This secondary analysis of a randomized clinical trial examines whether a germline mutation in a microRNA-binding site in KRAS is a predictive biomarker of cetuximab response and altered immunity following radiotherapy and cisplatin treatment for head and neck squamous cell cancer.
Key Points
Question
Can an inherited microRNA-disrupting variant, called the Kirsten rat sarcoma viral oncogene homolog (KRAS)–variant, predict cetuximab response in head and neck cancer?
Findings
In an analysis of a phase 3 randomized clinical trial of 891 patients with head and neck squamous cell carcinoma, the KRAS-variant predicted a significant, positive response to cetuximab. In addition, the KRAS-variant was found to interact with p16 status and was associated with significantly elevated transforming growth factor β1 levels in these patients.
Meaning
The KRAS-variant appears to be an inherited biomarker of cetuximab response in patients with head and neck squamous cell carcinoma.
Abstract
Importance
There is a significant need to find biomarkers of response to radiotherapy and cetuximab in locally advanced head and neck squamous cell carcinoma (HNSCC) and biomarkers that predict altered immunity, thereby enabling personalized treatment.
Objectives
To examine whether the Kirsten rat sarcoma viral oncogene homolog (KRAS)–variant, a germline mutation in a microRNA-binding site in KRAS, is a predictive biomarker of cetuximab response and altered immunity in the setting of radiotherapy and cisplatin treatment and to evaluate the interaction of the KRAS-variant with p16 status and blood-based transforming growth factor β1 (TGF-β1).
Design, Setting, and Participants
A total of 891 patients with advanced HNSCC from a phase 3 trial of cisplatin plus radiotherapy with or without cetuximab (NRG Oncology RTOG 0522) were included in this study, and 413 patients with available samples were genotyped for the KRAS-variant. Genomic DNA was tested for the KRAS-variant in a CLIA-certified laboratory. Correlation of the KRAS-variant, p16 positivity, outcome, and TGF-β1 levels was evaluated. Hazard ratios (HRs) were estimated with the Cox proportional hazards model.
Main Outcomes and Measures
The correlation of KRAS-variant status with cetuximab response and outcome, p16 status, and plasma TGF-β1 levels was tested.
Results
Of 891 patients eligible for protocol analyses (786 male [88.2%], 105 [11.2%] female, 810 white [90.9%], 81 nonwhite [9.1%]), 413 had biological samples for KRAS-variant testing, and 376 had plasma samples for TGF-β1 measurement. Seventy patients (16.9%) had the KRAS-variant. Overall, for patients with the KRAS-variant, cetuximab improved both progression-free survival (PFS) for the first year (HR, 0.31; 95% CI, 0.10-0.94; P = .04) and overall survival (OS) in years 1 to 2 (HR, 0.19; 95% CI, 0.04-0.86; P = .03). There was a significant interaction of the KRAS-variant with p16 status for PFS in patients treated without cetuximab. The p16-positive patients with the KRAS-variant treated without cetuximab had worse PFS than patients without the KRAS-variant (HR, 2.59; 95% CI, 0.91-7.33; P = .07). There was a significant 3-way interaction among the KRAS-variant, p16 status, and treatment for OS (HR, for KRAS-variant, cetuximab and p16 positive, 0.22; 95% CI, 0.03-1.66; HR for KRAS-variant, cetuximab and p16 negative, 1.43; 95% CI, 0.48-4.26; HR for KRAS-variant, no cetuximab and p16 positive, 2.48; 95% CI, 0.64-9.65; and HR for KRAS-variant, no cetuximab and p16 negative, 0.61; 95% CI, 0.23-1.59; P = .02). Patients with the KRAS-variant had significantly elevated TGF-β1 plasma levels (median, 23 376.49 vs 18 476.52 pg/mL; P = .03) and worse treatment-related toxic effects.
Conclusions and Relevance
Patients with the KRAS-variant with HNSCC significantly benefit from the addition of cetuximab to radiotherapy and cisplatin, and there is a significant interaction between the KRAS-variant and p16 status. Elevated TGF-β1 levels in patients with the KRAS-variant suggests that cetuximab may help these patients by overcoming TGF-β1–induced suppression of antitumor immunity.
Trial Registration
clinicaltrials.gov Identifier: NCT00265941
Introduction
The current standard of care for the nonsurgical management of locally advanced head and neck squamous cell carcinoma (HNSCC) is concurrent platinum-based chemotherapy and radiotherapy. Unfortunately, however, this approach results in an approximately 50% treatment failure rate in unselected patients. Cetuximab is an anti–epidermal growth factor receptor (EGFR) monoclonal antibody previously reported to be beneficial for such patients, improving survival when given with definitive radiotherapy or with cisplatin and fluorouracil in recurrent or metastatic HNSCC. However, a randomized phase 3 clinical trial (NRG Oncology RTOG 0522) failed to show any improvement in survival with the addition of 8 weeks of cetuximab to cisplatin and radiotherapy. To date, beyond EGFR surface expression, no predictive biomarker of cetuximab response has been identified for these patients.
Recently, human papillomavirus, reliably measured by p16 expression, has been established as a prognostic biomarker in HNSCC, with p16-positive patients having a favorable outcome compared with p16-negative patients. One reason for the better prognosis in p16-positive disease is thought to be attributable to immune-mediated mechanisms because cisplatin and radiotherapy expose p16-associated antigens, which are immunogenic, enabling host-mediated immune tumor killing. Consistent with a prior report, p16-positive patients in NRG Oncology RTOG 0522 had improved outcome compared with p16-negative patients. However, there was no benefit of cetuximab treatment when p16 status was considered in NRG Oncology RTOG 0522. One hypothesis put forward is lower EGFR tumor cell staining in p16-positive tumors in NRG Oncology RTOG 0522 vs p16-negative tumors (22% vs 30%).
