Abstract
A novel gene was identified in the Treponema denticola prcA-prtP protease operon. Strains with mutations in either the prcA-prtP or the msp region showed altered expression of a product(s) of the other locus. Together, these results provide information on the assembly of outer membrane complexes involved in T. denticola interaction with host cells and tissue.
Two Treponema denticola surface components with a wide range of cytopathic activities have been reported: the oligomeric major surface protein (Msp) (7, 13) and a protease complex (20) encoded by the prcA-prtP locus (17, 18). The Msp has functional characteristics of an outer membrane porin (6, 7). The protease complex consists of the PrtP protease (dentilisin) and two proteins (PrcA1 and PrcA2) that result from posttranslational processing of the PrcA polypeptide (18). Both Msp and PrcA-PrtP exhibit adhesin-like binding activity and are cytotoxic to epithelial cells (7). Recent reports implicate Msp in the disruption of fibroblast calcium responses and cytoskeletal assembly (1, 3). PrtP activity contributes to T. denticola tissue penetration (4, 12) and modulation of inflammatory cytokine responses (2, 5). PrtP is reported to either contribute to (15) or interfere with (21) T. denticola binding to Porphyromonas gingivalis. Using isogenic mutants, we showed that the production of native Msp and PrtP are related (10, 18) and that PrtP is required for PrcA processing (18). An msp mutant producing a C-terminally truncated Msp makes no detectable protease proteins, while an Msp-deficient mutant produces wild-type PrcA-PrtP protease. Strains mutated in the protease locus demonstrate reduced expression and oligomerization of Msp. The complex phenotypes of mutant strains must be taken into account when using mutants to study T. denticola interaction with host cells and tissue. The present study was undertaken to extend understanding of the relationship between these membrane complexes. Here, we further characterize the expression of the protease activity and Msp in isogenic mutant strains and identify a novel open reading frame in the protease operon whose expression is required for outer membrane complex formation.
Growth behavior of T. denticola strains.
To investigate the role of PrtP activity in anaerobic growth in NOS medium (14), parent strain 35405 and isogenic strains with mutations in either the protease locus or the msp locus (Table 1) were monitored for 7 days following 1:20 dilution of actively growing cultures (optical density at 600 nm [OD600] of 0.35; triplicate samples; four replicates). The parent strain reached stationary phase within 100 h. All of the mutant strains tested had a slightly slower growth rate, reaching stationary phase at 120 to 140 h (data not shown). Final optical densities of protease-deficient strains, including strain MPE, were consistently lower than that of strain 35405, but differences were statistically insignificant. High-molecular-weight media constituents were not degraded by the protease-deficient strains (data not shown), consistent with the hypothesis that PrtP activity contributes to nutrient processing or acquisition. Interestingly, strain MHE, which produces no Msp but has wild-type levels of protease activity, also grew at a slower rate than 35405. This result supports the hypothesis that Msp is essential for proper outer membrane function and is suggestive of functional interactions between PrtP protease activity and Msp porin activity in nutrient acquisition and uptake.
TABLE 1.
Treponema denticola strains used in this study
| Straina | Phenotypeb | Source or reference |
|---|---|---|
| Parent 35405 | SAAPFNA+, PrcA+, PrtP+, oligomeric Msp | ATCC |
| MHE (msp) | SAAPFNA+, PrcA+, PrtP+, Msp− | 10 |
| MPE (msp) | SAAPFNA−, PrcA−, PrtP−, truncated monomeric Msp | 10 |
| CKE (prcA-prtP) | SAAPFNA−, truncated PrcA, PrtP−, reduced Msp, mostly monomeric | 10 |
| CCE (prtP) | SAAPFNA−, unprocessed PrcA, PrtP−, reduced Msp, mostly monomeric | 18 |
| PNE (prcA) | SAAPFNA−, PrcA−, PrtP−, reduced Msp, mostly monomeric | 18 |
| P0760 (TDE0760) | SAAPFNA−, PrcA−, PrtP−, reduced Msp, mostly monomeric | This study |
The gene disrupted by ermF-ermB insertion is given in parentheses.
