Abstract
Background/Aims:
Hepatitis D virus (HDV) is a defective RNA virus that is dependent on hepatitis B surface antigen (HBsAg) for transmission and replication. HDV significance arises from the possibility of poor prognosis of hepatitis B virus (HBV) infection. In Saudi Arabia, HDV prevalence varied from 8 to 32% before the HBV vaccination program and ranged from 0 to 14.7% after the vaccination program was started. The last study, performed in 2004, showed a prevalence of 8.6% in hospital-based HBV cases and 3.3% in healthy donors. The aim of this study was to investigate the prevalence and molecular characterization of HDV in chronic hepatitis B (CHB) patients at the King Abdulaziz University Hospital in Jeddah, Saudi Arabia by molecular and serological techniques. To the best of our knowledge, this is the first study to detect HDV at the molecular level in Saudi Arabia.
Patients and Methods:
The study included samples from 182 CHB patients from Jeddah; 13 samples with HBsAg negative were excluded. Samples were tested for HDV-Ab, viral RNA by reverse transcriptase–polymerase chain reaction (RT-PCR) in the HDV L-Ag region and sequence analysis.
Results:
The mean age of the participants was 44.36 years; 75.1% of the participants were Saudi nationals, 58% were males. Nine samples were positive for HDV-Ab and four were borderline; all were subjected to RT-PCR amplification. Three of the positive HDV-Ab cases and 1 borderline case were positive by RT-PCR. All the positive cases had HBV genotype D, and the positive RT-PCR cases were positive for HBV DNA. One of the HDV viremic samples was of genotype 1 by sequencing. The prevalence of HDV in the study was 7.7%, which was lower in Saudis (6.3%) than in non-Saudis (11.9%).
Conclusion:
HDV coinfection does not seem to have an effect on the clinical status of the recruited CHB cases in this study. More studies are needed to investigate the genetic diversity in other areas such as the southern parts of the Kingdom.
Keywords: Chronic HBV, hepatitis D virus, prevalence, Saudi Arabia
INTRODUCTION
Viral hepatitis is a significant public health problem, which affects hundreds of millions worldwide with significant morbidity and mortality via acute and chronic infections. Chronic viral hepatitis infections are associated with the development of cirrhosis and hepatocellular carcinoma (HCC).[1]
Hepatitis Delta virus (HDV) was first detected by Rizzetto[2] among patients with a severe form of infection with hepatitis B virus (HBV). HDV is a defective RNA virus that is dependent on hepatitis B surface antigen (HBsAg) for transmission and replication.[3,4] HDV infection is significant because it can cause severe liver disease that includes fulminant liver failure and rapid progression to cirrhosis, as well as increased risk of HCC.[5]
HDV has a small circular RNA genome of approximately 1700 bases (smallest human RNA virus); the genome is single-stranded negative sense and forms a covalently closed circle.[6] The RNA encodes a protein called the delta antigen, which is subsequently encased in an envelope embedded with HBsAg.[6] The viral RNA genome encodes single open reading frame (ORF) that codes for the small delta antigen (SDAg) and large delta antigen (LDAg). Two major specific patterns of HDV/HBV infection have been described – coinfection and superinfection; while coinfection is the concurrent infection with both HBV and HDV, superinfection is the infection by HDV in chronic HBV cases. Both coinfection and superinfection with HDV result in more severe complications compared to infection with HBV alone.[7,8]
In the last few years, a study estimated that there were 18 million individuals who were infected with HDV from the 350 million HBV carriers worldwide.[9] The study showed that countries such as Pakistan and Iran had an increase in HDV prevalence whereas countries such as Japan, Australia, Turkey, China, India, and Taiwan have shown a decline in the incidence rate, although some of them have high prevalence rates.[9] HDV infection affects all age groups. The highest prevalence is found in the Middle East, Mediterranean basin, western and central Africa, central and northern Asia, the Amazon basin, the Pacific islands,[10,11] and Vietnam.[12] Similar to HBV, HDV infection can be acquired parenterally, sexually, or vertically from positive mothers.[13]