Beyond inhibition of EGFR signaling, however, cetuximab has also been found to work through immune-mediated mechanisms by enhancing adaptive immunity via antibody-dependent cell-mediated cytotoxicity and by better enabling cytotoxic T-lymphocyte cross-priming by dendritic cells. Recently, cetuximab combined with chemotherapy was further confirmed to trigger immunogenic cell death in mouse models through increased phagocytosis by dendritic cells. These immune-based mechanisms of cetuximab efficacy have not yet been studied in the setting of radiotherapy, although radiotherapy also enables antitumor immunity, with the release of danger signals, and dendritic and T-cell activation. Recently, a biomarker of altered immune function, FCGR2A, a polymorphism in an Fcy receptor, was found to influence the response rate to cetuximab for patients with Kirsten rat sarcoma viral oncogene homolog (KRAS) (OMIM 190070)–mutant tumors, indicating that inherited polymorphisms can affect cetuximab response.
In colon cancer, response to cetuximab is associated with an inherited biomarker disrupting KRAS, referred to as the KRAS-variant. The KRAS-variant is the first microRNA-binding site mutation discovered in cancer and is a functional germline mutation in the 3′ untranslated region of KRAS. Although the KRAS-variant is rare in tumors with acquired KRAS mutations, it leads to gene expression patterns consistent with KRAS addiction across tumor types. The KRAS-variant is associated with increased cancer risk and the development of multiple cancers and more recently was found to be a predictive biomarker of response to cancer treatment irrespective of tumor type or tumor-acquired mutations. In HNSCC, the KRAS-variant is found in 15% to 32% of patients and is therefore highly relevant. Patients with HNSCC and the KRAS-variant have previously been reported to have a poor response to platinum therapy, similar to findings for patients with the KRAS-variant and other cancer types.
Although the mechanisms leading to the predictive power of the KRAS-variant in treatment response have not been fully elucidated, one could hypothesize that the KRAS-variant identifies patients with altered immunity based on the fact that KRAS signaling and let-7 microRNA expression, both universally abnormal in KRAS-variant–associated tumors, are critical in systemic immunity. In addition, mutant KRAS directly inhibits antitumor immunity through induction of transforming growth factor β1 (TGF-β1) secretion, which is known to lead to conversion of conventional T cells into regulatory T cells or type 17 helper T cells. In the setting of radiotherapy, TGF-β1 is proposed to be a master regulator of treatment response, with high levels preventing radiation-induced antitumor immunity. Interestingly, in cells with high TGF-β1 levels, cetuximab has been found to restore the killing activity of patient natural killer cells against primary HNSCC cells via enhanced antibody-dependent cell-mediated cytotoxicity in a dose-dependent manner. Cetuximab response has also been found to overcome TGF-β1–expressing regulatory T cells, known to inhibit natural killer cell killing, providing a potential mechanism of action. To date, the role of altered immunity and TGF-β1 levels as potential mechanisms of cetuximab response in patients with the KRAS-variant has not been evaluated.
Thus, we sought to test the predictive role of the KRAS-variant for outcomes in patients with HNSCC in the setting of radiotherapy, cisplatin, and cetuximab, considering p16 status. We further evaluated immunity in patients with the KRAS-variant through plasma TGF-β1 levels.
Methods
Protocol and Patients
NRG Oncology RTOG 0522 was a phase 3 trial that tested the addition of cetuximab to radiotherapy with concurrent cisplatin for patients with advanced HNSCC. The study was approved by all institutional review boards where patients were enrolled, and oral informed consent was obtained from all study participants. Race and sex data were collected and are reported in eTable 1 in the Supplement. Eligible patients had pathologically proven squamous cell carcinoma of the oropharynx, hypopharynx, or larynx, with selected stage III or IV disease (T2N2-3M0 or T3-4NM0 [any N]), Zubrod performance status of 0 to 1, age of 18 years or older, and adequate bone marrow, hepatic, and renal function. Human papillomavirus status was evaluated by p16 expression as previously described. A total of 940 patients were enrolled into NRG Oncology RTOG 0522, of whom 891 (94.8%) were eligible for protocol analyses. A total of 413 of 891 patients had biological samples available for KRAS-variant analysis (46.4%) (eFigure 1 in the Supplement). Patients not tested for the KRAS-variant had significantly younger age (median, 56 vs 58; P = .02) than patients tested for the KRAS-variant, but the difference in medians was only 2 years (56 vs 58 years) (eTable 1 in the Supplement). Progression-free survival (PFS) and overall survival (OS) were similar for the patients tested and not tested for the KRAS-variant (PFS hazard ratio [HR], 0.92; 95% CI, 0.76-1.13; P = .44; OS HR, 0.99; 95% CI, 0.78-1.25; P = .93). DNA genotyping was performed from January 2015 to May 2015. At the time of analysis, median follow-up for surviving patients was 4.8 years (range, 0.2-6.9 years). Of the 413 study participants tested for the KRAS-variant, 5 (1.2%) were homozygous (GG) and 65 (15.7%) were heterozygous (TG), resulting in an overall prevalence of 16.9%. The prevalence of the KRAS-variant was not significantly different in p16-negative (17.4%) vs p16-positive (16.0%) patients, in agreement with a prior study.
