Abstract
The Burkholderia pseudomallei K96243 genome contains multiple type IV pilin-associated loci, including one encoding a putative pilus structural protein (pilA). A pilA deletion mutant has reduced adherence to human epithelial cells and is less virulent in the nematode model of virulence and the murine model of melioidosis, suggesting a role for type IV pili in B. pseudomallei virulence.
Burkholderia pseudomallei is the causative agent of melioidosis, a disease endemic to southeast Asia and northern Australia (9, 21, 42). Infections occur via inhalation or percutaneous inoculation, and clinical manifestations include subclinical infections, acute septicemia, and chronic disease (42). B. pseudomallei can infect almost any host organ, is resistant to many antibiotics, and can persist for long periods (6). Both B. pseudomallei and its close relative Burkholderia mallei are potential bioterrorist agents listed by the Centers for Disease Control and Prevention (17, 42).
A number of factors are associated with B. pseudomallei virulence, including products secreted by the general secretory pathway, type III secretory systems, flagella, lipopolysaccharide, and capsule (1, 7, 11-13, 29, 30, 35, 36). Despite these studies, little is known about how B. pseudomallei causes disease.
Adherence is an important virulence mechanism mediated by carbohydrate molecules, pilus, and nonpilus adhesins (14, 16, 20, 34, 37). Type IV pili (TFP) are important for virulence in many gram-negative bacteria and are divided into two subclasses, IVA and IVB, based on the presence of conserved motifs (38). The Flp subgroup of type IVB pili are shorter than other pilins and have a characteristic Flp motif (18).
B. pseudomallei adheres to human epithelial cell lines, but the molecular basis for this adherence is unknown (5). We describe the identification of multiple TFP-associated loci in B. pseudomallei and show that a TFP gene homologue is required for efficient adherence of B. pseudomallei to cultured cells and for virulence in vivo.
B. pseudomallei TFP genes.
We identified eight TFP-associated loci (designated TFP1 to TFP8) in the B. pseudomallei K96243 genome (http://www.sanger.ac.uk/Projects/B_pseudomallei/) by in silico probing with multiple pilin homologues and biogenesis proteins; five such loci contain one or more type IV pilin subunits (Table 1). The presence of two type IVB subunits in TFP7 and TFP8 suggests that B. pseudomallei K96243 may synthesize pili with a composite architecture (41, 43).
TABLE 1.
Summary of TFP loci identified in the B. pseudomallei K96243 genome
| Locus | Gene | Amino acidsa | Subunit/other homologue (organism; GenBank accession no.)b | % Similarity/% identity (amino acidsc) |
|---|---|---|---|---|
| Chromosome 1 | ||||
| TFP1 | BPSL0782 | 207 | Type IVA pilus subunit PilA (Pseudomonas aeruginosa; AAL12242) | 49/32 (217) |
| TFP2 | BPSL1821 | 63 | Type IVB pilus subunit Flp1 (Actinobacillus actinomycetemcomitans; AAN75204) | 42/24 (76) |
| TFP3 | BPSL1899 | 56 | Type IVB pilus subunit Flp1 (Actinobacillus actinomycetemcomitans; AAN75204) | 46/28 (75) |
| TFP4 | BPSL2752d | 150 | Type IV prepilin leader protein PilE (Pseudomonas aeruginosa; AAA79363) | 46/26 (157) |
| BPSL2756d | 186 | Type IV prepilin leader protein FimT (Pseudomonas aeruginosa; AAB39270) | 38/24 (189) | |
| TFP5 | BPSL3008e | 419 | Type IV pilus biogenesis protein PilB (Pseudomonas aeruginosa; A35384) | 47/34 (569) |
| TFP6 | BPSL3170e | 442 | Type IV pilus biogenesis protein PilQ (Pseudomonas aeruginosa; S37345) | 36/22 (714) |
| Chromosome 2 | ||||
| TFP7 | BPSS1593 | 557 | Type IVB minor pilus subunit PilV (Escherichia coli; AAL05526) | 33/21 (561) |
| BPSS1595 | 184 | Type IVB major pilus subunit PilS (Escherichia coli; BAA77979) | 43/24 (206) | |
| TFP8 | BPSS2185 | 56 | Type IVB pilus subunit Flp1 (Actinobacillus actinomycetemcomitans; AAN75204) | 43/22 (75) |
| BPSS2186 | 72 | Type IVB pilus subunit Flp1 (Actinobacillus actinomycetemcomitans; AAN75204) | 48/28 (82) |
Number of amino acids in the predicted protein.
