Abstract
Staphylococcus aureus is an important human pathogen that is also able to kill the model nematode Caenorhabditis elegans. We constructed a 2,950-member Tn917 transposon insertion library in S. aureus strain NCTC 8325. Twenty-one of these insertions exhibited attenuated C. elegans killing, and of these, 12 contained insertions in different genes or chromosomal locations. Ten of these 12 insertions showed attenuated killing phenotypes when transduced into two different S. aureus strains, and 5 of the 10 mutants correspond to genes that have not been previously identified in signature-tagged mutagenesis studies. These latter five mutants were tested in a murine renal abscess model, and one mutant harboring an insertion in nagD exhibited attenuated virulence. Interestingly, Tn917 was shown to have a very strong bias for insertions near the terminus of DNA replication.
In addition to being a common commensal bacterial species of humans, Staphylococcus aureus is a remarkably versatile pathogen, capable of causing a broad range of human disease (44). The diverse range of S. aureus disease has been attributed to its ability to produce an array of virulence factors, the expression of which is tightly regulated by global regulatory systems in response to cell density, energy availability, oxygen tension, and other environmental signals (for review, see references 8 and 32).
It has been difficult to identify S. aureus virulence factors due to the unwieldiness of mammalian pathogenesis models. As an alternative to traditional mammalian pathogenesis models, our laboratory and others have utilized a strategy for studying host-pathogen interactions, using the nematode Caenorhabditis elegans as a simple surrogate model host (3). A variety of human bacterial and fungal pathogens kill C. elegans, and there is a remarkably high degree of correlation between pathogen virulence factors required for nematode killing and virulence in vertebrate models (39). This allows genome-wide screening for pathogen virulence factors by a relatively high-throughput screening procedure (15, 16, 23, 26, 29, 42, 43).
Recently, we showed that a variety of S. aureus strains efficiently kill C. elegans and that S. aureus genes known to be important in mammalian pathogenesis are also required for nematode killing (40). As described here, we generated a Tn917 transposon insertion library of 2,950 mutants in S. aureus NCTC 8325 and screened the library for attenuated killing of C. elegans. We identified 10 genes important for virulence in C. elegans, one of which appears to encode a previously undescribed virulence factor important for pathogenicity in a murine renal abscess model.
(This work was presented in part at the 103rd General Meeting of the American Society for Microbiology, Washington, D.C., 18 to 22 May 2003.)
MATERIALS AND METHODS
Bacterial and C. elegans strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. Bacterial strains and the Bristol N2 strain of C. elegans were grown and maintained as described previously (5, 27).
TABLE 1.
Strains and plasmid used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | Restriction-deficient derivative of NCTC 8325-4 | 33 |
| NCTC 8325 | Wild-type strain, rsbU mutant | 21; The Staphylococcal Genetic Stock Center |
| Newman | ATCC 25904; high level of clumping factor, σB+ | 14; laboratory collection |
| ALC638 | agr sarA mutant in Newman background | A. L. Cheung |
| E. coli | ||
| OP50 | Uracil auxotrophy | 6; laboratory collection |
| DH5α | Invitrogen | |
| C. elegans | ||
| Bristol N2 | Wild-type strain | Caenorhabditis Genetics Center |
| Plasmid | ||
| pLTV1 | Temperature-sensitive plasmid harboring Tn917 transposon | 7 |
Construction of an S. aureus Tn917 library.
Plasmid pLTV1 (7) was electroporated into S. aureus strain RN4220 (38), purified (QIAprep spin mini kit; QIAGEN), and then electroporated into S. aureus strain 8325. Strain 8325(pLTV1) was grown overnight in tryptic soy (TS) broth containing 5-μg/ml tetracycline at 30°C, 10-fold dilutions were plated on TS agar plates containing 2-μg/ml erythromycin, and the plates were incubated overnight at 43°C. A total of 2,950 colonies were picked into TS medium containing 5-μg/ml erythromycin and grown overnight at 37°C in 96-well plates. Glycerol was added (final concentration, 15%), and the library was stored at −80°C.
