Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that causes frequently nosocomial infections, currently becoming more difficult to treat due to the various resistance mechanisms and different virulence factors. The purpose of this study was to determine the risk factors independently associated with the development of bacteremia by carbapenem-resistant P. aeruginosa, the frequency of virulence genes in metallo-β-lactamases producers and to evaluate their ability to produce biofilm. We conducted a case–control study in the Uberlândia Federal University – Hospital Clinic, Brazil. Polymerase Chain Reaction was performed for metallo-β-lactamases and virulence genes. Adhesion and biofilm assays were done by quantitative tests. Among the 157 strains analyzed, 73.9% were multidrug-resistant, 43.9% were resistant to carbapenems, 16.1% were phenotypically positive for metallo-β-lactamases, and of these, 10.7% were positive for blaSPM gene and 5.3% positive for blaVIM. The multivariable analysis showed that mechanical ventilation, enteral/nasogastric tubes, primary bacteremia with unknown focus, and inappropriate therapy were independent risk factors associated with bacteremia. All tested strains were characterized as strongly biofilm producers. A higher mortality was found among patients with bacteremia by carbapenem-resistant P. aeruginosa strains, associated independently with extrinsic risk factors, however it was not evident the association with the presence of virulence and metallo-β-lactamases genes.
Keywords: Pseudomonas aeruginosa, Resistance, Metallo-β-lactamases, Virulence, Biofilm
Introduction
Healthcare-associated infections (HAIs) like bacteremia caused by multidrug-resistant Pseudomonas aeruginosa strains result in an increased morbidity and mortality, prolonging the hospitalization, and higher costs compared to those infections caused by susceptible strains.1, 2, 3 Infections caused by these resistant microorganisms are often associated with age, cancer, heart disease, diabetes, intensive use of antibiotics, and invasive procedures such as hemodialysis, mechanical ventilation catheter, tracheostomy, and others.4
The increasing incidence of multidrug-resistant P. aeruginosa as a cause of nosocomial infection is a global problem, a consequence of the ability of this microorganism to develop resistance to almost all other antimicrobial agents during antimicrobial chemotherapy, either by selection of mutations in chromosomal genes or by horizontal gene transfer.5, 6 In Brazil, this problem is even more significant, since there is a very high density of antibiotic use, especially β-lactams, carbapenems, and fluoroquinolones.7, 8 The resistance in P. aeruginosa to carbapenems is up to 60% in some Brazilian hospitals9, 10 and mainly occurs by production of metallo-β-lactamases (MBL).
Ten subclasses of the MBL enzymes are known: IMP (Imipenemase), VIM (Verona Imipenemase), SPM-1 (São Paulo MBL), GIM (German Imipenemase), SIM-1 (Seul Imipenemase),11 AIM-1 (Australian Imipenemase),12 KHM (Kyorin University Hospital),13 NDM-1 (New Delhi MBL),14 DIM-1 (Dutch Imipenemase),15 and TMB (Tripoli MBL).16 In Brazil, the most prevalent subclasses are IMP-1 and SPM-1.11
Several other virulence genes as well as biofilm formation may contribute to the pathogenicity of severe infections, particularly including those associated with invasive procedures.17 Among the major virulence factors described in the literature, we highlight those related to the adherence of microorganisms to host cells through the flagella, fimbriae, and alginate18 and those that facilitate the disruption of epithelial integrity and interfere with the immune system, such as elastase, phospholipase C and protease alkaline, further exotoxin A, pyocyanin, and pyoverdine.19 The biofilm production is particularly associated with the difficulty of antibiotics to penetrate the cells, since them secrete a polymeric matrix composed of polysaccharides, proteins, and DNA.20, 21
The aim of this study was to identify the risk factors associated with bacteremia caused by carbapenem-resistant P. aeruginosa as well as the production of MBL. Additionally, we investigated the frequency of virulence genes and their ability to form biofilm.
Materials and methods
Patients and hospital
The P. aeruginosa strains were recovered from patients admitted to the Uberlandia University Hospital (Brazil), Federal University of Uberlândia (HC-UFU), and obtained from the Microbiology Laboratory of the HC-UFU, during the period from May 2009 to December 2012, considering only the first episode of infection.
Study design
We conducted a case (patients with bacteremia due to P. aeruginosa resistant to carbapenems) versus control (patients with bacteremia caused by P. aeruginosa susceptible to carbapenems) study to identify risk factors among patients infected with P. aeruginosa resistant to carbapenems. Additionally, we evaluated secondary outcomes, including periods of hospitalization, admission to the intensive care unit (ICU), and use of invasive procedures. Demographic, clinical, and epidemiological characteristics of each patient included in the study were recovered from the clinical records.
