Abstract
Enterohemorrhagic Escherichia coli, enteropathogenic E. coli, and Citrobacter rodentium are highly adapted enteropathogens that successfully colonize their host's gastrointestinal tract via the formation of attaching and effacing (A/E) lesions. These pathogens utilize a type III secretion system (TTSS) apparatus, encoded by the locus of enterocyte effacement, to translocate bacterial effector proteins into epithelial cells. Here, we report the identification of EspJ (E. coli-secreted protein J), a translocated TTSS effector that is carried on the 5′ end of the cryptic prophage CP-933U. Infection of epithelial cells in culture revealed that EspJ is not required for A/E lesion activity in vivo and ex vivo. However, in vivo studies performed with mice demonstrated that EspJ possesses properties that influence the dynamics of clearance of the pathogen from the host's intestinal tract, suggesting a role in host survival and pathogen transmission.
Enterohemorrhagic Escherichia coli (EHEC), enteropathogenic E. coli (EPEC) (35), and Citrobacter rodentium (29) are highly adapted enteropathogens that successfully colonize their host's gastrointestinal tract via the formation of attaching and effacing (A/E) lesions (reviewed in references 13 and 28). These lesions are characterized by the localized destruction (effacement) of intestinal epithelial microvilli, an intimate attachment between the bacterium and the host cell apical membrane, and the formation of pedestal-like structures containing high concentrations of actin (26) and intermediate filaments (2) directly beneath sites of bacterial attachment. Formation of A/E lesions is mediated by a filamentous type III secretion system (FTTSS) apparatus (20, 42) responsible for the translocation of bacterial effectors into eukaryotic cells, where they subvert host cell signaling (13). The TTSS apparatus is encoded on a pathogenicity island called the locus of enterocyte effacement (LEE). The LEE was first described in EPEC O127:H6 strain E2348/69 by McDaniel et al. (31) but is also present in EHEC (36), rabbit-specific EPEC (44), and C. rodentium (10).
The LEE also encodes six effector proteins, Tir, Map, EspF, EspG, EspH, and SepZ, which are all translocated into the host cell via the LEE-encoded FTTSS (11, 22, 24, 32, 45; James Kaper, personal communication). Tir is inserted into the host cell plasma membrane (22), where it functions as a bacterial receptor for intimin (21), an outer-membrane bacterial adhesion molecule (reviewed in reference 13). In the plasma membrane, Tir adopts a hairpin-loop topology featuring a central extracellular domain that binds intimin (17). The amino- and carboxy-terminal domains of Tir are oriented in the host cell cytoplasm, where they interact with host cytoskeletal and signaling components (reviewed in reference 4). Map targets mitochondria and has been suggested to disrupt their normal function (24), and it also initiates filopodium formation immediately upon interaction with the host cell via the GTPase Cdc42 (23). In contrast, EspH represses filopodium formation and enhances the formation of actin-rich pedestals (45). EspG is a 44-kDa protein homologous to VirA, a translocated protein of Shigella flexneri that triggers host microtubule destabilization but has an unknown role in virulence (11). EspF disrupts intestinal barrier function (32) and therefore potentially contributes to EPEC diarrhea. EspF also plays a role in epithelial cell apoptosis (7). Importantly, all the LEE-encoded effectors except Tir are dispensable for A/E lesion formation.
In addition, three non-LEE-encoded proteins named Cif (cycle-inhibiting factor) (30), EspI/NleA (16, 34), and TccP (Tir cytoskeleton coupling protein)/EspFu (5, 15) are translocated via the LEE-encoded FTTSS. Cif is carried on a lambdoid phage and triggers an irreversible cytopathic effect in HeLa cells, which is characterized by the progressive recruitment of focal adhesions, assembly of stress fibers, and arrest of the cell cycle (30). EspI/NleA is carried within prophage CP-933P, localizes to the Golgi (16), and is required for full virulence in the C. rodentium model (16, 34). TccP/EspFU is an EHEC O157 effector that is carried on prophage CP-933U and translocated into host cells, where it displays an Nck-like activity (5, 15).
EHEC infection is associated with a range of symptoms from nonbloody diarrhea, fever, and vomiting to bloody diarrhea (hemorrhagic colitis) and hemolytic-uremic syndrome, a life-threatening condition. EPEC causes infantile nonbloody diarrhea in developing countries, and although EPEC and EHEC share many genes implicated in virulence, EHEC, but not EPEC, produces a potent cytotoxin, Shiga toxin (Stx), which is responsible for the severe complications that characterize its infection (reviewed in reference 35). EHEC and EPEC are human pathogens, and as such, they are poorly pathogenic in other animal species. At present, there is no natural small-animal model that allows in vivo study of EHEC or EPEC. However, C. rodentium, a mouse-specific pathogen that possesses the LEE pathogenicity island, causes transmissible colonic hyperplasia (1; reviewed in reference 28), and induces colonic A/E lesions indistinguishable from those caused by EHEC and EPEC (40, 41), provides an excellent small animal model to simulate in vivo infection for those two pathogens (16, 34). In addition, neonatal- and weaned-lamb infection models have recently been reported (48). Animals experimentally infected with EHEC O157:H7 exhibited cecal and rectal colonization with A/E, although the infection was not associated with clinical disease. Intimin has been shown to be essential for persistence of EHEC in lambs (48).