Homologues were assigned based on TBLASTN (24) searches in Artemis release 4 (31). Homologues reported are those that have been functionally characterized and published.
Number of residues including gaps in the full-length protein alignment.
Predicted peptides contain prepilin-like leader sequences.
ORF corresponds to the first gene in the predicted locus, as no subunit is present.
Analysis of TFP1 and PilA.
B. pseudomallei K96243 open reading frame (ORF) BPSL0782 was designated pilA because the full-length predicted gene product shares 49% similarity (32% identity) to PilA from Pseudomonas aeruginosa (GenBank accession no. AAL12242). This locus was designated TFP1 (Table 1). While ORFs downstream of pilA in P. aeruginosa are clearly pilus associated (reviewed in reference 10), ORFs flanking pilA in B. pseudomallei K96243 differ substantially and may not be involved with pilin biogenesis.
pilA is predicted to encode the only type IVA pilin in B. pseudomallei K96243; it possesses the conserved glycine and phenylalanine residues of type IVA pilins, between which is the predicted leader sequence cleavage site, and also the invariant glutamic acid residue 5 amino acids from the mature N terminus, associated with most pilin types (Fig. 1) (38). The PilA leader is predicted to be 40 amino acids long, based on an upstream Shine-Dalgarno sequence; this sets it apart from other IVA pilins, which have shorter leader sequences (Fig. 1). Alternative pilA start codons are not associated with Shine-Dalgarno sequences. Ralstonia solanacearum, also from the family Burkholderiaceae, has a type IVA pilin with a longer leader sequence (15 amino acid residues) (19).
FIG. 1.
Comparison of the N-terminal amino acid sequence of B. pseudomallei K96243 PilA with representative pilins from the type IVA and IVB subclasses. Species abbreviations: Bp, B. pseudomallei strain K96243 PilA (IVA); Pa, P. aeruginosa strain PAK PilA (IVA; P02973); Ng, Neisseria gonorrhoeae strain MS11 PilE (IVA; CAA47307); Ec, Escherichia coli strain O127:H6 BfpA (IVB; P33553); Vc, Vibrio cholerae strain classical O1 Z17561 TcpA (IVB; CAA45455). Conserved glycine (G) and phenylalanine (F) residues (F is conserved in type IVA pilins) are highlighted, between which the signal peptides are predicted to be cleaved. The invariant glutamic acid (E) residue associated with most pilins is also indicated.
A number of consecutive arginine residues occur in the putative signal sequence, reminiscent of a twin-arginine translocation secretion signal; however, a consensus motif (26) is not evident, or the arginine residues are too close to the putative cleavage site.
PCR with pilA-flanking primers amplified the gene in B. pseudomallei strains of diverse origin (Table 2 shows the strains studied). The predicted amino acid sequence was conserved in all strains where sequencing was undertaken (results not shown).
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| B. pseudomallei strains investigated for conservation of pilA by PCR and sequencinga | ||
| 576 (AY598724) | Clinical isolate, Thailand | PHLb |
| 2889 (AY598726) | Pneumonic clinical isolate, Thailand | PHLb |
| NCTC4845 (AY598725) | Monkey isolate | NCTCc |
| B. pseudomallei strains investigated for conservation of pilA by PCR | ||
| 08 | Clinical isolate | 4 |
| GCH | Clinical isolate | Gold Coast Hospitald |
| RBH | Clinical isolate | 4 |
| 06 | Environmental isolate | 4 |
| GF1 | Environmental isolate | 4 |
| GF2 | Environmental isolate | 4 |
| THP375 | Environmental isolate | 4 |
| B. pseudomallei | ||
| K96243 | Clinical isolate, Genr Strr Chls, Thailand | Siriraj Hospitale |
| JAB16.1x | K96243 derivative; merodiploid strain; ΔpilA (nucleotides 67-612)::pAEH16 (cat sacBR oriT oriR6K); Genr Strr Chlr | This work |
| JAB16 | K96243 derivative; unmarked deletion mutant strain; ΔpilA (nucleotides 67-612); Genr Strr Chls | This work |
| E. coli | ||
| OP50 | Uracil auxotroph | 3 |
| S17.1 (λpir) | RP4-2-Tc::Mu Km::Tn7 Tp Sm (λpir) phoA20 thi-1 rpsE rpoB | 28 |
| JABEC16 | S17.1 (λpir) containing pAEH16 | This work |
| Plasmids | ||
| pDM4 | Suicide vector, sacBR oriT oriR6K Chlr | 25 |
| pAEH16 | pDM4 containing the pilA deletion construct | This work |
Numbers in parentheses are GenBank accession numbers.
