Abstract
This study was conducted to investigate the occurrence of PAH degrading microorganisms in two river systems in the Western Cape, South Africa and their ability to degrade two PAH compounds: acenaphthene and fluorene. A total of 19 bacterial isolates were obtained from the Diep and Plankenburg rivers among which four were identified as acenaphthene and fluorene degrading isolates. In simulated batch scale experiments, the optimum temperature for efficient degradation of both compounds was determined in a shaking incubator after 14 days, testing at 25 °C, 30 °C, 35 °C, 37 °C, 38 °C, 40 °C and 45 °C followed by experiments in a Stirred Tank Bioreactor using optimum temperature profiles from the batch experiment results. All experiments were run without the addition of supplements, bulking agents, biosurfactants or any other form of biostimulants. Results showed that Raoultella ornithinolytica, Serratia marcescens, Bacillus megaterium and Aeromonas hydrophila efficiently degraded both compounds at 37 °C, 37 °C, 30 °C and 35 °C respectively. The degradation of fluorene was more efficient and rapid compared to that of acenaphthene and degradation at Stirred Tank Bioreactor scale was more efficient for all treatments. Raoultella ornithinolytica, Serratia marcescens, Bacillus megaterium and Aeromonas hydrophila degraded a mean total of 98.60%, 95.70%, 90.20% and 99.90% acenaphthene, respectively and 99.90%, 97.90%, 98.40% and 99.50% fluorene, respectively. The PAH degrading microorganisms isolated during this study significantly reduced the concentrations of acenaphthene and fluorene and may be used on a larger, commercial scale to bioremediate PAH contaminated river systems.
Keywords: Polycyclic aromatic hydrocarbons (PAHs), Bioremediation, Bioreactor, PAH degrading microorganisms
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are a group of recalcitrant, bioaccumulative, semi-volatile organic pollutants that are widespread in the environment and enter environmental matrices via natural (such as volcanoes) and anthropogenic sources (such as industrial activities).1 They are composed of two or more fused aromatics (benzene rings) and a system of hydrophobic and lipophilic double bonds throughout their hydrocarbon rings. These groups of compounds have potential harmful effects on ecosystems as well as human health because many of them have been shown to be carcinogenic, teratogenic and mutagenic.2
Several approaches and strategies including physical, chemical and biological strategies have been developed, optimised and utilised to ameliorate PAH contamination and replenish polluted sites. Some of the available physical and chemical (conventional) techniques have been shown to have significant drawbacks such as technological complexity, high cost and a general lack of acceptance. Bioremediation, which is the use of biological systems to control pollution, has been shown to be a cost effective and environmentally friendly approach to remediate contaminated sites.3 Several bacterial (Pseudomonas, Alcanivorax, Microbulbifer, Sphingomonas, Micrococcus, Cellulomonas, Dietzia, Gordonia, Marinobacter4) and fungal species (Aspergillus sp., Trichocladium canadense, and Fusarium oxysporum5) capable of degrading PAH compounds have been isolated and characterised. It is however imperative to investigate the PAH degrading capability of indigenous microorganisms in various ecosystems especially those that are subject to significant point sources of PAH pollution. This study isolated and identified four PAH degrading microorganisms from the Diep and Plankenburg rivers in the Western Cape, South Africa and also investigated their degradative potential of two PAH compounds (acenaphthene and fluorene) under optimum temperature conditions.
Acenaphthene and fluorene were selected as model compounds for this study, because of their relatively simple structure and solubility capacity in organic solvent (acetonitrile) which allowed easier simulation during laboratory experiments. Acenaphthene is a PAH compound consisting of naphthalene with an ethylene bridge connecting positions 1 and 8 while fluorene contains two benzene rings each of which is coplanar with the central Carbon 9.6
Materials and methods
Study area
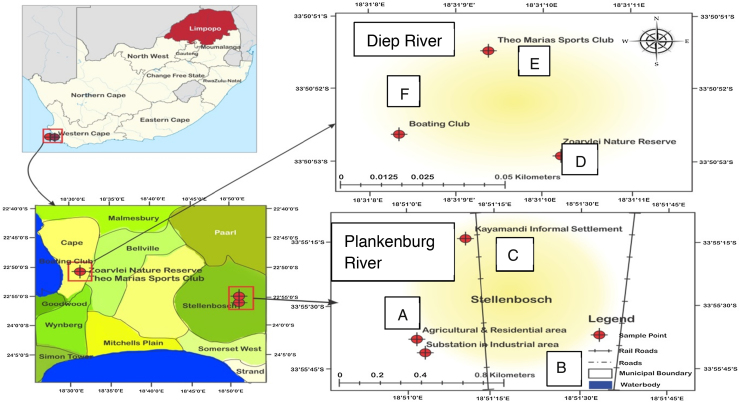
Three sampling points were identified along each of the two rivers studied. Along the Plankenburg River; Points A–C represents an agricultural farming and residential area, a substation in industrial area and the informal settlement of Kayamandi, respectively. For the Diep River, Points D–F represents the Zoarvlei nature reserve (industrial as well as residential), the Theo Marias Sports club (Industrial and Residential Area) and the Rietvlei Boating club respectively as shown in Fig. 1.
Fig. 1.
Map of the Diep and Plankenburg rivers showing locations of sampling sites (A – agricultural farming and residential area, B – substation in industrial area, C – informal settlement of Kayamandi, D – Zoarvlei nature reserve, E – Theo Marias sportsclub and F – Rietvlei boating club).
Sampling
Water and sediment was collected once a month over a period of one year. Water samples were collected in sterile amber bottles while sediment samples were collected using a sterile Ekman grab and placed in polypropylene containers which were wrapped in aluminium foil. Both the water and sediment samples were kept on ice at 4 °C during transport to the laboratory for analyses. During sampling, physicochemical parameters such as temperature, pH and conductivity were measured and recorded using a handheld PCS teslr 35 multi-parameter gauge (Wirsam, SA). All samples were analysed in not more than 90 h after collection.
Determination of the presence and concentration of acenaphthene and fluorene in the river systems
Acenaphthene and fluorene were extracted from the water samples with 50 ml 4:1 mixture of n-hexane and dichloromethane (DCM)7 in 500 ml separating funnels. The flask was left for 15 min at room temperature to allow for equilibration and efficient phase separation. This procedure was repeated thrice to ensure good PAH recovery (above 70%). The extracts were combined and dried in a water bath at 35 °C under a stream of nitrogen. The dried samples were reconstituted to 2 ml with n-hexane.
The extracts were cleaned using solid phase extraction (SPE) technique. The SPE glass tube frits were conditioned by eluting each one with 10 ml DCM, and 20 ml n-hexane at a flow rate of 1.0 ml/min. Each PAH concentrate extract was then loaded on the SPE/PTFE frits tubes and eluted with 70 ml of n-hexane. The eluates were dried in a water bath under a nitrogen stream. The resulting residues were re-dissolved in 3 ml methanol and dried again in a water bath under a nitrogen stream. Each of the dried concentrates was dissolved in 1 ml of n-hexane and filtered through 0.45 μm Millipore acrodisc membrane filters.8 The samples were run on GC/FID (gas chromatograph/flame ionisation detection).