Of the 70 patients with the KRAS-variant, the cetuximab-treated subset had fewer patients of white race (28 [87.5%] of 32 vs 38 [100%] of 38, P = .04). Within the nonvariant cohort, the cetuximab-treated subset had a significantly younger age than the subset not receiving cetuximab, but the difference in medians was only 2 years (59 vs 57 years, 174 cetuximab treated vs 169 not cetuximab treated, P = .05) (eTable 2 in the Supplement).
KRAS-Variant Testing
Genomic DNA from peripheral blood mononuclear cells or whole blood was isolated as previously described, and 100 ng was analyzed in a Clinical Laboratory Improvement Amendments–certified laboratory for the KRAS-variant (MiraDx). Patients that were homozygous (GG) were grouped with those that were heterozygous (TG) for these analyses.
Plasma TGF-β1 Levels
Plasma levels of TGF-β1 from 376 patients enrolled in NRG Oncology RTOG 0522 were measured using multiplex bead assays with the use of reagents from Millipore and the Luminex 100 system as previously described. The assay was run using manufacturer controls to ensure precision. The correlation between KRAS-variant status and plasma TGF-β1 levels was evaluated by Wilcoxon rank sum test.
Statistical Analysis
Locoregional failure (LRF), distant metastasis (DM), PFS, and OS were as defined in the NRG Oncology RTOG 0522 protocol. The LRF and DM rates were estimated by the cumulative incidence method. The PFS and OS rates were estimated by the Kaplan-Meier method. The HRs were estimated by the Cox proportional hazards model. Adverse events were graded by Common Terminology Criteria for Adverse Events, version 3.0. Odds ratios (ORs) were estimated by logistic regression. Patient characteristics were compared with the Fisher exact test (categorical variables) or Wilcoxon rank sum test (ordinal or continuous variables). All analyses were performed using SAS statistical software, version 9.4 (SAS Institute Inc). P < .05 was considered statistically significant.
Results
Prediction of Response to Cetuximab
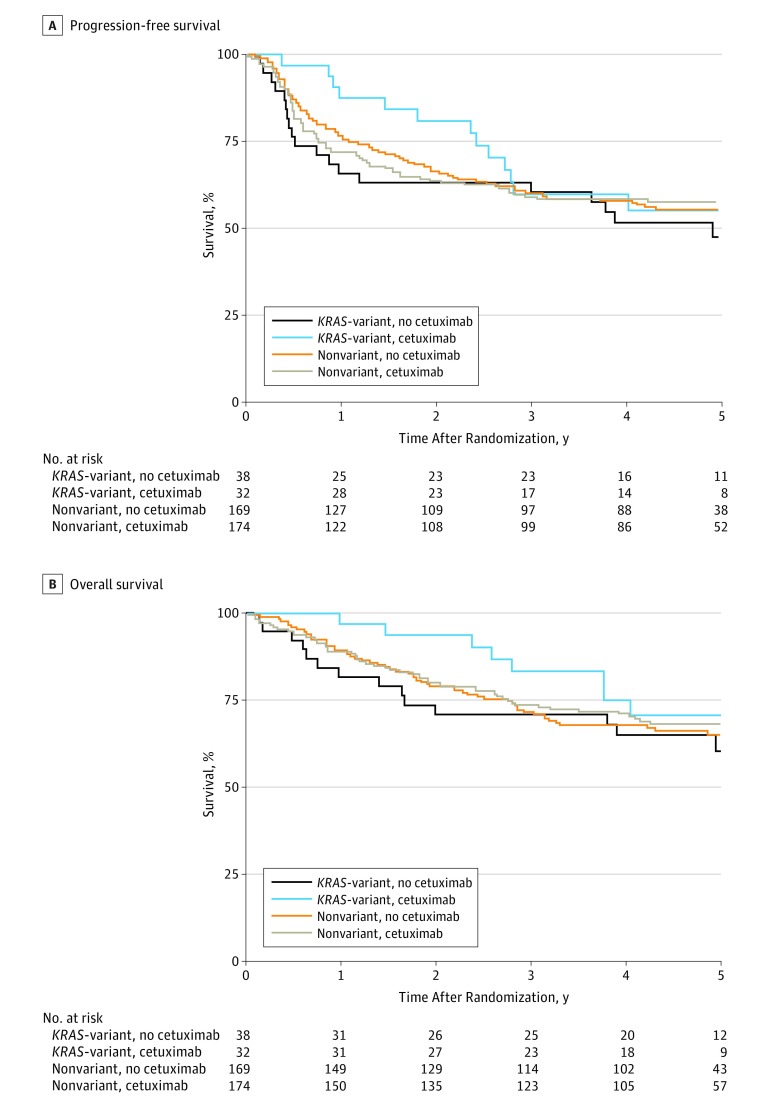
Of 891 patients eligible for protocol analyses (786 male [88.2%], 105 [11.2%] female, 810 white [90.9%], 81 nonwhite [9.1%]), 413 had biological samples for KRAS-variant testing, and 376 had plasma for TGF-β1 measurement. We investigated the association of the KRAS-variant with cetuximab response in HNSCC and found a significant, positive effect of 8 weeks of cetuximab treatment for all patients with the KRAS-variant. Patients with the KRAS-variant had significantly improved PFS in the first year (HR, 0.31; 95% CI, 0.10-0.94; P = .04) but not thereafter (HR, 1.76; 95% CI, 0.62-4.95; P = .28) (Figure 1A). The cetuximab treatment effect significantly varied over time (P = .02 for interaction between treatment and PFS time [>1 year]). In the nonvariant group, there was no effect of cetuximab on PFS with a treatment effect HR (cetuximab vs no cetuximab) of 1.00 (95% CI, 0.72-1.38; P = .98). In multivariate analysis, the interaction among treatment, time (>1 year), and KRAS-variant group remained significant (HR, 0.42; 95% CI, 0.14-1.26, for early treatment effect; HR, 1.22; 95% CI, 0.53-2.80, for late treatment effect; P = .02), indicating that there is an interaction between treatment and time in the KRAS-variant group (Table 1).
