Abstract
Salmonella enterica serovar Typhimurium has the fascinating ability to form tubular structures known as Salmonella-induced filaments (Sifs) in host cells. Here, we show that the prevalence of the Sif phenotype in HeLa cells is affected by host cell density, growth, and the multiplicity of infection. Sif formation was observed in cells that displayed rapid intracellular bacterial replication and was found to be dynamic, being maximal 8 to 10 h postinfection and declining thereafter. The virulence factors SpvB and SseJ were found to negatively modulate Sif formation. Our findings demonstrate the complex and dynamic nature of the Sif phenotype.
Salmonella enterica serovar Typhimurium is a gram-negative facultative intracellular pathogen that can cause a variety of diseases in different hosts (38). Both in vitro (4, 15, 18) and in vivo (23, 30, 32, 33) models have demonstrated the ability of serovar Typhimurium to inhabit and replicate within a membrane-bound compartment within host cells, the Salmonella-containing vacuole (SCV). From within this compartment, these bacteria can modulate trafficking of the SCV. For example, serovar Typhimurium is capable of blocking delivery of both the NADPH oxidase (13, 39, 40) and inducible nitric oxide synthase (iNOS) (9) to the SCV. Modulation of SCV trafficking is mediated in part by two different type III secretion systems (TTSS) encoded on the Salmonella chromosome. These specialized protein delivery systems mediate translocation of bacterial proteins called effectors into the host cell, where they modulate host cell machinery (16).
Late stages of SCV modification (6 to 8 h postinfection) include the formation of tubular membrane extensions known as Salmonella-induced filaments (Sifs). Sifs are thought to result from the fusion of late endocytic compartments with the SCV (6). Sif formation requires at least four effectors of the Salmonella pathogenicity island 2 (SPI-2)-encoded TTSS. SifA is essential for Sif formation and mutants lacking either this effector or regulators of its expression do not induce the Sif phenotype (28, 36). Deletion of sifA also leads to instability of the SCV, possibly through the actions of the putative glycerophospholipid-cholesterol acyltransferase SseJ, another SPI-2 effector (1, 31). The SPI-2 effectors SseF and SseG are also essential for Sif formation; sseF and sseG mutants form pseudo-Sif structures which do not acquire the late endosomal markers characteristic of Sifs (14, 20). Also, recent studies have shown that the SPI-2 effector SopD2 plays a role in promoting maximal Sif formation (16a). Regulation of the Sif phenotype by these and possibly other bacterial proteins is not yet understood.
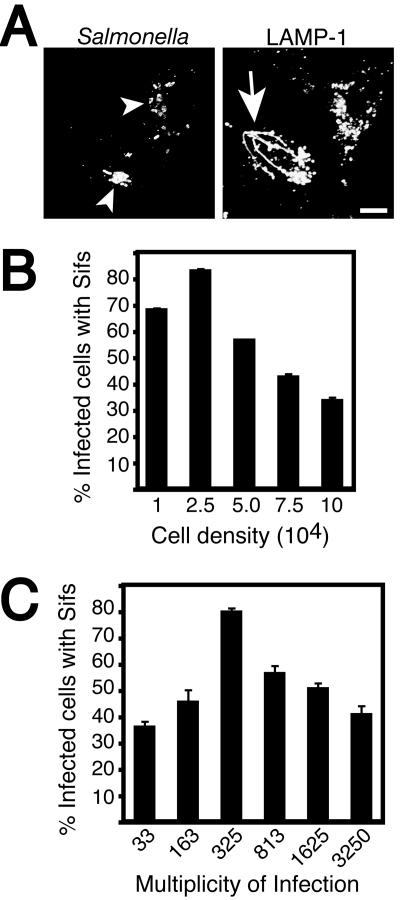
An intriguing question is why only some infected cells make Sifs. Previous studies demonstrated Sif formation in only 50 to 80% of infected cells (1, 2, 14, 19, 28, 31, 36). This result is exemplified in Fig. 1A, which shows that not all infected cells make Sifs in vitro. Recent observations in our laboratory suggested that the density of the host cell monolayer prior to infection affected the presentation of the Sif phenotype. To address the effect of host cell density on Sif formation, HeLa epithelial cells were plated at various densities prior to infection with serovar Typhimurium (Fig. 1B). For these studies, we used a method previously optimized for invasion by serovar Typhimurium (35). Cells were briefly (10 min) exposed to a high multiplicity of infection (MOI) of late log phase serovar Typhimurium. The bacteria invaded cells by using the SPI-1-encoded TTSS during this pulse, and extracellular bacteria were then removed by extensive washing and the addition of gentamicin to the media (35). As shown in Fig. 1B, cell density had a major influence on presentation of the Sif phenotype. Sifs were observed in up to 80% of infected cells at the optimal cell density of 2.5 × 104 cells/ml, but they were much less prevalent at other densities.