Isolation and identification of microorganisms from the Diep and Plankenburg river systems
Standard microbiological techniques including serial dilution, plating and culturing were employed to isolate microorganisms. Various general, selective and differential media including Nutrient agar, MacConkey agar (Merck, Germany) Mannitol Salt agar, Eosin Methylene Blue (EMB) agar, Pseudomonas isolation agar base (Oxoid, England), Aeromonas isolation agar and Glutamate Starch Phenol Red (GSP) agar (Fluka, India), amongst others were used to isolate bacterial species. Phenotypic identification techniques such as staining and biochemical tests were conducted for ‘tentative’ identification of the isolates. Isolates were selected on the basis of morphology; colour, cell shape and size, pigmentation and Gram reaction. Isolates were further identified using molecular techniques. DNA was extracted from the bacterial cultures using the ZR Fungal/Bacterial DNA kit™ (Zymo Research). The concentrated DNA samples of the bacterial strains were amplified by polymerase chain reaction (PCR) using a thermal cycler (Mastercycler ® personal, Eppendorf AG, Germany). The 16S target region was amplified using the universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-CGGTTACCTTACGACTT-3′).9, 10 The PCR reactions include an initial denaturation step at 94 °C for 10 min followed by 30 cycles of denaturation at 95 °C for 15 s, primer annealing at 60 °C for 1 min and primer extension at 72 °C for 1 min. In the final step, the samples were incubated at 72 °C for 10 min. PCR amplification was verified by gel electrophoresis, performed in horizontal submarine apparatus with 1% agarose gel, TAE buffer as the tank buffer. Electrophoresis was carried out for 2 h at 50 V and the gel visualised in an UV illuminator. The PCR amplified DNA was purified and sequenced in the forward and reverse directions on the ABI PRISM™ 3500 analyser. The nucleotide sequences obtained were analysed using CLC main workbench 7 followed by a BLAST (Basic Local Alignment Search Tool) search provided by NCBI (National Centre for Biotechnology Information) (www.ncbi.nlm.nih.gov) and identified.
Identification of potential PAH-degrading bacterial species using temperature optimisation screening
Each isolate designated DR, DB, PA, LC, LP, MA, DL, PY, PR, XP, RC, SE, AH, BA, TA, QO, ST, KP and DG was screened to determine which members of the bacterial species isolated are potential PAH degraders. In simulated experiments, 114 flasks (each containing 80 ml sterile distilled water, 10 ml of the analyte (acenaphthene, fluorene) in solution and 10 ml of bacterial culture) were used as starter medium. The isolates were cultured in liquid medium overnight and each PAH compound was dissolved first in 30% acetonitrile because of PAH poor aqueous solubility and subsequently taken into solution by gently shaking in amber bottles in a shaking incubator for three days at 70 rpm. The experiment was carried out in a shaking incubator over a period of four weeks at 25 °C, 30 °C, 35 °C, 37 °C, 38 °C, 40 °C and 45 °C,11, 12, 13, 14 and 150 rpm.15 Bacterial culturing was done daily in order to assess which microorganisms can withstand the PAH compounds and which isolates can utilise them as carbon sources. The culturing was done by taking 1 ml of the sample as rapidly as possible, diluting serially ten-fold and subsequently plating on media plates. Plate count, morphology, motility, Gram reaction and biochemical tests were all monitored throughout the experiments. The degradation potential of the identified isolates was determined using HPLC coupled with a dual wavelength absorbance detector.16, 17, 18 Only isolates that could degrade up to 75% acenaphthene and fluorene were selected for subsequent degradation studies.
Degradation study
The microorganisms that successfully degraded acenaphthene and fluorene up to 75% and above from the temperature optimisation experiments were selected as potential PAH degraders and used for degradation experiments at flask and Stirred Tank Bioreactor scale; where natural conditions were mimicked and no supplementary nutrients added in order to keep the system cost-effective. Samples were obtained daily to monitor bacterial growth, changes in morphology, Gram reaction and the number of cells was counted using plate counts. The degree of degradation was assessed using HPLC equipped with a dual wavelength absorbance detector16; the mobile phase was acetonitrile, the flow rate of the mobile phase was maintained at 1 ml/min, standard solutions of the compounds were used as reference/control, the samples were injected one after the other and the utilisation rates of the compounds were calculated based on the peak area per cent and retention time. The ratio of the amount of substrate degraded in test reactors to the amount of the substrate recovered in the control reactors was also calculated to determine the extent of degradation. The filters used to purify samples prior to running on the HPLC were of Polypropylene material to prevent adsorbance of the analyte unto the filters thereby leading to erroneous conclusions. Draw time was approximately one minute, as the sample collection was done as rapidly as possible to avoid disrupting the experiment and all experiments were run in triplicate.
Flask scale degradation
The methodology adopted for the flask scale experiment was similar to the screening protocols. The experiment was repeated for the selected PAH degrading microorganisms and the biodegradative potential of single species were compared with consortium (cocktail) flasks. Solutions containing 10% analyte, 10 ml overnight culture and made up to a final volume of 100 ml with sterile distilled water was placed in 36 separate conical flasks. Prior to inoculating with overnight cultures, the PAHs were dissolved in acetonitrile (30%) and taken into solution. For each compound, there were two flasks containing consortium (a combination of all isolates), as well as a flask that was not inoculated designated as sterile control for each compound. The sterile control was to account for PAH losses due to other factors apart from biological such as photooxidation and volatilisation amongst others and was also used to compare degradative capabilities of the isolated cultures. The experiment was carried out in a shaking incubator which was run at 25 °C, 30 °C, 35 °C, 37 °C, 38 °C, 40 °C and 45 °C and 150 rpm for 14 days (the 14 day period was selected because the PAH degrading microorganisms all degraded the compounds within that time range at the temperature optimisation screening scale). All flasks were capped with cotton wool and covered with aluminium foil to minimise losses due to photooxidation.
Stirred Tank Bioreactor scale
A total of 18 reactors were used during this study. The reactors were amber coloured glass containers with a working volume of 1 L (Glasschem, Stellenbosch, South Africa). All the reactors were equipped with over-head stirrers with flat-blade radial turbine impellers and were all run at 150 rpm. The flange of each reactor had five openings which were all capped with polypropylene plastic caps and fit to the reactor vessel by a wire spring. Each reactor was run for a total of four weeks with the same content as used in the flask scale reactors. Temperatures were maintained at optimum for each sample obtained from flask scale experiments using a hot water bath equipped with a thermometer to ensure temperature accuracy. One of the 18 Stirred Tank Bioreactors was left uninoculated and served as the sterile control, while the other 17 were inoculated with DR, DB, SE, AH and the cocktail respectively.
Data analysis
All data obtained from this study were analysed by an SPSS statistical package using repeated measures ANOVA. The means and standard deviations of triplicate treatments were also calculated and Microsoft EXCEL software was used to illustrate graphs.