Nematode killing assay and screening protocol.
S. aureus killing plates were prepared as previously described (40). Primary screening was carried out by transferring 20 to 30 L4-stage nematodes to each plate (27), incubating plates at 25°C, and scoring for live and dead worms after 40 h. Subsequent assays were performed with 25 to 35 L4-stage nematodes per plate in triplicate.
Determination of Tn917 insertion site.
The DNA segment adjacent to Tn917 insertion sites was determined with genomic DNA (DNeasy tissue kit; QIAGEN) and a nested “arbitrary” PCR technique using the transposon-specific primer Seq1 and the arbitrary primer Arb1a in the first round and the nested primers Arb2 and Int1 in subsequent rounds (primer sequences are shown in Table 2) (34). PCR products were purified (QIAquick PCR purification kit; QIAGEN), sequenced with primer Int2 (Taq DyeDeoxy Terminator Cycle sequencing kit; Applied Biosystems) in concert with an ABI 3700 PRISM automated sequencer, and analyzed with the Vector NTI Suite 7 software package (InforMax, Inc.).
TABLE 2.
Oligonucleotide primers used in this study
| Primer name | Primer sequence |
|---|---|
| Arbitrary PCR primers | |
| Int1 | CTATTCCTAAACACTTAAGAGAATTG |
| Int2 | CATTGGTTTAGTGGGAATTTGTACC |
| Seq1 | GGAGAGTATAAATTTGACTTG |
| Arb1a | GGCCACGCGTCGACTAGTACNNNNNNNNNNGTATA |
| Arb2 | GGCCACGCGTCGACTAGTAC |
| Int3 | CTCACAATAGAGAGATGTCACCGTC |
| Insertion-specific primers for transduction confirmation | |
| o3H1 | ATCAAGGCCGCAAGTAAATATG |
| o4D8 | TGCCTGTTGTTTCTTATC |
| o5F1 | AAATGGCTGAAACGAAGAAAGG |
| o6A5 | GATGCTCCTGGTGATTGTTCTG |
| o7A2 | GCATACATCATTCGTTTAG |
| o7G12 | TATCATTACCCATTACTTG |
| o15G12 | ATTCGCATACTAAGCATTG |
| o25G6 | GTCATGCCAGAGGTTAAAG |
| o28G12 | TTGCGCGTGATGATTATTAC |
| o29C3 | GTAGCCATTGCTGTAGGTATTG |
| o30A5 | TTAATTTAGAATACGGCAC |
| o31B11 | GCGCCACAAATTCGTATGATTC |
In the case of two mutants, 7G12 and 15G12, a plasmid marker rescue protocol was utilized to determine the location of the transposon insertions (7). Briefly, Tn917 was excised from genomic DNA using EcoRI (New England Biolabs), circularized with T4 DNA ligase (New England Biolabs), and electroporated into Escherichia coli DH5α (Invitrogen). Plasmid DNA was purified from these strains, and flanking DNA was sequenced with the transposon-specific outward-facing primers Int3 and Seq1.
Transduction of Tn917 insertions.
The general transducing phage φ80 was used as previously described (24), and transductants were confirmed by PCR with Seq1 and a mutant-specific primer listed in Table 2.
Murine kidney abscess infections.
Bacterial inocula were prepared from overnight cultures grown at 37°C in TS medium with the appropriate antibiotic and diluted to approximately 108 CFU/ml in TS medium. Appropriate dilutions were prepared and plated on TS agar plates to determine the precise inoculum. Female CD1 mice (Charles River Laboratories) were injected intraperitoneally with 0.2 ml of the bacterial suspension, and their health was monitored daily. Five days postchallenge, mice were sacrificed and both kidneys from each mouse were harvested, washed in phosphate-buffered saline, weighed, and homogenized in 2 ml of phosphate-buffered saline and colony counts were determined on TS agar plates with appropriate antibiotics. Animal study protocols were reviewed and approved by the Subcommittee on Animal Studies of the Massachusetts General Hospital.