PrtP protease activity was assayed by hydrolysis of SAAPFNA; PrcA, PrtP, and Msp production and forms are listed.
Gene expression and protease activity.
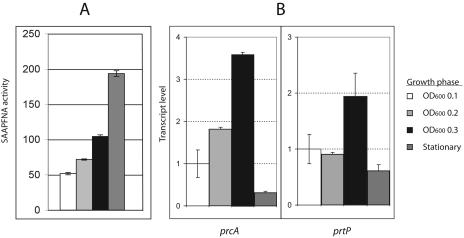
PrtP protease activity and transcription of prcA and prtP in parent and isogenic mutant strains were monitored throughout growth by quantitative reverse transcription-PCR (QRT-PCR). Protease activity in 35405, as measured by cleavage of succinyl-l-alanyl-l-alanyl-l-prolyl-l-phenylalanine-p-nitroanilide (SAAPFNA) (8), increasedduring active growth and persisted at high levels in stationary phase (Fig. 1A). Transcription of prcA and prtP increased during active growth but was greatly reduced at stationary phase (Fig. 1B). We detected no significant differences in prcA or prtP transcription during active growth between the parent strain and the msp mutants MHE and MPE (data not shown). This result is of particular interest due to the distinctly different Msp and PrcA-PrtP phenotypes of these strains (Table 1). The lack of PrcA-PrtP proteins and activity in strain MPE, but not MHE, is therefore due to posttranscriptional events and may be related to the presence of a defective, truncated Msp in MPE.
FIG. 1.
Protease activity and transcription of prcA-prtP in T. denticola 35405. (A) SAAPFNA activity (8) expressed in A405/h/ml/OD600. Error bars represent the range of values obtained with triplicate samples of a representative experiment. (B) QRT-PCR of prcA and prtP. Total RNA was extracted from cultures harvested at an OD600 of 0.1, 0.2, and 0.3 and at stationary phase (fifth day). DNase-treated RNA samples were reversed transcribed with random hexamer primers by using the SuperScript First-Strand synthesis system for RT-PCR (Invitrogen). One microliter of the resulting first-strand cDNA was amplified by using QuantiTect SYBR Green PCR (QIAGEN) in 25 μl of reaction buffer, with 16S rRNA serving as an internal control for normalization between samples. Gene-specific primers separated by approximately 80 to 100 bp were designed with Primer Express software (Perkin-Elmer Applied Biosystems). Thermal cycling was performed in an iCycler iQ Multi-Color Real Time PCR detection system (Bio-Rad) at 95°C for 15 min, followed by 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s. Threshold values were calculated by using baseline cycles 2 to 10. Data were analyzed by using software supplied by the manufacturer. The data are shown with the gene expression level defined as 1.0 at an OD600 of 0.1. Error bars represent standard deviations of the means of results from triplicate samples in two independent experiments.
Expression of msp in prcA and prtP mutants.
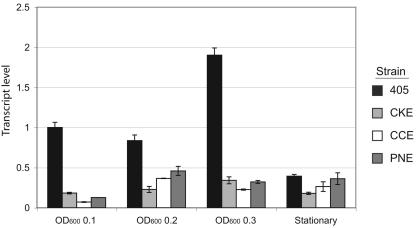
Total Msp and especially Msp oligomers were greatly reduced in strains with defined mutations in either prcA or prtP or both (10, 18). To determine whether this result was reflected in msp transcription, QRT-PCR assays were performed on parent and mutant strains. As shown in Fig. 2, msp transcription in isogenic prcA and prtP mutants was reduced by greater that 50% during active growth. This result is consistent with Msp protein expression results (10, 18) and suggests that protease gene expression might contribute to the regulation of msp transcription. However, reduced msp transcription does not account for the absence of Msp oligomers in protease mutants.