FIG. 1.
Effect of MOI on the formation of Sifs by serovar Typhimurium. (A) HeLa cells were infected for 8 h with serovar Typhimurium SL1344, fixed with 2.5% paraformaldehyde, and coimmunostained for LAMP-1 and Salmonella as indicated. Fixed cells were then analyzed by confocal microscopy. Arrowheads indicate LAMP-1+ bacteria within Salmonella-containing vacuoles. Tubular extensions of the bacterial vacuole, known as Sifs, are indicated with an arrow. Bar = 10 μm. (B) Asynchronous HeLa cells were plated at the indicated cell densities (cells/milliliter) prior to infection with a constant dilution of serovar Typhimurium for 8 h. Infected cells were then fixed and stained as for panel A, and Sif formation was enumerated by counting the numbers of infected cells which displayed Sif formation by using fluorescence microscopy. The averages ± the standard errors (SE) for at least three experiments are shown. A minimum of 100 infected cells were counted for each sample in a given experiment. (C) HeLa cells were plated at 2.5 × 104 cells/ml prior to infection with serovar Typhimurium for 8 h. For these experiments, the MOI was varied as indicated for each sample. Infected cells were then fixed and stained as described for panel A, and Sif formation was enumerated by fluorescence microscopy as described for panel B.
The experiments shown in Fig. 1B suggested that the ratio of invading bacteria to cell density affected Sif formation. To determine the effect of MOI on this phenotype, we plated cells at a constant density and infected serovar Typhimurium at various MOIs. As shown in Fig. 1C, an MOI of 325:1 led to optimal Sif formation, and lesser numbers of bacteria were unable to induce Sifs to the same extent. Surprisingly, higher MOIs also had an inhibitory effect on Sif formation. Together, these findings suggest that differences in host cell density and MOI are likely to explain the variability in results obtained by different investigators (1, 2, 14, 19, 31, 36).
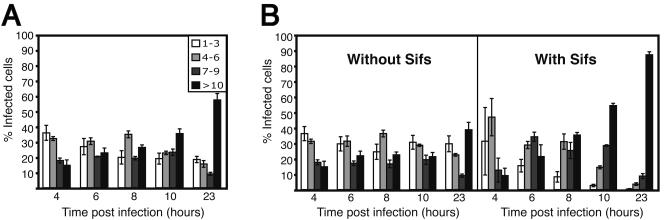
Despite many attempts to optimize conditions for Sif formation, we were unable to induce this phenotype in more than ∼80% of infected cells. This result may reflect at least two possibilities: (i) a population of infecting bacteria are unable to initiate the Sif phenotype, or (ii) a population of infected host cells have alterations to their endosomal systems that preclude the endocytic fusion events leading to Sif formation. To address the first possibility, we infected cells with serovar Typhimurium and counted the numbers of intracellular bacteria. Only lysosome-associated membrane protein 1-positive (LAMP-1+) bacteria were enumerated in order to exclude cytosolic bacteria (7). The numbers of intracellular bacteria were grouped into four categories and plotted for each time point. As shown in Fig. 2A, the majority of infected cells contain low numbers of bacteria 4 h postinfection, with very few (∼15%) cells containing >10 bacteria per cell. As time progressed during the experiment, the bacteria replicated within the SCV. As such, the numbers of infected cells with one to three bacteria decreased, while the numbers of infected cells with >10 bacteria increased over time, culminating with ∼60% of infected cells containing >10 bacteria by 23 h postinfection.