Recently, we monitored the global transcription profile of EHEC O157:H7 (Sakai strain) during attachment to eukaryotic plasma membranes (8). This has shown that the majority of the LEE-genes were down-regulated in attached bacteria. In addition, decreased levels of two mRNAs of adjacent genes, Z3071 (Ecs2714) and Z3072 (Ecs2715), carried within prophage CP-933U, were also detected (8). Importantly, Z3072 (TccP/EspFU) is present in EHEC but not in EPEC or C. rodentium (5, 15), while Z3071 is present in EHEC, EPEC, and C. rodentium, and its product is homologous to the type III effector HopF of Pseudomonas syringae (43). The aim of this study was to define the role of Z3071 in the pathogenesis of A/E pathogens in vitro, ex vivo, and in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Bacteria were grown in Luria-Bertani (LB) medium or in Dulbecco's modified Eagle's medium. When appropriate, additional antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 50 μg/ml; and tetracycline, 12.5 μg/ml. Minimal medium consisted of M9 salts supplemented with 1% glucose.
TABLE 1.
Strains and plasmids used in this study
| Name | Description | Reference or source |
|---|---|---|
| Strains | ||
| E2348/69 | Wild-type EPEC O127:H6 | 27 |
| ICC171 | ΔescF::Kan in E2348/69 | 47 |
| 85-170 | EHEC O157:H7 Δstx | 38 |
| ICC169 | Spontaneous naladixic acid-resistant derivative of wild-type C. rodentium | 46 |
| ICC188 | ΔespJ::Kan in 85-170 | This study |
| ICC189 | ΔespJ::Kan in ICC169 | This study |
| ICC190 | ΔespJ::Kan in E2348/69 | This study |
| Plasmids | ||
| pCX340 | pBR322 derivative; cloning vector used to fuse genes to blaM, which encodes the mature form of TEM-1 β-lactamase | 6 |
| pCX327 | Derivative of pCX340 encoding a fusion of residues 1 to 16 of Cif to TEM-1 | 6 |
| pICC283 | Derivative of pCX340 encoding a fusion of EspJ to TEM-1 | This study |
| pGEMT | Cloning vector | Promega |
| pSB315 | Source of aphT cassette | 14 |
| pKD46 | oriR101 repA101(Ts) blaM araBp-gam-bet-exo | 9 |
| pKD4 | Template plasmid conferring kanamycin resistance | 9 |
Translocation assay.
Translocation assays were performed as described by Charpentier and Oswald (6). Wild-type and ΔescF EPEC E2348/69 strains carrying derivatives of the pCX340 plasmid, a cloning vector encoding the mature form of TEM-1 β-lactamase, were subcultured in LB medium supplemented with tetracycline and incubated for 16 h at 37°C. The cultures were diluted 1/100 in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 2 mM glutamine at 37°C for 3.5 h (preactivation). HeLa cells grown on glass coverslips were infected with 1 ml of preactivated bacterial culture and incubated at 37°C in 5% CO2. After 30 min of infection, IPTG (isopropyl-β-d-thiogalactopyranoside) was added at a final concentration of 1 mM, and the coverslips were incubated for an additional hour. The cell monolayers were washed three times with phosphate-buffered saline (PBS) and covered with 100 μl of PBS plus 25 μl of 6× CCF2/AM solution freshly prepared from the CCF2/AM Loading kit (Invitrogen) (final concentration of CCF2/AM, 1 μM). The cells were incubated in darkness at room temperature for 2 h and then washed three times with PBS and observed under a Nikon Eclipse E600 fluorescence microscope using a UV-2A filter set (330- to 380-nm excitation). Pictures were taken using a Nikon DXM1200 digital camera.
Construction of nonpolar espJ mutations.
The EHEC 85-170 and C. rodentium espJ mutants were constructed by using a modification of the one-step method (9). In each case, a part of the gene and the flanking regions from both sides were amplified from the wild-type genomic DNA by PCR using the following pairs of primers (Table 2): for the EHEC mutant, EHEC-espJ-flank-f1 plus EHEC-espJ-flank-r1 (fragment 1) and EHEC-espJ-flank-f2 plus EHEC-espJ-flank-r2 (fragment 2); for the C. rodentium mutant, Citro-espJ-flank-f1 plus Citro-espJ-flank-r1 (fragment 1) and Citro-espJ-flank-f2 plus Citro-espJ-flank-r2 (fragment 2). For each mutant, the two PCR fragments (fragment 1 and fragment 2) were digested with BamHI, ligated to each other, amplified, and cloned into the cloning vector pGEMT. Then, in order to inactivate the gene and facilitate mutant strain identification, the nonpolar aphT cassette (14) conferring kanamycin resistance was inserted between the fused fragments (1 and 2) contained in pGEMT, at the BamHI site. The inserts containing the aphT cassette in the correct orientation were then amplified by PCR with primers EHEC-espJ-flank-f1 and EHEC-espJ-flank-r2 for the EHEC mutant and Citro-espJ-flank-f1 and Citro-espJ-flank-r2 for the C. rodentium mutant. In order to enhance allelic exchange, plasmid pKD46 was transformed into wild-type EHEC O157:H7 strain 85-170 and into wild-type C. rodentium strain ICC169 by electroporation, generating strains 85-170(pKD46) and ICC169(pKD46). The PCR product containing the flanking regions and the kanamycin resistance cassette was transformed by electroporation into each wild-type strain containing the pKD46 plasmid. Clones were grown on LB medium containing kanamycin to select for kanamycin resistance. pKD46 was cured by growth at 42°C. Mutations were verified by PCR.
TABLE 2.
