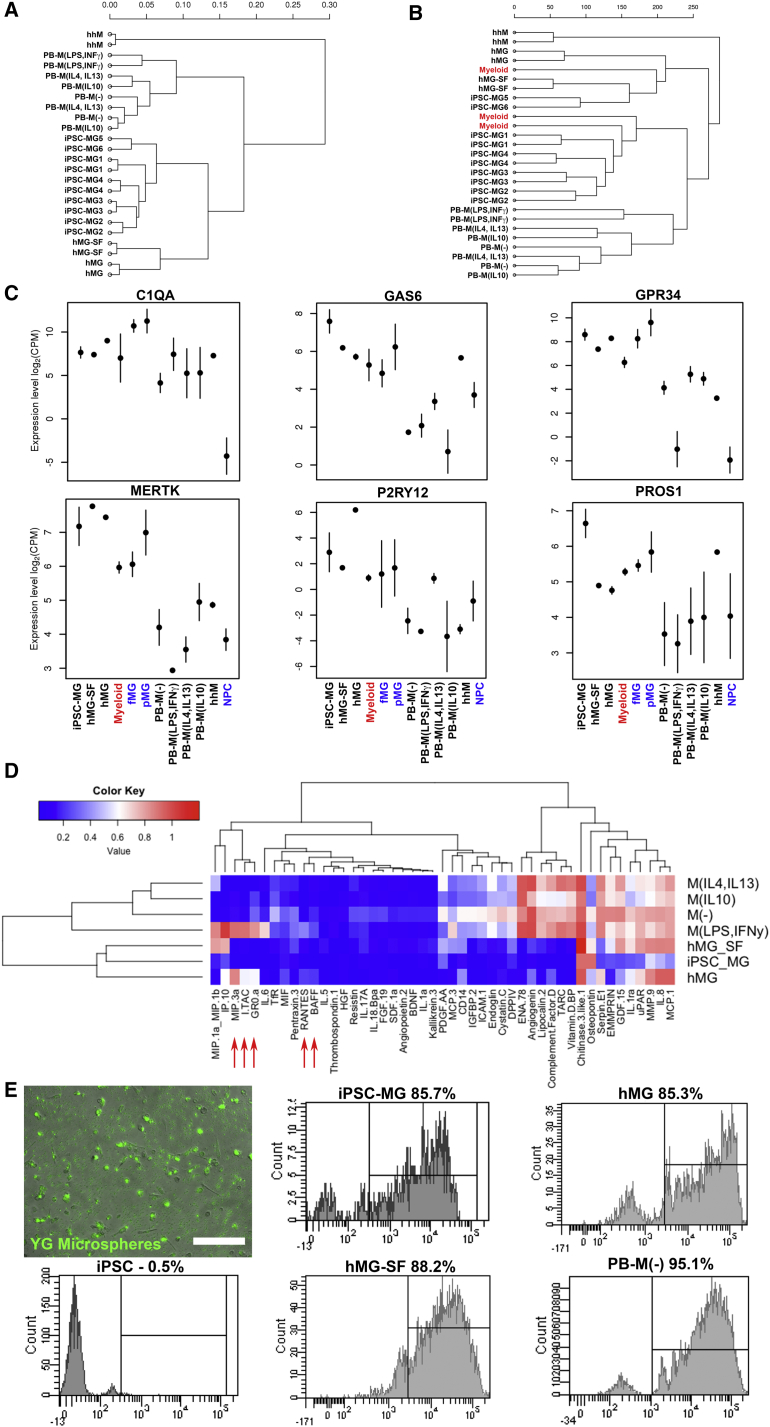
iPSC-MG Resemble Human Primary Microglia by Gene Expression
To further support the microglial identity of iPSC-MG, we performed whole-transcriptome analysis with next-generation deep RNA sequencing (RNA-seq). iPSC-MG from six unrelated healthy donors were compared with peripheral blood-derived macrophages (PB-M(−)) or polarized to M(lipopolysaccharide [LPS], interferon γ [IFNγ]), M(IL-4,IL-13), and M(IL-10), primary human hepatic macrophages (hhM), and primary human microglia cultured in serum-containing medium supplied by the provider (hMG) or in our serum-free medium (hMG-SF). We obtained high-quality reads (mean Phred quality score >38.4) and 86.2% mapped to the human genome; 18,516 genes were considered expressed and used for analysis.