FIG. 2.
Transcription of msp in protease mutant strains. T. denticola strains were harvested at an OD600 of 0.1, 0.2, and 0.3 and at stationary phase (fifth day). QRT-PCR was performed as described in the legend to Fig. 1. Data are shown with the gene expression level defined as 1.0 at an OD600 of 0.1. Error bars represent standard deviations of the means of results from triplicate samples in two independent experiments.
The prcA-prtP transcript includes an additional upstream gene.
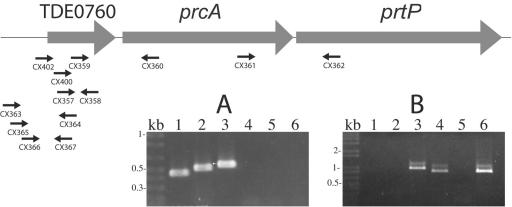
Expression of PrcA, which is encoded directly upstream of the gene encoding PrtP, is required for PrtP expression and activity (16, 18). A recent study reported that the prtP transcript is at least 5 kb, large enough to include both prcA and prtP (16). RT-PCR using primer pair CX361 and CX362 (Fig. 3A, lane 3) demonstrated that prcA and prtP are cotranscribed. Transcription of prtP was not detected in prcA mutant strain PNE (data not shown), and a typical rho-independent transcription termination sequence is found directly downstream of prtP. Taken together, these data demonstrate that prcA and prtP are cotranscribed. Because prcA and prtP together comprise only 4.2 kb, we examined the sequence upstream of prcA by RT-PCR and demonstrated that the prcA-prtP transcript extends upstream through the 5′ end of TDE0760 (Fig. 3B, lanes 1 and 4). TDE0760 encodes a conserved hypothetical protein of 18 or 21 kDa, depending on whether the translation initiates at nucleotide 799483 (ATG) or at 799576 (GTG), as indicated by the T. denticola genome annotation. Sequence analysis using PSORT (http://psort.nibb.ac.jp/) predicts that the TDE0760 protein, including the additional N-terminal region, is membrane associated and may be acylated.
FIG. 3.
RT-PCR analysis of prcA-prtP operon. DNase-treated RNA samples were reversed transcribed as described in the legend to Fig. 1 by using a gene-specific primer or random hexamer primers supplied with the SuperScript First-Strand synthesis system for RT-PCR (Invitrogen). One microliter of the resulting first-strand cDNA was amplified according to the manufacturer's instructions. The location and orientation of oligonucleotide primers used for RT-PCR are shown relative to TDE0760, prcA, and prtP in the graphic map. RT-PCR products, including positive (genomic DNA template) and negative (no RT enzyme) controls were analyzed by agarose gel electrophoresis. Panel A demonstrates continuity of transcription between TDE0760 and prtP. Lane 1, CX357-CX358; lane 2, CX359-CX360; lane 3, CX361-CX362; lane 4, CX363-CX364; lane 5, CX365-CX364; and lane 6, CX366-CX367. Panel B localizes the 5′ end of the transcript. CX402-CX360, lanes 1 to 3; CX400-CX360, lanes 4 to 6. Lanes 1 and 4, cDNA template; lanes 2 and 5, RNA template; lanes 3 and 6, DNA template.
Construction of a TDE0760 mutant.
To characterize the relationship between TDE0760 and the protease complex, we constructed a TDE0760 isogenic mutant by allelic replacement mutagenesis. A 1,475-bp PCR product containing all but the 3′ end of TDE0760 that was generated by using standard PCR conditions (9) with CX412 (GAGCTTTGTCTTCTACATTG) and CX413 (CAAATTGATGATCTTCCCTG) was cloned in pSTBlue-1 (Novagen). The ermF/ermB cassette (11) was inserted in opposite transcriptional orientation to TDE0760 at a pshA1 site 43 nucleotides 3′ to the putative ATG initiation codon at nucleotide 799483, yielding pCF353. Plasmid pCF353 was then digested with PvuII, releasing the vector sequences. T. denticola was electroporated with the resulting linear DNA, and transformants were selected for erythromycin resistance (10, 19). Genomic DNA from Emr colonies was screened by PCR for the presence of the ermF/ermB cassette in TDE0760 by using combinations of oligonucleotide primers specific for TDE0760, prcA, and ermF/ermB (data not shown). A single validated isolate was designated T. denticola P0760.