Statistical analysis.
Nematode survival was calculated by the Kaplan-Meier method, and survival differences were tested for significance by use of the log-rank test, comparing mutant and wild-type survival curves (GraphPad Prism, version 3.0). Differences in colonization of murine kidneys by S. aureus mutant strains and wild-type S. aureus were tested for significance with an unpaired two-tailed t test (GraphPad Prism, version 3.0). P values < 0.05 were considered significant.
RESULTS
Screening a Tn917 insertion library for attenuated clones.
A 2,950-member Tn917 transposon insertion library was generated by introducing the temperature-sensitive plasmid pLTV1 into S. aureus strain NCTC 8325 and selecting for erythromycin-resistant clones that grew at 43°C (see Materials and Methods). The library was screened for clones that showed a decreased ability to kill C. elegans after 40 h in comparison to worms feeding on wild-type S. aureus. Of 2,950 mutants screened, 145 exhibited attenuated virulence in the primary screen, and 21 of these, which consistently exhibited less-pathogenic phenotypes, were selected for further analysis.
Determination of insertion site and phenotypic confirmation by transduction.
The insertion sites of the Tn917 transposons in the 21 selected mutants were determined by an arbitrary PCR or a plasmid marker rescue method (7, 16, 34) as described in Materials and Methods. BLAST analysis (4) against the S. aureus 8325 genome and the annotated S. aureus N315 genome identified the insertion sites and associated disrupted open reading frames (ORFs) in all 21 mutants (22, 25). As shown in Table 3, eight distinct genes were directly disrupted, three mutants contained insertions within 200 bp of the 5′ end of three different genes, and one mutant contained a Tn917 insertion site in intergenic sequence that was downstream of two convergently transcribed genes. Twelve mutants (indicated in Table 3) with mutations corresponding to insertions in 12 different genes or chromosomal locations were chosen for further analysis. These clones resulted from Tn917 transposition and not whole plasmid integration, since all mutants were tetracycline sensitive. DNA blot analysis showed that each of these 12 mutants contained a single Tn917 insertion (data not shown).
TABLE 3.
Attenuated S. aureus mutants identified by screening in C. elegans
| Mutants and mutationa | N315 ORFb | Gene annotation | Gene name/function |
|---|---|---|---|
| TCA cycle genes | |||
| 3E1, 4D8, 8D9, 10B10, 22A5, 28C12, 28C11, 29E1 | SA1245 | odhAc,d,e | 2-Oxoglutarate dehydrogenase |
| 25G6, 29B8, 29G6 | SA1244 | odhBc,d,e | Dihydrolipoamide succinyltransferase |
| 7G12 | SA1669 | citG | Fumarate hydratase, class II |
| Transporters | |||
| 5F1 | SA1239 | 5′ braBc,d | Branched chain amino acid transporter |
| 7A2 | SA1214 | opp-2Cc,d,e | Oligopeptide transporter membrane permease |
| Nucleic acid metabolism/DNA replication | |||
| 30A5 | SA1045 | pyrAAe | Carbamoyl-phosphate synthase small chain (pyrimidine/arginine synthesis) |
| 3H1 | SA1288 | dinGc,d,e | DNA helicase |
| 28G12 | SA0467 | 5′ yacA/tilS | Lysidine biosynthesis (isoleucine tRNA synthesis) |
| Miscellaneous | |||
| 29C3 | SA2304 | 5′ fbp | Fructose-bisphosphatase |
| 6A5 | SA0790 | nagD | Conserved hypothetical protein |
| Mutants with no virulence-related phenotype after transduction | |||
| 15G12 | SA1241 | Similar to nitrogen oxide reductase | |
| 31B11 | 3′ SA1239 | 3′ braB | Downstream of braB and SA1238 |
Insertional mutants in boldface were transduced into 8325, and their C. elegans-killing phenotypes were confirmed.
ORF numbers from the S. aureus N315 genome sequencing project (25).