Supplied by Ty Pitt, Central Public Health Laboratory, Colindale, United Kingdom.
National Collection of Type Cultures.
Supplied by D. Alfredson, Gold Coast Hospital, Gold Coast, Australia.
Supplied by S. Songsivilai, Department of Immunology, Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok, Thailand.
Construction of an unmarked pilA deletion mutant strain, JAB16.
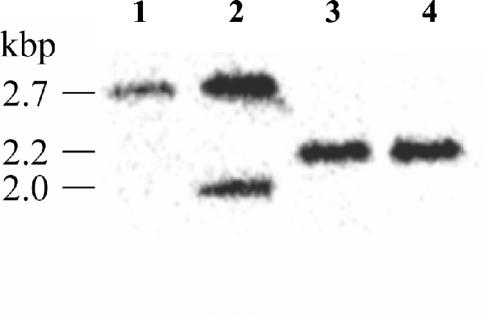
An allelic-exchange mutant (JAB16) was generated which contains an unmarked in-frame 546-bp deletion in the pilA gene (32). Briefly, the deleted pilA allele was constructed by PCR and transferred to the suicide vector pDM4 to give pAEH16. This was conjugated from JABEC16 to B. pseudomallei K96243, and merodiploid integrants were selected. One such colony, JAB16.1x, was cultured without selection and plated onto medium lacking sodium chloride but containing 15% sucrose to enrich for excision of integrated vector DNA (2), resulting in either a wild-type or deleted pilA allele. Chloramphenicol sensitivity (Cms) was assessed, and Cms colonies were analyzed by PCR (data not shown) and Southern blotting (Fig. 2) to distinguish pilA mutants from wild type. Three of the first eight Cms colonies screened contained the deleted pilA allele, and one was designated JAB16.
FIG. 2.
Southern hybridization of ClaI-digested genomic DNA with the use of a pilA-specific DNA probe. Lanes: 1, the hybridizing probe identified a 2.7-kbp fragment from K96243 DNA containing the wild-type pilA allele; 2, the hybridizing probe identified two fragments from JAB16.1x DNA, one fragment containing the wild-type pilA allele (2.7 kbp) and the other containing the deleted pilA allele (2.0 kbp); 3 and 4, the hybridizing probe identified only the DNA fragment containing the deleted pilA allele (2.2 kbp) from DNA isolated from two individual putative deletion mutants (one designated JAB16). The 2.2-kbp fragment in the deletion mutant strains is larger than the 2.0-kbp fragment in JAB16.1x, as the excised plasmid contained a ClaI site.
JAB16 has reduced adherence to human epithelial cell lines.
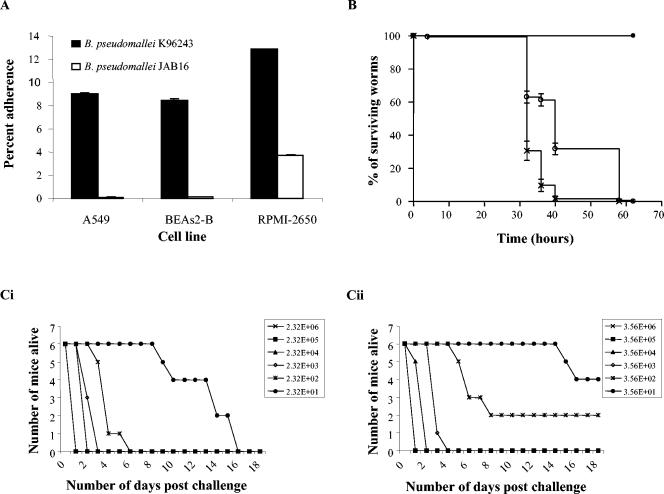
The interaction of B. pseudomallei K96243 or JAB16 with human respiratory cell lines was studied. Cell lines were cultured and prepared as previously described (40). Bacterial inocula were prepared from overnight cultures grown in nutrient broth, incubated statically for 16 h at 37°C. Monolayers were infected with diluted bacterial cultures (∼105 CFU/ml) for 1 h at 37°C, and inocula were enumerated by plate counts. Nonadherent bacteria were removed by five washes with phosphate-buffered saline. Monolayers were lysed with 0.1% (vol/vol) Triton X-100 for 30 min at 37°C, and adherent-cell-associated bacteria were enumerated by plate counts. JAB16 adhered significantly less than the K96243 parent to A549, BEAS-2B, and RPMI-2650 cell lines (P < 0.01) (Fig. 3A). These data suggest a role for pilA in the adherence of B. pseudomallei in vitro.