Definitions
Healthcare-associated infections (HAIs) are defined as any infection acquired after a patient's admission to the hospital. HAIs may manifest during hospitalization or after discharge, since they are related to hospitalization or procedures performed during the hospitalization.22 Bacteremia, according to the Centers for Disease Control and Prevention,23 can be defined as the presence of viable bacteria in the blood documented by a positive blood culture result. Bacteremia was classified as primary when it was unrelated to another focus of infection or when it was related to an intravenous catheter, and secondary when it was clinically related to infection in another anatomic site.9 Multidrug-resistance is defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories.24 The antimicrobial therapy was considered inappropriate when an isolated microorganism presented in vitro resistance to antimicrobials used for treating the patient and/or a lack of treatment for 24 h after diagnosis of microbial infections.25
Clinical microbiological and molecular testing
Cultures were collected according to the protocol used by the Microbiology Laboratory of the HC-UFU and were processed using the automated system BACT/Alert® (BioMérieux, Durham, USA). The identification and antimicrobial susceptibility tests were performed by automation using the VITEK II system and the strains that showed intermediate susceptibility were considered resistant. Quality-control protocols were used according to the standards of the Clinical and Laboratory Standard Institute.26, 27, 28 The carbapenem-resistant P. aeruginosa isolates were phenotypically screened for MBL production using double-disc synergy tests, as previously described.29, 30 In addition, to assess the presence of MBL genes in P. aeruginosa strains, a multiplex PCR was performed, as described previously.31 The cycling conditions were: 94 °C for 5 min, followed by 30 denaturation cycles at 94 °C for 30 s, annealing at 53 °C for 45 s and extension at 72 °C for 30 s, followed by final extension at 72 °C at 10 min, all in a MasterCycler personal (Eppendorf). Detection of virulence genes codifying alkaline protease (aprA), elastase A (lasA), elastase (lasB),32 haemolytic phospholipase C (plcH), non-haemolytic phospholipase C (plcN), exotoxin A (toxA) and alginate (algD)33 were determined by uniplex PCR, using the following protocol: 94 °C for 3 min, 30 cycles at 94 °C for 30 s, 55 °C for 1 min and 72 for 1 min and 30 s and 72 °C for 5 min.
Initial adhesion assay34
In order to evaluate the initial adhesion, 200 μL of a cell suspension containing 1 × 107 cells/mL prepared in TSB was added to 96-well polystyrene plates. Initial adhesion was allowed to occur for 2 h at 37 °C with rotation at 120 rpm. Bacteria adhered in 96-well polystyrene plates were washed twice with a 0.9% NaCl solution and harvested by scraping the wells for 90 s. The cell suspension obtained was plated on TSA for colony-forming unit (CFU) enumeration. All experiments were done in triplicate in three independent experiments. The strain ATCC15692 (PAO1) was used as a positive control and TSB without bacteria was used as a negative control.
Biofilm formation assay35
200 μL of a cell suspension containing 1 × 107 cells/mL prepared in TSB was added to 96-well polystyrene plates. Biofilm formation was allowed to occur for 24 h at 37 °C with rotation at 120 rpm. Bacteria grown in 96-well polystyrene plates were washed twice with a 0.9% NaCl solution and left to dry in an inverted position. The total biomass was measured by methanol (Merck) fixation, crystal violet (Merck) staining, and acid acetic (Merck) elution as previously described. The eluted dye was removed from each well and placed in a new 96-well microtiter plate, and its absorbance was read on an ELISA plate reader (BioTek Instruments Inc., Vermont, USA) at 570 nm. The experiments were done with eight replicates for each strain in three independent experiments. TSB without bacteria was used as a negative control. The optical density cut-off value (ODc) was established as three standard deviations (SD) above the mean of the optical density (OD) of the negative control: ODc = average OD of negative control + 3x SD of negative control. For easier interpretation of the results, strains were divided into the following categories according to optical density: (ODi): ODi ≤ ODc or ODc < ODi < 2x ODc = non-biofilm producer/weak biofilm producer; 2x ODc < ODi < 4x ODc = moderate biofilm producer; 4x ODc < ODi = strong biofilm producer.
Biofilm cell concentration36
The biofilm cell concentration was determined by CFU enumeration. After biofilm formation, the biofilms were washed twice with a 0.9% NaCl solution and harvested after scraping the wells for 90 s. The cell suspension obtained was plated onto TSA plates. All experiments were done in triplicate, on three independent occasions.
Statistical analysis
Statistical analysis was performed using GraphPad Prism v.5 (GraphPad Software, San Diego, CA). Quantitative assays were compared using the Kruskal–Wallis, applying Dunn's multiple comparison test. All tests were performed with a confidence level of 95% and statistical significance was defined as p < 0.05.