Results and discussion
The mean physicochemical parameters measured and recorded at sampling sites during winter and summer sampling time are recorded in Table 1.
Table 1.
Mean physicochemical parameters of river systems recorded during sampling time.
| Sampling points | Temperature (°C) | pH | Conductivity (mS/m) |
|---|---|---|---|
| A | 11.8 | 6.7 | 449 |
| 22.9 | 5.8 | 740 | |
| B | 12.1 | 6.9 | 668 |
| 22.3 | 6.1 | 711 | |
| C | 12.1 | 7.2 | 708 |
| 23.7 | 6.8 | 749 | |
| D | 11.4 | 7.3 | 715 |
| 27.8 | 8.0 | 761 | |
| E | 13.5 | 7.1 | 444 |
| 28.8 | 7.9 | 751 | |
| F | 13.9 | 7.0 | 589 |
| 28.5 | 7.8 | 773 |
PAHs in the river systems
The two PAHs investigated in this study were detected at varying concentrations in the collected sediment and water samples. They were detected at more elevated concentrations in sediment samples than in surface water samples (Table 2). This could be attributed to the capacity of PAH compounds to adsorb unto particulate matter.1 Higher concentrations of the compounds were detected during the winter months (May to September) compared to during the summer sampling time (December to March). This trend is comparable with results obtained by Zhang and Tao19 who reported higher PAH occurrence in winter compared to summer time in Beijing, China. This suggests that the most important source of PAH compounds into these river systems might be atmospheric deposition, because during winter there is increased vehicular activity and other fossil fuel combustion activities (such as the use of heating systems) which significantly deposits PAH compounds into environmental matrices.20, 21 The most contaminated site was Site F (Rietvlei boating club) with an average of 0.80 and 0.90 ppm acenaphthene and fluorene detected from sediments respectively during winter months and 0.6 and 0.7 respectively during summer sampling time. Therefore, in addition to atmospheric deposition, boating activities (emissions from which being a significant petrogenic PAH source), could also contribute to the input of the PAH compounds in the river.
Table 2.
Acenaphthene and fluorene concentrations detected at the sampling sites along the Diep and Plankenburg rivers.
| Sampling session/matrix | Sampling points |
|||||||
|---|---|---|---|---|---|---|---|---|
| Point A | Point B | Point C | Point D | Point E | Point F | |||
| Winter | Water | ND | 0.004 | 0.0008 | ND | ND | 0.004 | Ace (ppm) |
| 0.0009 | 0.20 | 0.0007 | ND | 0.0006 | 0.004 | Flu (ppm) | ||
| Sediment | 0.20 | 0.40 | 0.10 | ND | 0.07 | 0.80 | Ace(ppm) | |
| 0.60 | 0.90 | 0.60 | 0.40 | 0.80 | 0.90 | Flu (ppm) | ||
| Summer | Water | ND | 0.0006 | ND | ND | ND | 0.0006 | Ace(ppm) |
| ND | 0.003 | ND | ND | 0.0009 | 0.005 | Flu (ppm) | ||
| Sediment | 0.04 | 0.20 | 0.03 | ND | 0.004 | 0.6 | Ace(ppm) | |
| 0.04 | 0.70 | 0.002 | ND | 0.004 | 0.7 | Flu (ppm) | ||
Bacterial isolates identified from the Diep and Plankenburg river systems using conventional techniques
A total of 19 bacterial isolates were obtained from the sampling points along the Diep and Plankenburg river systems (surface water and sediments). The isolates’ DNA sequences queried on the NCBI database corresponded with the named biological sequences within the NCBI database. Table 3 presents the 19 bacterial species isolated from Diep and Plankenburg rivers.
Table 3.
Molecular (BLAST) identity of the bacterial species isolated from the Diep and Plankenburg rivers.
| S/N | Isolate | Percentage similarity (%) | BLAST ID |
|---|---|---|---|
| 1 | DR | 99 | Raoultella ornithinolytica |
| 2 | DB | 100 | Bacillus megaterium |
| 3 | PA | 99 | Pseudomonas aeruginosa |
| 4 | LC | 99 | Raoultella planticola |
| 5 | LP | 99 | Klebsiella oxytoca |
| 6 | MA | 99 | Escherichia coli |
| 7 | DL | 99 | Enterobacter cloacae |
| 8 | PY | 99 | Exiguobacterium acetylicum |
| 9 | PR | 99 | Acinetobacter sp. |
| 10 | XP | 99 | Exiguobacterium sp. |
| 11 | RC | 99 | Bacillus sp. |
| 12 | SE | 99 | Serratia marcescens |
| 13 | AH | 99 | Aeromonas hydrophila |
| 14 | BA | 99 | Bacillus aryabhattai |
| 15 | TA | 100 | Bacillus aquimaris |
| 16 | QO | 100 | Bacillus marisflavi |
| 17 | ST | 99 | Staphylococcus saprophyticus |
| 18 | KP | 99 | Citrobacter freundii |
| 19 | DG | 99 | Exiguobacterium undae |
Key: ID- Identity.
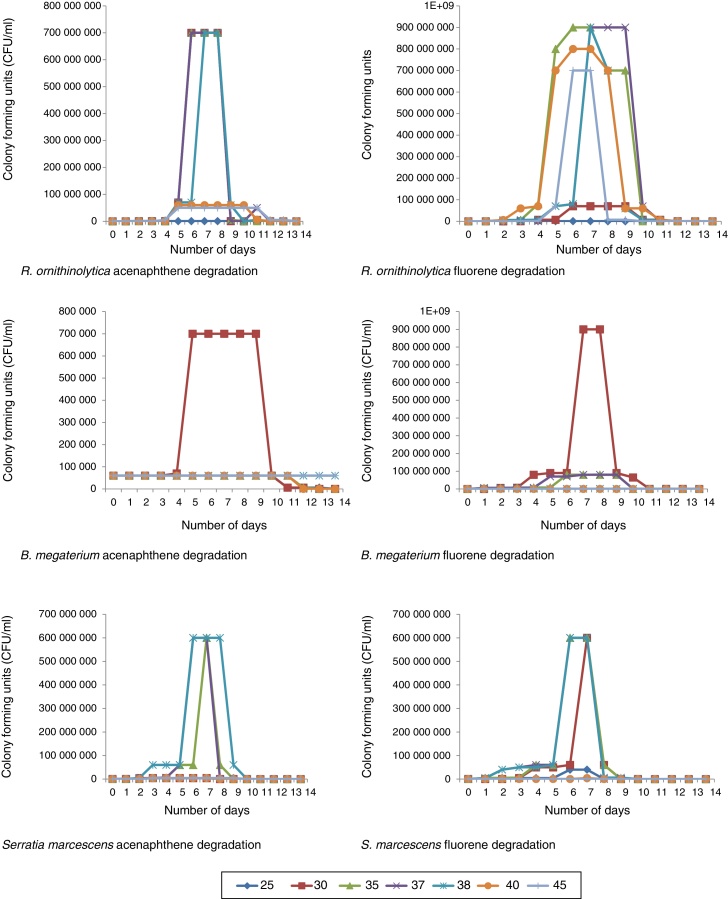
After all the isolated species were screened, four of the isolates successfully degraded acenaphthene and fluorene above 75% and were selected for further degradation studies. The microorganisms selected were: Raoultella ornithinolytica, Serratia marcescens, Bacillus megaterium and Aeromonas hydrophila. These microorganisms’ DNA sequences have been deposited in The National Centre for Biotechnology Information (NCBI) Genbank under the accession numbers; KT239136, KT239137, KT239139 and KT239138 respectively. Of the four selected microorganisms, three (R. ornithinolytica, S. marcescens and A. hydrophila) are Gram negative microorganisms. The reason why there are more Gram negative microorganisms able to ‘pass’ the screening compared to Gram positive microorganisms is because of the thicker peptidoglycan wall that Gram positive bacteria possess. The peptidoglycan wall of Gram positive bacteria absorbs the contaminants (PAHs) and the bacterial cell becomes overwhelmed, thus killing the cell.22 The percentage degradation recorded by all isolated bacterial species at the screening stage are shown in Table 4. Certain isolates such as Bacillus sp., B. aryabhattai, B. marisflavi and C. freundii successfully degraded up to and above 75% fluorene, but did not degrade acenaphthene accordingly (Table 4) and thus were not suitable candidates for degradation studies. The reason for this trend could not be verified as both compounds have the same number of rings (Fig. 1). Some bacterial species could degrade neither compound and some could not withstand the stress induced by the exposure to the compounds. This was deduced from the plate count, Gram reactions and biochemical tests monitoring during the experiments. Klebsiella oxytoca, E. coli, E. cloacae and S. saprophyticus did not grow on culture media plates for the duration of the experimentation. For species that successfully degraded the compounds, an increase in number of colonies was observed exponentially throughout the period of the experiment. This continued well until after the microorganisms had degraded a significant portion of the compounds, after which, a decline in colony growth was observed (Fig. 2).