Latest studies suggested at least eight HDV clades, which are phylogenetically distinct and have predominance in different regions worldwide.[14,15] The most prevalent HDV genotype worldwide is genotype 1,[16] which is related to a broad spectrum of pathogenicity and is predominant in the USA, Middle East, and Europe.[17,18] Genotype 2 is predominant in the Far East.[16] Genotype 3 is associated with severe forms of hepatitis and is predominant in Northern South America.[8] Genotype 4 in Taiwan and Japan and Genotypes 5 through to 8 are predominant in Africa.[19,20]
In Saudi Arabia, HDV frequency was found to be 8.0% in a sample of 36 HBsAg carriers in 1986.[21] Another large study conducted among 488 HBsAg positive cases in 1986[22] showed that the frequency in Riyadh area was 22.2% in patients with chronic HBV (superinfection), 7.9% in those with acute HBV (coinfection), and 6.7% in HBsAg inactive carriers. In Najran, the frequency was 9.6% and in Al-Hafouf it was 5.3%. This study showed that no HDV-Ab was detected in the areas of Khaiber and Jizan. In 1987, a high HDV prevalence of 32% was found in HBsAg-positive Saudi patients with liver disease in Riyadh.[23] In 1988, the prevalence was 8% in HBsAg carriers in Jizan.[24] The prevalence in HBsAg pregnant women was 9.7% in 1988; however, follow-up of 17 of their infants showed no evidence of perinatal transmission.[25] In 1991, HDV prevalence in HBsAg-positive patients was found to be 17.6%;[26] while a study in Jeddah performed in 1998 showed HDV prevalence to be 14.7% in intravenous drug abusers (IVDAs)/HBsAg carriers and 0.0% in non-IVDAs HBsAg positive cases.[27] In 2004, Al-Traif et al. showed that HDV prevalence among healthy HBsAg positive blood donors was 3.3% whereas the prevalence in hospital and clinic-based HBsAg patients was 8.6%, with expected decline in HDV infection in Saudi Arabia due to global vaccination.[28] All the studies conducted so far in Saudi Arabia were serological studies and did not attempt molecular detection of the viral genome.
PATIENTS AND METHODS
One hundred eighty-two blood samples were collected from chronically infected hepatitis B patients, including eight blood samples collected from HCC patients during their routine visit to King Abdulaziz University Hospital, Jeddah, Saudi Arabia. The samples were collected between March 2013 and December 2015. Blood samples were transferred to Special Infectious Agents Unit, King Fahd Medical Research Center, King Abdulaziz University for laboratory analysis and storage. Blood samples were centrifuged and separated and serum samples were kept at −80°C until laboratory analysis. Inclusion criteria were age >18 years, chronic HBV cases, no coinfection with HCV or HIV.
Serological markers
All serum samples were analyzed using commercial enzyme-linked immunosorbent assay (ELISA) kits for the detection of HBsAg (ABBOTT Murex HBsAg version 3, Germany), total anti-HBc (ABBOTT Murex HBcAb, Germany), HBeAg (ABBOTT Murex HBeAg, Germany), HCV Ab (ABBOTT Murex anti-HCV version 4.0, Germany), and HIV Ab (ABBOTT Murex HIV Ab, Germany) following the manufacturer's instructions. HBsAg positive serum samples were analyzed for anti-HD using commercial ELISA kit (ETI-AB-DELTAK-2 DiaSorin, Italy) following the manufacturer's instructions. This assay is a simultaneous competitive assay. Anti-HD present in the sample and a labeled anti-HD antibody compete for a fixed quantity of HDAg bound to the solid phase. The quantity of enzyme tracer bound to the solid phase and consequently the enzyme activity are inversely proportional to the anti-HD concentration present in samples or controls.
Molecular detection of hepatitis B virus infection
DNA was extracted from 200 μ l of serum sample using QIAamp® DNA Mini kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions. HBV genome was detected by nested-polymerase chain reaction (PCR) for amplification of HBV S-gene nucleotide (nt) 181-619, according to Kao et al.[29] HBV viral load was determined using Abbott RealTime HBV kit (Abbott, Germany).