Primers used in this study
| Name | Primer sequence (5′→3′) |
|---|---|
| EHEC-espJ-flank-f1 | GAATTCAAGGTAGTAGTACTTATCTGC |
| EHEC-espJ-flank-r1 | CGGGATCCCGATAATTGACATTATAAATGCCTT |
| EHEC-espJ-flank-f2 | CGGGATCCCGGATACATCATGCTCTCTGAG |
| EHEC-espJ-flank-r2 | CTGCAGTCATGCAGATTACCTTATAAG |
| Citro-espJ-flank-f1 | GAATTCTTCTAACACACCAGATCCTG |
| Citro-espJ-flank-r1 | CGGGATCCCGACAGTTTTCCTGTTTGTTCT |
| Citro-espJ-flank-f2 | CGGGATCCCGCATCATTACACTTCCAGAATC |
| Citro-espJ-flank-r2 | CTGCAGTTCACCAGGCATTGCGAAT |
| EPEC-espJ-pKD4-f | CCAATCATAAAGAACTGCTTATCATCAATTAGTAACATATTACGCAACGAGTGTAGGCTGGAGCTGCTTC |
| EPEC-espJ-pKD4-r | CATCCAGCCTGACTGTTTCTGGAAGTGTAATAATGAATGGTTCTCCCAGTCATATGAATATCCTCCTTAG |
The EPEC E2346/69 espJ mutant was constructed by using the one-step method (9). Briefly, a PCR product was generated by amplifying pKD4 template plasmid using long primers (EPEC-espJ-pKD4-f and EPEC-espJ-pKD4-r) that corresponded to 20 nucleotides of pKD4 and 50 nucleotides of flanking DNA regions homologous to the 5′ and 3′ ends of the espJ gene. The PCR product contains a kanamycin resistance gene (derived from pKD4) flanked by 50-bp sequences located upstream and downstream of the espJ gene. The PCR product was then transformed by electroporation into EPEC E2348/69 containing the pKD46 plasmid. Clones were grown on LB medium containing kanamycin to select for kanamycin resistance. pKD46 was cured by growth at 42°C. Mutation was verified by PCR.
FAS test and IVOC assay.
Fluorescent actin staining (FAS) testing was performed on infected HEp-2 cells as described by Knutton et al. (25). For the human intestinal in vitro organ culture (IVOC) assay, tissue was obtained with fully informed parental consent and local ethical committee approval using grasp forceps during routine endoscopic (Fujinon EG/EC-41 pediatric endoscope) investigation of intestinal disorders. Terminal ileal mucosal biopsy specimens from the Peyer's patch region that appeared macroscopically normal were taken for organ culture experiments. Light microscopy subsequently showed no histological abnormality. IVOC infections were performed as described previously (19). 85-170 ΔespJ (ICC188) was examined using tissue from three patients (aged 46, 110, and 141 months). In each experiment, an uninoculated sample (to exclude endogenous bacterial adhesion) and a positive control (IVOC with the parental strain 85-170 to exclude host factors) were included. Samples were fixed with 2.5% glutaraldehyde, postfixed in 1% aqueous osmium tetroxide, and processed for viewing by a JEOL JSM 5300 scanning electron microscope.
Mice.
Two murine models that differ in their susceptibility to C. rodentium were used in this study. Female 6- to 8-week-old C57BL/6J mice and male 5- to 6-week-old C3H/HeJ mice were purchased from Harlan Olac (Bichester, United Kingdom) and came from specific-pathogen-free stocks. During the course of these studies, sentinel animals were screened for common murine pathogens every 2 months. All animals were housed in individually HEPA-filtered cages with sterile bedding and free access to sterilized food and water.
Oral infection of mice.
Mice were orally inoculated using a gavage needle with 200 μl of bacterial suspension (≈1010 CFU for C57BL/6J mice (46) and ≈2 × 108 CFU for C3H/HeJ mice (34). The viable count of the inoculum was determined by retrospective plating on LB agar containing the appropriate antibiotic. Independent experiments were performed at least twice using groups of at least four mice per strain. Stool samples were recovered aseptically at various times after inoculation, and the number of viable bacteria per gram of stool was determined by plating the stool onto LB agar containing the appropriate antibiotics. At selected times postinfection, mice were killed by cervical dislocation. The colon and cecum were aseptically removed and weighed after the removal of fecal pellets and cecal contents. The organs were then homogenized mechanically in 5 ml of sterile PBS using a Seward (London, United Kingdom) 80 stomacher, and the number of viable bacteria per gram of organ homogenate was determined by plating the homogenate onto LB agar containing the appropriate antibiotics.
Oral inoculation of sheep.
Ten 6-week-old crossbred lambs were randomly divided into two equal groups, supplied with food and water ad libitum, and confirmed to be free of EHEC O157 by enrichment and O157 immunomagnetic separation. All of the lambs were housed in biosecure containment level 2 accommodations. Each group was housed in a separate room with its own air handling. The animals were visited only by experienced staff, who changed clothing between groups. Five lambs were each dosed orally with either 109 CFU of 85-170 Nalr or 85-170 ΔespJ::Kanr resuspended in 10 ml of PBS (pH 7.4). Approximately 24 h after the dosing, and as required thereafter for up to 27 days, rectal fecal samples from each lamb were collected for direct plating onto sorbitol-MacConkey (Oxoid) plates supplemented with either 15 μg of nalidixic acid/ml or 25 μg of kanamycin/ml (Sigma). Samples that were negative on direct plating were enriched in buffered peptone water for 6 h at 37°C and then plated onto sorbitol-MacConkey plates supplemented with the appropriate antibiotic. Representative colonies were confirmed to be E. coli O157 by latex agglutination (Oxoid). All animal experiments were performed in accordance with the Animals Scientific Procedures Act (1986) and were approved by the local ethical review committee.