Protein expression in mutant strain P0760.
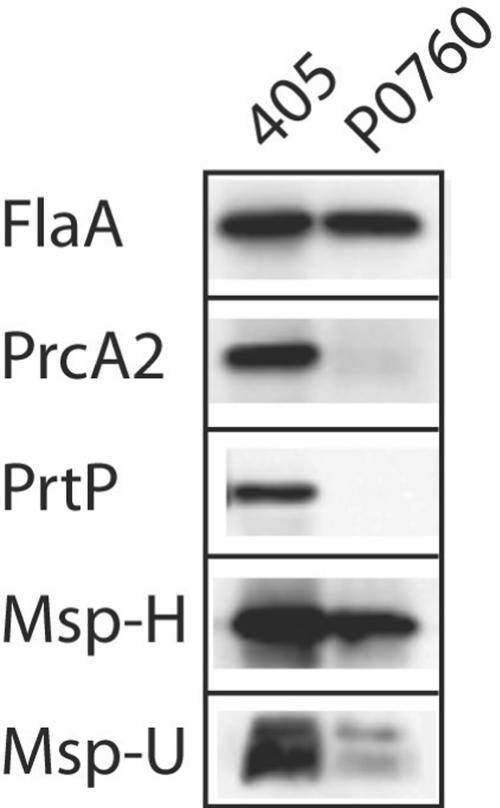
As shown in Fig. 4, mutagenesis of TDE0760 resulted in the loss of expression of both PrcA2 and PrtP. RT-PCR products of prtP, prcA, and the 3′ region of TDE0760 were present in 35405 but not in P0760 (data not shown). SAAPFNA activity was absent in P0760, essentially identical to that of CKE, PNE, CCE, and MPE (data not shown). While total Msp was considerably reduced in P0760 (Fig. 4, Msp-H), Msp oligomers (Fig. 4, Msp-U) were barely detectable in P0760 compared with 35405.
FIG. 4.
Western immunoassay of T. denticola parent and mutant strains. Equal amounts of whole-cell extracts of T. denticola 35405 (405) and P0760 were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to membranes, and probed with antibodies raised against recombinant FlaA, PrcA2, PrtP, or Msp polypetides. All samples were heated at 100°C for 5 min prior to electrophoresis, with the exception of Msp-U, which was held at 4°C.
While it has not yet been possible to determine the mechanism for the connection between the expression of Msp and the PrcA-PrtP protease complex, the present study further explored this relationship. We have presented new information on growth and transcription in T. denticola strains with mutations in genes encoding periodontal virulence factors. Transcriptional analysis supports the hypothesis that the production of active protease complex and Msp oligomers are closely related. While its function is not yet known, initial characterization of TDE0760 contributes to our understanding of protease complex expression. This report should help to provide a context for interpretation of studies using these and similar defined mutant strains to assay biological activities of Msp and PrtP.
Acknowledgments
This work was supported by Public Health Service grant DE13565 from the National Institute of Dental and Craniofacial Research (J.C.F.).