Molecular detection of hepatitis delta virus infection
Viral RNA was extracted from 140 μ l serum sample using QIAamp Viral RNA Mini Kit (QIAGEN GmbH, Hilden, Germany). Nested reverse transcriptase–polymerase chain reaction (RT-PCR) was performed to detect HDV genome (nt 877–1290). For the reverse transcription and first round of PCR, 10 μ l of extracted RNA were amplified using SuperScript® III One-Step RT-PCR/Platinum® Taq Mix (Invitrogen, life technologies, California, USA). Primers used for the first PCR round were 8531 U and 1302 D.[30] PCR conditions were 45°C for 30 min, followed by 2 min at 94°C, 40 cycles for 1 min at 94°C, 1 min at 56°C and 1 min at 68°C, and a final extension of 5 min at 68°C. For the second round PCR, the primers HDV-E and HDV-A were used.[30] The cycling conditions consisted of 2 min at 94°C, 40 cycles of 1 min at 94°C, 1 min at 54°C and 1 min at 72°C, and a final extension for 7 min at 72°C. PCR products were visualized by agarose gel electrophoresis after staining with ethidium bromide [Figure 1].
Figure 1.
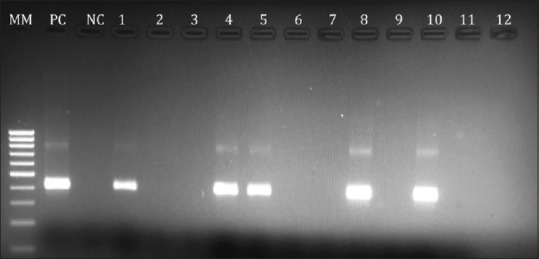
Agarose gel electrophoresis photo showing the HDV PCR products; 1st lane (MM) 100bp ladder marker; 2nd lane positive control (PC) for HDV; 3rd lane negative control (NC); lanes no. 1,4,5,8 and 10 are positive HDV PCR samples with product size 404 bp and lanes 2,3,6,7,9,11 and 12 are negative HDV PCR samples
Sequencing method
HBV was genotyped by amplifying the S-gene and sequencing of the amplified products according to Kao et al.[29] using an ABI 3500 Automatic Sequencer (Applied Biosystems, USA). Sequencing conditions were performed according to the instructions of the Bigdye Terminator V3.1 Reaction Cycle Kit (ABI, Germany).
For HDV genotyping, PCR bands (404 bp) were agarose gel purified using QIAquick Gel Extraction Kit (Qiagen, Germany) according to manufacturer's instructions. Cycle sequencing of the PCR products was performed on an ABI 3500 Automatic Sequencer (Applied Biosystems, USA) using the Bigdye Terminator V3.1 Reaction Cycle Kit (ABI, Germany) according to manufacturer's instructions. The viral sequence was assembled using Geneious software (version).[31] The assembled sequenced were initially searched for similarity using BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/). Genbank sequences with higher similarity were selected for further analysis. The HDV sequence was submitted to Genbank and was given the accession number KX219662.
Phylogenetic analysis
Phylogenetic analysis and distance calculations were performed using the MEGA6 software,[32] with the Neighbor-Joining method of the Maximum Composite Likelihood model, gamma-distributed rates among sites with 1000 bootstrap replicates [Figure 2].
Figure 2.

Phylogenetic tree of the positive HDV sample from this study represented by a red square. Bold face red font taxa represent isolates originating from the same country collapsed to save space. The evolutionary history was inferred using the Neighbor-Joining method. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site
Fibrosis score
We assessed the liver fibrosis using transient elastography (Fibroscan 502, Echosens, Paris, France 2005). The examination was performed by a well-trained certified technician, and each test result was verified by one of the two expert hepatologists. All included patients had to have at least 10 valid readings, success rate of at least 70%, and interquartile range (IQR) of less than 30%. The stiffness score was measured in Kappa. We defined the fibrosis stages as followoing: F1 <7, F2 7–9.5, F3 9.5–12.4, and F4 >12.5 kPa.[33]
Statistical analysis
Chi-square or Fisher's exact test were used to compare data. Stepwise logistic regression analysis was used to identify association of independent factors with HDV-Ab. Two-sided P value of <0.05 was used as a predictor for statistical significance. Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS) version 21 (IBM corporation, USA).