Statistics.
The unpaired Student t test was used to compare normally distributed values from groups of animals. The nonparametric Mann-Whitney and Kruskal-Wallis tests were used to compare nonnormally distributed values.
RESULTS
Z3071 is present in EHEC, EPEC, and C. rodentium, and its product is similar to HopF, an effector protein of P. syringae.
Z3071 (Ecs2714) is located within the 5′ end of the cryptic prophage CP-933U in EDL933 (37) and RIMD 0509952 Sakai (18), two clinical EHEC O157:H7 isolates that were recently sequenced. Z3071 is also present in EPEC O127:H6 strain E2348/69 (http://www.sanger.ac.uk/projects/Escherichia_Shigella) and C. rodentium (http://www.sanger.ac.uk/Projects/C_rodentium), but not in the E. coli K-12 genome (3), with 79.3 and 75.1% identity at the amino acid level, respectively. Z3071 encodes a 217-amino-acid protein with unknown function and is located upstream of Z3072 (tccP/espFu), encoding an EHEC type III effector protein that is required for EHEC-induced actin polymerization (5, 15). Similar to tccP and most of the LEE-located genes, Z3071 is also down-regulated in bacteria closely attached to plasma membranes (8). Importantly, a search of the database revealed that Z3071 has 22% identity with HopF, an effector protein of P. syringae (http://ca.expasy.org; 43). The data suggested that Z3071 might be a virulence factor of EHEC, EPEC, and C. rodentium, and it was therefore chosen for further investigation.
Z3071 is translocated into epithelial cells.
To determine whether Z3071 is translocated into epithelial cells, the novel TEM-1 translocation assay was applied (6). The system is based on a translational fusion of effector proteins with a mature TEM-1 β-lactamase and the detection of TEM-1 activity within eukaryotic cells by using the fluorescent β-lactamase substrate CCF2/AM. If translocation occurs, the translational fusion will be present in the eukaryotic cells, and the cells will appear blue. If translocation does not occur, the cells will appear green.
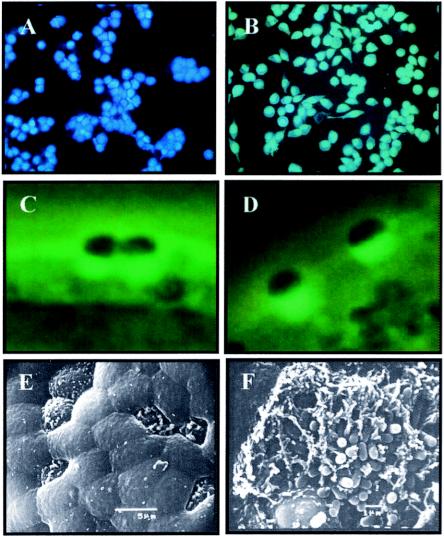
HeLa cell monolayers were infected with wild-type and ΔescF (ICC171) mutant (47) EPEC strains carrying the pICC283 plasmid encoding the TEM-1 fusion with Z3071. In addition, cells were infected with EPEC containing pCX340 (empty vector; negative control) or pCX327 (containing the first 16 residues of Cif; positive control) (6). Cells infected with EPEC(pCX340) appeared green (data not shown), indicating a lack of TEM-1 activity in these cells. Cells infected with EPEC(pCX327) appeared blue (data not shown), indicating that TEM-1 was translocated into the host cells. Cells infected with EPEC carrying the translation fusion Z3071-TEM-1 also appeared blue, indicating that Z3071 is a new translocated effector (Fig. 1A). The translocation of Z3071 was dependent on an active FTTSS, as a ΔescF EPEC strain carrying the EspJ-TEM-1 fusion appeared green, indicating that Z3071 is translocated via the LEE-encoded FTTSS (Fig. 1B). In accordance with the conventional nomenclature, we named Z3071 EspJ (for E. coli-secreted protein J).
FIG. 1.
In vitro analysis of EspJ. The translational fusion EspJ-TEM-1 is translocated into HeLa cells infected with wild-type EPEC(pICC283) (A) but not into HeLa cells infected with ICC171(pICC283) (B). EspJ is not required for A/E lesion activity in vitro, as indistinguishable actin-rich pedestals were observed underneath sites of bacterial adhesion in both the espJ mutant (C) and EHEC 85-170 (D). EspJ is not required for A/E lesions activity ex vivo. An uninfected follicle-associated epithelium devoid of bacteria is shown in panel E, and ICC188-induced A/E lesions are shown in panel F.
EspJ is not required for the formation of A/E lesions in vitro.
In order to characterize the EspJ function, an espJ mutant was generated in EHEC O157:H7 strain 85-170, EPEC strain E2348/69, and C. rodentium ICC169 by deleting the open reading frame. The resulting mutant strains were analyzed for growth in vitro in both rich and minimal media and were found to have growth rates identical to those of the parental strains, as measured by optical density (data not shown).
An espJ mutant and the wild-type EHEC and EPEC strains were assessed for A/E lesion formation in vitro. Infecting HEp-2 cells with the mutant and the wild-type strains and employing the FAS test (25) revealed actin-rich pedestals under adherent EHEC ΔespJ (strain ICC188) (Fig. 1C) and EPEC ΔespJ (strain ICC190) (data not shown) that were indistinguishable from those formed by the wild-type strains (Fig. 1D), indicating that espJ is not required for this activity in vitro.
EspJ is not required for the formation of A/E lesions ex vivo.
