Abstract
Importance
Castleman disease (CD) is an ultrarare, interleukin-6 (IL-6)–driven lymphoproliferative disorder whose underlying molecular alterations are unknown. Siltuximab (anti–IL-6 antibody) is approved for treatment of this disease. To our knowledge, genomic sequencing of CD has not been reported.
Objective
To investigate and identify molecular aberration(s) that help explain the exceptional response to siltuximab in a patient with cutaneous CD.
Design, Setting, and Participants
This case study examines data from comprehensive genomic profiling (using targeted next-generation sequencing) of tissue from a patient with cutaneous CD who demonstrated an exceptional response to siltuximab treated at a National Cancer Institute–designated Comprehensive Cancer Center.
Interventions
Intravenous siltuximab 12 mg/kg every 3 weeks. Tissue from the patient was interrogated by next-generation sequencing (405 genes). Serum was evaluated for IL-6 levels by enzyme-linked immunoassay.
Main Outcomes and Measures
Identification of pretreatment serum IL-6 levels and somatic variants that may explain the exceptional response to siltuximab in this patient with cutaneous CD.
Results
Patient pretreatment serum IL-6 levels were normal. Treatment with siltuximab resulted in a complete response lasting 7 years. Next-generation sequencing demonstrated a JAK1V310I missense mutation. Janus Kinase 1 (JAK1) is a crucial signaling component of the IL-6/IL-6 receptor/gp130 machinery. JAK1V310I may induce a conformation change with functional activation effect leading to enhanced sensitivity to the IL-6 ligand.
Conclusions and Relevance
Our observations suggest that a JAK1 alteration may explain the underlying biology of a patient’s cutaneous CD, as well as the patient’s exceptional response to siltuximab.
This case report investigates and identifies molecular aberrations to help explain the exceptional response to siltuximab in a patient with cutaneous Castleman disease.
Key Points
Question
What potential molecular aberration(s) may help explain the exceptional response observed in a patient with cutaneous Castleman disease treated with the anti–interleukin-6 (anti–IL-6) antibody siltuximab?
Findings
In this case report, a missense mutation in the Janus Kinase 1 gene (JAK1V310I) was identified, which is a crucial signaling component of the IL-6/IL-6 receptor/gp130 machinery. JAK1V310I may induce a conformational change that drives Castleman disease by enabling activation of the IL-6R/gp130/JAK1 complex even in the presence of normal levels of IL-6.
Meaning
Genomic profiling of Castleman disease may reveal important molecular drivers of disease, treatment resistance, and therapeutic targets.
Introduction
Interleukin 6 (IL-6) plays a central role in the pathogenesis of Castleman disease (CD), an ultrarare lymphoproliferative disorder. Castleman disease may involve a single lymph node (unicentric) or multiple sites (multicentric). Multicentric CD is associated with excessive release of proinflammatory cytokines, hyperproliferation of immune cells, cutaneous manifestations, ascites, pleural effusions, and can result in multiorgan system impairment.
Clinical and preclinical models have established a central role for IL-6 in symptomatology of multicentric CD. These observations have led to development of antibodies targeting IL-6 and IL-6 receptor (IL-6R). Siltuximab, a chimeric monoclonal antibody against human IL-6, has now been approved in the United States for treatment of CD.