Figure 1. Progression-Free Survival and Overall Survival for Patients With Head and Neck Squamous Cell Carcinoma by KRAS-Variant Status and Assigned Treatment.
A, Progression-free survival (PFS). In total, 179 of 413 patients (43.3%) experienced a PFS failure: 19 of 38 (50.0%) in the non–cetuximab-treated KRAS-variant group, 13 of 32 (40.6%) in the cetuximab-treated KRAS-variant group, 74 of 169 (43.8%) in the non–cetuximab-treated nonvariant group, and 73 of 174 (42.0%) in the cetuximab-treated nonvariant group. B, Overall survival. In total, 134 of 413 patients (32.4%) have died: 14 of 38 (36.8%) in the non–cetuximab-treated KRAS-variant group, 8 of 32 (25.0%) in the cetuximab-treated KRAS-variant group, 58 of 169 (34.3%) in the non–cetuximab-treated nonvariant group, and 54 of 174 (31.0%) in the cetuximab-treated nonvariant group.
Table 1. Multivariate Analysis of PFS and OS on the Basis of KRAS-Variant Status.
| Variable | PFS (n = 179 Events) |
OS (n = 134 Events) |
||
|---|---|---|---|---|
| HR (95% CI)a | P Value | HR (95% CI)a | P Value | |
| KRAS-variant | ||||
| Earlyb | 0.42 (0.14-1.26) | .12 | 0.27 (0.06-1.21) | .09 |
| Late | 1.22 (0.53-2.80) | .64 | 1.47 (0.53-4.03) | .46 |
| Nonvariant | ||||
| Earlyb | 1.20 (0.80-1.80) | .39 | 0.89 (0.56-1.41) | .62 |
| Late | 0.79 (0.47-1.33) | .38 | 0.81 (0.45-1.48) | .49 |
Abbreviations: HR, hazard ratio; OS, overall survival; PFS, progression-free survival.
The HRs were estimated from Cox proportional hazards models, including treatment (cetuximab vs no cetuximab), treatment × PFS and OS time interaction, KRAS (variant vs nonvariant), treatment × KRAS interaction, treatment × PFS and OS time × KRAS interaction, age, Zubrod performance status (1 vs 0), primary site (oropharynx vs others), T stage (T4 vs T2-3), and N stage (N2b-3 vs N0-2a).
First year for PFS and first 2 years for OS.
The pattern of failure in a multivariate analysis suggested that DM rather than LRF may be a more likely contributor to the difference in PFS for patients with the KRAS-variant: in the KRAS-variant group, the treatment effect is 0.45 (95% CI, 0.12-1.70) for DM and 0.84 (95% CI, 0.29-2.42) for LRF (eFigure 2 and eFigure 3 in the Supplement). In the nonvariant group, the treatment effects are 0.90 (95% CI, 0.48-1.70) for DM and 1.24 (95% CI, 0.80-1.92) for LRF.
Patients with the KRAS-variant also had significantly improved OS in the first 2 years (HR, 0.19; 95% CI, 0.04-0.86; P = .03) but not thereafter (HR, 2.34; 95% CI, 0.58-9.41; P = .23) (Figure 1B). Again, the cetuximab treatment effect significantly varied over time (P = .02 for interaction between treatment effect and survival time [>2 years]). In the nonvariant group, cetuximab treatment had no effect on OS (treatment effect HR [cetuximab vs no cetuximab], 0.90; 95% CI, 0.62-1.30; P = .56). In a multivariate analysis, the interaction among treatment, time (>2 years), and KRAS-variant group remained significant (Table 1), indicating that there is an interaction between treatment and time in the KRAS-variant group. The treatment effect in the first 2 years for the KRAS-variant group is 0.27 (95% CI, 0.06-1.21; P = .09) and 1.47 (95% CI, 0.53-4.03; P = .46) thereafter (P = .05 for the interaction).
Interaction With p16 Status for PFS
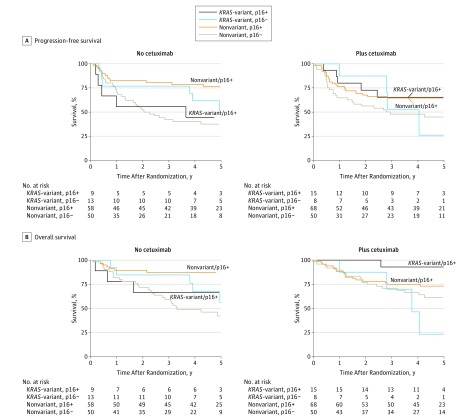
Because p16, a key cell cycle inhibitor, is a known prognostic biomarker in HNSCC, we evaluated outcome in patients with the KRAS-variant while considering p16 status, without or with cetuximab treatment. For PFS, in patients treated without cetuximab, the 2-way interaction between KRAS and p16 was significant (Table 2). The p16-positive patients with the KRAS-variant treated without cetuximab had worse PFS than the p16-positive patients without the KRAS-variant (HR, 2.59; 95% CI, 0.91-7.33; P = .07) (Figure 2A). However, in cetuximab-treated patients, there was no longer a difference in outcome between the p16-positive patients with the KRAS-variant vs without the KRAS-variant (HR, 0.89; 95% CI, 0.34-2.35; P = .82) (Figure 2A).