FIG. 2.
Sif formation is observed in cells that contain a rapidly replicating population of serovar Typhimurium. Asynchronous HeLa cells were infected with serovar Typhimurium SL1344 (MOI, ∼325:1) for the indicated times, fixed with 2.5% paraformaldehyde, and coimmunostained for LAMP-1 and Salmonella. Fixed cells were then analyzed by fluorescence microscopy, and the number of LAMP-1+ bacteria in each infected cell was enumerated. These numbers were grouped into one of four categories (see legend) and plotted for each time point. (A) The total population of infected cells. (B) Cells with Sifs have been plotted separately from those without Sifs. The average ± the SE for three separate experiments is shown. A minimum of 200 infected cells were counted for each time point in a given experiment.
The results shown in Fig. 2A represent the total population of intracellular bacteria in LAMP-1+ vacuoles. During the enumeration of intracellular bacteria, we also noted which cells contained Sifs and plotted them separately from those without Sifs (Fig. 2B). As shown, there was a dramatic difference between the two infected cell populations. Infected cells with Sifs displayed a rapid loss in the population of cells with one to three bacteria, concomitant with a rapid increase in the population of cells with higher bacterial numbers. By 23 h postinfection, nearly 90% of infected cells with Sifs had 10 or more bacteria, compared to 40% in those cells without Sifs. These findings suggest that the bacteria capable of inducing Sifs are undergoing rapid replication and that a static (or slow-growing) population of bacteria exists that is unable to induce the Sif phenotype. Therefore, Sifs appear to be associated with rapidly replicating serovar Typhimurium in vitro.
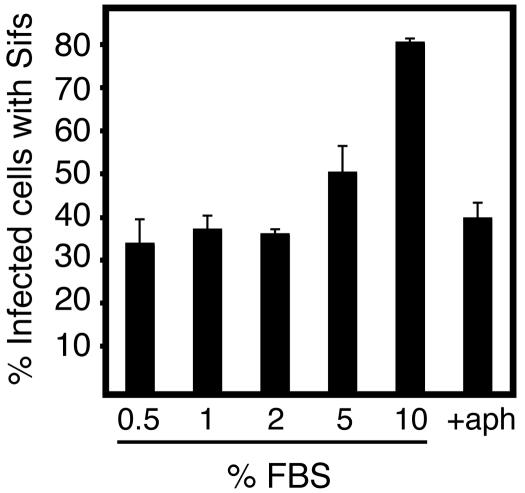
To address whether host cell growth affects Sif formation, HeLa cells were infected with wild-type serovar Typhimurium and then cultured in media containing different percentages of fetal bovine serum (FBS) (Fig. 3). Lowering serum concentrations is a commonly used method to slow cell growth and replication in culture. Normal culture conditions (10% FBS) showed maximal Sif formation 8 h postinfection, while suboptimal growth conditions exhibited a detrimental effect. This effect was also observed after treatment of cells with aphidicolin, a drug that blocks normal cell cycle progression by inhibiting DNA replication (34). As shown in Fig. 3, the addition of aphidicolin also reduced the numbers of Sifs formed by invading bacteria, although this drug did not inhibit bacterial replication on its own (data not shown). These results show that the growth rate of the host cell has an impact on Sif formation.
FIG. 3.