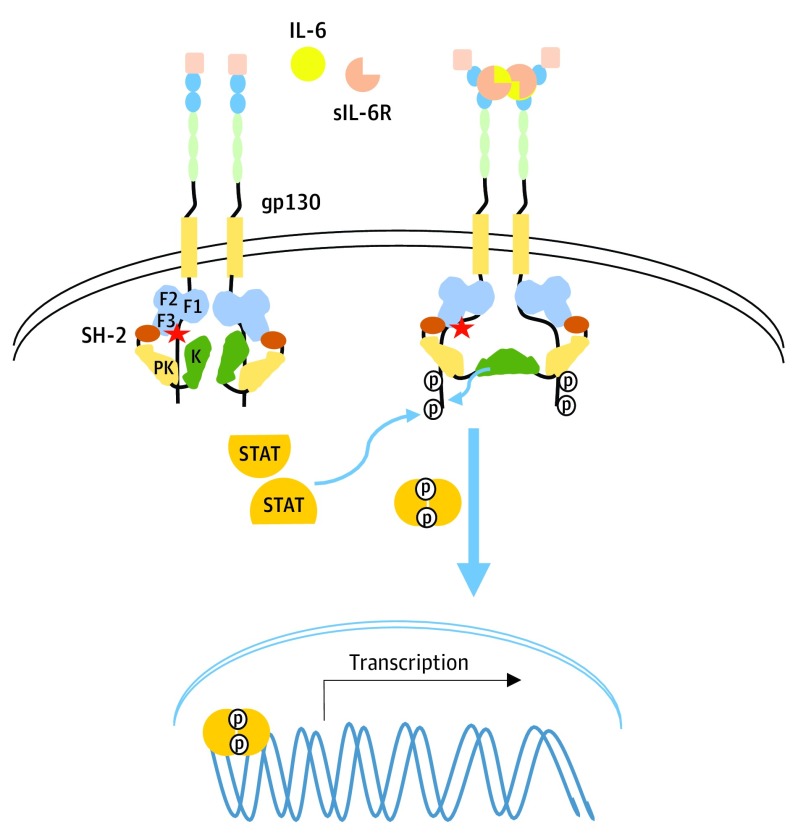
Even though IL-6 is a central figure in CD, the mechanisms by which IL-6 mediates the activation of the IL-6/IL-6R/gp130/JAK1 machinery are not completely understood. The current model proposes that the IL-6-mediated signaling cascade is initiated as 2 IL-6 molecules bind to either 2 membrane-bound IL-6Rs or 2 soluble IL-6Rs (sIL-6R). These receptor-ligand (or soluble receptor–ligand) complexes (acting as agonists) then bind to 2 gp130 signaling subunits to form a complete hexameric complex, to which Janus Kinases (JAKs) are constitutively bound. Through an unidentified mechanism, gp130 homodimerization leads to conformational changes and activation of JAK1, presumably through the release of inhibitory intramolecular interactions (Figure 1).
Figure 1. Proposed IL-6 Transsignaling Pathway and Proposed Impact of JAK1V310I Mutation.
JAK1V310I mutation in the F3 lobe of the FERM (4.1/ezrin/radixin/moesin) domain (indicated by the star) may release currently unidentified inhibitory intramolecular interactions resulting in full activation of JAK1 and signaling upon ligand-receptor complex engagement with gp130 and subsequent homodimerization (indicated with bold blue arrow). IL-6 indicates interleukin 6; K, kinase; PK, pseudokinase; sIL-6R, soluble interleukin 6 receptor; STAT, signal transducer and activator of transcription.
Herein, to our knowledge, we report for the first time genomic characterization of CD, identifying a somatic mutation in JAK1 in a patient with CD who attained a long-term complete remission (CR) after siltuximab treatment and discuss the mechanism by which this alteration may potentiate IL-6 signaling.
Methods
Genomic Sequencing
Targeted next-generation sequencing was performed (FoundationOne; Foundation Medicine) on a skin biopsy specimen. All exomes of 405 genes as well as introns of 31 cancer-related genes were analyzed using hybridization-based capture (https://www.foundationmedicine.com/). The study and treatment were conducted and informed consent obtained in accordance with the Declaration of Helsinki, UCSD Moores Cancer Center, and MD Anderson Cancer Center internal review board requirements.
IL-6 Quantitation
Interleukin 6 levels were assayed using a commercial enzyme linked immunoassay kit (ELISA; Quantikine R&D Systems) per manufacturer’s instructions.
Report of a Case
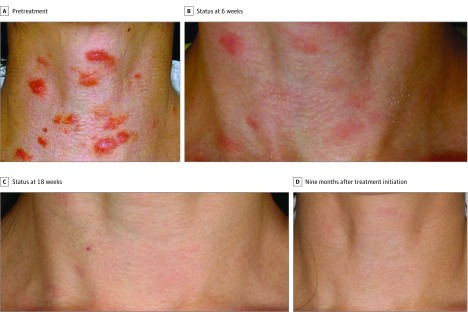
The patient is a female currently in her 50s, who was healthy until she developed multiple plaques on the face and neck. There was no disease on scans, nor any systemic symptoms. She was treated with rituximab, valacyclovir, azathioprine, plaquenil, minocycline, and steroids without salutary effects. Skin biopsy results, reviewed by a dermatopathologist, were diagnostic for cutaneous CD. Serum IL-6 levels were within normal range (0.9 pg/mL; lower limit of sensitivity, 0.7 pg/mL). Median levels for 118 patients with diffuse large-cell lymphoma were 4.6 pg/mL (range, undetectable to 225 pg/mL); median levels for 50 healthy volunteers were undetectable (range, undetectable to 4.3 pg/mL). The patient was both human immunodeficiency virus and human herpesvirus 8 negative. The patient was enrolled in a clinical trial with intravenous siltuximab 12 mg/kg administered every 3 weeks. As reported previously, her skin lesions improved within 24 hours (Figure 2) and she experienced no adverse effects. Patient attained a CR, which was durable on treatment for 7 years, despite increasing the time interval between treatment infusions to every 6 weeks. Treatment was then discontinued on her request and, within 1 year, she relapsed in the cutaneous area of the neck. She resumed intravenous siltuximab 12 mg/kg every 3 weeks and experienced rapid improvement of skin lesions. At the time of relapse, diagnosis was confirmed by repeat biopsy. Tissue was also sent for next-generation sequencing.