Table 2. Multivariate Analysis of PFS and OS on the Basis of p16, Cetuximab, and KRAS-Variant Status.
| Variable | PFS (n = 116 Events) |
OS (n = 85 Events) |
||
|---|---|---|---|---|
| HR (95% CI)a | P Value | HR (95% CI)a | P Value | |
| Treatment × KRAS | ||||
| × p16 | NA | .20 | NA | .02 |
| If p16-positive | NA | .14 | NA | .05 |
| Treatment × p16 | ||||
| If KRAS-variant | NA | .51 | NA | .15 |
| If KRAS-nonvariant | NA | .12 | NA | .01 |
| KRAS × p16 | ||||
| If cetuximab | NA | .84 | NA | .11 |
| If no cetuximab | NA | .04 | NA | .10 |
| Treatment effect | ||||
| If KRAS-variant and p16 positive | 0.60 (0.17-2.10) | .42 | 0.21 (0.02-2.04) | .18 |
| If KRAS nonvariant and p16 positive | 1.74 (0.88-3.42) | .11 | 2.36 (0.98-5.67) | .05 |
| KRAS effect | ||||
| If no cetuximab and p16 positive | 2.59 (0.91-7.33) | .07 | 2.48 (0.64-9.65) | .19 |
| If cetuximab and p16 positive | 0.89 (0.34-2.35) | .82 | 0.22 (0.03-1.66) | .14 |
| p16 effect | ||||
| If cetuximab and KRAS-variant | 0.73 (0.20-2.73) | .64 | 0.12 (0.01-1.11) | .06 |
| If cetuximab and KRAS nonvariant | 0.63 (0.36-1.11) | .11 | 0.80 (0.41-1.54) | .50 |
| If no cetuximab and KRAS-variant | 1.34 (0.40-4.44) | .64 | 0.86 (0.20-3.64) | .84 |
| If no cetuximab and KRAS nonvariant | 0.32 (0.17-0.61) | <.001 | 0.21 (0.09-0.49) | <.001 |
Abbreviations: HR, hazard ratio; NA, not applicable (HRs for interaction terms are not meaningful and are therefore note reported); OS, overall survival; PFS, progression-free survival.
The HRs were estimated from Cox proportional hazards models, including treatment (cetuximab vs no cetuximab), treatment × PFS and OS time interaction, KRAS (variant vs nonvariant), treatment × KRAS interaction, treatment × PFS and OS time × KRAS interaction, age, Zubrod performance status (1 vs 0), primary site (oropharynx vs others), T stage (T4 vs T2-3), and N stage (N2b-3 vs N0-2a).
Figure 2. Progression-Free Survival and Overall Survival by KRAS Genotype and p16 Status for Patients Treated Without or With Cetuximab Treatment.
A, Progression-free survival. There is a significant 2-way interaction between KRAS-variant status and p16 in patients treated without cetuximab (P = .04). B, Overall survival. There is a significant 3-way interaction among cetuximab treatment, KRAS-variant status, and p16 status (P = .02).
Interaction With p16 and Treatment for OS
For OS, there is a significant 3-way interaction among cetuximab treatment, KRAS-variant status, and p16 status. There is also a significant 2-way interaction between treatment and KRAS for p16-positive patients and between treatment and p16 status for patients with the KRAS-variant (Table 2). In patients treated without cetuximab, p16-positive patients with the KRAS-variant had worse OS than p16-positive patients without the KRAS-variant (HR, 2.48; 95% CI, 0.64-9.65; P = .19), which appeared to be nonsignificantly reversed with cetuximab treatment (HR, 0.22; 95% CI, 0.03-1.66; P = .14) (Figure 2B). In p16-positive patients, the cetuximab treatment effect is 0.21 (95% CI, 0.02-2.04; P = .18) for patients with the KRAS-variant compared with 2.36 (95% CI, 0.98-5.67; P = .05) for patients without the KRAS-variant.
Immunologic Landscape and Toxic Effects in Patients With the KRAS-Variant
On the basis of the differential effect of cetuximab, especially in p16-positive patients, we investigated immune factors in patients with the KRAS-variant. We measured plasma TGF-β1 (generally a negative regulator of local immune response) levels in patients from NRG Oncology RTOG 0522 and found that patients with the KRAS-variant had significantly higher plasma TGF-β1 levels (median, 23 376.49 pg/mL; interquartile range, 12 574.03-44 809.10 pg/mL; range, 5034.35-109 758.72 pg/mL) compared with patients without the KRAS-variant (median, 18 477 pg/mL; interquartile range, 8837.00-32 378.43 pg/mL; range, 2261.42-123 264.70 pg/mL) (Wilcoxon rank sum test, P = .03). The TGF-β1 levels were also compared by p16 and KRAS-variant status, but the differences were not statistically significant in this smaller cohort (eTable 3 in the Supplement).