Previously identified in the STM screen of Mei et al. (31).
Previously identified in the STM screen of Coulter et al. (11).
Upregulated by agr by transcriptional profiling (13).
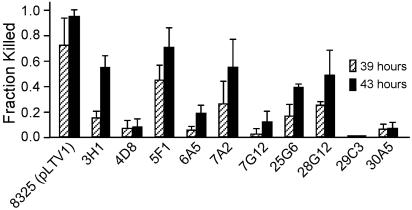
To determine whether the 12 transposon insertions chosen for further analysis were linked to the attenuated ability to kill C. elegans, the 12 transposon insertions were transduced into the parent S. aureus 8325 strain by using the generalized transducing phage φ80 (24). As shown in Table 3, among the 12 transduced insertions, 10 maintained their attenuation after transduction. Figure 1 shows that these 10 mutants have significantly decreased virulence toward C. elegans. All 10 mutants grew similarly to wild-type S. aureus in TS medium, except for mutant 30A5, which contains Tn917 inserted into the pyrAA gene (data not shown).
FIG. 1.
C. elegans killing by transduced S. aureus 8325 mutants. Shown are the fractions of nematodes killed after 39 and 43 h of feeding on S. aureus 8325(pLTV1) and transduced isogenic Tn917 insertion mutants. Following the growth of bacteria on TS agar plates, 25 to 35 L4-stage hermaphroditic N2 worms were transferred to each plate. Nematode death was determined after 39 and 43 h of feeding. The data represent the mean + standard deviation (error bar) of three sets of plates and are representative of three independent experiments.
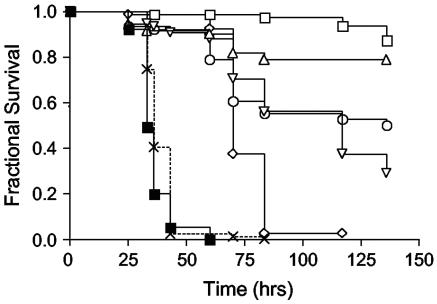
Five of the 10 genes were previously identified in signature-tagged mutagenesis (STM) screens carried out by Mei et al. (31) (odhA and opp-2C) or Coulter et al. (11) (odhA, odhB, braB, opp-2C, and dinG). To ensure that the attenuated phenotypes of insertions in the five previously unidentified virulence-related genes (citG, pyrAA, 5′ yacY/tilS, 5′ fbp, and nagD) (Table 3) were not strain specific, the insertions in these five strains were transduced into the S. aureus Newman strain, and the resulting transductants were tested for nematocidal activity. As shown in Fig. 2, the transposon insertion mutations that had been verified by transduction into 8325 also resulted in attenuated virulence compared to the wild-type strain when transduced into the Newman strain. Transduction of insertions that did not affect virulence, such as 15G12 (Fig. 2), did not reduce the virulence of the Newman strain, demonstrating that transduction of Tn917 itself does not affect C. elegans killing (not shown).
FIG. 2.
Survival of C. elegans on S. aureus Newman mutants. Shown is the survival of nematodes fed S. aureus Newman (wild type, closed squares; n = 77) and transduced isogenic mutants 6A5 (nagD, open diamonds; n = 79), 7G12 (citG, open squares; n = 80), 15G12 (SA1241, ×; n = 79), 28G12 (5′ yacA, open circles; n = 76), 29C3 (5′ fbp, open triangles; n = 71), and 30A5 (pyrAA, open inverted triangles; n = 76). Survival was plotted by the Kaplan-Meier method. Pairwise comparisons (log-rank test) by strain were as follows: Newman versus 6A5, P < 0.0001; Newman versus 7G12, P < 0.0001; Newman versus 15G12, P not significant; Newman versus 28G12, P < 0.0001; Newman versus 29C3, P < 0.0001; and Newman versus 30A5, P < 0.0001.
Distribution of insertion sites.