Host cell growth conditions affect Sif formation. Asynchronous HeLa cells were infected with serovar Typhimurium (MOI, ∼325:1) and then cultured in Dulbecco's modified Eagle medium containing the indicated concentration of FBS or with 1.25 μg of aphidicolin (aph)/ml and 10% FBS. At 8 h postinfection, cells were fixed and coimmunostained for LAMP-1 and Salmonella. Sif formation was then enumerated by fluorescence microscopy as described for Fig. 1.
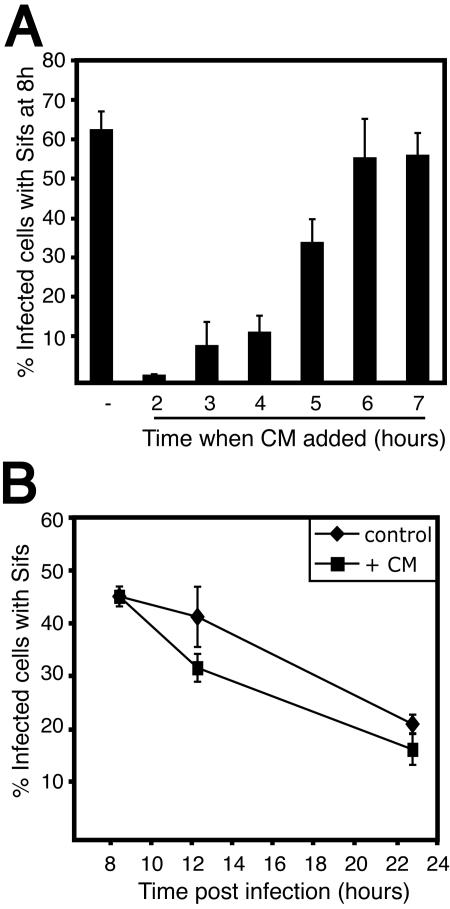
To further characterize the Sif phenotype, we analyzed the regulation of Sif formation. In Fig. 4A, chloramphenicol was added to stop bacterial protein synthesis at various times postinfection. In this way, bacterial effectors involved in Sif formation would no longer be synthesized. All cells were then fixed at 8 h to enumerate the extent of Sif formation. By 5 h postinfection, effectors were synthesized and Sif formation was maintained until the 8 h endpoint in ∼40% of the infected cells. By 6 h, the number of Sifs formed was equivalent to the control, in which no drug was added (Fig. 4A). These results suggest that the bacterial effectors necessary for Sif formation were translocated into host cells by 5 to 6 h postinfection and are consistent with previous studies demonstrating translocation of effector proteins (3, 5, 11, 12, 17, 19-21, 25-27).
FIG. 4.
Sif formation and stability during infection. (A) HeLa cells were infected with serovar Typhimurium SL1344 (MOI, ∼163:1), and 200 μg of chloramphenicol (CM)/ml was added to stop bacterial protein synthesis at the indicated times postinfection. Cells were fixed at 8 h and coimmunostained for LAMP-1 and Salmonella, and Sif formation was enumerated by fluorescence microscopy as described for Fig. 1. The control bar indicates Sif formation at 8 h postinfection with no chloramphenicol added. (B) HeLa cells were infected as described for panel A, and chloramphenicol was added to all samples at 8 h postinfection. Cells were fixed at the indicated time points and stained and analyzed as described for panel A. The control indicates normal Sif dynamics with no chloramphenicol added.
An interesting observation made during these studies was that Sifs are a dynamic phenotype. Maximal Sif formation is observed 8 to 10 h postinfection, but it then declines abruptly to ∼20% by 21 h (Fig. 5B, wild-type data). While there are many possibilities for this decline, we wished to determine whether effectors of the SPI-2 TTSS were responsible. In order to analyze the breakdown and stability of Sifs, HeLa cells were infected with serovar Typhimurium and chloramphenicol was added at 8 h. Cells were then fixed at 12 and 23 h to determine the role of bacterial protein synthesis in Sif stability and/or breakdown. As shown in Fig. 4B, antibiotic addition had no dramatic effect on Sif downregulation, suggesting that any bacterial factors required for Sif downregulation are stable or that the decline of Sifs is not dependent upon bacterial protein synthesis.