iPSC-MG, hMG, and hMG-SF clustered together in a hierarchical cluster analysis using all the expressed genes showing a high degree of similarity (Spearman's correlation coefficient 0.901–0.997) and were distinct from all macrophage subtypes (Figure 3A). When we included data obtained from an independent study (Zhang et al., 2016) that isolated primary CD45+ cells from human brain extracts (termed “Myeloid” in Figure 3B), iPSC-MG samples clustered together with the “Myeloid” samples, while PB-M created a distinct cluster and hhM clustered separately, appearing the most dissimilar.
Figure 3.
Gene Expression, Cytokine Release Profile, and Phagocytosis of Microglia and Macrophages
(A) Hierarchical clustering dendrogram of the RNA-seq data based on global mRNA expression. Sample distances were calculated from Pearson's correlation coefficient.
(B) Dendrogram showing hierarchical clustering of our RNA-seq data and data obtained from an independent study of human primary CD45+ cells in the brain (“Myeloid” samples in red, GEO: GSE73721). Analysis is based on transcriptome-wide expression.
(C) Graphs showing the expression levels of the six human microglial signature genes. Error bars are means ± SD. Colored samples correspond to data from independent studies (GEO: GSE73721 in red; GEO: GSE85839 in blue).
(D) Heatmap of the released cytokine profiles of five independent iPSC-MG runs from two lines, two independent hMG samples, and one hMG-SF sample compared with PB-M. Red arrows indicate the five proteins upregulated in hMG and M(LPS,IFNγ).
(E) Representative fluorescent image and flow-cytometry histograms showing phagocytosis of yellow-green (YG)-labeled microspheres. Scale bar, 200 μm. iPSC, undifferentiated iPSCs used as negative control.
Three recent studies (Bennett et al., 2016, Butovsky et al., 2014, Hickman et al., 2013) provided datasets with unique genes expressed in microglia from primary rodent cells. We selected genes that were identified in at least two of these studies and assessed their expression in our samples (Table S1). Of the 31 selected genes, 29 were expressed (at least 1 cpm) by hMG and 28 by iPSC-MG. Overall, their expression was comparable with the exception of P2RY13 (2.6-fold lower in iPSC-MG) and CYSLTR1 (3.8-fold lower in hMG). Moreover, LIPH had very low expression in all human cell types, while hMG showed low expression of CX3CR1, and iPSC-MG low expression of TMEM119 and LAG3. However, CX3CR1 and TMEM119 proteins were detected by FACS and immunofluorescent staining, respectively.
Comparison with hhM and PB-M(−) highlighted 11 genes that were consistently higher in both iPSC-MG and hMG (shown in red in Table S1).
Butovsky et al. proposed six genes, namely C1QA, GAS6, GPR34, MERTK, P2RY12, and PROS1, as unique signature in human primary microglia. Indeed, their high expression was consistent among all our microglial samples (iPSC-MG, hMG, hMG-SF) as well as in human microglial samples from two independent studies (Muffat et al., 2016, Zhang et al., 2016) (Figure 3C), whereas macrophages and neural progenitor cells (NPCs) showed lower levels (t test: p < 0.05 for microglia versus macrophages or NPCs in all six genes).