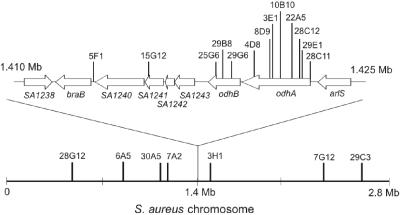
Multiple mutants were recovered from the screen with Tn917 insertions in the odhAB operon (Table 3). These mutants resulted from independent insertion events, since each insertion occurred at a different nucleotide position (data not shown). To investigate the nature of this apparent hot spot for transposition, the distribution of insertion sites within the S. aureus genome was examined. As shown in Fig. 3, there appears to be a strong bias for transposition within the region of the chromosome that lies in close proximity to the chromosome terminus, which was estimated to lie between 1.38 and 1.41 Mb in S. aureus strain N315 by GC-skew analysis (25). Similar results with Tn917 were obtained in our laboratory during mutagenesis of Enterococcus faecalis strain OG1RF (17).
FIG. 3.
Distribution of Tn917 insertion sites within the S. aureus genome. Shown is a schematic representation of the Tn917 insertion sites of those mutants with reduced virulence in the C. elegans infection model along a linearized S. aureus chromosome. Lines indicate nucleotide insertion sites. An enlarged depiction of the terminus region, containing a majority of the Tn917 insertion sites, is shown above the chromosome.
Examination of previously uncharacterized mutants in vivo.
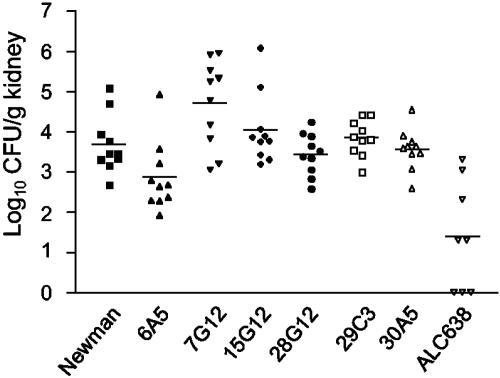
The five selected insertion mutants (transduced into the Newman strain) with mutations not previously shown to correspond to genes relevant for mammalian pathogenesis were tested in a modified murine renal abscess model (2). Mice were inoculated intraperitoneally with 0.2 ml of S. aureus culture, and after a 5-day incubation period, the kidneys were harvested and the bacterial loads were determined. As shown in Fig. 4, inoculation of 1 × 108 CFU of the wild-type Newman strain intraperitoneally resulted in an average of 5.5 × 103 CFU/kidney at day 5. Of the mutants tested, strain 6A5 (corresponding to an insertion in nagD) was significantly attenuated in this model (P < 0.05).
FIG. 4.
Select S. aureus Newman mutants tested in a murine renal abscess model. Groups of five CD1 mice were inoculated intraperitoneally with the following strains: Newman (wild type; 9.8 × 107 CFU), ALC638 (agr sarA; 1.4 × 108 CFU), 6A5 (nagD; 1.4 × 108 CFU), 7G12 (citG; 6.6 × 107 CFU), 28G12 (5′ yacA; 5.0 × 107 CFU), 29C3 (5′ fbp; 6.0 × 107 CFU), 30A5 (pyrAA; 1.4 × 108 CFU), and 15G12 (ORF SA1241, negative control; 1.6 × 108 CFU). Five days after inoculation, the mice were sacrificed and the kidneys were excised, homogenized, diluted, and plated. CFU of Newman, ALC638, and Tn917 mutants in renal tissue were calculated and plotted after log transformation. Statistical significance was determined using a two-tailed t test; compared to wild-type strain Newman, ALC638 and 6A5 were attenuated in renal tissue bacterial load (P < 0.05). Data are representative of two independent experiments.
DISCUSSION
Genome-wide approaches for studying S. aureus pathogenesis have recently involved the use of in vivo expression technology, which uses a promoter trap library to identify genes that are selectively transcriptionally active within the host (30), and signature tagged mutagenesis (STM), which compares the in vivo survival of a series of “tagged” mutants (19). Lowe et al. used in vivo expression technology to identify 45 S. aureus genes induced in a renal abscess model, including 6 previously identified virulence genes (28).