FIG. 5.
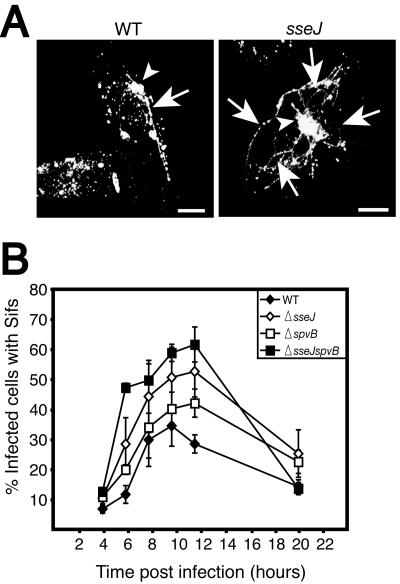
Sif formation is a dynamic phenotype under the regulation of multiple virulence proteins. (A) HeLa cells were infected with either wild-type (WT) serovar Typhimurium (CS401) or an isogenic mutant of this strain lacking sseJ (strain JAF43; see reference 11) (MOI, ∼325:1) as indicated. Infected cells were fixed at 10 h postinfection and immunostained for LAMP-1 and Salmonella. Confocal flat projections are shown for LAMP-1 staining. The arrowheads indicate LAMP-1+ bacteria within Salmonella-containing vacuoles. Sifs are indicated by arrows. As shown, the deletion of sseJ led to extensive Sif formation compared to that for wild-type serovar Typhimurium. Bars = 5 um. (B) HeLa cells were infected for the indicated times with either wild-type serovar Typhimurium (CS401) or isogenic mutants of this strain lacking sseJ, spvB (strain CS725; see reference 25), or a double mutant lacking sseJ and spvB (MBO60, created by transduction of a Kan insertion in the spvB gene from CS725 into JAF43) as described for panel A. Infected cells were fixed at the indicated time points and coimmunostained for LAMP-1, and Salmonella and Sif formation was enumerated by fluorescence microscopy as described for Fig. 1. The average result ± the SE from three separate experiments is shown.
A previous study by Ruiz-Albert et al. demonstrated that transfection of cells with the SPI-2 effector SseJ prior to infection inhibits Sif formation induced by wild-type serovar Typhimurium (31). To determine whether SseJ is involved in downregulating Sifs, we infected cells with mutants lacking this effector. For these experiments, the serovar Typhimurium CS401 wild-type strain was used. Optimization was carried out for this strain as it was for SL1344. An MOI of ∼325:1 was found to be optimal for CS401, as well, although Sif formation reached a maximum of only 30 to 50% by 8 h (data not shown). As illustrated in Fig. 5A, deletion of sseJ led to a dramatic increase in the amount of Sif tubules formed per infected cell. An increased percentage of infected cells had Sifs, as well (Fig. 5B). This increase was observed by 6 h postinfection and maintained at later times. However, Sif formation by this mutant still underwent a decline after peaking at 10 h postinfection. These findings demonstrate that SseJ antagonizes Sif formation during the first 8 to 10 h postinfection and that it does not mediate the downregulation of the Sif phenotype at later times postinfection.
Downregulation of the Sif phenotype correlated temporally with depolymerization of actin around the SCV, which was previously examined by us and others (3, 24, 25). Depolymerization of this actin is largely dependent upon the actions of SpvB, an ADP-ribosylating enzyme encoded on the virulence plasmid of serovar Typhimurium (10, 22, 25, 29, 37). The SPI-2 effectors SspH2 and SseI also contribute to remodeling of actin on the SCV, though their contribution is more subtle (25). To test whether actin depolymerization events are involved in the downregulation of Sifs, we infected cells with an isogenic mutant of serovar Typhimurium lacking spvB. This mutant displayed a slightly higher number of Sifs at 8 to 10 h; however, the breakdown of Sifs in the mutant was not affected compared to that in the wild type (Fig. 5B). Nonetheless, this is the first evidence that SpvB is involved in the Sif phenotype. We also infected cells with a double mutant lacking both sseJ and spvB. As expected, these bacteria induced Sifs in a larger fraction of infected cells at 10 h postinfection in a synergistic manner than the sseJ and spvB mutants alone (Fig. 5B).