RESULTS
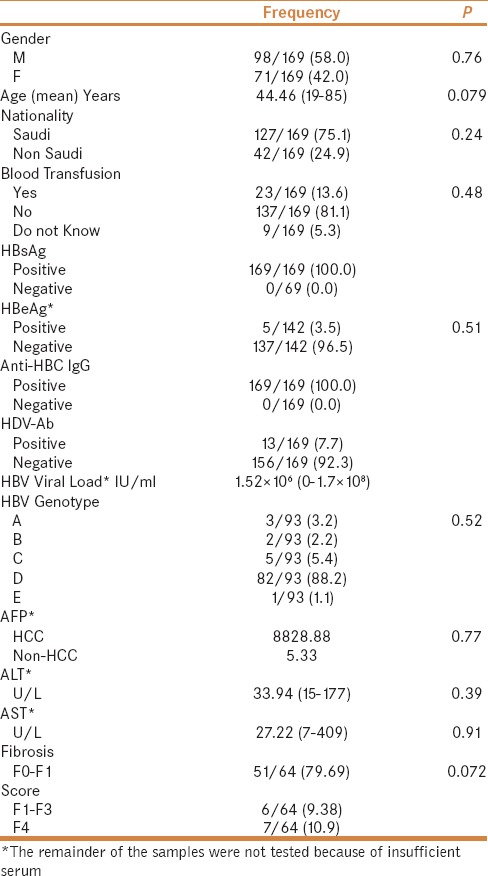
A total of 182 CHB patients including 8 HCC cases were recruited from the King Abdulaziz University Hospital, Jeddah. Of the recruited patients, 13 CHB patients were HBsAg negative and were excluded from the analysis. Of the recruited patients, 63 were under HBV antiviral treatment. The mean age of the participants was 44.46 years (19–85 years); 127 patients were Saudi nationals (75.1%) and 42 were non-Saudis (24.9%). Ninety-eight (58.0%) were males and 71 (42.0%) were females. One hundred and forty-two samples were tested for HBeAg, 137 (96.5%) of them were negative and only 5 (3.5%) samples were positive. Eighty samples were tested for HBeAb; 77 (96.3%) of them were positive and 3 (3.7%) were negative. All samples tested were positive for IgG class anti-HBc. The demographic, clinical, and laboratory data of the recruited patients are summarized in Table 1.
Table 1.
Characteristics of the recruited patients in the study
All the positive and negative samples for HBsAg were tested for anti-HDV antibodies. HBsAg negative samples were found to be negative for HDV-Ab and were excluded from the analysis. Of the 169 HBsAg positive samples, 9 samples representing 5.3% tested positive for anti-HDV with sample/cutoff (s/cutoff) values ranging from 0.23 to 0.86. Four other samples were borderline having s/cutoff ranging from 0.97 to 1.06; these samples when repeated gave comparable s/cutoff results. All the positive and borderline HDV-Ab samples were tested for HDV-RNA by RT-PCR; 3 of the positive HDV-Ab cases and 1 borderline cases were positive for HDV RNA. Because one of the borderline samples was positive for HDV RNA, we assumed the borderline cases to be positive for HDV-Ab and made the calculations on this assumption. The remaining samples were negative for HDV-Ab with s/cutoff ranging from 1.23–2.35.
HBV genotyping was done by phylogenetic analysis of the S-region of the HBV viral genome on 106 samples, which showed that the samples were 88.5% genotype D, 5.2% genotype C, 3.1% genotype A, 2.1% genotype B, and 1.0% genotype E. We were only able to sequence one HDV-RNA positive sample (CHB077) that clustered with genotype 1 sequences from Genbank; this sample had HBV genotype D.
Characteristics of the HDV-Ab positive patients
Positive cases for HDV-Ab (13 cases) were all positive for HBsAg and negative for HBeAg; these included 7 males and 6 females; there were 8 Saudi nationals, 3 Yemenis, and 2 Palestinians. The positive HDV-Ab cases had a fibrosis score ranging from F1–F2 to F2–F3, and one case with confirmed HCC. The mean value of ALT was 39.55 U/L which showed no significant difference compared to the mean of the negative cases (33.38). Liver function tests did not show a significant difference between HDV-Ab positive versus HDV-Ab negative cases (39.55 vs 33.38 for ALT, 25.91 vs 27.35 for AST, 36.45 vs 36.48). Of the positive HDV-Ab cases (1 sample positive for HDV RNA), 6 were under treatment with no significant difference between HDV-Ab positive and HDV-Ab negative cases.
No significant association was found with age, nationality, HBV genotype, AFP levels, duration of HBV infection, HBV DNA viral load, and fibrosis score [Table 1].
