Summary
Both the promises and pitfalls of the cell reprogramming research platform rest on human genetic variation, making the measurement of its impact one of the most urgent issues in the field. Harnessing large transcriptomics datasets of induced pluripotent stem cells (iPSC), we investigate the implications of this variability for iPSC-based disease modeling. In particular, we show that the widespread use of more than one clone per individual in combination with current analytical practices is detrimental to the robustness of the findings. We then proceed to identify methods to address this challenge and leverage multiple clones per individual. Finally, we evaluate the specificity and sensitivity of different sample sizes and experimental designs, presenting computational tools for power analysis. These findings and tools reframe the nature of replicates used in disease modeling and provide important resources for the design, analysis, and interpretation of iPSC-based studies.
Keywords: induced pluripotent stem cells, iPSC, disease modeling, human genetic variation, transcriptomics, power analysis, sample size, mixed models, spurious
Highlights
-
•
Differences between individuals dominate transcriptional variance
-
•
Using multiple clones per individual can be detrimental to differential analysis
-
•
We provide resources for the design, analysis, and interpretation of iPSC studies
Germain and Testa harness large public induced pluripotent stem cells (iPSC) transcriptomic datasets to assess the impact of human variability on iPSC-based disease modeling. They evaluate different experimental designs, showing among other things that using multiple clones per individual can be detrimental to the robustness of the results, establishing standards and informing decisions on the design of iPSC-based studies.
Introduction
Induced pluripotent stem cells (iPSCs), and more broadly the technologies emerging around cell reprogramming, have developed rapidly and are already transforming biomedical research. The defining, paradigm-shifting innovation of the iPSC-based research platform is the possibility of repeating development, which should be understood not merely as the in vitro reproduction of developmental processes, but as the feat of making it repeatable, i.e., transforming hitherto unique organismal processes—the development of a given individual—into something experimentally and statistically tractable. As such, cell reprogramming has made human genetic variability amenable to experimentation. Indeed, as was emphasized early on (Colman, 2008), a key asset of iPSCs is that, contrary to embryonic stem cells (ESCs), they are associated with medical histories and thus pave the way to what we previously described as the functional annotation of human genomes (Adamo et al., 2015), in terms of genotype-phenotype correlation and disease impact.
Yet at the same time the extent of human variability, of which we are only beginning to take the full measure, poses an important practical problem for the field of iPSC-based modeling. Given background genetic differences between individuals, the key question is which experimental designs and degrees of replication are necessary for robust and sensitive results, and what are the best measures to guide their selection. Here, we address such questions by harnessing the large datasets recently made available by the Human Induced Pluripotent Stem Cell Initiative (HipSci; Streeter et al., 2016, Kilpinen et al., 2016) and the National Heart, Lung, Blood Institute (NHLBI) NextGen consortium (Carcamo-Orive et al., 2016). We derive empirically grounded methodological recommendations for the design and analysis of iPSC-based studies.
Given the accessibility of expression profiling and the critical importance of gene expression for the regulation of cellular identity and function, transcriptomic dysregulations have emerged for their rapid and informative insight into the molecular underpinning of diseases, representing for many a first high-content in vitro phenotype. This is especially true for many diseases such as neurodevelopmental disorders, for which the iPSC technology is of prime relevance due to the inaccessibility of the relevant tissues, and whose main disease-associated genes are strongly enriched for transcription factors and chromatin regulators (De Rubeis et al., 2014). Transcription is therefore of key importance in itself, as well as representing a powerful stand-in for other, more directly functional assays.
Results
Differences between Individuals Dominate the iPSC Transcriptional Variance
In addition to the condition under study, transcriptomes from iPSC lines can vary because of several factors that can coarsely be boxed into genetic, epigenetic, and technical. Although skin biopsies harbor somatic mutations (Young et al., 2012) and a minority of epigenetic marks can be retained upon reprogramming (Kim et al., 2010), the majority of differences between individuals are most likely due to genetic variation. Conversely, while different iPSC clones from the same individual tend to have minor genetic differences chiefly due to mosaicism in the initial sample (Young et al., 2012, Ji et al., 2012) and ensuing selection (Kilpinen et al., 2016), most of the variability is likely related to differences in the epigenetic state of the cells (Salomonis et al., 2016). Finally, differences between RNA-sequencing (RNA-seq) libraries of the same clone are most likely to capture essentially technical sources of variation. As previously reported (Kilpinen et al., 2016, Carcamo-Orive et al., 2016), ANOVA reveals that transcriptomic variance is dominated by genetic differences, or at any rate differences between donors' specimens, which holds not only for the whole transcriptome but also for the majority of genes taken individually (Figure S1). The proportion of variance explained by differences between individuals increased with expression levels (Figures S1B and S1D), suggesting that genes not showing high susceptibility to genetic differences due to their low expression in iPSC are likely to do so in other tissues.
Spurious Differential Expression Is Exacerbated by Including Multiple iPSC Clones per Individual
Given the very large number of genes showing individual-dependent differential expression, the comparison of unrelated individuals—as in the traditional and currently most prevalent design of iPSC-based modeling studies—will inevitably lead to the identification of differentially expressed genes (DEGs) that are only spuriously associated to the comparison of interest. This risk is further compounded by the fact that, very often, in-house controls or standard ESC lines are compared with samples received from collaborating centers, introducing a further confounding factor for the conditions under study.