Ethical approval
The research Ethics Committee of the Federal University Uberlandia evaluated and approved our study design.
Results
From May 1, 2009, to December 31, 2012, a hospital surveillance of the incidence of P. aeruginosa bacteremia was performed in the microbiology laboratory of the University Hospital. In this period, 157 non-repetitive patients with P. aeruginosa bacteremia were included in the study. The detailed information on factors associated with the development of bacteremia and the clinical and demographic characteristics, as well the distribution of patients by wards are shown in Table 1, Table 2. The primary bacteremia occurred in 75.8% and those with unknown focus accounted in 62.4% of the cases. Secondary bacteremia accounted for 24.2% of patients, where the respiratory tract was the main focus of infection (17.2%). Furthermore, 43.9% of patients had bacteremia with carbapenem-resistant P. aeruginosa, and most of these patients were admitted to the ICU. Most patients were male (66.8%), with an average hospital stay of 63.2 ± 80.17 days and an average age of 52.01 ± 20.24 years.
Table 1.
Distribution of patients infected with Pseudomonas aeruginosa in different units of the Clinical Hospital of the Federal University of Uberlândia from May/2009 to December/2012.
| Clinics | Total | Carbapenem-resistant Pseudomonas aeruginosa |
|---|---|---|
| N = 157 (%) | N = 69 (%) | |
| Surgery | 35 (22.3) | 16 (23.2) |
| Medical clinic | 24 (15.3) | 10 (14.5) |
| Oncology | 14 (8.9) | 2 (2.9) |
| Emergency room | 9 (5.7) | 5 (7.3) |
| ICUa | 50 (31.9) | 31 (44.9) |
| Neonatal ICU | 2 (1.3) | 0 |
| Pediatric ICU | 2 (1.3) | 1 (1.5) |
| Transplant | 2 (1.3) | 0 |
| Othersb | 3 (1.9) | 0 |
Intensive care unit.
Burneds, thoracic pain unit.
Table 2.
Risk factors associated with carbapenem resistance in patients with bacteremia caused by Pseudomonas aeruginosa.
| Risk factors | Total | Carbapenem resistant | Multisensitivea | Univariate |
Multivariate |
||
|---|---|---|---|---|---|---|---|
| N = 157 (%) | N = 69 (%) | N = 65 (%) | ORb (CIc 95%) | pd | OR (CI 95%) | p | |
| Age-mean (years) | 52.01 ± 24.49 | 59.01 ± 20.24 | 43.18 ± 27.56 | – | 0.0009* | – | – |
| Male/female | 105/52 (66.88/33.12) | 45 (65.22)/24 (34.78) | 43 (66.15)/22 (33.85) | 0.9593 (0.4698–1.959) | 0.9092 | – | – |
| Time of hospitalization, mean (days) | 63.2 ± 80.17 | 60.01 ± 48.97 | 68.86 ± 110.4 | – | 0.3477 | – | – |
| Intensive care unit | 74 (47.13) | 38 (55.07) | 22 (33.85) | 2.396 (1.190–4.822) | 0.0135 | – | – |
| Surgery | 69 (43.95) | 37 (53.62) | 20 (30.77) | 2.602 (1.281–5.282) | 0.0075* | – | – |
| Invasive procedures (72 h) | 139 (88.53) | 64 (92.75) | 54 (83.08) | 2.607 (0.8527–7.973) | 0.0843 | – | – |
| Mechanical ventilation | 89 (56.69) | 48 (69.56) | 28 (43.08) | 3.020 (1.485–6.144) | 0.002* | 3.0674 (1.22–7.73) | 0.0175 |
| Tracheostomy | 70 (44.58) | 43 (62.32) | 17 (26.15) | 4.670 (2.234–9.759) | <0.0001* | – | – |
| Urinary catheter | 101 (64.33) | 52 (75.36) | 34 (52.31) | 2.789 (1.340–5.804) | 0.0054* | – | – |
| Venous central catheter | 125 (79.62) | 59 (85.51) | 48 (73.85) | 2.090 (0.8761–4.984) | 0.0926 | – | – |
| Surgical drain | 24 (15.29) | 14 (20.29) | 7 (10.77) | 2.109 (0.7918–5.618) | 0.1297 | – | – |
| Probes enteral/gastric nutrition | 109 (69.43) | 54 (78.26) | 40 (61.54) | 2.250 (1.053–4.810) | 0.0345* | 3.8347 (1.43–10.26) | 0.0074 |