Figure 2. Clinical Response to Siltuximab in a Patient With Cutaneous Castleman Disease and a JAK1 Mutation.
Photographs show a patient (A) pretreatment, (B) at 6 weeks after siltuximab initiation, (C) 18 weeks after siltuximab initiation, and (D) 9 months after siltuximab initiation.
Results and Discussion
This patient with cutaneous CD attained a durable CR on anti–IL-6 treatment despite having normal serum IL-6 levels, the latter being consistent with previous reports demonstrating that localized CD without systemic manifestations lacks increased IL-6 gene expression in lymphoid tissue. Next-generation sequence studies showing an alteration in JAK1 may explain these findings because this alteration could sensitize the IL-6/IL-6R/gp130/JAK1 machinery to normal levels of ligand.
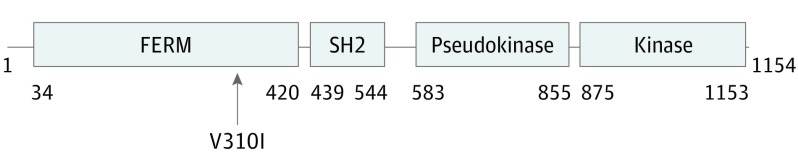
The patient harbored a JAK1V310I missense mutation, as well as a subclonal alteration in Notch1N390S. The JAK1 locus of V310 is found within the FERM (4.1/ezrin/radixin/moesin) domain. JAK family members share 4 common structural domains (Figure 3). JAKs and FAKs are the only tyrosine kinases with FERM domains, which are divergent from other ERM (ezrin, radixin, moesin) proteins. The FERM domain regulates protein localization, often at the plasma membrane, via intermolecular interactions. The most N-terminal region of the FERM domain of JAKs mediates interaction with type I and II cytokine receptors. Interaction of JAK1 with gp130 via the FERM domain is required for IL-6R–mediated signal transduction. Indeed, many of the gain-of-function mutations found within the JAK mutational hot-spot, located within pseudokinase region, require a functional FERM domain capable of mediating interactions with receptors in order for signaling events to occur.
Figure 3. Domain Structures of Janus Kinase Family Members.
JAKs contain 4 functional domains: (1) the FERM (4.1/ezrin/radixin/moesin) domain, (2) the Src homology 2 (SH2) domain, (3) the pseudokinase domain (PK), and (4) the kinase domain. Depicted here are domain structure boundaries for JAK1. The arrow shows the mutation identified in this patient.
JAK1V310I mutations have been reported previously in solid malignancies (http://cancer.sanger.ac.uk). Support for the functional significance of this amino acid substitution comes from the most common patient–derived gain-of-function mutations occurring in the pseudokinase domain observed in myeloproliferative neoplasms—the JAK1V658F and the corresponding JAK2V617F/V617I mutations. The substitution of V658F in JAK1, or other amino acid changes (eg, valine to isoleucine), resulting in increasing hydrophobic side chains, have a functional impact on JAK1 similar to that of the V658F mutation. Biochemical characterization of the JAK2V617I mutant identified in patients with myeloproliferative neoplasms also supports the hypothesis that this substitution impacts JAK2 conformation resulting in increasing JAK2 signaling.
The precise consequences of the JAK1V310I mutation are not known, due to the lack of structural information on the JAK1 FERM domain. However, single–particle electron microscopy studies with full-length JAK1 revealed significant conformational flexibility, and suggests that JAK1 makes multiple intermolecular interactions with gp130, along with multiple intramolecular interactions. Full-length JAK1 exists in a closed compact conformation where the FERM domain may make contacts with the kinase domain (Figure 1). This type of compact assembly due to intramolecular interactions between the FERM/kinase domains has also been demonstrated for FAK. We considered the detailed structural information of FAK, which reveals that FERM/kinase domain interactions regulate its activity. Current evidence demonstrates that FAK activation occurs in an orderly and sequential manner, whereby the release of inhibitory interactions between FERM and kinase domains are the initial key steps in full activation of FAK. Here we propose that the JAK1V310I mutation within the F3 lobe of the FERM domain may also lead to release of these presumed inhibitory interactions, with subsequent discharge of other, more well-established inhibitory interactions between the pseudokinase/kinase domains upon ligand–induced receptor complex conformation change. The loss of inhibitory interactions should allow JAK1 to take conformations that are more readily activated by ligand-induced gp130 dimerization, resulting in hyperresponsiveness to normal levels of cytokines (Figure 1), as has been demonstrated for many other activating mutations identified within JAKs. Clinical response to siltuximab observed in this individual, despite the lack of elevated IL-6, also supports this contention.