On the basis of the known role of TGF-β1 in normal tissue radiation toxicity, we evaluated toxic effects in patients with and without the KRAS-variant. Patients with the KRAS-variant appeared to have elevated levels of grade 3 to 4 mucositis even without cetuximab treatment (47.4% without cetuximab vs 50.0% with cetuximab; OR, 1.11; 95% CI, 0.43-2.85; P = .83). In contrast, in patients without the KRAS-variant, the addition of cetuximab significantly increased grade 3 to 4 mucositis (37.9% vs 50.6%; OR, 1.68; 95% CI, 1.09-2.58; P = .02) (eTable 4 in the Supplement). However, a test of the interaction between KRAS-variant status and cetuximab treatment was not significant (ORs, 1.11 and 1.68; P = .43). Patients with the KRAS-variant also appeared to have elevated levels of grade 3 to 4 skin reaction inside the portal even without cetuximab treatment (18.4% without cetuximab vs 15.6% with cetuximab; OR, 0.82; 95% CI, 0.23-2.89; P = .76). In contrast, in patients without the KRAS-variant, the addition of cetuximab significantly increased grade 3 to 4 skin reactions inside the portal (11.2% vs 21.8%; OR, 2.21; 95% CI, 1.21-4.01; P = .05), but again a test of interaction was not significant (P = .16) (eTable 5 in the Supplement). As expected, both patients with and without the KRAS-variant developed increased skin reaction outside the portal with cetuximab (nonvariant OR, 50.15; 95% CI, 6.81-369.54; P < .001; KRAS-variant OR, 8.54; 95% CI, 0.97-75.20; P = .05) (eTable 6 in the Supplement).
To further evaluate whether KRAS-variant status and TGF-β1 levels together predict toxic effects, we evaluated patients with the KRAS-variant with known TGF-β1 levels (n = 65) and looked at grade 3 to 4 mucositis or in-field skin toxic effects. We only included patients not treated with cetuximab because cetuximab is known to be a radiosensitizer. We found that patients with the KRAS-variant with TGF-β1 levels greater than vs at the median or below treated without cetuximab appeared to have an increase in risk of grade 3 to 4 toxic effects (OR, 2.31; 95% CI, 0.57-9.41). However, the interaction between the KRAS-variant and TGF-β1 was not significant (P = .40). In contrast, patients without the KRAS-variant with TGF-β1 levels greater than vs at the median or below treated without cetuximab had little correlation of TGF-β1 levels on toxic effects (OR, 1.19; 95% CI, 0.62-2.25).
Discussion
Patients with advanced HNSCC with the KRAS-variant significantly benefit from the addition of cetuximab to radiotherapy and cisplatin therapy, resulting in improved PFS and OS. We found that this benefit fades over time, possibly because of the short course of cetuximab delivered in this study. We further found that there is a significant interaction among the KRAS-variant, p16 status, and cetuximab, with p16-positive patients with the KRAS-variant doing poorly compared with p16-positive patients without the KRAS-variant, a deficit that seems to be overcome by cetuximab. Furthermore, we found that patients with the KRAS-variant have significantly elevated TGF-β1 levels, suggesting that they may have a baseline immunodeficient and/or anti–tumor immunodefective state, which cetuximab in combination with radiotherapy may be helping to overcome.
For HNSCC, although p16 is an important prognostic biomarker, it appears that knowledge of p16 status and KRAS-variant status may be important to determine optimal personalized treatment for these patients. It is particularly notable that p16-positive patients with the KRAS-variant had the worst outcomes of all subgroups of patients with HNSCC in our study, which appeared to be improved with a short course of cetuximab treatment given with radiotherapy and cisplatin. These patients should perhaps be considered a different entity from p16-positive patients without the KRAS-variant, who had excellent outcomes with radiotherapy and cisplatin alone. Work is ongoing to further validate these observations.
Our finding that patients with HNSCC and the KRAS-variant may have an altered immune state compared with patients without the KRAS-variant is novel but perhaps not surprising, considering that the KRAS-variant is germline and, thus, present in all cells, including immune cells. This inherited mutation is known to lead to globally altered gene expression profiles consistent with KRAS addiction, as well as altered expression of the let-7 microRNA, both of which have significant effects on the immune system. In addition, our findings of elevated TGF-β1 expression in these patients, which has long been known to be involved in immunity and immunosuppression, further supports our hypothesis that patients with the KRAS-variant are immunosuppressed. Baseline differences in immunity could also help explain the increased cancer risk found in patients with the KRAS-variant. Interestingly, cetuximab overcomes the impaired natural killer group 2D–dependent functionality of activated natural killer cells in the presence of TGF-β1, potentially explaining the global benefit of cetuximab treatment seen in patients with the KRAS-variant. Whether the altered TGF-β1 we see in these patients represents baseline perturbations in homeostasis or altered responses to tumor challenge and is a finding that will be found for all KRAS-variant patients with cancer remains unclear and is the subject of important ongoing investigations.
Although the sample size was small in this study and a larger study would strengthen the results, our current findings are consistent with prior work with KRAS-variant patients with recurrent or metastatic HNSCC treated with cisplatin with or without cetuximab, revealing that the addition of cetuximab improved median survival to 3.9 months; however, the interaction with p16 status could not be evaluated previously because of the small sample size. Our findings are also consistent with those found in KRAS-variant patients with colon cancer, in whom cetuximab monotherapy is clearly beneficial. What this study has additionally found is the potential importance of cetuximab delivered in conjunction with radiotherapy for patients with the KRAS-variant to seemingly enable them to mount appropriate immunologic responses after tumor-targeted irradiation, diminishing metastatic failures. We also find that patients with the KRAS-variant may have baseline normal tissue radiation sensitivity, possibly through their elevated TGF-β1 levels. Although cetuximab is a known radiosensitizer, there did not appear to be worsened radiosensitivity with the addition of cetuximab for patients with the KRAS-variant. Studies are ongoing to better understand the mechanisms of radiosensitivity in these patients to further personalize their treatment.