| Hemodialysis | 39 (24.84) | 27 (39.13) | 8 (12.31) | 4.580 (1.892–11.09) | 0.0004* | ||
| Parenteral nutrition | 25 (1.92) | 14 (20.29) | 8 (12.31) | 1.814 (0.7052–4.664) | 0.2125 | – | – |
| Comorbidity conditions | 122 (77.70) | 53 (76.81) | 49 (75.38) | 1.082 (0.4886–2.394) | 0.8465 | – | – |
| Heart failure | 35 (22.29) | 21 (30.43) | 12 (18.46) | 1.932 (0.8597–4.343) | 0.1079 | – | – |
| Cancer | 32 (20.38) | 8 (11.59) | 17 (26.15) | 0.3703 (0.1473–0.9308) | 0.0306 | – | – |
| Diabetes mellitus | 21 (13.37) | 9 (13.04) | 7 (10.75) | 1.243 (0.4341–3.558) | 0.6849 | – | – |
| Chronic renal failure | 40 (25.48) | 21 (30.43) | 16 (24.61) | 1.340 (0.6249–2.873) | 0.4514 | – | – |
| Human immunodeficiency virus | 10 (6.37) | 5 (7.25) | 3 (4.61) | 1.615 (0.3699–7.048) | 0.7191 | – | – |
| ASISescore ≥ 4 | 81 (51.59) | 35 (50.72) | 33 (50.77) | 0.9982 (0.5068–1.966) | 0.9959 | – | – |
| Primary bacteremia | 119 (75.79) | 45 (65.22) | 57 (87.69) | 0.2632 (0.1080–0.6413) | 0.0023* | – | – |
| Central line catheter related | 21 (13.37) | 11 (15.94) | 7 (10.75) | 1.571 (0.5693–4.338) | 0.3802 | – | – |
| Unknown | 98 (62.42) | 34 (49.27) | 50 (76.9) | 0.2914 (0.1383–0.6143) | 0.0009 | 0.2363 (0.08–0.73) | 0.0127 |
| Secondary bacteremia | 38 (24.20) | 24 (34.78) | 8 (12.31) | 3.800 (1.559–9.260) | 0.0023* | – | – |
| Respiratory tract | 27 (17.19) | 17 (24.64) | 5 (7.69) | 3.923 (1.353–11.37) | 0.0081* | – | – |
| Urinary tract | 6 (3.82) | 4 (5.79) | 1 (1.54) | 3.938 (0.4282–36.22) | 0.3666 | – | – |
| Inappropriate therapy | 49 (31.21) | 32 (46.37) | 8 (12.31) | 6.162 (2.560–14.83) | <0.0001* | 8.5534 (3.28–22.32) | <0.0001 |
| Previous use of antibiotics | 123 (78.34) | 59 (85.51) | 43 (66.15) | 3.019 (1.297–7.025) | 0.0086* | – | – |
| Mortality | 92 (58.59) | 48 (69.56) | 32 (49.23) | 2.357 (1.163–4.779) | 0.0165* | 2.3660 (1.15–4.85) | 0.0186 |
Multisensitive strains were considered as those who presented susceptibility to all antimicrobials tested.
Odds ratio.
Confidence interval.
p value.
Average severity of illness score.
Statistically significant (P≤0.05).
The previous use of antibiotics (78.3%), invasive procedures (88.5%), comorbidities (77.7%), and prior surgery (43.9%) were common. It was found that 31.2% of patients received inadequate treatment, and the death rate was 58.6%. Several intrinsic and extrinsic factors for the development of bacteremia by strains of P. aeruginosa resistant to carbapenems were detected by univariate analysis. However, only mechanical ventilation, enteral/nasogastric tubes, primary bacteremia with unknown focus, and inappropriate therapy were risk factors independently associated with the development of carbapenem-resistant P. aeruginosa bacteremia.
MBL production was analyzed for 56 carbapenem-resistant P. aeruginosa isolates. Nine (25.0%) isolates were phenotypically positive and a total of 16.1% (n= 9/56) were consistent with the amplicons MBL genes, being 10.71% blaSPM-1 and 5.3% blaVIM genes In general, the strains showed a multidrug resistance profile. For other virulence genes evaluated (aprA, plcH, plcN, lasA, lasB, toxA, and algD), all strains showed a high frequency (88%).
Among the 88 carbapenem-susceptible P. aeruginosa strains, 21 (23.9%) were resistant to other β-lactams. For 69 strains resistant to carbapenems, the resistance profile was high: β-lactam 50/69 (72.5%), fluoroquinolones 48/69 (69.6%), and aminoglycosides 50/69 (72.5%) (Table 3).
Table 3.