Cytokine Profiles of Human iPSC-Derived and Primary Microglia Are Similar, but Differ from Peripheral Blood-Derived Macrophages
We analyzed the proteins released by iPSC-MG, hMG, hMG-SF, and PB-M, including differentially polarized macrophages (Figure 3D). Interestingly, similarities between iPSC-MG and primary microglia drastically increased when hMG were cultured in our differentiation medium (hMG-SF), as Pearson's correlation coefficient increased from r = 0.473 to r = 0.824. Of note, hMG showed upregulation of cytokines, such as RANTES, I-TAC, BAFF, GR0-a, and MIP3a, which are typically released upon inflammation, and in fact were also expressed by M(LPS,IFNγ) macrophages (red arrows in Figure 3D). Nevertheless, microglial samples clustered together and away from all PB-M subtypes.
iPSC-MG Are Functional Phagocytes
iPSC-MG, hMG, hMG-SF, PB-M, and undifferentiated iPSCs were challenged with a given amount of fluorescently labeled latex microspheres per cell. Flow-cytometry analysis showed that the majority of iPSC-MG were able to phagocytose (90% ± 6%) as were both hMG and hMG-SF. As expected, PB-M(−) macrophages were also able to engulf microspheres while undifferentiated iPSCs were not (Figure 3E).
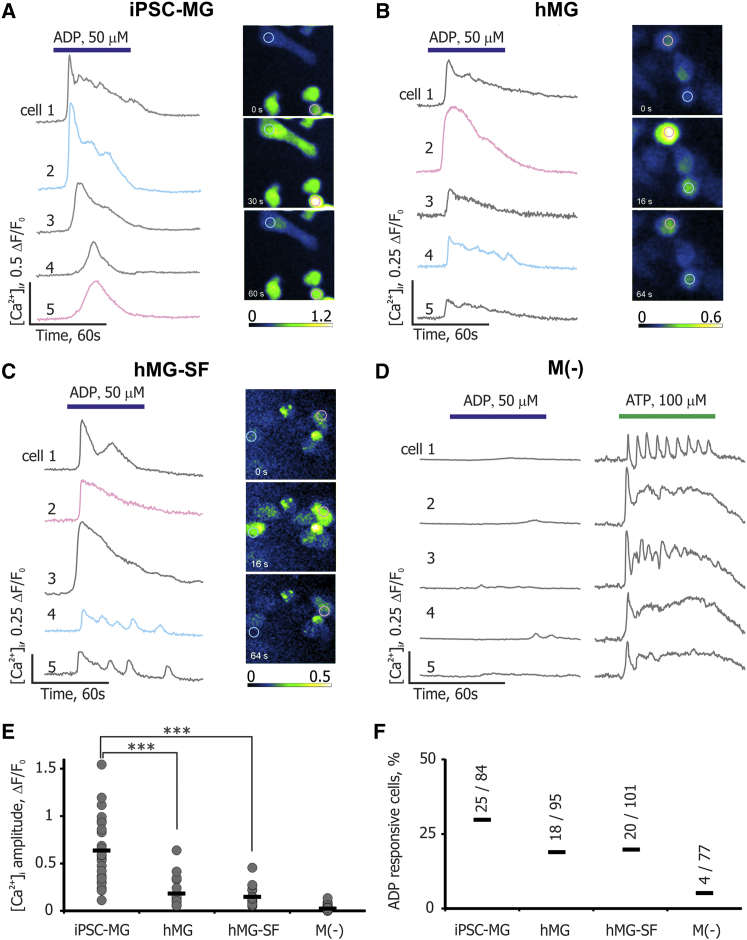
iPSC-MG Release Intracellular Ca2+ in Response to ADP
The microglial signature gene P2RY12 encodes a GI protein-coupled receptor (Haynes et al., 2006) that responds to ADP, resulting in intracellular Ca2+ ([Ca2+]i) transients, whereas PB-M that lack P2RY12 expression do not respond to ADP (Moore et al., 2015). Thus, ADP-induced [Ca2+]i transients can be used to differentiate between microglia and macrophages. When we stimulated iPSC-MG, hMG, hMG-SF, and PB-M with ADP, only microglial cells responded (Figures 4A–4C). The peak amplitude of ADP responses in iPSC-MG (Figure 4E) as well as the number of responsive cells (Figure 4F) were higher than either hMG or hMG-SF. On the contrary, none of the differentially polarized PB-M responded to ADP, but [Ca2+]i transients were reliably observed upon stimulation with ATP (Figures 4D and S4).
Figure 4.