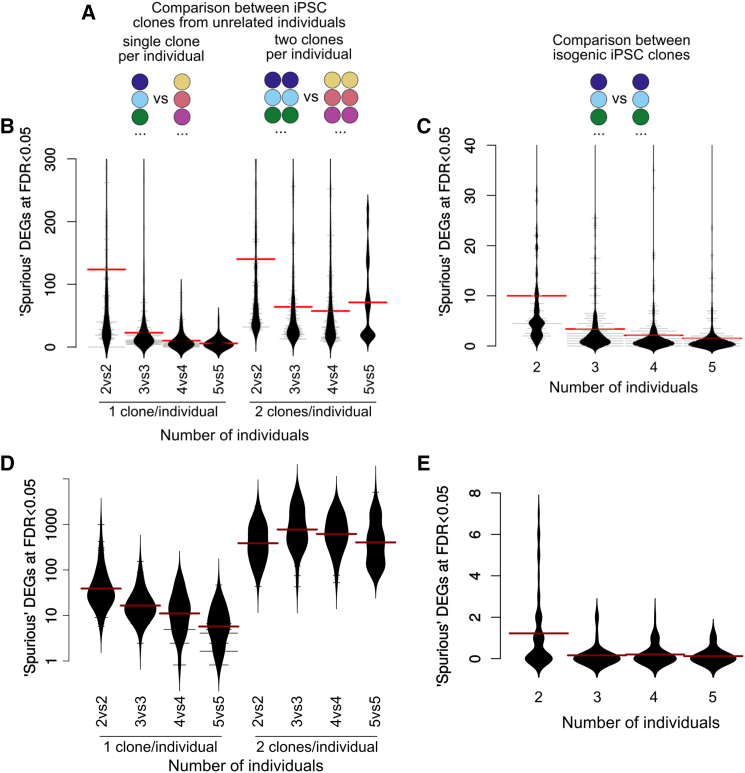
To assess the risk of such spurious results depending on different experimental designs, we performed a series of permutation analyses on both iPSC datasets. We performed more than 15,000 differential expression analyses (DEAs) between random sex-balanced groups of varying sizes and designs (illustrated in Figure 1A). In particular, we tested three types of experimental design: (1) comparisons between unrelated individuals using a single clone per individual, (2) comparison between unrelated individuals using two clones per individual, and (3) comparison between a group of individuals and different clones from the same individuals.
Figure 1.
Spurious Differential Expression between Groups of Random Individuals
Scheme of the types of permutations (A), below which are shown the corresponding distributions of the number of spurious DEGs in the HipSci (B and C; 500 permutations each) and NHLBI/GSE79636 (D and E; 50 permutations each) datasets. For reasons of visibility, the y axis of (D) is on a log scale. Red bars indicate the mean of each distribution. See also Figures S1–S4.
Random comparisons showed a surprisingly high frequency of “spurious” differentially expressed genes (Figures 1B and 1D). In contrast, basic measurements on cell morphology, which are available for a small subset of the HipSci samples (28 clones from 14 individuals), did not show such a dominant dependence on genetic differences (Figure S2B), although this can reflect the technical or intrinsic variability of the measurements (Figure S2A).
The fact that the comparisons between isogenic clones (Figures 1C and 1E) show very little spurious DEGs even at small sample sizes indicates that the much higher spurious results in the other two comparisons are related to genetic differences. In addition, it warrants the use of controls established by genetic correction of the patient-derived cells when this is cost-effectively commensurate to the specific objective.
Most notably, using more than one iPSC clone per individual results in a major increase in spurious DEGs (Figures 1B and 1D). Furthermore, and in contrast to comparisons involving single clones which show a clear downward trend upon increasing sample size, such a trend appears absent from the comparisons with two clones per individual. As expected given the much higher genetic heterogeneity of the NHLBI dataset (Figure S3), the distribution of “spurious” DEGs was considerably higher (Figure 1D). Nevertheless, we could observe the same pattern as in the HipSci dataset, albeit stronger, when using one versus two clones per individual. Importantly, when manually adding “true” differential expression (see below), using two clones per individual resulted in a dramatic increase of the false discovery rate (FDR) (Figure S4). Together, these results call into question the widespread habit (also entrenched thus far in the guidelines of this journal) of including more than one iPSC clone per patient.
This finding, however, should not come as a surprise, for the statistical methods commonly used in gene expression analysis assume that replicates are independent. For instance, the edgeR manual recommends summing the read counts of technical replicates before DEA instead of treating them as bona fide replicates. The reason is simple: treating technical replicates as independent artificially reduces the variability of the group's mean. The same applies to treating iPSC clones from the same individual as independent when they are clearly not (at least in terms of the overriding contribution of genetic makeup to the variance under study). While multiple clones per individual improve the internal robustness of the results, their inclusion hampers their generalizability to other individuals, unless appropriate methods are employed (see below).
Genes Recurrently Found Differentially Expressed across Random Individuals Are Related to Fundamental Developmental Processes
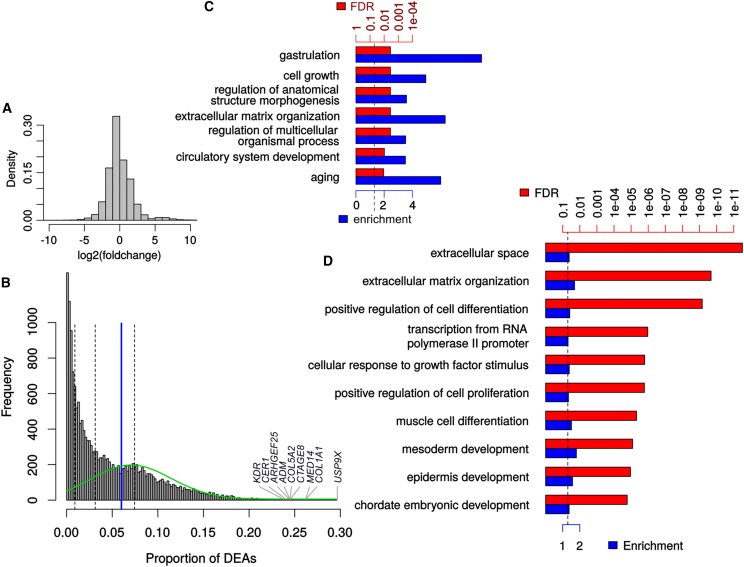
Importantly, the spurious DEGs are not limited to modest fold changes, and nearly half (48%) of them change by a factor of more than 2 (Figure 2A), indicating that a simple fold-change threshold cannot adequately address the problem. Some genes are found differentially expressed in the permutation DEAs much more often than others—in fact 98 genes are differentially expressed in more than 20% of the comparisons (Figure 2B)—which prompted us to investigate the nature of these recurrent DEGs in the HipSci dataset. Surprisingly, the top 100 DEGs show very strong enrichment for genes associated with both early developmental processes (nearly 9-fold enrichment for gastrulation-related genes) and aging (6-fold) (Figure 2C). In addition, and as previously reported (Adamo et al., 2015), genes related to the extracellular matrix (ECM) organization were also strongly enriched. The same enrichment for ECM-related genes appears in the larger set of all recurrent DEGs (Figure 2D), as well early developmental processes. Since the background used for the enrichment analysis excludes genes that are not expressed in iPSC, we can exclude that these enrichments are merely due to the pluripotent state, and instead must conclude that they reveal instead an unexpected degree of genetic variability in the expression of these genes, possibly linked to an evolutionarily selected increased robustness of the system to their variation. This result is in line with the enrichment, among iPSC-specific expression quantitative trait loci, for targets of pluripotency factors (Kilpinen et al., 2016).