The IVOC adhesion assay is based on the infection of human intestinal biopsy specimens with pathogens and offers a more physiological model for EHEC-EPEC infection. Using this model, Phillips et al. demonstrated that EHEC O157:H7 binds to the follicle-associated epithelium of Peyer's patches (39). In order to assess the contribution of EspJ to the colonization of human intestinal explants, ΔespJ (ICC188) and 85-170 wild-type strains were used to infect terminal ileal mucosal biopsy specimens, which contained both villous and lymphoid follicular areas. Bacterial adhesion was not seen on the uninoculated negative control samples (Fig. 1E). The infection showed that the ΔespJ mutant attaches to and causes A/E lesions on the follicle-associated epithelium of distal ileal samples (Fig. 1F), but not on ileal villous surfaces, a phenotype identical to that of the parent strain, 85-170 (data not shown). Therefore, EspJ is not required for A/E lesion activity ex vivo.
Contribution of EspJ to colonization of C3H/HeJ mice.
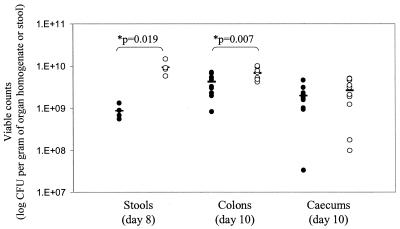
Infection experiments were performed in the murine model to determine the level of virulence of a C. rodentium ΔespJ mutant. The ability of the mutant strain to establish itself and expand in mice was investigated by monitoring the viable counts recovered from stools. In the sensitive C3H/HeJ mouse strain, the ΔespJ C. rodentium mutant (ICC189) produced the classic growth curve previously reported for the wild-type strain (1); however, the size of the challenging population increased steadily to peak at ca. 109 to 1010 CFU g of stool−1 at day 8, significantly higher than the levels for the wild-type strain (P = 0.019) (Fig. 2). Ten days postinfection, the mice had become too ill for us to continue (as determined by weight loss, piloerection, and lack of mobility), so the experiment was terminated. The pathogen burdens of the organs of C3H/HeJ mice (n = 12) infected with the wild type and with ICC189 (ΔespJ) were determined, as were the abilities of the strains to cause hyperplasia. Mice infected with ΔespJ had significantly higher pathogen burdens in the colon than mice infected with wild-type bacteria (6.86 × 109 ± 1.96 × 109 compared to 4.32 × 109 ± 1.92 × 109 CFU g of organ homogenate−1; P = 0.007) but similar burdens within the cecum (2.70 × 109 ± 1.67 × 109 compared to 2.01 × 109 ± 1.13 × 109 CFU g of organ homogenate−1; P = 0.273) (Fig. 2). However, there was no significant difference between the induced levels of colonic hyperplasia, as indicated by increased colon weight (data not shown).
FIG. 2.
Virulence of C. rodentium wild-type (ICC169) (solid symbols) and ΔespJ mutant (ICC189) (open symbols) strains in individual sensitive C3H/HeJ mice as determined by viable counts in stools and organ homogenates. *, significant differences were found in the viable counts obtained from ICC169- and ICC189-infected mice recovered from stools on day 8 postinfection and from the colons on day 10 postinfection.
Dynamics of clearance of C. rodentium ΔespJ in C57BL/6J mice.
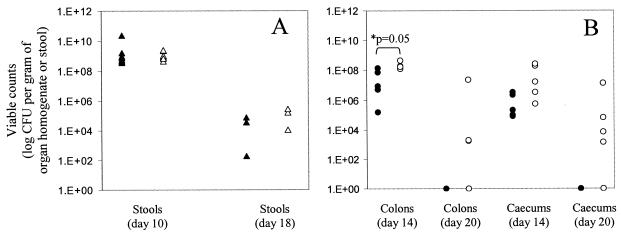
Since C3H/HeJ mice infected with ΔespJ and wild-type bacteria became too ill for us to follow the clearance of the pathogens, the more resistant C57BL/6J strain was used to follow the dynamics of colonization and clearance after infection (n = 10). The ability of the ΔespJ mutant strain (ICC189) to establish itself and expand in mice was investigated by monitoring the viable counts recovered from stools. For the first 10 days postinfection, the ΔespJ mutant produced the classic growth curve previously reported for the wild-type strain (46), with the size of the challenging population increasing steadily to peak at ca. 109 CFU g of stool−1 on day 10 (Fig. 3A). Fourteen days postinfection, the pathogen burdens of the organs of infected mice were determined, as were the abilities of the strains to cause hyperplasia. Mice infected with ΔespJ bacteria had significantly higher pathogen burdens in the colon than mice infected with wild-type bacteria (2.18 × 108 ± 1.22 × 108 compared to 4.80 × 107 ± 5.73 × 107 CFU g of organ homogenate−1; P = 0.05). Although the pathogen burdens within the cecum were higher for mice infected with ΔespJ bacteria, this difference was not significant (9.73 × 107 ± 1.13 × 107 compared to 1.19 × 106 ± 1.34 × 106 CFU g of organ homogenate−1; P = 0.163) (Fig. 3B). Twenty days postinfection, mice infected with wild-type bacteria had completely cleared infection in both the cecum and colon, as previously described (46). In contrast, three out of five mice infected with ΔespJ bacteria still had bacteria present within the colonic mucosa (ranging from 1.79 × 103 to 2.27 × 107 CFU g of organ homogenate−1). In addition, four out of five mice infected with ΔespJ still had bacteria present within the cecum (ranging from 1.41 × 103 to 1.42 × 107 CFU g of organ homogenate−1) (Fig. 3B). However, there was no significant difference between the induced levels of colonic hyperplasia, as indicated by increased colon weight (data not shown), on either day 14 or day 20 postinfection.