ADP-Evoked [Ca2+]i Transients in Microglia and Macrophages
(A) Left panel shows five example traces of [Ca2+]i transients following ADP application in iPSC-MG loaded with the Ca2+ indicator Fluo-4/AM. Right panel shows time lapse of changes in fluorescence intensity produced by ADP application. Magenta and cyan traces originate from cells indicated by same-colored regions of interest in the right panel. Bars represent duration of ADP or ATP application.
(B and C) Same data as in (A), obtained from primary human microglia (hMG) and hMG-SF correspondingly.
(D) ADP and ATP responses in PB-M(−). Note the absence of significant [Ca2+]i transients in response to ADP.
(E) Statistical analysis for the amplitudes of [Ca2+]i transients. Maximum amplitude of [Ca2+]i transient for each responsive cell is presented as a gray dot in the corresponding category (∗∗∗p < 0.001 by t test).
(F) Percentages of ADP-responsive cells among all different cell types analyzed. Ratios for each cell type indicate the number of responsive cells out of total number of cells analyzed. iPSC-MG data are pooled from three independent experiments.
See also Figure S4.
Discussion
As in vitro hematopoietic differentiation of PSCs resembles in vivo primitive hematopoiesis rather than definitive hematopoiesis (Vanhee et al., 2015), we reasoned that PSC-derived myeloid progenitors would resemble in vivo primitive yolk sac myeloid progenitors, and therefore could give rise to microglia in vitro. Stimulating PSCs with a myeloid inductive medium followed by treatment with microglia-promoting cytokines generated KDR+CD235a+ primitive hemangioblasts, which subsequently transitioned from CD45+CX3CR1− to CD45+CX3CR1+ microglial progenitors in vitro.
To ensure robustness and reproducibility of the protocol, we tested a panel of 16 PSC lines (Table S3) including iPSCs from individuals with varying disease status, age, and sex, generated using different reprogramming strategies (e.g., mRNA/microRNA, Sendai virus). We were able to obtain microglial progenitors from all lines, with an average yield of two to three progenitors per undifferentiated PSC. The yield of progenitors varied across the lines without correlation to a specific disease, reprogramming method, or sex and age of the donor. The resulting microglia expressed typical markers, were ramified with highly motile processes, and were able to phagocytose with efficiency equivalent to that of human primary microglia.
While the identity of human microglia has not been well established, recent genome-wide studies in mouse have provided datasets to facilitate the distinction of microglia from other myeloid or CNS cell types (Bennett et al., 2016, Butovsky et al., 2014, Hickman et al., 2013). Therefore, we compared global mRNA expression of iPSC-MG with primary microglia and both peripheral blood-derived and hepatic macrophages to evaluate the proposed “signature genes” in human microglia. As obtaining all these cell types from the same individual was not feasible, we included samples with different genetic backgrounds, which may increase the “noise” of the data and possibly mask differences between cell types. However, our analyses clearly showed that iPSC-MG were clustered away from both circulating and other tissue-specific macrophages, and together with primary microglia and CD45+ cells (called “myeloid”), isolated from human brains (Zhang et al., 2016). Furthermore, iPSC-MG expressed the six genes suggested as unique to human microglia (Butovsky et al., 2014) and many other genes enriched in mouse microglia (Table S1).
The cytokine profile of microglia was distinct from PB-M, independent of polarization status. iPSC-MG clustered together with hMG and tighter when hMG were cultured in our medium (hMG-SF), probably due to the absence of serum. In vivo, microglia reside behind the blood-brain barrier, and the presence of serum components triggers their activation (Ransohoff and Perry, 2009). Indeed, hMG cultured in serum showed increased levels of inflammatory molecules such as RANTES, GR0-A, I-TAC, BAFF, and MIP3a, similarly to M(LPS,IFNγ) pro-inflammatory macrophages.