Figure 2.
Description of the Genes Recurrently Found Differentially Expressed across iPSC from Groups of Random Individuals
(A) Distribution of fold changes across spurious DEGs of all HipSci permutations.
(B) Distribution of the proportion at which each gene appears differentially expressed. Dashed lines delineate the quartiles while the blue line indicates the threshold for “recurrent genes.” The green line represents the distribution of particularly recurrent genes found by expectation maximization-based mixture modeling.
(C) Most specific enriched gene ontology terms among the top 100 most recurrent genes. FDR, false discovery rate.
(D) Top most specific enriched gene ontology terms among the recurrent genes (>6% chance being found differentially expressed).
In (C) and (D) FDR is shown in red and fold enrichment in blue. See also Figure S5.
Despite the significance of these enrichments, it would be still theoretically possible that these genes are largely irrelevant to the fully formed organism. To assess this possibility, we tested whether the recurrent DEGs as well as the top 100 recurrent ones were enriched for genes found to be homozygously knocked out in healthy individuals (Sulem et al., 2015), but could find no significant relationship (Figure S5A). Nor was there any significant relationship between the frequency of a gene being differentially expressed across permutations and the prevalence of CNVs affecting that gene in the general population based on Exome Aggregation Consortium (ExAC; Kosmicki et al., 2017) data (Figures S5B and S5C).
Thus, the fact that some genes are recurrently found differentially expressed between random individuals does not necessarily mean that they are medically irrelevant. Indeed, pathogenic mechanisms might well be related to similar but subclinical phenotypic variations in healthy individuals. An interesting example is the recent finding that variation in the GTF2I gene, a transcription factor hemizygously deleted in Williams-Beuren syndrome and associated with its sociocognitive phenotype, is associated with anxiety in the general population (Jabbi et al., 2015). Therefore, we estimated the likelihood of recurrent genes being disease-significant by looking at their overlap with known disease genes from the database of Online Mendelian Inheritance in Man (OMIM). While OMIM genes were more likely to be differentially expressed between random individuals (p ∼ 3 × 10−16 by Mann-Whitney test), the enrichment was very small (Figure S5A). Together, these results suggest that genes recurrently found differentially expressed across individuals are neither depleted nor considerably enriched for genes more likely to be medically relevant.
Sensitivity across Experimental Designs and Sample Sizes
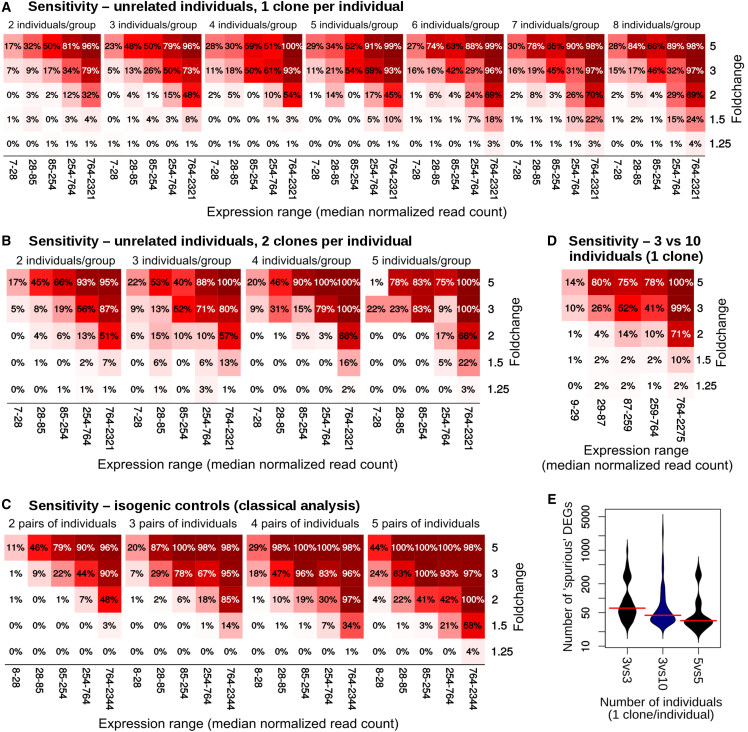
We next assayed to what extent different experimental designs and sample sizes could detect input differential expression. To this end we repeated the permutation analysis, each time introducing a total of 100 DEGs at five different fold changes, and distributed across different expression levels (see Experimental Procedures). Notably, the two datasets showed large differences in overall sensitivity (Figures 3 and S6), possibly owing in part to differences in coverage and different degrees of technical standardization.
Figure 3.
Sensitivity of Different Experimental Designs across Fold Change and Expression of the DEGs in the HipSci Dataset
(A–C) Using a single clone per individual (A), using two clones per individual (B), and comparing isogenic clones (C). Each square represents the average across 300 permutations.
(D) Sensitivity when comparing a small cohort with a large set of unrelated controls.
(E) Distribution of false positives when comparing a small cohort with a large set of unrelated controls.
See also Figure S6.
When comparing single clones from unrelated individuals, the sensitivity seemed to largely plateau after six individuals per group (Figure 3A). Using two clones per individual resulted in an increase in sensitivity, albeit at the cost of a massive loss in specificity, as shown above (Figures 1 and S4). Isogenic controls also showed a marked improvement in sensitivity in one of the datasets (Figure 3C).
In all cases, the sensitivity was quite good for high fold changes, but rapidly decreased with fold change and read count. Fold changes of 1.5, which are especially relevant in the context of gene duplications, were particularly difficult to detect, and most of them are unlikely to be detected unless the genes are very stable or highly expressed. This is particularly relevant given the importance of copy-number alterations for a variety of diseases (McCarroll and Altshuler, 2007, Cook and Scherer, 2008, Luo et al., 2012).