FIG. 3.
Virulence of C. rodentium wild-type (ICC169) (solid symbols) and ΔespJ mutant (ICC189) (open symbols) strains in individual C57BL/6J mice as determined by viable counts in stools (A) and organ homogenates (B). *, significant differences were found in the viable counts obtained from ICC169- and ICC189-infected mice recovered from colons on day 14 postinfection. Twenty days postinfection, mice infected with wild-type C. rodentium had completely cleared infection in both the cecum and colon, while three of five and four of five mice infected with ICC189 still had bacteria present within the colonic mucosa and cecum, respectively.
The contribution of EspJ to the colonization of conventional 6-week-old lambs.
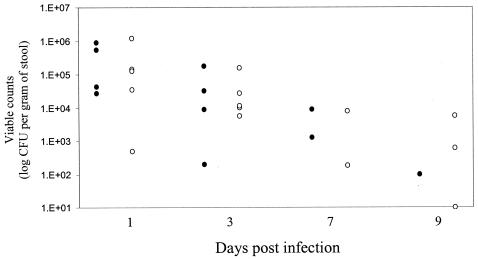
Infection experiments were performed in the conventional 6-week-old lamb model to determine the persistence of E. coli O157:H7 strain 85-170 and an espJ mutant. The abilities of the isolates to establish themselves and persist in lambs were investigated by monitoring the viable counts recovered in stools collected per rectum. Typically, wild-type EHEC produced the classic shedding pattern (48), persisting in relatively high numbers in the early stages of infection and then declining until it became detectable in only one lamb by day 9 postinfection (Fig. 4). EHEC was then undetectable by direct plating or enrichment for the duration of the study (27 days). In contrast, while EHEC ΔespJ behaved like the wild type in the early stages of infection, three lambs still had detectable EHEC ΔespJ on day 9 postinfection (Fig. 4) and one lamb continued to shed bacteria (as detected by enrichment) until the end of the study 27 days postinfection (data not shown). However, these differences were not statistically significant at any of the time points.
FIG. 4.
Shedding of EHEC O157 85-170 wild type (solid symbols) and a ΔespJ mutant (open symbols) from orally infected individual conventional lambs. By 9 days postinfection, the wild type was present in one out of five lambs, whereas the espJ mutant was present in three out of five lambs.
DISCUSSION
Using a DNA microarray specifically designed for E. coli O157:H7, we have recently identified two novel genes (Z3071 and Z3072) implicated in virulence that are located adjacent to each other within prophage CP-933U and are absent from the E. coli K-12 genome (8). Further studies demonstrated that Z3072 (TccP/EspFU) is a type III effector protein that has an Nck-like activity following translocation, recruiting N-WASP, Arp2/3 complex, and actin to the site of bacterial adhesion (5, 15). The purpose of this study was the characterization of the role of Z3071 (EspJ) in virulence.
Of particular interest is the fact that, while espJ is found in the genomes of EHEC O157:H7, EPEC, and C. rodentium, the gene downstream, tccP/espFu, is found only in EHEC O157:H7. Selective evolutionary pressures must have operated on espJ and tccP/espFu in the genetic backgrounds of EPEC and EHEC once prophage CP-933U was introduced which maintained espJ in EHEC O157:H7, EPEC, and C. rodentium and espFU/tccP in EHEC O157:H7 only.
In this study, we have demonstrated, by employing the TEM-1 reporter system, that EspJ is translocated into HeLa cells (6). Therefore, espJ encodes the fourth identified TTSS effector protein outside the LEE, after Cif (30), NleA/EspI (16, 34), and TccP/EspFU (5, 15). In addition, by carrying out in vitro and ex vivo adhesion assays, we have shown that EspJ does not affect the A/E lesion phenotype, as indistinguishable lesions were observed in both the espJ mutant and the wild-type strain.
An unexpected effect of EspJ on virulence was revealed in vivo. In the murine model of infection, higher levels of colonization by the ΔespJ mutant (ICC189) were found in the gastrointestinal tract sites than for the wild-type strain in both susceptible and more resistant mouse strains. As infection of C3H/HeJ mice rendered the animals too ill to be followed for extended periods, the dynamics of colonization by and clearance of ΔespJ were studied in the more resistant C57BL/6J mice. This model indicates that the dynamics of clearance of the ΔespJ mutant and the wild type are very different. While mice infected with wild-type bacteria had completely cleared infection at 20 days postinfection, C. rodentium was still recovered from the colon and cecum of mice infected with the ΔespJ mutant at the same time. It therefore appears that the ΔespJ mutant exhibits a more persistent colonization phenotype than the wild-type strain. It is worth noting that the wild-type and mutant strains grew equally well on LB and M9 minimal media, indicating that the phenotype obtained is not caused by a higher growth rate. In addition, for both the wild-type and ΔespJ mutant strains, the symptoms of disease, including levels of colonic hyperplasia, were identical. This suggests that the phenotype of the ΔespJ mutant may be due to a difference in the dynamics of clearance of the colonizing bacteria and not the initial inoculum or rates of progression of infection. Recently, the dynamics of colonization and the subsequent clearance of C. rodentium from orally infected mice were followed by using bioluminescence imaging. In this study, it was shown that the cecal patch is the first site to be colonized and the first to be cleared, followed by colonization and clearance of the colon. It has been suggested that this organ acts as a reservoir, shedding bacteria into the colon (46).