Two STM experiments have been carried out with S. aureus. Mei et al. screened a tagged library in a murine bacteremia model and identified 50 mutants harboring insertions in mainly biosynthetic loci (31). Coulter et al. used three different murine infection models (cutaneous abscess, bacteremia, and wound infection) to identify 237 genes important for in vivo survival, the majority of which also code for biosynthesis-related enzymes (11). Genome-wide transcriptional profiling has also been used to identify virulence-related genes. Dunman et al. identified the set of genes regulated by the important regulatory systems agr and sar, which included many previously identified putative virulence factors, a range of metabolic genes, and many hypothetical genes (13). Similarly, Saïd-Salim et al. defined the regulon of the transcriptional regulator rot, which was found to negatively regulate many secreted proteins and to positively regulate many cell wall-associated proteins (37).
Screening a Tn917 insertion library in a pathogenicity model.
In this study, we constructed a Tn917 transposon insertion mutation library in S. aureus NCTC 8325. Screening the library resulted in the identification of 10 unique attenuated mutants (Table 3). Five of these genes (odhA, odhB, braB, opp-2C, and dinG) had previously been identified in the STM screens of Coulter et al. and Mei et al. (11, 31). Additionally, five of these genes (odhA, odhB, opp-2C, pyrAA, and dinG) were found to be upregulated by the agr system based on transcriptional profiling (13). While the genes identified in our screen are associated with metabolic functions in S. aureus, all mutants were able to grow well in TS medium, with the exception of the pyrAA mutant. The disrupted genes can be placed into four functional categories based on the proteins they encode: tricarboxylic acid (TCA) cycle components, nucleic acid metabolism/DNA replication, transporters, and miscellaneous proteins. The same classes of genes were identified previously in the STM screens (11, 31).
The TCA cycle appears to play several critical roles in S. aureus pathogenesis. The TCA cycle is critical for the switch from exponential growth to postexponential growth and the subsequent production of capsule (12) and secretion of extracellular toxins such as α-hemolysin and staphylococcal enterotoxin C (10, 41). In addition, many TCA cycle elements are upregulated by the agr global regulatory system (13). As in other bacterial pathogens, nucleic acid biosynthesis is important for survival within hosts, likely due to the poor availability of purines and pyrimidines. The two transporters identified in this study have been previously identified as being important for in vivo survival in the two STM screens performed in S. aureus (11, 31) and likely reflect the importance of amino acid and peptide uptake in the setting of infection.
Importantly, 5 out of the 10 genes (odhA, odhB, braB, opp-2C, and dinG) identified in our in vivo screen in C. elegans were shown previously to have reduced virulence in murine infection models (11, 31). We employed a murine kidney abscess model to investigate the role in mammalian virulence of the remaining five mutants that had not previously been characterized in a murine infection model. Of these, the nagD mutant was attenuated at the P = 0.03 level. The role of nagD (SA0790) in S. aureus virulence is not clear. It shares a high degree of homology with B. subtilis yutF (50% identity, 68% similarity) and E. coli nagD (31% identity, 50% similarity). In E. coli, Peri et al. and Plumbridge independently cloned a locus involved in N-acetylglucosamine catabolism, termed nag (35, 36). However, nagD, the 3′-most member of the nagBACD operon, is not required for N-acetylglucosamine catabolism nor does it appear to contribute to the regulation of the operon (35, 36). In S. aureus and B. subtilis, the N-acetylglucosamine catabolism operon consists of nagAB only and the B. subtilis nagD homologue yutF is in a distinct three-gene operon of unknown function (yutDEF).
Although four of the five mutants that we tested were not attenuated in the kidney abscess model, this does not exclude the possibility that they may be attenuated in other animal models. For example, of the 237 mutants Coulter et al. found that were attenuated in at least one infection model, only 10% of the recovered mutants were attenuated in all three models (11). Similarly, inactivation of the aconitase gene does not affect mouse mortality when S. aureus is inoculated intraperitoneally but does cause attenuation in a subcutaneous infection model (41).