All of the positive cases had HBV infection of genotype D. The HDV RNA positive cases were also positive for HBV DNA. Compared to the closest strain, AM779584, the Saudi strain (KX219662) showed 8 (NT) mutations, 5 of which were non-synonymous and resulted in amino acid substitution at positions K5E, R11G, A42D, E46D, and I89M of the HDV L-Ag.
DISCUSSION
HDV has gained importance in the clinical and epidemiological setting since it was reported to contribute to severe liver diseases in HBsAg carriers. The virus has different prevalence rates in different geographical locations.[34] In Saudi Arabia, several reports have investigated the seroprevalence of HDV in different clinical and epidemiological settings. These studies were old and most were performed before the introduction of the mandatory HBV vaccination program in 1990. Results of the studies performed so far showed a difference in prevalence in the different populations and different time periods; while in the 1980s[21,22,23,24,25] studies showed a prevalence ranging from 5.4% in apparently healthy individuals to 22.2% in CHB cases in Riyadh; in 2004, a study showed a decline in HDV prevalence to 3.3% in HBsAg healthy blood donors and 8.6% in chronic HBV hospital-based patients with expected decline due to mandatory HBV vaccination.[28] The present study, to the best of our knowledge, is the first to attempt molecular detection and genotyping of HDV in Saudi Arabia.
This study was performed to investigate the prevalence of HDV in CHB cases in Jeddah, Saudi Arabia, 12 years after the last study by Al-Traif et al.[28] This study showed the prevalence of HDV-Ab in CHB cases with positive HBsAg in Jeddah, Saudi Arabia to be 7.7%. This indicates a slight decline in the prevalence of HDV compared to the results reported by Al-Traif et al. in 2004,[28] which showed a prevalence of 8.6% in hospital- and clinic-based HBsAg positive cases and 3.3% prevalence in HBsAg positive healthy blood donors. All the positive cases in our study were older than 30 years indicating that they might not have been vaccinated against HBV in the mandatory vaccination program. We have included HDV-Ab borderline cases as positive since one of them was positive by RT-PCR, which might be due to lack of sensitivity of the ELISA assay to detect them or these cases could be at an early stage of infection and the level of HDV-Ab was very low for the assay to detect. Another possible explanation is that the epitopes in the ELISA assay were not specific enough to detect these samples. There was not enough serum to test for the HDV-IgM antibodies in these cases.
Since Jeddah is an open city with a large population of expatriates, this study included both Saudi nationals (75.1%) and expatriates working in Saudi Arabia (24.9%). The overall prevalence in the studied population is 7.7%; we found that the prevalence among Saudi nationals (6.3%) was lower than non-Saudis (11.9%); the higher prevalence among non-Saudis might be because 3 of the non-Saudi cases were from Yemen, which is a hyperendemic area of HBV. In this study, HDV coinfection did not have an effect on the clinical status of the CHB cases, as indicated by the lack of significant statistical association with liver function tests, HBV DNA viral load, AFP level, and fibrosis score. This indicates that HDV coinfection has a limited role in the progression of HBV infection in the studied population.
Phylogenetic analysis of the sequenced sample showed that it belongs to HDV genotype 1 and has highest NT similarities (94.9–94.1%) with strains from Iran (KJ744243, FJ233056) followed by Turkey (AM779584, HQ005368). The Saudi strain clustered away from the other 7 clades of HDV, as reported by Foupouapouognigni et al.[14]
CONCLUSION
This study provides more information regarding the HDV prevalence in Jeddah, Saudi Arabia, showing a prevalence of 7.7% in our study population. Phylogenetic analysis of the sequenced sample showed that it is closely related to strains from Iran and Turkey of genotype 1. The coinfection with HDV in CHB cases of this study did not seem to have an effect on the clinical status of the cases. More studies are needed to investigate the prevalence and genetic diversity of HDV in other parts of Saudi Arabia including high-risk populations such as intravenous drug abusers and hemodialysis patients.