It is important to note that C. rodentium mutants with different colonization and clearance dynamics in the murine model have been previously reported (34). C. rodentium strains lacking the genes espI (strain ICC179) and map (strains P6C6 and P10H2) exhibited a different colonization phenotype in mice than wild-type bacteria, with viable counts recovered from the stools over an extended period. However, these strains also differ in their initial colonization dynamics, with the mutant strains being slower to become established and hence reaching a peak several days behind that of the wild-type strain. In contrast, the rates of progression of colonization of mice were identical for ΔespJ and wild-type bacteria. In addition, the mutant map and espI strains also produced lower levels of hyperplasia, whereas the level observed with the ΔespJ mutant was indistinguishable from that of the wild type. This strengthens the observation that the ΔespJ mutant exhibits a clearance defect phenotype unrelated to its in vivo growth rate.
Colonization of the cecum and the colon is also a characteristic of the infection of neonatal and weaned lambs with EHEC O157:H7. Although this infection is not associated with clinical disease, A/E lesions were detected in the cecum and rectum in 6-week-old lambs infected with EHEC O157:H7 (48). Previous studies of 6-week-old lambs have confirmed a role for intimin in the persistence of EHEC O157:H7 in the ovine intestine (48). In order to confirm the findings observed in the murine infection model, we also infected 6-week-old conventional lambs with the wild-type or ΔespJ mutant E. coli O157:H7 strain 85-170. In this model, the ΔespJ mutant was found to behave like the wild type in the early stages of infection but was recoverable from three out of five lambs on day 9 postinfection and persisted for 27 days postinfection in one lamb, whereas the wild type was found in only one out of five lambs 9 days postinfection. These data strengthen the observation that the espJ mutant exhibits a clearance defect phenotype.
Here, we report a TTSS effector that possesses properties that influence the dynamics of clearance of the pathogen from the host's intestinal tract in a novel manner. Our data indicate that the espJ mutant persists longer in mammalian hosts; hence, the espJ gene exhibits “antivirulence” properties. Antivirulence genes have been described in other pathogens, including Salmonella enterica (33) and Streptococcus pyogenes (12), and two main hypotheses have been put forward for their existence: they may favor host survival and thereby aid pathogen transmission, or they may contribute to pathogen fitness in nonhost environments. Indeed, the Salmonella antivirulence gene pcgL was found to be required for survival in nutrient-limited environments (33). However, we found no such requirement for espJ for growth in nutrient-poor media, and the presence of C. rodentium ΔespJ in the organs of mice after the wild-type strain had been cleared suggests that in the case of espJ a role in host survival, and hence in aiding pathogen transmission, may be more likely.
Acknowledgments
We thank Eric Oswald for pCX340 and pCX327 and Junkal Garmendia and Oliver Marches for technical help.
This study was supported by the Wellcome Trust and the BBSRC.
Editor: J. T. Barbieri
REFERENCES
- 1.Barthold, S. W., G. L. Coleman, R. O. Jacoby, E. M. Livstone, and A. M. Jonas. 1976. Transmissible murine colonic hyperplasia. Vet. Pathol. 15:223-236. [DOI] [PubMed] [Google Scholar]
- 2.Batchelor, M., J. Guignot, A. Patel, N. Cummings, J. Cleary, S. Knutton, D. W. Holden, I. Connerton, and G. Frankel. 2004. Involvement of the intermediate filament protein cytokeratin-18 in actin pedestal formation during EPEC infection. EMBO Rep. 5:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Campellone, K. G., and J. M. Leong. 2003. Tails of two Tirs: actin pedestal formation by enteropathogenic E. coli and enterohemorrhagic E. coli O157:H7. Curr. Opin. Microbiol. 6:82-90. [DOI] [PubMed] [Google Scholar]
- 5.Campellone, K. G., D. Robbins, and J. M. Leong. 2004. EspFU Is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7:1-12. [DOI] [PubMed] [Google Scholar]
- 6.Charpentier, X., and E. Oswald. 2004. Analysis of type III translocation signals of enteropathogenic and enterohemorrhagic Escherichia coli effectors using TEM-1 beta-lactamase as a fluorescence-based reporter. J. Bacteriol. 186:5486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crane, J. K., B. P. McNamara, and M. S. Donnenberg. 2001. Role of EspF in host cell death induced by enteropathogenic Escherichia coli. Cell Microbiol. 3:197-211. [DOI] [PubMed] [Google Scholar]
- 8.Dahan, S., S. Knutton, R. K. Shaw, V. E. Crepin, G. Dougan, and G. Frankel. 2004. Transcriptome of enterohemorrhagic Escherichia coli O157 adhering to eukaryotic plasma membranes. Infect. Immun. 72:5452-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, W., Y. Li, B. A. Vallance, and B. B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, S. J., E. O. Krejany, J. L. Mellies, R. M. Robins-Browne, C. Sasakawa, and J. B. Kaper. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 69:4027-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engleberg, N. C., A. Heath, A. Miller, C. Rivera, and V. J. DiRita. 2001. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J. Infect. Dis. 183:1043-1054. [DOI] [PubMed] [Google Scholar]
- 13.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 14.Galan, J. E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J. Bacteriol. 174:4338-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garmendia, J., A. Phillips, Y. Chong, S. Schuller, O. Marches, S. Dahan, E. Oswald, R. K. Shaw, S. Knutton, and G. Frankel. 2004. TccP is an enterohaemorrhagic E. coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. EMBO J. 6:1167-1183. [DOI] [PubMed] [Google Scholar]