Finally, since it is relatively common for laboratories specialized in cell reprogramming to have assembled banks of control iPSC lines against which disease-specific lines can be compared, we also evaluated the sensitivity of designs comparing only three patient-specific lines with a larger set of controls (n = 10). While such design provided fair sensitivity (Figure 3D), it could not achieve the same degree of spurious DEG minimization as more balanced groups (Figure 3E).
A good experimental design should optimize both sensitivity and specificity. While the ideal tradeoff between the two ultimately depends on the context, specificity (type I error) is most often considered at least as important, if not more so, than sensitivity (type II error). Therefore, while using multiple clones per individual increased sensitivity (albeit not necessarily more so than using more individuals), it did so at a much larger cost in specificity. This favors increasing the number of individuals instead of the number of clones per individual. Whenever this is not possible, or when the data have already been generated, analysis methods ought to be used that take into account the samples' interdependence.
Comparison of Methods for Dealing with Multiple Clones
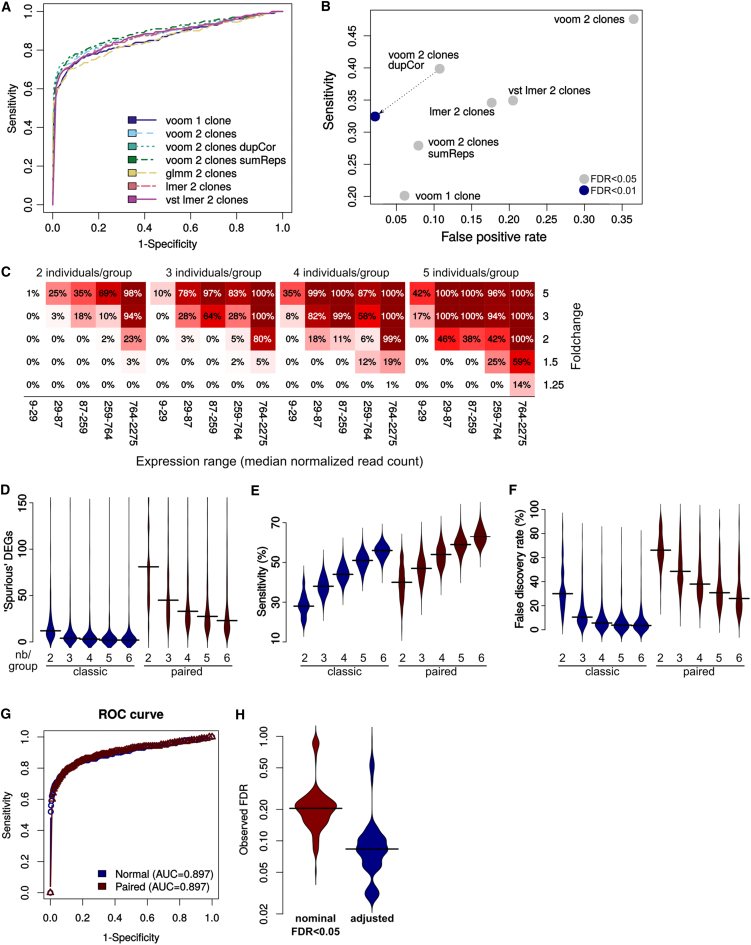
Although multiple iPSC clones per individual cannot be considered full replicates, they nevertheless provide useful information on some aspects of the variability. We therefore tested whether more appropriate statistical methods, in particular approaches based on mixed models and treating the individual as a random-effect variable, could harness the availability of additional clones while keeping good control of false positives. Since such modeling methods are not typically implemented in RNA-seq analysis packages, we tested alternative methods of applying or approximating them in this context (see Experimental Procedures), and compared their ability to distinguish “true” inputted DEGs from “spurious” ones (Figures 4A and 4B). The first implementation relies on the duplicateCorrelation function of the limma R package (Law et al., 2014), which approximates mixed models, in combination with a voom-based analysis (Ritchie et al., 2015). This was compared with a standard voom-based analysis of one or two clones per individual, as well as to a voom-based analysis previously summing the clones of each individual (sumReps), as recommended for technical replicates. Finally, we also tried alternative implementations of mixed models, in particular: (1) a glmm-based application of mixed models using the quasi-Poisson distribution (the negative binomial not being implemented in the glmmML package) on normalized counts; (2) a lme4-based application of mixed models on voom-transformed normalized counts; and (3) a lme4-based application of mixed models on DESeq2's variance-stabilizing transformation (Love et al., 2014). As expected, using one clone per patient resulted in the smallest spurious/FDR at a nominal FDR < 0.05, which was instead very high when using two clones per patient without special treatment. This effect could be mitigated to different extents, and with different impacts on sensitivity, by the different approaches. In particular, limma's duplicateCorrelation appeared to offer the best performance at nominal FDR < 0.05 (Figure 4B) and the best receiver-operating characteristic (ROC) curve at high specificity (Figure 4A), followed by the sum of replicates. While none of these approaches yielded a specificity equivalent to the use of single clones per individual at nominal FDR < 0.05, the last two came very close, enabling a considerable gain in sensitivity and having a superior net area under the ROC curve, indicating that they are advisable, especially with a slightly more stringent threshold. Indeed, using limma's duplicateCorrelation approach with an FDR threshold of 0.01 retained much of the increase in sensitivity of using two clones with a specificity superior to using a single clone (Figures 4B and 4C). We therefore recommend this approach, i.e., limma's duplicateCorrelation with a slightly more stringent significance threshold, for analyses of datasets involving more than one clone per individual.
Figure 4.
Comparison of Analysis Methods
(A and B) Sensitivity and specificity of analysis methods when dealing with multiple clones per patient (three individuals per group). Shown are the averages of 30 permutations for each method. The glmm-based method is not shown in (B) because it led to no positive at this FDR threshold.
(C) Sensitivity when using the dupCor approach and an FDR < 0.01 threshold.
(D–G) Comparison of edgeR's classic (exact test) and paired analysis (GLM-based) for paired experimental designs. At a q < 0.05 threshold, paired analysis leads to a major increase in “spurious” DEGs (D) for a very modest increase in sensitivity (E), resulting in a massive increase in the FDR (F) but little impact on the ROC curve (G).