Editor: J. T. Barbieri
REFERENCES
- 1.Amin, M., A. C. S. Ho, J. Y. Lin, A. P. Batista da Silva, M. Glogauer, and R. P. Ellen. 2004. Induction of de novo subcortical actin filament assembly by Treponema denticola major outer sheath protein. Infect. Immun. 72:3650-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, Y., T. Jinno, and T. Ogawa. 2003. Oral treponemes and their outer membrane extracts activate human gingival epithelial cells through Toll-like receptor 2. Infect. Immun. 71:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batista da Silva, A. P., W. Lee, E. Bajenova, C. A. McCulloch, and R. P. Ellen. 2004. The major outer sheath protein of Treponema denticola inhibits the binding step of collagen phagocytosis in fibroblasts. Cell. Microbiol. 6:485-498. [DOI] [PubMed] [Google Scholar]
- 4.Chi, B., M. Qi, and H. K. Kuramitsu. 2003. Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res. Microbiol. 154:637-643. [DOI] [PubMed] [Google Scholar]
- 5.Deng, Q. D., Y. Han, X. Xia, and H. K. Kuramitsu. 2001. Effects of the oral spirochete Treponema denticola on interleukin-8 expression from epithelial cells. Oral Microbiol. Immunol. 16:185-187. [DOI] [PubMed] [Google Scholar]
- 6.Egli, C., W. K. Leung, K. H. Müller, R. E. Hancock, and B. C. McBride. 1993. Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola. Infect. Immun. 61:1694-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenno, J. C., P. M. Hannam, W. K. Leung, M. Tamura, B. C. McBride, and V.-J. Uitto. 1998. Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect. Immun. 66:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenno, J. C., S. Y. Lee, C. H. Bayer, and Y. Ning. 2001. The opdB locus encodes the trypsin-like peptidase activity of Treponema denticola. Infect. Immun. 69:6193-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenno, J. C., K.-H. Müller, and B. C. McBride. 1996. Sequence analysis, expression and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J. Bacteriol. 178:2489-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenno, J. C., G. W. K. Wong, P. M. Hannam, and B. C. McBride. 1998. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol. Lett. 163:209-215. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher, H. M., H. A. Schenkein, R. M. Morgan, K. A. Bailey, C. R. Berry, and F. L. Macrina. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenier, D., V.-J. Uitto, and B. C. McBride. 1990. Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect. Immun. 58:347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haapasalo, M., K.-H. Müller, V.-J. Uitto, W. K. Leung, and B. C. McBride. 1992. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect. Immun. 60:2058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haapasalo, M., U. Singh, B. C. McBride, and V.-J. Uitto. 1991. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect. Immun. 59:4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto, M., S. Ogawa, Y. Asai, Y. Takai, and T. Ogawa. 2003. Binding of Porphyromonas gingivalis fimbriae to Treponema denticola dentilisin. FEMS Microbiol. Lett. 226:267-271. [DOI] [PubMed] [Google Scholar]
- 16.Ishihara, K., H. K. Kuramitsu, and K. Okuda. 2004. A 43-kDa protein of Treponema denticola is essential for dentilisin activity. FEMS Microbiol. Lett. 232:181-188. [DOI] [PubMed] [Google Scholar]
- 17.Ishihara, K., T. Miura, H. K. Kuramitsu, and K. Okuda. 1996. Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin). Infect. Immun. 64:5178-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, S. Y., X.-L. Bian, G. W. K. Wong, P. M. Hannam, B. C. McBride, and J. C. Fenno. 2002. Cleavage of Treponema denticola PrcA polypeptide to yield protease complex-associated proteins PrcA1 and PrcA2 is dependent on PrtP. J. Bacteriol. 184:3864-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, H., J. Ruby, N. Charon, and H. Kuramitsu. 1996. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J. Bacteriol. 178:3664-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uitto, V.-J., D. Grenier, E. C. S. Chan, and B. C. McBride. 1988. Isolation of a chymotrypsinlike enzyme from Treponema denticola. Infect. Immun. 56:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vesey, P. M., and H. K. Kuramitsu. 2004. Genetic analysis of Treponema denticola ATCC 35405 biofilm formation. Microbiology 150:2401-2407. [DOI] [PubMed] [Google Scholar]