Recently it was shown that S. aureus strain 8325, in which we constructed the Tn917 library, harbors an 11-bp deletion of the rsbU gene that decreases expression of the alternate sigma factor σB, which affects the expression of a variety of virulence-related genes (18). Indeed reconstruction of rsbU+ results in diminished agr expression and diminished expression of the secreted proteins SspA and Hla but increased nematocidal activity (20, 40). Therefore, to ensure that any transposon insertions showing attenuated C. elegans killing were not specific to the 8325 background, we confirmed the phenotype of selected mutants in the S. aureus Newman strain.
If the Tn917 insertion sites in our library were distributed randomly, then our library of 2,950 mutants would represent ∼1.5 times the coverage of the nonessential regions of the genome. However, as shown in Fig. 3, Tn917 appears to have a strong insertion bias near the terminus of the S. aureus chromosome. Assuming that the distribution of Tn917 insertions as a whole is similar to that found among attenuated mutants, we estimate that we have screened less than one-third of the nonessential genes in S. aureus. Future screens may benefit from the development of an alternative gram-positive transposon system, such as a mariner-based system, which has been shown to insert at random in a variety of organisms (1, 9).
In summary, we have shown that C. elegans can be used relatively efficiently to identify S. aureus virulence factors. Six out of 10 genes identified in C. elegans are relevant for mammalian pathogenesis, including the nagD gene, which was not previously known to encode a virulence factor. Future screens in C. elegans using a fully saturated mutant library would likely identify additional genes involved in mammalian pathogenesis.
ADDENDUM IN PROOF
A recent publication (T. Bae, A. K. Banger, A. Wallace, E. M. Glass, F. Aslund, O. Schneewind, and D. M. Missiakas, Proc. Natl. Acad. Sci. USA 101:12312-12317, 2004) describes the construction of an ordered library of Staphylococcus aureus insertion mutants by using a mariner-based transposon and the results of a screen for decreased virulence in the Caenorhabditis elegans system.
Acknowledgments
We thank Ambrose Cheung, Dartmouth Medical School, for providing strain ALC638, Edward Kazyanskaya for assistance with library screening, and Andrea Bernal for assistance with the murine model. C. elegans strains were originally obtained from the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health National Center of Research Resources.
This work was supported by a postdoctoral fellowship from the Howard Hughes Medical Institute and NIAID grant K08 AI053677 to C.D.S. and by a research grant from Aventis Pharmaceuticals to F.M.A. and S.B.C.
Editor: A. D. O'Brien
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, A. Camilli, D. J. Lampe, H. M. Robertson, and J. J. Mekalanos. 1998. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. USA 95:8927-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albus, A., R. D. Arbeit, and J. C. Lee. 1991. Virulence of Staphylococcus aureus mutants altered in type 5 capsule production. Infect. Immun. 59:1008-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alegado, R. A., M. C. Campbell, W. C. Chen, S. S. Slutz, and M. W. Tan. 2003. Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell Microbiol. 5:435-444. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M. 1988. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 6.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilli, A., A. Portnoy, and P. Youngman. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J. Bacteriol. 172:3738-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 9.Chiang, S. L., and E. J. Rubin. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296:179-185. [DOI] [PubMed] [Google Scholar]
- 10.Collins, F. M., and J. Lascelles. 1962. The effect of growth conditions on oxidative and dehydrogenase activity in Staphylococcus aureus. J. Gen. Microbiol. 29:531-535. [DOI] [PubMed] [Google Scholar]
- 11.Coulter, S. N., W. R. Schwan, E. Y. Ng, M. H. Langhorne, H. D. Ritchie, S. Westbrock-Wadman, W. O. Hufnagle, K. R. Folger, A. S. Bayer, and C. K. Stover. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30:393-404. [DOI] [PubMed] [Google Scholar]