Table 4.
Percentage degradation achieved by bacterial isolates at the temperature optimisation screening scale.
| Isolate | Percentage degradation (%)/temperature (°C) |
Analyte | ||||||
|---|---|---|---|---|---|---|---|---|
| 25 | 30 | 35 | 37 | 38 | 40 | 45 | ||
| R. ornithinolytica | 55.20 | 92.00 | 95.00 | 95.00 | 98.50 | 73.20 | 62.90 | Ace |
| 91.20 | 90.00 | 96.50 | 99.00 | 99.60 | 91.60 | 91.60 | Flu | |
| B. megaterium | 64.80 | 88.40 | 73.20 | 62.80 | 62.80 | 61.50 | 60.90 | Ace |
| 92.30 | 95.40 | 94.60 | 92.30 | 92.30 | 71.40 | 93.80 | Flu | |
| P. aeruginosa | 12.02 | 25.00 | 38.56 | 46.89 | 45.23 | 10.00 | 10.00 | Ace |
| 66.00 | 68.00 | 70.00 | 70.90 | 70.90 | 55.00 | 47.00 | Flu | |
| R. planticola | ND | ND | ND | ND | ND | ND | ND | Ace |
| ND | ND | ND | 5.01 | 3.02 | 3.02 | ND | Flu | |
| K. oxytoca | ND | ND | ND | ND | ND | ND | ND | Ace |
| ND | ND | ND | ND | ND | ND | ND | Flu | |
| E. coli | ND | ND | ND | ND | ND | ND | ND | Ace |
| ND | ND | ND | ND | ND | ND | ND | Flu | |
| E. cloacae | ND | ND | ND | ND | ND | ND | ND | Ace |
| ND | ND | ND | ND | ND | ND | ND | Flu | |
| E. acetylicum | ND | ND | ND | 11.09 | 18.66 | ND | 55.62 | Ace |
| 45.00 | 69.58 | 69.71 | 72.89 | 72.00 | 71.00 | 71.00 | Flu | |
| Acinetobacter sp. | ND | ND | ND | ND | ND | ND | ND | Ace |
| ND | ND | 19.28 | 56.32 | ND | ND | ND | Flu | |
| Exiguobacterium sp. | 52.00 | 12.00 | 54.00 | 55.00 | 66.21 | 55.89 | 64.00 | Ace |
| 66.09 | 68.21 | 60.23 | 60.23 | ND | 63.20 | 70.23 | Flu | |
| Bacillus sp. | 31.20 | 45.09 | 16.30 | ND | ND | ND | ND | Ace |
| 88.90 | 93.20 | 91.02 | 87.74 | 25.32 | 18.00 | 07.00 | Flu | |
| Serratia marcescens | 15.00 | 5.80 | 77.30 | 91.90 | 91.70 | 75.40 | 73.70 | Ace |
| 62.30 | 92.30 | 95.40 | 97.90 | 97.90 | 73.40 | 23.20 | Flu | |
| Aeromonas hydrophila | 75.40 | 99.40 | 99.50 | 99.20 | 89.88 | 62.80 | 55.20 | Ace |
| 74.10 | 95.80 | 99.23 | 99.80 | 74.40 | 64.60 | 57.30 | Flu | |
| Bacillus aryabhattai | ND | ND | ND | ND | ND | ND | ND | Ace |
| 88.00 | 99.00 | 91.00 | 88.32 | 84.20 | 63.20 | ND | Flu | |
| Bacillus aquimaris | ND | ND | ND | ND | ND | ND | ND | Ace |
| ND | ND | ND | ND | ND | ND | ND | Flu | |
| Bacillus marisflavi | ND | ND | ND | ND | ND | ND | ND | Ace |
| 78.00 | 98.00 | 90.00 | 72.00 | 71.00 | 13.20 | 05.78 | Flu | |
| S. saprophyticus | ND | ND | ND | ND | ND | ND | ND | Ace |
| ND | ND | ND | ND | ND | ND | ND | Flu | |
| Citrobacter freundii | ND | ND | ND | ND | ND | ND | ND | Ace |
| ND | 77.00 | 79.30 | 94.32 | 88.56 | 45.23 | ND | Flu | |
| E. undae | 13.02 | ND | ND | ND | 12.01 | 36.01 | 61.32 | Ace |
| 70.00 | 69.00 | 58.00 | 23.00 | 73.00 | 73.89 | 74.23 | Flu | |
Key: Ace: acenaphthene; Flu: fluorine; ND: no degradation.
Fig. 2.
Microbial plate counts from first to fourteenth day during biodegradation experiments.