Finally, we showed that iPSC-MG express functional P2RY12 at both transcript and protein levels. This receptor distinguishes rodent and human microglia from other myeloid cells (Butovsky et al., 2014) and its activation via ADP results in [Ca2+]i transients (Moore et al., 2015). All microglial samples (iPSC-MG, hMG, and hMG-SF) showed ADP-evoked [Ca2+]i transients, while PB-M were unresponsive to ADP but showed [Ca2+]i upon exposure to ATP, indicating that they were healthy and functional.
While this manuscript was under review, Muffat et al. (2016) published a microglial differentiation protocol, and we are providing a comparison with their RNA-seq data (Figures 3C and S3). Both approaches mimic embryonic development (derived via CD235a+ yolk sac progenitors), use IL-34 as the main driver to microglial lineage commitment and maturation in chemically defined media, and generate microglia with motile processes that express typical makers and are able to perform phagocytosis. Our strategy, based on monolayer cultures instead of embryoid bodies (EBs), is comparable in efficiency but requires fewer starting PSCs. Importantly, our protocol does not require manual selection of specific EBs. We isolate microglial progenitors via FACS or magnetic beads, enabling high-throughput applications such as compound and genetic screens.
While the global expression profile of iPSC-MG strongly resembles that of human primary microglia, discrepancies were found for some microglia markers including TMEM119, LAG3, and CX3CR1. Surprisingly, mRNA levels of TMEM119 were very low in iPSC-MG compared with hMG, even though the protein was detected by immunofluorescence. TMEM119 mRNA levels in the study by Muffat et al. (2016) were variable across different iPSC lines and there was a trend for downregulation when cells were cultured in “NPC-conditioned medium.” Another recent differentiation protocol (Pandya et al., 2017) describes the generation of iPSC-derived microglia from hematopoietic progenitors but requires astrocyte co-culture in the presence of serum. Under these experimental conditions, microglia differentiation is accomplished within 4 weeks with a yield comparable with both our and the Muffat protocols. Although their gene expression analysis was performed using microarrays, limiting direct comparison of expression data, TMEM119 was also not highly expressed and not significantly different from iPSC or fetal MG expression in their dataset. On the other hand, serum exposure upregulated TMEM119 in our hMG, suggesting that culture environment can alter expression. As studies in rodents suggest that Tmem119 is developmentally regulated (Bennett et al., 2016), variable expression could also reflect immaturity. In addition, minimal levels of LAG3 mRNA were detected in human primary and iPSC-derived microglia, unless they were exposed to either “NPC-conditioned medium” (in Muffat et al., 2016) or serum, despite previous studies in rodents (Butovsky et al., 2014, Hickman et al., 2013) that identified Lag3 as a key microglial gene. As microglia are particularly sensitive to exogenous stimuli, different culture conditions are likely responsible for the subtle discrepancies in transcript levels observed between different protocols. Furthermore, microglia are known to express the chemokine receptor CX3CR1 early in development, and it was detected by flow cytometry in both hMG and iPSC-MG derived through our protocol and from Pandya et al. (2017). In contrast, mRNA was not considered expressed in human primary microglia (Muffat et al., 2016 and our study) or iPSC-derived microglia (Muffat et al., 2016) and varied in our iPSC-MG. Transcript levels of CX3CR1 were also relatively low and variable in the Pandya et al. (2017) study despite detectable protein expression by FACS. Further work will be needed to address these discrepancies and distinguish culture-related regulation from developmental regulation of markers.
Overall, our protocol provides iPSC-MG as a new source of human microglial cells, which will complement studies in mouse models to better understand the role of microglia in health and disease. We have shown that the protocol is highly reproducible, and we anticipate that it will become a critical tool to investigate microglial dysfunction in CNS disorders. The inclusion of microglia in co-culture or three-dimensional systems, involving iPSC-derived neurons and other glial cell types, will be crucial to advance in vitro disease modeling and better recapitulate the complexity of the in vivo environment.
Experimental Procedures
Pluripotent Stem Cell Lines
Two human embryonic stem cell lines (RUES1 and H9, both NIH approved) and 13 iPSC lines reprogrammed at New York Stem Cell Foundation upon institutional review board approvals and informed consent and one iPSC line from Dr. Ricardo Feldman were used in this study (Supplemental Experimental Procedures and Table S3). Human primary microglia and hepatic macrophages were purchased from ScienCell Research Laboratories.