Conclusions.
We have shown that many conditions affect Sif formation in vitro, which may explain the variance of this phenotype among different investigators. Additionally, we report a number of novel observations relating to the Sif phenotype. First, Sif formation was found to correlate with cells infected with rapidly replicating bacteria. This observation is consistent with previous evidence suggesting that static (or dying) and rapidly growing populations of serovar Typhimurium exist in host cells (8). Second, bacterial numbers per infected cell do not appear to correlate directly with Sifs, as initial small numbers of intracellular bacteria (1-2) are sufficient to initiate Sif formation (see Fig. 2B). Also, the sseJ mutant, which is attenuated for intracellular replication (11, 31), can produce more Sifs than wild-type bacteria. Finally, we show that Sifs are a dynamic phenotype and are downregulated after 8 to 10 h in a manner that does not appear to require bacterial protein synthesis. While it remains unclear how the Sif phenotype benefits intracellular bacteria, this phenotype is tightly controlled by both positive (SifA, SseF, SseG, SopD2) (14, 36) and negative (SseJ and SpvB) (31) regulators. For future studies of the Sif phenotype, the double sseJ spvB mutant will provide a valuable reagent.
Acknowledgments
This work was supported by grant funding and a New Investigator Award from the Canadian Institutes of Health Research to J.H.B. Infrastructure for the Brumell laboratory was provided by a New Opportunities Fund from the Canadian Foundation for Innovation and the Ontario Innovation Trust. C.L.B. is the recipient of a University of Toronto Open Fellowship Award and a studentship from the Natural Sciences and Engineering Research Council of Canada.
We thank Michael Woodside for assistance with confocal microscopy and Brett Finlay, Nat Brown, Andrew Perrin, and members of the Brumell laboratory for helpful discussions and critical reading of the manuscript.
Editor: A. D. O'Brien
REFERENCES
- 1.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucrot, E., C. R. Beuzón, D. W. Holden, J.-P. Gorvel, and S. Méresse. 2003. Salmonella typhimurium SifA effector protein requires its membrane-anchoring C-terminal hexapeptide for its biological function. J. Biol. Chem. 278:14196-14202. [DOI] [PubMed] [Google Scholar]
- 3.Brumell, J. H., D. L. Goosney, and B. B. Finlay. 2002. SifA, a type III secreted effector of Salmonella typhimurium, directs Salmonella-induced filament (Sif) formation along microtubules. Traffic 3:407-415. [DOI] [PubMed] [Google Scholar]
- 4.Brumell, J. H., and S. Grinstein. 2004. Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 7:78-84. [DOI] [PubMed] [Google Scholar]
- 5.Brumell, J. H., S. Kujat-Choy, N. F. Brown, B. A. Vallance, L. A. Knodler, and B. B. Finlay. 2003. SopD2 is a novel type III secreted effector of Salmonella typhimurium that targets late endocytic compartments upon delivery into host cells. Traffic 4:36-48. [DOI] [PubMed] [Google Scholar]
- 6.Brumell, J. H., P. Tang, S. D. Mills, and B. B. Finlay. 2001. Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic 2:643-653. [DOI] [PubMed] [Google Scholar]
- 7.Brumell, J. H., P. Tang, M. L. Zaharik, and B. B. Finlay. 2002. Disruption of the Salmonella-containing vacuole leads to increased replication of Salmonella enterica serovar Typhimurium in the cytosol of epithelial cells. Infect. Immun. 70:3264-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchmeier, N. A., and S. J. Libby. 1997. Dynamics of growth and death within a Salmonella typhimurium population during infection of macrophages. Can. J. Microbiol. 43:29-34. [DOI] [PubMed] [Google Scholar]