Conclusions
Our report describes a novel genomic alteration within the IL-6/sIL-6R/gp130/JAK1 signaling axis that may in part explain the hyperactivation of this pathway in CD. This is an important step in understanding the molecular pathogenesis of the disease. Genomic profiling of CD is currently being pursued by the Castleman Disease Collaborative Network, and it will be important to determine if mutations within the IL-6/IL-6R/gp130/JAK1/STAT signaling pathway are a common occurrence.
References
- 1.Fajgenbaum DC, van Rhee F, Nabel CS. HHV-8-negative, idiopathic multicentric Castleman disease: novel insights into biology, pathogenesis, and therapy. Blood. 2014;123(19):2924-2933. [DOI] [PubMed] [Google Scholar]
- 2.Munoz J, Naing A, Qi M, Kurzrock R. Cutaneous Castleman disease. Br J Haematol. 2012;157(6):652-652. [DOI] [PubMed] [Google Scholar]
- 3.van Rhee F, Casper C, Voorhees PM, et al. . A phase 2, open-label, multicenter study of the long-term safety of siltuximab (an anti-interleukin-6 monoclonal antibody) in patients with multicentric Castleman disease. Oncotarget. 2015;6(30):30408-30419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurzrock R, Voorhees PM, Casper C, et al. . A phase I, open-label study of siltuximab, an anti-IL-6 monoclonal antibody, in patients with B-cell non-Hodgkin lymphoma, multiple myeloma, or Castleman disease. Clin Cancer Res. 2013;19(13):3659-3670. [DOI] [PubMed] [Google Scholar]
- 5.Garbers C, Aparicio-Siegmund S, Rose-John S. The IL-6/gp130/STAT3 signaling axis: recent advances towards specific inhibition. Curr Opin Immunol. 2015;34:75-82. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed B, Tschen JA, Cohen PR, et al. . Cutaneous castleman’s disease responds to anti interleukin-6 treatment. Mol Cancer Ther. 2007;6(9):2386-2390. [DOI] [PubMed] [Google Scholar]
- 7.Kurzrock R. Cytokine deregulation in hematological malignancies: clinical and biological implications. Clin Cancer Res. 1997;3(12 Pt 2):2581-2584. [PubMed] [Google Scholar]
- 8.Leger-Ravet MB, Peuchmaur M, Devergne O, et al. . Interleukin-6 gene expression in Castleman’s disease. Blood. 1991;78(11):2923-2930. [PubMed] [Google Scholar]
- 9.Lietha D, Cai X, Ceccarelli DF, Li Y, Schaller MD, Eck MJ. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129(6):1177-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilkens CM, Is’harc H, Lillemeier BF, et al. . A region encompassing the FERM domain of Jak1 is necessary for binding to the cytokine receptor gp130. FEBS Lett. 2001;505(1):87-91. [DOI] [PubMed] [Google Scholar]
- 11.Hornakova T, Chiaretti S, Lemaire MM, et al. . ALL-associated JAK1 mutations confer hypersensitivity to the antiproliferative effect of type I interferon. Blood. 2010;115(16):3287-3295. [DOI] [PubMed] [Google Scholar]
- 12.Gordon GM, Lambert QT, Daniel KG, Reuther GW. Transforming JAK1 mutations exhibit differential signalling, FERM domain requirements and growth responses to interferon-γ. Biochem J. 2010;432(2):255-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou YJ, Chen M, Cusack NA, et al. . Unexpected effects of FERM domain mutations on catalytic activity of Jak3: structural implication for Janus kinases. Mol Cell. 2001;8(5):959-969. [DOI] [PubMed] [Google Scholar]
- 14.Ma W, Kantarjian H, Zhang X, et al. . Mutation profile of JAK2 transcripts in patients with chronic myeloproliferative neoplasias. J Mol Diagn. 2009;11(1):49-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupardus PJ, Skiniotis G, Rice AJ, et al. . Structural snapshots of full-length Jak1, a transmembrane gp130/IL-6/IL-6Rα cytokine receptor complex, and the receptor-Jak1 holocomplex. Structure. 2011;19(1):45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]