Microglial Differentiation Protocol
PSC differentiation was induced with mTeSR Custom medium (STEMCELL Technologies) containing 80 ng/mL BMP4. At day 4 cells were induced with 25 ng/mL basic fibroblast growth factor, 100 ng/mL stem cell factor (SCF), and 80 ng/mL vascular endothelial growth factor in StemPro-34 SFM (with 2 mM GutaMAX, Life Technologies). Two days later, the medium was supplemented with 50 ng/mL SCF, 50 ng/mL IL-3, 5 ng/mL thrombopoietin, 50 ng/mL macrophage CSF (M-CSF) and 50 ng/mL Flt3l, and from day 14 with 50 ng/mL M-CSF, 50 ng/mL Flt3l, and 25 ng/mL GM-CSF. Between days 25 and 50, CD14+ or CD14+CX3CR1+ progenitors were isolated and plated onto tissue culture-treated dishes or Thermanox plastic coverslips (all from Thermo Fisher Scientific) in Microglial Medium (RPMI-1640 [Life Technologies] with 2 mM GlutaMAX-I, 10 ng/mL GM-CSF, and 100 ng/mL IL-34). Medium was replenished every 3–4 days for at least 2 weeks.
Other experimental procedures are described in the Supplemental Experimental Procedures.
Author Contributions
P.D., D.O.F., V.F., and S.N. designed the study and reviewed the manuscript with input from S.G. and E.S., and all authors. P.D. contributed to every assay and analysis. B.S. and G.L. performed differentiation experiments, I.K. and C.T. Ca2+ imaging and analysis, and M.Z. flow cytometry. M.W. and B.Z. analyzed the RNA-seq data. D.O.F. analyzed the cytokine release data. P.D. and V.F. wrote the paper.
Acknowledgments
We thank Dr. Isabella Pallotta and Emily Wrona for help with PB-M, and Drs. Ricardo Feldman, Aiqun Li, and Daniel Paul for the iPSC lines. This work was supported by the New York Stem Cell Foundation, the NIA/NIH grant U01AG046170 (to B.Z., M.W., E.S., S.G., and S.N.) a component of the AMP-AD Target Discovery and Preclinical Validation Project, the ORIO-13-051 grant (to D.O.F.) from the Oak Foundation, and by the 20140243 grant (to V.F.) from the Conrad N. Hilton Foundation. P.D. is an NYSCF-Druckenmiller fellow.
Published: May 18, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, three tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.04.023.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE97744.
Supplemental Information
References
- Aarum J., Sandberg K., Haeberlein S.L., Persson M.A. Migration and differentiation of neural precursor cells can be directed by microglia. Proc. Natl. Acad. Sci. USA. 2003;100:15983–15988. doi: 10.1073/pnas.2237050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajami B., Bennett J.L., Krieger C., Tetzlaff W., Rossi F.M. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- Bennett M.L., Bennett F.C., Liddelow S.A., Ajami B., Zamanian J.L., Fernhoff N.B., Mulinyawe S.B., Bohlen C.J., Adil A., Tucker A. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA. 2016;113:E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton C.A., Gilbert D.L. Production of superoxide anions by a CNS macrophage, the microglia. FEBS Lett. 1987;223:284–288. doi: 10.1016/0014-5793(87)80305-8. [DOI] [PubMed] [Google Scholar]
- Davalos D., Grutzendler J., Yang G., Kim J.V., Zuo Y., Jung S., Littman D.R., Dustin M.L., Gan W.B. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- Fenn A.M., Henry C.J., Huang Y., Dugan A., Godbout J.P. Lipopolysaccharide-induced interleukin (IL)-4 receptor-alpha expression and corresponding sensitivity to the M2 promoting effects of IL-4 are impaired in microglia of aged mice. Brain Behav. Immun. 2012;26:766–777. doi: 10.1016/j.bbi.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch U.K., Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 2007;10:1387–1394. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- Haynes S.E., Hollopeter G., Yang G., Kurpius D., Dailey M.E., Gan W.B., Julius D. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci. 2006;9:1512–1519. doi: 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- Hickman S.E., Kingery N.D., Ohsumi T.K., Borowsky M.L., Wang L.C., Means T.K., El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf K., Erny D., Goldmann T., Sander V., Schulz C., Perdiguero E.G., Wieghofer P., Heinrich A., Riemke P., Holscher C. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- Luheshi N.M., Kovacs K.J., Lopez-Castejon G., Brough D., Denes A. Interleukin-1alpha expression precedes IL-1beta after ischemic brain injury and is localised to areas of focal neuronal loss and penumbral tissues. J. Neuroinflammation. 2011;8:186. doi: 10.1186/1742-2094-8-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C.S., Ase A.R., Kinsara A., Rao V.T., Michell-Robinson M., Leong S.Y., Butovsky O., Ludwin S.K., Seguela P., Bar-Or A. P2Y12 expression and function in alternatively activated human microglia. Neurol. Neuroimmunol. Neuroinflamm. 2015;2:e80. doi: 10.1212/NXI.0000000000000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J., Li Y., Yuan B., Mitalipova M., Omer A., Corcoran S., Bakiasi G., Tsai L.H., Aubourg P., Ransohoff R.M. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I., Neumann H. Microglial clearance function in health and disease. Neuroscience. 2009;158:1030–1038. doi: 10.1016/j.neuroscience.2008.06.046. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A. Two-photon imaging of microglia in the mouse cortex in vivo. Cold Spring Harb. Protoc. 2012;2012 doi: 10.1101/pdb.prot069294. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A., Kirchhoff F., Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Ohgidani M., Kato T.A., Setoyama D., Sagata N., Hashimoto R., Shigenobu K., Yoshida T., Hayakawa K., Shimokawa N., Miura D. Direct induction of ramified microglia-like cells from human monocytes: dynamic microglial dysfunction in Nasu-Hakola disease. Sci. Rep. 2014;4:4957. doi: 10.1038/srep04957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya H., Shen M.J., Ichikawa D.M., Sedlock A.B., Choi Y., Johnson K.R., Kim G., Brown M.A., Elkahloun A.G., Maric D. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat. Neurosci. 2017;20:753–759. doi: 10.1038/nn.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli R.C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., Giustetto M., Ferreira T.A., Guiducci E., Dumas L. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Ransohoff R.M. How neuroinflammation contributes to neurodegeneration. Science. 2016;353:777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- Ransohoff R.M., Perry V.H. Microglial physiology: unique stimuli, specialized responses. Annu. Rev. Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- Schulz C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Smith A.M., Dragunow M. The human side of microglia. Trends Neurosci. 2014;37:125–135. doi: 10.1016/j.tins.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Sturgeon C.M., Ditadi A., Awong G., Kennedy M., Keller G. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat. Biotechnol. 2014;32:554–561. doi: 10.1038/nbt.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M., Fujita Y., Tanaka T., Nakamura Y., Kikuta J., Ishii M., Yamashita T. Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci. 2013;16:543–551. doi: 10.1038/nn.3358. [DOI] [PubMed] [Google Scholar]
- Vanhee S., De Mulder K., Van Caeneghem Y., Verstichel G., Van Roy N., Menten B., Velghe I., Philippe J., De Bleser D., Lambrecht B.N. In vitro human embryonic stem cell hematopoiesis mimics MYB-independent yolk sac hematopoiesis. Haematologica. 2015;100:157–166. doi: 10.3324/haematol.2014.112144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagimachi M.D., Niwa A., Tanaka T., Honda-Ozaki F., Nishimoto S., Murata Y., Yasumi T., Ito J., Tomida S., Oshima K. Robust and highly-efficient differentiation of functional monocytic cells from human pluripotent stem cells under serum- and feeder cell-free conditions. PLoS One. 2013;8:e59243. doi: 10.1371/journal.pone.0059243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.