Antimicrobial resistance profiles in samples of Pseudomonas aeruginosa isolated from blood.
| Antimicrobial | Carbapenems |
p (OR) | |
|---|---|---|---|
| Resistant group N = 69 (%) |
Susceptible group N = 88 (%) |
||
| Ceftazidime | 51 (73.9) | 10 (11.4) | <0.0001 (9.446–51.71) |
| Cefepime | 45 (65.2) | 17 (19.3) | <0.0001 (3.792–16.17) |
| Piperacillin/tazobactam | 37 (53.6) | 12 (13.6) | <0.0001 (3.386–15.84) |
| Fluorquinolones (ciprofloxacin and levofloxacin) | 48 (69.6) | 17 (19.3) | <0.0001 (4.568–19.95) |
| Aminoglicosides (amikacin and gentamicin) | 50 (72.5) | 12 (13.6) | <0.0001 (7.442–37.32) |
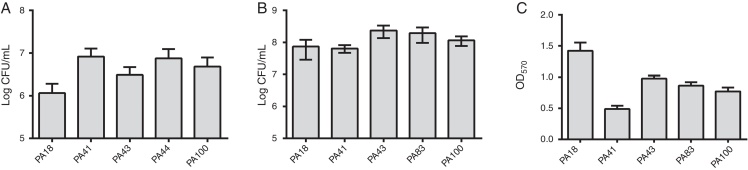
In addition to MBL research, five strains were selected (two containing SPM-1, one containing VIM, and two phenotypically positive for MBL) to the evaluation of the biofilm formation using quantitative assays of total biomass by staining with crystal violet. Considering the presence of MBL genes, all strains were identified as strongly biofilm producers with an average of 0.90 ± 0.61 of biomass. Differences in initial adhesion of individual strains were not observed. However, there was a lower OD for the strain containing the blaVIM gene (Fig. 1).
Fig. 1.
(A) Number of cells adhered to a polystyrene surface after 2 h, expressed as log CFU/mL for 5 samples of Pseudomonas aeruginosa isolated from blood. (B) Number of viable cells in the biofilm (log CFU/mL). (C) Biomass of biofilm expressed as optical density of crystal violet (OD570nm).
Discussion
Hospital bacteremia caused by multiresistant microorganisms, whether Gram-positive or Gram-negative, has often been described as a significant health problem that increases hospital costs and makes it difficult to establish an appropriate antimicrobial therapy, which results in a worse prognosis.37 The high proportion of hospital bacteremia caused by P. aeruginosa resistant to carbapenems indicates the importance of this organism as a significant cause of this infection in our hospital. The high proportion of hospital bacteremia caused by P. aeruginosa resistant to carbapenems indicates the importance of this organism as a significant cause of this infection in our hospital. The results of different studies have suggested that intrinsic risk factors such as mechanical ventilation, use of a nasogastric tube, and prior use of antibiotics increase the risk of bacteremia development caused by P. aeruginosa resistant to carbapenems, as well as the risk of morbidity and mortality.38
In this study, more than 50% of the risk factors were associated with infection by this microorganism according to univariate analysis. However, only mechanical ventilation, enteral/gastric feeding tubes, primary bacteremia of unknown origin, and inappropriate treatment were considered independent risk factors by multivariate analysis.
A central venous catheter is one of the most significant risk factors for acquired bacteremia in the hospital, as it was associated with more than 90% of these infections.39 Although it was not a significant risk factor in this study, 85.5% of patients with bacteremia with carbapenem-resistant P. aeruginosa had this invasive procedure.
Often, bacteremia caused by P. aeruginosa resistant to antibiotics has a higher mortality rate due, in particular, to the administration of inappropriate antibiotic therapy.40 In this study, we investigated a cohort of 157 patients with bacteremia caused by P. aeruginosa strains, with 69 of them infected by carbapenem-resistant P. aeruginosa isolates. Total mortality was higher in the carbapenem-resistant group; however, the presence of resistant strains was significantly associated with inappropriate antimicrobial therapy, proving that this group usually has a poorer prognosis. Similar to our findings, previous studies showed that P. aeruginosa isolated from patients who received inappropriate therapy had a worse prognosis, with rates of 46.1%41 and 53.8%.42
Besides to be associated with severe infections, resistance to carbapenems in P. aeruginosa often results in the production of MBL.43 Our results showed that the carbapenem-resistant P. aeruginosa were prevalent throughout the hospital with genes that encoding these enzymes in 16.1% of the strains, being 10.7% of genotype blaSPM-1 and 5.4% of genotype blaVIM. Since the frequency of multidrug-resistant strains was high, these results suggest that other resistance mechanisms coexist in these strains, such as efflux pumps and impermeability of the membrane.44
The prevalence of MBL as a resistance mechanism has increased, particularly in Latin America.45 In Brazil, the prevalence of MBL-producing P. aeruginosa varies between different regions and between hospitals, with rates ranging from 7.5% to 44%.11
SPM-1 enzyme is considered the most common in Brazil, followed by IMP-1,11, 46 however, there has also been an increase in the frequency of P. aeruginosa isolates containing VIM enzyme.45, 47 In our study, the SPM-1 enzyme was detected in 16.7% of phenotypically producing samples of MBL, followed by VIM enzyme, which was detected in 8.3% of cases. These results indicate a significant spread of MBL-encoding genes in our region.