- 16.Gruenheid, S., I. Sekirov, N. A. Thomas, W. Deng, P. O'Donnell, D. Goode, Y. Li, E. A. Frey, N. F. Brown, P. Metalnikov, T. Pawson, K. Ashman, and B. B. Finlay. 2004. Identification and characterization of NleA, a non-LEE encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 51:1233-1249. [DOI] [PubMed] [Google Scholar]
- 17.Hartland, E. L., M. Batchelor, R. M. Delahay, C. Hale, S. Matthews, G. Dougan, S. Knutton, I. Connerton, and G. Frankel. 1999. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol. 32:151-158. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 28:11-22. [DOI] [PubMed] [Google Scholar]
- 19.Hicks, S., G. Frankel, J. B. Kaper, G. Dougan, and A. D. Phillips. 1998. Role of intimin and bundle foming pili in enteropathgenic Escherichia coli adhesion to pediatric intestine in vitro. Infect. Immun. 66:1570-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 23.Kenny, B., S. Ellis, A. D. Leard, J. Warawa, H. Mellor, and M. A. Jepson. 2002. Co-ordinate regulation of distinct host cell signalling pathways by multifunctional enteropathogenic Escherichia coli effector molecules. Mol. Microbiol. 44:1095-1107. [DOI] [PubMed] [Google Scholar]
- 24.Kenny, B., and M. Jepson. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell Microbiol. 2:579-590. [DOI] [PubMed] [Google Scholar]
- 25.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knutton, S., D. R. Lloyd, and A. S. McNeish. 1987. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect. Immun. 55:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine, M. M., E. J. Berquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, S. Stoman, and B. Rowe. 1978. Escherichia coli that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:119-122. [DOI] [PubMed] [Google Scholar]
- 28.Luperchio, S. A., and D. B. Schauer. 2001. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 3:330-340. [DOI] [PubMed] [Google Scholar]
- 29.Luperchio, S. A., J. V. Newman, C. A. Dangler, M. D. Schrenzel, D. J. Brenner, A. G. Steigerwalt, and D. B. Schauer. 2000. Citrobacter rodentium, the causative agent of transmissible murine colonic hyperplasia, exhibits clonality: synonymy of C. rodentium and mouse-pathogenic Escherichia coli. J. Clin. Microbiol. 38:4343-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marches, O., T. N. Ledger, M. Boury, M. Ohara, X. Tu, F. Goffaux, J. Mainil, I. Rosenshine, M. Sugai, J. De Rycke, and E. Oswald. 2003. Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol. Microbiol. 50:1553-1567. [DOI] [PubMed] [Google Scholar]
- 31.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNamara, B. P., A. Koutsouris, C. B. O'Connell, J. P. Nougayrede, M. S. Donnenberg, and G. Hecht. 2001. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Investig. 107:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouslin, C., F. Hilbert, H. Huang, and E. A. Groisman. 2002. Conflicting needs for a Salmonella hypervirulence gene in host and non-host environments. Mol. Microbiol. 45:1019-1027. [DOI] [PubMed] [Google Scholar]
- 34.Mundy, M., L. Petrovska, K. Smollett, N. Simpson, R. K. Wilson, J. Yu, X. Tu, I. Rosenshine, S. Clare, G. Dougan, and G. Frankel. 2004. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and the roles of this and other secreted proteins in infection. Infect. Immun. 72:2288-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perna, N. T., G. R. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 38.Phillips, A. D., and G. Frankel. 2000. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J. Infect. Dis. 181:1496-1500. [DOI] [PubMed] [Google Scholar]
- 39.Phillips, A. D., S. Navabpor, S. Hicks, G. Dougan, T. Wallis, and G. Frankel. 2000. Enterohaemorrhagic Escherichia coli O157:H7 targets Peyer's patches in man and causes attaching-effacing lesions in both human and bovine intestine. Gut 47:377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schauer, D. B., and S. Falkow. 1993. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schauer, D. B., and S. Falkow. 1993. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect. Immun. 61:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 98:11638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shan, L., H. S. Oh, J. Chen, M. Guo, J. Zhou, J. R. Alfano, A. Collmer, X. Jia, and X. Tang. 2004. The HopP to F locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol. Plant-Microbe Interact. 17:447-455. [DOI] [PubMed] [Google Scholar]
- 44.Tauschek, M., R. A. Strugnell, and R. M. Robins-Browne. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44:1533-1550. [DOI] [PubMed] [Google Scholar]
- 45.Tu, X., I. Nisan, C. Yona, E. Hanski, and I. Rosenshine. 2003. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol. Microbiol. 47:595-606. [DOI] [PubMed] [Google Scholar]
- 46.Wiles, S., S. Clare, J. Harker, A. Huett, D. Young, G. Dougan, and G. Frankel. 2004. Organ-specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell Microbiol. 6:963-972. [DOI] [PubMed] [Google Scholar]
- 47.Wilson, R. K., R. K. Shaw, S. Daniell, S. Knutton, and G. Frankel. 2001. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell Microbiol. 3:753-762. [DOI] [PubMed] [Google Scholar]
- 48.Woodward, M. J., A. Best, K. A. Sprigings, G. R. Pearson, A. M. Skuse, A. Wales, C. M. Hayes, J. M. Roe, J. C. Low, and R. M. La Ragione. 2003. Non-toxigenic Escherichia coli O157:H7 strain NCTC12900 causes attaching-effacing lesions and eae-dependent persistence in weaned sheep. Int. J. Med. Microbiol. 293:299-308. [DOI] [PubMed] [Google Scholar]