Ethical approval
Ethical approval was obtained from the ethical research committee of King Fahd Medical Research Center, King Abdulaziz University (approval#: 011-CEGMR-06-ETH) and the study was conducted in accordance with the Helsinki Declaration of 1975, as revised in 2008. Informed consent was obtained from all patients for being included in the study. All procedures performed in this study were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Financial support and sponsorship
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH) – King Abdulaziz City for Science and Technology, the Kingdom of Saudi Arabia – award number (12-BIO2247-03). The authors also acknowledge with thanks, the Science and Technology Unit, King Abdulaziz University for technical support.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Thomas E, Yoneda M, Schiff ER. Viral hepatitis: Past and future of HBV and HDV. Cold Spring Harb Perspect Med. 2015;5:a021345. doi: 10.1101/cshperspect.a021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizzetto M, Canese MG, Arico S, Crivelli O, Trepo C, Bonino F, et al. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut. 1977;18:997–1003. doi: 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koytak ES, Yurdaydin C, Glenn JS. Hepatitis D. Curr Treat Options Gastroenterol. 2007;10:456–63. doi: 10.1007/s11938-007-0045-8. [DOI] [PubMed] [Google Scholar]
- 4.Eskandar H, SeyedJalal H, Fariba J. Seroprevalence of Delta Hepatitis in Patients with Chronic Hepatitis B and its Clinical Impact in Khuzestan Province, Southwest Iran. Hepatitis Monthly. 2009;9:287–92. [Google Scholar]
- 5.Noureddin M, Gish R. Hepatitis Delta: Epidemiology, Diagnosis and Management 36 Years After Discovery. Curr Gastroenterol Rep. 2014;16 doi: 10.1007/s11894-013-0365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flores R, Serra P, Minoia S, Di Serio F, Navarro B. Viroids: From genotype to phenotype just relying on RNA sequence and structural motifs. Front Microbiol. 2012;3:217. doi: 10.3389/fmicb.2012.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascarella S, Negro F. Hepatitis D virus: An update. Liver Int. 2011;31:7–21. doi: 10.1111/j.1478-3231.2010.02320.x. [DOI] [PubMed] [Google Scholar]
- 8.Casey JL, Brown TL, Colan EJ, Wignall FS, Gerin JL. A genotype of hepatitis D virus that occurs in northern South America. Proc Natl Acad Sci U S A. 1993;90:9016–20. doi: 10.1073/pnas.90.19.9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbas Z, Jafri W, Raza S, Hepatitis D. Scenario in the Asia-Pacific region. World J Gastroenterol. 2010;16:554–62. doi: 10.3748/wjg.v16.i5.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alavian S, Assari S, Manzoori-Joybari H, MoghaniLankarani M, Doroudi T, Haji-Beigi B, et al. Frequency and Risk Factors of Hepatitis D Virus in Hepatitis B Patients. Govaresh. 2005;10:21–6. [Google Scholar]
- 11.Alvarado-Mora MV, Locarnini S, Rizzetto M, Pinho JR. An update on HDV: Virology, pathogenesis and treatment. Antivir Ther. 2013;18(3 Pt B):541–8. doi: 10.3851/IMP2598. [DOI] [PubMed] [Google Scholar]
- 12.Dunford L, Carr MJ, Dean J, Nguyen LT, Ta Thi TH, Nguyen BT, et al. A Multicentre Molecular Analysis of Hepatitis B and Blood-Borne Virus Coinfections in Viet Nam. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davaalkham D, Ojima T, Uehara R, Watanabe M, Oki I, Nymadawa P, et al. Hepatitis delta virus infection in mongolia: Analyses of geographic distribution, risk factors, and disease severity. Am J Trop Med Hyg. 2006;75:365–9. [PubMed] [Google Scholar]
- 14.Foupouapouognigni Y, Noah DN, Sartre MT, Njouom R. High Prevalence and Predominance of Hepatitis Delta Virus Genotype 1 Infection in Cameroon. J Clin Microbiol. 2011;49:1162–4. doi: 10.1128/JCM.01822-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deny P. Hepatitis delta virus genetic variability: From genotypes I, II, III to eight major clades? Curr Top Microbiol Immunol. 2006;307:151–71. doi: 10.1007/3-540-29802-9_8. [DOI] [PubMed] [Google Scholar]