Limitations
This was an ad hoc analysis of a prospective study with a limited number of KRAS-variant patients. Additional prospective trials are planned to directly confirm the findings. End points to be further validated include the findings that the KRAS-variant identifies patients with altered immunity and differential sensitivity to radiotherapy and immune-modulating therapies.
Conclusions
Although a germline biomarker that can identify individuals with altered immunity that can be used to direct the best therapeutic combinations is a bold new concept, the KRAS-variant is unfolding as a true candidate. On the basis of the findings of this study, defining how anticancer agents affecting the tumor-host-immune association differently affect patients with the KRAS-variant is a critical and pressing need because this knowledge could help in personalizing developing immune-based cancer therapies for all patients.
eTable 1. Pretreatment Characteristics by Whether or Not KRAS Genotype Is Known
eTable 2. Pretreatment Characteristics by KRAS Genotype and Assigned Treatment
eTable 3. TGFB1 by p16 and KRAS-Variant Status
eTable 4. Grade 3-4 Treatment-Related [1] Radiation Mucositis by KRAS-Variant and Assigned Treatment
eTable 5. Grade 3-4 Treatment-Related [1] Skin Reaction Inside Portal [2] by KRAS-Variant and Assigned Treatment
eTable 6. Grade 3-4 Treatment-Related [1] Skin Reaction Outside Portal [2] by KRAS-Variant and Assigned Treatment
eFigure 1. CONSORT Flow Diagram RTOG 0522
eFigure 2. Distant Metastasis
eFigure 3. Local Regional Failure
References
- 1.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ang KK, Zhang Q, Rosenthal DI, et al. Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J Clin Oncol. 2014;32(27):2940-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354(6):567-578. [DOI] [PubMed] [Google Scholar]
- 4.Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359(11):1116-1127. [DOI] [PubMed] [Google Scholar]
- 5.Boeckx C, Baay M, Wouters A, et al. Anti-epidermal growth factor receptor therapy in head and neck squamous cell carcinoma: focus on potential molecular mechanisms of drug resistance. Oncologist. 2013;18(7):850-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261-269. [DOI] [PubMed] [Google Scholar]
- 7.Chung CH, Zhang Q, Kong CS, et al. p16 protein expression and human papillomavirus status as prognostic biomarkers of nonoropharyngeal head and neck squamous cell carcinoma. J Clin Oncol. 2014;32(35):3930-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spanos WC, Nowicki P, Lee DW, et al. Immune response during therapy with cisplatin or radiation for human papillomavirus-related head and neck cancer. Arch Otolaryngol Head Neck Surg. 2009;135(11):1137-1146. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava RM, Lee SC, Andrade Filho PA, et al. Cetuximab-activated natural killer and dendritic cells collaborate to trigger tumor antigen-specific T-cell immunity in head and neck cancer patients. Clin Cancer Res. 2013;19(7):1858-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephenson RM, Lim CM, Matthews M, Dietsch G, Hershberg R, Ferris RL. TLR8 stimulation enhances cetuximab-mediated natural killer cell lysis of head and neck cancer cells and dendritic cell cross-priming of EGFR-specific CD8+ T cells. Cancer Immunol Immunother. 2013;62(8):1347-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Zhang X, Mortenson ED, Radkevich-Brown O, Wang Y, Fu Y-X. Cetuximab-mediated tumor regression depends on innate and adaptive immune responses. Mol Ther. 2013;21(1):91-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pozzi C, Cuomo A, Spadoni I, et al. The EGFR-specific antibody cetuximab combined with chemotherapy triggers immunogenic cell death. Nat Med. 2016;22(6):624-631; Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharyya T, Purushothaman K, Puthiyottil SS, Bhattacharjee A, Muttah G. Immunological interactions in radiotherapy-opening a new window of opportunity. Ann Transl Med. 2016;4(3):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derer A, Frey B, Fietkau R, Gaipl U. Immune-modulating properties of ionizing radiation: rationale for the treatment of cancer by combination radiotherapy and immune checkpoint inhibitors. Cancer Immunol Immunother. 2016;65(7):779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frey B, Rubner Y, Kulzer L, et al. Antitumor immune responses induced by ionizing irradiation and further immune stimulation. Cancer Immunol Immunother. 2014;63(1):29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kjersem JB, Skovlund E, Ikdahl T, et al. FCGR2A and FCGR3A polymorphisms and clinical outcome in metastatic colorectal cancer patients treated with first-line 5-fluorouracil/folinic acid and oxaliplatin +/− cetuximab. BMC Cancer. 2014;14:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saridaki Z, Weidhaas J, Lenz H-J, et al. A let-7 microRNA-binding site polymophism is KRAS predicts improved outcome in metastatic colorectal cancer (mCRC) patients treated with salvage cetuximab/panitumumab monotherapy. Clin Cancer Res. 2014;20(17):4499-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin LJ, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68(20):8535-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paranjape T, Heneghan H, Lindner R, et al. A 3′-untranslated region KRAS variant and triple-negative breast cancer: a case-control and genetic analysis. Lancet Oncol. 2011;12(4):377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratner E, Lu L, Boeke M, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70(16):6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pilarski R, Patel DA, Weitzel J, et al. The KRAS-variant is associated with risk of developing double primary breast and ovarian cancer. PLoS One. 2012;7(5):e37891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McVeigh TP, Jung S-Y, Kerin MJ, et al. Estrogen withdrawal, increased breast cancer risk and the KRAS-variant. Cell Cycle. 2015;14(13):2091-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratner E, Keane F, Lindner R, et al. A KRAS-variant is a biomarker of poor outcome, platinum chemotherapy resistance and a potential target for therapy in ovarian cancer. Oncogene. 2012;31(42):4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weidhaas J, Kim E, Herbst R, et al. The KRAS-variant and treatment response in BATTLE-1 [abstract]. J Clin Oncol. 2014;32(5)(suppl):8135. [Google Scholar]