Considering the presence of MBL and biofilm production, 100% of the strains were classified as strong producers. Another study in Brazil found that 40% of P. aeruginosa classified as strong biofilm-producing were also MBL producers.35 We believe that these results are important once these characteristics (biofilm production and MBL) overlap and infections caused by these bacteria are difficult to treat. This is justified, in part, because the growth of bacteria in the biofilm is about 64 times more resistant to antimicrobial.48
P. aeruginosa has a large number of extracellular virulence factors that also contribute to the pathogenicity and severity of these infections, such as that encoded by aprA, plcH, plcN, lasA, lasB, toxA, and algD genes.32, 33 Our data showed that the presence of a multiresistant profile, in the most of the strains (88%) were positive for the aprA, plcH, plcN, lasA, lasB, toxA, and algD virulence genes, except in one strain that did not show the toxA and plcH genes, which can further contribute to a worse prognosis associated with severe infections.
Our results confirm previous findings regarding risk factors for the development of P. aeruginosa carbapenem-resistant bacteremia, as well as the dissemination of MBL-producing strains of SPM-1 type. However, this study contributes for further evidence of the spread of MBL-producing strains, particularly the VIM type, in highly virulent strains and strongly biofilm producers.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgment
The autors would like to thank the financial support by FAPEMIG (Fundação de Amparo à Pesquisa de Minas Gerais).
Associate Editor: Elizabeth de Andrade Marques
References
- 1.Hirsch E.B., Tam V.H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res. 2010;10:441–451. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morales E., Cots F., Sala M. Hospital costs of nosocomial multi-drug resistant Pseudomonas aeruginosa acquisition. BMC Health Serv Res. 2012;12:122–129. doi: 10.1186/1472-6963-12-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang C.I., Kim S.H., Park W.B. Bloodstream infections caused by antibiotic-resistant Gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother. 2005;49:760–766. doi: 10.1128/AAC.49.2.760-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shorr A.F., Tabak Y.P., Killian A.D. Healthcare-associated bloodstream infection: a distinct entity? Insights from a large U.S. database. Crit Care Med. 2006;34:2588–2595. doi: 10.1097/01.CCM.0000239121.09533.09. [DOI] [PubMed] [Google Scholar]
- 5.Xavier D.E., Picao R.C., Girardello R. Efflux pumps expression and its association with porin down-regulation and b-lactamase production among Pseudomonas aeruginosa causing bloodstream infections in Brazil. BMC Microbiol. 2010;10:1–7. doi: 10.1186/1471-2180-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zavascki A.P., Gaspareto P.B., Martins A.F. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing SPM-1 metallo-β-lactamase in a teaching hospital in southern Brazil. J Antimicrob Chemother. 2005;56:1148–1151. doi: 10.1093/jac/dki390. [DOI] [PubMed] [Google Scholar]
- 7.Moreira M.R., Guimarães M.P., Rodrigues A.A.A. Antimicrobial use, incidence, etiology and resistance patterns in bacteria causing ventilator-associated pneumonia in a clinical–surgical intensive care unit. Rev Soc Bras Med Trop. 2013;246:39–44. doi: 10.1590/0037-868216722013. [DOI] [PubMed] [Google Scholar]
- 8.Porto J.P., Santos R.O., Gontijo-Filho P.P. Active surveillance to determine the impact of methicillin resistance on mortality in patients with bacteremia and influences of the use of antibiotics on the development of MRSA infection. Rev Soc Bras Med Trop. 2013;46:713–718. doi: 10.1590/0037-8682-0199-2013. [DOI] [PubMed] [Google Scholar]
- 9.Baumgart A.M., Molinari M.A., Silveira A.C. Prevalence of carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii in high complexity hospital. Braz J Infect Dis. 2010;14:433–436. doi: 10.1590/s1413-86702010000500002. [DOI] [PubMed] [Google Scholar]
- 10.Kiffer C., Hsiung A., Oplustil C. Antimicrobial susceptibility of Gram-negative bacteria in Brazilian hospitals: the MYSTIC Program Brazil 2003. Braz J Infect Dis. 2005;9:216–224. doi: 10.1590/s1413-86702005000300004. [DOI] [PubMed] [Google Scholar]
- 11.Mendes R.E., Castanheira M., Pignatari A.C. Metallo-β-lactamases. Arch Pathol Lab Med. 2006;42:103–113. [Google Scholar]