(H) Adjusting the FDR of differentially expressed genes on the basis of their frequency in the permutations results in a considerable improvement in the FDR (based on 300 permutations of comparisons between two groups of four individuals, using two clones per individual, HipSci dataset).
Paired Analysis in the Context of Isogenic Controls Results in Increased False Discoveries
Popular differential expression methods, such as edgeR (Robinson et al., 2010), DESeq2 (Anders and Huber, 2010, Love et al., 2014), and limma/voom (Law et al., 2014, Ritchie et al., 2015), implement generalized linear models to provide the possibility to analyze more complex experimental designs than a binary comparison between two groups. In the context of studies involving isogenic or half-matched controls (i.e., unaffected sibling or parent), this permits comparison of each patient-derived line with its matched control and the gauging of consistent relative changes (i.e., ∼background + condition). Such a paired analysis naturally increases sensitivity, but at the cost of increasing the degrees of freedom of the tested model. We therefore compared the sensitivity and occurrence of spurious DEGs in paired versus classical analysis (Figures 4D–4G). Of note, while paired analysis did indeed result in an increase in sensitivity (Figure 4E), it also massively increased the detection of spurious DEGs (Figure 4D), leading to an important net increase in the FDR (Figure 4F). This, however, had no noticeable impact on the ROC curve (Figure 4G), indicating that the paired analysis can be used provided that an accordingly more stringent FDR threshold is adopted.
Knowledge of Variable Genes Can Improve the False Discovery Rate
Since genes are variously likely to be found differentially expressed across random groups of individuals, we assessed whether the knowledge of this probability for each gene could help adjust the FDR of differential expression analysis. Using a very simple approach based on the frequency of the gene and fold change among spurious differential expression across permutations (see Experimental Procedures), we adjusted the nominal FDR of the differential expression analysis on the basis of the results of our permutation analysis, which resulted in a considerable improvement of the effective FDR (Figure 4H). This approach thus offers a proof of concept of the utility of such large datasets in the interpretation of iPSC-based studies, paving the way to more complex approaches, for instance involving hypothesis weighting or Bayesian updating, which can probably make even greater use of this information on variable genes. This being said, such corrections should be interpreted with care given that, as emphasized earlier, it could well be that genes whose expression is highly variable in the general population are nevertheless associated with the conditions under study or related subclinical traits.
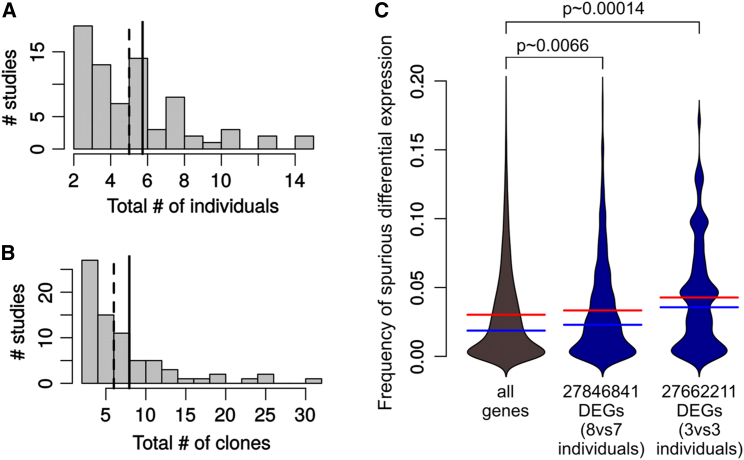
A Considerable Proportion of Published iPSC-Based Disease-Modeling Studies Has Insufficient Precision
To assess the impact of our observations for the iPSC modeling field, we sampled the most recent iPSC disease-modeling literature, selecting 77 studies published in 2016 (see Experimental Procedures and Table S1). The vast majority (79%) of the studies relied on unrelated controls, with 10% using a combination of unrelated and half-matched controls and 10% using isogenic controls. The distribution of the total number of individuals (all conditions pooled) from which the iPSC lines were derived and the total number of iPSC clones used are plotted in Figures 5A and 5B. Importantly, 41% of the studies used more than one clone per individual (the proportion increases to 51% in studies involving at least six individuals in total) without adopting appropriate statistical methods, which as we have shown can artificially inflate significance and lead to many false positives.
Figure 5.
Survey of the 2016 iPSC Disease-Modeling Studies
(A) Total number of individuals (all conditions pooled) from which the studied iPSC lines were derived.
(B) Total number of iPSC clones studied (all conditions pooled). The dashed lines in (A) and (B) indicate the median and the solid lines indicate the mean.
(C) Distribution of the probability of genes to be found differentially expressed in random permutations, comparing all genes (left) with the DEGs of two sample iPSC-based studies from our survey (right). The red line indicates the mean and the blue line indicates the median. The p values were calculated from a Mann-Whitney test comparing the distributions.
As an example, we took two studies focusing on the transcriptome as relevant endophenotype and that reported lists of DEGs (several studies involving transcriptomics did not report such a list, and some did not even make the data available). The first (PubMed ID 27846841) involved a rather large number of individuals per group (eight versus seven), while the second (PubMed ID 27662211) included a fairly small number of individuals (three per group). Despite the difference in cell type, the genes reported as differentially expressed by these studies had a significantly higher probability of being differentially expressed in our random permutations (Figure 5C), particularly for the study involving a small sample size, which questions the specificity of at least a subset of these genes for the disease under study.
This being said, it must be emphasized that many of the studies reviewed were not exclusively based on (and in some cases not even involving) transcriptional phenotypes. In light of some of the results described here (Figure S2) as well as evolutionary considerations, it is possible that lower-content assays measuring, for instance, cellular phenotypes could show a lower rate of spurious results due to genetic differences. Moreover, the combination of independent lines of evidence can often strengthen results obtained from weak (from an intrinsic statistical point of view) studies. Nevertheless, the present results indicate that a considerable proportion of iPSC-based disease-modeling studies, especially when based on next-generation sequencing experiments, may present a high rate of false positives and (unless one focuses on very highly expressed genes and/or fairly large fold changes, i.e., >2) be underpowered.