Degradation efficiencies
Flask scale degradation
For all flask scale degradation experiments, the optimum temperature ranged between 30 °C and 38 °C which are comparable with results obtained by Antizar-Ladislao et al.23 and Moscoso et al.15 For most experiments, there was a sharp decline in degradation efficiency at 40 °C and 45 °C (Table 5), this could be due to the increase in temperatures coupled with a reduction in oxygen in the reactors at higher temperature profiles.24 For R. ornithinolytica and S. marcescens on acenaphthene and fluorene, the most efficient degradation was observed at 37 °C with a mean percentage degradation of 97.80% and 99.90% achieved by R. ornithinolytica and degradation percentages of 91.90% and 97.90% achieved for S. marcescens (Table 5). This might be due to the fact that both organisms grow optimally at 37 °C.25, 26 Both organisms also showed good degradation efficiencies at 38 °C, which are also comparable to results obtained by Antizar-Ladislao et al.23, 27 B. megaterium efficiently degraded both compounds at 30 °C with mean degradation percentages of 88.40% and 95.40% for acenaphthene and fluorene respectively (Table 5). This trend could also be attributed to optimum growth temperature for B. megaterium as shown by Logan and De Vos28 who compared growth temperature between 3 °C and 45 °C and determined 30 °C to be the optimum temperature for growth of the organism. For A. hydrophila, optimum degradation was observed at 35 °C with percentage degradation of 99.50% and 99.10% achieved for acenapthene and fluorene respectively. At temperature values of 30 °C, 35 °C and 37 °C efficient degradation was achieved for acenaphthene (99.40%, 99.50%, 99.20%) and fluorene (95.20%, 99.10%, 98.50%) respectively (Table 5).
Table 5.
Mean biodegradation values of acenaphthene and fluorene recorded at flask scale after 14 days.
| Flasks | Temperature (°C) | Initial concentration (ppm) | Residual concentration (ppm) | SD (±) | Percentage degradation (%) |
|---|---|---|---|---|---|
| A (Ace + R. ornithinolytica) | 25 | 50 | 22.38 | 0.54 | 55.2 |
| 30 | 50 | 3.72 | 0.8 | 92.6 | |
| 35 | 50 | 2.31 | 1.5 | 95.4 | |
| 37 | 50 | 1.09 | 0.2 | 97.8 | |
| 38 | 50 | 0.21 | 0.3 | 95.8 | |
| 40 | 50 | 13.41 | 1.23 | 73.2 | |
| 45 | 50 | 18.56 | 0.9 | 62.9 | |
| B (Ace + S. marcescens) | 25 | 50 | 27.49 | 0.02 | 45 |
| 30 | 50 | 22.38 | 0.89 | 55.2 | |
| 35 | 50 | 11.33 | 0.96 | 77.3 | |
| 37 | 50 | 4 | 1.63 | 91.9 | |
| 38 | 50 | 4.14 | 0.64 | 91.7 | |
| 40 | 50 | 12.29 | 0.45 | 75.4 | |
| 45 | 50 | 13.16 | 1.19 | 73.7 | |
| C (Ace + B. megaterium) | 25 | 50 | 17.58 | 0.63 | 64.8 |
| 30 | 50 | 5.82 | 0.32 | 88.4 | |
| 35 | 50 | 13.41 | 0.74 | 73.2 | |
| 37 | 50 | 18.62 | 1.19 | 62.8 | |
| 38 | 50 | 18.57 | 1.6 | 62.8 | |
| 40 | 50 | 19.25 | 3.56 | 61.5 | |
| 45 | 50 | 19.5 | 3.36 | 60.9 | |
| D (Ace + A. hydrophila) | 25 | 50 | 12.29 | 3.98 | 75.4 |
| 30 | 50 | 0.28 | 0.23 | 99.4 | |
| 35 | 50 | 0.24 | 0.06 | 99.5 | |
| 37 | 50 | 0.42 | 1.75 | 99.2 | |
| 38 | 50 | 5.06 | 1.97 | 89.88 | |
| 40 | 50 | 18.62 | 2.67 | 62.8 | |
| 45 | 50 | 22.38 | 0.35 | 55.2 | |
| (Ace + cocktail) | 25 | 50 | 13.1 | 0.03 | 73.8 |
| 30 | 50 | 0.28 | 0.65 | 99.4 | |
| 35 | 50 | 0.3 | 0.54 | 99.4 | |
| 37 | 50 | 0.42 | 0.34 | 99.2 | |
| 38 | 50 | 0.02 | 0.74 | 99.9 | |
| 40 | 50 | 0.42 | 0.56 | 91.5 | |
| 45 | 50 | 4.78 | 0.21 | 90.4 | |
| J (fluorene + R. ornithinolytica) | 25 | 50 | 4.22 | 1.23 | 91.6 |
| 30 | 50 | 1.96 | 1.12 | 96.1 | |
| 35 | 50 | 0.75 | 1.69 | 98.5 | |
| 37 | 50 | 0.02 | 0.65 | 99.9 | |
| 38 | 50 | 0.18 | 0.36 | 99.6 | |
| 40 | 50 | 4.22 | 0.39 | 91.6 | |
| 45 | 50 | 4.33 | 0.98 | 91.6 | |
| K (fluorene + S. marcescens) | 25 | 50 | 18.87 | 0.36 | 62.3 |
| 30 | 50 | 3.65 | 0.21 | 92.3 | |
| 35 | 50 | 2.28 | 0.14 | 95.4 | |
| 37 | 50 | 1.06 | 0.47 | 97.9 | |
| 38 | 50 | 1.07 | 0.13 | 97.7 | |
| 40 | 50 | 13.29 | 0.36 | 73.4 | |
| 45 | 50 | 38.4 | 0.97 | 23.2 | |
| L (fluorene + B. megaterium) | 25 | 50 | 2.68 | 1.16 | 94.6 |
| 30 | 50 | 2.28 | 1.23 | 95.4 | |
| 35 | 50 | 3.85 | 4.56 | 92.3 | |
| 37 | 50 | 3.87 | 4.41 | 92.3 | |
| 38 | 50 | 3.87 | 2.13 | 92.3 | |
| 40 | 50 | 14.2 | 0.12 | 71.4 | |
| 45 | 50 | 3.08 | 0.96 | 93.8 | |
| M (fluorene +A. hydrophila) | 25 | 50 | 12.7 | 1.1 | 74.4 |
| 30 | 50 | 2.39 | 0.36 | 95.2 | |
| 35 | 50 | 0.44 | 0.21 | 99.1 | |
| 37 | 50 | 0.73 | 0.15 | 98.5 | |
| 38 | 50 | 12.79 | 0.92 | 74.4 | |
| 40 | 50 | 14.71 | 0.89 | 64.6 | |
| 45 | 50 | 21.36 | 6.39 | 57.3 | |
| N (fluorene + cocktail) | 25 | 50 | 0.69 | 1.69 | 98.6 |
| 30 | 50 | 0.25 | 2.36 | 99.5 | |
| 35 | 50 | 0.31 | 3.35 | 99.4 | |
| 37 | 50 | 0.29 | 2.13 | 99.4 | |
| 38 | 50 | 0.3 | 0.36 | 99.4 | |
| 40 | 50 | 0.25 | 0.36 | 99.5 | |
| 45 | 50 | 0.25 | 0.38 | 99.5 |
Key: Ace: acenaphthene, Cocktail: R. ornithinolytica, S. marcescens, B. megaterium, A. hydrophila.