- 12.Dassy, B., and J.-M. Fournier. 1996. Respiratory activity is essential for post-exponential-phase production of type 5 capsular polysaccharide by Staphylococcus aureus. Infect. Immun. 64:2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 15.Gan, Y. H., K. L. Chua, H. H. Chua, B. Liu, C. S. Hii, H. L. Chong, and P. Tan. 2002. Characterization of Burkholderia pseudomallei infection and identification of novel virulence factors using a Caenorhabditis elegans host system. Mol. Microbiol. 44:1185-1197. [DOI] [PubMed] [Google Scholar]
- 16.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garsin, D. A., J. Urbach, J. C. Huguet-Tapia, J. E. Peters, and F. M. Ausubel. 2004. Construction of an Enterococcus faecalis Tn917-mediated gene disruption library offers insight into Tn917 insertion patterns. J. Bacteriol. 186:7280-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 20.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iandolo, J. J. 2000. Genetic and physical map of the chromosome of Staphylococcus aureus 8325, p. 317-325. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. A. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 22.Iandolo, J. J., V. Worrell, K. H. Groicher, Y. Qian, R. Tian, S. Kenton, A. Dorman, H. Ji, S. Lin, P. Loh, S. Qi, H. Zhu, and B. A. Roe. 2002. Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene 289:109-118. [DOI] [PubMed] [Google Scholar]
- 23.Joshua, G. W., A. V. Karlyshev, M. P. Smith, K. E. Isherwood, R. W. Titball, and B. W. Wren. 2003. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology 149:3221-3229. [DOI] [PubMed] [Google Scholar]
- 24.Kasatiya, S. S., and J. N. Baldwin. 1967. Nature of the determinant of tetracycline resistance in Staphylococcus aureus. Can. J. Microbiol. 13:1079-1086. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 26.Kurz, C. L., S. Chauvet, E. Andres, M. Aurouze, I. Vallet, G. P. Michel, M. Uh, J. Celli, A. Filloux, S. De Bentzmann, I. Steinmetz, J. A. Hoffmann, B. B. Finlay, J. P. Gorvel, D. Ferrandon, and J. J. Ewbank. 2003. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 22:1451-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis, J. A., and J. T. Fleming. 1995. Basic culture methods. Methods Cell Biol. 48:3-29. [PubMed] [Google Scholar]
- 28.Lowe, A. M., D. T. Beattie, and R. L. Deresiewicz. 1998. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol. Microbiol. 27:967-976. [DOI] [PubMed] [Google Scholar]
- 29.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 30.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 31.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 32.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 33.Novick, R. P. 1990. The Staphylococcus as a molecular genetic system, p. 1-37. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, Inc., New York, N.Y.
- 34.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 35.Peri, K. G., H. Goldie, and E. B. Waygood. 1990. Cloning and characterization of the N-acetylglucosamine operon of Escherichia coli. Biochem. Cell Biol. 68:123-137. [DOI] [PubMed] [Google Scholar]
- 36.Plumbridge, J. A. 1989. Sequence of the nagBACD operon in Escherichia coli K12 and pattern of transcription within the nag regulon. Mol. Microbiol. 3:505-515. [DOI] [PubMed] [Google Scholar]
- 37.Saïd-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 39.Sifri, C. D., J. Begun, and F. M. Ausubel. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol., in press. [DOI] [PubMed]
- 40.Sifri, C. D., J. Begun, F. M. Ausubel, and S. B. Calderwood. 2003. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71:2208-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somerville, G. A., M. S. Chaussee, C. I. Morgan, J. R. Fitzgerald, D. W. Dorward, L. J. Reitzer, and J. M. Musser. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70:6373-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenor, J. L., B. A. McCormick, F. M. Ausubel, and A. Aballay. 2004. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr. Biol. 14:1018-1024. [DOI] [PubMed] [Google Scholar]
- 44.Waldvogel, F. A. 2000. Staphylococcus aureus (including staphylococcal toxic shock), p. 2069-2092. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, Pa.