- 9.Chakravortty, D., I. Hansen-Wester, and M. Hensel. 2002. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 195:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fierer, J., L. Eckmann, F. Fang, C. Pfeifer, B. B. Finlay, and D. Guiney. 1993. Expression of the Salmonella virulence plasmid gene spvB in cultured macrophages and nonphagocytic cells. Infect. Immun. 61:5231-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman, J. A., M. E. Ohl, and S. I. Miller. 2003. The Salmonella enterica serovar Typhimurium translocated effectors SseJ and SifB are targeted to the Salmonella-containing vacuole. Infect. Immun. 71:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freeman, J. A., C. Rappl, V. Kuhle, M. Hensel, and S. I. Miller. 2002. SpiC is required for translocation of Salmonella pathogenicity island 2 effectors and secretion of translocon proteins SseB and SseC. J. Bacteriol. 184:4971-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallois, A., J. R. Klein, L. A. Allen, B. D. Jones, and W. M. Nauseef. 2001. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J. Immunol. 166:5741-5748. [DOI] [PubMed] [Google Scholar]
- 14.Guy, R. L., L. A. Gonias, and M. A. Stein. 2000. Aggregation of host endosomes by Salmonella requires SPI2 translocation of SseFG and involves SpvR and the fms-aroE intragenic region. Mol. Microbiol. 37:1417-1435. [DOI] [PubMed] [Google Scholar]
- 15.Holden, D. W. 2002. Trafficking of the Salmonella vacuole in macrophages. Traffic 3:161-169. [DOI] [PubMed] [Google Scholar]
- 16.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Jiang, X., O. W. Rossanese, N. F. Brown, S. Kujat-Choy, J. E. Galan, B. B. Finlay, and J. H. Brumell. 2004. The related effector proteins SopD and SopD2 from Salmonella enterica serovar Typhimurium contribute to virulence during systemic infection of mice. Mol Microbiol. 54:1186-1198. [DOI] [PubMed] [Google Scholar]
- 17.Knodler, L. A., J. Celli, W. D. Hardt, B. A. Vallance, C. Yip, and B. B. Finlay. 2002. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol. Microbiol. 43:1089-1103. [DOI] [PubMed] [Google Scholar]
- 18.Knodler, L. A., and O. Steele-Mortimer. 2003. Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4:587-599. [DOI] [PubMed] [Google Scholar]
- 19.Knodler, L. A., B. A. Vallance, M. Hensel, D. Jackel, B. B. Finlay, and O. Steele-Mortimer. 2003. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol. Microbiol. 49:685-704. [DOI] [PubMed] [Google Scholar]
- 20.Kuhle, V., and M. Hensel. 2002. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell. Microbiol. 4:813-824. [DOI] [PubMed] [Google Scholar]
- 21.Kuhle, V., D. Jackel, and M. Hensel. 2004. Effector proteins encoded by Salmonella pathogenicity island 2 interfere with the microtubule cytoskeleton after translocation into host cells. Traffic 5:356-370. [DOI] [PubMed] [Google Scholar]
- 22.Lesnick, M. L., N. E. Reiner, J. Fierer, and D. G. Guiney. 2001. The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Mol. Microbiol. 39:1464-1470. [DOI] [PubMed] [Google Scholar]
- 23.Mastroeni, P., and M. Sheppard. 2004. Salmonella infections in the mouse model: host resistance factors and in vivo dynamics of bacterial spread and distribution in the tissues. Microbes Infect. 6:398-405. [DOI] [PubMed] [Google Scholar]