- 12.Yong D., Bell J., Ritchie B. A novel sub group metallo-β-lactamase (MBL), AIM-1 emerges in Pseudomonas aeruginosa (PSA) from Australia, abstr. C1-593. 47th Annual Interscience Conference on Antimicrobial Agents Chemotherapy (ICAAC); Chicago; 2007. [Google Scholar]
- 13.Sekiguchi J., Morita K., Kitao T. KHM-1, a novel plasmid-mediated metallo-beta-lactamase from a Citrobacter freundii clinical isolate. Antimicrob Agents Chemother. 2008;52:4194–4197. doi: 10.1128/AAC.01337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yong D., Toleman M.A., Giske G.C. Characterization of a new metallo-beta-lactamase gene, bla (NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel L., Rodruigues-Martinez J.M., Naiemi N. Characterization of DIM-1, an integron-encoded metallo-beta-lactamase from a Pseudomonas stutzeri clinical isolate in the Netherlands. Antimicrob Agents Chemother. 2009;54:2420–2424. doi: 10.1128/AAC.01456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Salabi A., Borra P.S., Toleman M.A. Genetic and biochemical characterization of a novel metallo-β-lactamase, TMB-1, from an Achromobacter xylosoxidans strain isolated in Tripoli, Libya. Antimicrob Agents Chemother. 2012;56:2241–2245. doi: 10.1128/AAC.05640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todar K. University of Wisconsin-Madison. Department of Bacteriology; 2004. Pseudomonas aeruginosa. Todar's Online Textbook of Bacteriology.http://textbookofbacteriology.net/pseudomonas.html [Google Scholar]
- 18.Kipnis E., Sawa T., Wiener-Kronish J. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med et. Maladies Infect. 2006;36:78–91. doi: 10.1016/j.medmal.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 19.Jaffar-Bandjee M.C., Lazdunski A., Bally M. Production of elastase, exotoxin A, and alkaline protease in sputa during pulmonary exacerbation of cystic fibrosis in patients chronically infect by Pseudomonas aeruginosa. J Clin Microbiol. 1995;33:924–929. doi: 10.1128/jcm.33.4.924-929.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 21.Høiby N., Bjarnsholt T., Givskov M. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Rodrigues E., Richtmann R. Sarvier; São Paulo: 2008. IRAS: Infecção relacionada à assistência à saúde-Orientações Práticas. [Google Scholar]
- 23.Centers for Disease Control and Prevention Guidelines for the prevention of intravascular catheter-related infections. Morb Mortal Wkly Rep. 2002;51(RR-10):1–36. [PubMed] [Google Scholar]
- 24.Magiorakos A.P., Srinivasan A., Carey R.B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert D.N., Sande M.A., Moellering R.C. Antimicrobial Therapy Incorporated; Sperryville, VA: 2007. The Sanford Guide to Antimicrobial Therapy 2007. [Google Scholar]
- 26.Clinical and Laboratory Standard Institute Performance standards for antimicrobial susceptibility testing. 19th Informational Supplement M100-S19; Wayne, PA; 2009. [Google Scholar]
- 27.Clinical and Laboratory Standard Institute Performance standards for antimicrobial susceptibility testing. 20th Informational Supplement M100-S20; Wayne, PA; 2010. [Google Scholar]
- 28.Clinical and Laboratory Standard Institute Performance standards for antimicrobial susceptibility testing. 21st Informational Supplement M100-S21; Wayne, PA; 2011. [Google Scholar]
- 29.Arakawa Y., Shibata N., Shibayama K. Convenient test for screening metallo-beta-lactamase-producing bacteria by using thiol compounds. J Clin Microbiol. 2000;38:40–43. doi: 10.1128/jcm.38.1.40-43.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee K., Chong Y., Shin H.B., Kim Y.A., Yong D., Yum J.H. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase producing strains of Pseudomonas aeruginosa and Acinetobacter species. Clin Microbiol Infect. 2001;7:88–91. doi: 10.1046/j.1469-0691.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- 31.Woodford N. Rapid characterization of β-lactamases by multiplex PCR. In: Gillespie S.H., McHugh T.D., editors. Antibiotic Resistance Protocols. 2nd ed. 2010. pp. 181–192. [DOI] [PubMed] [Google Scholar]
- 32.Lomholt J.A., Poulsen K., Kilian M. Epidemic population structure of Pseudomonas aeruginosa: evidence for a clone that is pathogenic to the eye and that has a distinct combination of virulence factors. Infect Immun. 2001;69:6284–6295. doi: 10.1128/IAI.69.10.6284-6295.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanotte P., Watt S., Mereghetti L. Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J Med Microbiol. 2004;53:73–81. doi: 10.1099/jmm.0.05324-0. [DOI] [PubMed] [Google Scholar]