- 16.Moatter T, Abbas Z, Shabir S, Jafri W. Clinical presentation and genotype of hepatitis delta in Karachi. World J Gastroenterol. 2007;13:2604–7. doi: 10.3748/wjg.v13.i18.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farci P. Delta hepatitis: An update. J Hepatol. 2003;39:212–9. doi: 10.1016/s0168-8278(03)00331-3. [DOI] [PubMed] [Google Scholar]
- 18.Shih HH, Shih C, Wang HW, Su CW, Sheen IJ, Wu JC. Pro-205 of large hepatitis delta antigen and Pro-62 of major hepatitis B surface antigen influence the assembly of different genotypes of hepatitis D virus. J Gen Virol. 2010;91(Pt 4):1004–12. doi: 10.1099/vir.0.017541-0. [DOI] [PubMed] [Google Scholar]
- 19.Le Gal F, Gault E, Ripault MP, Serpaggi J, Trinchet JC, Gordien E, et al. Eighth major clade for hepatitis delta virus. Emerg Infect Dis. 2006;12:1447–50. doi: 10.3201/eid1209.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radjef N, Gordien E, Ivaniushina V, Gault E, Anais P, Drugan T, et al. Molecular phylogenetic analyses indicate a wide and ancient radiation of African hepatitis delta virus, suggesting a deltavirus genus of at least seven major clades. J Virol. 2004;78:2537–44. doi: 10.1128/JVI.78.5.2537-2544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashraf SJ, Arya SC, Arendrup M, Krogsgaard K, Parande CM, Orskov B, et al. Frequencies of hepatitis B, delta and HTLV-III virus markers in Saudi Arabia. Liver. 1986;6:73–7. doi: 10.1111/j.1600-0676.1986.tb00271.x. [DOI] [PubMed] [Google Scholar]
- 22.el-Hazmi MA, Ramia S. Epidemiology of delta agent infection in Arabia: Geographical distribution and prevalence of anti-delta. Vox Sang. 1986;50:216–9. doi: 10.1111/j.1423-0410.1986.tb04884.x. [DOI] [PubMed] [Google Scholar]
- 23.Ramia S, el-Hazmi MA, Vivian PA, Waller DK, Mushahwar IK, Frosner GG. Delta agent infection in Riyadh, Saudi Arabia. Trans R Soc Trop Med Hyg. 1987;81:317–8. doi: 10.1016/0035-9203(87)90251-3. [DOI] [PubMed] [Google Scholar]
- 24.Arya SC, Ashraf SJ, Parande CM, Tobeiqi MS, Ageel AR. Hepatitis B and delta markers in primary hepatocellular carcinoma patients in the Gizan area of Saudi Arabia. APMIS Suppl. 1988;3:30–4. [PubMed] [Google Scholar]
- 25.Ramia S, Bahakim H. Perinatal transmission of hepatitis B virus-associated hepatitis D virus. Ann Inst Pasteur Virol. 1988;139:285–90. doi: 10.1016/s0769-2617(88)80041-8. [DOI] [PubMed] [Google Scholar]
- 26.Massoud M HO, Saleh WA. Hepatitis D anti-bodies in some HBs Ag positive in Saudis at Riyad. J Egyptian Soc Parasitol. 1991;21:561–5. [PubMed] [Google Scholar]
- 27.Njoh J, Zimmo S. Prevalence of antibody to hepatitis D virus among HBsAg-positive drug-dependent patients in Jeddah, Saudi Arabia. East Afr Med J. 1998;75:327–8. [PubMed] [Google Scholar]
- 28.Al-Traif I, Ali A, Dafalla M, Al-Tamimi W, Qassem L. Prevalence of hepatitis delta antibody among HBsAG carriers in Saudi Arabia. Ann Saudi Med. 2004;24:343–4. doi: 10.5144/0256-4947.2004.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao JH, Chen PJ, Lai MY, Chen DS. Sequence analysis of pre-S/surface and pre-core/core promoter genes of hepatitis B virus in chronic hepatitis C patients with occult HBV infection. J Med Virol. 2002;68:216–20. doi: 10.1002/jmv.10188. [DOI] [PubMed] [Google Scholar]
- 30.Villa DdF, Cortes-Mancera F, Payares E, Montes N, Hoz Fdl, Arbelaez MP, et al. Hepatitis D virus and hepatitis B virus infection in Amerindian communities of the Amazonas state, Colombia. Virol J. 2015;12:1. doi: 10.1186/s12985-015-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castera L, Forns X, Alberti A. Non-invasive evaluation of liver fibrosis using transient elastography. J Hepatol. 2008;48:835–47. doi: 10.1016/j.jhep.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Romeo R, Perbellini R. Hepatitis delta virus: Making the point from virus isolation up to 2014. World J Hepatol. 2015;7:2389–95. doi: 10.4254/wjh.v7.i22.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]