FIG. 3.
Characterization of JAB16. (A) Adherence of B. pseudomallei K96243 and JAB16 to human epithelial cell lines. Percent adherence was determined by dividing the number of adherent bacteria by the inoculum and multiplying by 100. Percent adherence for the RPMI-2650 cell line was multiplied by 100. Data represent the means ± standard errors of the means for triplicate wells of single representative experiments. Each experiment was performed at least three times. The independent sample t test was used to analyze differences in cell-associated bacteria between K96243 and JAB16. (B) Killing of C. elegans by B. pseudomallei K96243 and JAB16. Between 10 and 20 C. elegans L4-stage worms were seeded onto nematode growth medium agar, which was inoculated with 10 μl of an overnight culture of B. pseudomallei K96243 (×) or B. pseudomallei JAB16 (○). E. coli OP50 (•) was used as a negative control. Each value is the mean ± standard error of the mean of six replicates. The survival data were plotted using the Kaplan-Meier method and analyzed using the Mantel-Haenszel log rank test in the statistical package Prism, version 3.02. (C) Survival curves of BALB/c mice challenged with different doses of B. pseudomallei K96243 (Ci) and JAB16 (Cii), by the intranasal route of infection. Infected BALB/c mice (groups of six) were monitored for 18 days after bacterial challenge. By the use of regression with life data B. pseudomallei JAB16 is attenuated with respect to B. pseudomallei K96243 (P = 0.012).
JAB16 exhibits reduced virulence in nematode worms.
The soil nematode Caenorhabditis elegans is susceptible to B. pseudomallei (15). C. elegans strain N2 nematodes which had been synchronized to the L4 stage (39) and suspended in K medium (33) were exposed to a lawn of JAB16 or K96243 at 25°C on nematode growth medium agar, and nematode survival was recorded. C. elegans started to die by 32 h postinfection, regardless of the infecting strain, but worms infected with JAB16 survived significantly longer than K96243-infected worms (Fig. 3B), 99% of which were dead by 58 and 40 h, respectively (P < 0.001). A P. aeruginosa pilA mutant displayed reduced piliation and was also deficient for secretion of proteins by the general secretory pathway (23). While this could be the case with JAB16, O'Quinn et al. showed that a B. pseudomallei general secretory pathway mutant, deficient in secretion, was unable to delay the time to death of C. elegans (27). Since JAB16 adheres less in cell culture, we suggest that the mechanism by which JAB16 delays the time to death of nematodes is due to a reduced-adherence phenotype.
JAB16 is attenuated in a murine model of infection.
Groups of six BALB/c mice were challenged with different doses of JAB16 and K96243, by either the intraperitoneal route or the intranasal route (22). JAB16 was not attenuated compared to K96243 via the intraperitoneal challenge route (data not shown). In contrast, JAB16 was less virulent than K96243 via the intranasal route but only at low challenge doses (P < 0.05) (Fig. 3C). This may be due to expression of other pili (Table 1) or other putative adhesins present in the genome (results not shown). Differences in the attenuation of pilus mutants according to the route of challenge have been reported previously with Yersinia pseudotuberculosis (8). As bacterial adhesins often recognize specific receptors (20, 34, 37), attenuation differences for JAB16 due to route of challenge may be due to differential distribution and/or expression of the cognate host receptor.