An R Package to Estimate the Power of iPSC Study Designs
Lastly, we provide the R package iPSCpoweR, available on github, to reproduce all of the analyses performed in the present study (including multi-threaded DEA permutations, plots, etc.) and conduct further power analysis. We hope that this resource can help scientists in the field to make more informed decisions regarding the design of future iPSC-based studies.
Discussion
The predominance of inter-individual differences in explaining transcriptional variability has already been reported (Rouhani et al., 2014, Burrows et al., 2016, Carcamo-Orive et al., 2016). These datasets are likely to underestimate human genetic variability due to the populations sampled (Figure S3); in particular, all individuals from which the HipSci lines used were derived are described by the consortium as “white,” and most of them are labeled as being of English origin. The predominance of inter-individual differences is in line with the surprising degree of genetic variation in human gene expression regulation (Kasowski et al., 2013, Melé et al., 2015, Barrera et al., 2016). Indeed, Barrera et al. (2016) showed that the median human genome harbors 60 heterozygous and 20 homozygous missense SNPs that change the amino acid sequence of transcription factor (TF) DNA binding domains, resulting in changes in affinity and/or specificity in at least 75% of the cases. Together with variations in TF target sites and other changes affecting DNA conformation, these can lead to wide differences in gene expression.
While the present results were obtained from the transcriptome of pluripotent cells, the impact is unlikely to be smaller in differentiated cell types. Indeed, Banovich et al. (2016) recently reported that regulatory variation between individuals is lower in iPSCs than in two differentiated cell types. Furthermore, given the difficulty in obtaining samples from patients harboring rare mutations, it is not uncommon for probands' and control lines to have different origins, and hence potentially confounding background genomic differences. It is therefore likely that the “spurious” differences observed here in the permutation DEAs are relatively conservative estimates. On the other hand, while the present study aimed at measuring the impact of spurious differential expression, it is possible that spurious differences passing multiple testing correction in this context, namely with little real differences between groups, might not necessarily pass it if the two groups show overriding transcriptional differences related to the condition of interest. This is due to the fact that popular correction methods (e.g., Benjamini-Hochberg) are rank based; hence the larger the transcriptional effect of the studied condition, the smaller the number of spurious DEGs will be. Finally, for specific cellular or functional assays with a proven more direct relationship to the ultimate traits under fitness selection, it is plausible that compared with transcription they are more robust and less influenced by genetic variation.
A key observation of the present study is that, contrary to a widespread practice, using multiple clones per individual can be very detrimental to the robustness of the study unless appropriate statistical methods are adopted. This represents a critical warning for the iPSC modeling field, since most leading studies, including our own work (Adamo et al., 2015), in a large number of publications have embraced the thus far current standard of including multiple clones per individual without accounting for their interdependence in the statistical analysis. One way of doing so, harnessing the additional power of multiple clones while keeping a good control of false positives, was through the combination of the voom and duplicateCorrelation functions implemented in the limma package (Law et al., 2014, Ritchie et al., 2015) used with a slightly more stringent significance threshold (here FDR < 0.01). This approach dramatically reduces the rate of false or spurious discovery, making the design equivalent to an isogenic experimental design, albeit with a much larger total number of clones profiled.
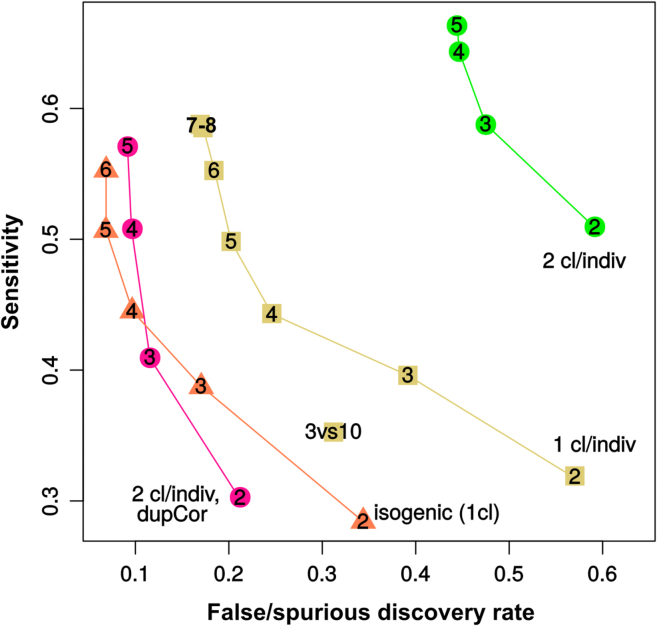
On the basis of our results (summarized in Figure 6), we propose the following recommendations. Isogenic controls corrected for the mutation, which we modeled here using different clones from the same individual, represent an ideal albeit cost- and time-intensive experimental design. Using two clones per individual with the aforementioned mixed-models approach offered a comparable performance. In both these cases, using at least three individuals per group (ideally four or more) offered a decent control of false positives, with additional individuals offering increases in sensitivity. When single clones from unrelated individuals are used, a strict minimum of four individuals per group should be used. In general (and to the extent that such comparisons were made possible by the current datasets), more than six to seven individuals per group offered only marginal improvements. Finally, the use of multiple clones per individual without adequate statistical treatment of their interdependence is strongly discouraged. Just like technical replicates, different clones from the same individuals capture only a portion of the biological variability, and treating them as statistically independent artificially reduces the variability of the group's mean.
Figure 6.
Summary of the Precision and Sensitivity of the Main Experimental and Computational Designs
The numbers in the data points represent the number of individuals per group. Triangles indicate isogenic lines, squares indicate unrelated individuals with one clone per individual, and circles indicate unrelated individuals with two clones per individual, with standard (green) or mixed-models (pink) analysis (through limma's duplicateCorrelation). With a standard analysis, using two clones per individual has a catastrophic impact on the FDR. Using mixed models, however, dramatically improves the FDR, making the design equivalent (albeit with a much larger total number of clones profiled) to an isogenic experimental design.