- 24.Méresse, S., K. E. Unsworth, A. Habermann, G. Griffiths, F. Fang, M. J. Martinez-Lorenzo, S. R. Waterman, J.-P. Gorvel, and D. W. Holden. 2001. Remodelling of the actin cytoskeleton is essential for replication of intravacuolar Salmonella. Cell. Microbiol. 3:567-577. [DOI] [PubMed] [Google Scholar]
- 25.Miao, E. A., M. Brittnacher, A. Haraga, R. L. Jeng, M. D. Welch, and S. I. Miller. 2003. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol. Microbiol. 48:401-415. [DOI] [PubMed] [Google Scholar]
- 26.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 28.Mills, S. D., S. R. Ruschkowski, M. A. Stein, and B. B. Finlay. 1998. Trafficking of porin-deficient Salmonella typhimurium mutants inside HeLa cells: ompR and envZ mutants are defective for the formation of Salmonella-induced filaments. Infect. Immun. 66:1806-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otto, H., D. Tezcan-Merdol, R. Girisch, F. Haag, M. Rhen, and F. Koch-Nolte. 2000. The spvB gene-product of the Salmonella enterica virulence plasmid is a mono(ADP-ribosyl)transferase. Mol. Microbiol. 37:1106-1115. [DOI] [PubMed] [Google Scholar]
- 30.Richter-Dahlfors, A., A. M. J. Buchan, and B. B. Finlay. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz-Albert, J., X. J. Yu, C. R. Beuzón, A. N. Blakey, E. E. Galyov, and D. W. Holden. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 32.Salcedo, S. P., M. Noursadeghi, J. Cohen, and D. W. Holden. 2001. Intracellular replication of Salmonella typhimurium strains in specific subsets of splenic macrophages in vivo. Cell. Microbiol. 3:587-597. [DOI] [PubMed] [Google Scholar]
- 33.Sheppard, M., C. Webb, F. Heath, V. Mallows, R. Emilianus, D. Maskell, and P. Mastroeni. 2003. Dynamics of bacterial growth and distribution within the liver during Salmonella infection. Cell. Microbiol. 5:593-600. [DOI] [PubMed] [Google Scholar]
- 34.Spadari, S., G. Pedrali-Noy, M. C. Falaschi, and G. Ciarrocchi. 1984. Control of DNA replication and cell proliferation in eukaryotes by aphidicolin. Toxicol. Pathol. 12:143-148. [DOI] [PubMed] [Google Scholar]
- 35.Steele-Mortimer, O., S. Méresse, J.-P. Gorvel, B.-H. Toh, and B. B. Finlay. 1999. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell. Microbiol. 1:33-51. [DOI] [PubMed] [Google Scholar]
- 36.Stein, M. A., K. Y. Leung, M. Zwick, F. Garcia-del Portillo, and B. B. Finlay. 1996. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 20:151-164. [DOI] [PubMed] [Google Scholar]
- 37.Tezcan-Merdol, D., T. Nyman, U. Lindberg, F. Haag, F. Koch-Nolte, and M. Rhen. 2001. Actin is ADP-ribosylated by the Salmonella enterica virulence-associated protein SpvB. Mol. Microbiol. 39:606-619. [DOI] [PubMed] [Google Scholar]
- 38.Tsolis, R. M., R. A. Kingsley, S. M. Townsend, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Of mice, calves, and men. Comparison of the mouse typhoid model with other Salmonella infections. Adv. Exp. Med. Biol. 473:261-274. [PubMed] [Google Scholar]
- 39.Vázquez-Torres, A., G. Fantuzzi, C. K. Edwards III, C. A. Dinarello, and F. C. Fang. 2001. Defective localization of the NADPH phagocyte oxidase to Salmonella-containing phagosomes in tumor necrosis factor p55 receptor-deficient macrophages. Proc. Natl. Acad. Sci. USA 98:2561-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vázquez-Torres, A., Y. Xu, J. Jones-Carson, D. W. Holden, S. M. Lucia, M. C. Dinauer, P. Mastroeni, and F. C. Fang. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655-1658. [DOI] [PubMed] [Google Scholar]