Concluding remarks.
The identification of eight loci encoding a total of seven putative type IV pilin subunits, and many accessory genes, may be related to the capacity of B. pseudomallei to exist and replicate in the environment and infect various animal hosts and tissues. It is possible that different pili are required to mediate interactions with specific host receptors or that the expression of B. pseudomallei pili is regulated in a complex manner. Work to further characterize the role of TFP loci is in progress.
An unmarked in-frame deletion of pilA in B. pseudomallei decreases adherence to cultured respiratory cell lines, decreases the time to death of C. elegans, and reduces the killing of BALB/c mice. Since the mutation in JAB16 is an in-frame deletion, it should not affect the expression of downstream genes, making it highly likely that the phenotypes observed for JAB16 are due to the absence of pilA. Our data suggest that PilA may be an important mediator of the pathogenic process in humans and should be considered as a target in future attempts to generate a protective vaccine against melioidosis.
Acknowledgments
We thank Bryan Lingard for technical assistance and Thomas Laws for helpful discussion and statistical analysis.
The nematode strains used in this work were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH National Center for Research Resources (NCRR). Justin Boddey and Nat Brown acknowledge Australian Postgraduate Awards.
Editor: J. B. Bliska
REFERENCES
- 1.Atkins, T., R. Prior, K. Mack, P. Russell, M. Nelson, J. Prior, J. Ellis, P. C. F. Oyston, G. Dougan, and R. W. Titball. 2002. Characterisation of an acapsular mutant of Burkholderia pseudomallei identified by signature tagged mutagenesis. J. Med. Microbiol. 51:539-547. [DOI] [PubMed] [Google Scholar]
- 2.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, N. F., and I. R. Beacham. 2000. Cloning and analysis of genomic differences unique to Burkholderia pseudomallei by comparison with B. thailandensis. J. Med. Microbiol. 49:993-1001. [DOI] [PubMed] [Google Scholar]
- 5.Brown, N. F., J. A. Boddey, C. P. Flegg, and I. R. Beacham. 2002. Adherence of Burkholderia pseudomallei cells to cultured human epithelial cell lines is regulated by growth temperature. Infect. Immun. 70:974-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaowagul, W. 2000. Recent advances in the treatment of severe melioidosis. Acta Trop. 74:133-137. [DOI] [PubMed] [Google Scholar]
- 7.Chua, K. L., Y. Y. Chan, and Y. H. Gan. 2003. Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 71:1622-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collyn, F., M. A. Lety, S. Nair, V. Escuyer, A. Ben Younes, M. Simonet, and M. Marceau. 2002. Yersinia pseudotuberculosis harbors a type IV pilus gene cluster that contributes to pathogenicity. Infect. Immun. 70:6196-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dance, D. A. 2000. Melioidosis as an emerging global problem. Acta Trop. 74:115-119. [DOI] [PubMed] [Google Scholar]
- 10.Darzins, A., and M. A. Russell. 1997. Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system—a review. Gene 192:109-115. [DOI] [PubMed] [Google Scholar]
- 11.DeShazer, D., P. J. Brett, M. N. Burtnick, and D. E. Woods. 1999. Molecular characterization of genetic loci required for secretion of exoproducts in Burkholderia pseudomallei. J. Bacteriol. 181:4661-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeShazer, D., P. J. Brett, R. Carlyon, and D. E. Woods. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeShazer, D., P. J. Brett, and D. E. Woods. 1998. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 30:1081-1100. [DOI] [PubMed] [Google Scholar]
- 14.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gan, Y. H., K. L. Chua, H. H. Chua, B. P. Liu, C. S. Hii, H. L. Chong, and P. Tan. 2002. Characterization of Burkholderia pseudomallei infection and identification of novel virulence factors using a Caenorhabditis elegans host system. Mol. Microbiol. 44:1185-1197. [DOI] [PubMed] [Google Scholar]