Fluorene degradation was generally more efficient and more rapid than acenaphthene degradation (Table 5, Table 6) as evidenced by the higher degradation percentages obtained over a wider range of temperatures. R. ornithinolytica degraded above 91% of fluorene at all temperature values tested (25–45 °C). This is more efficient compared to acenaphthene degradation at sub-optimal temperatures (55.20%, 73.20%, 62.90% at 25 °C, 40 °C and 45 °C respectively). B. megaterium also degraded above 92% at all temperature profiles except at 40 °C where 71.40% degradation was achieved. These values are better compared to 88.40% achieved at optimum temperature and between 73.20% and 64.80% achieved at sub-optimal temperatures during acenaphthene degradation studies. For most treatments, more than half of the compound had been degraded by the fifth day of treatment. The reason for this trend could not be verified.
Table 6.
Mean biodegradation percentages of acenaphthene and fluorene by the PAH degrading microorganisms at the Stirred Tank Bioreactor scale.
| Reactor | Initial concentration (ppm) | Residual concentration (ppm) | SD | Percentage degradation (%) |
|---|---|---|---|---|
| O (Ace + R. ornithinolytica) | 50.00 | 0.68 | 0.01 | 98.60 |
| P (Ace + S. marcescens) | 50.00 | 2.15 | 0.30 | 95.70 |
| Q (Ace + B. megaterium) | 50.00 | 4.91 | 0.90 | 90.20 |
| R (Ace + A. hydrophila) | 50.00 | 0.03 | 0.06 | 99.90 |
| S (ace + cocktail) | 50.00 | 0.15 | 0.93 | 99.60 |
| T (fluorene + R. ornithinolytica) | 50.00 | 0.02 | 0.09 | 99.90 |
| U (fluorene + S. marcescens) | 50.00 | 0.99 | 0.36 | 97.90 |
| V (fluorene + B. megaterium) | 50.00 | 0.82 | 0.23 | 98.40 |
| W (fluorene +A. hydrophila) | 50.00 | 0.24 | 0.36 | 99.50 |
| X (fluorene + cocktail) | 50.00 | 0.39 | 1.6 | 99.20 |
Key: Ace- acenaphthene, Cocktail: Raoultella ornithinolytica, Serratia marcescens, Bacillus megaterium and Aeromonas hydrophila.
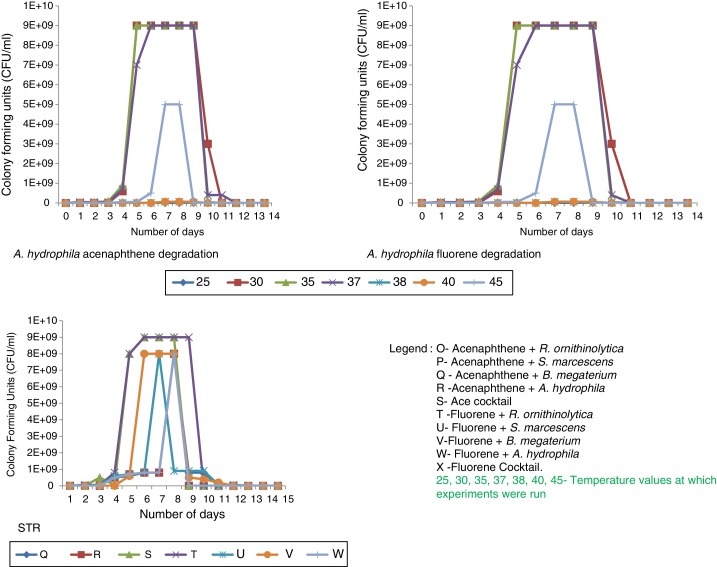
The results obtained showed that for the pure strain experiments, A. hydrophila degraded acenaphthene best (99.50%) while R. ornithinolytica degraded fluorene most efficiently (99.90%). The poorest degradation was achieved in reactor B (acenaphthene and B. megaterium) for which only 88.40% degradation percentage was achieved at optimum temperature (Table 5). After the first few days of the experiment, biodegradation profiles became static in reactor B, factors influencing this occurrence could include (i) a decrease in the bioavailability of the compound; (ii) accumulation of toxic metabolites; or (iii) the enrichment of more recalcitrant compounds.24 For the cocktail experiments, a slight improvement in degradation efficiency was observed for both compounds especially fluorene with degradation percentages within the range of 98.60% and 99.50% recorded at all temperature profiles. This could be due to multiple, co-operative metabolic capacities which could in turn improve the efficiency of the bioremediation processes.24 At temperatures higher than 60 °C the isolates could not survive and thus degradation percentages significantly declined.
Stirred Tank Bioreactor scale
At the Stirred Tank Bioreactor scale mean percentage degradation achieved ranged between 90.20% and 99.90% for all experiments (Table 6). R. ornithinolytica, S. marcescens, B. megaterium, A. hydrophila and the cocktail showed a 98.60%, 95.70%, 90.20%, 99.90% and 99.60% degradation efficiency for acenaphthene and a 99.90%, 97.90%, 98.40%, 99.50% and 99.20% degradation efficiency for fluorene. The improved efficiency at STR scale compared to flask scale could be attributed to improved oxygenation and mixing afforded the system by the overhead stirrers and impellers as opposed to the gentle agitation available in the shaking incubator at the flask scale. Agitation has been shown to significantly improve the dissolution rate of PAH particles as PAH biodegradation is known to be inhibited by slow dissolution rates thus contributing to increased efficiency of STRs.29 A study by Vinas et al.,24 showed significant increase in degradation efficiency rates with improved agitation and stirring compared to gentle agitation. The increased dissolution rate is influenced by two factors both of which enhance the volumetric mass transfer co-efficient. These factors include: reduced film thickness due to intense turbulence generated by the agitation inside the bioreactor and the increased interfacial surface area of the PAH particles resulting from collisions between particles and particles as well as between particles and the impeller.29 The mean biodegradation percentages achieved by the microorganisms at the Stirred Tank Bioreactor scale are represented in Table 6.
Microbial cell count during and after degradation
During R. ornitinolytica treatment of acenaphthene, at optimal temperature range (30–38 °C) there was an increase in cell count from 5 × 104 CFU/ml to 7 × 108 CFU/ml by the seventh day of the experiment (Fig. 2). However, by the 14th day of the experiment, there was a drastic decrease in cell count to around 5 × 103 CFU/ml. At 25 °C, 40 °C and 45 °C there was an increase in cell count from 5 × 104 CFU/ml to 5 × 105 CFU/ml, 6 × 107 CFU/ml, 5 × 107 CFU/ml respectively by day seven (Fig. 2). For fluorene, at 25 °C, 30 °C, 35 °C, 37 °C, 38 °C, 40 °C and 45 °C there was an increase in cell count from 5 × 103 CFU/ml to 6 × 105 CFU/ml, 7 × 107 CFU/ml, 9 × 108 CFU/ml, 9 × 108 CFU/ml, 9 × 108 CFU/ml, 8 × 108 CFU/ml and 7 × 108 CFU/ml respectively by the seventh day of the experiment (Fig. 2).