- 34.Cerca N., Pier G.B., Vilanova M. Quantitative analysis of adhesion and biofilm formation on hydrophilic and hydrophobic surfaces of clinical isolates of Staphylococcus epidermidis. Res Microbiol. 2005;156:506–514. doi: 10.1016/j.resmic.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez L.R., Antunes A.L., Freitas A.L. When the resistance gets clingy: Pseudomonas aeruginosa harboring metallo-β-lactamase gene shows high ability to produce biofilm. Eur J Clin Microbiol Infect Dis. 2012;31:11–714. doi: 10.1007/s10096-011-1363-2. [DOI] [PubMed] [Google Scholar]
- 36.Cassat J.E., Semltzer M.S., Lee C.Y. Investigation of biofilm formation in clinical isolates of Staphylococcus aureus. Methods Mol Biol. 2014;1085:195–211. doi: 10.1007/978-1-62703-664-1_12. [DOI] [PubMed] [Google Scholar]
- 37.Fernandes T.A., Pereira A.C.P., Petrili A.S. Caracterização molecular de Pseudomonas aeruginosa resistentes a carbapenêmicos e produtoras de metalo-β-lactamase isoladas em hemoculturas de crianças e adolescentes com câncer. Rev Soc Bras Med Trop. 2010;43:372–376. doi: 10.1590/s0037-86822010000400007. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed S.H., Daef E.A., Badary M.S. Nosocomial blood stream infection in intensive care units at Assiut University Hospitals (Upper Egypt) with special reference to extended spectrum β-lactamase producing organisms. BMC Res Notes. 2009;2:76–86. doi: 10.1186/1756-0500-2-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jamal M.A., Rosenblatt J., Jiang Y. Prevention of transmission of multi-drug resistant organisms during catheter exchange using antimicrobial catheters. Antimicrob Agents Chemother. 2014 doi: 10.1128/AAC.02886-14. AAC-02886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y.J., Jun Y.H., Kim Y.R. Risk factors for mortality in patients with Pseudomonas aeruginosa bacteremia; retrospective study of impact of combination antimicrobial therapy. BMC Infect Dis. 2014;14:161. doi: 10.1186/1471-2334-14-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joo E.J., Kang C.I., Ha Y.E. Impact of inappropriate empiric antimicrobial therapy on outcome in Pseudomonas aeruginosa bacteraemia: a stratified analysis according to sites of infection. Infection. 2011;39:309–318. doi: 10.1007/s15010-011-0124-6. [DOI] [PubMed] [Google Scholar]
- 42.Morata L., Cobos-Trigueros N., Martínez J.A. Influence of multidrug resistance and appropriate empirical therapy on the 30-day mortality rate of Pseudomonas aeruginosa bacteremia. Antimicrob Agents Chemother. 2012;56:4833–4837. doi: 10.1128/AAC.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavalcanti F.L.D.S., Almeida A.C.S., Vilela M.A. Changing the epidemiology of carbapenem-resistant Pseudomonas aeruginosa in a Brazilian teaching hospital: the replacement of São Paulo metallo-β-lactamase-producing isolates. Mem Inst Oswaldo Cruz. 2012;107:420–423. doi: 10.1590/s0074-02762012000300019. [DOI] [PubMed] [Google Scholar]
- 44.Manoharan A., Chatterjee S., Mathai D. Detection and characterization of metallo beta lactamases producing Pseudomonas aeruginosa. Indian J Med Microbiol. 2010;28:241–244. doi: 10.4103/0255-0857.66486. [DOI] [PubMed] [Google Scholar]
- 45.Sader H.S., Castanheira M., Mendes R.E. Dissemination and diversity of metallo-b-lactamases in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Int J Antimicrob Agents. 2005;25:57–61. doi: 10.1016/j.ijantimicag.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 46.Gales A.C., Menezes L.C., Silbert S. Dissemination in distinct Brazilian regions of an epidmic carbapenem-resistant Pseudomonas aeruginosa producing SPM metalo-β-lactamases. J Antimicrob Chemother. 2003;52:699–702. doi: 10.1093/jac/dkg416. [DOI] [PubMed] [Google Scholar]
- 47.Franco M.R., Caiaffa-Filho H.H., Burattini M.N. Metallo-beta-lactamases among imipenem-resistant Pseudomonas aeruginosa in a Brazilian university hospital. Clinics. 2010;65:825–829. doi: 10.1590/S1807-59322010000900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antunes A.L., Bonfanti J.W., Perez L.R. High vancomycin resistance among biofilms produced by Staphylococcus species isolated from central venous catheters. Mem Inst Oswaldo Cruz. 2011;106:51–55. doi: 10.1590/s0074-02762011000100008. [DOI] [PubMed] [Google Scholar]