Part of the problem therefore lies in deep ambiguities regarding the meaning of technical and biological replicates. Indeed, whether a replicate is biological or technical has become an increasingly vague notion that thrives in the diversity of possible answers, and partly for good reasons: there are not two but diverse possible degrees of replication, each capturing different layers of variability. Moreover, as biological processes have gradually been mastered in the laboratory, they have increasingly ceased to be in themselves objects of investigation and have joined the scientist's toolkit, so that in many contexts biological processes have become purely technical operations. In the language of the historian of science Hans-Jörg Rheinberger, they went from “epistemic things” to “technical objects” (Rheinberger, 1997). It might therefore be useful to replace the distinction between technical and biological replicates for one inspired by these categories. In this view, technical replicates are replicates capturing the variability of (all or some of) the processes through which we study a phenomenon, whereas epistemic replicates capture, in addition to this technical variability, the variability in the phenomenon itself under study. The concepts are therefore relative to the questions being asked. Insofar as the aim of a study is to uncover disease-relevant effects of, say, a pathogenic mutation or copy-number alteration, epistemic replicates should capture the variability of the patients harboring the disease, and hence iPSC clones represent technical replicates. If, instead, the aim is to learn about the effects a given genetic lesion against a specific background genome, or about the impact of reprogramming-induced changes on the expressivity of that lesion, iPSC clones represent epistemic replicates.
More clarity in choosing and reporting the type and degree of replication would greatly help to improve the robustness of findings in iPSC-based studies.
Experimental Procedures
Samples Used
The description of the HipSci samples used can be obtained using the getSamplesInfo function of the package. A summary of the samples is shown in Figure S3, and the SRA-run IDs are listed in Table S2. All the samples not cultivated on feeders, not associated to a disease, and available on open access at the time of this study were used (a total of 62 lines from 47 individuals, including 15 individuals with two clones).
From the NHLBI NextGen consortium dataset, we included only lines that have been reprogrammed from the same type of cells (Erythroblasts) using the same rna method (Life-Tech GITC) and cultured in the same Matrigel preparation (a total of 107 lines from 37 individuals were used). These are listed in Table S2, and are available (both counts and annotation) in the R package (data(“GSE79636”)).
RNA-Seq Quantification
For the HipSci data, RNA-seq quantification was performed using Salmon v6.1 (Patro et al., 2016), using FMD indexes and the Refseq transcript annotation. The expression matrix is available through the getTxExpr and getGeneExpr functions of the package. For all analyses, the TMM normalization was used (Robinson and Oshlack, 2010). For the NHLBI NextGen data, we used the quantification made available by the authors on the GEO (GEO: GSE79636).
ANOVA
The analysis of transcriptional variance (Figure S1) was performed on log-transformed transcripts per million (TPM) values aggregated at the gene level. Both linear models (Figures S1A and S1B) and mixed random-effect models implemented by the lme4 R package (Figures S1C–S1E) were used. The analysis can be reproduced using the transcriptionalVarianceExplained function of the package.
For the ANOVA in cell morphology, we used only the data collected on the middle concentration of fibronectin. The data can be accessed from the cellpheno data object of the package, and the ANOVA can be reproduced using the cellphenoVarianceExplained function.
Differential Expression Analysis
With the exception of the mixed-models approaches (Figures 4A, 4B, and 6), all differential expression analysis was performed with edgeR v.3.12.1, which has among the most robust performances (Germain et al., 2016). For testing, only genes for which at least two samples had more than ten reads were considered. For the paired analysis, generalized linear models and the likelihood-ratio test was used; otherwise the exact test was used. See the edgeRwrapper function for the exact code.
In the comparison of approaches to dealing with multiple clones per individual (Figures 4A and 4B, pink data points in Figure 6), we used limma v.3.28.5. The dupCor method, as implemented in the voomWrapper function of the iPSCpoweR package, uses a two-step voom transformation (before calling duplicateCorrelation, and after, including the output of the function). The SumReps method simply sums, for each individual, the read counts of all clones, and is implemented in the voomWrapperSumReps function. The lmer- and glmm-based methods are respectively implemented in the voomLmerWrapper, vstLmerWrapper, and glmmWrapper functions, and use the individual as a random-effect variable (i.e., ∼1 + [1|individual] + group). The lmer-based methods rely on the drop1 approach with a chi-squared test for statistical significance.
Input/“True” Differential Expression
To assess sensitivity, we introduced differences between the groups by enabling the addDE option of the package's permutation functions, with default settings. The differences are introduced multiplying a gene's read count, in each sample of one of the groups, by a predefined fold change. For each fold change (1.25, 1.5, 2, 3, 5, and their inverse), ten genes were randomly selected spanning the range of expression levels of the tested transcriptome, for a total of 100 introduced DEGs in each analysis. Importantly, this is making the assumption that, all other things being equal, “real” differentially expressed genes have an intra-group variability comparable with other genes.
Permutation Analysis
Permutation DEA analyses were performed using the DEA.permutateIndividuals and DEA.permutateClones functions of the package, using the default setting except for the addition of the expression filter (at least two samples having more than ten reads). Permutations that were not balanced for sex were discarded. For the NHLBI dataset, we instead ensured that the comparisons were balanced for technical batches. For comparisons involving 12 or fewer total individuals, only the individuals with more than one available iPSC clone were used in order to maximize the comparability with other experimental designs. For comparisons involving more than 12 individuals in total it was, however, necessary to use the whole cohort of samples to have a sufficient number of valid comparisons.
The permutation analysis of cellular morphology (Figure S2) can be reproduced with the cellpheno.permutateIndividuals function.
Data-Informed Correction of the Q Values
For Figure 4H, the nominal FDR values reported by edgeR were corrected using the multiProbSpurious function of the package. The function reports, for each tested gene, the frequency of the gene's absolute fold change or higher in the distribution of fold changes of the spurious DEG, as well as the frequency at which the given gene was found differentially expressed, in the union of all permutations of the HipSci dataset. For genes whose fold change was plausible in the distribution of spurious fold changes (p > 0.05), we then replaced edgeR's FDR by the frequency of the given gene in the permutations, if greater.
Gene Ontology Enrichment Analysis
Gene Ontology enrichment analyses were performed using the goseq R package (Young et al., 2010) against the background of tested genes (passing the filter of >10 reads in >2 samples), correcting for eventual RNA-seq transcript length bias, and excluding genes without annotation. Categories with at least ten genes and maximum 1,000 genes were considered. To increase the informativeness of the terms, we removed terms with significantly enriched children terms.