- 16.Hultgren, S. J., S. Abraham, M. Caparon, P. Falk, J. W. St. Geme III, and S. Normark. 1993. Pilus and nonpilus bacterial adhesins: assembly and function in cell recognition. Cell 73:887-901. [DOI] [PubMed] [Google Scholar]
- 17.Jeddeloh, J. A., D. L. Fritz, D. M. Waag, J. M. Hartings, and G. P. Andrews. 2003. Biodefense-driven murine model of pneumonic melioidosis. Infect. Immun. 71:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542-554. [DOI] [PubMed] [Google Scholar]
- 19.Kang, Y. W., H. L. Liu, S. Genin, M. A. Schell, and T. P. Denny. 2002. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46:427-437. [DOI] [PubMed] [Google Scholar]
- 20.Klemm, P., and M. A. Schembri. 2000. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 290:27-35. [DOI] [PubMed] [Google Scholar]
- 21.Leelarasamee, A., and S. Bovornkitti. 1989. Melioidosis: review and update. Rev. Infect. Dis. 11:413-425. [DOI] [PubMed] [Google Scholar]
- 22.Liu, B. P., G. C. Koo, E. H. Yap, K. L. Chua, and Y. H. Gan. 2002. Model of differential susceptibility to mucosal Burkholderia pseudomallei infection. Infect. Immun. 70:504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, H. M., S. T. Motley, and S. Lory. 1997. Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol. Microbiol. 25:247-259. [DOI] [PubMed] [Google Scholar]
- 24.Madden, T. L., R. L. Tatusov, and J. H. Zhang. 1996. Applications of network BLAST server. Methods Enzymol. 266:131-141. [DOI] [PubMed] [Google Scholar]
- 25.Milton, D. L., R. O'Toole, P. Horstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Quinn, A. L., E. M. Wiegand, and J. A. Jeddeloh. 2001. Burkholderia pseudomallei kills the nematode Caenorhabditis elegans using an endotoxin-mediated paralysis. Cell. Microbiol. 3:381-393. [DOI] [PubMed] [Google Scholar]
- 28.Penfold, R. J., and J. M. Pemberton. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145-146. [DOI] [PubMed] [Google Scholar]
- 29.Rainbow, L., C. A. Hart, and G. Winstanley. 2002. Distribution of type III secretion gene clusters in Burkholderia pseudomallei, B. thailandensis and B. mallei. J. Med. Microbiol. 51:374-384. [DOI] [PubMed] [Google Scholar]
- 30.Reckseidler, S. L., D. DeShazer, P. A. Sokol, and D. E. Woods. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J. F., D. W. Russell, and N. Irwin. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Smith, M. P., T. R. Laws, T. P. Atkins, P. C. F. Oyston, D. I. de Pomerai, and R. W. Titball. 2002. A liquid-based method for the assessment of bacterial pathogenicity using the nematode Caenorhabditis elegans. FEMS Microbiol. Lett. 210:181-185. [DOI] [PubMed] [Google Scholar]
- 34.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens, M. P., A. Friebel, L. A. Taylor, M. W. Wood, P. J. Brown, W. D. Hardt, and E. E. Galyov. 2003. A Burkholderia pseudomallei type III secreted protein, BopE, facilitates bacterial invasion of epithelial cells and exhibits guanine nucleotide exchange factor activity. J. Bacteriol. 185:4992-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens, M. P., M. W. Wood, L. A. Taylor, P. Monaghan, P. Hawes, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649-659. [DOI] [PubMed] [Google Scholar]
- 37.St. Geme, J. W., III. 1997. Bacterial adhesins: determinants of microbial colonization and pathogenicity. Adv. Pediatr. 44:43-72. [PubMed] [Google Scholar]
- 38.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 39.Sulston, J., and J. Hodgkin. 1988. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Thomas, R. J., and T. J. Brooks. 2004. Oligosaccharide receptor mimics inhibit Legionella pneumophila attachment to human respiratory epithelial cells. Microb. Pathog. 36:83-92. [DOI] [PubMed] [Google Scholar]
- 41.Toma, C., H. Kuroki, N. Nakasone, M. Ehara, and M. Iwanaga. 2002. Minor pilin subunits are conserved in Vibrio cholerae type IV pili. FEMS Immun. Med. Microbiol. 33:35-40. [DOI] [PubMed] [Google Scholar]
- 42.White, N. J. 2003. Melioidosis. Lancet 361:1715-1722. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida, T., N. Furuya, M. Ishikura, T. Isobe, K. Haino-Fukushima, T. Ogawa, and T. Komano. 1998. Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili. J. Bacteriol. 180:2842-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]