- 25.Graziano F, Canestrari E, Loupakis F, et al. Genetic modulation of the Let-7 microRNA binding to KRAS 3′-untranslated region and survival of metastatic colorectal cancer patients treated with salvage cetuximab-irinotecan. Pharmacogenomics J. 2010;10(5):458-464. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W, Winder T, Ning Y, et al. A let-7 microRNA-binding site polymorphism in 3′-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. 2011;22(1):104-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganzinelli M, Rulli E, Caiola E, et al. Role of KRAS-LCS6 polymorphism in advanced NSCLC patients treated with erlotinib or docetaxel in second line treatment (TAILOR). Sci Rep. 2015;5:16331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sclafani F, Chau I, Cunningham D, et al. Prognostic role of the LCS6 KRAS variant in locally advanced rectal cancer: results of the EXPERT-C trial. Ann Oncol. 2015;26(9):1936-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen BC, Moyer BJ, Avissar M, et al. A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis. 2009;30(6):1003-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung CH, Lee JW, Slebos RJ, et al. A 3′-UTR KRAS-variant is associated with cisplatin resistance in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol. 2014;25(11):2230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Ruyck K, Duprez F, Ferdinande L, et al. A let-7 microRNA polymorphism in the KRAS 3′-UTR is prognostic in oropharyngeal cancer. Cancer Epidemiol. 2014;38(5):591-598. [DOI] [PubMed] [Google Scholar]
- 32.Smakman N, Veenendaal LM, van Diest P, et al. Dual effect of Kras(D12) knockdown on tumorigenesis: increased immune-mediated tumor clearance and abrogation of tumor malignancy. Oncogene. 2005;24(56):8338-8342. [DOI] [PubMed] [Google Scholar]
- 33.Sathe A, Ayyar K, Reddy K. MicroRNA let-7 in the spotlight: role in innate immunity. Inflamm Cell Signal. 2014;1:e109. doi: 10.14800/ics.109 [DOI] [Google Scholar]
- 34.Zdanov S, Mandapathil M, Abu Eid R, et al. Mutant KRAS conversion of conventional T cells into regulatory T cells. Cancer Immunol Res. 2016;4(4):354-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99-146. [DOI] [PubMed] [Google Scholar]
- 36.Gutcher I, Donkor MK, Ma Q, Rudensky A, Flavell R, Li M. Autocrine transforming growth factor-β1 promotes in vivo Th17 cell differentiation. Immunity. 2011;34(3):396-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanpouille-Box C, Diamond JM, Pilones KA, et al. TGFβ is a master regulator of radiation therapy-induced antitumor immunity. Cancer Res. 2015;75(11):2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klöss S, Chambron N, Gardlowski T, et al. Cetuximab reconstitutes pro-inflammatory cytokine secretions and tumor-infiltrating capabilities of sMICA-inhibited NK cells in HNSCC tumor spheroids. Front Immunol. 2015;6:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jie HB, Schuler PJ, Lee SC, et al. CTLA-4+ regulatory T cells increased in cetuximab-treated head and neck cancer patients suppress NK cell cytotoxicity and correlate with poor prognosis. Cancer Res. 2015;75(11):2200-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin L, Ratner E, Leng S, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3′UTR increases non-small cell cancer risk. Cancer Res. 2008;68:8535-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byers LA, Holsinger FC, Kies MS, et al. Serum signature of hypoxia-regulated factors is associated with progression after induction therapy in head and neck squamous cell cancer. Mol Cancer Ther. 2010;9(6):1755-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalbfleisch J, Prentice R. The Statistical Analysis of Failure Time Data. New York, NY: Wiley; 1980. [Google Scholar]
- 43.Kaplan E, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457-481. [Google Scholar]
- 44.Yoshimura A, Muto G. TGF-β function in immune suppression. Curr Top Microbiol Immunol. 2011;350:127-147. [DOI] [PubMed] [Google Scholar]
- 45.Guo SW, Du Y, Liu X. Platelet-derived TGF-β1 mediates the down-modulation of NKG2D expression and may be responsible for impaired natural killer (NK) cytotoxicity in women with endometriosis. Hum Reprod. 2016;31(7):1462-1474. [DOI] [PubMed] [Google Scholar]
- 46.Klöß S, Chambron N, Gardlowski T, et al. Increased sMICA and TGFβ1 levels in HNSCC patients impair NKG2D-dependent functionality of activated NK cells. Oncoimmunology. 2015;4(11):e1055993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Pretreatment Characteristics by Whether or Not KRAS Genotype Is Known
eTable 2. Pretreatment Characteristics by KRAS Genotype and Assigned Treatment
eTable 3. TGFB1 by p16 and KRAS-Variant Status
eTable 4. Grade 3-4 Treatment-Related [1] Radiation Mucositis by KRAS-Variant and Assigned Treatment
eTable 5. Grade 3-4 Treatment-Related [1] Skin Reaction Inside Portal [2] by KRAS-Variant and Assigned Treatment
eTable 6. Grade 3-4 Treatment-Related [1] Skin Reaction Outside Portal [2] by KRAS-Variant and Assigned Treatment
eFigure 1. CONSORT Flow Diagram RTOG 0522
eFigure 2. Distant Metastasis
eFigure 3. Local Regional Failure