For B. megaterium an increase was observed in plate count from 6 × 104 CFU/ml to 7 × 105 CFU/ml at 30 °C during acenaphthene treatment by the fourth day of the experiment (Fig. 2). At all other temperature profiles, there was no significant increase in plate count. For fluorene however, at 25 °C, 30 °C, 35 °C, 37 °C, 38 °C, 40 °C and 45 °C there was an increase in plate count from 6 × 104 /ml to 9 × 105 CFU/ml, 9 × 108 CFU/ml, 8 × 107 CFU/ml, 8 × 107 CFU/ml, 8 × 105 CFU/ml, 7 × 104 CFU/ml, 7 × 104 CFU/ml respectively after 7 days (Fig. 2).
There was an increase in average plate count ranging from 4 × 105 CFU/ml to 6 × 108 CFU/ml after seven days for S. marcescens degradation experiments within the optimum temperature range for acenaphthene (35–38 °C) and fluorene (30–38 °C). At 25 °C, 40 °C and 45 °C there was no significant increase in plate count for both compounds (Fig. 2).
At Stirred Tank Bioreactor scale, an increase was also observed in plate count ranging from 4 × 104 CFU/ml to 5 × 106 CFU/ml to 8 × 109 CFU/ml to 9 × 109 CFU/ml by the seventh day for all experiments (Fig. 2). The reason for the increase in plate count was associated with rapid cell proliferation because of the supply of adequate carbon sources.30, 31 However, by the end of the experiments, there was either a drastic decline in cell growth or the microorganisms were dead evidenced by no cell growth on culture media plates. This was attributed to the possibility that the microorganisms had utilised all the contaminants (serving as carbon sources) and hence, stopped replicating abundantly. Apart from substrate depletion, another plausible explanation for the drastic decline in microbial numbers is that toxic intermediates and by-products such as salicylate as well as oxy-PAHs; including PAH-ketones, quinones and coumarins32, 33, 34, 35 might have been produced and accumulated in the reactors thereby causing the death of the bacterial cells.
The indigenous microorganisms used in this study have great hydrocarbonoclastic potential since natural attenuation occurred without any nutrient supplementation or any other sort of biostimulation. They cannot be described as obligate hydrocarbonoclastic microorganisms (OHCBs) since they were successfully cultured on undefined growth media.
The temperature conditions under which the microorganisms were isolated are not best suitable for the biodegradative potential of the isolates to be optimally expressed. This is shown by the varying optimum temperature profiles encountered during the flask scale experiments (none of which corresponds with the temperature conditions at sampling time). Each microorganism investigated had a particular optimum temperature requirement which directly influenced treatment efficiency of the PAH compounds. However, for the likely adoption of these microorganisms and temperature protocols for restoration of PAH contaminated river systems, the temperature of river systems cannot be controlled in order to achieve successful bioremediation, but bioremediation efforts can be made on warm days when the average temperature ranges between 30 °C and 38 °C.
Under appropriate temperature conditions, the microorganisms studied can utilise acenaphthene and fluorene as carbon or energy sources therefore might be capable of efficiently remediating PAH polluted environments, as evidenced by the increase in cell numbers recorded during bioremediation studies.
The hydrocarbonoclastic microorganisms identified in this study could possibly be utilised in the remediation of PAH polluted river systems on a commercial scale. In addition, industries in the vicinity of the river systems can use these microorganisms to pre-treat their wastes/effluents prior to release into the environment or waste disposal systems. Furthermore, it would be a more efficient, cost-effective and environmental friendly approach to pre-treat effluents and waste waters compared to many other available techniques, such as activated sludge systems, desalination, distillation, dark fermentation and wet oxidation, among others.
Attempts can possibly be made to use the identified hydrocarbonoclastic microorganisms for degradation of other polycyclic aromatic hydrocarbon compounds. The structure and toxicity of the by-products and intermediates produced during the microbial metabolism of acenaphthene and fluorene should be investigated in further studies. The factors responsible for the more proficient degradation of fluorene compared to acenaphthene should also be investigated.
Raoultella ornithinolytica, S. marcescens and B. megaterium can be used on a larger, commercial scale to restore polluted aquatic ecosystems. However, A. hydrophila has been shown to cause diseases in commercially important aquatic species such as fish36, 37 and therefore, cannot be used on a larger, commercial scale to replenish PAH contaminated river systems due to safety reasons.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The authors wish to acknowledge the financial support of the National Research Foundation (NRF), South Africa, through the Thuthuka Research Grant No. 84185 awarded to Prof B.O. Opeolu. They also wish to acknowledge Dr. O. Oguntoke for his technical assistance.
Associate Editor: Lucy Seldin
References
- 1.Wick AF, Haus NW, Sukkariyah BF, Hearing KC, Lee Daniels W. Remediation of PAH-contaminated soils and sediments: a literature review. Department of Crop and Soil Environmental Sciences Blacksburg, VA 2011:24061 Virginia Polytechnic Institute and State University.
- 2.Quinn L., Pieters R., Nieuwoudt C., Borgen A., Kylin H., Bouwman H. Distribution profiles of selected organic pollutants in soils and sediments of industrial, residential and agricultural areas of South Africa. J Environ Monit. 2009;9:1647–1657. doi: 10.1039/b905585a. [DOI] [PubMed] [Google Scholar]
- 3.Soleimani M. Bioremediation: an environmental friendly solution for oil contaminated soils. J Bioremed Biodegrad. 2012;3:2155–6199. [Google Scholar]
- 4.Wu M., Chen L., Tian Y., Dinga Y., Dick W.A. Degradation of polycyclic aromatic hydrocarbons by microbial consortia enriched from three soils using two different culture media. Environ Pollut. 2013;178:152–158. doi: 10.1016/j.envpol.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Silva I.S., Grossman M., Durranta L.R. Degradation of polycyclic aromatic hydrocarbons (2–7 rings) under microaerobic and very-low-oxygen conditions by soil fungi. Int Biodeterior Biodegrad. 2009;63:224–229. [Google Scholar]
- 6.Skupinska K., Mislewicz I., Kasprzycka-Guttman T. Polycyclic aromatic hydrocarbons: environmental appearance and impact on living organisms. Acta Pol Pharma-Drug Res. 2004;61:233–240. [PubMed] [Google Scholar]
- 7.Guerin T.F. The extraction of aged polycyclic aromatic hydrocarbon (PAH) residues from clay soil using sonification and soxhlet extraction procedure: a comparative study. J Environ Monit. 1999;1:63–67. doi: 10.1039/a807307d. [DOI] [PubMed] [Google Scholar]