Selection of iPSC-Based Disease-Modeling Studies
For the brief review of experimental designs used in recent iPSC disease-modeling studies, a PubMed search for “Induced pluripotent stem cells” OR “iPSC” was performed and publications published in 2016 were considered. Only studies performing human iPSC-based modeling of specific diseases were considered (reviews, studies involving therapeutic application of iPSC derivatives; using direct conversion or reporting the development/improvement of technologies were discarded), and reports of the mere generation of single lines were discarded. For a large number of studies, the number of individuals/lines had to be inferred from the figures (lines were considered if at least some of the assays were performed on them beyond the validation of their pluripotency). Of the 81 publications meeting these criteria (see Table S1), three were excluded as not indicating the number of lines involved and four more were considered ambiguous in this respect, so that 74 studies were finally used to compile the number of clones/individuals (see Table S1).
Author Contributions
P.L.G. and G.T. conceived and designed the study; P.L.G. performed the analyses and made the figures; and P.L.G. and G.T. wrote the paper.
Acknowledgments
This work was supported by the European Research Council (ERC) grant DISEASEAVATARS 616441 (G.T.); the Telethon Foundation (grant GGP14265 to G.T.); the Italian Association for Cancer Research (AIRC) (IG grant to G.T.); the Umberto Veronesi Foundation (P.L.G.); the ERANET-Neuron grant from the Italian Ministry of Health (P.L.G.); The Italian Ministry of Health (Ricerca Corrente grant to G.T.); and the Regione Lombardia (grant Ricerca Indipendente 2012) (G.T.).
Published: June 6, 2017
Footnotes
Supplemental Information includes six figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.05.012.
Contributor Information
Pierre-Luc Germain, Email: pierre.germain@ieo.it.
Giuseppe Testa, Email: giuseppe.testa@ieo.it.
Supplemental Information
References
- Adamo A., Atashpaz S., Germain P., Zanella M., D'Agostino G., Albertin V., Testa G. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat. Genet. 2015;47:132–141. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banovich, N.E., Li, Y.I., Raj, A., Ward, M.C., Greenside, P., Calderon, D., Gilad, Y. (2016). Impact of regulatory variation across human iPSCs and differentiated cells. http://dx.doi.org/10.1101/091660. [DOI] [PMC free article] [PubMed]
- Barrera L.A., Vedenko A., Kurland J.V., Rogers J.M., Gisselbrecht S.S., Rossin E.J., Bulyk M.L. Survey of variation in human transcription factors reveals prevalent DNA binding changes. Science. 2016;351:1450–1454. doi: 10.1126/science.aad2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows C.K., Banovich N.E., Pavlovic B.J., Patterson K., Gallego Romero I., Pritchard J.K., Gilad Y. Genetic variation, not cell type of origin, underlies the majority of identifiable regulatory differences in iPSCs. PLoS Genet. 2016;12:1–18. doi: 10.1371/journal.pgen.1005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo-Orive I., Hoffman G.E., Cundiff P., Beckmann N.D., D’Souza S.L., Knowles J.W., Lemischka I. Analysis of transcriptional variability in a large human iPSC library reveals genetic and non-genetic determinants of heterogeneity. Cell Stem Cell. 2016;20:518–532. doi: 10.1016/j.stem.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman A. Induced pluripotent stem cells and human disease. Cell Stem Cell. 2008;3:236–237. doi: 10.1016/j.stem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Cook E.H., Jr., Scherer S.W. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Ercument Cicek A., Buxbaum J.D. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain P.L., Vitriolo A., Adamo A., Laise P., Das V., Testa G. RNAontheBENCH: computational and empirical resources for benchmarking RNAseq quantification and differential expression methods. Nucleic Acids Res. 2016;44:5054–5067. doi: 10.1093/nar/gkw448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbi M., Chen Q., Turner N., Kohn P., White M., Kippenhan J.S., Berman K.F. Variation in the Williams syndrome GTF2I gene and anxiety proneness interactively affect prefrontal cortical response to aversive stimuli. Transl. Psychiatry. 2015;5:e622. doi: 10.1038/tp.2015.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Ng S.H., Sharma V., Neculai D., Hussein S., Sam M., Batada N.N. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435–440. doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- Kasowski M., Kyriazopoulou-panagiotopoulou S., Grubert F., Zaugg J.B., Kundaje A., Liu Y., Snyder M. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H., Goncalves A., Leha A., Afzal V., Danecek P., Faulconbridge A., Kolb-Kokocinski A. Common genetic variation drives molecular heterogeneity in human iPSCs. bioRxiv. 2016 [Google Scholar]
- Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Daley G.Q. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmicki J.A., Samocha K.E., Howrigan D.P., Sanders S.J. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet. 2017;49:504–510. doi: 10.1038/ng.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.W., Chen Y., Shi W., Smyth G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–34. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Sanders S., Tian Y., Voineagu I. Genome-wide transcriptome profiling reveals the functional impact of rare de novo and recurrent CNVs in autism spectrum disorders. Am. J. Hum. Genet. 2012;91:38–55. doi: 10.1016/j.ajhg.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S.A., Altshuler D.M. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- Melé M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides accurate, fast, and bias-aware transcript expression estimates using dual-phase inference. bioRxiv. 2016 [Google Scholar]
- Rheinberger H.J. Stanford University Press; 1997. Toward a History of Epistemic Things: Synthesizing Proteins in the Test Tube. [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhani F., Kumasaka N., de Brito M.C., Bradley A., Vallier L., Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10:e1004432. doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N., Dexheimer P.J., Omberg L., Schroll R., Bush S., Huo J., Lutzko C. Integrated genomic analysis of diverse induced pluripotent stem cells from the progenitor cell biology consortium. Stem Cell Rep. 2016;7:110–125. doi: 10.1016/j.stemcr.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter I., Harrison P.W., Faulconbridge A., The HipSci Consortium. Flicek P., Parkinson H., Clarke L. The human-induced pluripotent stem cell initiative-data resources for cellular genetics. Nucleic Acids Res. 2016;45:D691–D697. doi: 10.1093/nar/gkw928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P., Helgason H., Oddson A., Stefansson H., Gudjonsson S.A., Zink F., Stefansson K. Identification of a large set of rare complete human knockouts. Nat. Genet. 2015;47:448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.A., Larson D.E., Sun C.-W., George D.R., Ding L., Miller C.A., Ley T.J. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.