- 8.Olatunji O.S., Fatoki O.S., Opeolu B.O., Xhimba B.J. Determination of polycyclic aromatic hydrocarbons [PAHs] in processed meat products using gas chromatography – flame ionization detector. Food Chem. 2014;156:296–300. doi: 10.1016/j.foodchem.2014.01.120. [DOI] [PubMed] [Google Scholar]
- 9.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons; New York: 1991. pp. 21–32. [Google Scholar]
- 10.Turner S., Pryer K.M., Miao V.P., Palmer J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 11.Pathak H., Kantharia D., Malpani A., Madamwar D. Naphthalene degradation by Pseudomonas sp. HOB1: in vitro studies and assessment of naphthalene degradation efficiency in simulated microcosms. J Hazard Mater. 2009;166:1466–1473. doi: 10.1016/j.jhazmat.2008.12.074. [DOI] [PubMed] [Google Scholar]
- 12.Chen J., Wong M.H., Wong Y.S., Tam N.F.Y. Multi-factors on biodegradation kinetics of polycyclic aromatic hydrocarbons (PAHs) by Sphingomonas sp. a bacterial strain isolated from mangrove sediment. Marine Poll Bull. 2008;57:695–702. doi: 10.1016/j.marpolbul.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Antizar-Ladislao B., Lopez-Real J., Beck A.J. Degradation of polycyclic aromatic hydrocarbons (PAHs) in an aged coal tar contaminated soil under in-vessel composting conditions. Environ Pollut. 2006;141(3):459–468. doi: 10.1016/j.envpol.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 14.Amenu D. Isolation of poly aromatic hydrocarbons (PAHs) degrading bacteria. Landmark Res J. 2014;1:1–3. [Google Scholar]
- 15.Moscoso F., Ferreira L., Deive F.J., Morán P., Sanromán M.A. Viability of phenanthrene biodegradation by an isolated bacterial consortium: optimization and scale-up. Bioprocess Biosyst Eng. 2012;36(2):133–141. doi: 10.1007/s00449-012-0768-3. [DOI] [PubMed] [Google Scholar]
- 16.Dodor D.E., Hwang H.M., Ekunwe S.I. Oxidation of anthracene and benzo(a)pyrene by immobilized laccase from Trametes versicolor. Enzyme Microb Technol. 2004;35:210–217. [Google Scholar]
- 17.Bordoloi N.K., Konwar B.K. Bacterial biosurfactant in enhancing solubility and metabolism of petroleum hydrocarbons. J Hazard Mater. 2009;170(1):495–505. doi: 10.1016/j.jhazmat.2009.04.136. [DOI] [PubMed] [Google Scholar]
- 18.Vo-Dinh T., Fetzer J., Campligia A.D. Monitoring and characterization of polyaromatic compounds in the environment. Talanta. 1998;47(4):943–969. doi: 10.1016/s0039-9140(98)00162-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Tao S. Global atmospheric emission inventory of polycyclic aromatic hydrocarbons (PAHs) for 2004. Atmos Environ. 2009;43:812–819. [Google Scholar]
- 20.Liang Y., Fung P.K., Tse M.F., Hong H.C., Wong M.H. Sources and seasonal variation of PAHs in the sediments of drinking water reservoirs in Hong Kong and the Dongjiang River (China) Environ Monit Assess. 2008;146:41–50. doi: 10.1007/s10661-007-0058-8. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H., Sun L., Sun T., Li H., Luo Q. Spatial distribution and seasonal variation of polycyclic aromatic hydrocarbons (PAHs) contaminations in surface water from the Hun River, Northeast China. Environ Monit Assess. 2013;185:1451–1462. doi: 10.1007/s10661-012-2644-7. [DOI] [PubMed] [Google Scholar]
- 22.Silhavy T.J., Kahne D., Walker S. The bacterial cell envelope. Cold Spring Harbor Persp Biol. 2010 doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antizar-Ladislao B., Spanova K., Beck A.J., Russell N.J. Microbial community structure changes during bioremediation of PAHs in an aged coal-tar contaminated soil by in-vessel composting. Int Biodeterior Biodegr. 2008;61:357–364. [Google Scholar]
- 24.Vinas M., Sabate J., Espuny M., Solanas A. Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol. 2005;71:7008–7018. doi: 10.1128/AEM.71.11.7008-7018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdou A.M. Purification and partial characterisation of psychrotrophic Serratia marcescens lipase. J Dairy Sci. 2003;86:127–132. doi: 10.3168/jds.S0022-0302(03)73591-7. [DOI] [PubMed] [Google Scholar]
- 26.Lin C.S., Kung H.F., Lin C.M., Tsai H.C., Tsai Y.H. Histamine production by Raoultella ornithinolytica in mahi-mahi meat at various storage temperatures. J Food Drug Anal. 2016;24(2):305–310. doi: 10.1016/j.jfda.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antizar-Ladislao B., Lopez-Real J., Beck A.J. Laboratory studies of the remediation of polycyclic aromatic hydrocarbon contaminated soil by in-vessel composting. Waste Manage. 2005;25(3):281–289. doi: 10.1016/j.wasman.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Logan N.A., De Vos P. Genus Bacillus Cohn 1872. In: Vos P.D., Garrity G., Jones D., Krieg N.R., Ludwig W., Rainey F.A., Schleifer K.H., Whitman W.B., editors. Bergey's Manual of Systematic Bacteriology. Springer; New York: 2009. pp. 3–9. [Google Scholar]
- 29.Ruihong Y. University of Saskatchewan; 2006. Bioremediation of Polycyclic Aromatic Hydrocarbon (PAH)-Contaminated Soils in a Roller Baffled Bioreactor. PhD thesis. [Google Scholar]
- 30.Johnsen A.R., Bendixen K., Karlson U. Detection of microbial growth on polycyclic aromatic hydrocarbons in microtiter plates by using the respiration indicator WST-1. Appl Environ Microbiol. 2002;68:2683–2689. doi: 10.1128/AEM.68.6.2683-2689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnsen A.R., Schmidt S., Hybholt T.K., Henriksen S., Jacobsen C.S., Andersen O. Strong impact on the polycyclic aromatic hydrocarbon (PAH)-degrading community of a PAH-polluted soil but marginal effect on PAH degradation when priming with bioremediated soil dominated by mycobacteria. Appl Environ Microbiol. 2007;73(5):1474–1480. doi: 10.1128/AEM.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bamforth S.M., Singleton I. Bioremediation of polycyclic aromatic hydrocarbons: current knowledge and future directions. J Chem Technol Biotechnol. 2005;80:723–736. [Google Scholar]
- 33.Mrozik A., Piotrowska-Seget S., Labuzek S. Bacterial degradation and bioremediation of polycyclic aromatic hydrocarbons. Polish J Environ Stud. 2003;12:15–25. [Google Scholar]
- 34.Moscoso F., Tejiz I., Deive F.J., Sanroman M.A. Efficient PAHs biodegradation by a bacterial consortium at flask and bioreactor scale. Bioresour Technol. 2012;119:270–276. doi: 10.1016/j.biortech.2012.05.095. [DOI] [PubMed] [Google Scholar]
- 35.Moscoso F., Deive F.J., Longo M.A., Sanromán M.A. Insights into polyaromatic hydrocarbon biodegradation by Pseudomonas stutzeri CECT 930: operation at bioreactor scale and metabolic pathways. Int J Environ Sci Technol. 2015;12:1243–1252. [Google Scholar]
- 36.Janda J.M., Abott S.L. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23(1):35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Austin B., Austin D.A. Bacterial fish pathogens: disease of farmed and wild fish. In: Dobbins P., editor. Bacterial Fish Pathogens. 3rd ed. Springer-Verlag KG; Berlin, Germany: 1999. [Google Scholar]